Abstract
Within the last decade, active research on circular RNAs (circRNAs) has dramatically improved our understanding of the expression and function of these non-coding RNAs. While several mechanisms for circRNA function have been proposed, including sequestration of microRNAs and regulation of cellular proteins, studies provide evidence that circRNAs can regulate transcription and may also serve as biomarkers. Due to the heterogeneous nature of the brain, and the dynamic transcriptional mechanisms that support neurobiological pathways, the influence of circRNAs is potentially extensive. Understanding how circRNAs contribute to key neurological pathways will fill gaps in our understanding of brain function and provide valuable insight into novel therapeutic approaches to treat neurological diseases. Here, we review recent research on circRNA expression in the brain, describe the proposed functions of circRNAs, and evaluate the role of circRNAs in neurological diseases.
Keywords: circRNA, Non-coding RNA, Brain, Transcriptional regulation, Neurological disease, microRNA
Abbreviations: PSD, post-synaptic density
1. An overview of circRNAs
The growth of biotechnology has facilitated new discoveries in transcriptomics and RNA biology. Over the past few decades, our awareness of the complexities of RNAs and transcriptional regulation has expanded significantly, first with the identification of microRNAs (miRs) and piwi RNAs and more recently with a newly recognized class of non-coding RNAs (ncRNA) known as circular RNAs (circRNAs). Although these species were originally described in the 1970s [1] and again decades later [[2], [3], [4]], research in the past decade has greatly improved our understanding of circRNA features and functions. Here, we first provide an overview on what is currently understood about circRNAs followed by a discussion on circRNA expression and function in the brain.
CircRNAs are different from other non-coding RNAs due to their circular secondary structure, which is created by the back-splicing of a single-stranded linear transcript and the formation of a covalent bond. The outcome is an enclosed non-polyadenylated circular transcript [5,6]. As a result, circRNAs are estimated to be 2.5 to 5 times more stable than linear transcripts due to the absence of exposed ends that may be targeted by 3′ or 5’ exoribonucleases [2,[7], [8], [9], [10]]. In eukaryotes, the lengths of circRNAs are heterogeneous, ranging from approximately 100 to 4000 base pairs long [11,12], and are estimated to include 0.1–10% as many molecules as linear transcripts [8,11]. CircRNA sequences are also well conserved across the mouse, rat, and human genomes [10,13], and as well as between humans and primates [14].
1.1. CircRNA biogenesis
Multiple mechanisms have been proposed for circRNA biogenesis. These models propose that circRNAs form during RNA splicing, and include spliceosome-mediated backsplicing [11,15], spliceosome-mediated intronic circularization [16,17], and exon skipping, driven by lariat or intron-pairing mechanisms [18]. Although circRNAs contain primarily exonic sequences [6,19], they may also contain non-coding sequences, including those derived from antisense strands, introns, untranslated sequences, and intergenic regions [6,20], whereas intronic RNA lariats are formed by intronic circularization. Also, in silico analysis of read directionality from paired-end sequencing data from the Encyclopedia of DNA Elements (ENCODE) Consortium shows that most circRNAs are transcribed from the same strand as their linear counterparts [8]. This finding was validated in Drosophila [21], whereby biogenesis of circRNAs competes with pre-mRNA splicing and whereby the rate of circRNAs synthesis is heavily influenced by flanking introns. However, although circRNAs and linear RNAs have common junction sequences when they are derived from the same host gene, they may have different exon and intron compositions [8]; this feature enables the formation of a potentially diverse circRNA repertoire for individual genes.
1.2. CircRNAs and host gene expression
Multiple studies on mammalian cells have reported an inverse relationship between the abundance of circRNAs and their respective linear mRNAs [6,13,[21], [22], [23]], potentially due to competition from concurrent transcription. This correlation was also observed in the aging mouse brain, for which elevated circRNA expression was observed alongside low linear mRNA expression [24]. On the other hand, a positive correlation was observed for brain circRNAs and their corresponding linear transcripts during neuronal differentiation [13]. However, in rhesus macaque brains, approximately 80% of identified circRNAs showed no correlation with host gene expression to provide evidence of the complexity of competitive and cooperative expression between circRNAs and linear mRNAs [14]. The relationship between circRNA and linear transcript expression is also cell-specific [8], and cellular metabolic demands may influence circRNA expression. Given the energetic needs of the brain, it is likely that circRNAs regulate the function of healthy brains and thus may have roles in diseases of the brain. In this review, we will discuss recent insights into the role of circRNAs in the brain with respect to expression levels, putative functions, and roles in neurological diseases.
2. CircRNA expression in the brain
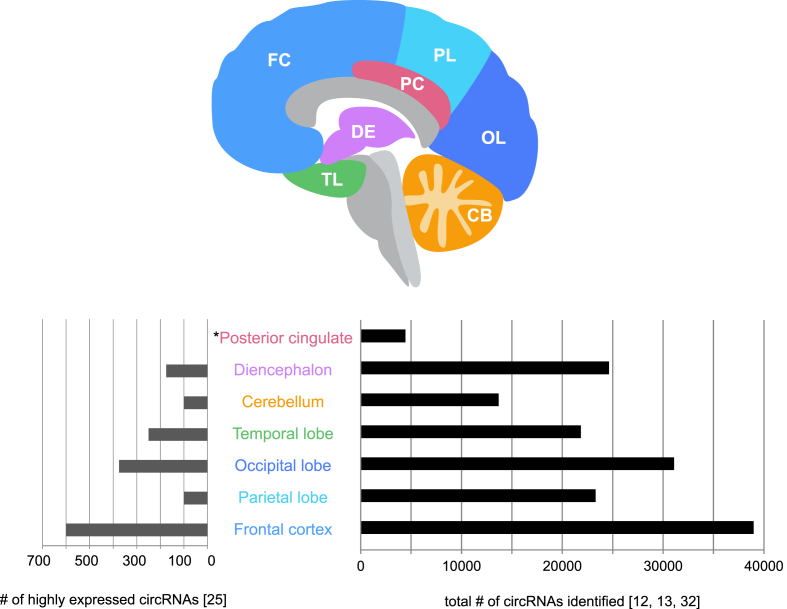
Recent studies have reported preferential back-splicing of neural genes and increased expression of circRNAs in the mammalian brain compared to other tissues [10,13,25]. Analysis of human and mouse neuronal cell lines, as well as ENCODE datasets, identified more circRNAs in the mammalian brain compared to other tissues, including the lungs, heart, kidney, testis and spleen, such that 20% of the protein coding genes in the brain produced circRNAs [10]. In a separate landscape analysis of clinically-relevant tissues from a single human donor, 339 circRNAs were identified in the cerebral cortex, with 141 being unique to the cortex compared to 19 other tissues [26]. Several of these circRNAs are derived from host genes that regulate brain function. These genes include neurotrophic receptor tyrosine kinase 2 (NTRK2), reticulon 4 (RTN4), and homer scaffold protein 1 (HOMER1) [26]. NTRK2 encodes a membrane receptor that binds brain-derived neurotrophic factor (BDNF), which regulates neuronal morphology, synaptic plasticity [27], and the synaptic dysfunction of striatal neurons in Huntington's disease [28]. RTN4 encodes an inhibitor of axonal sprouting and regulates disease progression in an amyloid precursor protein (APP)-expressing transgenic mouse model of Alzheimer's disease (AD) [29]. Lastly, HOMER1, which encodes a post-synaptic density (PSD) protein, regulates neuronal survival following injury [30] and in AD [31]. Fig. 1 summarizes the landscape of circRNA expression in the human brain based on tissue analyses [12,13,25,32].
Fig. 1.
