Abstract
In this study, we investigated low frequency and rare variants associated with blood pressure (BP) by focusing on a linkage region on chromosome 16p13. We used whole genome sequencing (WGS) data obtained through the NHLBI Trans-Omics for Precision Medicine (TOPMed) program on 395 Cleveland Family Study (CFS) European Americans (CFS-EA). By analyzing functional coding variants and non-coding rare variants with CADD score > 10 residing within the chromosomal region in families with linkage evidence, we observed 25 genes with nominal statistical evidence (burden or SKAT p < 0.05). One of the genes is RBFOX1, an evolutionarily conserved RNA-binding protein that regulates tissue-specific alternative splicing that we previously reported to be associated with BP using exome array data in CFS. After follow-up analysis of the 25 genes in 10 independent TOPMed studies with individuals of European, African, and East Asian ancestry, and Hispanics (N = 29,988), we identified variants in SLX4 (p = 2.19 × 10−4) to be significantly associated with BP traits when accounting for multiple testing. We also replicated the associations previously reported for RBFOX1 (p = 0.007). Follow-up analysis with GTEx eQTL data shows SLX4 variants are associated with gene expression in coronary artery, multiple brain tissues, and right atrial appendage of the heart. Our study demonstrates that linkage analysis of family data can provide an efficient approach for detecting rare variants associated with complex traits in WGS data.
Keywords: Linkage analysis, whole genome sequencing, blood pressure, rare variants
Introduction
Blood pressure (BP) is a complex trait that has been widely studied in genome-wide association studies (GWAS) (International Consortium for Blood Pressure Genome-Wide Association et al. 2011; Levy et al. 2009; Liang et al. 2018; Sung et al. 2018; Warren et al. 2017; Zhu and Cooper 2007; Zhu et al. 2015; Zhu et al. 2005; Zhu et al. 2011). High blood pressure or hypertension is a major modifiable risk factor for cardiovascular disease and an important risk factor for stroke and kidney disease. Family and twin studies suggest 30–50% of the variation in BP is attributable to genetic heritability (Cooper et al. 2002; Kupper et al. 2005; Miall and Oldham 1963; van Rijn et al. 2007). To date, over 900 loci have been identified to be associated with BP, accounting for nearly 6% of the heritability of this trait (Evangelou et al. 2018; Hoffmann et al. 2017; Liu et al. 2016a). However, rare variants are not well examined by GWAS due to their poor tagging by common variants. When multiple rare variants contribute to inter-individual trait variation, these rare variants can be enriched through ascertainment of families (Jun et al. 2018; Zhu et al. 2010). Correspondingly, linkage analysis of family data is a valid and promising approach for detecting genetic signals because it is insensitive to allelic heterogeneity and facilitates the discovery of missing heritability due to rare variants (Ott et al. 2015). Using this approach, we identified multiple low frequency and rare variants in several genes on chromosome 16 contributing to BP variation using exome array data (He et al. 2017), further demonstrating that family-based study designs are valuable for identifying rare variants.
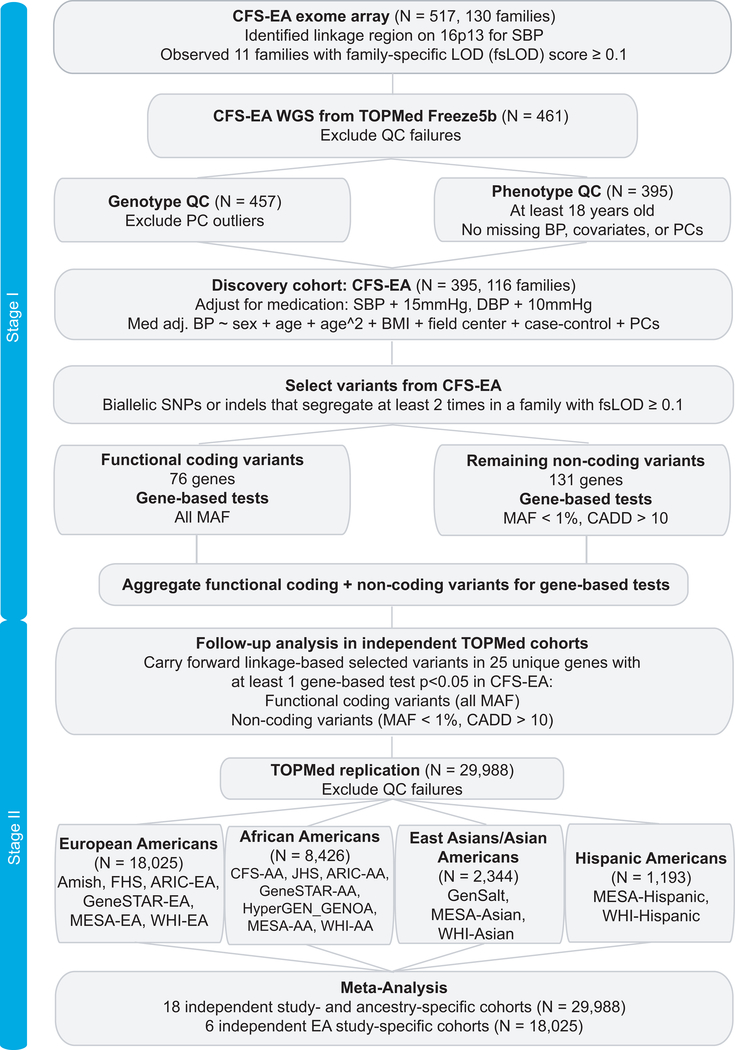
We reexamined a previously identified BP linkage region on chromosome 16 using whole genome sequencing (WGS) data from the National Heart, Lung, and Blood Institute’s (NHLBI) Trans-Omics for Precision Medicine (TOPMed) program. The 16p13 linkage region was initially identified in European Americans from the Cleveland Family Study (CFS), which included 517 individuals in 130 families, genotyped with the Illumina OmniExpress Exome array (focused on protein-coding regions of the genome) (He et al. 2017). We utilized a two-stage approach in this study (Fig. 1): stage I is the discovery of single variants as well as signals from gene-based tests in the discovery cohort and stage II is the independent external replication. Here, we report association analysis in 30,383 individuals aged 18–91 years at collection, with deep coverage WGS and harmonized BP measurements.
Fig. 1. Study design for discovery and replication data.
Figure was produced in Adobe Illustrator CS6.
