Although phylogenetically ancient, the olfactory system has received less attention than other sensorial systems. However, olfactory dysfunction is considered an early prodromal event in neurodegenerative diseases (NDs) (Doty, 2012; Attems et al., 2014), which may vary from severe smell loss (e.g., Alzheimer’s and Parkinson’s diseases) to relatively moderate loss (e.g., progressive supranuclear palsy) (Doty, 2017). Recently, a cluster of neuropathological and functional discoveries has evidenced the relevant role of the olfactory bulb (OB) during the neurodegenerative process (Attems et al., 2014; Rey et al., 2018). For instance, the double-transgenic APP/PS1 mouse model of Alzheimer’s disease (AD) develops early proteomic disturbances accompanied by a specific modulation of the focal adhesion kinase (FAK) and mitogen-activated protein kinase (MAPK) dynamics at the level of the OB, demonstrating that olfactory molecular alterations occur prior to β-amyloid plaque appearance and memory impairments in APP/PS1 transgenic mice (Lachen-Montes et al., 2016). Nevertheless, there are currently specific questions which should be addressed in human neurodegeneration: Does the loss of smell precede the onset of ND-specific neuropathological features? What is the metabolic impairment induced by protein inclusions in the OB? Is it disease-specific or tauopathy/synucleinopathy-dependent? How the neuropathological substrates modulate their constitutive interactome at olfactory level? Are there common olfactory targets across different neurological backgrounds?
In this perspective, we briefly summarize the current knowledge generated by the application of neuroproteomic strategies for in-depth molecular characterization of the OB in AD and Parkinson’s disease (PD), and discuss challenges to move the field forward.
Neuroproteomics as a tool to characterize the molecular landscape of the olfactory bulb in proteinopathies: During decades, the molecular complexity of the mammalian olfactory system has impeded its systemic characterization, but shotgun “omic” approaches together with bioinformatics developments are now pushing forward the frontiers of olfaction research at an increasing pace (Zelaya et al., 2015; Lachen-Montes et al., 2017a). Specifically, neuroproteomics is being applied to identify and quantify olfactory-related proteomes in different biological contexts such as olfactory learning, ageing and neurodegeneration (Lachen-Montes et al., 2016). Using protein and peptide fractionation strategies followed by liquid chromatography tandem mass spectrometry (LC-MS/MS), the first proteomic atlas of the human OB was characterized in 2012, identifying 1466 proteins involved in diverse biological functions highlighting nucleotide, RNA binding, hydrolase and phosphatase activities (Fernandez-Irigoyen et al., 2012). Along with recent exhaustive proteomic descriptions performed in human olfactory structures (Lachen-Montes et al., 2016), proteomic workflows are also being used to quantitatively monitor OB molecular derangements in human subjects with different neurological syndromes to discover novel potential therapeutic targets and biomarkers.
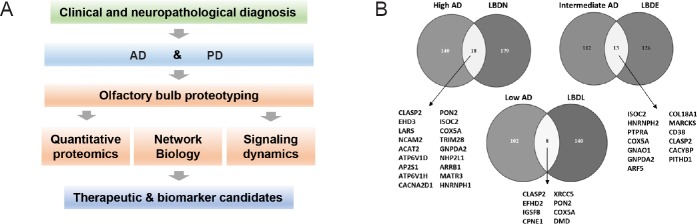
Olfactory proteotyping: commonalities and differences across human tauopathies and synucleinopathies: Most of the neuroproteomics workflows focused on the characterization of human AD and PD neurodegeneration, although informative, have ignored the neuropathological progression of the disease across AD/PD related-brain structures. We consider that the elucidation of the progressive proteome-wide alterations that occurs in a stage-dependent manner in early-affected OB region, may help to unveil the biochemical routes affected during the olfactory pathophysiology of AD and PD. Using a discovery pipeline combining neuropathological examination, quantitative proteomics, network biology and biochemical approaches, we have partially characterized the proteostatic imbalance present in the OB from AD and PD subjects (Lachen-Montes et al., 2017b, 2018) (Figure 1). In particular: i) around 20% of the quantified proteome differs between AD and PD phenotypes respect to nondemented controls, ii) differential functional interactors for neuropathological substrates (APP, Tau, and alpha-synuclein) have been detected in the OB, iii) 24 OB proteins are commonly deregulated across AD grading (Braak I–II, Braak III–IV and Braak V–VI stages), and iv) 65 OB proteins are commonly deregulated across Lewy body pathology (limbic, early-neocortical and neocortical stages) (Lachen-Montes et al., 2017b, 2018).
Figure 1.
Olfactory proteotyping across Alzheimer’s disease (AD) and Parkinson’s disease (PD).