CircRNA expression in adult human brain tissue. Left plot: total number of circRNAs detected that demonstrated greater expression than their mRNA counterparts (>50% reads supporting circRNAs, P < 0.05) [25]. Right plot: total number of circRNAs detected. Data for the cerebellum were derived from two studies, and the median number of circRNAs is shown. *For the posterior cingulate (PC), the number of circRNAs identified in astrocytes is shown [32]. Evaluation of the number of circRNAs with greater expression than their mRNA counterparts was not performed for the PC data set. Diencephalon (DE) [12,13,25]; cerebellum (CB) [13,25]; temporal lobe (TL) [13,25]; occipital lobe (OL) [13,25]; parietal lobe (PL) [13,25]; frontal cortex (FC) [13,25].
2.1. CircRNA expression during brain development and aging
Brain circRNA expression levels are altered in development and aging. During embryonic development [25] and in the developing human foetal neocortex [33], over 6,000 total circRNAs were detected during the early and late stages of gestation, with 39% of genes expressing linear transcripts also expressing circRNAs [33]. Furthermore, 89,137 circRNAs were found in the human foetal brain, compared to less than 13,400 circRNAs identified in other foetal tissue types and compared to 65,731 circRNAs in the adult human brain [13], suggesting that circRNAs may have a significant role in the developing brain [19]. Differences in circRNA expression have also been observed in the brains of adult (10 years old; n = 4, two females and two males) and aged (20 years old; n = 4, two females and two males) rhesus macaques [14]. From analysis of eight brain regions including the prefrontal cortex, posterior cingulate cortex, temporal cortex, parietal cortex, occipital cortex, hippocampus, dentate gyrus, and cerebellum, 17,050 circRNAs were identified. Of these, 3% were found to be aging-related based on differential expression between adult and aged brains. In addition to age-specific expression, gender-specific expression changes was also observed with the expression of circRNAs derived from male-specific host genes including circCD99 [34], circPREX1, and circTSPAN15 [35]. Additional gender-specific circRNAs included circEFCAB2 for males and circPCTP and circZNF484 for females.
In other species, 4,634 unique circRNAs have been identified in the porcine embryonic brain, with 20% of the splice sites involved in circRNA biogenesis being conserved in mice and humans [36]. Although as of yet, circRNAs have not been reported to be profiled in other porcine tissues or in the adult porcine brain, regulated patterns of circRNA expression were identified in the fetal porcine brain, with elevated circRNA abundance occurring in the cortex at day 60 of gestation [36]. A similar regulated pattern was also observed with aging in Drosophila heads for which there was an age-associated increase in the expression of over 250 circRNAs [37]. Similarly, elevated and dynamic expression of circRNAs has been observed during neuronal differentiation in human forebrain neuron progenitor cells [38]. Along these lines, with the role of senescence in aging pathways, the circRNA circPVT1 was found to be required for the expression of proteins that prevent senescence [39] to suggest a possible regulatory role of this circRNA in aging processes. These reports of evolutionarily conserved, dynamically regulated circRNA expression levels in the brain suggest that circRNAs are important for brain development, possibly with respect to formation of neural structures and cell growth and distribution, and as well as aging and normal brain functions by way of intricate transcriptional programs.
2.2. Cell-specificity of circRNA expression
While more research is necessary, circRNAs have demonstrated variable expression in different brain cell types. We previously described the expression of 4,438 circRNAs in posterior cingulate astrocytes in the brains of elderly subjects [32]. In silico analysis revealed that these circRNAs are derived from host genes that are involved with regulating immune function, a known role of astrocytes. In a separate study, a similar number of unique circRNAs was identified in primary mouse cortical neurons (n = 5,265), with 797 demonstrating decreased expression and 1,926 demonstrating increased expression during neuronal maturation [13]. The magnitude of circRNA up-regulation was lower in cultures containing large numbers of glial cells, suggesting that the observed up-regulation may be specific to neurons [13], or may be associated with neuron-neuron interactions. In other analyses, the ratio of circRNA expression levels to the levels of the corresponding linear transcript, as well as splice donor and acceptor usage, have been reported to be cell-specific [8,15,40]. However, circRNA expression was found to not be more cell-specific than expression of the corresponding linear transcript [41].
Neuronal studies further show specificity of circRNA expression in cellular compartments unique to this cell type. In synaptosomes and microdissected neuropil from mouse hippocampal slices, enrichment of circRNAs relative to host gene linear transcripts has been observed [10], however analyses in other brain regions are needed to determine if this relationship expands beyond the hippocampus. Furthermore, circRNAs have been identified in subcellular compartments including the cell body and dendrites [10], or more broadly in the cytoplasm [6,42]. This finding is not surprising given local and activity-dependent translation of mRNA in neurons and the potential role that circRNAs may have in regulating or influencing mRNA levels.
CircRNAs may also regulate the formation of neuron-specific subcellular structures. In murine hippocampal neurons, a postnatal shift in circRNA expression, independent of expression of host transcripts, was observed with the onset of synaptogenesis [10]. During this transition, expression of circRNAs derived from host genes encoding synaptic function proteins such as DLG associated protein 1 (Dlgap1), K(lysine) acetyltransferase 6B (Myst4), and kelch like family member 2 (Klhl2) were found to be up-regulated [10]. For Dlgap1, circRNA expression increased 20 fold between embryonic and post-natal stages, while only a 4 fold increase was observed for Dlgap1 mRNA. On the other hand, Myst4 and Klhl2 circRNAs demonstrated increased expression during development alongside decreased expression of their respective host mRNAs.
2.3. CircRNA expression outside of the brain
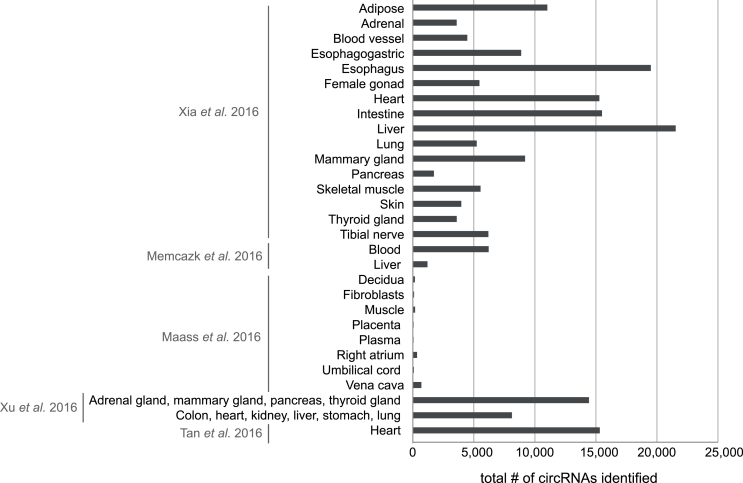
CircRNAs are also found outside the brain. They have been detected in heart, colon, kidney, liver, lung, and glandular tissues such as the adrenal gland, mammary gland, pancreas and thyroid gland (Fig. 2) [12,19,26,43,44]. Additionally, extracellular fluids such as plasma and serum contain circRNAs [12] (Fig. 2). Across tissue types, the number of circRNAs detected ranges from 57 in plasma [26] to 21,536 in the liver [19]. In comparison, in the adult human brain, anywhere from 6,289 to 38,983 circRNAs have been reported, highlighting the variability in circRNA expression across organs. Across all studies, both unique, tissue-specific circRNAs, and as well as common circRNAs, were identified with 33 ubiquitously expressed circRNAs identified in one tissue landscape analysis [44]. This study additionally observed that 36%–50% of circRNAs identified in each tissue were specific to that tissue, and that fetal tissue displayed up-regulated expression of circRNAs compared to adult tissue.
Fig. 2.
CircRNA expression in adult human non-brain tissues. Shown are the total numbers of circRNAs reported for each tissue using data from the indicated studies [12,19,26,43,44].
