Abstract
The metabolic instability of mRNA currently limits its utility for gene therapy. Compared to plasmid DNA, mRNA is significantly more susceptible to digestion by RNase in the circulation following systemic dosing. To increase mRNA metabolic stability, we hybridized a complementary reverse mRNA with forward mRNA to generate double stranded mRNA (dsmRNA). RNase A digestion of dsmRNA established a 3000-fold improved metabolic stability compared to single stranded mRNA (ssmRNA). Formulation of a dsmRNA polyplex using a PEG-peptide further improved the stability 3000-fold. Hydrodynamic dosing and quantitative bioluminescence imaging of luciferase expression in the liver of mice established the potent transfection efficiency of dsmRNA and dsmRNA polyplexes. However, hybridization of the reverse mRNA against the 5’ and 3’ UTR of forward mRNA resulted in UTR denaturation and a 10-fold loss in expression. Repeat dosing of dsmRNA polyplexes produced equivalent transient expression, suggesting the lack of an immune response in mice. Co-administration of excess uncapped dsmRNA with a dsmRNA polyplex failed to knockdown expression, suggesting dsmRNA is not a Dicer substrate. Maximal circulatory stability was achieved using a fully complementary dsmRNA polyplex. The results established dsmRNA as a novel metabolically stable and transfection competent form of mRNA.
Introduction
The liver has been the focus of numerous investigations of non-viral DNA and mRNA gene delivery systems1–21 because of its unique fenestrated endothelium cells that allow nanoparticles to escape the circulation and contact hepatocytes22–23. An efficient systemically dosed DNA delivery system to transfect hepatocytes has eluded the field24. Even though DNA delivery offers several advantages over mRNA delivery, which include greater metabolic stability, ease of preparation, and greater amplification, delivery of DNA to the nucleus of quiescent hepatocytes remains a major hurdle25–26. The delivery of mRNA to the cytosol of hepatocytes circumvents the need for nuclear delivery, resulting in efficient but short lived transient expression27.
There have been several investigations demonstrating the efficacy of systemically delivered mRNA to achieve expression in liver hepatocytes17–18, 20–21, 28. Systemically delivered lipid nanoparticles produced appreciable luciferase expression in mouse liver following administration of a 1 μg mRNA dose18. A lipid nanoparticle was able to deliver human erythropoietin mRNA resulting in physiologically relevant expression28. Likewise, a LUNAR lipid nanoparticle containing human factor IX mRNA was efficacious when delivered to hemophilia mice21. Therapeutic efficacy against HIV challenge was achieved by systemic delivery of mRNA encoding an anti-HIV antibody in a lipid nanoparticle17. However, in the examples above, despite high expression efficiency, lipid nanoparticle doses of 10–15 μg of mRNA were required to achieve efficacy in mice.
The metabolic instability of mRNA remains a significant obstacle toward improving the efficiency and persistence of expression27. Naked mRNA is completely digested in human plasma in 15 seconds29. A study of polyethylene glycol (PEG)-polylysine mRNA polyplex resulted in only 1% of dose recovery after 5 min, whereas a more stabilized formulation improved recovery to 25%30. By comparison, an optimized PEG-peptide DNA polyplex remains fully stable in the circulation for up to four hrs31–33. A key element of this polyplex is the incorporation and spacing of four Acr-Lys residues (acridine attached to the ɛ-amine of Lys) to increase binding affinity for DNA by polyintercalation31. PEGylated polyacridine peptides also delayed the metabolism of single stranded (ss) mRNA due to intercalation into the double stranded stem loop native structure resulting in increased serum stability34.
In the present study, we demonstrate an approach to further improve the metabolic stability of mRNA by developing and testing double stranded (ds) mRNA PEGylated polyacridine peptide polyplexes. Hybridization of a partially complementary reverse RNA strand mRNA results in dsmRNA that demonstrates increased RNase stability and remains fully translationally competent in mice when dosed hydrodynamically, but rapidly loses activity when dosed systemically. Elongation of the reverse RNA strand to be fully complementary with mRNA diminished dsmRNA polyplex translational competency but increased circulatory stability. Unlike viral dsmRNA which is highly immunogenic35, capped and poly(A) tailed dsmRNA appears to be well-tolerated upon repeated hydrodynamic dosing with no apparent adverse effects. These results suggest that dsmRNA may improve the efficacy of other systemically delivered non-viral gene delivery vectors.
Materials and Methods
Rev Luc-UTR pcDNA 3.1 (-)
Template DNA was prepared by incorporating a reverse-oriented T7 promoter (3’ to 5’ relative to the coding sequence of luciferase) along with an M13 sequencing primer into Luc-UTR pcDNA 3.1 (-) between the BamHI and HindIII cut sites on the 3’ end of the 3’ UTR (Supplemental Fig. S1). Luc-UTR pcDNA 3.1 (-) has been described previously34. Insert DNA was purchased from Integrated DNA Technologies, Coralville, IA. The insert was digested with HindIII-HF and BamHI-HF (New England Biolabs; NEB, Ipswich, MA, USA). HindIII-HF and BamHI-HF linearized plasmid DNA was treated with alkaline phosphatase (Calf Intestinal Alkaline Phosphatase; CIP, NEB) to prevent self-ligation, followed by phenol-chloroform extraction and alcohol precipitation. Linear DNA and insert were ligated with T4 DNA ligase (NEB), then used to transform DH5-α E. coli., resulting in positive colonies, from which plasmid DNA was isolated using a Qiagen Miniprep kit (Qiagen). Sequences were confirmed by Sanger Sequencing at the Iowa Institute of Human Genetics using an M13 sequencing primer. The schematic of Rev Luc-UTR pcDNA 3.1 (-) is illustrated in supplemental figure S1.
Luc-UTR 80A pc DNA3.1 (-)
An 81-adenosine encoded stretch was installed into Rev Luc-UTR pcDNA 3.1 (-)[9]. Two DNA inserts were restricted along with Rev Luc-UTR pcDNA 3.1 (-) as described in supplemental figure S2. The two inserts and linear DNA vector were ligated and transformed into E. coli as described above. A 33A insert was isolated and confirmed by Sanger Sequencing. An internal BsmBI restriction site was removed by site directed mutagenesis. Two additional poly(A) insertion steps of 24 A’s were installed as indicated in supplemental figure S2. The resulting plasmid, Luc-UTR 80A pc DNA 3.1 (-), was verified by Sanger sequencing (Supplemental Fig. S2).
Rev Luc-UTR 80A pcDNA 3.1 (-)
Luc-UTR 80A pcDNA 3.1 (-) was digested sequentially with HindIII-HF and BsmBI and dephosphorylated using recombinant shrimp alkaline phosphatase (rSAP). A hybridized T7 promoter DNA insert (IDT) was annealed and ligated to linearized Luc-UTR 80A pcDNA 3.1 (-) using T4 DNA ligase. The ligated plasmid was transformed into DH5-α E. coli and the plasmid was isolated and subjected to Sanger Sequencing. The resulting plasmid (Rev Luc-UTR 80A pc DNA3.1 (-) is described in supplemental figure S3.
PCR Generation of Luc-DNA.
Luc-UTR pcDNA 3.1 (-) was linearized with AseI and BsaI to disrupt the T7 promoter. A reverse primer containing a T7 promoter was annealed upstream of the 3’ UTR and a forward primer was annealed at the start codon immediately downstream of the 5’ UTR (Supplemental Fig. S4). The DNA was amplified by PCR using PfuUltra High-Fidelity DNA Polymerase (Agilent Technologies, Santa Clara, CA, USA). The PCR product (Luc-DNA) was purified by phenol-chloroform extraction and verified by Sanger sequencing (Supplemental Fig. S4).
