Abstract
Transcriptomics has been widely applied to study grape berry development. With few exceptions, transcriptomic studies in grape are performed using the available genome sequence, PN40024, as reference. However, differences in gene content among grape accessions, which contribute to phenotypic differences among cultivars, suggest that a single reference genome does not represent the species’ entire gene space. Though whole genome assembly and annotation can reveal the relatively unique or “private” gene space of any particular cultivar, transcriptome reconstruction is a more rapid, less costly, and less computationally intensive strategy to accomplish the same goal. In this study, we used single molecule-real time sequencing (SMRT) to sequence full-length cDNA (Iso-Seq) and reconstruct the transcriptome of Cabernet Sauvignon berries during berry ripening. In addition, short reads from ripening berries were used to error-correct low-expression isoforms and to profile isoform expression. By comparing the annotated gene space of Cabernet Sauvignon to other grape cultivars, we demonstrate that the transcriptome reference built with Iso-Seq data represents most of the expressed genes in the grape berries and includes 1,501 cultivar-specific genes. Iso-Seq produced transcriptome profiles similar to those obtained after mapping on a complete genome reference. Together, these results justify the application of Iso-Seq to identify cultivar-specific genes and build a comprehensive reference for transcriptional profiling that circumvents the necessity of a genome reference with its associated costs and computational weight.
Keywords: transcriptome reference, RNA-seq, Vitis vinifera, berry ripening
Grape berries undergo a series of complex physiological and biochemical changes during their development that determine their characteristics at harvest (Kuhn et al. 2014). Genome-wide expression studies using microarray and, more recently, RNA sequencing (RNA-seq) revealed that berry development involves the expression and modulation of approximately 23,000 genes (Massonnet et al. 2017a) and that the ripening transition is associated with a major transcriptome shift (Fasoli et al. 2012). Transcriptomic studies characterized the ripening program across grapevine cultivars (Venturini et al. 2013; Da Silva et al. 2013; Jiao et al. 2015; Massonnet et al. 2017a), identifying key ripening-related genes (Palumbo et al. 2014; Massonnet et al. 2017a) and determining the impact of stress and viticultural practices on ripening (Deluc et al. 2009; Pastore et al. 2013; Xi et al. 2014; Amrine et al. 2015; Blanco-Ulate et al. 2015, 2017; Corso et al. 2015; Hopper et al. 2016; Savoi et al. 2016; Zenoni et al. 2017; Lecourieux et al. 2017; Massonnet et al. 2017b). This knowledge increases the possibility of exerting control over the ripening process, improving fruit composition under suboptimal or adverse conditions, and enhancing desirable traits in a crop with outstanding cultural and commercial significance (Savoi et al. 2016, 2017; Serrano et al. 2017; Zenoni et al. 2017).
These genome-wide expression analyses were possible because a highly contiguous assembly for the species was produced (Jaillon et al. 2007); this first effort used a grape line (PN40024) created by several rounds of backcrossing to reduce heterozygosity, facilitating genome assembly (Jaillon et al. 2007). Though poor by current standards, this pioneering, chromosome-resolved assembly served as the basis for numerous publications. However, the structural diversity of grape genomes makes using a single one-size-fits-all reference genome inappropriate (Golicz et al. 2016a, 2016b). There is substantial unshared gene content between cultivars, with 8–10% of the genes missing when two cultivars are compared (Da Silva et al. 2013). Although many of these genes are not essential for plant survival, they can account for 80% of the expression within their respective families and expand key gene families possibly associated with cultivar-specific traits (Da Silva et al. 2013).
Assembling genome references for all interesting cultivars is impractical, in part because its cost remains prohibitive and because of genomic features that impede the development of high-quality genome assemblies for any grape cultivar. Although the V. vinifera genome is relatively small (∼487Mb) (Lodhi and Reisch 1995; Jaillon et al. 2007) and as repetitive as other plant genomes of similar size (41.4%) (Jaillon et al. 2007; Michael and Jackson 2013), it is highly heterozygous (∼13%) (Jaillon et al. 2007; Velasco et al. 2007). Most domesticated grape cultivars are crosses between distantly related parents; this and clonal propagation cause the high heterozygosity observed in the species (Strefeler et al. 1992; Ohmi et al. 1993; Bowers and Meredith 1997; Sefc et al. 1998; Lopes et al. 1999; Di Gaspero et al. 2005; Tapia et al. 2007; Ibáñez et al. 2009; Cipriani et al. 2010; Myles et al. 2011; Lacombe et al. 2013; Chin et al. 2016; Minio et al. 2017; Zhou et al. 2017). Earlier attempts using short reads struggled to resolve complex, highly heterozygous genomes (Gnerre et al. 2011; Huang et al. 2012; Di Genova et al. 2014; Kajitani et al. 2014; Safonova et al. 2015). A limited ability to call consensus polymorphic regions yields highly fragmented assemblies where structural ambiguity occurs and alternative alleles at heterozygous sites are excluded altogether (Velasco et al. 2007). Single Molecule Real Time (SMRT) DNA sequencing (Pacific Biosciences, California, USA) has emerged as the leading technology for reconstructing highly contiguous, diploid assemblies of long, repetitive genomes that include phased information about heterozygous sites (Chin et al. 2013, 2016; Doi et al. 2014; Gordon et al. 2016; Pryszcz and Gabaldón 2016; Ricker et al. 2016; Seo et al. 2016; Vij et al. 2016; Huddleston et al. 2017). Recently, we used Vitis vinifera cv. Cabernet Sauvignon to test the ability of SMRT reads and the FALCON-Unzip assembly pipeline to resolve both alleles at heterozygous sites in the genome (Chin et al. 2016). The assembly produced was significantly more contiguous (contig N50 = 2.17 Mb) than the original PN40024 assembly (contig N50 = 102.7 kbp) and provided the first phased sequences of the diploid V. vinifera genome (Minio et al. 2017).
Despite recent advances in genome reconstruction methodologies, assembling a complex plant genome is still costly. Transcriptome reconstruction is the only alternative strategy to depict known and unknown gene content information (Venturini et al. 2013; Da Silva et al. 2013; Jiao et al. 2015). De novo assembly of RNA-seq reads is widely used for this purpose (Grabherr et al. 2011; Ashrafi et al. 2012; Venturini et al. 2013; Bellucci et al. 2014). SMRT technology was recently deployed to investigate expressed gene isoforms (Iso-Seq) in a variety of organisms, including a handful of plant species (Liu et al. 2017; Zulkapli et al. 2017; Filichkin et al. 2018). Long reads delivered by this methodology report full-length transcripts sequenced from their 5′-ends to polyadenylated tails (Dong et al. 2015; Weirather et al. 2015; Gao et al. 2016; Tombácz et al. 2016; Kuo et al. 2017; Price and Gibas 2017; Workman et al. 2017), making Iso-Seq an ideal technology for reconstructing a transcriptome without a reference genome sequence and without assembling fragments to resolve the complete isoform sequence (Honaas et al. 2016; Ju et al. 2017). Moreover, alternative transcripts that contribute to the gene space complexity (Brett et al. 2002) and vary with cell type (Swarup et al. 2016), developmental stage (Thatcher et al. 2016), and stress (Yan et al. 2012; Liu et al. 2016) cannot be definitively characterized without full-length transcript information.
The objective of this study was to test whether full-length cDNA sequencing with Iso-Seq technology is a suitable alternative to traditional genome sequencing, assembly, and annotation for reconstructing a grape transcriptome reference for transcriptional profiling. We compared how Cabernet Sauvignon’s Iso-Seq transcriptome fares as a reference for RNA-seq analysis vs. its annotated genome. We sequenced the full-length transcripts of ripening berries with Iso-Seq and Illumina RNA-seq reads. The high-coverage short-read data were used to profile gene expression and to error-correct low-expression isoforms that would have been otherwise lost by the standard Iso-Seq pipeline. The transcriptome reference built with Iso-Seq data represented most of the expressed genes in the grape berries and included cultivar-specific or “private” genes. When used as the reference for RNA-seq, Iso-Seq generated transcriptome profiles quantitatively similar to those obtained by mapping on a complete genome reference. These results support using Iso-Seq to capture the gene space of a plant and build a comprehensive reference for transcriptional profiling without a pre-defined reference genome.
Methods
Plant material and RNA isolation
Grape berries from Cabernet Sauvignon FPS clone 08 were collected in Summer 2016 from vines grown in the Foundation Plant Services (FPS) Classic Foundation Vineyard (Davis, CA, USA). Between 10 and 15 berries were sampled at pre-véraison, véraison, post-véraison, and at commercial maturity. Table S1 provides weather information for the sampling days. The ripening stages were visually assessed based on color development and confirmed by measurements of soluble solids (Figure S1; Table S2). On the day of sampling, berries were deseeded, frozen in liquid nitrogen, and ground to powder (skin and pulp). Total RNA was isolated using a Cetyltrimethyl Ammonium Bromide (CTAB)-based extraction protocol as described in Blanco-Ulate et al. (2013). RNA purity was evaluated with a Nanodrop 2000 spectrophotometer (Thermo Scientific, Hanover Park, IL, USA). RNA was quantified with a Qubit 2.0 Fluorometer using the RNA broad range kit (Life Technologies, Carlsbad, CA, USA). RNA integrity was assessed using electrophoresis and an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). Only RNA with integrity number (RIN) greater than 8.0 was used for SMRTbell library preparation.