3. The spectrum of circRNA functions
Given the high abundance of circRNAs in the mammalian brain, circRNAs may play important roles in healthy brain function, aging, and neurological disease. Because they are found in synaptosomes, circRNAs may have roles in synaptic functions, or may be released into the synaptic cleft, possibly through vesicles, to influence neighbouring cells [45]. Further, both up-regulation and down-regulation of circRNA expression, without changes in host linear transcript expression, occurs in hippocampal neurons following excitatory neural activity [10]. Thus, circRNAs expression may have roles in healthy brain function, particularly learning and memory, due to their localization to synapses and their observed changes at excitatory synapses. Here we describe known and proposed molecular mechanisms for circRNA functions in the brain.
3.1. Transcriptional regulation
The most frequently reported functions of circRNAs are binding and sequestering miRs [20,46], thereby acting as “miR sponges”. Of note, in silico analyses suggest that circRNAs do not bind miRs or RNA binding proteins (RBPs) more efficiently than long non-coding RNAs [20,46] to suggest that any binding competition may not be influenced by differences in binding capacity. Despite these findings, experimental evidence shows that circRNAs regulate miR activity. For example, the circRNA sponge for miR-7 (CDR1as, or ciRS-7), which is elevated in the mammalian brain, is a known miR regulator [10,41]. Using photoactivatable-ribonucleoside-enhanced crosslinking, immuno-precipitation, and miRNA seed matching analyses, CDR1as was observed to harbour over 70 binding sites for the miR-7 effector protein argonaute (AGO), and may bind up to 20,000 miR-7 transcripts per cell [20,46,47]. Notably, the interaction between CDR1as and miR-7 has been proposed to be brain-specific and both are co-expressed in the mesencephalon [20]. Further, overexpression of CDR1as in zebrafish decreased midbrain size, while CDR1as knockout down-regulated miR-7 targets in HEK293 cells [20].
In the context of brain aging, Xu et al. reported expression of specific circRNAs involved in aging processes with evidence of circRNA transcriptional regulation in rhesus macaques [14]. Seventy-three percent of circRNAs derived from host genes involved in calcium homeostasis and synaptic plasticity functions demonstrated aging-specific differential expression. Deeper investigation led to the observation that knocking down expression of two highly expressed circRNAs whose host genes are involved in calcium signalling (circ-CACNA2D1 [L-type Ca2+ channel], circ-CACNA1E [R-type Ca2+ channel]) resulted in increased expression of the respective calcium channel genes in cultured rhesus macaque hippocampal neurons to suggest that the circRNAs may directly regulate mRNAs. Due to the central role of calcium signalling in neuronal function, circRNA regulation of calcium signalling gene expression may significantly impact both normal brain functions and as well as aging processes. Such findings provide further credence towards circRNA transcriptional regulation and the potential role of circRNAs in the aging brain.
In other studies, the molecular mechanisms linking other circRNAs to transcriptional regulation are less clear. For instance, circRNAs encoded in the sex-determining region Y gene (circSRY) [41,46] and the E3 ubiquitin-protein ligase gene (ciR-ITCH) [41,46], as well as circ-001569 [48], and circRNA-CER [49], have been shown to bind miRs to provide evidence that they may act as “miR sponges.” However, it is not clear if such interactions impact transcriptional regulation. Despite this gap, these circRNAs have been found to impact multiple pathways including chondrocyte extracellular matrix degradation [49], cell proliferation and invasion in colorectal cancer [48], and the Wnt/Beta-catenin pathway in cancers [50,51].
Multiple mechanisms have been proposed for circRNA function independent of miR or mRNA regulation. Possible mechanisms supported by experimental data include binding and regulating RNA polymerase II (Pol II) in the nucleus [16,22], binding small nuclear ribonucleoprotein complexes to up-regulate transcription of specific genes [22,52], and enabling the formation of a diverse set of transcripts during circRNA back-splicing [[21], [22], [23],53]. Based on demonstrated interactions between circRNAs and Pol II, circRNAs may also bind proteins outside the transcriptional machinery to regulate other cellular functions. For example, intronic circRNAs were found to bind and sequester TDP-43, a protein that accumulates in degenerating motor neurons in amyotrophic lateral sclerosis, to prevent the pathogenic effects of TDP-43 aggregation [54]. As circRNAs have been identified in various subcellular compartments, it has been theorized that circRNAs may bind RBPs to reach their destination in the cell [55], or they may help to organize RNA-protein complexes [13,55]. In addition, the circRNA circ-Foxo3 has been found to bind and sequester stress proteins in the cytoplasm to influence senescence and stress pathways [56]. Lastly, as circRNAs have been reported to be co-expressed alongside linear transcripts, their transcription alone may introduce competition and suppress the formation of mRNAs from the respective host gene.
3.2. CircRNA translation
One controversial potential mechanism of circRNA function is through direct protein translation of the circRNA. Evaluation of rat brains revealed that no ribosomal-bound RNA sequences mapped to circRNA junctions and polysome profiling of mouse brains revealed that circRNAs were depleted in ribosome/polysome-bound fractions and enriched in non-ribosomal RNA fractions [10]. Similarly, analysis of human fibroblasts revealed that exonic circRNAs containing the translation start site sequence did not bind ribosomes or polysomes, suggesting that exonic circRNAs are not translated [6]. Also, in silico analysis of RNAseq data from a human osteosarcoma cell line showed that ribosome-protected fragments did not align to circRNA junctions [41]. Despite these findings, evidence for circRNA translation has been observed in HEK293 cells [57], E. coli [58], and HeLa cells [59]. In the latter two studies, circRNAs were synthesized with an infinite open reading frame and resulted in the translation of proteins. The E. coli study further revealed that the synthesized circRNA generated more protein compared to its linear counterpart by two orders of magnitude [58]. While more evidence of circRNA translation is needed, it remains a possibility that translation may occur despite the absence of a cap and internal ribosome entry site [60], creating a putative non-canonical mechanism for circRNA translation. Although evidence for circRNA translation has yet to be demonstrated in the mammalian brain, given the localization of circRNAs to synapses and activity-dependent translation that occurs at that location, the occurrence of such processing remains a possibility.
4. CircRNAs in neurological diseases and conditions
Additional studies using animal models and human tissues support a role for circRNAs in neurological disease. For instance, the circRNA CDR1as may play roles in neuropsychiatric and neurodegenerative diseases, and as well as in brain tumours. In a recent study, Piwecka et al. showed that Cdr1as knockout mice display behavioural phenotypes associated with neuropsychiatric disorders [84]. Furthermore, the expression of CDR1as was reduced in hippocampal CA1 samples from sporadic AD patients compared to controls [85]. This reduction in CDR1as levels was hypothesized to increase miR-7 levels and reduce expression of miR-7 targets such as ubiquitin protein ligase A (UBE2A), which is involved in clearing amyloid in AD brains [86]. In further support of a role for CDR1as in AD, in vitro analysis showed that CDR1as, or ciRS-7, drives degradation of APP and beta-secretase 1 (BACE1), and that APP decreases expression of CDR1as [87]. While miR-7 was not specifically evaluated in this study, miR-7 regulation by CDR1as may have roles in other neurological diseases. For instance, miR-7 has been suggested to play a role in Parkinson's disease by decreasing expression of alpha-synuclein [88], and miR-7 was reported to suppress glioblastoma growth by regulating the PI3K/ATK and Raf/MEK/ERK pathways [89]. However, the role of circRNAs in glioblastoma is further complicated by evidence showing decreased circRNA expression in human glioblastoma compared to healthy controls [90]. Lastly, in multiple system atrophy, another alpha-synucleinopathy for which alpha-synuclein accumulates in oligodendrocytes, RNAseq analysis of the frontal cortex revealed five differentially expressed circRNAs that were not coupled to expression changes in the linear host transcript [91]. These circRNAs were derived from the IQ motif containing K (IQCK), mitogen-activated protein kinase kinase kinase kinase 3 (MAP4K3), EF-hand calcium binding domain 11 (EFCAB11), dystrobrevin alpha (DTNA), and multiple C2 and transmembrane domain containing 1 (MCTP1) host genes, all of which may be associated with neural processes or conditions [91].