In Vitro Transcription of ssmRNA
Rev Luc-UTR pcDNA 3.1 (-) was linearized with BamHI-HF to produce a plasmid encoding a translatable forward strand of ssmRNA. Briefly, 12 μg of plasmid was linearized with 50 U of BamHI-HF 37°C for 1 hr, then digested with 0.6 U of proteinase K (Thermo Fisher Scientific, Pittsburgh, PA, USA) in 0.5% SDS (Research Products International; RPI, Mt. Prospect, IL, USA) to remove residual RNase A. The plasmid was purified by phenol-chloroform extraction and isopropanol precipitation, and quantified by absorbance to yield 90%, with an absorbance 260/280 ratio of 1.8. BamHI-linearized DNA template was used to prepare pre-mRNA by in vitro transcription (IVT) using the Ambion MEGAscript T7 Kit (Life Technologies, Grand Island, NY, USA) according to manufacturer’s instructions. One μg of linearized template was combined with 7.5 mM ATP, GTP, CTP, and ΨTP (TriLink Biotechnologies, San Diego, CA, USA), 10× reaction buffer, and T7 RNA polymerase in a total volume of 20 μl, then incubated at 37°C for 4 h. Two units of TURBO DNase (Ambion) was added and incubated for 15 min at 37°C to remove the DNA template. The reaction was diluted with water (30 μl) and terminated with the addition of 30 μl of 7.5 M lithium chloride and 50 mM EDTA (pH 8.0) and chilled to precipitate the pre-mRNA. The pellet was recovered by centrifugation at 16,000 × g for 30 min, washed with 70% ethanol, re-centrifuged, dissolved in nuclease free water (RPI) and quantified by absorbance. One 20 μl IVT reaction resulted in a yield of 190 μg of pre-mRNA with a 260/280 nm absorbance ratio of 1.9. A 5’ cap and poly(A) tail was added to the pre-mRNA using the Vaccinia Capping System, mRNA Cap 2’-O-Methyltransferase, and E. coli Poly(A) Polymerase (NEB) according to an adaptation from the manufacturer’s protocol. Pre-mRNA (180 μg in 240 μl) was heat-denatured at 65°C for 5 min and chilled on ice for 5 min, followed by the addition of 9 μl of 40 U/μl RNase OUT (Life Technologies), 36 μl of 10× capping buffer, 18 μl of 10 mM GTP, 18 μl of 2 mM SAM, 18 μl of 10 U/μl Vaccinia Capping Enzyme, and 18 μl of 50 U/μl mRNA Cap 2´-O-Methyltransferase, incubated at 37°C for 1 h. Capped mRNA was diluted in 1× E. coli poly(A) polymerase reaction buffer, and tailed by reaction with 1 mM ATP and 100 U of E. coli poly(A) polymerase. The resulting capped and tailed mRNA of approximately 1990 bases was purified by lithium chloride precipitation to yield 220 μg of ssmRNA with a 260/280 nm absorbance ratio of 2.2 (Supplemental Figure S1). Alternatively, Luc-UTR 80A pcDNA 3.1 (-) was linearized by digestion with BsmBI (NEB) to yield a 1,974-base luciferase transcript with a poly(A) tail of 80 nucleotides without a non-A overhang. IVT and capping were performed as described above to yield 150 μg of capped and tailed ssmRNA(80A) (Supplemental Figure S2). ssmRNA(80A) was also prepared by individual substitution of ATP, GTP, CTP, and ΨTP with phosphorothioate nucleotides ATPαS, UTPαS, GTPαS and CTPαS during in vitro transcription36.
In Vitro Transcription of Rev-mRNA
Rev Luc-UTR pcDNA 3.1 (-) was linearized with BsaI-HF and used to produce a 1903 base reverse mRNA1 (Rev-mRNA1) by IVT. Rev-mRNA1 was purified and quantified as above to yield 130 μg (Supplemental Fig. S1). A shorter 1,656-base Rev-mRNA2 was prepared by subjecting Luc-DNA to IVT. Rev-mRNA2 was purified and quantified as described above to yield 130 μg (Supplemental Fig. S4). Rev Luc-UTR 81A pcDNA 3.1 (-) was linearized by digestion with BsaI-HF. IVT followed by purification on RNeasy MinElute (Qiagen) resulted in a 17 μg yield of a 1980 base Rev-mRNA(80A) (Supplemental Fig. S3). Positioning the T7 promoter directly in front of a poly(U) stretch, as well as the first nucleotides being GGU instead of GGGAGA, accounted for the low yield.
Preparation and Characterization of dsmRNA
dsmRNA was prepared by hybridizing equimolar quantities of forward ssmRNA or ssmRNA(80A) with Rev-mRNA1, Rev-mRNA2, or Rev-mRNA(80A) at a concentration 0.2 μg/μl in a total volume 25 μl of SSC (150 mM sodium chloride, 15 mM sodium citrate, pH 7) by thermocycler heating at 85°C for 5 min, then cooling to 25°C over 10 min. dsmRNA (0.5 μg, 15μl) was combined with 3 μl 5× LB loading medium (FBM; Faster Better Media, Hunt Valley, MD, USA) and electrophoresed for 30 min at 145 V on a 1% agarose gel, containing 2 μl of 5 mg/ml ethidium bromide, in 250 ml of 0.5× lithium boric acid buffer (FBM). Agarose gels were imaged with the UVP BioSpectrum Imaging System and VisionWorks LS software (UVP, Upland, CA, USA) using an ethidium bromide filter. Hybridized dsmRNA was identified by band shift relative to heat cycled ssmRNA and Rev-mRNA.
Formulation of ssmRNA and dsmRNA Polyplexes
Peptide synthesis and PEGylation was accomplished as previously described34. To establish the stoichiometry of PEGylated polyacridine peptide binding to ss and dsmRNA, an agarose gel electrophoretic band mobility shift assay was conducted. Heat denatured ssmRNA (0.5 μg/7.5 μl in water) or dsmRNA1 (0.5 μg/7.5 μl in SSC) were combined with an equal volume of 20, 50, 100, 400, or 800 pmol of PEGylated polyacridine peptide in 5 mM HEPES pH 7 in a total volume of 15 μl. Agarose gel electrophoresis was performed as described above. The particle size and zeta potential of mRNA nanoparticles were determined at a concentration of 50 μg/ml in 5 mM HEPES by dynamic light scattering on a Brookhaven ZetaPlus (Holtsville, NY). The values reported are the intensity averaged unimodal distribution mean and standard error from 10 replicate measurements.
ss and dsmRNA Metabolic Stability
The metabolic stability of dsmRNA1, ssmRNA, and polyplexes were compared by RNase digestion as described previously20. Polyplexes were formed at a stoichiometry of 0.8 nmol of PEG-peptide per 1 μg of ss or dsmRNA. RNase A (0, 10 fg, 300 fg, 10 pg, 300 pg, 10 ng, 300 ng, or 10 μg, 50 U) was added to digest 1 μg of ssmRNA, dsmRNA1, ssmRNA polyplex, or dsmRNA1 polyplex in 15 μl of 3 mM HEPES pH 7.4 / 0.3× SSC at 37°C for 15 min. RNase A digestion was stopped by 1 hr digestion at 37°C with 5 U of proteinase K in 400 μl RNase extraction buffer (50 mM Tris, 1% SDS, 100 mM NaCl, pH 8). mRNA was extracted with phenol-chloroform to remove PEG-peptide, precipitated in ethanol containing 20 μg of glycogen, and reconstituted in 15 μl RNase-free water (RPI). Isolated mRNA was heat denatured and loaded onto a 1% agarose gel and electrophoresed as described above.
The metabolic stability of ss and dsmRNA were also compared by digestion with mouse serum. dsmRNA1 and heat-denatured ssmRNA (1 μg) where combined with 0 to 1 v/v% mouse serum (Sigma-Aldrich, St. Louis, MO, USA) in a total volume of 15 μl of 5 mM HEPES pH 7.4. The serum digestions were incubated at 37°C for 15 min then combined with 3 μl 5× loading buffer and analyzed by agarose gel electrophoresis as described above.
Hydrodynamic Dosing of ss and dsmRNA
mRNA (1 μg) and mRNA polyplexes were diluted in 1.8 ml of saline and hydrodynamically dosed in triplicate mice by rapid 5 sec administration via the tail vein as previously described34. Bioluminescence imaging (BLI) was performed at 4, 24, 48 and 72 h post-dosing as previously described34. Hydrodynamic stimulation was performed by tail vein administration of 1 μg of mRNA or mRNA polyplex in 100 μl of saline. At circulation delay times of 5 to 20 min, a hydrodynamic stimulatory dose of 1.8 ml of saline (without mRNA) was rapidly administered in 5 sec via the tail vein followed by BLI quantification of luciferase expression at 24 h post hydrodynamic-dosing.
siRNA silencing of dsmRNA
A dicer substrate siRNA (DsiRNA) was designed to target the 5’ coding region of the luciferase mRNA beginning three nucleotides after the AUG start codon. The hybridized sense sequence, 5’GGACGCCAAGAAUAUCAAGAAAGGC 3’, (GC represents DNA bases) and antisense sequence, 5’GCCUUUCUUGAUAUUCUUGGCGUCCUC 3’, obtained from Integrated DNA Technology (Coralville IA) was reconstituted in water. Translationally non-functional dsmRNA was also evaluated as a dicer substrate. Uncapped ssmRNA was prepared and hybridized with Rev-mRNA2. A 10 μg dose of DsiRNA or uncapped dsmRNA2 were combined with a 1 μg dose of translationally active ssmRNA or dsmRNA2 in 1.8 ml of saline and administered hydrodynamically in triplicate mice. The level of luciferase expression was quantified at 24 hrs by BLI.