Library preparation and sequencing
RNAs from four biological replicates per developmental stage were pooled in equal amounts. One µg of the pooled RNA was used for cDNA synthesis and for SMRTbell library construction using the SMARTer PCR cDNA synthesis kit (Clontech Laboratories, Inc. Mountain View, CA, USA). First-strand cDNA synthesis was performed using the SMRTScribe Reverse Transcriptase (Clontech Laboratories, Inc. Mountain View, CA, USA). Each developmental stage was individually barcoded (Table S3). To minimize artifacts during large-scale amplification, a cycle optimization step was performed by collecting five 5 µl aliquots at 10, 12, 14, 16, and 18 PCR cycles. PCR reaction aliquots were loaded on an E-Gel pre-cast agarose gel 0.8% (Invitrogen, Life Technologies, Carlsbad, CA, USA) to determine the optimal cycle number. Second-strand cDNA was synthesized and amplified using the Kapa HiFi PCR kit (Kapa Biosystems, Wilmington, MA, USA) with the 5′ PCR primer IIA (Clontech Laboratories, Inc. Mountain View, CA, USA) following the manufacturer’s instructions. Large-scale PCR was performed using the number of cycles determined during the optimization step (14 cycles). Barcoded double-stranded cDNAs were pooled at equal amounts and used for size selection. Size selection was carried out with a BluePippin (Sage Science, Beverly, MA, USA) and 1-2 kbp, 2-3 kbp, 3-6 kbp, and 5-10 kbp fractions were collected. After size selection, each fraction was PCR-enriched prior to SMRTbell template library preparation. cDNA SMRTbell libraries were prepared using 1-3 µg of PCR enriched size-selected samples, followed by DNA damage repair and SMRTbell ligation using the SMRTbell Template Prep Kit 1.0 (Pacific Biosciences, Menlo Park, CA, USA). A second size selection was performed on the 3-6 kbp and 5-10 kbp fractions to remove short contaminating SMRTbell templates. A total of 8 SMRT cells were sequenced on a PacBio Sequel system (DNA Technologies Core, University of California, Davis, USA). Demultiplexing, filtering, quality control, clustering and polishing of the Iso-Seq sequencing data were performed using SMRT Link ver. 4.0.0 (Table S4). Iso-Seq read error rates were estimated using the identity of the best alignment on the diploid Cabernet Sauvignon genomic assembly (Chin et al. 2016). Alignment was performed with GMAP ver. 2015-09-29 (Wu and Watanabe 2005) using the parameters “-K 20000 -B 4 -f 2”. Coding sequences (CDS) were identified using Transdecoder (Haas et al. 2013) as implemented in the PASA ver. 2.1.0 (Haas et al. 2003).
RNA-seq libraries were prepared using the Illumina TruSeq RNA sample preparation kit v2 (Illumina, San Diego, CA, USA), following the low-throughput protocol. Each biological replicate was barcoded individually. Libraries were evaluated for quantity and quality with the High Sensitivity chip on a Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA, USA) and sequenced on an Illumina HiSeq4000 (DNA Technologies Core Facility, University of California, Davis, USA; Table S5). Quality filtering and adapter trimming were performed with Trimmomatic ver. 0.36 (Bolger et al. 2014) using the following parameters: “ILLUMINACLIP:2:30:10 LEADING:7 TRAILING:7 SLIDINGWINDOW:10:20 MINLEN:36” (Table S5). Error correction of the Full-length Non-Chimeric (FLNC) Iso-Seq reads was carried out using high-quality Illumina reads and LSC ver. 2.0 (Au et al. 2012) with a minimum coverage threshold of 5 reads (“–short_read_coverage_threshold 5”).
Transcriptome reconstruction and annotation
Isoforms identified by the SMRT Link pipeline and error-corrected Iso-Seq reads were merged using EvidentialGene (Gilbert 2013) in order to obtain a non-redundant transcriptome (ISNT). Contaminant sequences were searched by parsing blastn (Altschul et al. 1990) alignments against the NCBI NT database (ftp://ftp.ncbi.nlm.nih.gov/blast/db/, retrieved January 17th, 2017) using Megan ver. 6.6.5 (Huson et al. 2007) with default parameters. Sequences detected as non-viridiplantae were removed. Isoforms with no RNA-seq read mapping on their sequence over the 16 samples (662 isoforms) were considered as putative artifacts and were also discarded. ISNT sequences are provided in File S1. Functional annotation was performed with blastx (Altschul et al. 1990) using RefSeq plant proteins as database (ftp://ftp.ncbi.nlm.nih.gov/refseq, retrieved January 17th, 2017) imposing an HSP length cutoff of 50 amino acids. Functional domains were identified with InterProScan ver. 5.28-68.0 (Jones et al. 2014; File S2). Tree view of identified GO terms was generated using WEGO ver. 2.0 (Ye et al. 2006). Iso-Seq reads were considered derived from repetitive regions when showing a RepeatMasker (Smit et al. 2013) hit with coverage ≥ 75% and an identity ≥ 50% using the custom-created repeat library (see below). Non-coding RNAs were identified with Infernal ver. 1.1.2 (Nawrocki et al. 2009) using the Rfam database ver. 12.2 (Nawrocki et al. 2015). Secondary overlapping alignments and structures with an e-value ≥ 0.01 were rejected. Hits on the minus strand of the Iso-Seq reads were rejected as well as matches that were truncated or covering less than 80% of the entire read.
Cabernet Sauvignon genome annotation
Cabernet Sauvignon primary contigs and haplotigs (Chin et al. 2016) were used as genomic reference. Primary contigs are the backbone sequence of the genome assembly and haplotigs are the alternative haplotype sequences where the assembler could phase the two haplotypes. A repeat library was created ad hoc for Cabernet Sauvignon. MITEs were identified with MITEHunter ver. 11.2011 (Han and Wessler 2010); LTRs and TRIMs were identified with LTRharvest (GenomeTools ver. 1.5.7; Ellinghaus et al. 2008) and LTRdigest (GenomeTools ver. 1.5.7; Steinbiss et al. 2009). RepeatModeler ver. 1.0.8 (Smit and Hubley 2008), and RepeatMasker ver. open-4.0.6 (Smit et al. 2013) were used to generated a custom library of repeat models. Repetitive elements in Cabernet Sauvignon genome were then identified with RepeatMasker (Smit et al. 2013) using both custom and plant repeat models altogether (Table S6).
Ab initio trainings and predictions were carried out with SNAP ver. 2006-07-28 (Korf 2004), Augustus ver. 3.0.3 (Stanke et al. 2006), GeneMark-ES ver. 4.32 (Lomsadze et al. 2005), GlimmerHMM ver. 3.0.4 (Majoros et al. 2004), GeneID ver. 1.4.4 (Parra et al. 2000) and Twinscan ver. 4.1.2 (Korf et al. 2001; Brent 2008) using TAIR10 annotation for Arabidopsis as informant species. MAKER-P ver. 2.31.3 (Campbell et al. 2014a) was used to integrate the ab initio predictions with the experimental evidence listed in Table S7 using the parameters reported in File S3. Only MAKER-P models showing an Annotation Edit Distance (AED) < 0.5 were kept.
Gene structure refinement was carried out with PASA ver. 2.1.0 (Haas et al. 2003), parameters can be found in File S4. As transcriptomic evidence, we used the Iso-Seq data as well all the publicly available grape transcriptomic data (Table S7). Public Cabernet Sauvignon RNA-seq data (Table S7) were de novo assembled separately using HISAT2 and Stringtie ver. 1.1.3 (Pertea et al. 2015). De novo transcript sequences were then clustered in a non-redundant dataset using CD-HIT-EST ver. 4.6 (Li and Godzik 2006) with an identity threshold of 99%. Genome annotation summary is reported in Table S8. Alternative splicing forms were classified using AStalavista ver. 3.0 (Foissac and Sammeth 2007).
Non-coding RNAs were searched with Infernal ver. 1.1.2 (Nawrocki et al. 2009) as described above (Table S9). The predicted transcripts’ functional annotation was made with blastp search using the RefSeq plant protein database. Functional domain identification was done with InterProScan as described above for the ISNT. For each gene locus, a non-redundant list of the GO terms attributed to all the alternative transcripts was generated.
Gene expression analysis with RNA-seq
For expression profiling, short reads were aligned on transcript sequences using Bowtie2 ver. 2.26 (Langmead and Salzberg 2012) with options “–sensitive–dpad 0–gbar 99999999–mp 1,1–np 1–score-min L,0,-0.1”. Evaluation of expression at isoform and gene locus levels was carried out using RSEM ver. 1.1.14b3 (Li and Dewey 2011) with default parameters. Differential expression analysis was performed using EBSeq ver. 1.16.0 (Leng et al. 2013). For each pairwise comparison of consecutive growth stages, size factors were calculated with median normalization using five iterations of the EM algorithm. Genes were considered as significantly differentially expressed if they had a minimum posteriori estimate probability (PPDE) threshold of 0.95 and mean RPKM greater than 1 in at least one of two growth stages. Pearson correlation matrices and PCAs were performed using the log2-transformed RPKM values. Heatmaps of the Pearson correlation matrices and dendrograms were generated using heatmap.2 function from the gplots R package ver. 3.0.1 (Warnes et al. 2016). PCAs were carried out in R with the FactoMineR package ver. 1.41 (Lê et al. 2008).
In order to compare expression values of ISNT transcripts and gene loci in the Cabernet Sauvignon genes, ISNT transcripts were aligned to the Cabernet Sauvignon genome using GMAP as described above. ISNT transcripts were associated with gene loci annotated on the Cabernet Sauvignon genome if they aligned at least for 66% of their length. Alignments with translocation were excluded (Table S10). For each ISNT-gene locus association, gene expression was calculated as the sum of the mean expression values of all ISNT transcripts and all Cabernet Sauvignon gene loci, separately, using RSEM ver. 1.1.14b3 (Li and Dewey 2011). Differential expression analysis at cluster level was performed using EBSeq ver. 1.16.0 (Leng et al. 2013).