Material and Methods
Study Population
The TOPMed program is sponsored by the NHLBI and generates data from multiple omics platforms aimed to improve our understanding of the underlying biological mechanisms for heart, lung, blood, and sleep disorders. TOPMed generated WGS data on all contributed samples with a target of 30x coverage on average. WGS provides a comprehensive view of the human genome; thus, these data offer an unprecedented resource to study the genetic architecture of many heart, lung, blood, and sleep disorders. We used the CFS European American samples (CFS-EA; N = 395; 116 families) for discovery analysis in stage I. All of these families were also in the exome array data for the original linkage analysis and 390 subjects were in both exome array and TOPMed WGS. In stage II, independent replication was performed in 10 TOPMed studies (18 ancestry- and study-specific cohorts). These 10 studies (N = 29,988) contain European Americans (EA), African Americans (AA), individuals of East Asian ancestry as well as Hispanic Americans from the following studies: Atherosclerosis Risk in Communities Study from the Venous Thromboembolism (VTE) project (ARIC; EA and AA), Cleveland Family Study African Americans (CFS; AA), Framingham Heart Study (FHS; EA), Genetic Epidemiology Network of Salt Sensitivity (GenSalt; East Asian), Genetics of Cardiometabolic Health in the Amish (Amish; EA), Genetic Studies of Atherosclerosis Risk (GeneSTAR; EA and AA), Hypertension Genetic Epidemiology Network and Genetic Epidemiology Network of Arteriopathy (HyperGEN_GENOA; AA), Jackson Heart Study (JHS; AA), Multi-Ethnic Study of Atherosclerosis (MESA; EA, AA, Asian American, and Hispanic), and the Women’s Health Initiative (WHI; EA, AA, Asian American, and Hispanic). These studies vary in design: ARIC, JHS, and MESA are community-based studies; Amish, CFS, FHS, GeneSTAR, GenSalt, and HyperGEN_GENOA are family-based studies; and WHI is a population-based cohort study in which a case-control sample was selected for TOPMed. The study was approved by the institutional review board (IRB) at Case Western Reserve University. Each individual cohort study was approved by the appropriate IRB in the corresponding institute and appropriate informed consent was obtained from human subjects for participation in the study.
Quality Control
We included only biallelic single nucleotide polymorphisms (SNPs) and insertion-deletion polymorphisms (indels) that passed all filters and had a Phred-scaled quality score (QUAL) > 127, following the quality control (QC) procedures performed centrally by the TOPMed Sequencing Centers, the Informatics Research Center (IRC), and the Data Coordinating Center (DCC). Genotypes for all individuals at all sites passing QC have a minimum 10x sequencing depth. We included participants from 10 TOPMed studies from the freeze_5b release (aligned to GRCh38) and retained only unique subjects that the DCC reported to have no currently known identity problems, reflecting the December 1st, 2017 sample annotation. We further restricted our analyzed sample to individuals who were at least 18 years old at time of measurement and excluded principal component (PC) outliers. The PCs were calculated by the TOPMed DCC using the PC-AiR method, which makes robust population structure inference in the presence of known or cryptic relatedness (Conomos et al. 2015). Lastly, we included only individuals whose harmonized blood pressure measurements were available for our analysis, resulting in a combined sample size of N = 30,383.
Phenotype Harmonization
Phenotype data were collectively harmonized by members of the TOPMed Blood Pressure Working Group. Inclusion criteria for phenotype harmonization include: 1) resting/sitting systolic blood pressure (SBP) and diastolic blood pressure (DBP) recorded as part of a research examination, 2) at least 2 BP measurements were made, and 3) availability of information on the use of antihypertensive medication. With the exception of CFS that utilized data from its last longitudinal examination, when measurements were most comprehensive, all other studies reported measurements from their baseline examinations. For studies with 2 BP measurements at the baseline visit, the average of the first and second measurement was reported. Studies with 3 or more BP measurements at baseline reported the average of the second and third measurement. For each of the 19 ancestry- and study-specific cohorts analyzed, we used harmonized SBP and DBP values and for those reporting current use of antihypertensive medication (32% of study subjects), we added 15 mmHg to their SBP and 10 mmHg to their DBP (Law et al. 2009). Pulse pressure (PP) was calculated as the difference between the (medication-adjusted) SBP and DBP. Covariates used in the analyses were measured at the same visit as the blood pressure measurements.
We calculated regression residuals for medication-adjusted SBP, DBP, and PP after adjusting for age, age2, sex, body mass index (BMI), field center (if data within a study were collected from multiple centers), case-control status (WHI only; grouped all stroke and VTE cases together), and principal components (3 PCs for individuals of European ancestry and 10 PCs for individuals of African or East Asian ancestry, or Hispanics). Because the residuals are approximately normally distributed, no phenotype transformation was performed on any of the studies (Online Resource Figures 1-4). Residuals of these regressions were used as the phenotype for association analysis.
Statistical Analyses
Instead of pooling all the data together, we analyzed each of the 19 ancestry- and study-specific cohorts separately and meta-analyzed the results using Fisher’s method to reduce potential bias with study design heterogeneity (Fig. 1). We conducted single SNP and gene-based associations for all protein-coding genes within the linkage region on 16p13 (chr16:2737103–16223464) using the software EPACTS (EPACTS: Efficient and Parallelizable Association Container Toolbox). A kinship matrix was generated for each of the 10 TOPMed studies analyzed using EPACTS (EPACTS: Efficient and Parallelizable Association Container Toolbox) and these were incorporated into all of the association analyses to adjust for within-study relatedness.
In the stage I discovery analysis, using TOPMed CFS-EA WGS data, we selected for variants in protein-coding genes that segregate at least 2 times in at least 1 of the 11 identified families contributing to the linkage evidence (He et al. 2017). Within the linkage region, we first filtered the variants by protein-coding genes and consequently excluded the intergenic regions. The gene region is defined by Ensembl Variant Effect Predictor (Ensembl Variation - Calculated variant consequences) as a part of the functional annotations curated by WGSA (Liu et al. 2016b), which is provided by the TOPMed DCC. These variants are hereafter referred to as linkage-based selected variants and they were divided into 2 groups using WGSA (Liu et al. 2016b) functional annotations for single variant and gene-based association tests: a) functional coding variants that result in an amino acid change and b) the remaining non-coding variants. The functional coding variants include the following classifications: inframe deletions/insertions, exon loss variant (deletion of an exon), frameshift variant, initiator codon variant non-canonical start codon, splice acceptor variant, splice region variant, start lost variant, stop lost/gained variant, and missense variant. The remaining non-coding variants include any classifications that are not listed previously except for intergenic variants; however, this subset of variants also includes synonymous variants, which although coding, do not lead to amino acid changes. The majority of the non-coding variants are intronic variants.
Single variant association tests for SNPs and indels were performed for all linkage-based selected variants in the linkage region using the Efficient Mixed-Model Association eXpedited (EMMAX) test for quantitative traits (Kang et al. 2010). Gene-based tests for SNPs and indels were performed using the variable-threshold burden test (burdenVT) (Price et al. 2010), combined multivariate and collapsing burden test (burdenCMC) (Li and Leal 2008), and mixed-model sequence kernel association test (SKAT) (Wu et al. 2011). We incorporated linkage-based selected functional coding variants with any MAF and non-coding rare variants (MAF < 1%). Although the focus of our study is on rare and low frequency variants, we included common functional coding variants in the analysis as they may have important biological implications. We imposed an additional filter for non-coding variants using the CADD Phred-like score (Liu et al. 2016b). We used a threshold of CADD Phred-like score > 10 to retain the top 10% most deleterious variants for analysis. In the stage II replication analysis, we replicated variants across studies using the same set of variants identified from CFS-EA, regardless of their availability or study-specific MAF in the independent replication studies. For the gene-based tests, meta-analyses of 6 EA replication cohorts and 18 multi-ancestry replication cohorts were calculated using Fisher’s combined P-value method. Initially, we analyzed functional coding and rare non-coding variants separately under the assumption that the functional coding variants are more likely to have a unidirectional effect, whereas non-coding variants are likely to have bi-directional effects on the BP traits. However, to minimize potentially issues with multiple comparisons, in the stage II independent replication analysis, we further combined functional coding and non-coding variants for gene-based analysis and the final reported genes were based on this analysis. The significance level for stage II p-values was determined by two independent traits for SBP, DBP, and PP, 25 unique genes tested for the analysis, and three gene-based tests, resulting in a conservative threshold (p = 3.3 × 10−4). While the genetic correlation between SBP and DBP is high (0.93–0.98), the genetic correlation between DBP and PP is low (0.05) (van Rijn et al. 2007).
