Abstract
The obesity epidemic is developing into the most costly health problem facing the world. Obesity, characterized by excessive adipogenesis and enlarged adipocytes, promotes morbidities, such as diabetes, cardiovascular disease, and cancer. Regulation of adipogenesis is critical to our understanding of how fat cell formation causes obesity and associated health problems. Thy1 (also called CD90), a widely used stem cell marker, blocks adipogenesis and reduces lipid accumulation. Thy1-knockout mice are prone to diet-induced obesity. Although the importance of Thy1 in adipogenesis and obesity is now evident, how its expression is regulated is not. We hypothesized that DNA methylation has a role in promoting adipogenesis and affects Thy1 expression. Using the methylation inhibitor 5-aza-2′-deoxycytidine (5-aza-dC), we investigated whether DNA methylation alters Thy1 expression during adipogenesis in both mouse 3T3-L1 preadipocytes and mouse mesenchymal stem cells. Thy1 protein and mRNA levels were decreased dramatically during adipogenesis. However, 5-aza-dC treatment prevented that phenomenon. Methylation-sensitive pyrosequencing analysis showed that CpG sites at the Thy1 locus have increased methylation during adipogenesis, as well as increased methylation in adipose tissue from diet-induced obese mice. These new findings highlight the potential role of Thy1 and DNA methylation in adipogenesis and obesity.—Flores, E. M., Woeller, C. F., Falsetta, M. L., Susiarjo, M., Phipps, R. P. Thy1 (CD90) expression is regulated by DNA methylation during adipogenesis.
Keywords: 3T3-L1, obesity, mesenchymal stem cells, 5-aza-dC, epigenetics
Obesity rates have risen markedly in the past 30 yr; >700 million people worldwide are clinically obese (1, 2). Obesity promotes type 2 diabetes, fatty liver disease, and cardiovascular disease and is linked with certain cancers (3–5). Health care costs associated with obesity and its comorbidities are enormous and will continue to rise as currents trends continue and the population ages (6). Thus, a further understanding of obesity and its underlying mechanisms and causes are urgently needed.
Obesity results from a positive energy balance when more calories are consumed than are used. The surplus energy is packaged into lipid-based storage molecules and sent to fat storage cells called adipocytes. In obesity, there is both an increase in adipocyte size and an increase in adipocyte number to accommodate the lipid (7). Adipocytes are formed during the process of adipogenesis and arise from stem cells, fibroblasts, or other progenitor cells, when appropriately programmed (8). Adipogenesis is a highly regulated process that requires the activation of several key signaling pathways, including STAT5 and Fyn, and activation of the transcription factors peroxisome proliferator activated receptor-γ (PPARγ) and C/EBPα. Numerous genes involved in fatty acid transport and storage, such as fatty acid binding protein 4 (Fabp4) are induced during adipogenesis to promote lipid accumulation in adipocytes.
Several proteins, including Pref-1, Wnt, TGF-β, and Thy1 (formally called CD90), have been shown to inhibit adipogenesis by blocking proadipogenic signaling (9–13). Our recent study (14) showed that Thy1 blocked the activity of the Src family kinase Fyn in preadipocytes and human fibroblasts. Thy1-mediated inhibition of Fyn activity prevented adipocyte formation. Interestingly, although preadipocytes expressed high levels of Thy1, its expression was lost during adipogenesis, and mature adipocytes expressed almost no Thy1. Thy1 is a member of the Ig supergene family and is a glycophosphatidylinositol-linked surface protein. Although Thy1 is expressed on preadipocytes and subsets of fibroblasts, neurons, and stem cells, little is known about how its expression is controlled. We recently showed that Thy1 levels can be regulated by microRNAs (miRs). Specifically, the miR-103/107 family of miRNAs can target Thy1 mRNA and reduce its expression (15). Furthermore, the Thy1 gene contains several CpG-rich elements, termed CpG islands, which are hotspots for cytosine methylation and gene regulation (16, 17). To date, there have been no reports studying DNA methylation of Thy1 during adipogenesis. However, Thy1 methylation has been studied in lung fibroblasts and in T cells, in which they show an increase in Thy1 methylation correlated with a decrease in Thy1 expression (18–20). Therefore, we investigated the same CG-rich region within intron 1 (termed Thy1-CGI1), which is part of the promoter (17, 20–23).
Although changes in DNA methylation patterns at the Thy1 locus have not been characterized in the context of adipogenesis, recent reports have shown that DNA methylation changes are an integral part of adipocyte formation (24, 25). In the most widely used and well-accepted model of in vitro adipogenesis, the murine 3T3-L1 preadipocyte line, global DNA methylation has been shown to increase during adipogenesis (26, 27). Interestingly, addition of the DNA methylation inhibitor 5-aza-2′-deoxycytidine (5-aza-dC) at the onset of adipogenic differentiation can completely block adipocyte formation, suggesting that DNA methylation changes are essential for adipogenesis (27, 28). Specific changes in regional DNA methylation are also critical for adipogenesis, and blocking those patterns can alter the differentiation pathway of precursor cells toward osteoblastogenesis, rather than adipogenesis (24, 29, 30). For example, when blocking DNA methylation with 5-aza-dC in 3T3-L1 cells, Wnt10a expression increased through the hypomethylation of several of its CpG sites. The increase in Wnt10a expression helped steer the differentiation program toward osteoblastogenesis and away from adipogenesis (30).
Because Thy1 protein and mRNA levels are rapidly suppressed during adipogenic differentiation, we hypothesized that it’s reduced expression was linked to hypermethylation of the Thy1 locus. Excitingly, we are the first, to our knowledge, to show methylation-sensitive pyrosequencing data of the Thy1-CGI1 region and to report that DNA methylation is one of the essential regulators of Thy1 expression during adipogenesis.
MATERIALS AND METHODS
Chemicals
5-Aza-dC, 3-isobutyl-1-methylxanthine, dexamethasone, and human recombinant insulin were all purchased from MilliporeSigma (Burlington, MA, USA).
Animals
C57BL/6 and Thy1-knockout mice (on a matched C57BL/6 background) were housed in colony cages with a 12/12-h light/dark cycle and fed ad libitum. Male mice were fed a standard chow diet or a high-fat diet (HFD) consisting of 60% kcal/fat (D12492; Research Diets, New Brunswick, NJ, USA) for 8 wk, as previously described (14). Mice were weighed at time of euthanization. Fat tissue was isolated from mice for pyrosequencing assays. All animal experiments were performed with approval of the University Committee on Animal Resources at the University of Rochester School of Medicine and Dentistry, and all methods were performed in accordance with the relevant guidelines and regulations.
Cell culture
All cells were incubated at 37°C with 7% humidified CO2. 3T3-L1 cells were maintained in 10% calf serum supplemented with DMEM medium. C57BL/6 mouse bone marrow mesenchymal stem cells (MSCs) were purchased from Cell Biologics (Chicago, IL, USA) and maintained in 10% MSC-qualified fetal bovine serum in supplemented MEM medium from Thermo Fisher Scientific (Waltham, MA, USA). Cells were plated at 60% confluence and treated at 80% confluency. To induce adipogenesis, a medium containing an adipogenic cocktail (ACT) was added to confluent cells, which consisted of 0.5 mM 3-isobutyl-1-methylxanthine, 0.5 μM dexamethasone, and 2 μg/ml insulin. Fresh ACT was added every 2 d. To inhibit methylation, cells were treated daily with 0.5 μM 5-aza-dC or DMSO as a control. Cells were then harvested on days indicated for each experiment.
Real-time quantitative PCR detection of mRNA
RNA was extracted with a miRNeasy Kit (Qiagen, Hilden, Germany) and was quantified with a NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific). An iScript Reverse Transcription Kit (Bio-Rad Laboratories, Hercules, CA, USA) was used to make cDNA from 150 ng RNA. Quantitative Real-Time PCR assays were then performed using SsoAdvanced Universal SYBR Green Supermix (Bio-Rad Laboratories), according to the manufacturer’s instructions. All genes of interest were normalized to 18S rRNA, and the relative percentages were normalized to 100% of medium + DMSO for Thy1 mRNA or 100% of ACT + DMSO for Fabp4 mRNA levels. Primer sequences were as follows: Thy1, (forward) 5′‑CCTTACCCTAGCCAACTTCAC‑3′, (reverse) 5′‑AGGATGTGTTCTGAACCAGC‑3′; Fabp4, (forward) 5′‑ATGTGTGATGCCTTTGTGGGAAC‑3′, (reverse) 5′‑TCATGTTGGGCTTGGCCATG‑3′; and 18s rRNA, (forward) 5′‑GTAACCCGTTGAACCCCATT‑3′, (reverse) 5′‑CCATCCAATCGGTAGTAGCG‑3′.