Data Availability
Sequencing data are accessible through NCBI (PRJNA433195) and other relevant datasets, such as protein-coding gene and repeat coordinates, can be retrieved from the Cantu lab github repository (http://cantulab.github.io/data.html). Supplemental material available at Figshare: https://doi.org/10.25387/g3.7291943.
Results
Isoform sequencing of the grape transcriptome during berry development
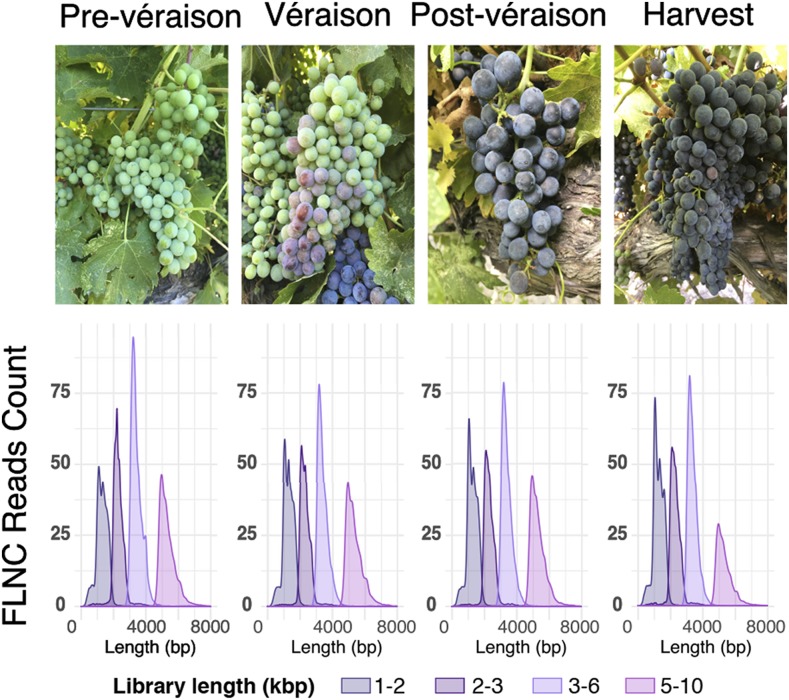
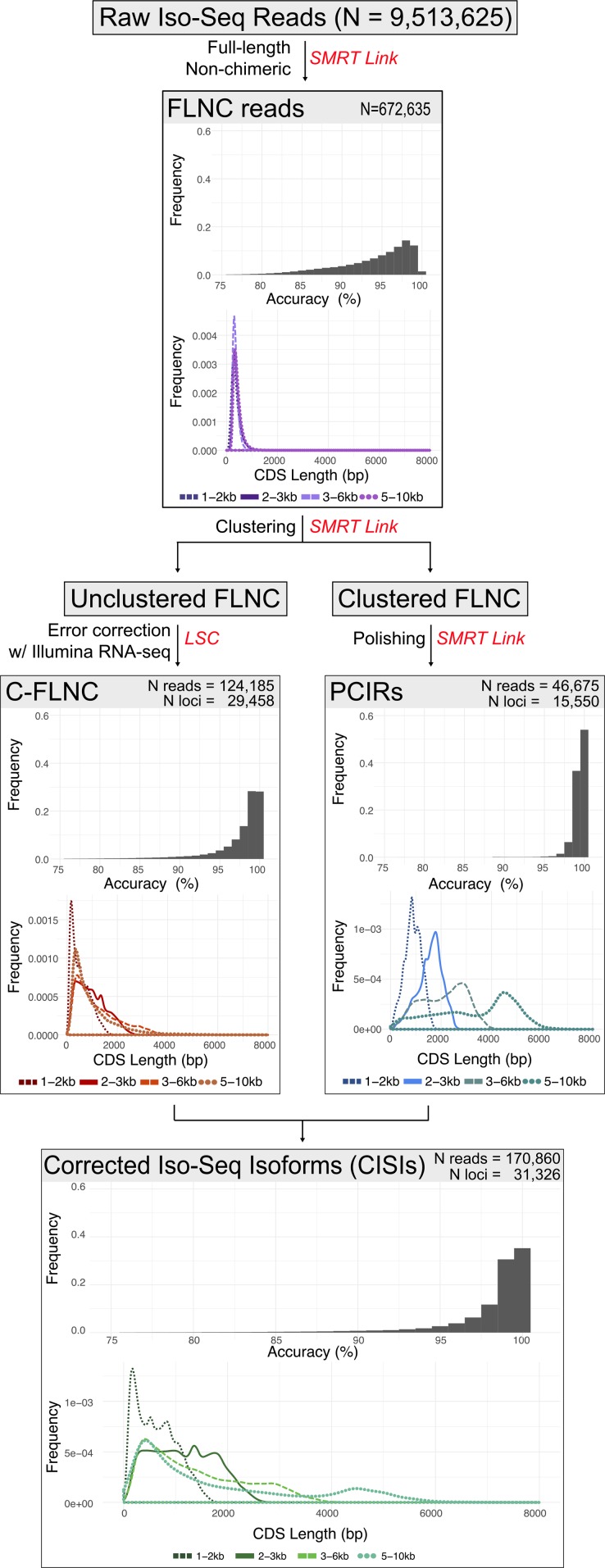
To obtain a comprehensive representation of the transcripts expressed during berry development, we isolated RNA from Cabernet Sauvignon berries (Figure 1) before the onset of ripening (4.35 ± 0.39 °Brix), at véraison (10.94 ± 0.26 °Brix), after véraison (18.38 ± 0.61 °Brix), and at commercial ripeness (20.33 ± 0.76 °Brix). To avoid loading bias, cDNAs were fractionated based on their length to produce four libraries at each developmental stage in size ranges of 1-2 kbp, 2-3 kbp, 3-6 kbp, or 5-10 kbp (Figure 1). Libraries derived from different developmental stages were barcoded and libraries with similar cDNA size were pooled together. Each library pool was sequenced independently on two SMRT cells of a Pacific Biosciences Sequel system generating a total of 23.6 Gbp. In parallel, the same samples were sequenced using Illumina technology (25,655,771 ± 3,512,980 high-quality reads per sample) to provide high-coverage sequence information for error correction and for gene expression quantification (Table S5). Demultiplexing, filtering and quality control of SMRT sequencing data were performed using SMRT Link as described in the Methods section. A total of 672,635 full-length non-chimeric (FLNC; Figure 2) reads with a maximum length of 14.6 kbp and a N50 of 3.5 kbp were generated (Table S4). FLNC reads were further polished and clustered into 46,675 single representatives of expressed transcripts (henceforth, polished-clustered Iso-Seq reads or PCIRs) ranging from 400 bp to 8.8 kbp with a N50 of 3.6 kbp (Table S4). The alignment of FLNC reads and PCIRs to the genomic DNA contigs of the same Cabernet Sauvignon clone (Chin et al. 2016; Minio et al. 2017) confirmed that sequence clustering and polishing successfully increased sequence accuracy, whose median values were 95.4% in FLNC reads and 99.6% in the PCIRs. The increase in sequence accuracy was also reflected by the significantly longer detectable coding sequences (CDS) in the PCIRs compared to the short and fragmented CDS found in the FLNC reads (Figure 2). The residual sequence discrepancy between PCIRs and the genomic contigs could be explained by heterozygosity and/or sequencing errors, but unexpectedly not by expression level (Figure S2).
Figure 1.
Full-length cDNA sequencing (Iso-Seq) of Cabernet Sauvignon berries. Representative pictures of berry clusters at the four growth stages used in this study and the read length distribution of the Iso-Seq libraries by cDNA size fraction. FLNC, Full-Length Non-Chimeric.
Figure 2.
Diagram depicting the main steps of Iso-Seq read analysis. Raw Iso-Seq reads were processed to obtain Full-Length Non-Chimeric (FLNC) reads and clustered isoform reads (PCIRs). FLNC reads that did not cluster in any of the PCIRs were error-corrected using RNA-seq data (C-FLNC). The final dataset described in this study included PCIRs and C-FLNC reads. For each step, sequencing accuracy and predicted coding sequence (CDS) length distributions are reported.
Over 18.5% of the FLNC reads did not cluster with any other reads and were discarded by the SMRT Link pipeline. When mapped on the genomic contigs, the uncorrected reads displayed a sequence accuracy that reflected the typical error rate of 10–20% of the technology (Figure 2) (Giordano et al. 2017; Zimin et al. 2017; Koren et al. 2017). High error rates also resulted in short and fragmented detectable CDS (Figure 2). To recover the information carried by these 124,185 uncorrected FLNCs, which represented an important fraction of the transcriptome (see below), we error-corrected their sequences with LSC (Au et al. 2012) using the short reads generated with Illumina technology. As for the PCIRs, error correction resulted in greater sequence accuracy and longer CDS (Figure 2). PCIRs and error-corrected FLNC (C-FLNC) reads were finally combined into a single dataset of 170,860 corrected Iso-Seq isoforms (CISIs). As low as 1.7% (2,826) of the CISIs showed significant homology with interspersed repeats. LTRs and LINEs were the most abundant orders with 778 and 729 representatives, respectively. Chloroplast and mitochondria genes represented a small fraction of the CISIs with only 89 isoforms (0.05%) having a significant match (50% identity and mutual alignment coverage). CISIs were also searched for non-coding RNA (ncRNA) using the covariance models of the Rfam database; only 182 isoforms were annotated as ncRNAs and were all ribosomal RNA (145 attributed to the large subunit, clan CL00112, and 37 to the small subunit, clan CL00111). Excluding these transcribed isoforms, only 164 CISIs (0.1%) failed to align to the Cabernet Sauvignon genomic contigs, confirming the completeness of the genome assembly and the negligible biological contamination of the berry samples.
