Abstract
Background:
The SH-group at Cys-34 of human serum albumin (HSA) is a unique and accessible functional group that can be exploited for efficient linkage of a maleimide containing cytotoxic drug derivative to albumin. The specific maleimide chemistry used for production of the maleimide-linked albumin drug (MAD) is critical, however, to minimize the plasma concentration of “free” cytotoxic drug spontaneously released from albumin carrier thus decreasing dose-limiting host toxicity while enhancing the plasma half-life from minutes to days (ie, pharmacokinetic effect) and tissue concentration of the MAD in the extracellular cellular fluid at sites of cancer (ie, EPR effect).
Methods:
To accomplish this goal, a chemical synthesis was developed using 2-fluoro-5-maleimidobenzoic acid to stably link the potent cytotoxic chemically modified analogue of the naturally occurring sesquiterpene γ-lactone, thapsigargin, 8-O-(12-aminododecanoyl)-8-O-debutanoyl thapsigargin (12ADT), to Cys-34 of albumin to produce 12ADT-MAD.
Results:
Using FITC-labeling, LC/MS analysis, and in vitro growth and clonogenic survival assays on a series of 6 human prostate cancer lines (LNCaP, LAPC-4, VCap, CWR22Rv1, PC3, and Du145), we documented that 12ADT-MAD is endocytosed by prostate cancer cells where it is degraded into its amino acids liberating cysteinyl-maleimide-12ADT which is both chemically stable at the acidic pH of 5.5 present in the endosome while retaining its high killing ability (IC50 50 nM) via SERCA inhibition.
Conclusions:
Based upon these positive in vitro validation results, the in vivo efficacy versus host toxicity of this 12-ADT-MAD approach is presently being evaluated against a series of patient derived androgen responsive and castration resistant human xenografts in immune-deficient mice.
Keywords: albumin based drug uptake, EPR effect, maleimide linkage, prostate cancer
1 |. INTRODUCTION
Cancer cells are “obligate parasites” which absolutely depend on systemic nutritional support as well as metabolic cooperation with host cells in order to produce a tumor microenvironment that can sustain their lethal growth.1 While evolution has produced a physiology to continuously provide adequate systemic supply of nutrients through the blood to normal tissue, as cancer cells grow, they eventually exceed the existing local blood supply, which results in the tumor microenvironment becoming acidic, hypoxic, and low in nutrients. Continued malignant growth in such a compromised (ie, stressful) microenvironment requires cancer cells to initiate autophagy to parasitize the host’s energy and nutrients. In the 1920s, Warburg and the Cori’s demonstrated the importance of carbohydrate metabolism and lactate generation by cancers.2,3 Since then, a variety of potential low molecular weight nutrients, such as fatty acids, ketone bodies, and amino acids have been identified. The first indications that plasma proteins, like albumin, might be involved in tumor autophagic nutrition were reported by Mider in the late 1940’s. This report concluded that tumors act as “nitrogen traps” after comparing the protein metabolism of normal tissue and of tumor tissue.4 These studies demonstrate one-way passage of amino acids from the body pool to the tumor without any appreciable return and that even the stress of starvation does not release tumor protein for the body. In 1954, Babson and Winnick reported that cancer cells, unlike normal cells, utilized plasma proteins more efficiently than free leucine.5 Based on these findings, they concluded that tumor growth in the starving animal and in the cachectic human is driven by the cancer cell’s acquired ability to trap plasma proteins and to use the degradation products (amino acids) for proliferation, a process that normal cells do not do. Later, it was shown that albumin, in order to be utilized for its amino acid content, enters cancer cells by an active process of endocytosis.6,7
More recent experiments using IV injected [111In]DTPA labeled albumin documented that large amounts of labeled albumin metabolites are retained within sites of cancer (ie, more than 20% of total injected albumin is concentrated in Dunning rat prostatic adenocarcinomas by 3 days following a single dose of radiolabeled albumin).8 This is significant because cancer cells must tap the energy and nitrogen resources of its host with high efficiency in order to proliferate. The required substrates, oxygen, glycolytic energy, and nitrogen must be transferred to the tumor by blood and plasma. Proteins account for 70% of the solubilized substances in plasma. Another 20% consist of a multitude of low molecular weight organic substrates (glucose, amino acids, fatty acids, urea, creatine, uric acid, glycerol) and the remaining 10% are inorganic ions. Plasma proteins are high quality nutrient and carry most of the readily available nitrogen and energy reserves of the host.9 Albumin is the most abundant among these proteins accounting for about 50–70% of the human plasma protein reserve. Its relevance as a metabolic reserve is demonstrated by comparing the millimolar concentrations of the amino acids in albumin with the concentration of free amino acids in plasma. Assuming a physiologic albumen concentration of 40 g/L, the concentration of amino acids that are released after complete lysosomal degradation of albumin (ie, 317 mM) exceeds the free amino acids in the plasma (ie, 2.7 mM) 117-fold.9 Besides providing amino acids, albumin catabolism also provides a considerable source of energy, providing a rationale for why autophagic uptake and degradation of albumin is a fundamental acquired characteristic of cancer cells.6,7