(A) Olfactory proteotyping approach to identify potential therapeutic targets and biomarkers. (B) Common deregulated targets across AD and PD stages. High AD: Braak V–VI stages; intermediate AD: Braak III–IV stages; low AD: Braak I–II stages; LBDN: Lewy body neocortical stage; LBDE: Lewy body early-neocortical stage; LBDL: Lewy body limbic stage.
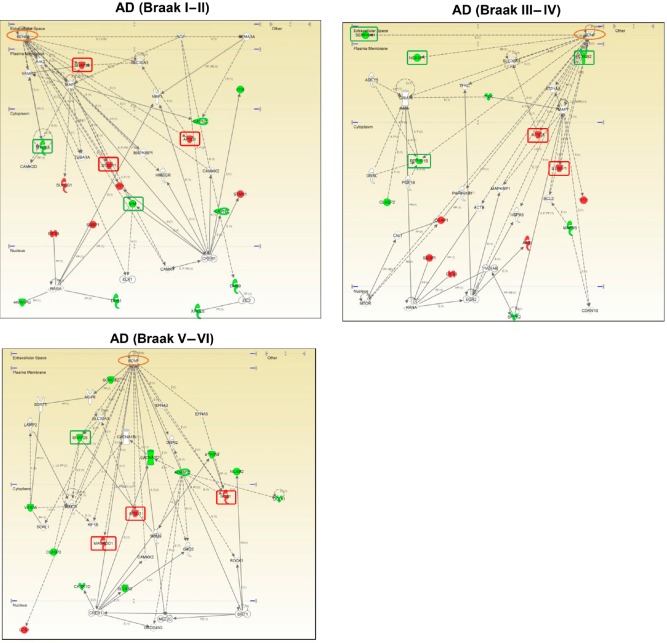
It has been hypothesized that differential disturbance of common pathological substrates (e.g., cholinergic damage) may be part of the molecular mechanisms that cause olfactory dysfunction across NDs (Doty, 2017). As brain-derived neurotrophic factor (BDNF) signalling is directly involved in the survival of cholinergic neurons, the characterization of the functional BDNF interactome at the level of the OB might provide mechanistic clues in the elucidation of olfactory impairments in AD and PD. In this context, network-driven proteomics has allowed to detect some differential functional interactors of BDNF in AD and PD subjects at the level of the OB (Lachen-Montes et al., 2017b, 2018) (Figure 2), suggesting that these protein alterations may differentially compromised the integrity of cholinergic pathways in both neurological syndromes. In addition, our cross-disease study has allowed the detection and identification of a subset of common protein mediators in PD and AD subjects with respect to nondemented controls (Lachen-Montes et al., 2017b, 2018) (Figure 1), suggesting that this shared OB proteome might be part of a common pathological disarrangement during the olfactory neurodegeneration in both neurological disorders. In particular, the mitochondrial cytochrome c oxidase subunit 5A (COX5A) and CLIP-associating protein 2 (CLASP2) were commonly deregulated across all AD and PD stages (Lachen-Montes et al., 2017b, 2018). COX5A is the terminal oxidase in mitochondrial electron transport, and the up-regulation observed in AD and PD suggests an early impairment in mitochondrial function and redox signaling at olfactory level. In the case of CLASP2, its down-regulation indicates that neurite outgrowth, synaptic function and neuronal polarity is compromised in AD and PD olfactory neurons. Considering that cell survival routes have been proposed as neuroprotective targets for slow the progression or delay onset the neurodegenerative process, the cross-disease monitorization of specific OB survival mechanisms also generated important differences in the activation dynamics between AD and PD. Specifically, the activation state of signaling axes such as MEK/ERK, and MKK3/6-p38 MAPK was clearly different between AD and PD phenotypes, whereas OB PDK1/PKC axis was similarly deregulated across AD and PD stages (Lachen-Montes et al., 2017b, 2018). Since no research has been specifically focused on the specific role of this shared protein/kinase panel between AD and PD across olfactory structures, further targeted experiments are needed to elucidate whether they could be potential mediators of the Aβ and α-synuclein spreading or are secondary manifestations of previous damage associated with olfactory deficit.
Figure 2.
Modulation of the brain-derived neurotrophic factor (BDNF) functional interactome across Alzheimer’s disease (AD) stages at the level of olfactory bulb (OB).