In addition to CDR1as, other circRNAs have been reported to be associated with AD. Huang et al. used microarray technology to identify circRNA expression changes in the hippocampus of five- and ten-month-old senescence-accelerated mice P8 (SAMP8) mice, an AD mouse model [92]. Through this analysis, they identified over 300 dysregulated circRNAs and also identified the mmu_circRNA_017963 circRNA, which is involved in multiple processes including apoptosis and synaptic function [92]. Microarray technology was also used in a separate study to simultaneously characterize circRNAs, miRs, and mRNAs in the hippocampus of an AD rat model [93]. In this study, 555 circRNAs were identified and potential regulatory networks constructed using the identified circRNAs, miRs, and mRNAs, were used to create a mechanistic model for AD pathogenesis in the hippocampus. Although these investigations were performed in AD models and human tissue evaluation is still needed, they are the first to describe hippocampal circRNA changes in AD, and thereby establishes a framework for interpreting the impact of circRNAs in AD.
Another circRNA with a possible role in brain function and neurological disease is circHomer1. In one study, circHomer1 expression was elevated after synaptic plasticity induction in primary hippocampal neurons [10]. Since circRNA transcription may compete for transcription with host gene mRNA, over-expression of circHomer1 may decrease expression of Homer 1b/c, whose expression normally inhibits plasticity [10,94]. With this loss of function, circHomer1 may putatively facilitate synaptic plasticity. Also, in rat cortical neuron cultures, knockdown of Homer1b/c expression decreased apoptosis following neuronal injury [30]. Thus, expression of circHomer1 and its impact on Homer1b/c expression may influence plasticity and neuroprotective mechanisms.
5. CircRNAs as neurological disease biomarkers
CircRNAs are promising biomarkers due to their stability, cell-specific expression patterns, and capacity for conditional expression [61]. CircRNAs have been identified in various biofluids that may be collected non-invasively, including blood [12,[62], [63], [64], [65], [66], [67], [68]], saliva [69,70], and plasma [71], and are also present in exosomes [[72], [73], [74]]. Therefore, they have been proposed to serve as biomarkers for multiple illnesses including lupus erythematosus [71], tuberculosis [[63], [64], [65]], rheumatoid arthritis [66], diabetes [67,68,75], and cancer [67,68,75]. Additionally, analysis of peripheral blood mononuclear cells in patients with major depressive disorder revealed differential expression of has_circRNA_103636 following an eight week antidepressant treatment regimen, suggesting that this circRNA could be a treatment biomarker [76]. Aside from this study, the potential uses of circRNAs as biomarkers in neurological diseases have not been well explored. Identifying biomarkers of brain disease is important due to the inaccessibility of brain tissue for biopsy. The potential for development of circRNA biomarkers for neurological disease is also compelling due to the presence of the blood-brain barrier, which regulates the movement of molecules between the peripheral circulation and the brain. In a disease state such as in AD, stroke, and ALS, the blood-brain barrier may be compromised [[77], [78], [79], [80], [81], [82]], thereby enabling free circRNAs or exosome-encapsulated circRNAs [83] to escape from the brain and be detected peripherally. Thus, brain-specific circRNAs may be released into the circulation during neurological disease. As a result, it may be possible to correlate the blood levels of these circRNAs with disease progression or response to treatment.
6. Future directions in circRNA research
Studies in the past few years have significantly increased our understanding of circRNA expression and function. Technical advances, such as the development of RNAseq and the creation of bioinformatics approaches to detect circRNAs, have significantly facilitated these discoveries. Opportunities for further investigation include analysing human samples to identify circRNAs associated with neurological disease and performing functional experiments, such as through knockdown of expression of specific circRNAs, to determine how circRNAs regulate cellular pathways. It will also be important to determine if RNA modification of circRNAs occurs and how such features may impact circRNA function, and to develop new bioinformatics approaches since existing algorithms are based on currently known models of molecular mechanisms, and such models may change over time. For example, it is possible that circRNAs biogenesis does not universally follow canonical splicing principles such that existing bioinformatics algorithms for circRNA detection would need to be revised. Continued exploration into circRNAs will help us to better understand the heterogeneous and dynamic aspects of brain function and design new strategies for treating neurological disease.
Acknowledgements
We thank the Translational Genomics Research Institute for support, as well as the Arizona Alzheimer's Disease Core Center through the National Institute on Aging (NIA) of the National Institutes of Health (NIH) under award number P30AG019610, the state of Arizona, and the Arizona Department of Health Services (ADHS14-052688). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. The funders had no role in the data collection and analysis, decision to publish, or preparation of the manuscript. Nancy Linford, PhD provided editing services.
Contributor Information
Shobana Sekar, Email: ssekar@tgen.org.
Winnie S. Liang, Email: wliang@tgen.org.
References
- 1.Sanger H.L., Klotz G., Riesner D., Gross H.J., Kleinschmidt A.K. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc. Natl. Acad. Sci. U. S. A. 1976;73(11):3852–3856. doi: 10.1073/pnas.73.11.3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cocquerelle C., Mascrez B., Hetuin D., Bailleul B. Mis-splicing yields circular RNA molecules. FASEB J. 1993;7(1):155–160. doi: 10.1096/fasebj.7.1.7678559. [DOI] [PubMed] [Google Scholar]
- 3.Nigro J.M., Cho K.R., Fearon E.R., Kern S.E., Ruppert J.M., Oliner J.D., Kinzler K.W., Vogelstein B. Scrambled exons. Cell. 1991;64(3):607–613. doi: 10.1016/0092-8674(91)90244-s. [DOI] [PubMed] [Google Scholar]
- 4.Kos A., Dijkema R., Arnberg A.C., van der Meide P.H., Schellekens H. The hepatitis delta (delta) virus possesses a circular RNA. Nature. 1986;323(6088):558–560. doi: 10.1038/323558a0. doi: 510.1038/323558a323550. [DOI] [PubMed] [Google Scholar]