While the half-life of albumin in the body is ~19 days in humans, its plasma half-life is only about 1 day.10 This is because despite tight endothelial barrier function, albumin constantly leaves the plasma via endothelial transcytosis and enters un-degraded into the extracellular fluid throughout the body. Due to this transcytosis, albumin makes ~15 around trips from the extracellular fluid returning back to the blood via the lymphatic thoracic duct during its life-span.10 Besides transcytosis, albumin also extravasates into the extracellular fluid within sites of metastatic cancer due to a characteristic decrease in tumor endothelial barrier function which, coupled with a lack of a functional lymphatic drainage at these sites, produces a cancer specific process known as the enhanced permeability and retention (ie, EPR) effect.11 We have documented that due to this malignancy driven EPR effect, a small molecule therapeutic drug bound to albumin in the blood stream is concentrated appropriately sixfold within sites of prostate cancer within 24 h of dosing.12
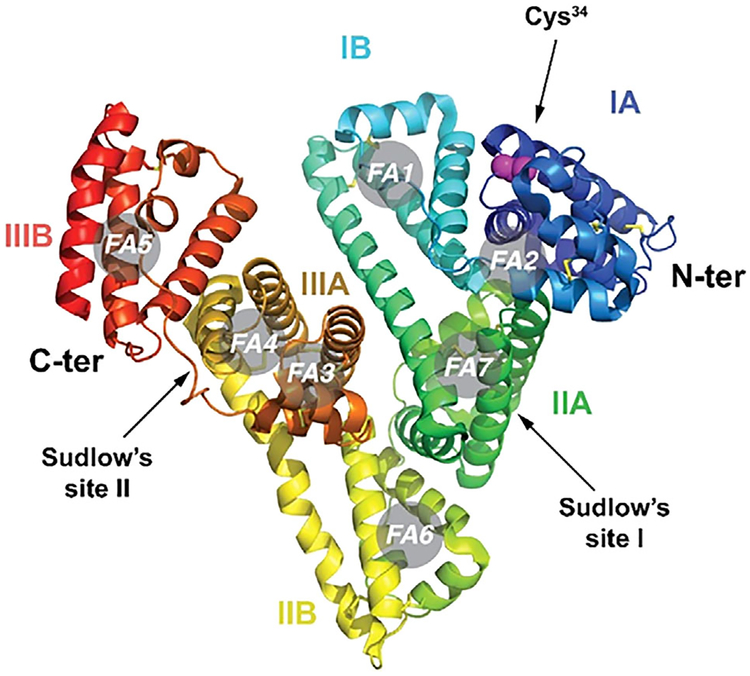
Coupling drugs to albumin in the blood is quite straightforward.13 This is because albumin has a modular structural organization producing a heart-shaped molecule composed of three homologous domains, each containing an A and B subdomain.14 The 1A domain contains a surface accessible cysteine at amino acid position 34 (Cys-34), Figure 1. Approximately 70% of circulating albumin in the blood stream contains the accessible Cys-34, not blocked by endogenous sulfhydryl scavenger compounds such as cysteine, homocysteine, glutathione, or nitric oxide. The concentration of these low molecular weight sulfhydryl compounds in human blood plasma in their reduced form, that is, cysteine (10–12 μM), homocysteine (0.15–0.25 μM), cysteinylglycine (3–4 μM), glutathione (4–5 μM) is low when compared to the total thiol concentration in human plasma, which is in the range 400–500 μM.15 Thus, the free thiol group (SH) at the Cys-34 position of HSA accounts for 80–90% of the total thiol concentration in blood plasma.16 Importantly, the SH group of Cys-34 of HSA is the most reactive thiol group in human plasma because of the low pKa of Cys-34 in HSA, which is approximately seven compared to 8.5 and 8.9 for cysteine and glutathione, respectively15 at physiological pH. Taken together, the SH group of Cys-34 of HSA is a unique and accessible functional group of a plasma protein that can be exploited for in situ coupling of circulating albumin following intravenous administration of a thiol-reactive prodrug.13 The X-ray structure of the defatted protein structure reveals that cysteine-34 is located in a hydrophobic crevice on the surface of the protein that is approximately 10–12 Å deep.14 When HSA is complexed with long-chain fatty acids as in the X-ray structure in which seven molecules of myristic acid (ie, denoted in gray circles in Figure 1) are bound, the crevice opens up, exposing the sulfhydryl group Cys-34.17 This allows efficient coupling of this Cys-34 sulfhydryl to small molecular cytotoxic compounds containing a thiol-reactive (eg, maleimide) group which follows second-order kinetics with second-order rate constants in the range 600–2250 L mol−1 min−1, with the highest rate constant occurring when all of the myristic acid binding sites of albumin are saturated, as occurs in the blood.15
FIGURE 1.
Molecular structure of human serum albumin (HSA) with an indication of its subdomains (IA, IB, IIA, IIB, IIIA, an IIIB), of the N and C termini, of Sudlow’s sites I and II and of the seven fatty acid binding sites (FA1 to FA7). The side chain residues of Cys-34 are shown as purple spheres.
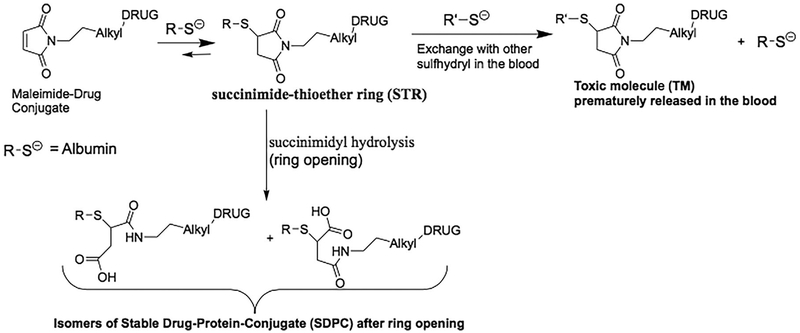
Thus, the SH-group at Cys-34 of HSA is a unique and accessible functional group that can be exploited for efficient linkage of a maleimide containing cytotoxic drug derivative to albumin.13,15 The specific maleimide chemistry used for production of the maleimide-linked albumin drug (MAD) conjugate is critical in order to minimize the plasma concentration of “free” cytotoxic drug spontaneously released from albumin carrier thus suppressing its acute host toxicity while enhancing both the plasma half-life (ie, from minutes to days) and tissue concentration of the MAD at sites of cancer (ie, EPR effect). N-alkyl maleimides are commonly used as linkers for the conjugation of the sulfhydryl group in cysteine of proteins to other therapeutic molecules to form succinimide-thioether ring. This succinimide thioether ring (STR) linkage, however, undergoes significant spontaneous cleavage by a retro-Michael reaction under physiologic conditions, Figure 2.18,19 When this reaction occurs in-vivo, it results in dose-limiting systemic toxicity as the therapeutic agent is spontaneously released to circulate in the blood forming adduct with other sulfhydryl containing species like gluthathione, cysteine etc. generating a systemically toxic molecule TM, Figure 2. In contrast, it has been documented that opening of the succinimide-thioether ring (succinimidyl hydrolysis) to form succinamic acid thioether results in forming a stable drug-protein-conjugate (SDPC) with a very long half-life of up to 2 years, Figure 2.20 Thus, if succinimide-thioether that is formed upon conjugation of sulfhydryl group of protein to maleimide is hydrolysed to the succinamic acid thioethers to form the SDPC, this would eliminate the problem of poor in-vivo stability of the protein-drug conjugate. Further investigations have shown that the electron withdrawing inductive effect of the N-substituent on the nitrogen of the succinimide-thioether plays a major mechanistic role in determining the rate of the succinimidy hydrolysis to succinamic acid thioethers.20 Thus, in the present study, 2-fluoro-5-maleimidobenzoic acid (compound 3 in Figure 3) was used for stably linking a potent cytotoxic drug to Cys-34 of albumin since it possesses the required electron withdrawing inductive ability needed by combining the resonance effect of the aromatic ring and the electron withdrawing property of fluorine at para position to the nitrogen of the succinimide-thioether ring.20–22
FIGURE 2.