Statistical Analysis
The base-10 logarithm of the raw luminescent output from BLI results were analyzed for statistically significant differences by t-test or one-way analysis of variance (ANOVA) with Dunnet’s Multiple Comparisons Test on GraphPad Prism version 7.01 (GraphPad Software, La Jolla, CA, USA).
Results
In an effort to increase the metabolic stability of mRNA we prepared Rev-mRNA and hybridized it with ssmRNA to form dsmRNA (Scheme 1). We predicted that our dsmRNA would be more resistant to metabolism than ssmRNA. However, since dsmRNA has not been previously described in the literature, it was not clear if this novel form of mRNA would be translationally active.
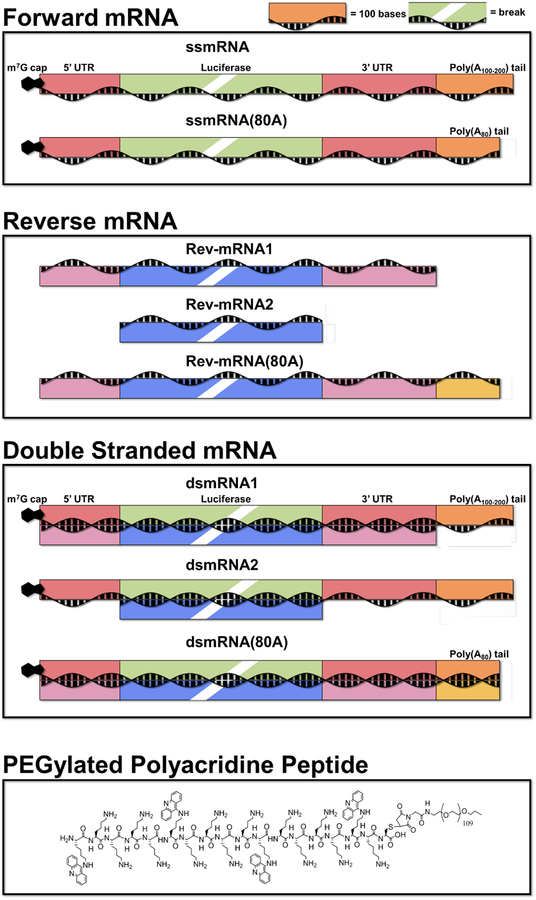
Scheme 1. Structure of ssmRNA, dsmRNA1, ssmRNA(80A), dsmRNA2, and dsmRNA(80A).
The schematic drawing of forward and reverse mRNA used to prepare dsmRNA is illustrated. Reverse strand hybridization of 3’ and 5’ UTRs distinguishes dsmRNA1 from dsmRNA2. Reverse strand hybridization with the poly(A) tail is included in dsmRNA(80A).
This study was further motivated by the finding that a previously described PEGylated polyacridine peptide bound to double stranded stem-loop regions in ssmRNA resulting in polyplexes with improved metabolic stability34 (Scheme 1). It was anticipated that the PEGylated polyacridine peptide would possess even greater affinity for dsmRNA due to increased polyintercalative binding, and that dsmRNA polyplexes would demonstrate improved metabolic stability.
In vitro transcription (IVT) was used to prepare reverse mRNA by installation of a reverse oriented T7 promoter on the 5’ end of the antisense strand of Luc-UTR pcDNA 3.1 (-)34. Restriction digestion of Luc-UTR pcDNA 3.1 (-) interrupted the forward T7 promoter, providing a linear DNA that transcribed a full length Rev-mRNA1 that was complementary to the 5’ UTR, luciferase transgene and 3’ UTR of capped and tailed mRNA (Scheme 1). Hybridization of equal-mol amounts of Rev-mRNA1 and ssmRNA resulted in the generation of a single band of higher molecular weight dsmRNA1 (Fig. 1).
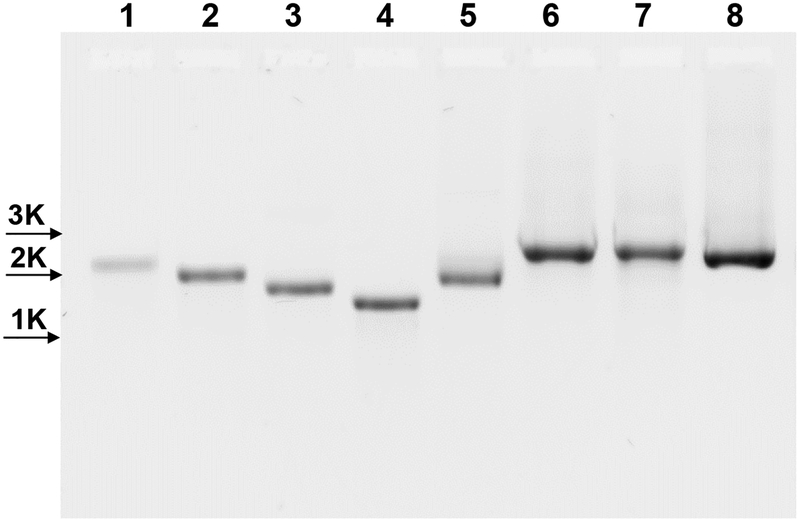
Figure 1. Agarose Gel Electrophoresis of dsmRNA Hybridization.
The electrophoretic migration of ssmRNA (1), ssmRNA(80A) (2), Rev-mRNA1 (3), Rev-mRNA2 (4), and Rev-mRNA(80A) (5) (0.5 μg) are compared to dsmRNA1 (6), dsmRNA2 (7) and dsmRNA(80A) (8) (0.5 μg total mRNA) on a 1% agarose gel detected by ethidium bromide. ssmRNAs were denatured at 65°C for 5 min and cooled at 4°C for 5 min prior to electrophoresis. dsmRNAs were annealed by heating equimolar amounts of ssmRNA and Rev-mRNA at 85°C for 5 min following by cooling to 25°C over ten min in 150 mM sodium chloride, 15 mM sodium citrate, pH 7.
Truncated Rev-mRNA was prepared by IVT from PCR-generated linear DNA. A primer containing a T7 promoter was hybridized near the 5’ end of the antisense strand along with a second 3’ primer to generate truncated DNA templates of precise length containing a reverse T7 promoter. These experiments led to the development of Rev-mRNA2 that hybridizes with ssmRNA while leaving the 5’ and 3’ UTR native (Scheme 1). Hybridization of Rev-mRNA2 with ssmRNA resulted in a single higher molecular weight band for dsmRNA2 (Fig. 1).