Adaptation of BDNF functional network in initial AD (Braak I–II) (left), intermediate AD (Braak III–IV) (right) and advanced AD (Braak V–VI) (down). Continuous lines represent direct interactions, while discontinuous lines correspond to indirect functional interactions. Up-regulated molecules in red, and down-regulated molecules in green. (See complete legend in Additional file 1 (158.3KB, pdf) ). Deregulated proteins are mainly involved in regulation of neurotransmitter release (synaptosomal associated protein-25; SNAP25), dendrite formation (unconventional myosin Va; MYO5A), synaptic vesicle docking (syntaxin-binding protein 1; STXBP1, AP3-complex subunit delta-1; AP3D1), endocytic trafficking (Nck-associated protein 1; NCKAP1) and axon guidance (Plexin-B2; PLXNB2). In the case of PD, the altered BDNF interactors were polypyrimidine tract-binding protein 1 (PTBP1; common to limbic and neocortical stages), and Laminin subunit beta-2 (LAMB2), agrin (AGRN), interferon-induced, double-stranded RNA-activated protein kinase (EIF2AK2) and Guanine deaminase (GDA) (specifically modulated in neocortical stage).
Olfactory proteomics as a novel strategy to detect secretable biomarker candidates: Based on the relevance of the smell loss in neurodegeneration, we consider that the application of proteomics in primary olfactory structures is an ideal approach to explore early pathophysiological changes, detecting olfactory proteins that might be tested in biofluids as potential biomarkers. Data-mining of mass-spectrometry-generated datasets has revealed that 30% of the OB proteome is also localized in cerebrospinal fluid. One of these secretable proteins over-expressed in the OB from PD subjects is Glucosamine-6-phosphate isomerase 2 (GNPDA2), an allosteric enzyme that catalyzes the reversible reaction converting D-glucosamine-6-phosphate into D-fructose-6-phosphate and ammonia (Lachen-Montes et al., 2018). Due to ammonia product causes deficits in excitatory glutamatergic and GABAergic neurotransmission, inflammatory responses, and memory impairments, we focused our attention into the GNPDA2 protein biofluid profile in patients with PD. Interestingly, GNPDA2 showed an inverse correlation with respect to α-synuclein protein levels detected in cerebrospinal fluid whereas serum levels were decreased in PD population respect to nondemented controls. Although these data may be a consequence of the damaged blood-brain barrier (BBB) previously report in PD, additional experiments are needed to clinically validate the monitoring of the GNPDA2 biofluid profile across different tauopathies and synucleinopathies, in order to evaluate the specificity and sensitivity of this metabolic protein as a biomarker.
Conclusion and future perspective: Based on the assumption that AD and PD share common underlying mechanisms due to the overlap in clinical presentation and brain neuropathology, cross-disease olfactory proteomic analyses have demonstrated pathology-specific molecular pathways and protein signatures that are common in AD and PD (Lachen-Montes et al., 2017b, 2018). It is evident that data from more neurodegenerative diseases are needed to complete the olfactory proteotyping across taupathies, synucleinopathies and tardopathies. However, the extensive dynamic range in protein concentration and the proteome heterogeneity originated by diverse interconnected neuronal microenvironments, cause difficulties to globally interpret the outputs from olfactory high-throughput molecular maps. In the near future, the acquisition of high-throughput qualitative and quantitative molecular data by technological developments in systems proteomics will allow the precise dissection of unanticipated peptide and protein regulators present in neurodegenerative primary olfactory structures. The application of MALDI imaging (Lachen-Montes et al., 2018) will offer the great advantage to investigate the neuropathological changes taking place directly in olfactory areas, enabling the so-called “molecular histology”. Technological advancements in the field of laser capture microdissection coupled to mass spectrometry will allow the targeted molecular characterization of specific olfactory cell layers present in the OB, providing novel insights regarding the molecular mechanisms that govern the olfactory dysfunction in a cell-type dependent manner. Based on the differences and similarities observed in the activation state of OB survival kinases, future phosphoproteomic studies will allow to precisely decipher novel cell-signaling networks disrupted during olfactory neurodegeneration across tauopathies and synucleinopathies.
Due to the olfactory neuroepithelium is the only portion of the central nervous system that is exposed to the external environment, intranasal therapies based on enzyme replacement or specific drug delivery are emerging as an alternative route to overpass the BBB and deliver potential therapeutic agents to the brain (Agrawal et al., 2018). Said that, we consider olfactory proteomics as a novel source of pathological olfactory substrates to develop unanticipated preclinical studies in order to potentially treat or cure the neurodegenerative process in its early phases.
This work was funded by grants from the Spanish Ministry of Economy and Competitiveness (MINECO) (No. SAF2014-59340-R), Department of Economic Development from Government of Navarra (No. PC023-PC024, PC025, PC081-82 and PI059) and Obra Social la Caixa to ES. The Proteomics Unit of Navarrabiomed is a member of Proteored, PRB3-ISCIII, and was supported by grant PT17/0019 to JFI, of the PE I+D+i 2013-2016, funded by ISCIII and ERDF.
This project is part of the HUPO Brain Proteome Project and these results are lined up with the Spanish Initiative on the Human Proteome Project (SpHPP). Part of this work was presented in the 27th HUPO Brain Proteome Project Workshop (May 9th–10th, 2017, Ruhr-University Bochum, Germany) and the XII European Proteomics Association (EUPA) Congress (June 16th–20th, Santiago de Compostela, Spain).