Rationale for chemical instability of N-alkyl linked maleimide-drug coupling to albumin
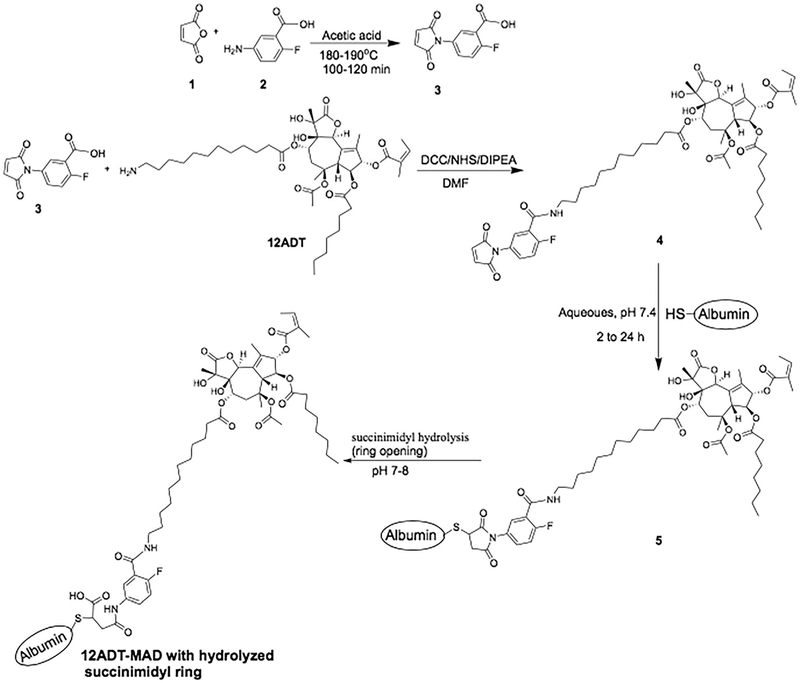
FIGURE 3.
Chemical scheme for synthesizing 2-fluoro-5-maleimidobenzoic acid (compound 3), 2-fluoro-5-maleimidobenzoic acid-12ADT (compound 4), and albumin coupling to produce 12ADT-MAD (compound 5)
Selection of an “appropriate” cytotoxic drug for such 2-fluoro-5-maleimidobenzoic acid coupling to albumin is based upon the following requirements: (i) it must be chemically linkable via an acid stable bond to 2-fluoro-5-maleimidobenzoic acid; (ii) it must itself be chemically stable at the acidic pH 5.5 environment of the endosome; and (iii) it must retain its potent cytotoxicity when liberated in the endosome coupled to free cysteine. Based upon these requirements, a chemically modified analogue of the naturally occurring sesquiterpene γ-lactone, thapsigargin, 8-O-(12-aminododecanoyl)-8-O-debutanoyl thapsigargin (12ADT), Figure 3, was chosen as the initial cytotoxic drug stably coupled to albumin. This is because 12ADT is acid stable and contains a primary amine allowing its covalent coupling via formation of a peptide bond.23 12ADT is a potent inhibitor (IC50 10 nM) of their endoplasmic reticulum (ER) calcium ATPase (ie, SERCA 2b) pumps and thus induces depletion of the high (ie, >500 μM) Ca+2 in the ER, inducing both an ER stress response and a “capacitance entrance” of extracellular Ca+2 in human mCRPCs producing a sustained increase in intracellular Ca+2 (Cai) to >1 μM over the next 18–36 h.23 The combination of ER stress and a sustained elevation of Cai, eventually results in the apoptotic death of mCRPC with an LD50 of <50 nM.23 As part of this death response, AR protein expression is rapidly decreased in AR positive mCRPC cells.24 In addition, 12ADT kills even multidrug resistant cells lacking apoptotic Bak and Bax proteins.25 An additional factor is that 12ADT’s potent killing ability is retained when coupled to single amino-acids.23,24,26
2 |. MATERIAL AND METHODS
2.1 |. General information
All solvents and reagents used were obtained from commercial sources and used without further purification. The 1H- and 13C—NMR spectra were obtained on a Bruker Avance III 500 MHz NMR spectrometer at 500 and 125 MHz, respectively in deuterated dimethyl sulfoxide (DMSO-d6). Chemical shifts are in δ units (ppm). MALDI-MS was done using Voyager DE-STR MALDI-TOF. Purity of the compounds was determined with reverse phase-HPLC. The purity of all the compounds was determined to be >95%.