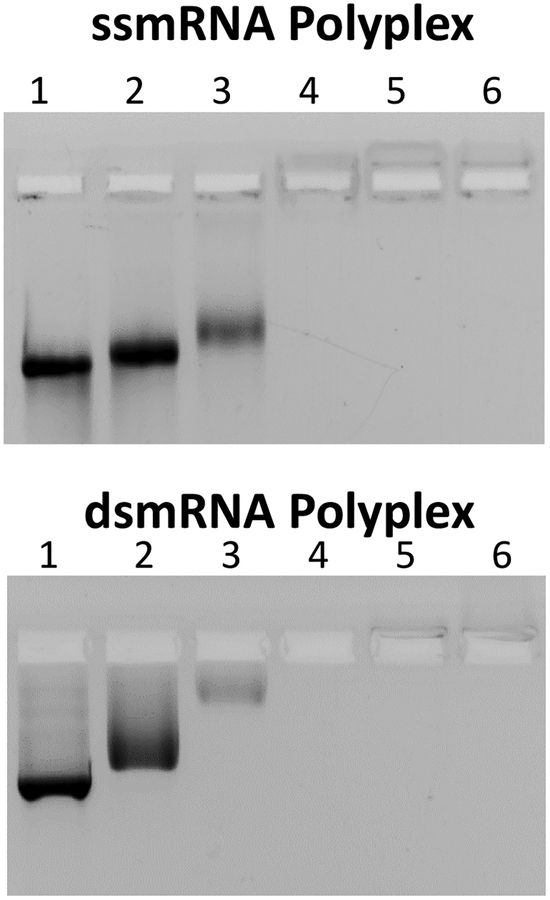
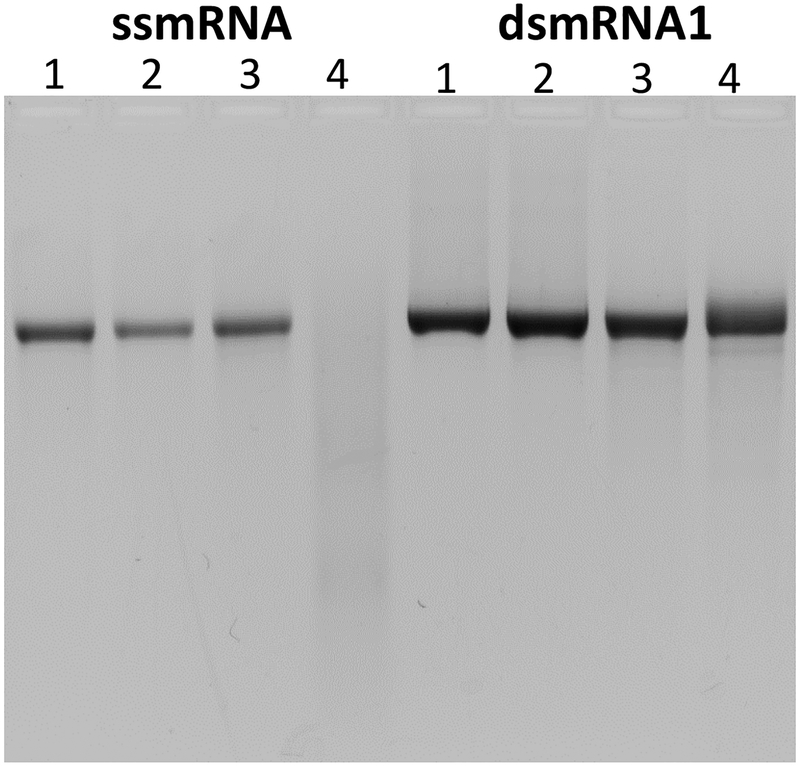
In a previous study, the affinity of a PEGylated polyacridine peptide for binding DNA was optimized to increase the circulatory stability of PEGylated DNA polyplexes31 (Scheme 1). The addition of 0.2 nmol of PEG-peptide to 1 μg of DNA resulted in the spontaneous formation of polyplexes31–33. The circulatory stability is dependent on the number and spacing of Lys-Acr (acridine) residues that bind to DNA by polyintercalation31. To determine the stoichiometry at which mRNA forms polyplexes, the amount of PEGylated polyacridine peptide was varied from 0 to 0.8 nmol combined with a fixed 0.5 μg of ss and dsmRNA and analyzed by agarose gel electrophoresis band shift assay. Complete band shift of ss and dsmRNA occurred at a stoichiometry of 0.2 nmol of PEG-peptide per μg of mRNA (Fig. 2). To ensure complete mRNA polyplex formation, a PEG-peptide stoichiometry of 0.8 nmol per μg of dsmRNA was used throughout the remainder of the study. Dynamic light scattering established a particle diameter for ssmRNA(80A) PEG-peptide polyplexes of 62 nm, compared to 167 nm for dsmRNA(80A), 185 nm for dsmRNA1 and 200 nm for dsmRNA2. The zeta potential ranged from +2 to +8 mV for all double stranded mRNA polyplexes.
Figure 2. Agarose Gel Electrophoresis Band Shift of ssmRNA and dsmRNA PEG-peptide Polyplexes.
Panel A and B illustrate the band shift for ssmRNA (0.5 μg) and dsmRNA1 (0.5 μg) respectively, when combined with 0, 20, 50, 100, 400, or 800 pmol (lanes 1-6) of PEG-peptide and electrophoresed on a 1% agarose gel detected by ethidium bromide.
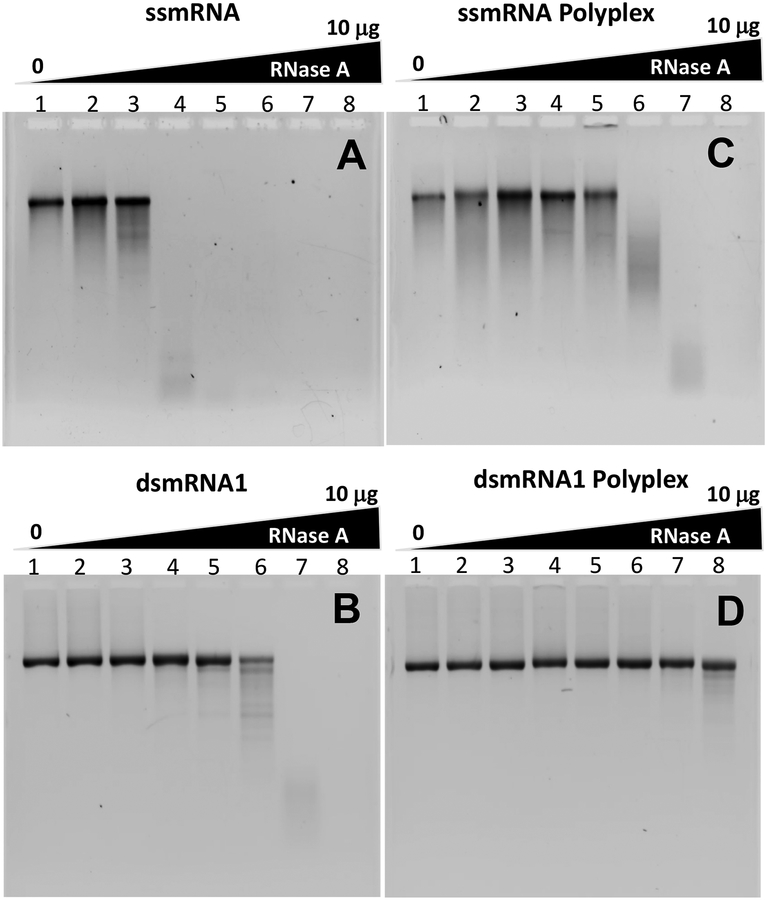
The relative metabolic stability of ssmRNA and dsmRNA to digestion with RNase A was evaluated by agarose gel electrophoresis. ssmRNA proved to be completely metabolized when digested with 10 pg (0.05 mU) of RNase A for 15 min (Fig. 3A). By comparison, dsmRNA was stable when digested with up to 300 ng (1.5 U) of RNase A, resulting in 3000-fold improved stability over ssmRNA (Fig. 3B). ssmRNA polyplexes were also stable when digested with up to 300 ng (1.5 U) of RNase A, demonstrating an improvement in metabolic stability of 3000-fold over ssmRNA (Fig. 3A & C). However, the metabolic stability of ssmRNA polyplexes were only equal to dsmRNA (Fig. 3B & C) whereas dsmRNA polyplexes resisted digestion with 10 μg (50 U) of RNase A (Fig. 3D). The overall improvement in RNase A stability was 1,000,000-fold when comparing dsmRNA polyplex with ssmRNA, which compares favorably with prior published results on mRNA stabilization by lipid nanoparticles20. To establish the relative stability of mRNA to mouse serum RNase34, ssmRNA and dsmRNA were digested with an increasing volume percent of mouse serum. Agarose gel electrophoresis established that dsmRNA possessed approximately 10-fold greater stability to mouse serum RNases compared ssmRNA (Fig. 4). The metabolic stability of dsmRNA1, dsmRNA 2 and dsmRNA(80A) were all similarly improved by relative to ssmRNA. Likewise, each dsmRNA PEG-peptide polyplex also possessed a similar improved protection to RNAse digestion. However, as demonstrated below, the circulatory stability of ssmRNA, dsmRNA1, dsmRNA2 and dsmRNA(80A) polyplexes were significantly different.
Figure 3. RNase Digestion of ssmRNA, dsmRNA, and Polyplexes.
Panel A-D illustrate the result of digestion of 1 μg of ssmRNA, dsmRNA1, ssmRNA polyplex and dsmRNA1 polyplex with 0 (1), 10 fg (2), 300 fg (3), 10 pg (4), 300 pg (5), 10 ng (6), 300 ng (7), or 10 μg (50 U) (8) of RNase A for 15 min in 15 μl 5 mM HEPES, 25 mM sodium chloride, and 2.5 mM sodium citrate. ssmRNA and dsmRNA were recovered by Proteinase K digestion followed by phenol-chloroform extraction and ethanol precipitation, then analyzed by agarose gel electrophoresis detected by ethidium bromide.
Figure 4. Serum Digestion of ssmRNA and dsmRNA.
Panel A and B illustrate the result of digestion of ssmRNA and dsmRNA1 (1 μg) digested with 0, 0.01, 0.1 or 1 vol% of mouse serum (lanes 1–4) for 15 min at 37°C, then analyzed by agarose gel electrophoresis detected by ethidium bromide.
T7 RNA polymerase was used to synthesize ssmRNA containing substituted phosphorothioate linkages for each base in an attempt to further improved the stability of dsmRNA. While these substitutions result in translationally active mRNA in prokaryotes36, hydrodynamic dosing of 1 μg of phosphorothioate A, U, C or G substituted ssmRNA(80A) in mice resulted in complete loss of luciferase expression relative to an unsubstituted control.