To reconstruct an Iso-Seq non-redundant transcriptome (ISNT) that would be tested as a reference for expression profiling, CISIs were clustered with EvidentialGene (Gilbert 2013) to reduce the redundancy between stages and libraries. One representative for each expressed transcript was retained for a total of 28,721 isoforms (File S1). Less than four percent of these isoforms did not contain a complete open reading frame, likely due to residual errors in their sequence. Mapping on the Cabernet Sauvignon genome showed that they potentially derive from 24,378 gene loci (i.e., protein-coding gene sequences) over both primary contigs (13,583 loci) and haplotigs (10,795 loci; File S5). Most isoforms (61.2%) aligned to both a locus on a primary contig and a locus on its associated haplotig, suggesting that despite the differences between haplotypes the mapping could not differentiate between alleles. For 10,727 loci (44%), multiple isoforms (2.03 ± 3.49 isoforms/locus) mapped on the same locus possibly due to alternative splicing and structural differences between alleles. ISNT included 77.6% of the BUSCO (Simão et al. 2015) conserved orthologous; while far from representing the complete grape transcriptome, the ISNT dataset included a remarkably high fraction of these conserved genes, particularly considering that ISNT was constructed using expression data from berries only. Interestingly, 301 BUSCO complete gene models (20%) were found in multiple copies in the ISNT, suggesting that alternative isoforms of these highly conserved genes are expressed during ripening. Putative functions were assigned to the ISNT transcripts as described in the Methods section (Table S11). Only four sequences did not match any sequence in the databases used. Gene Ontology (GO) terms were assigned to 23,386 transcripts with an average of ∼6.9 GO terms per transcript (Table S11, File S6-S8).
Isoform sequencing allows the discovery of private Cabernet Sauvignon genes
Previous analyses of gene content in a limited number of grape cultivars showed that up to 10% of grape isoforms were not shared between genotypes. Some of these “dispensable” genes were associated with cultivar-specific characteristics (Da Silva et al. 2013). To identify protein-coding transcripts characteristic of Cabernet Sauvignon (i.e., private genes), we looked for homologous sequences among the ISNT transcripts in the PN40024 genome (Jaillon et al. 2007; Vitulo et al. 2014) and in the transcriptomes of Corvina (Venturini et al. 2013), Tannat (Da Silva et al. 2013), and Nebbiolo (Gambino et al. 2017). Approximately five percent of the ISNT (1,501 isoforms) did not have a homologous copy in any of the four datasets (Table S12). These putative Cabernet Sauvignon private isoforms were involved in various biological processes of berry development and ripening like phenylpropanoid/flavonoid biosynthesis (a chalcone synthase, a flavanone 3-hydroxylase, and a flavonoid 3′-hydroxylase (Falginella et al. 2012)), sugar accumulation and transport (ten sucrose-phosphate synthases, a phosphofructokinase, a glucose-6-phosphate dehydrogenase, a sucrose transport SUC4-like, a polyol transporter, and an inositol transporter (Afoufa-Bastien et al. 2010; Xin et al. 2013)), water transport (eleven aquaporins), and cell wall metabolism and loosening (six cellulose synthases, a xyloglucan galactosyltransferase, one xyloglucan glycosyltransferase 9-like, two expansins, two xyloglucan endotransglucosylase/hydrolases, two pectinesterases, a pectin methylesterase/invertase inhibitor, seven polygalacturonases, and two β-galactosidases (Carey et al. 1995; Cosgrove 2000, 2005)).
Protein-coding gene models in the Cabernet Sauvignon genomic assembly
To evaluate the non-redundant Iso-Seq transcriptome’s completeness and usefulness as a reference for RNA-seq analysis, the protein-coding genes in the Cabernet Sauvignon genome were predicted as described in Figure S3. First, the repetitive regions of the genome were masked using a custom-made library of Cabernet Sauvignon MITE, LTR, and TRIM information. Overall, ∼51% of the assembly consisted of repetitive elements (Table S6), with 412,994 repetitive elements on the primary assembly (313 Mb) and 274,123 on the haplotigs (177Mb), LTRs were the most abundant class, covering over 335 Mb of the genomic sequences, with Gypsy and Copia families accounting for 201 Mb and 104 Mb, respectively. Next, MAKER-P (Campbell et al. 2014b) identified putative protein-coding loci, combining the results of six ab initio predictors trained ad hoc with publicly available experimental evidences. Ab initio predictors were trained using a custom set of 4,000 randomly selected gene models out of the 5,636 high-quality, non-redundant, and highly conserved gene models of the PN40024 V1 transcriptome (4,459 multiexonic and 1,177 monoexonic). Experimental evidence from public databases (Table S7) was incorporated and used to validate the predicted models. The final MAKER-P prediction included 38,227 high-quality gene models (AED < 0.5) on the primary contigs and 26,789 on the associated haplotigs. Using the covariance models from the Rfam database, 5,780 non-overlapping putative long non-coding RNAs (ncRNAs; 3,239 on primary contigs and 2,541 on haplotigs) belonging to 275 different families were annotated (Table S9). Gene models were further improved using the information from all Iso-Seq full-length datasets (PCIRs, C-FLNC, FLNC), RNA-seq, and the publicly available grapevine transcriptome assemblies. This final refinement improved the annotation of the UTRs and added isoform information. PCIRs helped identify 155 new loci not detected by MAKER-P, update the structure of 10,801 gene models, and add 2,712 alternative transcripts. C-FLNC reads introduced 830 additional missing loci and added 3,738 alternative transcripts to the annotation. Together, 14,388 gene models were updated. FLNC reads introduced 14 new loci and 20,493 alternative transcripts, bringing the number of updated model structures to 24,945. Predicted genes without similarity to known proteins in the RefSeq database and without any functional domains identified by InterProScan (Jones et al. 2014) were removed. The final predicted transcriptome included 55,887 transcripts on 36,689 loci on primary contigs and 40,444 transcripts on 25,479 loci on haplotigs (Table S8). GO terms were assigned to 80,752 transcripts (83.8%) based on homology with protein domains in RefSeq and InterPro databases (Table S13, File S9-S11). A genome browser for Cabernet Sauvignon, its annotation, and an associated blast tool are available at http://cantulab.github.io/data.
For 2,995 (11%) of the 25,479 protein-coding gene loci identified on the haplotigs, we could not find a corresponding homolog in the primary contigs, likely reflecting the diversity in gene content between parental genomes (Cabernet Franc and Sauvignon Blanc (Bowers and Meredith 1997)), as in Corvina (2,321 transcripts; Venturini et al. 2013), Tannat (1,873 transcripts; Da Silva et al. 2013), and Nebbiolo (3,961 transcripts; Gambino et al. 2017). We could not find homologous genes in PN40024, Corvina, Tannat, and Nebbiolo for 1,714 protein-coding gene loci (Table S14). Those genes included likely members of the phenylpropanoid/flavonoid biosynthesis pathway: four phenylalanine ammonia-lyase, three chalcone synthases, a chalcone isomerase, a dihydroflavonol-4-reductase, a flavanone 3-hydroxylase, a flavonoid 3′,5′-hydroxylase, a leucoanthocyanidin dioxygenase, and an anthocyanin acyltransferase. Other private Cabernet Sauvignon genes were associated with terpenoid biosynthesis, including six valencene synthases that may play a role in grapevine flower aroma, three vinorine synthases (Lücker et al. 2004; Martin et al. 2009), and a (E,E)-geranyllinalool synthase.
The incorporation of Iso-Seq data in the gene prediction pipeline also allowed the structural annotation of alternative transcripts. Twenty five percent (15,509) of the 62,168 annotated gene loci had two or more alternative isoforms, an average of 1.55 ± 1.29 alternative transcripts per locus, confirming previous reports in PN40024 (Vitulo et al. 2014). The frequency of splicing variant types was similarly observed in other plant species (Reddy et al. 2013). Intron retention was the most abundant type, accounting for over 44% (File S12), similar to what was observed for rice (45-55%; Zhang et al. 2015), Arabidopsis (30-64%; Marquez et al. 2012; Reddy et al. 2013; Zhang et al. 2015) and maize (40-58%; Zhang et al. 2015; Wang et al. 2016). Alternative acceptor sites (13%), alternative donor sites (10%), and exon skipping (8%) were the other most abundant types of alternative splicing found in the Cabernet Sauvignon genome; a full description of the selected splicing events is reported in File S12.