- 5.Chen I., Chen C.Y., Chuang T.J. vol. 6. Wiley Interdiscip Rev RNA; 2015. Biogenesis, Identification, and Function of Exonic Circular RNAs; pp. 563–579. (5). doi: 510.1002/wrna.1294. Epub 2015 Jul 1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jeck W.R., Sorrentino J.A., Wang K., Slevin M.K., Burd C.E., Liu J., Marzluff W.F., Sharpless N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. Rna. 2013;19(2):141–157. doi: 10.1261/rna.035667.112. doi: 110.1261/rna.035667.035112. Epub 032012 Dec 035618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Enuka Y., Lauriola M., Feldman M.E., Sas-Chen A., Ulitsky I., Yarden Y. Circular RNAs are long-lived and display only minimal early alterations in response to a growth factor. Nucleic Acids Res. 2016;44(3):1370–1383. doi: 10.1093/nar/gkv1367. doi: 1310.1093/nar/gkv1367. Epub 2015 Dec 1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Salzman J., Chen R.E., Olsen M.N., Wang P.L., Brown P.O. Cell-type specific features of circular RNA expression. PLoS Genet. 2013;9(9) doi: 10.1371/journal.pgen.1003777. e1003777. doi: 1003710.1001371/journal.pgen.1003777. Epub 1002013 Sep 1003775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Suzuki H., Zuo Y., Wang J., Zhang M.Q., Malhotra A., Mayeda A. Characterization of RNase R-digested cellular RNA source that consists of lariat and circular RNAs from pre-mRNA splicing. Nucleic Acids Res. 2006;34(8):e63. doi: 10.1093/nar/gkl151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.You X., Vlatkovic I., Babic A., Will T., Epstein I., Tushev G., Akbalik G., Wang M., Glock C., Quedenau C. Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat. Neurosci. 2015;18(4):603–610. doi: 10.1038/nn.3975. doi: 610.1038/nn.3975. Epub 2015 Feb 1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lasda E., Parker R. Circular RNAs: diversity of form and function. Rna. 2014;20(12):1829–1842. doi: 10.1261/rna.047126.114. doi: 1810.1261/rna.047126.047114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Memczak S., Papavasileiou P., Peters O., Rajewsky N. Identification and characterization of circular RNAs as a new class of putative biomarkers in human blood. PLoS One. 2015;10(10) doi: 10.1371/journal.pone.0141214. e0141214. doi: 0141210.0141371/journal.pone.0141214. eCollection 0142015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rybak-Wolf A., Stottmeister C., Glazar P., Jens M., Pino N., Giusti S., Hanan M., Behm M., Bartok O., Ashwal-Fluss R. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol. Cell. 2015;58(5):870–885. doi: 10.1016/j.molcel.2015.03.027. doi: 810.1016/j.molcel.2015.1003.1027. Epub 2015 Apr 1023. [DOI] [PubMed] [Google Scholar]
- 14.Xu K., Chen D., Wang Z., Ma J., Zhou J., Chen N., Lv L., Zheng Y., Hu X., Zhang Y. Annotation and functional clustering of circRNA expression in rhesus macaque brain during aging. Cell Discov. 2018;4:48. doi: 10.1038/s41421-018-0050-1. eCollection 42018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Starke S., Jost I., Rossbach O., Schneider T., Schreiner S., Hung L.H., Bindereif A. Exon circularization requires canonical splice signals. Cell Rep. 2015;10(1):103–111. doi: 10.1016/j.celrep.2014.12.002. doi: 110.1016/j.celrep.2014.1012.1002. Epub 2014 Dec 1024. [DOI] [PubMed] [Google Scholar]
- 16.Zhang Y., Zhang X.O., Chen T., Xiang J.F., Yin Q.F., Xing Y.H., Zhu S., Yang L., Chen L.L. Circular intronic long noncoding RNAs. Mol. Cell. 2013;51(6):792–806. doi: 10.1016/j.molcel.2013.08.017. doi: 710.1016/j.molcel.2013.1008.1017. Epub 2013 Sep 1012. [DOI] [PubMed] [Google Scholar]
- 17.Floris G., Zhang L., Follesa P., Sun T. Regulatory role of circular RNAs and neurological disorders. Mol. Neurobiol. 2017;54(7):5156–5165. doi: 10.1007/s12035-016-0055-4. doi: 5110.1007/s12035-12016-10055-12034. Epub 12016 Aug 12024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ebbesen K.K., Kjems J., Hansen T.B. Circular RNAs: identification, biogenesis and function. Biochim. Biophys. Acta. 2016;1859(1):163–168. doi: 10.1016/j.bbagrm.2015.07.007. doi: 110.1016/j.bbagrm.2015.1007.1007. Epub 2015 Jul 1011. [DOI] [PubMed] [Google Scholar]
- 19.Xia S., Feng J., Lei L., Hu J., Xia L., Wang J., Xiang Y., Liu L., Zhong S., Han L. Comprehensive characterization of tissue-specific circular RNAs in the human and mouse genomes. Briefings Bioinf. 2017;18(6):984–992. doi: 10.1093/bib/bbw081. doi: 910.1093/bib/bbw1081. [DOI] [PubMed] [Google Scholar]
- 20.Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A., Maier L., Mackowiak S.D., Gregersen L.H., Munschauer M. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338. doi: 10.1038/nature11928. doi: 310.1038/nature11928. Epub 12013 Feb 11927. [DOI] [PubMed] [Google Scholar]
- 21.Ashwal-Fluss R., Meyer M., Pamudurti N.R., Ivanov A., Bartok O., Hanan M., Evantal N., Memczak S., Rajewsky N., Kadener S. circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell. 2014;56(1):55–66. doi: 10.1016/j.molcel.2014.08.019. Epub 2014 Sep 1018. [DOI] [PubMed] [Google Scholar]
- 22.Chen L.L. The biogenesis and emerging roles of circular RNAs. Nat. Rev. Mol. Cell Biol. 2016;17(4):205–211. doi: 10.1038/nrm.2015.32. doi: 210.1038/nrm.2015.1032. Epub 2016 Feb 1024. [DOI] [PubMed] [Google Scholar]
- 23.Kelly S., Greenman C., Cook P.R., Papantonis A. Exon skipping is correlated with exon circularization. J. Mol. Biol. 2015;427(15):2414–2417. doi: 10.1016/j.jmb.2015.02.018. doi: 2410.1016/j.jmb.2015.2402.2018. Epub 2015 Feb 2426. [DOI] [PubMed] [Google Scholar]
- 24.Gruner H., Cortes-Lopez M., Cooper D.A., Bauer M., Miura P. CircRNA accumulation in the aging mouse brain. Sci. Rep. 2016;6:38907. doi: 10.1038/srep38907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Szabo L., Morey R., Palpant N.J., Wang P.L., Afari N., Jiang C., Parast M.M., Murry C.E., Laurent L.C., Salzman J. Statistically based splicing detection reveals neural enrichment and tissue-specific induction of circular RNA during human fetal development. Genome Biol. 2015;16 doi: 10.1186/s13059-015-0690-5. 126-015-0690-0695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maass P.G., Glazar P., Memczak S., Dittmar G., Hollfinger I., Schreyer L., Sauer A.V., Toka O., Aiuti A., Luft F.C. A map of human circular RNAs in clinically relevant tissues. J. Mol. Med. (Berl.) 2017;95(11):1179–1189. doi: 10.1007/s00109-017-1582-9. doi: 1110.1007/s00109-00017-01582-00109. Epub 02017 Aug 00125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang J., Harte-Hargrove L.C., Siao C.J., Marinic T., Clarke R., Ma Q., Jing D., Lafrancois J.J., Bath K.G., Mark W. proBDNF negatively regulates neuronal remodeling, synaptic transmission, and synaptic plasticity in hippocampus. Cell Rep. 2014;7(3):796–806. doi: 10.1016/j.celrep.2014.03.040. doi: 710.1016/j.celrep.2014.1003.1040. Epub 2014 Apr 1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Plotkin J.L., Day M., Peterson J.D., Xie Z., Kress G.J., Rafalovich I., Kondapalli J., Gertler T.S., Flajolet M., Greengard P. Impaired TrkB receptor signaling underlies corticostriatal dysfunction in Huntington's disease. Neuron. 2014;83(1):178–188. doi: 10.1016/j.neuron.2014.05.032. doi: 110.1016/j.neuron.2014.1005.1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Masliah E., Xie F., Dayan S., Rockenstein E., Mante M., Adame A., Patrick C.M., Chan A.F., Zheng B. Genetic deletion of Nogo/Rtn4 ameliorates behavioral and neuropathological outcomes in amyloid precursor protein transgenic mice. Neuroscience. 2010;169(1):488–494. doi: 10.1016/j.neuroscience.2010.04.045. doi: 410.1016/j.neuroscience.2010.1004.1045. Epub 2010 Apr 1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fei F., Rao W., Zhang L., Chen B.G., Li J., Fei Z., Chen Z. Downregulation of Homer1b/c improves neuronal survival after traumatic neuronal injury. Neuroscience. 2014;267:187–194. doi: 10.1016/j.neuroscience.2014.02.037. Epub 2014 Mar 1015. [DOI] [PubMed] [Google Scholar]