GTEx V6p cis-eQTL gene expression data and covariates were downloaded from the GTEx Portal (https://www.gtexportal.org/home/datasets). Imputed genotype data (N = 450) were downloaded from dbGaP. From this dataset, we performed gene-based association analysis between gene expression and linkage-based selected variants in a corresponding gene in the currently available 44 tissues (including 2 cell lines). We used the residual of the gene expression level as the phenotype, after adjusting for sex, platform, PCs 1–3, and tissue-specific latent factors inferred by GTEx using the PEER method (Stegle et al. 2012). The analyzed variants were limited to variants replicated across studies, where we aggregated linkage-based selected functional coding variants and rare non-coding variants identified from CFS-EA. For the imputed genotype data, the rare variant filters imposed by GTEx (maf05, maf01) were removed in order to include as many linkage-based selected rare variants as possible for this analysis. Imputation info score and Hardy-Weinberg Equilibrium filters were kept for QC.
While some variants in the promoter and non-coding regulatory region can be captured by our defined gene units, it’s possible that many functionally important intergenic variants are left out by our variant selection criteria and must be supplemented with regulatory annotations. Enhancer elements were defined by GeneHancer, a database that integrates enhancers reported from the Encyclopedia of DNA Elements (ENCODE), the Ensembl regulatory build, and Functional ANnoTation Of the Mammalian genome (FANTOM) project, and the VISTA Enhancer Browser (Fishilevich et al. 2017). We only included enhancer elements that were denoted as “elite” on GeneCards (http://www.genecards.org/), defined as enhancer-gene relations reflecting both a high-likelihood enhancer definition and a strong enhancer-gene association. Gene-enhancer associations were generated by integrating multiple sources of information, including expression quantitative trait loci (eQTLs), enhancer RNA (eRNA) co-expression, transcription factor (TF) co-expression, capture Hi-C (CHi-C), and gene target distance. The analysis groups were aggregated by gene units and each group consists of linkage-based selected functional coding variants and rare non-coding variants within the reported enhancer element. We performed gene-based association analysis using our defined variant sets and BP traits for each gene.
Results
Descriptive characteristics are provided for all subjects (Online Resource Table 1). Our previous study identified a linkage region on 16p13 with a maximum LOD score of 2.81 (He et al. 2017). In children, blood pressure increases with age and the normal blood pressure ranges are different from those of adults. Thus, we updated the 2 LOD-score drop region after removing individuals under 18 years old (MLOD = 2.54), resulting in a targeted region chr16:2737103–16223464.
It is important to verify whether common variants in the linkage region could be driving the linkage or association analysis. To the best of our knowledge, only 3 common variants have been identified through BP GWAS in the linkage region on 16p13: rs35450617 (g.6839674T>G), rs12921187 (g.4893018T>G), rs3915425 (g.15818687T>C) (Evangelou et al. 2018). These 3 variants reside within RBFOX1, PPL, and MYH11, respectively. We performed conditional association analysis in the discovery cohort by including these 3 variants as covariates. These 3 SNPs (MAFs > 0.27 for all) and were not associated with any BP trait in CFS-EA, suggesting that they have minimal effect on the linkage and association analyses in this study.
In CFS-EA linkage analysis with exome array data, we observed 11 families with family-specific LOD score (fsLOD) ≥ 0.1 at the most significant SNP (rs6501060; g.8041950T>C), regardless of the inclusion of participants under 18. In CFS-EA TOPMed WGS data, we used the same 11 families and selected for variants in protein-coding genes that segregate at least twice in at least one of these families. The size of these 11 families ranges from 2 to 11 individuals, with a total of 72 individuals. Our selection of variants using family information resulted in 76 genes with functional coding variants and 131 genes with remaining non-coding variants. From the CFS-EA discovery cohort, we observed 20 genes containing functional coding variants with at least 1 gene-based test p-value < 0.05 (Online Resource Table 2) and 8 genes containing non-coding variants (MAF < 1%, CADD > 10) with at least 1 gene-based test p-value < 0.05. Three genes overlap between these 2 groups. The linkage-based selected functional coding and non-coding variants from these 25 unique candidate genes (burden or SKAT p < 0.05) in CFS-EA (stage I) were carried forward for stage II replication analysis in 10 independent, multi-ancestry TOPMed studies (Online Resource Tables 3, 4, and 5).
We initially analyzed functional coding and rare non-coding variants separately in the replication gene-based analysis. Our previously identified RBFOX1 gene association remains nominally significant (SBP, p = 0.021 for burdenCMC) in the meta-analysis of 6 European-American cohorts involving functional variants within 20 genes. We observed 3 additional genes (CLUAP1, TRAP1, and SLX4) with functional coding variants that are nominally associated with BP traits (Online Resource Table 3). The association evidence became less significant when cohorts of African and East Asian ancestry and Hispanics were included (Online Resource Table 3), which is expected given many of these variants are not present in those cohorts. For non-coding variants in the 8 genes carried forward for independent replication, we observed 4 unique genes (MYH11, MTRNR2L4, RBFOX1, and SLX4) that were nominally significant in the meta-analysis of 6 EA cohorts (Online Resource Table 4), but none of the four genes pass multiple testing. Again, adding cohorts other than European Americans weakened the association evidence for the same reason as before.
Next, we aggregated linkage-based selected functional coding variants and non-coding variants in all 25 unique genes carried forward for replication (Online Resource Table 5). In particular, we focused on MTRNR2L4, RBFOX1, and SLX4 because they showed the most significant association evidence (Table 1). Since there were no linkage-based selected functional coding variants in MTRNR2L4, the aggregated gene-based test had the same results as the non-coding variant gene-based test for this gene. We observed an improvement in the association evidence for both RBFOX1 and SLX4 after aggregating both functional coding and non-coding variants for gene-based analysis. In the EA replication cohorts, SLX4 variants are significantly associated with PP (p = 2.19 × 10−4 for burdenVT), after Bonferroni correction, which accounts for 25 genes, 3 tests, and 2 independent traits for SBP, DBP, and PP.
Table 1.