HPLC Method 1 was used for analytical reversed-phase HPLC to determine the purity of the synthetic compounds using a Waters Delta 600 Controller equipped with a variable wavelength UV-vis detector (Waters 2487 Dual λ Absorbance Detector) set to detect at 215 and 285 nm and a Vydac 218TP54 (C18, 5 μm, 4.6 mm. i.d. X 250 mm) analytical column. The flow rate was 1.3 mL/min; mobile phase A: 5% MeCN, 95% water and 0.1% TFA. Mobile phase B: 100% MeCN, 0.1% TFA. Gradient: 0–2 min 100% mobile phase A; 2–20 min gradual change to 100% mobile phase B; 20–25, 100% mobile phase B; 25–27 min gradual change to 100% mobile phase A; 27–30 min, 100% mobile phase A. Injection volume: 100 μL.
HPLC Method 2 was used for preparative purification of compounds 4 and 6. Flow rate: 25 mL/min; mobile phase A: 5% MeCN, 95% water and 0.1% TFA. Mobile phase B: 100% MeCN, 0.1% TFA. Gradient: 0–4.0 min, 100% mobile phase A; 4.0–20.0 min gradual change to 100% mobile phase B; 20.0–20.5 min 100% mobile phase B; 20.5–21.0 min, sharp change to mobile phase A; 21.0–24.0 min mobile phase A. Injection volume: 1.0 mL.
2.2 |. Synthesis of 2-fluoro-5-maleimidobenzoic acid-12ADT (compound 4)
Thapsigargin was purified from Thapsia garganica seeds and 12 ADT analogue synthesized as described in detail previously.22 Synthesis was performed as summarized in the synthetic scheme shown in Figure 3. To obtain a stable HSA coupled 12ADT through the Cys-34 of albumin, we utilized 2-fluoro-5-maleimidobenzoic acid derivative, compound 3. This compound was synthesized as previously reported,23,24 but with modifications that facilitate a more efficient isolation and purification for large scale synthesis, as presented in Figure 3. To accomplish this synthesis, 5-amino-2-fluorobenzoic acid (5.0 g, 1 molar equivalent) and maleic anhydride (4.7 g, 1.5 molar equivalent) were separately dissolved in 100 mL acetic acid. The maleic anhydride clear solution was gradually added to the clear solution of 5-amino-2-fluorobenzoic acid and a slurry was formed. The mixture was placed in oil bath for refluxing and it became a clear, homogenous brown liquid upon heating. It was then refluxed at 180–190°C for 2 h. Solvent and all volatiles were evaporated from the mixture by using rotary evaporator to give a pale brown powdery precipitate. This crude product precipitate was then triturated in water, filtered and residue washed with water. The residue was air dried to afford 2-fluoro-5-maleimidobenzoic acid, compound 3 (5.94 g, 78.4% yield). 1H NMR (500 MHz, DMSO-d6) 7.20 (1H, s), 7.46 (1H, t, J = 10.0 Hz), 7.60–7.65 (1 H, m), 7.87 (1H, dd, J = 3.0 Hz, 6.5 Hz), 13.35–13.65 (1H, br s); 13C NMR (125 MHz, CDCl3) 118.5, 120.5, 127.9, 130.5, 133.5, 135.1 (2C), 159.2, 161.5 (2C), 164.8. HPLC purity 98.4%.
To obtain compound 4, purified compound 3 (300 mg, 1.277 mmol, 2.5 molar equivalent to 12ADT) was dissolved in 20 mL DMF. This was followed by the addition of N-hydroxysuccinimide (NHS) (293.7 mg, 2.554 mmol; 2 molar equivalent to 3) and dicyclohexylcarbodiimide (DCC) (527 mg, 2.558 mmol; 2 molar equivalent to 3). The reaction mixture was stirred at room temperature for 3 h and then filtered. The filtrate was added to a solution of 12ADT (397 mg, 0.5103 mmol; 1 molar equivalent to compound 3) in DMF and 5 molar equivalent of N,N-diisopropylethylamine (DIPEA). The reaction mixture was stirred at room temperature and monitored with MALDI-MS for indication of the mass of the desired product. The reaction was stopped after stirring for 2 h and solvent was evaporated completely. The crude product was then purified using preparative HPLC (method 2) to give N-aryl-maleimide-12ADT, compound 4. Yield: 57%, brown powder; MALDI-MS m/z 1018.33, ([C53H71FN2O15 + Na]+, calcd. 1018.14), HPLC tR is 12.5 min and purity is 97.8%.
2.3 |. Kinetics of 2-fluoro-5-maleimidobenzoic acid-12ADT (compound 4) coupling to HSA
To determine its coupling kinetics, a mixture of 10 mM of compound 4 in phosphate buffered saline (PBS) pH 7.4 containing 100 μM solution of HSA was incubated for various time spans up to 24 h at 37°C. The mixture was then centrifuged at 18 000g for 15 min to remove any precipitate. Supernatants were then assayed using Ellman’s reagent (G-Biosciences cat# BC87) according to manufacturer’s instructions to determine the amount of free HSA cysteine in each solution. Samples were read at 412 nm and free thiol concentration was determined using an extinction coefficient of 14,140 M−1 cm−1.
2.4 |. Production of 12ADT-MAD (compound 5)
A total of 10 mM solution of compound 4 was prepared in DMSO and an aliquot of this was diluted with fetal bovine serum (FBS) to 100 μM. This was incubated at 4°C, room temperature, and 37°C to obtain the optimum reaction condition. Reactions at room temperature and 37°C were monitored over a 2 h period and the reaction at 4°C were monitored over a 24 h period. Since FBS contains ~400 μM albumin, all compound 4 added was expected to covalently react with albumin in FBS. To determine this, at each time point 100 μL aliquot was taken and added to 300 μL of acetonitrile and the mixture was vortexed and centrifuged at 13 000 rpm for 6 min. The supernatant obtained was analyzed by analytical HPLC and MALDI-MS to determine the presence of unreacted compound 4.