- 31.Luo P., Li X., Fei Z., Poon W. Scaffold protein Homer 1: implications for neurological diseases. Neurochem. Int. 2012;61(5):731–738. doi: 10.1016/j.neuint.2012.06.014. doi: 710.1016/j.neuint.2012.1006.1014. Epub 2012 Jun 1027. [DOI] [PubMed] [Google Scholar]
- 32.Sekar S., Cuyugan L., Adkins J., Geiger P., Liang W.S. Circular RNA expression and regulatory network prediction in posterior cingulate astrocytes in elderly subjects. BMC Genomics. 2018;19(1):340. doi: 10.1186/s12864-018-4670-5. doi: 310.1186/s12864-12018-14670-12865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen B.J., Huang S., Janitz M. Changes in circular RNA expression patterns during human foetal brain development. Genomics. 2018 doi: 10.1016/j.ygeno.2018.04.015. [epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 34.Weickert C.S., Elashoff M., Richards A.B., Sinclair D., Bahn S., Paabo S., Khaitovich P., Webster M.J. Transcriptome analysis of male-female differences in prefrontal cortical development. Mol. Psychiatr. 2009;14(6):558–561. doi: 10.1038/mp.2009.5. doi: 510.1038/mp.2009.1035. [DOI] [PubMed] [Google Scholar]
- 35.Kang H.J., Kawasawa Y.I., Cheng F., Zhu Y., Xu X., Li M., Sousa A.M., Pletikos M., Meyer K.A., Sedmak G. Spatio-temporal transcriptome of the human brain. Nature. 2011;478(7370):483–489. doi: 10.1038/nature10523. doi: 410.1038/nature10523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Veno M.T., Hansen T.B., Veno S.T., Clausen B.H., Grebing M., Finsen B., Holm I.E., Kjems J. Spatio-temporal regulation of circular RNA expression during porcine embryonic brain development. Genome Biol. 2015;16:245. doi: 10.1186/s13059-015-0801-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Westholm J.O., Miura P., Olson S., Shenker S., Joseph B., Sanfilippo P., Celniker S.E., Graveley B.R., Lai E.C. Genome-wide analysis of drosophila circular RNAs reveals their structural and sequence properties and age-dependent neural accumulation. Cell Rep. 2014;9(5):1966–1980. doi: 10.1016/j.celrep.2014.10.062. doi: 1910.1016/j.celrep.2014.1910.1062. Epub 2014 Nov 1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Y., Xue W., Li X., Zhang J., Chen S., Zhang J.L., Yang L., Chen L.L. The biogenesis of nascent circular RNAs. Cell Rep. 2016;15(3):611–624. doi: 10.1016/j.celrep.2016.03.058. doi: 610.1016/j.celrep.2016.1003.1058. Epub 2016 Apr 1017. [DOI] [PubMed] [Google Scholar]
- 39.Panda A.C., Grammatikakis I., Kim K.M., De S., Martindale J.L., Munk R., Yang X., Abdelmohsen K., Gorospe M. Identification of senescence-associated circular RNAs (SAC-RNAs) reveals senescence suppressor CircPVT1. Nucleic Acids Res. 2017;45(7):4021–4035. doi: 10.1093/nar/gkw1201. doi: 4010.1093/nar/gkw1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liang D., Wilusz J.E. Short intronic repeat sequences facilitate circular RNA production. Genes Dev. 2014;28(20):2233–2247. doi: 10.1101/gad.251926.114. doi: 2210.1101/gad.251926.251114. Epub 252014 Oct 251923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guo J.U., Agarwal V., Guo H., Bartel D.P. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014;15(7):409. doi: 10.1186/s13059-014-0409-z. doi: 410.1186/s13059-13014-10409-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Salzman J., Gawad C., Wang P.L., Lacayo N., Brown P.O. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One. 2012;7(2) doi: 10.1371/journal.pone.0030733. e30733. doi: 30710.31371/journal.pone.0030733. Epub 0032012 Feb 0030731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tan W.L., Lim B.T., Anene-Nzelu C.G., Ackers-Johnson M., Dashi A., See K., Tiang Z., Lee D.P., Chua W.W., Luu T.D. A landscape of circular RNA expression in the human heart. Cardiovasc. Res. 2016;113(3):298–309. doi: 10.1093/cvr/cvw250. [DOI] [PubMed] [Google Scholar]
- 44.Xu T., Wu J., Han P., Zhao Z., Song X. Circular RNA expression profiles and features in human tissues: a study using RNA-seq data. BMC Genomics. 2017;18(Suppl 6):680. doi: 10.1186/s12864-017-4029-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Rossum D., Verheijen B.M., Pasterkamp R.J. Circular RNAs: novel regulators of neuronal development. Front. Mol. Neurosci. 2016;9:74. doi: 10.3389/fnmol.2016.00074. eCollection 02016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hansen T.B., Jensen T.I., Clausen B.H., Bramsen J.B., Finsen B., Damgaard C.K., Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388. doi: 10.1038/nature11993. doi: 310.1038/nature11993. Epub 12013 Feb 11927. [DOI] [PubMed] [Google Scholar]
- 47.Hansen T.B., Wiklund E.D., Bramsen J.B., Villadsen S.B., Statham A.L., Clark S.J., Kjems J. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J. 2011;30(21):4414–4422. doi: 10.1038/emboj.2011.359. doi: 4410.1038/emboj.2011.4359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xie H., Ren X., Xin S., Lan X., Lu G., Lin Y., Yang S., Zeng Z., Liao W., Ding Y.Q. Emerging roles of circRNA_001569 targeting miR-145 in the proliferation and invasion of colorectal cancer. Oncotarget. 2016;7(18):26680–26691. doi: 10.18632/oncotarget.8589. doi: 26610.18632/oncotarget.28589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu Q., Zhang X., Hu X., Dai L., Fu X., Zhang J., Ao Y. Circular RNA related to the chondrocyte ECM regulates MMP13 expression by functioning as a MiR-136 'sponge' in human cartilage degradation. Sci. Rep. 2016;6:22572. doi: 10.1038/srep22572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li F., Zhang L., Li W., Deng J., Zheng J., An M., Lu J., Zhou Y. Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/beta-catenin pathway. Oncotarget. 2015;6(8):6001–6013. doi: 10.18632/oncotarget.3469. doi: 6010.18632/oncotarget.13469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huang G., Zhu H., Shi Y., Wu W., Cai H., Chen X. cir-ITCH plays an inhibitory role in colorectal cancer by regulating the Wnt/beta-catenin pathway. PLoS One. 2015;10(6) doi: 10.1371/journal.pone.0131225. e0131225. doi: 0131210.0131371/journal.pone.0131225. eCollection 0132015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hindi N., Casali P.G., Morosi C., Messina A., Palassini E., Pilotti S., Tamborini E., Radaelli S., Gronchi A., Stacchiotti S. Imatinib in advanced chordoma: a retrospective case series analysis. Eur. J. Cancer. 2015;51(17):2609–2614. doi: 10.1016/j.ejca.2015.07.038. doi: 2610.1016/j.ejca.2015.2607.2038. Epub 2015 Aug 2614. [DOI] [PubMed] [Google Scholar]