Gene-Based association p-values of linkage-based selected functional coding and non-coding variants in MTRNR2L4, RBFOX1, and SLX4
| SBP | DBP | PP | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Gene a | NVARb | BurdenVT c | BurdenCMC c | SKAT c | BurdenVT | BurdenCMC | SKAT | BurdenVT | BurdenCMC | SKAT |
| CFS-EA Coding, N = 395 | ||||||||||
| RBFOX1 | 3 | 0.012 | 0.456 | 0.024 | 0.11 | 0.342 | 0.064 | 0.001 | 0.042 | 0.014 |
| SLX4 | 10 | 0.12 | 0.851 | 0.056 | 0.16 | 0.569 | 0.035 | 0.52 | 0.687 | 0.572 |
| CFS-EA Non-coding, N = 395 | ||||||||||
| MTRNR2L4 | 2 | 0.05 | 0.048 | 0.101 | 0.012 | 0.01 | 0.027 | 0.55 | 0.576 | 0.8 |
| RBFOX1 | 86 | 0.95 | 0.71 | 0.036 | 0.29 | 0.213 | 0.257 | 0.59 | 0.566 | 0.114 |
| SLX4 | 3 | 0.22 | 0.11 | 0.107 | 0.022 | 0.009 | 0.007 | 0.99 | 0.931 | 1 |
| CFS-EA Coding + Non-coding, N = 395 | ||||||||||
| MTRNR2L4 | 2 | 0.045 | 0.048 | 0.101 | 0.013 | 0.010 | 0.027 | 0.550 | 0.576 | 0.800 |
| RBFOX1 | 89 | 0.890 | 0.323 | 0.036 | 0.340 | 0.571 | 0.266 | 0.230 | 0.044 | 0.087 |
| SLX4 | 13 | 0.034 | 0.851 | 0.047 | 0.006 | 0.569 | 0.003 | 0.690 | 0.687 | 0.861 |
| EA Meta-Analysis Coding, N = 18,025 | ||||||||||
| RBFOX1 | 3 | 0.051 | 0.021 | 0.478 | 0.042 | 0.025 | 0.308 | 0.554 | 0.320 | 0.546 |
| SLX4 | 10 | 0.086 | 0.069 | 0.071 | 0.599 | 0.901 | 0.301 | 0.004 | 0.001 | 0.021 |
| EA Meta-Analysis Non-coding, N = 18,025 | ||||||||||
| MTRNR2L4 | 2 | 0.002 | 0.685 | 0.085 | 0.013 | 0.853 | 0.446 | 0.037 | 0.531 | 0.075 |
| RBFOX1 | 86 | 0.384 | 0.926 | 0.032 | 0.712 | 0.98 | 0.091 | 0.624 | 0.94 | 0.493 |
| SLX4 | 3 | 0.379 | 0.493 | 0.649 | 0.995 | 0.908 | 0.986 | 0.046 | 0.091 | 0.151 |
| EA Meta-Analysis Coding + Noncoding, N = 18,025 | ||||||||||
| MTRNR2L4 | 2 | 0.002 | 0.685 | 0.085 | 0.013 | 0.853 | 0.446 | 0.035 | 0.531 | 0.075 |
| RBFOX1 | 89 | 0.222 | 0.007 | 0.009 | 0.078 | 0.037 | 0.013 | 0.630 | 0.126 | 0.598 |
| SLX4 | 13 | 0.071 | 0.093 | 0.086 | 0.917 | 0.554 | 0.621 | 2.19E-04 | 0.006 | 0.009 |
Gene: only genes with at least 1 gene-based test p < 0.05 in stage I discovery analysis were moved forward for stage I replication
NVAR: number of passed variants; only include linkage-based selected functional coding variants and rare non-coding variants with MAF < 1%, CADD > 10
BurdenVT, BurdenCMC, and SKAT are gene-based tests described in the Methods section.
We further explored the tissue-specific gene expression associations using GTEx V6p data for the RBFOX1, SLX4, and MTRNR2L4. We investigated the gene expression levels of these 3 genes among individuals with imputed genotyping array data. Although we analyzed all of the currently available tissues, there are many systems of the human body that affect BP so we only presented the association evidence for a few tissues that may be relevant to BP, including the brain, heart, and blood vessels (Table 2). For tissue-specific gene-based analysis, we analyzed functional coding and non-coding variants replicated across studies and tested for gene-based association in all available tissues. Gene expression of RBFOX1 is available in 29 tissues. The linkage-based selected variants of RBFOX1 were associated with tissues in the nervous system, including the hypothalamus (p = 0.040 for burdenCMC), putamen (p = 0.014 for burdenVT), and tibial nerve (p = 4.35 × 10−5 for burdenCMC). SLX4 gene expression levels are available in 44 tissues and linkage-based selected variants within this gene are associated with several brain and heart tissues, including the cerebellar hemisphere (p = 0.027 for SKAT), hypothalamus (p = 0.033 for SKAT), coronary artery (p = 0.011 for burdenCMC), and heart atrial appendage (p = 0.028 for burdenCMC). For MTRNR2L4, none of the linkage-based selected variants can be found in the imputed genotyping array data; thus, there were no eligible variants for analysis.
Table 2.
GTEx gene expression association p-values for RBFOX1 and SLX4
| Gene | Tissue | NSa | NVARb | BurdenVT | BurdenCMC | SKAT |
|---|---|---|---|---|---|---|
| RBFOX1 | Brain_Hypothalamus | 81 | 5 | 0.109 | 0.041 | 0.102 |
| Brain_Putamen_basal_ganglia | 82 | 5 | 0.014 | 0.701 | 0.060 | |
| Nerve_Tibial | 256 | 8 | 3.00E-04 | 4.35E-05 | 0.069 | |
| SLX4 | Artery_Coronary | 118 | 11 | 0.018 | 0.011 | 0.060 |
| Brain_Cerebellar_Hemisphere | 89 | 11 | 0.220 | 0.071 | 0.027 | |
| Brain_Hypothalamus | 81 | 11 | 0.130 | 0.054 | 0.033 | |
| Heart_Atrial_Appendage | 159 | 11 | 0.130 | 0.028 | 0.081 |
NS: number of subjects
NVAR: number of passed variants; only include linkage-based selected functional coding variants and rare non-coding variants with MAF < 1%, CADD > 10
Note: Out of the 3 genes included in this analysis, MTRNR2L4 did not have any variants eligible for analysis in the GTEx V6p imputed array data.
In order to identify potential regulatory variants that fall outside of the genes of interest (e.g. intergenic variants), we used GeneHancer to detect candidate enhancer elements for RBFOX1, SLX4, and MTRNR2L4 (Fishilevich et al. 2017). For each gene, we aggregated linkage-based selected functional coding variants of the enhancer gene target with available non-coding variants (MAF < 5%; CADD > 10) within these enhancer elements. Then we conducted group-wise burden and SKAT tests for each gene (Table 3). As expected, most of the non-coding variants in enhancer elements were intergenic or intragenic enhancers. For these 3 genes, there was no overlap between variants within enhancer elements and linkage-based selected non-coding variants identified from CFS-EA. Because the GeneHancer data are based on multiple tissues, tissue-specific results are unavailable. We focused on the meta-analysis for all 7 European-American cohorts (N = 18,420). We found that variants in RBFOX1, SLX4, and MTRNR2L4 were associated with all 3 BP traits (p < 0.05), before but not after adjusting for multiple comparisons. The association evidence weakens after adding cohorts of African and East Asian ancestry and Hispanics (results not shown).