The in vitro translation of dsmRNA1 using the rabbit reticulocyte lysate assay was examined37. The results established that ssmRNA was translated with 100-fold greater efficiency compared to dsmRNA1. These results are consistent with a previous report that established this assay is inhibited by dsRNA37.
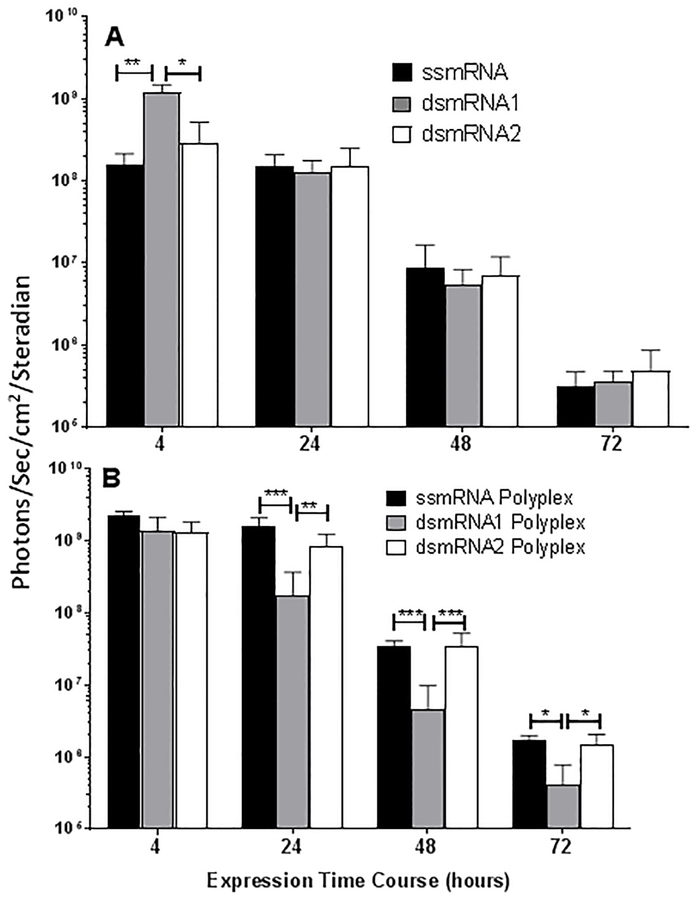
The translational competency of ssmRNA relative to dsmRNA1 and dsmRNA2, with and without PEG-peptide, was determined by luciferase expression in liver measured by bioluminescence imaging at 4–72 hrs following hydrodynamic-dosing34. Direct hydrodynamic-dosing of ssmRNA, dsmRNA1, and dsmRNA2 resulted in equivalent luciferase expression in liver at 24, 48, and 72 hr, thereby establishing that the hybridized reverse strand did not impede translation (Fig. 5A). dsmRNA1 produced significantly higher expression at 4 hrs relative to ssmRNA and dsmRNA2, which is likely due to the greater metabolic instability of ssmRNA and dsmRNA2 during hydrodynamic dosing. Likewise, the transient expression profiles for ssmRNA and dsmRNA1 and dsmRNA2 were equivalent, establishing that the increased metabolic stability of dsmRNA1 and dsmRNA2 failed to increase the persistence of luciferase expression in liver. Comparison of the result of hydrodynamic-dosing ssmRNA, dsmRNA1 and dsmRNA2 PEG-peptide polyplexes revealed a 10-fold increase in luciferase expression for ssmRNA and dsmRNA2 polyplex at 4 hrs, whereas the expression from dsmRNA1 polyplex was not increased (Fig. 5B). The significant increase in expression for ssmRNA and dsmRNA2 polyplexes relative to dsmRNA1 polyplex persisted for 24, 48 and 72 hrs (Fig. 5B). We hypothesize that dsmRNA1 polyplexes are expressed less efficiently due to denaturation of 5’ and 3’ UTR secondary structures. The shorter reverse RNA strand in dsmRNA2 preserves the native structure of the 5 and 3’ UTRs to maintain proper interaction of cis-regulatory elements34.
Figure 5. Transient Gene Expression of ssmRNA, dsmRNA and Polyplexes.
The luciferase expression in liver measured at 4–72 hrs is illustrated following hydrodynamic delivery of a 1 μg dose of mRNA to triplicate mice. Panel A illustrates the result of dosing ssmRNA, dsmRNA1, and dsmRNA2, and panel B illustrates the result of dosing PEG-peptide polyplexes. For significance, *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001.
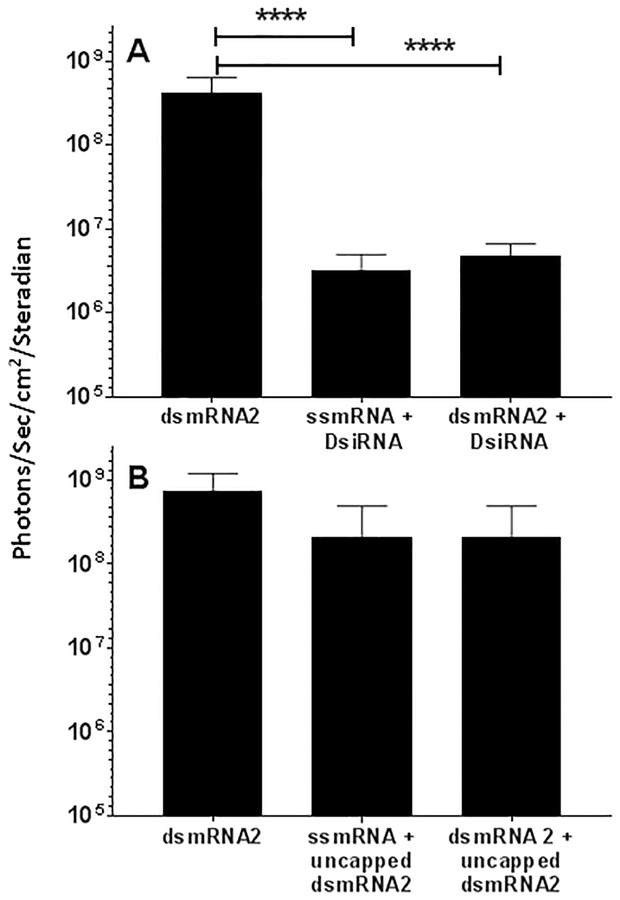
dsmRNA could potentially act as a Dicer substrate which, upon loading onto the RNA-induced silenceing complex (RISC), might silence dsmRNA mediated expression. To determine if dsmRNA was a Dicer substrate, we first attempted to establish if dsmRNA was susceptible to siRNA knockdown. We have previously established that co-hydrodynamic-dosing of an siRNA directed against a luciferase expressing plasmid (pGL3) resulted in dose and sequence-dependent knockdown in liver38. Consequently, co-hydrodynamic-dosing of DsiRNA with either ssmRNA polyplex or dsmRNA2 polyplex also resulted in the 100-fold knockdown of luciferase expression at 24 hrs for both ssmRNA polyplex and dsmRNA polyplexes (Fig. 6A). Surprisingly, the siRNA loaded RISC complex was able to act on both ssmRNA and dsmRNA with equal efficiency. However, co-administration of 10 μg of uncapped dsmRNA2 with 1 μg of capped dsmRNA2 resulted in no significant loss of luciferase expression from ssmRNA polyplex or dsmRNA2 polyplex (Fig. 6B). This result indicates that uncapped dsmRNA2 is not a Dicer substrate and, at the 10 μg dose tested, dsmRNA does not participate in RNAi mediated knockdown (Fig. 6B).
Figure 6. RNAi Knockdown of dsmRNA.
Panel A illustrates the result of hydrodynamic-dosing of 1 μg of ssmRNA and dsmRNA1 combined with 10 μg of DsiRNA in triplicate mice, followed by measuring luciferase expression in the liver by bioluminescence imaging at 24 hrs. Panel B illustrates the same experiment in which 10 μg of uncapped dsmRNA1 was substituted for DsiRNA. ****p ≤ 0.0001.