Western blot analysis
Cells were lysed with 60 mM Tris, 2% SDS, and protease inhibitor cocktail (MilliporeSigma); 10 μg of protein was loaded/lane and run on SDS-PAGE gels. Protein gels were transferred to 0.45 μm Immobilon-PVDF membranes (MilliporeSigma) and blocked with 5% bovine serum albumin (BSA) in 0.1% Tween 20 in PBS. Primary antibodies, sheep anti-mouse Thy1 (R&D Systems, Minneapolis, MN, USA), rabbit anti-mouse Fabp4 (Cell Signaling Technology, Danvers, MA, USA), and rabbit anti-mouse β-tubulin (Cell Signaling Technology) were diluted 1:5000, 1:500, and 1:5000, respectively, and incubated for 1 h. Membranes were washed in 0.1% Tween 20 in PBS then incubated in anti-sheep or anti-rabbit horseradish peroxidase–conjugated secondary antibodies at 1:5000 or 1:20,000 dilution, respectively. Protein was visualized with Immobilon Western chemiluminescent horseradish peroxidase substrate (MilliporeSigma). The MagicMark XP (Thermo Fisher Scientific) protein standard protocol was used for the ladder. Blots were developed by X-ray film. All blots are provided as uncropped images in the Supplemental Data.
Flow cytometry
Cells were trypsinized and washed in PBS, then fixed with 2% paraformaldehyde and blocked with 1:50 human Fc receptor blocker (Miltenyi Biotec, Bergisch Gladbach, Germany) in PBS. The cells were then incubated with anti-mouse Thy1.2-PE conjugated antibody, 1:500, (BD Biosciences, San Jose, CA, USA) for 1 h on ice. Cells were washed and resuspended in PBS. Cells were analyzed on a LSR II Flow Cytometer running FACSDiva software (BD Biosciences). Analysis of fluorescence data was performed with FlowJo software (v.10.1; Tree Star, Ashland, OR, USA).
Immunofluorescent staining
Cells treated in 12-well plates were washed with 1× PBS and fixed with 2% paraformaldehyde for 10 min and washed 3 times with PBS. Cells were blocked in 1% BSA and 0.1% Triton X-100 in PBS with normal donkey serum (Jackson ImmunoResearch Laboratories, West Grove, PA, USA) and Fc blocker 1:50 (BD Biosciences). The primary antibodies used were Thy1.2-PE–conjugated antibody, (BD Biosciences) and Fabp4 (Cell Signaling Technology), which were diluted 1:500 in 1% BSA and incubated for 2 h at room temperature in the dark. After removal of primary antibody and 3 washes, secondary antibody (donkey anti-rabbit AF647) was applied at a 1:2000 dilution for 1 h. Cells were then washed and visualized on an Evos-Fl Cell Imaging System (Thermo Fisher Scientific).
DNA extraction and bisulfite conversion
Genomic DNA was isolated from cells with a DNeasy DNA Extraction Kit (Qiagen), and quantified with the NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific). Genomic DNA (1000 ng) was then bisulfite converted with an Epitect Plus Bisulfite Conversion Kit (Qiagen) to be analyzed by a pyrosequencer.
Pyrosequencing assays
Bisulfite-treated DNA was amplified with the PyroMark PCR Kit (Qiagen) and conditions of 95°C for 5 min, then 45 cycles of 95°C for 30 s and an annealing temperature of 58°C for 30 s, then 72°C for 30 s, followed by 72°C for 10 min. The PCR product sizes were then verified with electrophoresis on a 2% agarose gel. Ten microliters of the biotinylated PCR products were mixed with 1 μl streptavidin-coated Sepharose beads, 40 μl PyroMark binding buffer (Qiagen), and 29 μl RNAse-free water for a total volume of 80 μl. That mixture was then run on a PyroMark Vacuum Workstation (Qiagen). The purified PCR products were then added to the annealing buffer, which contained the corresponding sequencing primer. After annealing, the plate was loaded into the PyroMark Q96 MD instrument (Qiagen). PyroMark-CpG software automatically generated a dispensation order of deoxynucleoside triphosphates and controlled dispensations based on the sequence to analyze. Controls were included in the dispensation order to check the performance of the reactions. All runs also included a no-template control. We analyzed the data with the PyroMark software (Qiagen) for quantification of the percentage of DNA CpG methylation.
Pyrosequencing primers
Methylation levels were measured in the first CpG island of intron 1, which is part of the promoter (17, 20–23) of mouse Thy1 (chr9:44,043,384–44,048,579; GRCm38/mm10) (94–349 bp) using the pyrosequencing assay. Gene-specific primers for Thy1 were designed with the Pyro-Mark assay design software (v.2.0; Qiagen). The program automatically generated primer sets that included both PCR and sequencing primers, based on selected target sequences. One of the primers was biotinylated to enable immobilization to streptavidin-coated beads. The sequences were as follows: (forward) 5′‑TTTAGTTATAGTTTTGGGAAAGGATAT‑3′, (reverse) biotinylated primer 5′‑CCACCTCCTCCCTCTATT‑3′, and sequencing primer: 5′‑ATATTAGGGAGTTTTTATAT‑3′.
Global CpG methylation analysis
Total DNA was extracted from adipose tissue or bone marrow–derived MSCs with a Tissue DNA Isolation Kit from Qiagen. Total DNA (50–100 ng) was analyzed for the presence of 5-methyl CpG levels by ELISA using the 5-mC DNA ELISA Kit from Zymo Research (Irvine, CA, USA) following the manufacturer’s instruction. Results were normalized to the wild-type tissue or cells.
Statistical analysis
All values are presented as means ± sem. Experiments were conducted in triplicate at separate times. A 2-way ANOVA was used for statistical analysis with Prism 6 (GraphPad Software, La Jolla, CA, USA). Values of P < 0.05 were considered significant.
RESULTS
Reduced Thy1 expression during adipogenesis is partially attenuated by 5‑aza-dC in bone marrow–derived mouse MSCs
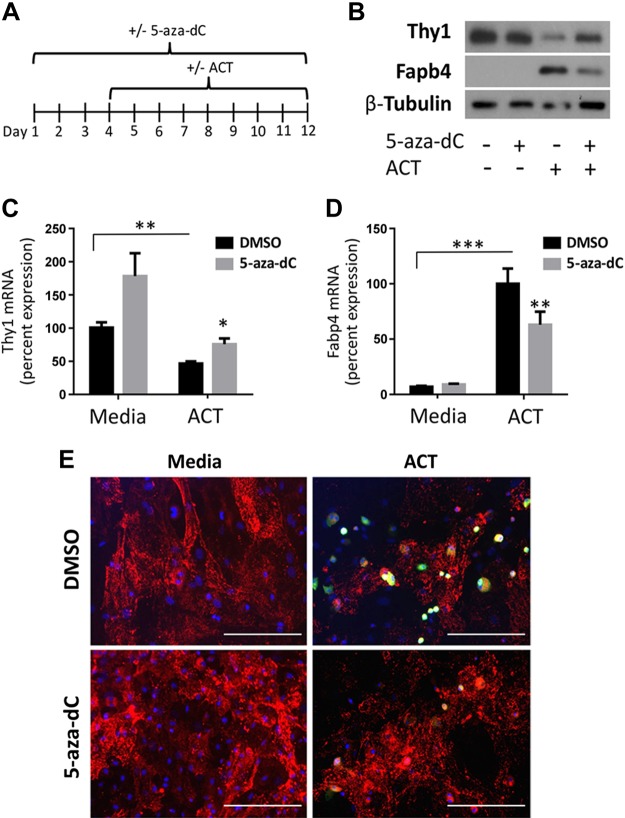
Previously, we demonstrated (14) that preadipocytes and MSCs must down-regulate Thy1 expression to differentiate into adipocytes. To determine whether DNA methylation regulates Thy1 expression during adipogenesis, we first pharmacologically inhibited DNA methyltransferase in bone marrow–derived mouse MSCs (mMSCs) before differentiation into adipocytes (28). mMSCs were treated daily with either DMSO (vehicle) or the DNA methyltransferase inhibitor, 5-aza-dC, for 12 d and were concurrently treated with either medium alone or the ACT medium, starting at d 3 (Fig. 1A). mMSCs typically differentiate into adipocytes in 2 wk when exposed to ACT (31). Therefore, we examined an intermediate time point to examine Thy1 levels. As expected, when given ACT, Thy1 levels decreased, whereas Fabp4 levels increased compared with medium alone (Fig. 1B–D); Fabp4 is highly expressed by adipocytes and serves as an adipogenic marker. However, inhibiting DNA methylation with 5-aza-dC increased Thy1 protein and mRNA levels vs. medium alone and attenuated the down-regulation of Thy1 in ACT samples (Fig. 1B, C). We also determined that protein and mRNA levels of Fabp4 were decreased in mMSCs treated with 5-aza-dC (Fig. 1B, D). Immunofluorescence staining also revealed that 5-aza-dC–treated mMSCs had increased surface expression of Thy1 and decreased Fabp4 levels (Fig. 1E). These results indicate that inhibition of DNA methylation impairs adipogenesis.