Table 3.
Gene-Based association p-values for GeneHancer elements
| SBP | DBP | PP | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Enhancer Target | NVARa | BurdenVT | BurdenCMC | SKAT | BurdenVT | BurdenCMC | SKAT | BurdenVT | BurdenCMC | SKAT |
| All European-American Cohorts, N = 18,420 | ||||||||||
| MTRNR2L4 | 20 | 0.028 | 0.015 | 0.144 | 0.216 | 0.007 | 0.356 | 0.014 | 0.032 | 0.113 |
| RBFOX1 | 3 | 0.005 | 0.032 | 0.148 | 0.025 | 0.031 | 0.144 | 0.027 | 0.120 | 0.137 |
| SLX4 | 26 | 0.046 | 0.047 | 0.026 | 0.199 | 0.365 | 0.046 | 0.081 | 0.005 | 0.055 |
NVAR: number of passed variants; only include linkage-based selected functional coding variants and available rare non-coding variants with MAF < 5% and CADD > 10
Discussion
This study demonstrates the added power and promise of linkage evidence when investigating low frequency and rare variants in complex diseases like hypertension. Previously, another study (Roeder et al. 2006) presented a method that uses linkage data to weight the association p-values. They implemented an exponential weighting scheme or cumulative weighting scheme using LOD scores. The samples used in linkage analysis and association analysis are independent. However, the weighting approach by Roeder et al. does not work for rare variant analysis within a gene or locus because there is no variation of the LOD scores. In our study, we weight the contribution of a rare variant to linkage evidence rather than directly weighting the LOD scores. Thus, our approach can be applied to rare variant analysis in a gene or locus.
Overall, our association evidence is stronger in European ancestry cohorts compared to African and East Asian ancestry cohorts and Hispanics. This finding is not surprising given rare variants were initially identified from European-American families, increasing the probability that linkage-based selected variants will be monomorphic or extremely rare in other ancestries or ethnicity. The majority of our study subjects are of European ancestry (61%). We were able to verify the association between RBFOX1 and BP traits as well as identify a novel gene SLX4.
For RBFOX1, SLX4, and MTRNR2L4, we looked at characteristics of CFS-EA subjects and families who carried the linkage-based selected variants used in this study. Our previous analysis of RBFOX1 was limited to linkage-based selected functional coding variants in the exome array data (He et al. 2017). By using TOPMed WGS data, we were able to further investigate the non-coding variants in RBFOX1. Summary characteristics for CFS-EA carriers of linkage-based selected variants are provided (Online Resource Table 6). Using family information, we identified two rare (MAF < 5%) and one common functional coding variants. The common variant is a splice region variant and the two rare variants are missense variants. For both functional coding rare variants of RBFOX1 (rs149974858 [p.Pro38Ala] and rs145873257 [p.Gly374Ser]), the directions of effect from meta-analysis of all 19 cohorts are negative for all 3 BP traits (Online Resource Table 7), consistent with a protective effect. All 3 carriers of rs149974858 reside within 1 family and were morbidly obese with a median BMI of 62.37. They had a median BP measure (SBP/DBP) of 115/84 mm Hg and median residuals of −27.05 and −2.38 for SBP and DBP, respectively. Given their elevated BMI, the blood pressure of these 3 carriers were lower than expected, which deviate from the positive correlation between BMI and BP found in previous studies (Droyvold et al. 2005; Dua et al. 2014). Nine carriers of rs145873257 reside in 3 families. They were overweight (median BMI 28.35) and had negative median residuals for BP (−15.83 and −12.26 for SBP and DBP, respectively). Again, this variant showed a protective effect on BP given their high BMI. On the other hand, non-coding variants had bidirectional effects with nearly half of the variants having a protective effect and the rest having a deleterious effect for all 3 BP traits (Online Resource Table 7). This is consistent with our gene-based meta-analysis, which shows a significant association in SKAT for non-coding variants (Online Resource Table 4).
In CFS-EA, we identified 10 functional coding variants and 3 non-coding rare variants in SLX4 (Online Resource Tables 6 and 9). All 10 functional variants are missense variants and only 1 of them is rare (rs140051968; p.Ser1342Gly; MAF = 0.0038). Out of the 9 common functional coding variants, 7 variants are in high LD (r2 > 0.9) with each other. However, excluding variants in high LD in the gene-based tests barely changes the EA meta-analysis association results (functional coding variants only: p = 0.005 for burdenVT in PP; functional coding and rare non-coding variants together: p = 2.5 × 10−4 for burdenVT in PP. In the single SNP meta-analysis, rs140051968 is presented in 4 EA cohorts and it had a positive direction of effect for all 3 BP traits, suggesting a deleterious effect that elevates BP. In CFS-EA, 3 subjects within the same family carry rs140051968. They have a median BP of 136/80 mmHg with a median BMI of 25, which indicates the carriers have an elevated BP without being overweight or obese. For the 3 non-coding variants in SLX4, the carriers are obese (median BMI > 30 for carriers of each SNP) and have an elevated BP (median SBP/DBP > 146/88 mmHg), which is expected based on published literature (Droyvold et al. 2005; Dua et al. 2014). The gene SLX4 plays an important role in DNA double-strand break repair (Yamamoto et al. 2011). Based on GTEx gene expression data, it is highly expressed in the cerebellum, a region implicated in BP control through the cerebellar adrenomedullary system (Figueira and Israel 2018).
We identified 2 non-coding rare variants in MTRNR2L4 (rs146514363 [n.84+2329C>T] and rs540895452 [n.84+7135G>A]) from CFS-EA. The SNP rs146514363 (MAF = 0.0038) is presented in 3 subjects (2 families) and can be found in 11 out of 16 replication cohorts and all 6 EA replication cohorts. There are 3 carriers for rs540895452 (MAF = 0.0038) and all of them are in the same family. This variant is only presented in 4 out of 16 replication cohorts, all of which are European. Because each gene-based test must have at least 2 variants, gene-based meta-analysis can only be done in EA cohorts. Similar to rs140051968 in SLX4, carriers of either rs146514363 or rs540895452 have elevated BP (median BP > 132/80) without being obese (median BMI < 27). MTRNR2L4 plays a role in neuroprotective and anti-apoptotic factor (Bodzioch et al. 2009), but its role in BP regulation remains unclear.
Although our statistical evidence for rare variants in MYH11 is modest, there is biological and clinical evidence that MYH11 could impact the risk of cardiovascular disease. MYH11 encodes the protein myosin-11, which is a component of the myosin heavy chain in smooth muscle. Mutations in MYH11 have been reported to cause thoracic aortic aneurysms and/or dissections (Takeda et al. 2015; Zhu et al. 2006). High blood pressure and high cholesterol are both risk factors for atherosclerosis and consequently may lead to thoracic aortic aneurysm.