2.5 |. Synthesis of cysteinyl-2-fluoro-5-maleimidobenzoic acid-12ADT (compound 6)
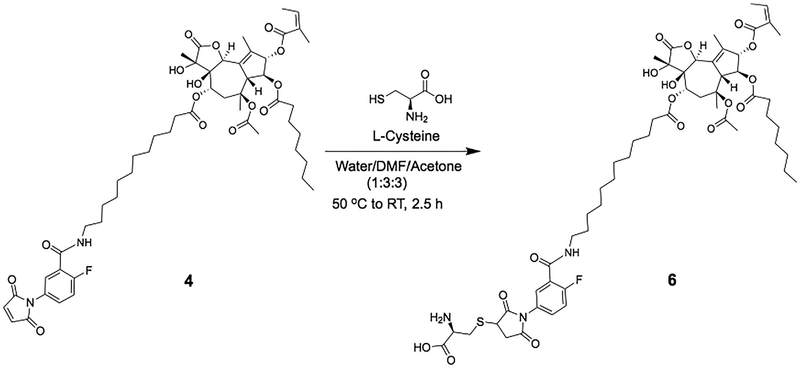
Synthesis was performed as summarized in the synthetic scheme shown in Figure 4. To obtain compound 6, 30 mg (0.0301 mmol) of compound 4 was dissolved in 3.5 mL DMF and to this was added3.5 mL acetone and 50 μL of DIPEA. This clear solution was then added gradually to a solution of L-cysteine (43.7 mg, 0.3612 mmol) in 2.5 mL PBS, pH 8 buffer solution. The homogenous mixture was incubated at 40–50°C for 5 min and then stirred at room temperature for 2 h. The crude product was purified using preparative HPLC (method 2) to give compound 6. Yield: 82%, pale brown powder; MALDI-MS m/z 1139.94, ([C56H78FN3O17S + Na]+, calcd. 1139.29), HPLC tR is 15.5 min and purity is 96.7%.
FIGURE 4.
Chemical scheme for synthesizing cysteinyl-2-fluoro-5-maleimidobenzoic acid-12ADT (compound 6)
2.6 |. Fluorescent-labeling of human serum albumin (HSA)
Human serum albumin (recombinant expressed in rice) was purchased from Sigma-Aldrich, Saint Louis, MO (Sigma cat#A9731) and reconstituted in PBS. To covalently couple a fluorescent label to the reactive lysines of HSA, NHS-fluorescein (Thermo Fisher, Rockford, IL, cat# 46410), dissolved in DMSO, was mixed in 10-fold excess with the HSA at a pH of 7.4. This reaction took place at room temperature for 2 h in the dark. To quickly assess the efficiency of the reaction, the labeled protein was passed through a dye removal column (Thermo Fisher cat# 22858) according to manufacturer’s recommendations and the effluent was analyzed by absorbance at 450 and 280 nm. For microscopy and cell based assays, all labeled HSA preparations were purified using a AKTAprime Plus FPLC System (GE 11001313) using a HiLoad Superdex 200 PG (GE 28989335) column with PBS pH 7.4 as the buffer. One milliliter fractions were collected and screened using A280, A450, and SDS PAGE for the presence of relevant amounts of labeled HSA. Positive fractions were pooled and sterile filtered for downstream purposes.
2.7 |. Cell lines and cell culture assays
The culture conditions and media for all of the human prostate cancer lines used in these studies (ie, LNCaP, LAPC-4, VCaP, CWR22Rv1, PC-3, DU-145) are as described previously.23,24 All lines were mycoplasma negative using the MycoSensor PCR Assay kit (Agilent Technologies, La Jolla, CA) and genetically authenticated within the last 6 months using STR profiling performed by the Johns Hopkins Genetic Resource Core Facility. Cell viability at various times post exposure to test compounds was determined using trypan blue exclusion as previously described.23 The dose-response ability of test compounds to kill prostate cancer cells was determined using a clonogenic survival assay as described previously,23 with the results expressed as the nmol/L concentration needed to lose 50% of the clonogenic ability (ie, LD50) after 48 h of exposure to the test compound. In addition, MTT growth assays performed as described previously27 were used to confirm the clonogenic results. SERCA pump inhibition, expressed as the concentration needed to inhibit 50% of the enzymatic activities (IC50 value) and the change in the intracellular Ca+2 concentration were determined as previously described.23
2.8 |. Fluorescence imaging
To assess cell uptake of HSA, cells were plated at confluency in 6-well plates and treated with 500 nM fluorescent HSA constructs for various time points. The media was then removed and cells were washed with PBS three times. Cells were then fixed with methanol and stained with ProLong™ Diamond Antifade Mountant with DAPI (Thermo Fisher cat# P36962). Cells were then imaged using a Nikon C1si True Spectral Imaging Confocal Laser Scanning Microscope System. All images were analyzed using ImageJ software.
2.9 |. Cellular production of cysteinyl-2-fluoro-5-maleimidobenzoic acid-12ADT (compound 6) from 12ADT-MAD
LNCaP cells were incubated with 500 nM 12ADT-MAD in RPMI-1640 media containing 10% fetal bovine serum for 24 h and then the media removed and the cells washed, trypsinized, and centrifuged. Cell pellets containing 107 cells were homogenized using a hand held glass homogenizer (Kimble Kontes size 22) in 1 mL of Complete Protease inhibitor (Roche Diagnostics, Indianapolis, IN) buffer. This whole homogenate was deproteinated with the addition of 3 volumes of acetonitrile containing 0.1% Formic acid (CH3CN/0.1% FA) and after mixing, samples were centrifuged at 15 000 RPM × 5 min. Resulting supernatant and calibration standards of Cysteinyl-maleimide-12ADT were separately analyzed by liquid chromatography coupled to a quadripole mass spectrometer (LC/MS/MS) (Applied Biosystems ABI 3000 triple quadrapole MS).