Figure 1.
The reduction of Thy1 expression during adipogenesis is attenuated by 5-aza-dC in mMSCs. mMSCs were treated daily with either DMSO or 5-aza-dC for 12 d continually with basal medium alone or with the introduction of the ACT starting at d 3. A) Timeline diagram of treatment for mMSCs. B) Western blot shows treatment with 5-aza-dC increases Thy1 protein levels and decreases protein levels of the adipogenic marker Fabp4 vs. cells only receiving the ACT. C, D) Quantitative real-time PCR results show that treatment with 5-aza-dC significantly increased Thy1 mRNA and decreased Fabp4 mRNA levels. Relative percentages were normalized to medium DMSO for Thy1 mRNA and to ACT DMSO for Fabp4 mRNA levels. E) Immunofluorescent images depict reduced FAPB4 (green) and increased Thy1 (red) expression in cells receiving 5-aza-dC vs. those receiving ACT alone. Cell nuclei are stained with DAPI and are depicted in blue. Scale bars, 200 μm. *P < 0.05, **P < 0.01, ***P < 0.001.
Reduced Thy1 expression, which is necessary for adipogenesis, is prevented by inhibition of global DNA methylation in 3T3-L1 cells
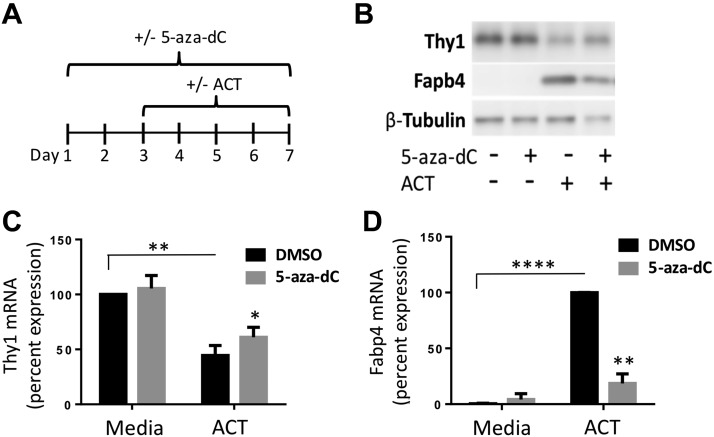
Primary MSCs are heterogeneous with only some cells fully differentiating into adipocytes (32). Thus, we next used the well-established preadipocyte murine cell line, 3T3-L1. Previously, we showed (33) that fully differentiated adipocytes treated with ACT no longer expressed Thy1 protein and mRNA. To observe changes in Thy1 expression, we examined an intermediate point in adipogenesis. At only 4 d of treatment with ACT, the cells were only partially differentiated, Thy1 was decreased, and Fabp4 expression was induced. Cells were treated daily for 7 d with either DMSO or 5-aza-dC and, starting at d 3, were either given ACT or continued with medium alone (Fig. 2A). As expected, Thy1 decreased at both the protein and mRNA levels in cells given ACT compared with medium alone (Fig. 2B, C). However, we show that treatment with 5-aza-dC prevented the decrease in Thy1 mRNA levels in ACT-treated cells, whereas medium alone with 5-aza-dC caused no significant changes (Fig. 2C). 5-Aza-dC also caused a decrease in Fabp4 levels in ACT-treated cells, indicative of decreased adipogenesis (Fig. 2B, D). Inhibiting global methylation blunted adipogenesis, which was reflected by the decreased levels of Fabp4 expression, and it, in part, sustained the Thy1 levels.
Figure 2.
Inhibiting methylation increases Thy1 expression in 3T3-L1 during adipogenesis. 3T3-L1 cells were treated daily with either DMSO or 5-aza-dC for 7 d continually with basal medium alone or with the introduction of ACT starting at d 3. A) Timeline diagram of treatment for 3T3-L1s. B) Western blot showing treatment with 5-aza-dC resulted in an increase in Thy1 total protein levels in both medium alone and ACT-exposed samples. ACT samples also had a decrease in Fabp4 protein levels when treated with 5-aza-dC, shown with a representative Western blot. C, D) Quantitative real-time PCR shows treatment with 5-aza-dC increases Thy1 mRNA and decreases Fabp4 mRNA levels in ACT samples. Relative percentages were normalized to 100% of medium DMSO for Thy1 mRNA or 100% of ACT DMSO for Fabp4 mRNA levels. *P < 0.05, **P < 0.01, ****P < 0.0001.
5-aza-dC partially restores Thy1 cell surface expression in 3T3-L1 cells when exposed to ACT
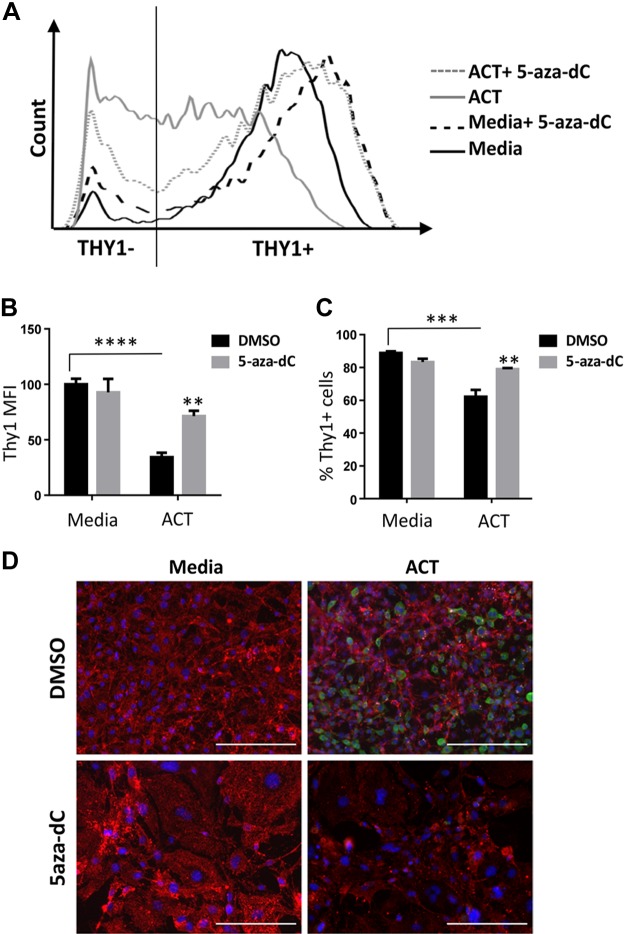
We next examined Thy1 cell surface expression on 3T3-L1 cells because Thy1 is a known cell marker on preadipocytes and is readily detected via flow cytometry and immunofluorescence. Cells were treated for 7 consecutive days with either DMSO or 5-aza-dC, whereas ACT samples were given the ACT starting at d 3, as previously described. The representative histogram in Fig. 3A shows cells treated with ACT shift out of the Thy1+ gate into Thy1− during differentiation, whereas cells treated with ACT and 5-aza-dC mostly remain in the Thy1+ gate. As expected, preadipocytes treated with ACT expressed significantly less surface Thy1 than did cells with medium alone, as evidenced by a lower mean fluorescence intensity (MFI) (Fig. 3B). However, preadipocytes cultured with ACT and 5-aza-dC showed a significant increase in Thy1 MFI compared with that of ACT alone, which occurred in tandem with an increase in the percentage of Thy1+ cells (Fig. 3B, C). Using immunofluorescence, we confirmed that Thy1 expression was sustained in ACT-treated preadipocytes also treated with 5-aza-dC, whereas Fabp4 expression decreased compared with cells treated with ACT alone (Fig. 3D).
Figure 3.
Thy1 surface protein levels attenuated by 5-aza-dC. 3T3-L1 cells were treated daily with either DMSO or 5-aza-dC for 7 d with the introduction of the ACT starting at d 3. A) Representative histogram of Thy1 surface levels measured by flow cytometry using a CD90.2-PE conjugated antibody. B) ACT samples had a decrease in Thy1 MFI, which was partially attenuated by 5-aza-dC treatment. C) ACT samples had a decrease in Thy1+ cells compared with medium alone, whereas ACT cells treated with 5-aza-dC had an increase in the Thy1+ population. D) Immunofluorescent images show decreased staining of Thy1 (red) with ACT compared with preadipocytes (medium alone) and increased Fabp4 (green). Thy1 staining was sustained when ACT samples were treated with 5-aza-dC, along with decreased staining by Fabp4. Cell nuclei were stained with DAPI and are depicted in blue. Scale bars, 200 μm. **P < 0.01, ***P < 0.001, ****P < 0.0001.