Iso-Seq transcriptome as reference for RNA-seq analysis
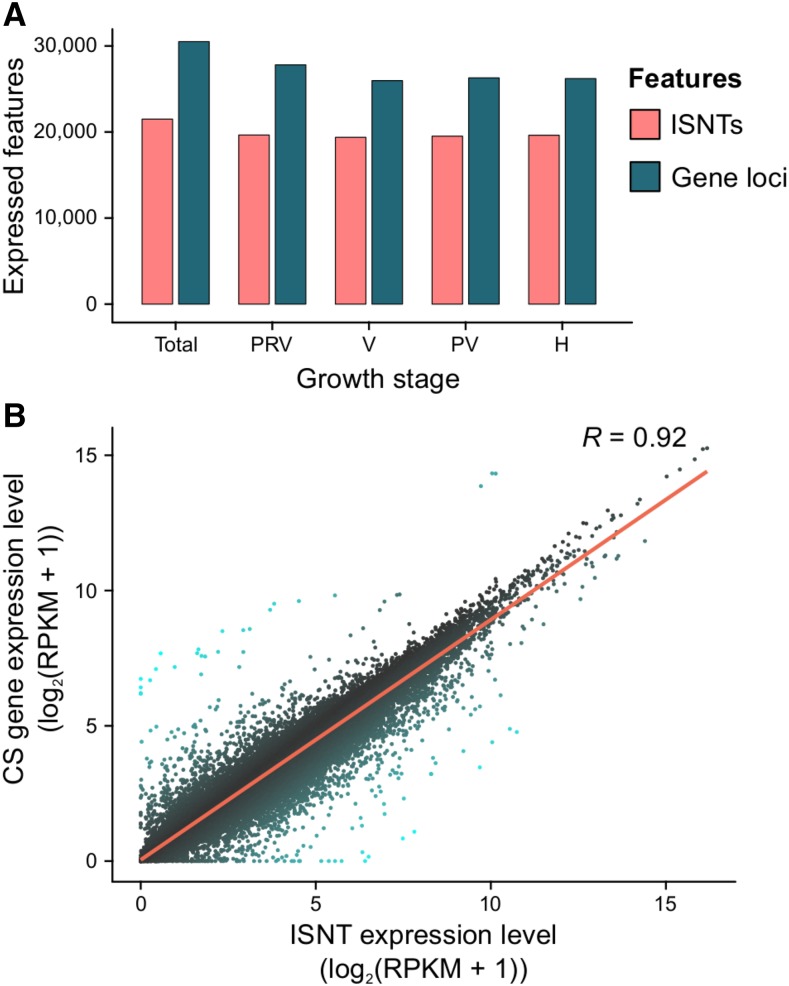
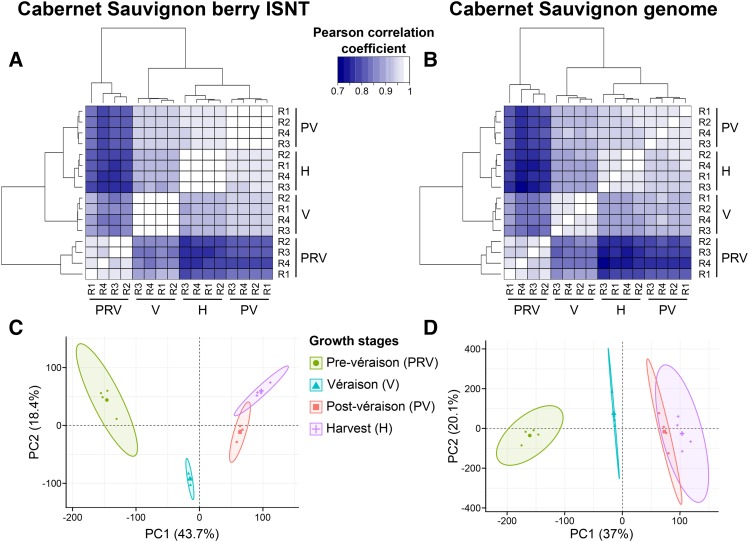
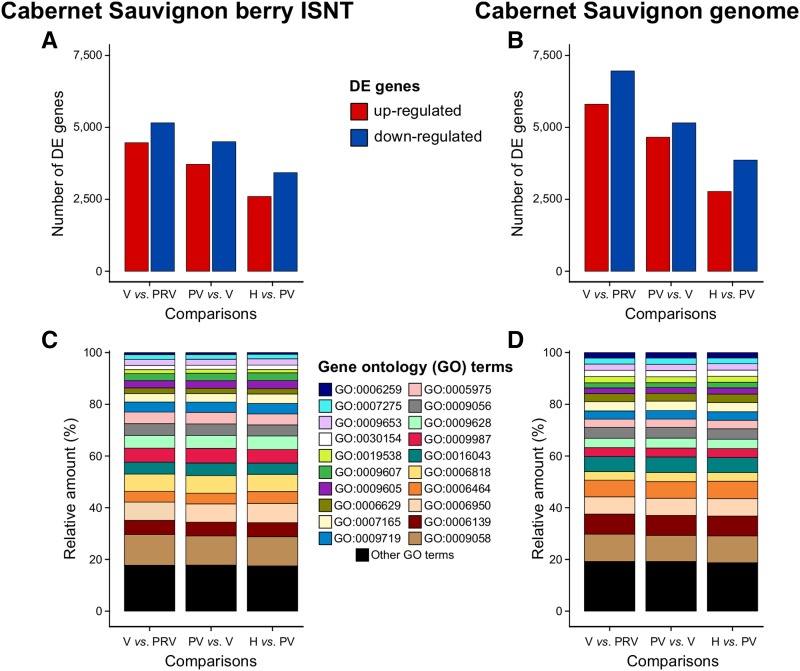
The final step of the analysis was to evaluate the effectiveness of the reconstructed ISNT as reference for RNA-seq analysis of berry development compared to the gene space predicted on the Cabernet Sauvignon genome. Comparisons between the predicted transcripts and the reconstructed ISNT as references for RNA-seq are summarized in Table 1. Only about three percent more RNA-seq reads mapped on the Cabernet Sauvignon predicted transcriptome (90.6 ± 0.8%) than on the ISNT (87.2 ± 0.8%), suggesting that Iso-Seq reconstructed most of the transcripts detectable by RNA-seq at a coverage of ∼26 M reads/sample. Approximately 75% of the ISNT (21,494 transcripts) and ∼49% of the predicted gene space (30,501 gene loci) was detected as expressed (mean RPKM ≥ 1) in at least one stage (Figure 3A, Table S15-S16). In both datasets, the number of expressed genes was slightly higher at pre-véraison stage than at later developmental stages, consistent with previous observations of ripening Cabernet Sauvignon berries (Fasoli et al. 2018). For both datasets, Pearson’s correlation matrix and Principal Component Analysis (PCA) showed a clear distinction between pre-véraison stage and the three ripening stages, as well as a stronger correlation between post-véraison and full-ripe berry transcriptomes (Figure 4), confirming the well-known transcriptional reprogramming associated with the onset of ripening (Fasoli et al. 2012; Massonnet et al. 2017a) and suggesting that similar global transcriptomic dynamics of berry development can be obtained using either Iso-Seq or the whole genome as reference. We then applied a sequence clustering approach to define associations between ISNT isoforms and gene loci to directly compare the expression values of each gene in the two transcriptomes. Based on reciprocal overlap of the alignment, we were able to associate 25,306 ISNT transcripts with 26,873 gene loci in the Cabernet Sauvignon genome (Table S10). Gene expression levels measured on the two transcriptomes were well-correlated (R = 0.92; P-value < 2.2 e-16; Figure 3B; Tables S17-18). Differential gene expression analysis identified 14,477 ISNT transcripts and 18,600 Cabernet Sauvignon genes significantly differentially expressed (PPDE ≥ 0.95) at least once during berry development (Table S15-S16). More genes were differentially regulated between pre-véraison and véraison than during ripening for both transcriptomes, (Figure 5A & B) as previously observed (Palumbo et al. 2014; Massonnet et al. 2017a). Ninety one percent of the differentially expressed ISNT isoforms were also differentially expressed when RNA-seq data were mapped on genomic loci. Similar relative amounts of Biological Process GO terms among differentially expressed genes were observed between the two transcriptomes (Figure 5C & D). Interestingly, 302 Cabernet Sauvignon private isoforms (transcripts not found in other cultivars) were differentially expressed during berry development, including transcripts encoding a polyol transporter, an inositol transporter, and five aquaporins.
Table 1. Summary of the RNA-seq results when Cabernet Sauvignon berry ISNT and the Cabernet Sauvignon genome were used as reference for short-read mapping.
| Iso-Seq transcriptome | Cabernet Sauvignon genome | |
|---|---|---|
| Mapping rate | 87.2 ± 0.8% | 90.6 ± 0.8% |
| Expressed features (mean RPKM ≥ 1 at least at one growth stage) | 21,494 isoforms (74.8%)* | 30,501 gene loci (49%)** |
| Expressed features with BP GO term annotation | 14,431 isoforms | 21,588 gene loci |
| Total BP GO terms among expressed features | 61,603 GO terms | 80,103 GO terms |
| DE features (PPDE ≥ 0.95 and mean RPKM ≥ 1 at least at one growth stage) | 14,477 isoforms (50.4%)* | 18,600 gene loci (29.9%)** |
| DE features with BP GO term annotation | 10,237 isoforms | 14,026 gene loci |
| Total BP GO terms among DE features | 44,179 GO terms | 53,349 GO terms |
Biological process, BP; differentially expressed, DE.
Percentage of ISNT.
Percentage of Cabernet Sauvignon predicted transcriptome.
Figure 3.
Gene expression analysis during berry development using the ISNT as reference and comparison with the Cabernet Sauvignon genome. (A) Number of ISNT transcripts (ISNTs) and gene loci expressed overall and at each ripening stage. Abbreviation of berry growth stages: PRV, Pre-véraison; V, Véraison; PV, Post- véraison; H, harvest. (B) Scatterplot showing the correlation between the gene expression level (log2(RPKM + 1)) of ISNT isoforms and Cabernet Sauvignon (CS) gene loci. Line of best fit and correlation coefficient factor (R) are provided.
Figure 4.