- 53.Zhang X.O., Wang H.B., Zhang Y., Lu X., Chen L.L., Yang L. Complementary sequence-mediated exon circularization. Cell. 2014;159(1):134–147. doi: 10.1016/j.cell.2014.09.001. doi: 110.1016/j.cell.2014.1009.1001. Epub 2014 Sep 1018. [DOI] [PubMed] [Google Scholar]
- 54.Armakola M., Higgins M.J., Figley M.D., Barmada S.J., Scarborough E.A., Diaz Z., Fang X., Shorter J., Krogan N.J., Finkbeiner S. Inhibition of RNA lariat debranching enzyme suppresses TDP-43 toxicity in ALS disease models. Nat. Genet. 2012;44(12):1302–1309. doi: 10.1038/ng.2434. doi: 1310.1038/ng.2434. Epub 2012 Oct 1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cortes-Lopez M., Miura P. Emerging functions of circular RNAs. Yale J. Biol. Med. 2016;89(4):527–537. eCollection 2016 Dec. [PMC free article] [PubMed] [Google Scholar]
- 56.Du W.W., Yang W., Chen Y., Wu Z.K., Foster F.S., Yang Z., Li X., Yang B.B. Foxo3 circular RNA promotes cardiac senescence by modulating multiple factors associated with stress and senescence responses. Eur. Heart J. 2017;38(18):1402–1412. doi: 10.1093/eurheartj/ehw001. doi: 1410.1093/eurheartj/ehw1001. [DOI] [PubMed] [Google Scholar]
- 57.Li X.F., Lytton J. A circularized sodium-calcium exchanger exon 2 transcript. J. Biol. Chem. 1999;274(12):8153–8160. doi: 10.1074/jbc.274.12.8153. [DOI] [PubMed] [Google Scholar]
- 58.Abe N., Hiroshima M., Maruyama H., Nakashima Y., Nakano Y., Matsuda A., Sako Y., Ito Y., Abe H. Rolling circle amplification in a prokaryotic translation system using small circular RNA. Angew Chem. Int. Ed. Engl. 2013;52(27):7004–7008. doi: 10.1002/anie.201302044. doi: 7010.1002/anie.201302044. Epub 201302013 May 201302028. [DOI] [PubMed] [Google Scholar]
- 59.Abe N., Matsumoto K., Nishihara M., Nakano Y., Shibata A., Maruyama H., Shuto S., Matsuda A., Yoshida M., Ito Y. Rolling circle translation of circular RNA in living human cells. Sci. Rep. 2015;5:16435. doi: 10.1038/srep16435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shatsky I.N., Dmitriev S.E., Terenin I.M., Andreev D.E. Cap- and IRES-independent scanning mechanism of translation initiation as an alternative to the concept of cellular IRESs. Mol. Cell. 2010;30(4):285–293. doi: 10.1007/s10059-010-0149-1. doi: 210.1007/s10059-10010-10149-10051. Epub 12010 Oct 10014. [DOI] [PubMed] [Google Scholar]
- 61.Abu N., Jamal R. Circular RNAs as promising biomarkers: a mini-review. Front. Physiol. 2016;7:355. doi: 10.3389/fphys.2016.00355. eCollection 02016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang Y.G., Yang H.L., Long Y., Li W.L. Circular RNA in blood corpuscles combined with plasma protein factor for early prediction of pre-eclampsia. Bjog. 2016;123(13):2113–2118. doi: 10.1111/1471-0528.13897. doi: 2110.1111/1471-0528.13897. Epub 12016 Feb 13895. [DOI] [PubMed] [Google Scholar]
- 63.Huang Z.K., Yao F.Y., Xu J.Q., Deng Z., Su R.G., Peng Y.P., Luo Q., Li J.M. Microarray expression profile of circular RNAs in peripheral blood mononuclear cells from active tuberculosis patients. Cell. Physiol. Biochem. 2018;45(3):1230–1240. doi: 10.1159/000487454. doi: 1210.1159/000487454. Epub 000482018 Feb 000487459. [DOI] [PubMed] [Google Scholar]
- 64.Qian Z., Liu H., Li M., Shi J., Li N., Zhang Y., Zhang X., Lv J., Xie X., Bai Y. Potential diagnostic power of blood circular RNA expression in active pulmonary tuberculosis. EBioMedicine. 2018;27:18–26. doi: 10.1016/j.ebiom.2017.12.007. Epub 2017 Dec 1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhuang Z.G., Zhang J.A., Luo H.L., Liu G.B., Lu Y.B., Ge N.H., Zheng B.Y., Li R.X., Chen C., Wang X. The circular RNA of peripheral blood mononuclear cells: hsa_circ_0005836 as a new diagnostic biomarker and therapeutic target of active pulmonary tuberculosis. Mol. Immunol. 2017;90:264–272. doi: 10.1016/j.molimm.2017.08.008. Epub 2017 Aug 1030. [DOI] [PubMed] [Google Scholar]
- 66.Ouyang Q., Wu J., Jiang Z., Zhao J., Wang R., Lou A., Zhu D., Shi G.P., Yang M. Microarray expression profile of circular RNAs in peripheral blood mononuclear cells from rheumatoid arthritis patients. Cell. Physiol. Biochem. 2017;42(2):651–659. doi: 10.1159/000477883. doi: 610.1159/000477883. Epub 000472017 Jun 000477815. [DOI] [PubMed] [Google Scholar]
- 67.Fang Y., Wang X., Li W., Han J., Jin J., Su F., Zhang J., Huang W., Xiao F., Pan Q. Screening of circular RNAs and validation of circANKRD36 associated with inflammation in patients with type 2 diabetes mellitus. Int. J. Mol. Med. 2018;42(4):1865–1874. doi: 10.3892/ijmm.2018.3783. doi: 1810.3892/ijmm.2018.3783. Epub 2018 Jul 1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhao Z., Li X., Jian D., Hao P., Rao L., Li M. Hsa_circ_0054633 in peripheral blood can be used as a diagnostic biomarker of pre-diabetes and type 2 diabetes mellitus. Acta Diabetol. 2017;54(3):237–245. doi: 10.1007/s00592-016-0943-0. doi: 210.1007/s00592-00016-00943-00590. Epub 02016 Nov 00523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lin X., Lo H.C., Wong D.T., Xiao X. Noncoding RNAs in human saliva as potential disease biomarkers. Front. Genet. 2015;6:175. doi: 10.3389/fgene.2015.00175. eCollection 02015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bahn J.H., Zhang Q., Li F., Chan T.M., Lin X., Kim Y., Wong D.T., Xiao X. The landscape of microRNA, Piwi-interacting RNA, and circular RNA in human saliva. Clin. Chem. 2015;61(1):221–230. doi: 10.1373/clinchem.2014.230433. doi: 210.1373/clinchem.2014.230433. Epub 232014 Nov 230436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li H., Li K., Lai W., Li X., Wang H., Yang J., Chu S., Kang C., Qiu Y. Comprehensive circular RNA profiles in plasma reveals that circular RNAs can be used as novel biomarkers for systemic lupus erythematosus. Clin. Chim. Acta. 2018;480:17–25. doi: 10.1016/j.cca.2018.01.026. Epub 2018 Feb 1020. [DOI] [PubMed] [Google Scholar]
- 72.Li J., Li Z., Jiang P., Peng M., Zhang X., Chen K., Liu H., Bi H., Liu X., Li X. Circular RNA IARS (circ-IARS) secreted by pancreatic cancer cells and located within exosomes regulates endothelial monolayer permeability to promote tumor metastasis. J. Exp. Clin. Canc. Res. 2018;37(1):177. doi: 10.1186/s13046-018-0822-3. doi: 110.1186/s13046-13018-10822-13043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li Z., Yanfang W., Li J., Jiang P., Peng T., Chen K., Zhao X., Zhang Y., Zhen P., Zhu J. Tumor-released exosomal circular RNA PDE8A promotes invasive growth via the miR-338/MACC1/MET pathway in pancreatic cancer. Cancer Lett. 2018;432:237–250. doi: 10.1016/j.canlet.2018.04.035. Epub 2018 Apr 1028. [DOI] [PubMed] [Google Scholar]