Our study illustrates some important implications. We observed that rare variants are more likely to be ancestry-specific. In the stage II replications, we found that many variants identified from the European-American discovery cohort are monomorphic in other ancestries. It is possible that different variants within the same genes may be associated with BP traits in different ancestral populations. Thus, we did gene-based analyses for MTRNR2L4, RBFOX1, and SLX4 in the replication cohorts using variants passing the following criteria in each cohort: any functional coding variants and rare non-coding variants with cohort-specific MAF < 1% and CADD > 10. All singletons (i.e. allele count = 1 within the cohort) have been removed from each cohort prior to gene-based analysis. We used these criteria as they are similar to the selection criteria we used previously with CFS-EA for association analysis. Based on these results, we noticed an improvement in the association evidence between RBFOX1 variants and SBP in the EA (burdenCMC p = 3.7 × 10−4) and multi-ethnic (burdenCMC p = 0.004) meta-analysis (Online Resource Table 8). The association evidence in EA is driven by FHS (SKAT p = 0.010), GeneSTAR-EA (burdenCMC p = 0.001), and WHI-EA (burdenCMC p = 0.007), which account for over half of the EA replication samples (12,106 out of 18,025 individuals). FHS and GeneSTAR-EA are both family studies, in which rare variants can be enriched. There are a number of coding variants identified in these 3 cohorts that are not found in CFS-EA: 18 for FHS, 6 for GeneSTAR, and 22 for WHI. Overall, these findings suggest that there are BP-associated variants within RBFOX1 that are absent in CFS-EA. This conclusion provides further evidence supporting our previous study (He et al. 2017), which states that RBFOX1 variants are associated with BP traits in European Americans. In non-European samples, SLX4 variants are associated with PP (SKAT p = 0.037) in African Americans before but not after Bonferroni correction.
When we looked at characteristics of CFS-EA carriers for the analyzed variants, individuals who are overweight or obese did not consistently have elevated BP or more severe hypertension, which departs from the expected positive correlation between BMI and BP. Due to limited information on comorbidities, we were unable to further examine the clinical characteristics of these carriers.
There are a few limitations in this study. First, BP measurement method and procedure varied among studies. Second, the gene expression data for TOPMed subjects were unavailable at the time of the study. Therefore, the gene expression analysis was done using GTEx subjects with a relatively small sample size. Gene expression data for TOPMed cohorts may become available in the future and allow us to conduct further analysis on the cellular transcriptome. Third, the discovery sample size is very small (N=395 in CFS-EA), although the exome array data with the linkage evidence had more complete families with additional individuals (N = 517). Carrying out the same study in much larger discovery studies may lead to additional discoveries. Lastly, we also had limited data on Hispanics and individuals of African and East Asian ancestry.
In summary, we performed association analysis of functional coding and rare non-coding variants within the 16p13 region using 30,383 subjects from the TOPMed WGS project. To improve power, we utilized linkage findings, enabling the discovery of a novel gene SLX4 and replication of a previously identified gene RBFOX1 for BP traits. While these variants may only explain a small proportion of BP variation at the population level, some variants could substantially impact blood pressure in individual carriers and may help identify pharmacological targets; thus, potentially making it a promising approach for personalized medicine. This study has shown that family information can be used to help discover genes and variants that may be missed by GWAS in isolation.
Supplementary Material
Online Resource Figure 1. Phenotypic characteristics of European-American cohorts
Online Resource Figure 2. Phenotypic characteristics of African-American cohorts
Online Resource Figure 3. Phenotypic characteristics of East Asian and Asian-American cohorts
Online Resource Figure 4. Phenotypic characteristics of Hispanic-American cohorts
Online Resource Note. TOPMed Consortium Banner Authors
Online Resource Table 1. Descriptive characteristics of all analyzed samples
Online Resource Table 2. Annotation of linkage-based selected functional coding variants identified in CFS-EA
Online Resource Table 3. Gene-Based analysis p-values of linkage-based selected functional coding variants in 20 genes
Online Resource Table 4. Gene-Based analysis p-values of linkage-based selected non-coding variants in 8 genes
Online Resource Table 5. Gene-Based analysis of linkage-based selected functional coding and non-coding variants in 25 unique genes carried forward for replication
Online Resource Table 6. Characteristics of CFS-EA (discovery) carriers for linkage-based selected variants in MTRNR2L4, SLX4, and RBFOX1
Online Resource Table 7. Meta-Analysis of single SNP association for functional coding variants in MTRNR2L4, SLX4, and RBFOX1
Online Resource Table 8. Meta-Analysis p-values using all available passed variants* in MTRNR2L4, SLX4, and RBFOX1
Acknowledgements
This work was supported by grants T32 HL007567, HL113338 and HL086694 from the National Heart, Lung, and Blood Institute (NHLBI) and HG003054 from the National Human Genome Research Institute (NHGRI). Whole genome sequencing (WGS) for the Trans-Omics in Precision Medicine (TOPMed) program was supported by the National Heart, Lung and Blood Institute (NHLBI). WGS for “NHLBI TOPMed: Atherosclerosis Risk in Communities” (phs001211.v1.p1) was performed at the Baylor College of Medicine Human Genome Sequencing Center (HHSN268201500015C, 3U54HG003273-12S2). WGS for “NHLBI TOPMed: The Framingham Heart Study” (phs000974.v2.p2) was performed at the Broad Institute of MIT and Harvard (HHSN268201500014C). WGS for “NHLBI TOPMed: Genetics of Cardiometabolic Health in the Amish” (phs000956.v2.p1) was performed at the Broad Institute of MIT and Harvard (3R01HL121007-01S1). WGS for “NHLBI TOPMed: The Jackson Heart Study” (phs000964.v2.p1) was performed at the University of Washington Northwest Genomics Center (HHSN268201100037C). WGS for “NHLBI TOPMed: The Cleveland Family Study” (phs000954.v1.p1) was performed at the University of Washington Northwest Genomics Center (3R01HL098433-05S1). WGS for “NHLBI TOPMed: Multi-Ethnic Study of Atherosclerosis (MESA)” (phs001416.v1.p1) was performed at the Broad Institute of MIT and Harvard (3U54HG003067-13S1). WGS for “NHLBI TOPMed: Hypertension Genetic Epidemiology Network and Genetic Epidemiology Network of Arteriopathy” (phs001293.v1.p1) was performed at the University of Washington Northwest Genomics Center (3R01HL055673-18S1). WGS for “NHLBI TOPMed: Genetic Epidemiology Network of Arteriopathy” (phs001345.v1.p1) was performed at the University of Washington Northwest Genomics Center (3R01HL055673-18S1). WGS for “NHLBI TOPMed: Genetic Studies of Atherosclerosis Risk” (phs001218.v1.p1) was performed at the Macrogen and the Broad Institute of MIT and Harvard (HHSN268201500014C). WGS for “NHLBI TOPMed: Genetic Epidemiology Network of Salt Sensitivity” (phs001217.v1.p1) was performed at the Baylor College of Medicine Human Genome Sequencing Center (HHSN268201500015C). WGS for “NHLBI TOPMed: Women’s Health Initiative” (phs001237.v1.p1) was performed at the Broad Institute of MIT and Harvard (HHSN268201500014C). Centralized read mapping and genotype calling, along with variant quality metrics and filtering were provided by the TOPMed Informatics Research Center (3R01HL-117626-02S1). Phenotype harmonization, data management, sample-identity QC, and general study coordination, were provided by the TOPMed Data Coordinating Center (3R01HL-120393-02S1). We gratefully acknowledge the studies and participants who provided biological samples and data for TOPMed. The Atherosclerosis Risk in Communities study has been funded in whole or in part with Federal funds from the National Heart, Lung, and Blood Institute, National Institutes of Health, Department of Health and Human Services (contract numbers HHSN268201700001I, HHSN268201700002I, HHSN268201700003I, HHSN268201700004I and HHSN268201700005I). The authors thank the staff and participants of the ARIC study for their important contributions. The Amish studies upon which these data are based were supported by NIH grants R01 AG18728, U01 HL072515, R01 HL088119, R01 HL121007, and P30 DK072488. See publication: PMID: 18440328. Support for the Cleveland Family Study were provided by NIH grants HL 046389, HL113338, and 1R35HL135818. The Framingham Heart Study has been supported by contracts N01-HC-25195 and HHSN268201500001I and grant R01 HL092577. The Framingham Heart Study thanks the study participants and the multitude of investigators who over its 70 year history continue to contribute so much to further our knowledge of heart, lung, blood and sleep disorders and associated traits. GeneSTAR was supported by grants from the National Institutes of Health/National Heart, Lung, and Blood Institute (U01 HL72518, HL087698, HL49762, HL58625, HL071025, HL112064), the National Institutes of Health/National Institute of Nursing Research (NR0224103), and by a grant from the National Institutes of Health/National Center for Research Resources (M01-RR000052) to the Johns Hopkins General Clinical Research Center. Support for GENOA was provided by the National Heart, Lung and Blood Institute (HL054457, HL054464, HL054481, and HL087660) of the National Institutes of Health. The Genetic Epidemiology Network of Salt-Sensitivity (GenSalt) was supported by research grants (U01HL072507, R01HL087263, and R01HL090682) from the National Heart, Lung and Blood Institute, National Institutes of Health, Bethesda, MD. The Jackson Heart Study (JHS) is supported and conducted in collaboration with Jackson State University (HHSN268201800013I), Tougaloo College (HHSN268201800014I), the Mississippi State Department of Health (HHSN268201800015I/HHSN26800001) and the University of Mississippi Medical Center (HHSN268201800010I, HHSN268201800011I and HHSN268201800012I) contracts from the National Heart, Lung, and Blood Institute (NHLBI) and the National Institute for Minority Health and Health Disparities (NIMHD). The authors also wish to thank the staffs and participants of the JHS. MESA and the MESA SHARe project are conducted and supported by the National Heart, Lung, and Blood Institute (NHLBI) in collaboration with MESA investigators. Support for MESA is provided by contracts HHSN268201500003I, N01-HC-95159, N01-HC-95160, N01-HC-95161, N01-HC-95162, N01-HC-95163, N01-HC-95164, N01-HC-95165, N01-HC-95166, N01-HC-95167, N01-HC-95168, N01-HC-95169, UL1-TR-000040, UL1-TR-001079, UL1-TR-001420, UL1-TR-001881, and DK063491. The WHI program is funded by the National Heart, Lung, and Blood Institute, National Institutes of Health, U.S. Department of Health and Human Services through contracts HHSN268201600018C, HHSN268201600001C, HHSN268201600002C, HHSN268201600003C, and HHSN268201600004C. Dr. Bress is supported by K01HL133468 from the NHLBI. Dr. Franceschini is supported by DK117445 from the National Institute of Diabetes and Digestive Kidney Diseases (NIDDK) and MD012765 from the National Institute on Minority Health and Health Disparities (NIMHD). The authors would like to acknowledge contributions from the investigators of the NHLBI TOPMed Consortium (https://www.nhlbiwgs.org/topmed-banner-authorship).
Footnotes
Declaration of Interests
The authors declare no competing interests.
Disclaimer
The views expressed in this manuscript are those of the authors and do not necessarily represent the views of the National Heart, Lung, and Blood Institute; the National Institutes of Health; or the U.S. Department of Health and Human Services.
Data Availability
The datasets analyzed during the current study are available in the dbGaP repository. Instructions for accessing TOPMed data can be found on: https://www.nhlbiwgs.org/topmed-data-access-scientific-community
Online Resource
Online Resource include seven tables, four figures, and a note containing consortium authors and affiliations. Tables are included in the Excel spreadsheet. Figures and note are included in the PDF file.
References
- Bodzioch M, Lapicka-Bodzioch K, Zapala B, Kamysz W, Kiec-Wilk B, Dembinska-Kiec A (2009) Evidence for potential functionality of nuclearly-encoded humanin isoforms Genomics 94:247–256 doi: 10.1016/j.ygeno.2009.05.006 [DOI] [PubMed] [Google Scholar]
- Conomos MP, Miller MB, Thornton TA (2015) Robust inference of population structure for ancestry prediction and correction of stratification in the presence of relatedness Genet Epidemiol 39:276–293 doi: 10.1002/gepi.21896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper RS et al. (2002) Genome Scan Among Nigerians Linking Blood Pressure to Chromosomes 2, 3, and 19 Hypertension 40:629–633 doi: 10.1161/01.hyp.0000035708.02789.39 [DOI] [PubMed] [Google Scholar]
- Droyvold WB, Midthjell K, Nilsen TI, Holmen J (2005) Change in body mass index and its impact on blood pressure: a prospective population study Int J Obes (Lond) 29:650–655 doi: 10.1038/sj.ijo.0802944 [DOI] [PubMed] [Google Scholar]
- Dua S, Bhuker M, Sharma P, Dhall M, Kapoor S (2014) Body mass index relates to blood pressure among adults N Am J Med Sci 6:89–95 doi: 10.4103/1947-2714.127751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ensembl Variation - Calculated variant consequences. https://useast.ensembl.org/info/genome/variation/prediction/predicted_data.html.
- EPACTS: Efficient and Parallelizable Association Container Toolbox. http://genome.sph.umich.edu/wiki/EPACTS.