Additional files:
Additional file 1 (158.3KB, pdf) : Ingenuity pathway analysis legend.
IPA 8 Legend
Additional file 2: Open peer review reports 1 (102.7KB, pdf) and 2 (99.1KB, pdf) .
Footnotes
Copyright license agreement: The Copyright License Agreement has been signed by both authors before publication.
Plagiarism check: Checked twice by iThenticate.
Peer review: Externally peer reviewed.
Open peer reviewers: Dolores E. Lopez, University of Salamanca, Spain; Cristina Maccallini, University G. d’Annunzio, Italy.
P-Reviewers: Lopez DE, Maccallini C; C-Editors: Zhao M, Liu WJ; T-Editor: Liu XL
References
- 1.Agrawal M, Saraf S, Antimisiaris SG, Chougule MB, Shoyele SA, Alexander A. Nose-to-brain drug delivery: An update on clinical challenges and progress towards approval of anti-Alzheimer drugs. J Control Release. 2018;281:139–177. doi: 10.1016/j.jconrel.2018.05.011. [DOI] [PubMed] [Google Scholar]
- 2.Attems J, Walker L, Jellinger KA. Olfactory bulb involvement in neurodegenerative diseases. Acta Neuropathol. 2014;127:459–475. doi: 10.1007/s00401-014-1261-7. [DOI] [PubMed] [Google Scholar]
- 3.Doty RL. Olfactory dysfunction in Parkinson disease. Nat Rev Neurol. 2012;8:329–339. doi: 10.1038/nrneurol.2012.80. [DOI] [PubMed] [Google Scholar]
- 4.Doty RL. Olfactory dysfunction in neurodegenerative diseases: is there a common pathological substrate? Lancet Neurol. 2017;16:478–488. doi: 10.1016/S1474-4422(17)30123-0. [DOI] [PubMed] [Google Scholar]
- 5.Fernandez-Irigoyen J, Corrales FJ, Santamaria E. Proteomic atlas of the human olfactory bulb. J Proteomics. 2012;75:4005–4016. doi: 10.1016/j.jprot.2012.05.011. [DOI] [PubMed] [Google Scholar]
- 6.Lachén-Montes M, González-Morales A, de Morentin XM, Pérez-Valderrama E, Ausín K, Zelaya MV, Serna A, Aso E, Ferrer I, Fernández-Irigoyen J, Santamaría E. An early dysregulation of FAK and MEK/ERK signaling pathways precedes the β-amyloid deposition in the olfactory bulb of APP/PS1 mouse model of Alzheimer’s disease. J Proteomics. 2016;148:149–158. doi: 10.1016/j.jprot.2016.07.032. [DOI] [PubMed] [Google Scholar]
- 7.Lachen-Montes M, Zelaya MV, Segura V, Fernandez-Irigoyen J, Santamaria E. Progressive modulation of the human olfactory bulb transcriptome during Alzheimer s disease evolution: novel insights into the olfactory signaling across proteinopathies. Oncotarget. 2017a;8:69663–69679. doi: 10.18632/oncotarget.18193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lachen-Montes M, Gonzalez-Morales A, Zelaya MV, Perez-Valderrama E, Ausin K, Ferrer I, Fernandez-Irigoyen J, Santamaria E. Olfactory bulb neuroproteomics reveals a chronological perturbation of survival routes and a disruption of prohibitin complex during Alzheimer’s disease progression. Sci Rep. 2017b;7:9115. doi: 10.1038/s41598-017-09481-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lachen-Montes M, Gonzalez-Morales A, Iloro I, Elortza F, Ferrer I, Gveric D, Fernandez-Irigoyen J, Santamaria E. Unveiling the olfactory proteostatic disarrangement in Parkinson’s disease by proteome-wide profiling. Neurobiol Aging. 2018;73:123–134. doi: 10.1016/j.neurobiolaging.2018.09.018. [DOI] [PubMed] [Google Scholar]
- 10.Rey NL, Wesson DW, Brundin P. The olfactory bulb as the entry site for prion-like propagation in neurodegenerative diseases. Neurobiol Dis. 2018;109:226–248. doi: 10.1016/j.nbd.2016.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zelaya MV, Perez-Valderrama E, de Morentin XM, Tunon T, Ferrer I, Luquin MR, Fernandez-Irigoyen J, Santamaria E. Olfactory bulb proteome dynamics during the progression of sporadic Alzheimer’s disease: identification of common and distinct olfactory targets across Alzheimer-related co-pathologies. Oncotarget. 2015;6:39437–39456. doi: 10.18632/oncotarget.6254. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
IPA 8 Legend