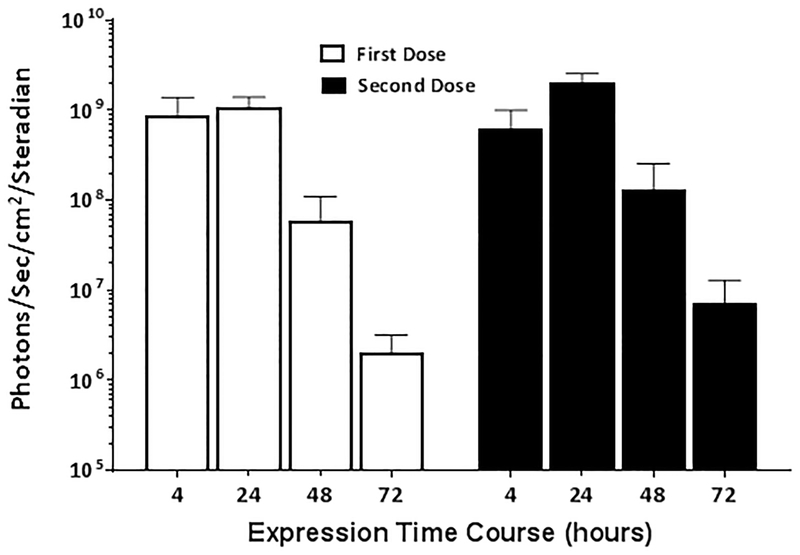
The potential for an innate immune response against dsmRNA is another concern that we addressed. A primary dose of dsmRNA2 polyplex delivered hydrodynamically resulted in a transient expression of luciferase over three days with peak expression occurring at 24 hrs post hydrodynamic delivery (Fig. 7). Administration of a second identical dose of dsmRNA2 polyplex to the same mice after one week resulted in an equivalent transient expression profile (Fig. 7). While these results do not definitively rule out any dose, or route-of-delivery dependent adverse immune responses, they do establish that dsmRNA polyplexes are well tolerated without adverse effects in a repeated hydrodynamic-dosing regimen.
Figure 7. dsmRNA Repeated Dosing.
The level of luciferase expression was measured over time following hydrodynamic-dosing of 1 μg of dsmRNA2 polyplex to triplicate mice. After one week, a second identical hydrodynamic-dose of 1 μg of dsmRNA2 polyplex was administered to the same mice and monitored for expression over time.
To further evaluate the influence of Rev-mRNA complementarity with ssmRNA, a plasmid encoding a fixed 81 poly(A) sequence was generated (Supplemental S2). IVT resulted in ssmRNA(80A) possessing 5’ and 3’ UTRs and an 80 base poly(A) tail. Direct hydrodynamic-dosing of ssmRNA(80A) confirmed luciferase expression equal to ssmRNA poly(A) tailed enzymatically. Installation of a reverse T7 promoter in the plasmid also allowed the IVT of RevmRNA(80A) that was fully complementary to ssmRNA(80A), including the 80-A tail (Supplemental Fig. S5).
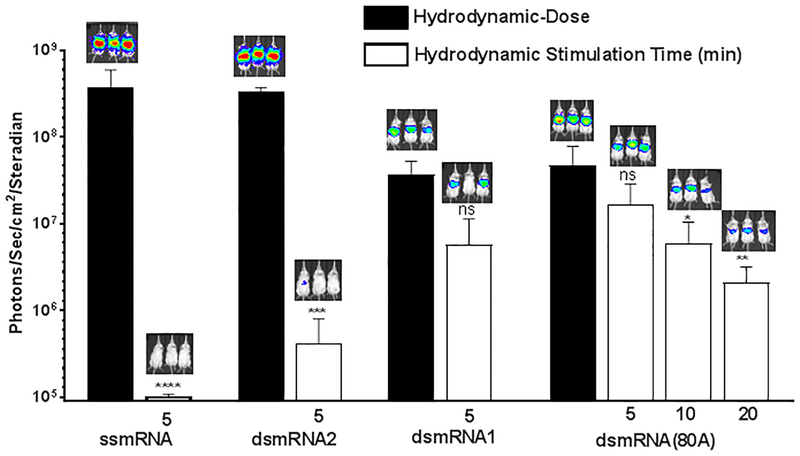
To determine if the metabolic stability of dsmRNA corresponded to increased circulatory stability, the luciferase expression of a 1 μg dose of ssmRNA polyplex, dsmRNA1 polyplex, dsmRNA2 polyplex and dsmRNA(80A) polyplex were compared by hydrodynamic stimulation. dsmRNA polyplexes were administered to triplicate mice in a small volume (100 μl) tail vein injection, followed by a hydrodynamic stimulatory dose of saline at 5 min. The luciferase expression was quantified by bioluminescence imaging at 24 hrs and compared to a benchmark control of direct hydrodynamic-dosing of 1 μg of each mRNA construct. The results established that a 1 μg dose of ssmRNA polyplex was completely inactivated during the 5 min circulation time, resulting in a greater than 4,000-fold loss in expression relative to a direct hydrodynamic-dosing control (Fig. 8). Similar results were obtained when dosing dsmRNA2 polyplex, in which a 5 min circulation time resulted in an 800-fold loss in luciferase expression relative to direct hydrodynamic dosing control (Fig. 8). In contrast, hybridization of the reverse strand with the 5’ and 3’ UTRs in dsmRNA1 decreased the level of expression 10-fold upon direct hydrodynamic dosing relative to dsmRNA2, but simultaneously improved the circulatory stability, resulting in a 7-fold loss in expression during a 5 min circulation (Fig. 8). Substitution with a polylysine PEG-peptide that lacked acridine-Lys residues resulted in the complete loss of 5 min circulatory stability34. These result suggest that the native 5’and 3’ UTRs are vulnerable to metabolism, which would result in removal of either the 5’ cap or poly(A) tail or both, accounting for a loss in luciferase expression. Similarly, direct hydrodynamic-dosing of dsmRNA(80A) polyplex also established a 10-fold loss in expression relative to dsmRNA2 polyplex due to 5’ and 3’ UTR denaturation (Fig. 8). However, the increased metabolic stability afforded by hybridizing against both the 5’ and 3’ UTR, and the poly (A) tail further improved the circulatory stability as determined by a 3-fold loss in luciferase expression in 5 min, 8-fold loss in 10 min and 20-fold loss in 20 min, relative to direct hydrodynamic dosing of dsmRNA(80A) (Fig. 8). These results establish the need to protect the 5’ and 3’ UTR, and the poly(A) tail from metabolism in the circulation. The decrease in expression resulting from denaturing the 3’ and 5’ UTR is offset by a large increase in the circulatory stability.
Figure 8. Circulatory Stability of dsmRNA Polyplexes.
The circulatory stability of a 1 μg dose of dsmRNA polyplexes was determined by comparison of luciferase expression at 24 hrs following a hydrodynamic-dose versus hydrodynamic stimulation at 5, 10, and 20 min. The loss in expression were either non-significant (ns), or significant with *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001 when comparing hydrodynamic-dosing with hydrodynamic stimulation.
Discussion
mRNA has been under investigation as a substitute for plasmid DNA in non-viral gene delivery for nearly 30 years39. The primary focus has been on using mRNA as a vaccine due to its ability to express an immunogenic protein and to serve as an adjuvant in muscle40. There have been an increasing number of reports that describe the systemic delivery of mRNA14, 17–18, 21, 28, 41–42. An early 2002 study described the hydrodynamic delivery of 50 μg of mRNA in mice resulting in the detection of low level (106 photons/sec/cm2/steradian) luciferase expression in the liver by bioluminescence imaging41. Peak expression occurred at 3 hrs post-mRNA delivery and required co-administration with 30 μg of decoy RNA and 400 U of RNase inhibitor41. A 2006 study used hydrodynamic delivery to administer a 50 μg dose of mRNA containing 5’ and 3’ Xenopus laevis β-globin UTRs flanking a luciferase gene. The addition of UTRs increased expression 15-fold at 3 hrs relative to mRNA lacking UTRs however, even with co-administration of decoy mRNA and RNase inhibitor, peak expression was not increased beyond 12 hrs43. A 2015 study further improved the level and persistence of mRNA mediated expression in the liver by installing a human β-globin 3’ and 5’ UTRs into a codon optimized mRNA34. Following a 1 μg dose administered hydrodynamically, the luciferase expression (108) peaked at 24 hrs and persisted at detectable levels for 3 days. A PEGylated polyacridine peptide was used to form ssmRNA polyplexes to resist metabolism during hydrodynamic-dosing, resulting in an additional 10-fold improvement in luciferase expression in the liver of mice. A seminal study in 2015 demonstrated delivery of a conventional i.v. dose of 1 μg of mRNA formulated in lipid nanoparticles resulting in peak luciferase expression (109) at 4 hrs18.
Despite these advances in mRNA potency and delivery, it has been much more challenging to develop mRNA formulations that are metabolically stable in the circulation27. In contrast, DNA can be formulated into stable nanoparticles with a long circulatory half-life27. Likewise, metabolically stabilized synthetic siRNA has been formulated into lipid nanoparticles44 and also conjugated directly to a targeting ligand45. To increase the metabolic stability of mRNA, we rationalized that a PEGylated polyacridine peptide would bind dsmRNA with higher affinity compared to ssmRNA and could potentially result in dsmRNA polyplexes that were metabolically stable in the circulation. The relative susceptibility to RNase digestion demonstrates an increase in the intrinsic metabolic stability for dsmRNA polyplex relative to ssmRNA polyplex (Fig. 3). Despite this, achieving in vivo metabolic stability is more demanding since polyplexes bind to serum proteins46, PEG-peptides can be stripped in the circulation47, and liver macrophages phagocytose and metabolize polyplexes48.