Wild-type and Thy1-null adipose tissue and MSCs have similar global methyl-CpG levels
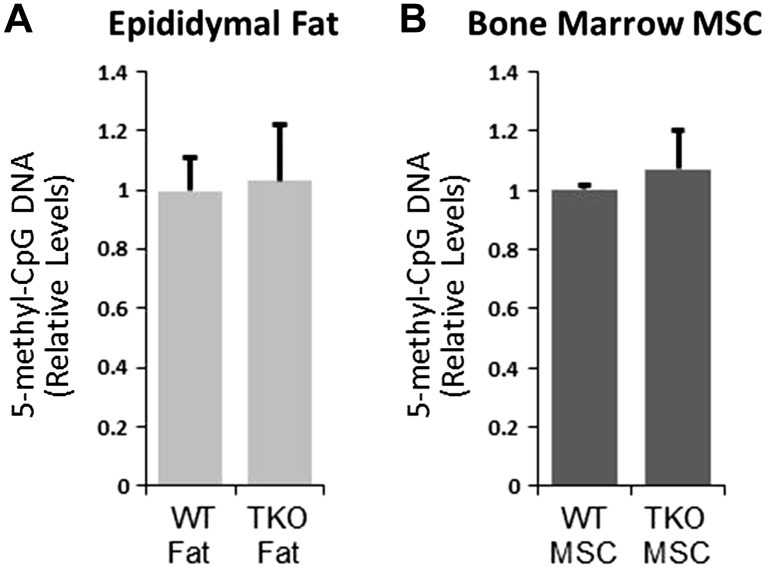
We previously demonstrated that Thy1 expression is lost during adipogenesis. Here, we show that, by blocking global CpG DNA methylation with 5-aza-dC, both adipogenesis and the loss of Thy1 expression are prevented. However, it was unknown whether Thy1 itself has a role in altering DNA methylation. To test whether Thy1 altered global CpG DNA methylation, we analyzed 5m-CpG DNA levels from epididymal fat pads of wild-type, and matched Thy1-knockout, mice (Fig. 4A). 5m-CpG DNA levels were similar in both wild-type and Thy1-knockout mice. As another test, bone marrow derived MSCs from wild-type and Thy1-knockout mice were isolated and analyzed for global 5m-CpG levels (Fig. 4B). Results showed that 5m-CpG levels are also similar in MSCs from both wild-type and Thy1-knockout mice. These results suggested that Thy1 does not alter global CpG methylation. Therefore, we further pursued the concept that Thy1 is regulated by changes in DNA methylation and that this may be key to the loss of Thy1 expression during adipogenesis.
Figure 4.
Thy1-null mice and MSCs do not have altered global methylation levels. A) Epididymal adipose tissue from wild-type C57BL/6 mice and Thy1-knockout (TKO) mice was analyzed for global CpG DNA methylation levels by ELISA. Results from adipose tissue from wild-type mice were normalized to 1. B) Bone marrow–derived MSCs from wild-type C57BL/6 and Thy1-knockout mice were analyzed for global DNA methylation as described; n = 5 animals/strain.
Thy1 (CD90) gene expression is regulated by DNA methylation during adipogenesis
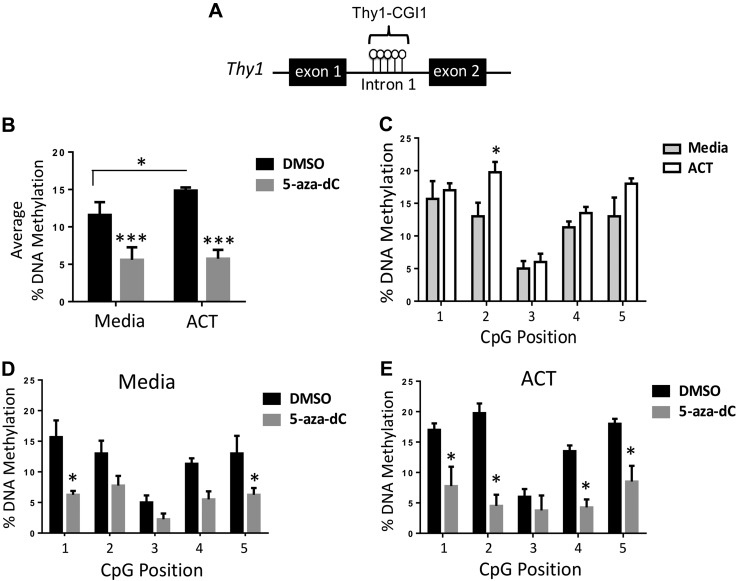
We went on to examine Thy1 DNA methylation, which typically occurs in cytosine (CG)-rich regions and is commonly associated with gene silencing (34, 35). As the transcriptional activation of the Thy1 gene involves both the upstream promoter and intron 1 (21, 23) and previous studies have shown methylation differences in that region of the Thy1 gene (17, 19–23), we focused on the most CpG-rich region within intron 1, which we term Thy1-CGI1 (Fig. 5A). Using bisulfite treatment and methylation-sensitive pyrosequencing, we analyzed methylation levels of 5 consecutive CpG sites within Thy1-CGI1. 3T3-L1 preadipocytes were treated as previously described. We found that Thy1-CGI1 was hypermethylated during differentiation comparing the average DNA methylation of ACT-treated samples to those treated with medium alone. Treatment with 5-aza-dC resulted in reduced methylation of those CpG sites in the Thy1 gene in samples treated both with medium alone and with ACT (Fig. 5B). Furthermore, individual CpG positions within Thy1-CGI1 showed an increase in DNA methylation when treated with the ACT vs. medium alone, with a significant increase at CpG position 2 (Fig. 5C). Methylation decreased across all 5 CpG sites when treated with 5-aza-dC, whether the cells were treated with medium alone or with ACT (Fig. 5D, E). Four of the 5 CpG sites examined had significantly reduced methylation levels when cells were treated with ACT and 5-aza-dC. Our data indicate that this region is methylation sensitive and can influence Thy1 expression during adipogenesis.
Figure 5.
Thy1 is hypermethylated during adipogenesis and demethylated by 5-aza-dC. 3T3-L1 cells were treated daily with either DMSO or 5-aza-dC for 7 d with the introduction of the ACT starting on d 3. A) Schematic of Thy1 gene showing Thy1-CGI1 and CpG sites between exon 1 and 2. B) Average percentage of DNA methylation by Thy1-CGI1 showed an increase in methylation in ACT cells compared with medium alone, whereas treatment with the methylation inhibitor 5-aza-dC resulted in a decrease in overall methylation in both groups. C) Individual CpG position sites were measured across Thy1-CGI1 and showed increases in methylation with ACT compared with medium alone. D, E) The medium alone and ACT groups had decreases in methylation in all individual CpG sites when treated with 5-aza-dC. *P < 0.05, ***P < 0.001.