Global transcriptomic changes during berry development ripening using the ISNT and Cabernet Sauvignon gene space as references. (A, B) Pearson correlation matrices of the 16 berry transcriptomes. (C, D) PCA plots of the 16 berry transcriptomes. Each point represents a biological replicate and ellipses define confidence areas (95%) for each berry growth stage. All analyses were performed using the log2-transformed RPKM values of expressed features (mean RPKM ≥ 1 at least at one stage).
Figure 5.
Comparison of the differentially expressed (DE) ISNT transcripts and Cabernet Sauvignon gene loci during berry development. (A, B) Number of up- and down-regulated genes between each pairwise comparison of developmental stages (PPDE ≥ 0.95 and mean RPKM ≥ 1 at least at one stage). (C, D) Relative amount of Biological Process Gene Ontology (GO) terms among differentially expressed genes. Abbreviation of berry growth stages: PRV, Pre-véraison; V, Véraison; PV, Post- véraison; H, harvest.
Discussion
Full-length cDNA sequencing with SMRT technology (Iso-Seq) can be used to rapidly reconstruct the grape berry transcriptome, enabling the identification of cultivar-specific isoforms, refinement of the Cabernet Sauvignon genome annotation, and the creation of a reference for transcriptome-wide expression profiling. In contrast to transcriptome reconstruction using short-read sequencing that requires de novo assembly, Iso-Seq delivers full-length transcripts that eliminate the introduction of assembly errors and artifacts like chimeric transcripts and incomplete fragments due to PolyA capture (Chang et al. 2014; Huang et al. 2016; Moreton et al. 2016; Smith-Unna et al. 2016; Geniza and Jaiswal 2017; Ungaro et al. 2017). The incorporation of high-coverage short-read sequencing is still necessary to benefit from the complete transcript sequencing enable by Iso-Seq. Although Iso-Seq provides much longer reads than second-generation sequencing platforms and as a result is excellent in resolving transcript structure, its sequencing error rate is high (10–20%) and throughput is still relatively low (Giordano et al. 2017; Zimin et al. 2017; Koren et al. 2017). Here we show that combining Iso-Seq with Illumina sequencing at high coverage enables expression profiling and sequence error correction of Iso-Seq reads, particularly those derived from low-expression genes. The clustering analysis of the SMRT link pipeline discarded ∼18.5% of the FLNC reads, likely caused by low sequence accuracy. To overcome this technical issue, we applied a hybrid error correction pipeline consisting in performing the error correction of the unclustered FLNC reads, followed by an additional clustering step of both to resolve redundancies. Error correction with Illumina reads recovered a significant amount of Iso-Seq reads that would have otherwise been removed by the standard Iso-Seq pipeline, highlighting the importance of integrating multiple sequencing technologies with complementary features (Koren et al. 2012; Au et al. 2012; Salmela and Rivals 2014; Hu et al. 2016).
Transcriptome reconstruction has been widely used to develop references for genome-wide expression profiling in the absence of an annotated genome assembly (Simon et al. 2009; Garber et al. 2011; Martin and Wang 2011; Yang and Kim 2015). Though a genome reference is available for grape, transcriptome reconstruction overcomes the limitations of a cultivar-specific reference that lacks the gene content of other cultivars. Although cultivar-specific genes appear nonessential for berry development, those private genes could contribute to cultivar characteristics. For example, the wine grape Tannat accumulates unusually high levels of polyphenols in the berry; its cultivar-specific genes account for more than 80% of the expression of phenolic and polyphenolic compound biosynthetic enzymes (Da Silva et al. 2013). De novo transcriptome assembly from short RNA-seq reads has been used to explore the gene content diversity in Tannat (Da Silva et al. 2013), Corvina (Venturini et al. 2013), and Nebbiolo (Gambino et al. 2017). Iso-Seq identified 1,501 Cabernet Sauvignon transcripts expressed during berry development that were found in neither the genome of PN40024 nor the transcriptomes of Tannat, Nebbiolo and Corvina. Some private Cabernet Sauvignon transcripts have functions potentially associated with traits characteristic of Cabernet Sauvignon grapes and wines like their color and sugar content. These transcripts included three sugar transporter-coding genes, which could be involved in the accumulation of glucose and fructose during berry ripening (Lecourieux et al. 2014), and a chalcone synthase, a flavanone 3-hydroxylase, and a flavonoid 3′-hydroxylase, all involved in the flavonoid pathway. Chalcone synthases catalyze the first committed step of the flavonoid biosynthesis pathway (Sparvoli et al. 1994), which produces different classes of metabolites in grape berry, including flavonols (yellow pigments), flavan-3-ols and proanthocyanidins (mouth-feel and smooth sensory perceptions), and anthocyanins (red, purple, and blue pigments). In addition, products of the flavonoid 3′-hydroxylase can lead to the synthesis of cyanidin-3-glucoside, a red anthocyanin (Castellarin et al. 2012). The analysis of the gene space in the genome assembly showed that private Cabernet Sauvignon genes identified using Iso-Seq are only a fraction of the private Cabernet Sauvignon transcriptome. As in other transcriptome reconstruction methods, Iso-Seq can only identify transcripts that are expressed in the organs and developmental stages used for RNA sequencing. Obtaining the full set of private transcripts without genome assembly would require sequencing additional organs and developmental stages. In addition, it is challenging to differentiate isoforms derived from close paralogous genes, alleles of the same gene, and alternative splicing variants, in any transcriptome obtained by RNA sequencing (including Iso-Seq); this potentially leads to an overestimation of the genes in the final transcriptome reference. This study could not resolve isoform redundancy in the final transcriptome for about 37% of the gene loci in the Cabernet Sauvignon genome. This is a limitation of Iso-Seq as well as of all transcriptome references that cannot be overcome without a complete genome assembly.
In this study, we tested whether the transcriptome reconstructed using Iso-Seq can be used for expression profiling. Only an approximately 3% difference in read alignment between ISNT and the genome reference was observed, implying that at high coverage, ISNT detects almost all genes expressed during berry development. The slight difference in mapping rate between the two references can be explained by either the absence of some low-expression transcripts in the ISNT or the residual error rate in isoform sequences. Gene expression analysis using the ISNT as reference showed similar results compared to the Cabernet Sauvignon genome assembly, with a very high correlation of expression level and differential gene expression, and with similar global transcriptomic changes. However, we observed differences in the number of expressed and differentially expressed features that depend on the reference used. Those differences could be explained by the diploid phasing of the Cabernet Sauvignon genome assembly and that multiple ISNT transcripts might correspond to a single gene locus. Nonetheless, similar relative amounts of Biological Process GO terms were found among the differentially expressed genes, confirming that the transcriptome obtained using Iso-Seq captured the transcriptional reprogramming underlying the main physiological and biochemical changes during grape berry development. In addition, gene expression analysis revealed that some private isoforms (20%) are significantly modulated during berry development, indicating that in addition to identifying the private gene space, the ISNT reference makes it possible to observe its expression.
In conclusion, this study demonstrates that Iso-Seq data can be used to create and refine a comprehensive reference transcriptome that represents most genes expressed in a tissue undergoing extensive transcriptional reprogramming during development. In grapes, this approach can aid developing transcriptome references and is particularly valuable given diverse cultivars with private transcripts and accessions that are genetically distant from available genome references, like the non-vinifera Vitis species used as rootstocks or for breeding. The pipeline described here can be useful in efforts to reconstruct the gene space in plant species with large and complex genomes still unresolved.
Acknowledgments
This work was supported by J. Lohr Vineyards and Wines, E. & J. Gallo Winery, the Louis P. Martini Endowment in Viticulture, and the NSF grant #1741627. Part of this work was carried out in collaboration with UC Davis Chile and funded by the Chilean Economic Development Agency (CORFO).
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25387/g3.7291943.