- Evangelou E et al. (2018) Genetic analysis of over 1 million people identifies 535 new loci associated with blood pressure traits Nature Genetics doi: 10.1038/s41588-018-0205-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueira L, Israel A (2018) Cerebellar adrenomedullin: A new target for blood pressure regulation Therapeutic Targets for Neurological Diseases 4 doi:: \\ 10.14800/ttnd.1039 [DOI] [Google Scholar]
- Fishilevich S et al. (2017) GeneHancer: genome-wide integration of enhancers and target genes in GeneCards Database (Oxford; ) 2017 doi: 10.1093/database/bax028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He KY et al. (2017) Rare variants in fox-1 homolog A (RBFOX1) are associated with lower blood pressure PLoS Genet 13:e1006678 doi: 10.1371/journal.pgen.1006678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann TJ et al. (2017) Genome-wide association analyses using electronic health records identify new loci influencing blood pressure variation Nat Genet 49:54–64 doi: 10.1038/ng.3715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Consortium for Blood Pressure Genome-Wide Association S et al. (2011) Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk Nature 478:103–109 doi: 10.1038/nature10405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun G et al. (2018) Evaluating the contribution of rare variants to type 2 diabetes and related traits using pedigrees Proc Natl Acad Sci U S A 115:379–384 doi: 10.1073/pnas.1705859115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HM et al. (2010) Variance component model to account for sample structure in genome-wide association studies Nat Genet 42:348–354 doi: 10.1038/ng.548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupper N, Willemsen G, Riese H, Posthuma D, Boomsma DI, de Geus EJ (2005) Heritability of daytime ambulatory blood pressure in an extended twin design Hypertension 45:80–85 doi: 10.1161/01.HYP.0000149952.84391.54 [DOI] [PubMed] [Google Scholar]
- Law MR, Morris JK, Wald NJ (2009) Use of blood pressure lowering drugs in the prevention of cardiovascular disease: meta-analysis of 147 randomised trials in the context of expectations from prospective epidemiological studies BMJ 338:b1665 doi: 10.1136/bmj.b1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy D et al. (2009) Genome-wide association study of blood pressure and hypertension Nat Genet 41:677–687 doi: 10.1038/ng.384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Leal SM (2008) Methods for detecting associations with rare variants for common diseases: application to analysis of sequence data Am J Hum Genet 83:311–321 doi: 10.1016/j.ajhg.2008.06.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J et al. (2018) Correction: Single-trait and multi-trait genome-wide association analyses identify novel loci for blood pressure in African-ancestry populations PLoS Genet 14:e1007345 doi: 10.1371/journal.pgen.1007345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C et al. (2016a) Meta-analysis identifies common and rare variants influencing blood pressure and overlapping with metabolic trait loci Nat Genet 48:1162–1170 doi: 10.1038/ng.3660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X et al. (2016b) WGSA: an annotation pipeline for human genome sequencing studies J Med Genet 53:111–112 doi: 10.1136/jmedgenet-2015-103423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miall WE, Oldham PD (1963) The hereditary factor in arterial blood-pressure British medical journal 1:75–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott J, Wang J, Leal SM (2015) Genetic linkage analysis in the age of whole-genome sequencing Nat Rev Genet 16:275–284 doi: 10.1038/nrg3908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AL, Kryukov GV, de Bakker PI, Purcell SM, Staples J, Wei LJ, Sunyaev SR (2010) Pooled association tests for rare variants in exon-resequencing studies Am J Hum Genet 86:832–838 doi: 10.1016/j.ajhg.2010.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder K, Bacanu S-A, Wasserman L, Devlin B (2006) Using linkage genome scans to improve power of association in genome scans American journal of human genetics 78:243–252 doi: 10.1086/500026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegle O, Parts L, Piipari M, Winn J, Durbin R (2012) Using probabilistic estimation of expression residuals (PEER) to obtain increased power and interpretability of gene expression analyses Nat Protoc 7:500–507 doi: 10.1038/nprot.2011.457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung YJ et al. (2018) A Large-Scale Multi-ancestry Genome-wide Study Accounting for Smoking Behavior Identifies Multiple Significant Loci for Blood Pressure Am J Hum Genet 102:375–400 doi: 10.1016/j.ajhg.2018.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda N, Morita H, Fujita D, Inuzuka R, Taniguchi Y, Nawata K, Komuro I (2015) A deleterious MYH11 mutation causing familial thoracic aortic dissection Hum Genome Var 2:15028 doi: 10.1038/hgv.2015.28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rijn MJ et al. (2007) Heritability of blood pressure traits and the genetic contribution to blood pressure variance explained by four blood-pressure-related genes J Hypertens 25:565–570 doi: 10.1097/HJH.0b013e32801449fb [DOI] [PubMed] [Google Scholar]
- Warren HR et al. (2017) Genome-wide association analysis identifies novel blood pressure loci and offers biological insights into cardiovascular risk Nat Genet 49:403–415 doi: 10.1038/ng.3768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu MC, Lee S, Cai T, Li Y, Boehnke M, Lin X (2011) Rare-variant association testing for sequencing data with the sequence kernel association test Am J Hum Genet 89:82–93 doi: 10.1016/j.ajhg.2011.05.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto KN et al. (2011) Involvement of SLX4 in interstrand cross-link repair is regulated by the Fanconi anemia pathway Proc Natl Acad Sci U S A 108:6492–6496 doi: 10.1073/pnas.1018487108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L et al. (2006) Mutations in myosin heavy chain 11 cause a syndrome associating thoracic aortic aneurysm/aortic dissection and patent ductus arteriosus Nat Genet 38:343–349 doi: 10.1038/ng1721 [DOI] [PubMed] [Google Scholar]
- Zhu X, Cooper RS (2007) Admixture mapping provides evidence of association of the VNN1 gene with hypertension PLoS One 2:e1244 doi: 10.1371/journal.pone.0001244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Feng T, Li Y, Lu Q, Elston RC (2010) Detecting rare variants for complex traits using family and unrelated data Genet Epidemiol 34:171–187 doi: 10.1002/gepi.20449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X et al. (2015) Meta-analysis of correlated traits via summary statistics from GWASs with an application in hypertension Am J Hum Genet 96:21–36 doi: 10.1016/j.ajhg.2014.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X et al. (2005) Admixture mapping for hypertension loci with genome-scan markers Nat Genet 37:177–181 doi: 10.1038/ng1510 [DOI] [PubMed] [Google Scholar]
- Zhu X et al. (2011) Combined admixture mapping and association analysis identifies a novel blood pressure genetic locus on 5p13: contributions from the CARe consortium Hum Mol Genet 20:2285–2295 doi: 10.1093/hmg/ddr113 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Online Resource Figure 1. Phenotypic characteristics of European-American cohorts
Online Resource Figure 2. Phenotypic characteristics of African-American cohorts
Online Resource Figure 3. Phenotypic characteristics of East Asian and Asian-American cohorts
Online Resource Figure 4. Phenotypic characteristics of Hispanic-American cohorts
Online Resource Note. TOPMed Consortium Banner Authors
Online Resource Table 1. Descriptive characteristics of all analyzed samples
Online Resource Table 2. Annotation of linkage-based selected functional coding variants identified in CFS-EA
Online Resource Table 3. Gene-Based analysis p-values of linkage-based selected functional coding variants in 20 genes
Online Resource Table 4. Gene-Based analysis p-values of linkage-based selected non-coding variants in 8 genes
Online Resource Table 5. Gene-Based analysis of linkage-based selected functional coding and non-coding variants in 25 unique genes carried forward for replication
Online Resource Table 6. Characteristics of CFS-EA (discovery) carriers for linkage-based selected variants in MTRNR2L4, SLX4, and RBFOX1
Online Resource Table 7. Meta-Analysis of single SNP association for functional coding variants in MTRNR2L4, SLX4, and RBFOX1
Online Resource Table 8. Meta-Analysis p-values using all available passed variants* in MTRNR2L4, SLX4, and RBFOX1