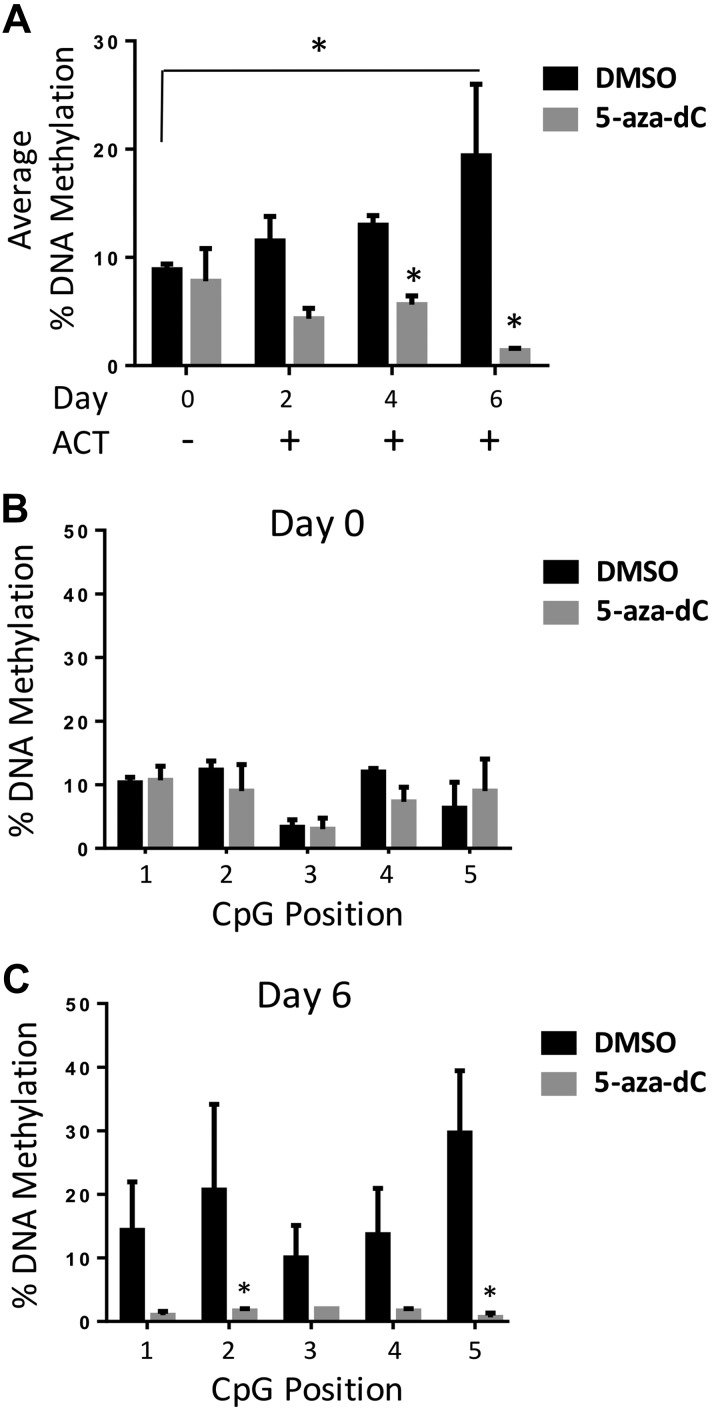
To test the methylation status of Thy1 over time during adipocyte differentiation, we examined methylation levels at those 5 sites on d 0 (before ACT), 2, 4, and 6. Cells were pretreated with DMSO or given 5-aza-dC for 24 h, then either harvested at d 0 or given ACT medium with treatment with DMSO (vehicle) or 5-aza-dC continued daily. Overall, average DNA methylation percentages increased in a time-dependent manner when exposed to the ACT, with the highest degree of 5m-CpG occurring on d 6 (Fig. 6A). The d-0 samples showed lower methylation levels compared with other time points in which cells had been exposed to ACT. Furthermore, there were no significant changes at any of the 5 CpG sites when given a single dose of 5-aza-dC (Fig. 6B). However, d-6 samples showed a significant 2-fold increase in methylation compared with d 0, along with a significant decrease in methylation when cells were exposed daily to 5-aza-dC at all 5 CpG sites (Fig. 6C). This implies that during the normal adipogenic process, CpG sites at the Thy1 locus become hypermethylated, which blunted Thy1 expression and allowed adipogenic differentiation.
Figure 6.
Thy1 is hypermethylated in a time-dependent manner during adipocyte differentiation and attenuated by 5-aza-dC. 3T3-L1 cells were pretreated for 24 h with either DMSO or 5-aza-dC, along with continuous daily treatment, and then harvested corresponding to days with ACT (d 0, no ACT). A) Average percentage of DNA methylation of Thy1-CGI1 showed an increase in methylation in normal adipogenesis differentiation process, whereas treatment with the methylation inhibitor 5-aza-dC resulted in a decrease in overall methylation. B, C) Individual CpG position sites were measured across Thy1-CGI1, and d 0 showed no relevant changes, whereas d 6 had a notable decrease in methylation when treated with 5-aza-dC. *P < 0.05.
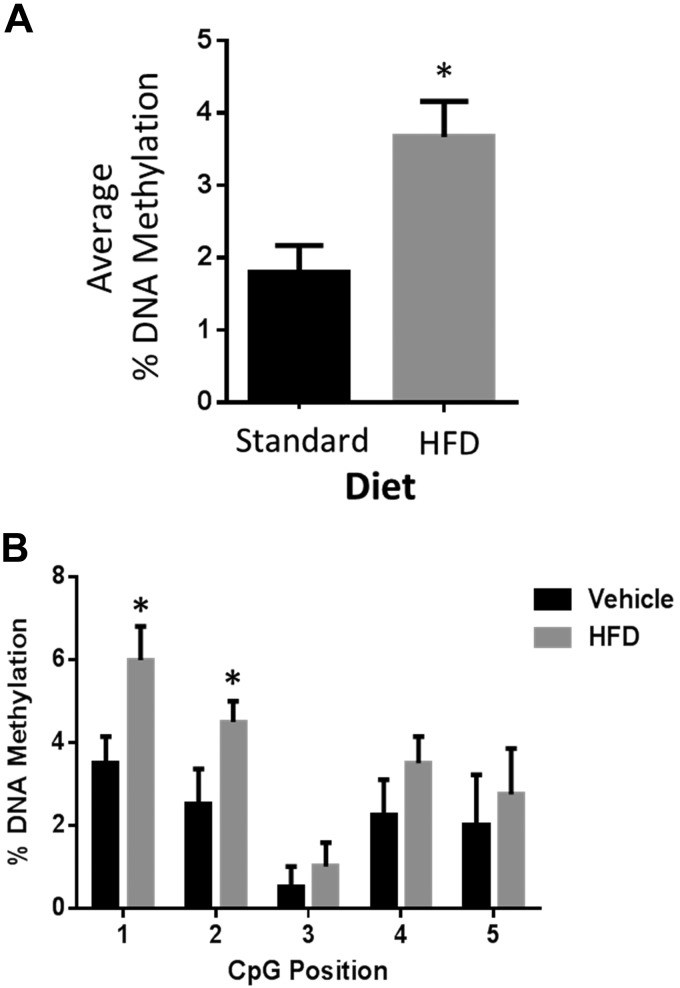
Although we found that Thy1 methylation was increased during adipogenesis in vitro, we wanted to test whether that occurred in vivo. To do that, we investigated Thy1 methylation in vivo by analyzing fat tissue from mice on either a standard chow diet (vehicle) or an HFD for 8 wk. As expected, mice gained more weight on an HFD compared with a standard diet (Supplemental Fig. S1). Interestingly, there was a significant 2-fold increase in the average percentage of DNA methylation in the Thy1-CGI1 region in adipose tissue from mice on an HFD compared with a standard diet (Fig. 7A). Additionally, we discovered that all 5 individual CpG sites analyzed within the Thy1-CGI1 region were hypermethylated in adipose tissue from mice given an HFD (Fig. 7B). These results indicate that diet-induced obesity results in hypermethylation of the Thy1 gene in vivo.
Figure 7.
DNA methylation is increased at the Thy1 gene in fat tissue of mice on an HFD. A) Average percentage of DNA methylation of Thy1-CGI1 showed an increase in methylation in fat tissue isolated from male mice on an HFD compared with mice on a standard chow diet. B) Individual CpG position sites were measured across the Thy1-CGI1 region and showed increases in CpG methylation at each site; n = 5 animals/group, *P < 0.05 (Student’s t test).
DISCUSSION
Excessive adipogenesis can lead to weight gain and obesity, which affects >700 million people worldwide. The consequences of obesity can be dire, including the development of cardiovascular or liver disease, diabetes, and other comorbidities, which result in significant morbidity and mortality. Therefore, understanding the adipogenic pathways and molecular changes that foster adipocyte differentiation will elucidate the mechanisms contributing to the pathogenesis of obesity. New understanding should lead to better solutions for this growing problem because lifestyle changes (e.g., improved diet and exercise) are often insufficient. Although genome-wide studies have shown changes in histone methylation/acetylation occur during adipogenesis (25, 36, 37), few specific adipogenesis-relevant genes have been identified as influenced by epigenetic changes (e.g., DNA methylation). In this study, we identify the Thy1 gene as being regulated by methylation during adipogenesis and demonstrate that DNA methylation has an active role in adipogenic differentiation. Ultimately, we found that inhibiting DNA methylation blunts adipogenesis and sustains Thy1 at levels that can suppress adipogenesis.
Thy1 is a cell-surface protein that is expressed on mouse thymocytes and on both mouse and human preadipocytes (38, 39). We have previously shown that Thy1 is down-regulated in a time-dependent manner during adipocyte differentiation, although cells overexpressing Thy1 no longer differentiate, even when given an ACT. However, here, we show that inhibiting DNA methylation blunts adipogenesis, even in the presence of ACT. These findings correlate with reduced levels of Fabp4 expression at both the protein and mRNA levels, along with partially attenuated levels of Thy1 when exposed to a global methylation inhibitor. Fabp4 levels inversely correlate with Thy1 levels, and Fabp4 expression increases, whereas Thy1 expression is suppressed during adipogenesis (14). We also demonstrated that during adipogenesis, Thy1-CGI1 methylation increases, which likely contributes to reduced Thy1 expression. Although we saw statistically significant changes at CpG position 2 in ACT-treated samples relative to untreated (medium alone) cells (Fig. 5C), we observed similar trends for the other CpG positions. Those results are consistent with previous studies showing alterations in methylation status at CpG sites causes significant changes in gene expression (40, 41). Treatment with the DNA methylation inhibitor, 5-aza-dC, resulted in hypomethylation of Thy1-CGI1, which in part may contribute to the attenuation of Thy1 expression. Because 5-aza-dC is a global DNA methylation inhibitor, it can affect other genes involved in adipogenesis in addition to Thy1. Although the Thy1 gene may be a key target for methylation during adipogenesis, there are undoubtedly other genes regulated by methylation during differentiation. Further investigation is necessary to identify other essential genes regulated through epigenetic means because that will increase our understanding of the adipogenic pathway. Our data suggest that DNA methylation is a necessary regulatory mechanism for preadipocytes to differentiate into fat cells, consistent with recent reports implying DNA methylation is involved in lineage-specific adipocyte development (24, 42).