Hydrodynamic dosing34 and hydrodynamic stimulation32 were used to investigate the transfection competency and circulatory stability of dsmRNA. Direct hydrodynamic dosing of ssmRNA, dsmRNA1, and dsmRNA2 revealed that reverse strand hybridization with the 5’ and 3’ UTR resulted in a 10-fold loss in expression (Fig. 7). However, the initial loss in expression efficiency is offset by increased circulatory stability when the reverse stand is hybridized against the 5’ and 3’ UTR. This is demonstrated by comparing the result of hydrodynamic stimulation of ssmRNA, dsmRNA1, dsmRNA2 and dsmRNA(80A) polyplexes (Fig. 8). The ability to slow the metabolism of a 1 μg i.v. dose of dsmRNA(80A) polyplex to retain 5% transfection efficiency following a 20 min circulation time demonstrates significant progress over prior formulations34, 43, 49.
The ability of dsmRNA to retain expression efficiency compared to ssmRNA suggests that the ribosome is capable of stripping the reverse strand during translation50. Stem-loop double stranded structures within mRNA are also similarly denatured by the ribosome during translation50. These double-stranded regions within translationally active mRNA do not serve as substrates for Dicer, and thereby, do not result in RNAi self-knockdown. Likewise, when examined in the present study, we found no evidence that dsmRNA participated in RNAi self-knockdown (Fig. 7). The finding that siRNA was equally efficient at knocking down gene expression from ssmRNA and dsmRNA suggests that dsmRNA is converted to ssmRNA before being acted upon by RISC51.
While the potential for immune response is a significant concern for any mRNA formulation, the present study only delivered a 1 μg dose of dsmRNA polyplexes systemically, without experiencing any adverse effects. Uridine was replaced with pseudouridine in both forward and reverse mRNA strands in an attempt to preemptively suppress immune activation52. Immune activation by viral dsRNA results from binding to cell surface toll-like receptor 3 and retinoic acid-inducible gene I-like receptors (RLRs)53, resulting in the activation of the type I interferon (IFN) pathway35. Infected cells enter an anti-viral state resulting in inhibition of protein translation54. Compared to viral dsRNA, mRNA has been reported to be immune privileged due to the presence of the unique cap(1) structure55–56. The finding that hydrodynamically dosed dsmRNA polyplexes were equally efficient in repeated administration suggests that hepatocytes avoided entering the anti-viral state (Fig. 7). This may be the result of the low dose used, the hydrodynamic route of delivery which avoids polyplex binding to cell surface receptors, or may be the result of masking dsmRNA within PEGylated polyplexes.
In conclusion, a metabolically stable and translationally active form of mRNA is described. dsmRNA is easily prepared in good yield and is much more metabolically stable. The complementarity of the reverse and forward strand inversely influence translation efficiency and circulatory stability in vivo. dsmRNA formulations avoid participation in RNAi and immune activation. Future studies will explore chemically functionalized dsmRNA.
Supplementary Material
Acknowledgements
The authors gratefully acknowledge support from NIH Grants GM117785 and T32 GM00865.
References
- 1.Wu GY; Wu CH, Receptor-mediated Gene Delivery and Expression In Vivo. J. Biol. Chem 1988, 263, 14621–14624. [PubMed] [Google Scholar]
- 2.Wu CH; Wilson JM; Wu GY, Targeting Genes: Delivery and Persistent Expression of a Foreign Gene Driven by Mammalian Regulatory Elements in Vivo. J. Biol. Chem 1989, 264, 16985–16987. [PubMed] [Google Scholar]
- 3.Wu GY; Wilson JM; Shalaby F; Grossman M; Shafritz DA; Wu CH, Receptor-mediated Gene Delivery In Vivo. Partial Correction of Genetic Analbuminemia in Nagase Rats. J. Biol. Chem 1991, 266, 14338–14342. [PubMed] [Google Scholar]
- 4.Wilson JM; Grossman M; Wu CH; Chowdhury NR; Wu GY; Chowdhury JR, Hepatocyte-directed Gene Transfer In Vivo Leads to Transient Improvement of Hypercholesterolemia in Low Density Lipoprotein Receptor-deficient Rabbits. J. Biol. Chem 1992, 267, 963–967. [PubMed] [Google Scholar]
- 5.Perales JC; Ferkol T; Beegen H; Ratnoff OD; Hanson RW, Gene transfer in vivo: Sustained expression and regulation of genes introduced into the liver by receptor-targeted uptake. Proc. Natl. Acad. Sci. USA 1994, 91, 4086–4090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stankovics J; Crane AM; Andrews E; Wu CH; Wu GY; Ledley FD, Overexpression of Human Methylmalonyl CoA Mutase in Mice after In Vivo Gene Transfer with Asialoglycoprotein Polylysine DNA Complexes. Hum. Gene. Ther 1994, 5, 1095–1104. [DOI] [PubMed] [Google Scholar]
- 7.Hara T; Tan Y; Huang L, In vivo gene delivery to the liver using reconstituted chylomicron remnants as a novel nonviral vector. Proceedings of the National Academy of Sciences of the United States of America 1997, 94, 14547–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nishikawa M; Takemura S; Takakura Y; Hashida M, Targeted delivery of plasmid DNA to hepatocytes in vivo: optimization of the pharmacokinetics of plasmid DNA/galactosylated Poly(L-lysine) complexes by controlling their physicochemical properties. JPET 1998, 287, 408–415. [PubMed] [Google Scholar]
- 9.Kwoh DY; Coffin CC; Lollo CP; Jovenal J; Banaszczyk MG; Mullen P, et al. , Stabilization of poly-L-lysine/DNA polyplexes for in vivo gene delivery to the liver. Biochimica et Biophysica Acta 1999, 1444, 171–190. [DOI] [PubMed] [Google Scholar]
- 10.Kawakami S; Fumoto S; Nishikawa M; Yamashita F; Hashida M, In vivo gene delivery to the liver using novel galactosylated cationic liposomes. Pharmaceutical Research 2000, 17, 306–13. [DOI] [PubMed] [Google Scholar]
- 11.Nishikawa M; Yamauchi M; Morimoto K; Ishida E; Takakura Y; Hashida M, Heptocyte-targeted in vivo gene expression by intraveneous injection of plasmid DNA complexed with synthetic multi-functional gene delivery system. Gene Therapy 2000, 7, 548–555. [DOI] [PubMed] [Google Scholar]
- 12.Al-Dosari MS; Knapp JE; Liu D, Hydrodynamic Delivery In Advances in Genetics, Academic Press: 2005; Vol. 54, pp 65–82. [DOI] [PubMed] [Google Scholar]
- 13.Phua KKL; Leong KW; Nair SK, Transfection efficiency and transgene expression kinetics of mRNA delivered in naked and nanoparticle format. Journal of controlled release: official journal of the Controlled Release Society 2013, 166, 227–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Y; Su H.-h.; Yang Y; Hu Y; Zhang L; Blancafort P, et al. , Systemic Delivery of Modified mRNA Encoding Herpes Simplex Virus 1 Thymidine Kinase for Targeted Cancer Gene Therapy. Molecular Therapy 2013, 21, 358–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li J; He Y; Wang W; Wu C; Hong C; Hammond PT, Polyamine-Mediated Stoichiometric Assembly of Ribonucleoproteins for Enhanced mRNA Delivery. Angew Chem Int Ed Engl 2017, 56, 13709–13712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li J; Wang W; He Y; Li Y; Yan EZ; Zhang K, et al. , Structurally Programmed Assembly of Translation Initiation Nanoplex for Superior mRNA Delivery. ACS Nano 2017, 11, 2531–2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pardi N; Secreto AJ; Shan X; Debonera F; Glover J; Yi Y, et al. , Administration of nucleoside-modified mRNA encoding broadly neutralizing antibody protects humanized mice from HIV-1 challenge. Nat Commun 2017, 8, 14630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pardi N; Tuyishime S; Muramatsu H; Kariko K; Mui BL; Tam YK, et al. , Expression kinetics of nucleoside-modified mRNA delivered in lipid nanoparticles to mice by various routes. J Control Release 2015, 217, 345–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thess A; Grund S; Mui BL; Hope MJ; Baumhof P; Fotin-Mleczek M, et al. , Sequence-engineered mRNA Without Chemical Nucleoside Modifications Enables an Effective Protein Therapy in Large Animals. Mol Ther 2015, 23, 1456–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kauffman KJ; Mir FF; Jhunjhunwala S; Kaczmarek JC; Hurtado JE; Yang JH, et al. , Efficacy and immunogenicity of unmodified and pseudouridine-modified mRNA delivered systemically with lipid nanoparticles in vivo. Biomaterials 2016, 109, 78–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ramaswamy S; Tonnu N; Tachikawa K; Limphong P; Vega JB; Karmali PP, et al. , Systemic delivery of factor IX messenger RNA for protein replacement therapy. Proc Natl Acad Sci U S A 2017, 114, E1941–E1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jacobs F; Wisse E; De Geest B, The Role of Liver Sinusoidal Cells in Hepatocyte-Directed Gene Transfer. The American Journal of Pathology 2010, 176, 14–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wisse E; Jacobs F; Topal B; Frederik P; De Geest B, The size of endothelial fenestrae in human liver sinusoids: implications for hepatocyte-directed gene transfer. Gene Ther 2008, 15, 1193–9. [DOI] [PubMed] [Google Scholar]