3 |. RESULTS
3.1 |. Kinetics of 2-fluoro-5-maleimidobenzoic acid 12ADT (compound 4) coupling with HSA
Initially, the kinetics of coupling 2-fluoro-5-maleimidobenzoic acid (compound 3) to the sulfhydryl group of a low molecular weight compound (ie, 2-mercatoethanol) was evaluated. When compound 3 was incubated with excess 2-mercaptoethanol (10 molar equivalent) in DMSO-d6 and the reaction was monitored using 1H-NMR. Within 30 min, complete disappearance of the peak corresponding to the maleimide peak at 7.20 and the presence of the peaks that indicate the expected Michael addition reaction and the formation of succinimide-thioether (indicated by peaks at 2.95 [1H, m], 3.31 [1H, m], 4.18 [1H, m]) were observed. These results prove that just like the common N-alkyl-maleimide derivative, 2-fluoro-5-maleimidobenzoic acid also undergo fast Michael addition reaction with sulfhydryl group to form succinimide-thioether. Based upon these positive results, the kinetics of 2-fluoro-5-maleimidobenzoic acid 12ADT (compound 4) coupling to HSA was tested. These studies documented that within 30 min, there was >95% loss of albumin associated free sulfhydryl.
3.2 |. Rapid cellular uptake of 2-fluoro-5-maleimidobenzoic acid coupled human serum albumin (HSA)
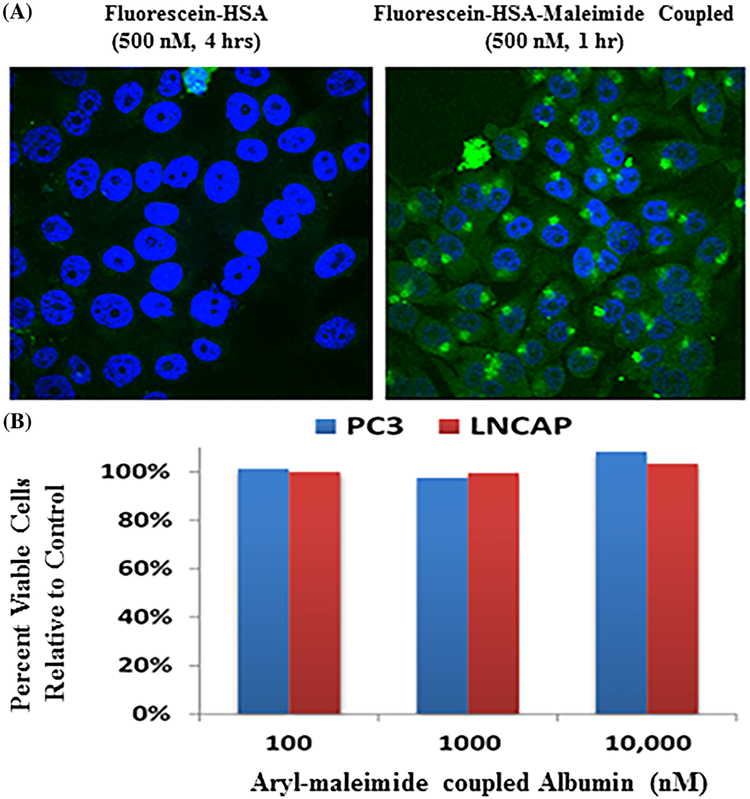
HSA was fluorescein-labeled via n-linkage to a series of lysines using standard techniques and FPLC purification. An aliquot of the fluorescently labeled HSA was coupled via Cys-34 to 2-fluoro-5-maleimidobenzoic acid (ie, compound 3). A variety of human prostate cancer cells were incubated in tissue culture with 500 nM of either the fluorescein-HSA with unreacted Cys-34 (ie, fluorescein-HSA) or fluorescein-HSA-maleimide coupled proteins and at various times, the cultures were washed, methanol fixed, counter-stained with DAPI to identify nuclei which are blue, and examined for green fluorescein-albumin using confocal microscopy. Unexpectedly, but significantly, only the 2-fluoro-5-maleimidobenzoic acid coupled fluorescein-labeled HSA is rapidly (ie within minutes) internalized producing both punctate endosomal and diffuse cytoplasmic staining, with little uptake of the green fluorescein-HSA with unreacted Cys-34 even after 4 h, Figure 5A. Western blot analysis demonstrated that >90% of the fluorescein-label in the cytoplasmic fraction is associated with small peptides and free fluorescein-labeled-lysine consistent with albumin degradation in the endosome. This is consistent with earlier studies documenting that endothelial cells transcytose unmodified albumin without degrading it, but albumin in which Cys-34 is capped with gold is taken up and degraded by the cells into its amino acids.28 Such selective cellular uptake of fluorescein-HSA-2-fluoro-5-maleimidobenzoic acid is not toxic to prostate cancer cells as documented by its lack of growth inhibitory ability even after 5 days of exposure at 10 μM, Figure 5B.
FIGURE 5.
A, Differential uptake by PC-3 human prostate cancer cells of 500nM FITC-HSA at 4 h vs 500 nM FITC-HSA-maleimide at 1 h. Blue fluorescence is due to DAPI staining of DNA and Green fluorescence is due to fluorescein-HSA-maleimide uptake. B, Dose-Response of LNCaP and PC-3 human prostate cancer cells to 5 day of fluorescein-HSA-maleimide as evaluate percent viability compared to untreated control cells.
These data document that when Cys-34 in albumin is “capped,” the protein is “recognized” differently than unmodified albumin and is phagocytized and degraded into its free amino-acids by cancer cells. Thus by capping albumin with a 2-fluoro-5-maleimidobenzoic acid coupled cytotoxic drug, we hypothesize that the resultant resulting maleimide-linked albumin drug (MAD) can selectively concentrate in the extracellular fluid (ECF) at sites of metastatic prostate cancer due to the combination of an enhanced serum half-life and tumor specific EPR effect. Once in the tumor ECF, MAD is taken up by the cancer cells and degraded within endosomes to liberate its cytotoxic moiety.