Adipogenesis is a complex process that is controlled by many factors, which include epigenetic and posttranscriptional modifications. Previous studies have established that the expression/activity of miRs, small RNAs ∼20–22 bp in length that bind to and block expression of specific, targeted genes, have a role adipogenesis (43). Furthermore, in obesity, it has been shown that there are significant changes in the expression of miRs, such as an increase in miR-103 levels (44, 45). We have recently shown that miR-103/107 levels increase during adipogenesis and that miR-103 can bind the 3′ UTR of Thy1 to blunt its expression (15). Here, we confirmed that the levels of miR-103 increased during adipogenesis and that those levels were unchanged by treatment with 5-aza-dC, suggesting that the regulatory effects of Thy1 methylation are separate from changes in miR expression (Supplemental Fig. S3). Thus, regulation of Thy1 expression by DNA methylation represents a novel insight into the process of adipogenic differentiation.
It is currently unclear the relative contribution that each pathway (miR vs. epigenetic) has in controlling Thy1 expression. One novel way to test the relative contribution of miR-103/107 vs. epigenetic regulation of Thy1 during adipogenesis would be to introduce single-nucleotide mutations in both the miR-103/107 seed sequence of the Thy1 3′-UTR and CpG sites in the Thy1-CGI1 using clustered regularly interspaced short palindromic repeats. Although that approach was beyond the scope of the current study, future work delineating the importance of those mechanisms is of great interest. Still, it is interesting to speculate that, based on results presented here, DNA methylation is more important than miR regulation of Thy1 during adipogenesis. Additionally, timing and stability of epigenetic vs. miR-mediated regulation of Thy1 are likely distinct. Methylation changes may be induced at early events of the adipogenic process and are likely passed on during proliferation of adipogenic progenitor cells (46). Indeed, in certain heterogeneous populations of fibroblasts, some are already prone to adipogenesis and are stably Thy1low/− (14, 33). Whether Thy1 methylation is different in Thy1hi/+ vs. Thy1low/− fibroblasts is unclear; however, given the relatively stable nature of epigenetic marks, that appears likely. In contrast, regulation of Thy1 by miRNA may occur at a more committed step of adipogenesis. MiR-103, which we previously reported targets Thy1 during adipogenesis, is a PPARγ-dependent miRNA (47). Because PPARγ is activated after adipogenic commitment, this is likely a later step in Thy1 regulation compared with methylation (48).
Another important finding in this study is that Thy1 CpG methylation is increased in an animal of diet-induced obesity. Showing that CpG methylation increases in both adipogenesis in vitro and obesity in vivo adds to the significance of this work and highlights the function of Thy1 in these processes. This has notable implications for heritable alterations in obesity. Increased Thy1 methylation levels may be inherited by offspring from obese parents. Interestingly, animal models of obesity have shown that maternal obesity epigenetically alters visceral fat progenitor cells in male offspring making them more prone to form adipocytes (49). Although Thy1 expression was not measured in those studies, it may be that Thy1 is lower in the more adipogenic progenitor cells. Interestingly, MSCs from obese human beings also show lower Thy1 levels compared with MSCs from nonobese individuals (50); however, whether or not DNA methylation has a role in that decrease is currently unknown.
Another possibility is that Thy1 DNA methylation, like miR regulation, is PPARγ dependent; however, few studies have shown that PPARγ itself can dictate DNA methylation. Future studies with PPARγ-null cells may shed light on that possibility. Interestingly, the PPARγ gene is also a target of CpG methylation (51). Methylation status of the PPARγ gene was inversely correlated with body mass index in this study. This suggests that, in contrast to Thy1, PPARγ gene methylation is lower in obesity.
Stem cells, such as MSCs, are distinguished and defined by various cell surface markers, including Thy1 (52). MSCs can differentiate into adipocytes, osteocytes, and chondrocytes (39). Recent studies have shown 5-aza-dC prevents adipogenesis and promotes osteoblastogenesis through the activation of Wnt10a (30). Wnt10a is known to be up-regulated and essential for bone formation (53–55). However, Thy1 also has a critical role (56, 57). It was recently shown that Thy1 is up-regulated during osteoblastogenesis and that Thy1− cells (knockdown and knockout) have an impaired ability to form osteoblasts (58). However, changes in Thy1 DNA methylation during osteoblast formation have not been analyzed.
Although we show that 5-aza-dC treatment decreased methylation in the Thy1-CGI1 region and blunted adipogenesis, it is possible that loss of methylation at the Thy1 gene could affect other cell types, such as fibroblasts or T lymphocytes, in a different manner. For example, it has been established that Thy1 expression is up-regulated in myofibroblasts from radiation-induced scarring compared with normal skin fibroblasts (59). Additionally, several types of malignant cells, including cancer stem cells, show increased expression of Thy1 (60, 61). Future studies are needed to determine whether DNA methylation regulates Thy1 expression in those cells and potentially in others.
In summary, our work shows for the first time that the Thy1 gene has increased DNA methylation during adipogenesis. We demonstrate herein that inhibiting DNA methylation attenuates the loss of Thy1 expression when cells are stimulated to differentiate into adipocytes. Blocking methylation leads to sustained Thy1 expression and prevents adipogenesis. These studies highlight the role that both Thy1 and CpG DNA methylation have during adipogenesis and, further, suggest these pathways may be dysregulated in metabolic diseases in which adipogenesis and adipocyte biology is altered, such as obesity.
Supplementary Material
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
ACKNOWLEDGMENTS
The authors thank Stephen Pollock (University of Rochester Medical Center) for assisting with figure preparation for the manuscript. This project is funded by U.S. National Institutes of Health (NIH) National Institute of Environmental Health Sciences Grants F31ES027767 (to E.F.), ES001247, and ES0023032; NIH National Heart, Lung, and Blood Institute Grant HL133761; and University of Rochester Clinical and Translational Science Award TL1-TR000096 from the NIH. The authors declare no conflicts of interest.
Glossary
- 5-aza-dC
5-aza-2′-deoxycytidine
- ACT
adipogenic cocktail
- BSA
bovine serum albumin
- Fabp4
fatty acid binding protein 4
- HFD
high-fat diet
- MFI
mean fluorescence intensity
- miR
microRNA
- mMSC
mouse mesenchymal stem cell
- MSC
mesenchymal stem cell
- PPARγ
peroxisome proliferator activated receptor-γ
- Thy1
thymocyte antigen-1
- Thy1-CGI1
Thy1 CpG island 1
Footnotes
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
AUTHOR CONTRIBUTIONS
E. M. Flores performed all experiments; E. M. Flores, C. F. Woeller, M. Susiarjo, and R. Phipps aided in experimental design and data analysis; E. M. Flores prepared figures and wrote the manuscript; and all authors edited and reviewed the manuscript.