- 24.Hu Y; Haynes MT; Wang Y; Liu F; Huang L, A Highly Efficient Synthetic Vector: Nonhydrodynamic Delivery of DNA to Hepatocyte Nuclei in Vivo. ACS Nano 2013, 7, 5376–5384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yao J; Fan Y; Li Y; Huang L, Strategies on the nuclear-targeted delivery of genes. Journal of Drug Targeting 2013, 21, 926–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lam AP; Dean DA, Progress and prospects: nuclear import of nonviral vectors. Gene Ther 2010, 17, 439–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guan S; Rosenecker J, Nanotechnologies in delivery of mRNA therapeutics using nonviral vector-based delivery systems. Gene Ther 2017, 24, 133–143. [DOI] [PubMed] [Google Scholar]
- 28.DeRosa F; Guild B; Karve S; Smith L; Love K; Dorkin JR, et al. , Therapeutic efficacy in a hemophilia B model using a biosynthetic mRNA liver depot system. Gene Ther 2016, 23, 699–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsui NB; Ng EK; Lo YM, Stability of endogenous and added RNA in blood specimens, serum, and plasma. Clin Chem 2002, 48, 1647–53. [PubMed] [Google Scholar]
- 30.Chen Q; Qi R; Chen X; Yang X; Wu S; Xiao H, et al. , A Targeted and Stable Polymeric Nanoformulation Enhances Systemic Delivery of mRNA to Tumors. Mol Ther 2017, 25, 92–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kizzire K; Khargharia S; Rice KG, High-affinity PEGylated polyacridine peptide polyplexes mediate potent in vivo gene expression. Gene Ther 2013, 20, 407–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fernandez CA; Baumhover NJ; Duskey JT; Khargharia S; Kizzire K; Ericson MD, et al. , Metabolically stabilized long-circulating PEGylated polyacridine peptide polyplexes mediate hydrodynamically stimulated gene expression in liver. Gene Ther 2011, 18, 23–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Khargharia S; Kizzire K; Ericson MD; Baumhover NJ; Rice KG, PEG length and chemical linkage controls polyacridine peptide DNA polyplex pharmacokinetics, biodistribution, metabolic stability and in vivo gene expression. J Control Release 2013, 170, 325–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Crowley ST; Poliskey JA; Baumhover NJ; Rice KG, Efficient expression of stabilized mRNA PEG-peptide polyplexes in liver. Gene Ther 2015, 22, 993–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nellimarla S; Mossman KL, Extracellular dsRNA: its function and mechanism of cellular uptake. J Interferon Cytokine Res 2014, 34, 419–26. [DOI] [PubMed] [Google Scholar]
- 36.Ueda T; Tohda H; Chikazumi N; Eckstein F; Watanabe K, Phosphorothioate-containing RNAs show mRNA activity in the prokaryotic translation systems in vitro. Nucleic Acids Res 1991, 19, 547–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hunter T; Hunt T; Jackson RJ; Robertson HD, The characteristics of inhibition of protein synthesis by double-stranded ribonucleic acid in reticulocyte lysates. J Biol Chem 1975, 250, 409–17. [PubMed] [Google Scholar]
- 38.McAnuff MA; Rettig GR; Rice KG, Potency of siRNA versus shRNA mediated knockdown in vivo. J Pharm Sci 2007, 96, 2922–30. [DOI] [PubMed] [Google Scholar]
- 39.Pardi N; Hogan MJ; Porter FW; Weissman D, mRNA vaccines - a new era in vaccinology. Nat Rev Drug Discov 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schlake T; Thess A; Fotin-Mleczek M; Kallen K-J, Developing mRNA-vaccine technologies. RNA Biology 2012, 9, 1319–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McCaffrey AP; Ohashi K; Meuse L; Shen S; Lancaster AM; Lukavsky PJ, et al. , Determinants of hepatitis C translational initiation in vitro, in cultured cells and mice. Mol Ther 2002, 5, 676–84. [DOI] [PubMed] [Google Scholar]
- 42.Phua KK; Leong KW; Nair SK, Transfection efficiency and transgene expression kinetics of mRNA delivered in naked and nanoparticle format. J Control Release 2013, 166, 227–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wilber A; Frandsen JL; Geurts JL; Largaespada DA; Hackett PB; McIvor RS, RNA as a Source of Transposase for Sleeping Beauty-Mediated Gene Insertion and Expression in Somatic Cells and Tissues. Molecular Therapy 2006, 13, 625–630. [DOI] [PubMed] [Google Scholar]
- 44.Tam YYC; Chen S; Cullis PR, Advances in Lipid Nanoparticles for siRNA Delivery. Pharmaceutics 2013, 5, 498–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nair JK; Willoughby JL; Chan A; Charisse K; Alam MR; Wang Q, et al. , Multivalent N-acetylgalactosamine-conjugated siRNA localizes in hepatocytes and elicits robust RNAi-mediated gene silencing. J Am Chem Soc 2014, 136, 16958–61. [DOI] [PubMed] [Google Scholar]
- 46.Oupický D; Koňák Č; Dash PR; Seymour LW; Ulbrich K, Effect of Albumin and Polyanion on the Structure of DNA Complexes with Polycation Containing Hydrophilic Nonionic Block. Bioconjugate Chemistry 1999, 10, 764–772. [DOI] [PubMed] [Google Scholar]
- 47.Khargharia S; Baumhover NJ; Crowley ST; Duskey J; Rice KG, The Uptake Mechanism of PEGylated DNA Polyplexes by the Liver Influences Gene Expression. Gene Therapy 2014, 21, 1021–1028. [DOI] [PubMed] [Google Scholar]
- 48.Baumhover NJ; Duskey JT; Khargharia S; White CW; Crowley ST; Allen RJ, et al. , Structure–Activity Relationship of PEGylated Polylysine Peptides as Scavenger Receptor Inhibitors for Non-Viral Gene Delivery. Molecular Pharmaceutics 2015, 12, 4321–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McCaffrey AP; Meuse L; Pham TT; Conklin DS; Hannon GJ; Kay MA, RNA interference in adult mice. Nature 2002, 418, 38–9. [DOI] [PubMed] [Google Scholar]
- 50.Qu X; Wen JD; Lancaster L; Noller HF; Bustamante C; Tinoco I Jr., The ribosome uses two active mechanisms to unwind messenger RNA during translation. Nature 2011, 475, 118–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ameres SL; Martinez J; Schroeder R, Molecular basis for target RNA recognition and cleavage by human RISC. Cell 2007, 130, 101–12. [DOI] [PubMed] [Google Scholar]
- 52.Karikó K; Muramatsu H; Welsh FA; Ludwig J; Kato H; Akira S, et al. , Incorporation of Pseudouridine Into mRNA Yields Superior Nonimmunogenic Vector With Increased Translational Capacity and Biological Stability. Molecular Therapy 2008, 16, 1833–1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chattopadhyay A; Raghuraman H, Application of fluorescence spectroscopy to membrane protein structure and dynamics In Curr Sci, 2004; Vol. 87, pp 175–180. [Google Scholar]
- 54.Stark GR; Kerr IM; Williams BR; Silverman RH; Schreiber RD, How cells respond to interferons. Annu Rev Biochem 1998, 67, 227–64. [DOI] [PubMed] [Google Scholar]
- 55.Ramanathan A; Robb GB; Chan SH, mRNA capping: biological functions and applications. Nucleic Acids Res 2016, 44, 7511–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zust R; Cervantes-Barragan L; Habjan M; Maier R; Neuman BW; Ziebuhr J, et al. , Ribose 2’-O-methylation provides a molecular signature for the distinction of self and non-self mRNA dependent on the RNA sensor Mda5. Nat Immunol 2011, 12, 137–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.