3.3 |. Kinetics of coupling of 2-fluoro-5-maleimidobenzoic acid-12ADT to albumin in situ in serum to produce 12ADT-MAD
There are two possible methods to produce 12ADT-MAD (compound 5) for therapeutic systemic delivery. The first is to couple 2-fluoro-5-maleimidobenzoic acid-12ADT (compound 4) with albumin directly ex vivo as presented in Figure 3 and then purify the 12ADT-MAD for subsequent intravenous injections. Alternatively, since we have documented that the rate of coupling to albumin is so rapid, compound 4 could be injected intravenously allowing coupling in the blood to the high (~400 μM) albumin concentration producing 12ADT-MAD in situ, as long as there are no serum components which either compete for or inhibit such coupling. To test for this possibility, the kinetics of such in situ coupling was evaluated by incubating 100 μM of compound 4 with fetal bovine serum containing ~400 μM albumin at 37°C. These results documented that coupling of compound 4 with albumin produces compound 5 rapidly in the serum (ie, >90% of the compound 4 is covalently linked to albumin within 5 min with essentially no free non-protein bound compound 4 being detectable after 30 min).
3.4 |. Therapeutic efficacy of 12ADT-MAD against prostate cancer cells
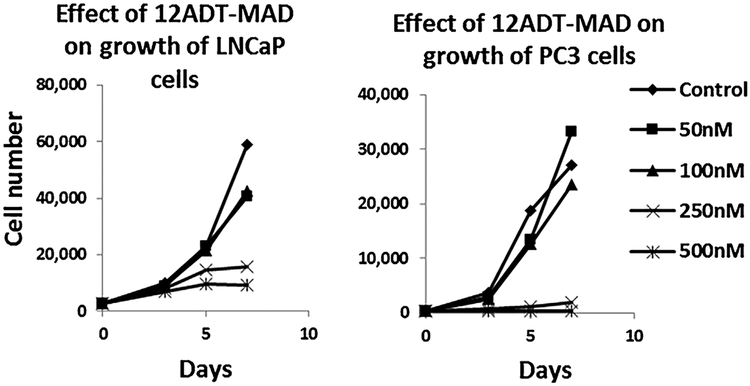
The dose response cytotoxicity of 12ADT-MAD (compound 5) was evaluated against a series of six human prostate cancer lines (LNCaP, LAPC-4, VCap, CWR22Rv1, PC3, and Du145). These results demonstrated that cell death is extensively induced in all lines with IC50 values ranging from 100 to 250 nM. Figure 6 presents representative data for the PC3 and LNCaP cell lines. Importantly, therapeutic response is totally dependent upon the 12ADT as part of the MAD since the albumin coupled aryl-maleimide without the 12ADT has no growth inhibitory ability, Figure 5B. Using fluorescein labeling and LC/MS, we documented that 12ADT-MAD is endocytosed by prostate cancer cells where it is degraded into amino acids liberating cysteinyl-maleimide-12ADT (compound 6). Compound 6 was separately documented to be chemically stable at the acidic pH of 5.5 in the endosome and to retain potent killing ability (IC50 <50 nM) for prostate cancer cells. This cell killing is due to its SERCA inhibition (IC50 20 nM) inducing an ER stress response and a “capacitance entrance” of extracellular Ca+2 in the prostate cancer cells causing an increase in intracellular Ca+2 (Cai) to >1 μM producing apoptotic death.
FIGURE 6.
Kinetics of the dose response toxicity of 12ADT-MAD to LNCaP and PC-3 human prostate cancer cells
4 |. DISCUSSION
Based upon these positive in vitro validation results, the in vivo dose-response efficacy versus host toxicity of this12ADT-MAD approach is presently being evaluated against a series of patient derived androgen responsive and castration resistant human xenografts in immune-deficient mice. As part of this testing, we are comparing the dose-response of anti-tumor efficacy versus host toxicity of intravenously injected ex vivo synthetically produced 12ADT-MAD versus in situ generated 12ADT-MAD produced by intravenous injection of 2-fluoro-5-maleimidobenzoic acid-12ADT. These future studies will thus define which approach is optimal for clinical translation.
ACKNOWLEDGMENTS
we would like to acknowledge the Department of Defense Prostate Cancer Research Program, Award No W81XWH-16–1-0410, NIH-Prostate SPORE; Grant number: P50 CA058236. Also we wish to thanks the Cell Imaging Facility supported by the SKCCC CCSG (P30 CA006973) for their services and assistance; the Mass Spectrometry and Proteomics Core facility of the Johns Hopkins School of Medicine for the Mass Spec analysis; and Johns Hopkins School of Medicine Pharmacology Department for the H and C-13 NMR analysis.