REFERENCES
- 1.Afshin A., Forouzanfar M. H., Reitsma M. B., Sur P., Estep K., Lee A., Marczak L., Mokdad A. H., Moradi-Lakeh M., Naghavi M., Salama J. S., Vos T., Abate K. H., Abbafati C., Ahmed M. B., Al-Aly Z., Alkerwi A., Al-Raddadi R., Amare A. T., Amberbir A., Amegah A. K., Amini E., Amrock S. M., Anjana R. M., Ärnlöv J., Asayesh H., Banerjee A., Barac A., Baye E., Bennett D. A., Beyene A. S., Biadgilign S., Biryukov S., Bjertness E., Boneya D. J., Campos-Nonato I., Carrero J. J., Cecilio P., Cercy K., Ciobanu L. G., Cornaby L., Damtew S. A., Dandona L., Dandona R., Dharmaratne S. D., Duncan B. B., Eshrati B., Esteghamati A., Feigin V. L., Fernandes J. C., Fürst T., Gebrehiwot T. T., Gold A., Gona P. N., Goto A., Habtewold T. D., Hadush K. T., Hafezi-Nejad N., Hay S. I., Horino M., Islami F., Kamal R., Kasaeian A., Katikireddi S. V., Kengne A. P., Kesavachandran C. N., Khader Y. S., Khang Y. H., Khubchandani J., Kim D., Kim Y. J., Kinfu Y., Kosen S., Ku T., Defo B. K., Kumar G. A., Larson H. J., Leinsalu M., Liang X., Lim S. S., Liu P., Lopez A. D., Lozano R., Majeed A., Malekzadeh R., Malta D. C., Mazidi M., McAlinden C., McGarvey S. T., Mengistu D. T., Mensah G. A., Mensink G. B. M., Mezgebe H. B., Mirrakhimov E. M., Mueller U. O., Noubiap J. J., Obermeyer C. M., Ogbo F. A., Owolabi M. O., Patton G. C., Pourmalek F., Qorbani M., Rafay A., Rai R. K., Ranabhat C. L., Reinig N., Safiri S., Salomon J. A., Sanabria J. R., Santos I. S., Sartorius B., Sawhney M., Schmidhuber J., Schutte A. E., Schmidt M. I., Sepanlou S. G., Shamsizadeh M., Sheikhbahaei S., Shin M. J., Shiri R., Shiue I., Roba H. S., Silva D. A. S., Silverberg J. I., Singh J. A., Stranges S., Swaminathan S., Tabarés-Seisdedos R., Tadese F., Tedla B. A., Tegegne B. S., Terkawi A. S., Thakur J. S., Tonelli M., Topor-Madry R., Tyrovolas S., Ukwaja K. N., Uthman O. A., Vaezghasemi M., Vasankari T., Vlassov V. V., Vollset S. E., Weiderpass E., Werdecker A., Wesana J., Westerman R., Yano Y., Yonemoto N., Yonga G., Zaidi Z., Zenebe Z. M., Zipkin B., Murray C. J. L.; GBD 2015 Obesity Collaborators (2017) Health effects of overweight and obesity in 195 countries over 25 years. N. Engl. J. Med. 377, 13–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Flegal K. M., Kruszon-Moran D., Carroll M. D., Fryar C. D., Ogden C. L. (2016) Trends in obesity among adults in the United States, 2005 to 2014. JAMA 315, 2284–2291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ahima R. S. (2009) Connecting obesity, aging and diabetes. Nat. Med. 15, 996–997 [DOI] [PubMed] [Google Scholar]
- 4.Smyth S., Heron A. (2006) Diabetes and obesity: the twin epidemics. Nat. Med. 12, 75–80 [DOI] [PubMed] [Google Scholar]
- 5.Heymsfield S. B., Wadden T. A. (2017) Mechanisms, pathophysiology, and management of obesity. N. Engl. J. Med. 376, 254–266 [DOI] [PubMed] [Google Scholar]
- 6.Cawley J., Meyerhoefer C. (2012) The medical care costs of obesity: an instrumental variables approach. J. Health Econ. 31, 219–230 [DOI] [PubMed] [Google Scholar]
- 7.Jo J., Gavrilova O., Pack S., Jou W., Mullen S., Sumner A. E., Cushman S. W., Periwal V. (2009) Hypertrophy and/or hyperplasia: dynamics of adipose tissue growth. PLOS Comput. Biol. 5, e1000324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Joe A. W., Yi L., Even Y., Vogl A. W., Rossi F. M. (2009) Depot-specific differences in adipogenic progenitor abundance and proliferative response to high-fat diet. Stem Cells 27, 2563–2570 [DOI] [PubMed] [Google Scholar]
- 9.Jing K., Heo J. Y., Song K. S., Seo K. S., Park J. H., Kim J. S., Jung Y. J., Jo D. Y., Kweon G. R., Yoon W. H., Hwang B. D., Lim K., Park J. I. (2009) Expression regulation and function of Pref-1 during adipogenesis of human mesenchymal stem cells (MSCs). Biochim. Biophys. Acta 1791, 816–826 [DOI] [PubMed] [Google Scholar]
- 10.Hudak C. S., Sul H. S. (2013) Pref-1, a gatekeeper of adipogenesis. Front. Endocrinol. (Lausanne) 4, 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim C. Y., Kim G. N., Wiacek J. L., Chen C. Y., Kim K. H. (2012) Selenate inhibits adipogenesis through induction of transforming growth factor-β1 (TGF-β1) signaling. Biochem. Biophys. Res. Commun. 426, 551–557 [DOI] [PubMed] [Google Scholar]
- 12.Kumar A., Ruan M., Clifton K., Syed F., Khosla S., Oursler M. J. (2012) TGF-β mediates suppression of adipogenesis by estradiol through connective tissue growth factor induction. Endocrinology 153, 254–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cawthorn W. P., Bree A. J., Yao Y., Du B., Hemati N., Martinez-Santibañez G., MacDougald O. A. (2012) Wnt6, Wnt10a and Wnt10b inhibit adipogenesis and stimulate osteoblastogenesis through a β-catenin-dependent mechanism. Bone 50, 477–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Woeller C. F., O’Loughlin C. W., Pollock S. J., Thatcher T. H., Feldon S. E., Phipps R. P. (2015) Thy1 (CD90) controls adipogenesis by regulating activity of the Src family kinase, Fyn. FASEB J. 29, 920–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Woeller C. F., Flores E., Pollock S. J., Phipps R. P. (2017) Editor’s highlight: Thy1 (CD90) expression is reduced by the environmental chemical tetrabromobisphenol-A to promote adipogenesis through induction of microRNA-103. Toxicol Sci. 157, 305–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Robinson C. M., Neary R., Levendale A., Watson C. J., Baugh J. A. (2012) Hypoxia-induced DNA hypermethylation in human pulmonary fibroblasts is associated with Thy-1 promoter methylation and the development of a pro-fibrotic phenotype. Respir. Res. 13, 74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kolsto A. B., Kollias G., Giguere V., Isobe K. I., Prydz H., Grosveld F. (1986) The maintenance of methylation-free islands in transgenic mice. Nucleic Acids Res. 14, 9667–9678 [PMC free article] [PubMed] [Google Scholar]
- 18.Mann V., Szyf M., Razin A., Chriqui-Zeira E., Kedar E. (1986) Characterization of a tumorigenic murine T-lymphoid-cell line spontaneously derived from an IL-2-dependent T-cell line. Int. J. Cancer. 37, 781–786 [DOI] [PubMed] [Google Scholar]
- 19.Neveu W. A., Mills S. T., Staitieh B. S., Sueblinvong V. (2015) TGF-β1 epigenetically modifies Thy-1 expression in primary lung fibroblasts. Am. J. Physiol. Cell Physiol. 309, C616–C626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sanders Y. Y., Pardo A., Selman M., Nuovo G. J., Tollefsbol T. O., Siegal G. P., Hagood J. S. (2008) Thy-1 promoter hypermethylation: a novel epigenetic pathogenic mechanism in pulmonary fibrosis. Am. J. Respir. Cell Mol. Biol. 39, 610–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Giguére V., Isobe K., Grosveld F. (1985) Structure of the murine Thy-1 gene. EMBO J. 4, 2017–2024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gundersen G., Kolstø A. B., Larsen F., Prydz H. (1992) Tissue-specific methylation of a CpG island in transgenic mice. Gene 113, 207–214 [DOI] [PubMed] [Google Scholar]
- 23.Spanopoulou E., Giguere V., Grosveld F. (1991) The functional domains of the murine Thy-1 gene promoter. Mol. Cell. Biol. 11, 2216–2228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lim Y. C., Chia S. Y., Jin S., Han W., Ding C., Sun L. (2016) Dynamic DNA methylation landscape defines brown and white cell specificity during adipogenesis. Mol. Metab. 5, 1033–1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Broholm C., Olsson A. H., Perfilyev A., Gillberg L., Hansen N. S., Ali A., Mortensen B., Ling C., Vaag A. (2016) Human adipogenesis is associated with genome-wide DNA methylation and gene-expression changes. Epigenomics 8, 1601–1617 [DOI] [PubMed] [Google Scholar]
- 26.Guo W., Chen J., Yang Y., Zhu J., Wu J. (2016) Epigenetic programming of Dnmt3a mediated by AP2α is required for granting preadipocyte the ability to differentiate. Cell Death Dis. 7, e2496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sakamoto H., Kogo Y., Ohgane J., Hattori N., Yagi S., Tanaka S., Shiota K. (2008) Sequential changes in genome-wide DNA methylation status during adipocyte differentiation. Biochem. Biophys. Res. Commun. 366, 360–366 [DOI] [PubMed] [Google Scholar]
- 28.Yang X., Wu R., Shan W., Yu L., Xue B., Shi H. (2016) DNA methylation biphasically regulates 3T3-L1 preadipocyte differentiation. Mol. Endocrinol. 30, 677–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao Q. H., Wang S. G., Liu S. X., Li J. P., Zhang Y. X., Sun Z. Y., Fan Q. M., Tian J. W. (2013) PPARγ forms a bridge between DNA methylation and histone acetylation at the C/EBPα gene promoter to regulate the balance between osteogenesis and adipogenesis of bone marrow stromal cells. FEBS J. 280, 5801–5814 [DOI] [PubMed] [Google Scholar]