- 74.Li Y., Zheng Q., Bao C., Li S., Guo W., Zhao J., Chen D., Gu J., He X., Huang S. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25(8):981–984. doi: 10.1038/cr.2015.82. doi: 910.1038/cr.2015.1082. Epub 2015 Jul 1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang S.J., Chen X., Li C.P., Li X.M., Liu C., Liu B.H., Shan K., Jiang Q., Zhao C., Yan B. Identification and characterization of circular RNAs as a new class of putative biomarkers in diabetes retinopathy. Invest. Ophthalmol. Vis. Sci. 2017;58(14):6500–6509. doi: 10.1167/iovs.17-22698. doi: 6510.1167/iovs.6517-22698. [DOI] [PubMed] [Google Scholar]
- 76.Cui X., Niu W., Kong L., He M., Jiang K., Chen S., Zhong A., Li W., Lu J., Zhang L. hsa_circRNA_103636: potential novel diagnostic and therapeutic biomarker in Major depressive disorder. Biomark. Med. 2016;10(9):943–952. doi: 10.2217/bmm-2016-0130. doi: 910.2217/bmm-2016-0130. Epub 2016 Jul 2212. [DOI] [PubMed] [Google Scholar]
- 77.Zipser B.D., Johanson C.E., Gonzalez L., Berzin T.M., Tavares R., Hulette C.M., Vitek M.P., Hovanesian V., Stopa E.G. Microvascular injury and blood-brain barrier leakage in Alzheimer's disease. Neurobiol. Aging. 2007;28(7):977–986. doi: 10.1016/j.neurobiolaging.2006.05.016. doi: 910.1016/j.neurobiolaging.2006.1005.1016. Epub 2006 Jun 1016. [DOI] [PubMed] [Google Scholar]
- 78.Zlokovic B.V. The blood-brain barrier in health and chronic neurodegenerative disorders. Neuron. 2008;57(2):178–201. doi: 10.1016/j.neuron.2008.01.003. doi: 110.1016/j.neuron.2008.1001.1003. [DOI] [PubMed] [Google Scholar]
- 79.Sandoval K.E., Witt K.A. Blood-brain barrier tight junction permeability and ischemic stroke. Neurobiol. Dis. 2008;32(2):200–219. doi: 10.1016/j.nbd.2008.08.005. doi: 210.1016/j.nbd.2008.1008.1005. Epub 2008 Aug 1027. [DOI] [PubMed] [Google Scholar]
- 80.Merali Z., Huang K., Mikulis D., Silver F., Kassner A. Evolution of blood-brain-barrier permeability after acute ischemic stroke. PLoS One. 2017;12(2) doi: 10.1371/journal.pone.0171558. e0171558. doi: 0171510.0171371/journal.pone.0171558. eCollection 0172017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sweeney M.D., Sagare A.P., Zlokovic B.V. Blood-brain barrier breakdown in Alzheimer disease and other neurodegenerative disorders. Nat. Rev. Neurol. 2018;14(3):133–150. doi: 10.1038/nrneurol.2017.188. doi: 110.1038/nrneurol.2017.1188. Epub 2018 Jan 1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Garbuzova-Davis S., Hernandez-Ontiveros D.G., Rodrigues M.C., Haller E., Frisina-Deyo A., Mirtyl S., Sallot S., Saporta S., Borlongan C.V., Sanberg P.R. Impaired blood-brain/spinal cord barrier in ALS patients. Brain Res. 2012;1469:114–128. doi: 10.1016/j.brainres.2012.05.056. Epub 2012 Jun 1027. [DOI] [PubMed] [Google Scholar]
- 83.Chen C.C., Liu L., Ma F., Wong C.W., Guo X.E., Chacko J.V., Farhoodi H.P., Zhang S.X., Zimak J., Segaliny A. Elucidation of exosome migration across the blood-brain barrier model in vitro. Cell. Mol. Bioeng. 2016;9(4):509–529. doi: 10.1007/s12195-016-0458-3. doi: 510.1007/s12195-12016-10458-12193. Epub 12016 Jul 12197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Piwecka M., Glazar P., Hernandez-Miranda L.R., Memczak S., Wolf S.A., Rybak-Wolf A., Filipchyk A., Klironomos F., Cerda Jara C.A., Fenske P. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science. 2017;357(6357) doi: 10.1126/science.aam8526. (pii):science.aam8526. doi: 8510.1126/science.aam8526. Epub 2017 Aug 8510. [DOI] [PubMed] [Google Scholar]
- 85.Lukiw W.J. Circular RNA (circRNA) in Alzheimer's disease (AD) Front. Genet. 2013;4:307. doi: 10.3389/fgene.2013.00307. eCollection 02013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhao Y., Alexandrov P.N., Jaber V., Lukiw W.J. Deficiency in the ubiquitin conjugating enzyme UBE2A in Alzheimer's disease (AD) is linked to deficits in a natural circular miRNA-7 sponge (circRNA; ciRS-7) Genes. 2016;7(12) doi: 10.3390/genes7120116. (pii):genes7120116. doi: 7120110.7123390/genes7120116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Shi Z., Chen T., Yao Q., Zheng L., Zhang Z., Wang J., Hu Z., Cui H., Han Y., Han X. The circular RNA ciRS-7 promotes APP and BACE1 degradation in an NF-kappaB-dependent manner. FEBS J. 2017;284(7):1096–1109. doi: 10.1111/febs.14045. doi: 1010.1111/febs.14045. Epub 12017 Mar 14013. [DOI] [PubMed] [Google Scholar]
- 88.Junn E., Lee K.-W., Jeong B.S., Chan T.W., Im J.-Y., Mouradian M.M. Repression of α-synuclein expression and toxicity by microRNA-7. Proc. Natl. Acad. Sci. Unit. States Am. 2009;106(31):13052–13057. doi: 10.1073/pnas.0906277106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Liu Z., Jiang Z., Huang J., Huang S., Li Y., Yu S., Yu S., Liu X. miR-7 inhibits glioblastoma growth by simultaneously interfering with the PI3K/ATK and Raf/MEK/ERK pathways. Int. J. Oncol. 2014;44(5):1571–1580. doi: 10.3892/ijo.2014.2322. [DOI] [PubMed] [Google Scholar]
- 90.Song X., Zhang N., Han P., Moon B.S., Lai R.K., Wang K., Lu W. Circular RNA profile in gliomas revealed by identification tool UROBORUS. Nucleic Acids Res. 2016;44(9):e87. doi: 10.1093/nar/gkw075. Epub 2016 Feb 1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chen B.J., Mills J.D., Takenaka K., Bliim N., Halliday G.M., Janitz M. Characterization of circular RNAs landscape in multiple system atrophy brain. J. Neurochem. 2016;139(3):485–496. doi: 10.1111/jnc.13752. doi: 410.1111/jnc.13752. Epub 12016 Sep 13755. [DOI] [PubMed] [Google Scholar]
- 92.Huang J.L., Qin M.C., Zhou Y., Xu Z.H., Yang S.M., Zhang F., Zhong J., Liang M.K., Chen B., Zhang W.Y. Comprehensive analysis of differentially expressed profiles of Alzheimer's disease associated circular RNAs in an Alzheimer's disease mouse model. Aging (Albany NY) 2018;10(2):253–265. doi: 10.18632/aging.101387. doi: 210.18632/aging.101387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wang Z., Xu P., Chen B., Zhang Z., Zhang C., Zhan Q., Huang S., Xia Z.A., Peng W. Identifying circRNA-associated-ceRNA networks in the hippocampus of Abeta1-42-induced Alzheimer's disease-like rats using microarray analysis. Aging (Albany NY) 2018;10(4):775–788. doi: 10.18632/aging.101427. doi: 710.18632/aging.101427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Xie L., Mao M., Xiong K., Jiang B. Circular RNAs: a novel player in development and disease of the central nervous system. Front. Cell. Neurosci. 2017;11:354. doi: 10.3389/fncel.2017.00354. eCollection 02017. [DOI] [PMC free article] [PubMed] [Google Scholar]