Funding information
DoD, Grant number: W81XWH-16-1-0410; NIH-Prostate SPORE, Grant number: P50 CA058236; Center for Scientific Review, Grant numbers: P50 CA058236, p30 CA006973
REFERENCES
- 1.Martinez-Outschoorn UE, Pestell RG, Howell A, et al. Energy transfer in “parasitic” cancer metabolism: mitochondria are the powerhouse and Achilles’ heel of tumor cells. Cell Cycle. 2011;10:4208–4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Warburg O, Wind F, Negelein E. The metabolism of tumors in the body. J Gen Physiol. 1927;8:519–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cori CA, Cori GT. The carbohydrate metabolism of tumours. J Biol Chem. 1925;65:397–405. [Google Scholar]
- 4.Mider GB. Some aspects of nitrogen and energy metabolism in cancerous subjects: a review. Cancer Res. 1951;11:821–829. [PubMed] [Google Scholar]
- 5.Babson AL, Winnick T. Protein transfer in tumor-bearing rats. Cancer Res. 1954;14:606–611. [PubMed] [Google Scholar]
- 6.Commisso C, Davidson SM, Soydaner-Azeloglu RG, et al. Macro-pinocytosis of protein is an amino acid supply route in Ras-transformed cells. Nature. 2013;497:633–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davidson SM, Jonas O, Keibler MA, et al. Direct evidence for cancer-cell-autonomous extracellular protein catabolism in pancreatic tumors. Nat Med. 2017;23:235–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stehle G, Wunder A, Schrenk HH, Hartung G, Heene DL, Sinn H. Albumin-based drug carriers: comparison between serum albumins of different species on pharmacokinetics and tumor uptake of the conjugate. Anticancer Drugs. 1999;10:785–790. [PubMed] [Google Scholar]
- 9.Stehle G, Sinn H, Wunder A, et al. Plasma protein (albumin) catabolism by the tumor itself-implications for tumor metabolism and genesis of cachexia. Crit Rev Oncol Hematol. 1997;26:77–100. [DOI] [PubMed] [Google Scholar]
- 10.Sleep D, Cameron J, Evans LR. Albumin as a versatile platform for drug half-life extension. Biochim Biophys Acta. 2013;1830:5526–5534. [DOI] [PubMed] [Google Scholar]
- 11.Matsumura Y, Meada H. A new concept for macromolecular therapeutics in cancer chemotherapy: mechanism of tumoritropic accumulation of proteins and antitumor agent Smancs. Cancer Res. 1986;46:6387–6392. [PubMed] [Google Scholar]
- 12.Isaacs JT, Dalrymple SL, Rosen DM, Hammers H, Olsson A, Leanderson T. Anti-cancer potency of tasquinimod is enhanced via albumin-binding facilitating increased uptake in the tumor microenvironment. Oncotarget. 2014;5:8093–8106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kratz F A clinical update of using albumin as a drug vehicle—a commentary. J Control Release. 2014;190:331–336. [DOI] [PubMed] [Google Scholar]
- 14.Bhattacharya AA, Grüne T, Curry S. Crystallographic analysis reveals common modes of binding of medium and long-chain fatty acids to HSA. J Mol Biol. 2000;303:721–732. [DOI] [PubMed] [Google Scholar]
- 15.Kratz F, Warnecke A, Scheuermann K, et al. Probing the cysteine-34 position of endogenous Serum albumin with thiol-binding doxorubicin derivatives: improved efficacy of an acid-sensitive doxorubicin derivative with specific albumin-binding properties compared to that of the parent compound. J Med Chem. 2002;45:5523–5533. [DOI] [PubMed] [Google Scholar]
- 16.Ogasawara Y, Mukai Y, Togawa T, Suzuki T, Tanabe S, Ishii K. Determination of plasma thiol bound to albumin using affinity chromatography and high performance liquid chromatography with fluorescence detection: ratio of cysteinyl albumin as a possible biomarker of oxidative stress. J Chromatogr B Anal Technol Biomed Life Sci. 2007;845:157–163. [DOI] [PubMed] [Google Scholar]
- 17.Gryzunov YA, Arroyo A, Vigne JL, et al. Binding of fatty acids facilitates oxidation of cysteine-34 and converts copper-albumin complexes from antioxidants to prooxidants. Arch Biochem Biophys. 2003;413:53–66. [DOI] [PubMed] [Google Scholar]
- 18.Alley SC, Benjamin DR, Jeffrey SC, Okeley NM, Meyer DL, Sanderson RJ. And Senter PD. Contribution of linker stability to the activities of anticancer immunoconjugates. Bioconjugate Chem. 2008;19:759–765. [DOI] [PubMed] [Google Scholar]
- 19.Shen BQ, Xu K, Liu L, et al. Conjugation site modulates the in vivo stability and therapeutic activity of antibody-drug conjugates. Nat Biotechnol. 2012;30:184–189. [DOI] [PubMed] [Google Scholar]
- 20.Fontaine SD, Reid R, Robinson L, Ashley GW, Santi DV. Long-term stabilization of maleimide-thiol conjugates. Bioconjugate Chem. 2015;26:145–152. [DOI] [PubMed] [Google Scholar]
- 21.Christie RJ, Fleming R, Bezabeh B, et al. Stabilization of cysteine-linked antibody drug conjugates with N-aryl maleimides. J Control Release. 2015;220:660–670. [DOI] [PubMed] [Google Scholar]
- 22.De Figueiredo RM, Oczipka P, Frohlich R, Christmann M. Synthesis of 4-maleimidobutyric acid and related maleimides. Synthesis. 2008;8: 1316–1318. [Google Scholar]
- 23.Denmeade SR, Jakobsen CM, Janssen S, et al. Prostate-specific antigen-activated thapsigargin prodrug as targeted therapy for prostate cancer. J Natl Cancer Inst. 2003;95:990–1000. [DOI] [PubMed] [Google Scholar]
- 24.Vander Griend DJ, Antony L, Dalrymple SL, et al. Amino acid containing thapsigargin analogues deplete androgen receptor protein via synthesis inhibition and induce the death of prostate cancer cells. Mol Cancer Ther. 2009;8:1340–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Janssen K, Horn S, Niemann MT, Daniel PT, Schulze-Osthoff K, Fischer U. Inhibition of ER Ca2+pump forces multidrug-resistant cells deficient in Bak and Bax into necrosis. J Cell Sci. 2009;122:4481–4491. [DOI] [PubMed] [Google Scholar]
- 26.Denmeade SR, Mhaka AM, Rosen DM, et al. Engineering a prostate-specific membrane antigen-activated tumor endothelial cell prodrug for cancer therapy. Sci Transl Med. 2012;4:140ra86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Uzgare AR, Isaacs JT. Enhanced redundancy in Akt and mitogen-activated protein kinase-induced survival of malignant versus normal prostate epithelial cells. Cancer Res. 2004;64:6190–6199. [DOI] [PubMed] [Google Scholar]
- 28.Schnitzer JE, Bravo J. High affinity binding, endocytosis, and degradation of conformationally modified albumins. J Biol Chem. 1993;268:7562–7567. [PubMed] [Google Scholar]