Communicating editor: T. Slotte
Literature Cited
- Afoufa-Bastien D., Medici A., Jeauffre J., Coutos-Thévenot P., Lemoine R., et al. , 2010. The Vitis vinifera sugar transporter gene family: Phylogenetic overview and macroarray expression profiling. BMC Plant Biol. 10: 245 10.1186/1471-2229-10-245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J., 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403–410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- Amrine K. C. H., Blanco-Ulate B., Riaz S., Pap D., Jones L., et al. , 2015. Comparative transcriptomics of Central Asian Vitis vinifera accessions reveals distinct defense strategies against powdery mildew. Hortic. Res. 2: 15037 10.1038/hortres.2015.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashrafi H., Hill T., Stoffel K., Kozik A., Yao J., et al. , 2012. De novo assembly of the pepper transcriptome (Capsicum annuum): a benchmark for in silico discovery of SNPs, SSRs and candidate genes. BMC Genomics 13: 571 10.1186/1471-2164-13-571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Au K. F., Underwood J. G., Lee L., Wong W. H., 2012. Improving PacBio long read accuracy by short read alignment. PLoS One 7: e46679 10.1371/journal.pone.0046679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellucci E., Bitocchi E., Ferrarini A., Benazzo A., Biagetti E., et al. , 2014. Decreased Nucleotide and Expression Diversity and Modified Coexpression Patterns Characterize Domestication in the Common Bean. Plant Cell 26: 1901–1912. 10.1105/tpc.114.124040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco-Ulate B., Amrine K. C., Collins T. S., Rivero R. M., Vicente A. R., et al. , 2015 Developmental and metabolic plasticity of white-skinned grape berries in response to Botrytis cinerea during noble rot. Plant Physiol. 169: pp.00852.2015. 10.1104/pp.15.00852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco-Ulate B., Hopfer H., Figueroa-Balderas R., Ye Z., Rivero R. M., et al. , 2017. Red blotch disease alters grape berry development and metabolism by interfering with the transcriptional and hormonal regulation of ripening. J. Exp. Bot. 68: 1225–1238. 10.1093/jxb/erw506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco-Ulate B., Vincenti E., Powell A. L. T., Cantu D., 2013. Tomato transcriptome and mutant analyses suggest a role for plant stress hormones in the interaction between fruit and Botrytis cinerea. Front. Plant Sci. 4: 1–16. 10.3389/fpls.2013.00142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger A. M., Lohse M., Usadel B., 2014. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers J. E., Meredith C. P., 1997. The parentage of a classic wine grape, Cabernet Sauvignon. Nat. Genet. 16: 84–87. 10.1038/ng0597-84 [DOI] [PubMed] [Google Scholar]
- Brent M. R., 2008. Steady progress and recent breakthroughs in the accuracy of automated genome annotation. Nat. Rev. Genet. 9: 62–73. 10.1038/nrg2220 [DOI] [PubMed] [Google Scholar]
- Brett D., Pospisil H., Valcárcel J., Reich J., Bork P., 2002. Alternative splicing and genome complexity. Nat. Genet. 30: 29–30. 10.1038/ng803 [DOI] [PubMed] [Google Scholar]
- Campbell M. S., Holt C., Moore B., Yandell M., 2014a Genome Annotation and Curation Using MAKER and MAKER-P, pp. 4.11.1–4.11.39 in Current Protocols in Bioinformatics, John Wiley & Sons, Inc., Hoboken, NJ, USA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell M. S., Law M., Holt C., Stein J. C., Moghe G. D., et al. , 2014b MAKER-P: A tool kit for the rapid creation, management, and quality control of plant genome annotations. Plant Physiol. 164: 513–524. 10.1104/pp.113.230144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey A. T., Holt K., Picard S., Wilde R., Tucker G. A., et al. , 1995. Tomato exo-(1–4)-b-D-galactanase. Isolation, changes during ripening in normal and mutant tomato fruit, and characterization of a related cDNA clone. Plant Physiol. 108: 1099–1107. 10.1104/pp.108.3.1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellarin S. D., Bavaresco L., Falginella L., Gonçalves M. I. V. Z., Di Gaspero G., 2012. Phenolics in grape berry and key antioxidants | BenthamScience. Biochem. Grape Berry 89–110. [Google Scholar]
- Chang Z., Wang Z., Li G., 2014. The impacts of read length and transcriptome complexity for de ovo assembly: A simulation study. PLoS One 9: 1–8. 10.1371/journal.pone.0094825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin C.-S., Alexander D. H., Marks P., Klammer A. A., Drake J., et al. , 2013. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods 10: 563–569. 10.1038/nmeth.2474 [DOI] [PubMed] [Google Scholar]
- Chin C.-S., Peluso P., Sedlazeck F. J., Nattestad M., Concepcion G. T., et al. , 2016. Phased diploid genome assembly with single-molecule real-time sequencing. Nat. Methods 13: 1050–1054. 10.1038/nmeth.4035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cipriani G., Spadotto A., Jurman I., Di Gaspero G., Crespan M., et al. , 2010. The SSR-based molecular profile of 1005 grapevine (Vitis vinifera L.) accessions uncovers new synonymy and parentages, and reveals a large admixture amongst varieties of different geographic origin. Theor. Appl. Genet. 121: 1569–1585. 10.1007/s00122-010-1411-9 [DOI] [PubMed] [Google Scholar]
- Corso M., Vannozzi A., Maza E., Vitulo N., Meggio F., et al. , 2015. Comprehensive transcript profiling of two grapevine rootstock genotypes contrasting in drought susceptibility links the phenylpropanoid pathway to enhanced tolerance. J. Exp. Bot. 66: 5739–5752. 10.1093/jxb/erv274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove D. J., 2005. Growth of the plant cell wall. Nat. Rev. Mol. Cell Biol. 6: 850–861. 10.1038/nrm1746 [DOI] [PubMed] [Google Scholar]
- Cosgrove D. J., 2000. Loosening of plant cell walls by expansins. Nature 407: 321–326. 10.1038/35030000 [DOI] [PubMed] [Google Scholar]
- Da Silva C., Zamperin G., Ferrarini A., Minio A., Dal Molin A., et al. , 2013. The high polyphenol content of grapevine cultivar tannat berries is conferred primarily by genes that are not shared with the reference genome. Plant Cell 25: 4777–4788 (erratum Plant Cell 29: 913). 10.1105/tpc.113.118810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deluc L. G., Quilici D. R., Decendit A., Grimplet J., Wheatley M. D., et al. , 2009. Water deficit alters differentially metabolic pathways affecting important flavor and quality traits in grape berries of Cabernet Sauvignon and Chardonnay. BMC Genomics 10: 212 10.1186/1471-2164-10-212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Gaspero G., Cipriani G., Marrazzo M. T., Andreetta D., Prado Castro M. J., et al. , 2005. Isolation of (AC)n-microsatellites in Vitis vinifera L. and analysis of genetic background in grapevines under marker assisted selection. Mol. Breed. 15: 11–20. 10.1007/s11032-004-1362-4 [DOI] [Google Scholar]
- Di Genova A., Almeida A. M., Muñoz-Espinoza C., Vizoso P., Travisany D., et al. , 2014. Whole genome comparison between table and wine grapes reveals a comprehensive catalog of structural variants. BMC Plant Biol. 14: 7 10.1186/1471-2229-14-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi K., Monjo T., Hoang P. H., Yoshimura J., Yurino H., et al. , 2014. Rapid detection of expanded short tandem repeats in personal genomics using hybrid sequencing. Bioinformatics 30: 815–822. 10.1093/bioinformatics/btt647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong L., Liu H., Zhang J., Yang S., Kong G., et al. , 2015. Single-molecule real-time transcript sequencing facilitates common wheat genome annotation and grain transcriptome research. BMC Genomics 16: 1039 10.1186/s12864-015-2257-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellinghaus D., Kurtz S., Willhoeft U., 2008. LTRharvest, an efficient and flexible software for de novo detection of LTR retrotransposons. BMC Bioinformatics 9: 18 10.1186/1471-2105-9-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falginella L., Di Gaspero G., Castellarin S. D., 2012. Expression of flavonoid genes in the red grape berry of “Alicante Bouschet” varies with the histological distribution of anthocyanins and their chemical composition. Planta 236: 1037–1051. 10.1007/s00425-012-1658-2 [DOI] [PubMed] [Google Scholar]
- Fasoli M., Dal Santo S., Zenoni S., Tornielli G. B., Farina L., et al. , 2012. The grapevine expression atlas reveals a deep transcriptome shift driving the entire plant into a maturation program. Plant Cell 24: 3489–3505. 10.1105/tpc.112.100230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasoli M., Richter C. L., Zenoni S., Bertini E., Vitulo N., et al. , 2018 Timing and order of the molecular events marking the onset of berry ripening in grapevine. Plant Physiol. 17: pp.00559.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filichkin S. A., Hamilton Mi., Dharmawardhana P. D., Singh S. K., Sullivan C., et al. , 2018. Abiotic stresses modulate landscape of poplar transcriptome via alternative splicing, differential intron retention, and isoform ratio switching. Front. Plant Sci. 9: 5 10.3389/fpls.2018.00005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foissac S., Sammeth M., 2007. ASTALAVISTA: Dynamic and flexible analysis of alternative splicing events in custom gene datasets. Nucleic Acids Res. 35: W297–W299. 10.1093/nar/gkm311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambino G., Dal Molin A., Boccacci P., Minio A., Chitarra W., et al. , 2017. Whole-genome sequencing and SNV genotyping of ‘Nebbiolo’ (Vitis vinifera L.) clones. Sci. Rep. 7: 17294 10.1038/s41598-017-17405-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao S., Ren Y., Sun Y., Wu Z., Ruan J., et al. , 2016. PacBio full-length transcriptome profiling of insect mitochondrial gene expression. RNA Biol. 13: 820–825 (erratum: RNA Biol. 13: 1323). 10.1080/15476286.2016.1197481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garber M., Grabherr M. G., Guttman M., Trapnell C., 2011. Computational methods for transcriptome annotation and quantification using RNA-seq. Nat. Methods 8: 469–477. 10.1038/nmeth.1613 [DOI] [PubMed] [Google Scholar]
- Geniza M., Jaiswal P., 2017. Tools for building de novo transcriptome assembly. Curr. Plant Biol. 11–12: 41–45. 10.1016/j.cpb.2017.12.004 [DOI] [Google Scholar]