- 30.Chen Y. S., Wu R., Yang X., Kou S., MacDougald O. A., Yu L., Shi H., Xue B. (2016) Inhibiting DNA methylation switches adipogenesis to osteoblastogenesis by activating Wnt10a. Sci. Rep. 6, 25283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu S., De Becker A., Van Camp B., Vanderkerken K., Van Riet I. (2010) An improved harvest and in vitro expansion protocol for murine bone marrow-derived mesenchymal stem cells. J. Biomed. Biotechnol. 2010, 105940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qian S. W., Li X., Zhang Y. Y., Huang H. Y., Liu Y., Sun X., Tang Q. Q. (2010) Characterization of adipocyte differentiation from human mesenchymal stem cells in bone marrow. BMC Dev. Biol. 10, 47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lehmann G. M., Woeller C. F., Pollock S. J., O’Loughlin C. W., Gupta S., Feldon S. E., Phipps R. P. (2010) Novel anti-adipogenic activity produced by human fibroblasts. Am. J. Physiol. Cell Physiol. 299, C672–C681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suzuki M. M., Bird A. (2008) DNA methylation landscapes: provocative insights from epigenomics. Nat. Rev. Genet. 9, 465–476 [DOI] [PubMed] [Google Scholar]
- 35.Hsieh C. L. (1994) Dependence of transcriptional repression on CpG methylation density. Mol. Cell. Biol. 14, 5487–5494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Keller M., Hopp L., Liu X., Wohland T., Rohde K., Cancello R., Klös M., Bacos K., Kern M., Eichelmann F., Dietrich A., Schön M. R., Gärtner D., Lohmann T., Dreßler M., Stumvoll M., Kovacs P., DiBlasio A. M., Ling C., Binder H., Blüher M., Böttcher Y. (2016) Genome-wide DNA promoter methylation and transcriptome analysis in human adipose tissue unravels novel candidate genes for obesity. Mol. Metab. 6, 86–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mikkelsen T. S., Xu Z., Zhang X., Wang L., Gimble J. M., Lander E. S., Rosen E. D. (2010) Comparative epigenomic analysis of murine and human adipogenesis. Cell 143, 156–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Haeryfar S. M., Hoskin D. W. (2004) Thy-1: more than a mouse pan-T cell marker. J. Immunol. 173, 3581–3588 [DOI] [PubMed] [Google Scholar]
- 39.Orbay H., Tobita M., Mizuno H. (2012) Mesenchymal stem cells isolated from adipose and other tissues: basic biological properties and clinical applications. Stem Cells Int. 2012, 461718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Attwood J. T., Yung R. L., Richardson B. C. (2002) DNA methylation and the regulation of gene transcription. Cell. Mol. Life Sci. 59, 241–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baylin S. B., Herman J. G., Graff J. R., Vertino P. M., Issa J. P. (1998) Alterations in DNA methylation: a fundamental aspect of neoplasia. Adv. Cancer Res. 72, 141–196 [PubMed] [Google Scholar]
- 42.Bowers R. R., Kim J. W., Otto T. C., Lane M. D. (2006) Stable stem cell commitment to the adipocyte lineage by inhibition of DNA methylation: role of the BMP-4 gene. Proc. Natl. Acad. Sci. USA 103, 13022–13027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Du J., Cheng X., Shen L., Tan Z., Luo J., Wu X., Liu C., Yang Q., Jiang Y., Tang G., Li X., Zhang S., Zhu L. (2016) Methylation of miR-145a-5p promoter mediates adipocytes differentiation. Biochem. Biophys. Res. Commun. 475, 140–148 [DOI] [PubMed] [Google Scholar]
- 44.Li M., Liu Z., Zhang Z., Liu G., Sun S., Sun C. (2015) miR-103 promotes 3T3-L1 cell adipogenesis through AKT/mTOR signal pathway with its target being MEF2D. Biol. Chem. 396, 235–244 [DOI] [PubMed] [Google Scholar]
- 45.Perri R., Nares S., Zhang S., Barros S. P., Offenbacher S. (2012) MicroRNA modulation in obesity and periodontitis. J. Dent. Res. 91, 33–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Noer A., Sørensen A. L., Boquest A. C., Collas P. (2006) Stable CpG hypomethylation of adipogenic promoters in freshly isolated, cultured, and differentiated mesenchymal stem cells from adipose tissue. Mol. Biol. Cell 17, 3543–3556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.John E., Wienecke-Baldacchino A., Liivrand M., Heinäniemi M., Carlberg C., Sinkkonen L. (2012) Dataset integration identifies transcriptional regulation of microRNA genes by PPARγ in differentiating mouse 3T3-L1 adipocytes. Nucleic Acids Res. 40, 4446–4460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lefterova M. I., Haakonsson A. K., Lazar M. A., Mandrup S. (2014) PPARgamma and the global map of adipogenesis and beyond. Trends Endocrinol. Metab. 25, 293–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liang X., Yang Q., Fu X., Rogers C. J., Wang B., Pan H., Zhu M. J., Nathanielsz P. W., Du M. (2016) Maternal obesity epigenetically alters visceral fat progenitor cell properties in male offspring mice. J. Physiol. 594, 4453–4466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Oñate B., Vilahur G., Ferrer-Lorente R., Ybarra J., Díez-Caballero A., Ballesta-López C., Moscatiello F., Herrero J., Badimon L. (2012) The subcutaneous adipose tissue reservoir of functionally active stem cells is reduced in obese patients. FASEB J. 26, 4327–4336 [DOI] [PubMed] [Google Scholar]
- 51.Volberg V., Yousefi P., Huen K., Harley K., Eskenazi B., Holland N. (2017) CpG Methylation across the adipogenic PPARγ gene and its relationship with birthweight and child BMI at 9 years. BMC Med. Genet. 18, 7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Alipour R., Sadeghi F., Hashemi-Beni B., Zarkesh-Esfahani S. H., Heydari F., Mousavi S. B., Adib M., Narimani M., Esmaeili N. (2010) Phenotypic characterizations and comparison of adult dental stem cells with adipose-derived stem cells. Int. J. Prev. Med. 1, 164–171 [PMC free article] [PubMed] [Google Scholar]
- 53.Kasaai B., Moffatt P., Al-Salmi L., Lauzier D., Lessard L., Hamdy R. C. (2012) Spatial and temporal localization of WNT signaling proteins in a mouse model of distraction osteogenesis. J. Histochem. Cytochem. 60, 219–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rudnicki M. A., Williams B. O. (2015) Wnt signaling in bone and muscle. Bone 80, 60–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yin X., Wang X., Hu X., Chen Y., Zeng K., Zhang H. (2015) ERβ induces the differentiation of cultured osteoblasts by both Wnt/β-catenin signaling pathway and estrogen signaling pathways. Exp. Cell Res. 335, 107–114 [DOI] [PubMed] [Google Scholar]
- 56.Chung M. T., Liu C., Hyun J. S., Lo D. D., Montoro D. T., Hasegawa M., Li S., Sorkin M., Rennert R., Keeney M., Yang F., Quarto N., Longaker M. T., Wan D. C. (2013) CD90 (Thy-1)-positive selection enhances osteogenic capacity of human adipose-derived stromal cells. Tissue Eng. Part A 19, 989–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bearden R. N., Huggins S. S., Cummings K. J., Smith R., Gregory C. A., Saunders W. B. (2017) In-vitro characterization of canine multipotent stromal cells isolated from synovium, bone marrow, and adipose tissue: a donor-matched comparative study. Stem Cell Res. Ther. 8, 218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Paine A., Woeller C. F., Zhang H., de la Luz Garcia-Hernandez M., Huertas N., Xing L., Phipps R. P., Ritchlin C. T. (2018) Thy1 is a positive regulator of osteoblast differentiation and modulates bone homeostasis in obese mice. FASEB J. 32, 3174–3183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hansen T. C., Woeller C. F., Lacy S. H., Koltz P. F., Langstein H. N., Phipps R. P. (2017) Thy1 (CD90) expression is elevated in radiation-induced periprosthetic capsular contracture: implication for novel therapeutics. Plast. Reconstr. Surg. 140, 316–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.He J., Liu Y., Zhu T., Zhu J., Dimeco F., Vescovi A. L., Heth J. A., Muraszko K. M., Fan X., Lubman D. M. (2012) CD90 is identified as a candidate marker for cancer stem cells in primary high-grade gliomas using tissue microarrays. Mol. Cell Proteomics 11, M111.010744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Parry P. V., Engh J. A. (2012) CD90 is identified as a marker for cancer stem cells in high-grade gliomas using tissue microarrays. Neurosurgery 70, N23–N24 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.