- Gilbert D. G., 2013. Gene-omes built from mRNA-seq not genome DNA, pp. 47405 in 7th Annual Arthropod Genomics Symposium. [Google Scholar]
- Giordano F., Aigrain L., Quail M. A., Coupland P., Bonfield J. K., et al. , 2017. De novo yeast genome assemblies from MinION, PacBio and MiSeq platforms. Sci. Rep. 7: 3935 10.1038/s41598-017-03996-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnerre S., Maccallum I., Przybylski D., Ribeiro F. J., Burton J. N., et al. , 2011. High-quality draft assemblies of mammalian genomes from massively parallel sequence data. Proc. Natl. Acad. Sci. USA 108: 1513–1518. 10.1073/pnas.1017351108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golicz A. A., Batley J., Edwards D., 2016a Towards plant pangenomics. Plant Biotechnol. J. 14: 1099–1105. 10.1111/pbi.12499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golicz A. A., Bayer P. E., Barker G. C., Edger P. P., Kim H. R., et al. , 2016b The pangenome of an agronomically important crop plant Brassica oleracea. Nat. Commun. 7: 13390 10.1038/ncomms13390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D., Huddleston J., Chaisson M. J., Hill C. M., Kronenberg Z. N., et al. , 2016. Long-read sequence assembly of the gorilla genome. Science 352: aae0344 10.1126/science.aae0344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr M. G., Haas B. J., Yassour M., Levin J. Z., Thompson D. A., et al. , 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29: 644–652. 10.1038/nbt.1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas B. J., Delcher A. L., Mount S. M., Wortman J. R., Smith R. K., Jr., et al. , 2003. Improving the Arabidopsis genome annotation using maximal transcript alignment assemblies. Nucleic Acids Res. 31: 5654–5666. 10.1093/nar/gkg770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas B. J., Papanicolaou A., Yassour M., Grabherr M., Blood P. D., et al. , 2013. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 8: 1494–1512. 10.1038/nprot.2013.084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y., Wessler S. R., 2010. MITE-Hunter: A program for discovering miniature inverted-repeat transposable elements from genomic sequences. Nucleic Acids Res. 38: e199 10.1093/nar/gkq862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honaas L. A., Wafula E. K., Wickett N. J., Der J. P., Zhang Y., et al. , 2016. Selecting superior de novo transcriptome assemblies: Lessons learned by leveraging the best plant genome. PLoS One 11: e0146062 10.1371/journal.pone.0146062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopper D. W., Ghan R., Schlauch K. A., Cramer G. R., 2016. Transcriptomic network analyses of leaf dehydration responses identify highly connected ABA and ethylene signaling hubs in three grapevine species differing in drought tolerance. BMC Plant Biol. 16: 118 10.1186/s12870-016-0804-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu R., Sun G., Sun X., 2016. LSCplus: a fast solution for improving long read accuracy by short read alignment. BMC Bioinformatics 17: 451 10.1186/s12859-016-1316-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Chen X. G., Armbruster P. A., 2016. Comparative performance of transcriptome assembly methods for non-model organisms. BMC Genomics 17: 523 10.1186/s12864-016-2923-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S., Chen Z., Huang G., Yu T., Yang P., et al. , 2012. HaploMerger: Reconstructing allelic relationships for polymorphic diploid genome assemblies. Genome Res. 22: 1581–1588. 10.1101/gr.133652.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huddleston J., Chaisson M. J., Steinberg K. M., Warren W., Hoekzema K., et al. , 2017. Discovery and genotyping of structural variation from long-read haploid genome sequence data. Genome Res. 27: 677–685. 10.1101/gr.214007.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson D. H., Auch A. F., Qi J., Schuster S. C., 2007. MEGAN analysis of metagenomic data. Genome Res. 17: 377–386. 10.1101/gr.5969107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibáñez J., Vargas A. M., Palancar M., Borrego J., De Andrés M. T., 2009. Genetic relationships among table-grape varieties. Am. J. Enol. Vitic. 60: 35–42. [Google Scholar]
- Jaillon O., Aury J.-M., Noel B., Policriti A., Clepet C., et al. , 2007. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449: 463–467. 10.1038/nature06148 [DOI] [PubMed] [Google Scholar]
- Jiao C., Gao M., Wang X., Fei Z., 2015. Transcriptome characterization of three wild Chinese Vitis uncovers a large number of distinct disease related genes. BMC Genomics 16: 223 10.1186/s12864-015-1442-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P., Binns D., Chang H.-Y., Fraser M., Li W., et al. , 2014. InterProScan 5: genome-scale protein function classification. Bioinformatics 30: 1236–1240. 10.1093/bioinformatics/btu031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju C., Zhao Z., Wang W., 2017. Efficient approach to correct read alignment for pseudogene abundance estimates. IEEE/ACM Trans. Comput. Biol. Bioinforma. 14: 522–533. 10.1109/TCBB.2016.2591533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajitani R., Toshimoto K., Noguchi H., Toyoda A., Ogura Y., et al. , 2014. Efficient de novo assembly of highly heterozygous genomes from whole-genome shotgun short reads. Genome Res. 24: 1384–1395. 10.1101/gr.170720.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren S., Schatz M. C., Walenz B. P., Martin J., Howard J. T., et al. , 2012. Hybrid error correction and de novo assembly of single-molecule sequencing reads. Nat. Biotechnol. 30: 693–700. 10.1038/nbt.2280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren S., Walenz B. P., Berlin K., Miller J. R., Bergman N. H., et al. , 2017. Canu: scalable and accurate long-read assembly via adaptive k -mer weighting and repeat separation. Genome Res. 27: 722–736. 10.1101/gr.215087.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korf I., 2004. Gene finding in novel genomes. BMC Bioinformatics 5: 59 10.1186/1471-2105-5-59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korf I., Flicek P., Duan D., Brent M. R., 2001. Integrating genomic homology into gene structure prediction. Bioinformatics 17: S140–S148. 10.1093/bioinformatics/17.suppl_1.S140 [DOI] [PubMed] [Google Scholar]
- Kuhn N., Guan L., Dai Z. W., Wu B. H., Lauvergeat V., et al. , 2014. Berry ripening: Recently heard through the grapevine. J. Exp. Bot. 65: 4543–4559. 10.1093/jxb/ert395 [DOI] [PubMed] [Google Scholar]
- Kuo R. I., Tseng E., Eory L., Paton I. R., Archibald A. L., et al. , 2017. Normalized long read RNA sequencing in chicken reveals transcriptome complexity similar to human. BMC Genomics 18: 323 10.1186/s12864-017-3691-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacombe T., Boursiquot J. M., Laucou V., Di Vecchi-Staraz M., Péros J. P., et al. , 2013. Large-scale parentage analysis in an extended set of grapevine cultivars (Vitis vinifera L.). Theor. Appl. Genet. 126: 401–414. 10.1007/s00122-012-1988-2 [DOI] [PubMed] [Google Scholar]
- Langmead B., Salzberg S. L., 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9: 357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lê S., Josse J., Husson F., 2008. FactoMineR : An R Package for Multivariate Analysis. J. Stat. Softw. 25: 1–18. 10.18637/jss.v025.i01 [DOI] [Google Scholar]
- Lecourieux F., Kappel C., Lecourieux D., Serrano A., Torres E., et al. , 2014. An update on sugar transport and signalling in grapevine. J. Exp. Bot. 65: 821–832. 10.1093/jxb/ert394 [DOI] [PubMed] [Google Scholar]
- Lecourieux F., Kappel C., Pieri P., Charon J., Pillet J., et al. , 2017. Dissecting the biochemical and transcriptomic effects of a locally applied heat treatment on developing Cabernet Sauvignon grape berries. Front. Plant Sci. 8: 53 10.3389/fpls.2017.00053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leng N., Dawson J. A., Thomson J. A., Ruotti V., Rissman A. I., et al. , 2013. EBSeq: An empirical Bayes hierarchical model for inference in RNA-seq experiments. Bioinformatics 29: 1035–1043. 10.1093/bioinformatics/btt087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Dewey C. N., 2011. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12: 323 10.1186/1471-2105-12-323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Godzik A., 2006. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22: 1658–1659. 10.1093/bioinformatics/btl158 [DOI] [PubMed] [Google Scholar]
- Liu J., Chen X., Liang X., Zhou X., Yang F., et al. , 2016 Alternative splicing of rice WRKY62 and WRKY76 transcription factor genes in pathogen defense. Plant Physiol. 171: pp.01921.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Mei W., Soltis P. S., Soltis D. E., Barbazuk W. B., 2017. Detecting alternatively spliced transcript isoforms from single-molecule long-read sequences without a reference genome. Mol. Ecol. Resour. 17: 1243–1256. 10.1111/1755-0998.12670 [DOI] [PubMed] [Google Scholar]
- Lodhi M. A., Reisch B. I., 1995. Nuclear DNA content of Vitis species, cultivars, and other genera of the Vitaceae. Theor. Appl. Genet. 90: 11–16. 10.1007/BF00220990 [DOI] [PubMed] [Google Scholar]
- Lomsadze A., Ter-Hovhannisyan V., Chernoff Y. O., Borodovsky M., 2005. Gene identification in novel eukaryotic genomes by self-training algorithm. Nucleic Acids Res. 33: 6494–6506. 10.1093/nar/gki937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes M. S., Sefc K. M., Eiras Dias E., Steinkellner H., Laimer Câmara Machado M., et al. , 1999. The use of microsatellites for germplasm management in a Portuguese grapevine collection. Theor. Appl. Genet. 99: 733–739. 10.1007/s001220051291 [DOI] [PubMed] [Google Scholar]
- Lücker J., Bowen P., Bohlmann J., 2004. Vitis vinifera terpenoid cyclases: Functional identification of two sesquiterpene synthase cDNAs encoding (+)-valencene synthase and (-)-germacrene D synthase and expression of mono- and sesquiterpene synthases in grapevine flowers and berries. Phytochemistry 65: 2649–2659. 10.1016/j.phytochem.2004.08.017 [DOI] [PubMed] [Google Scholar]
- Majoros W. H., Pertea M., Salzberg S. L., 2004. TigrScan and GlimmerHMM: Two open source ab initio eukaryotic gene-finders. Bioinformatics 20: 2878–2879. 10.1093/bioinformatics/bth315 [DOI] [PubMed] [Google Scholar]
- Marquez Y., Brown J. W. S., Simpson C., Barta A., Kalyna M., 2012. Transcriptome survey reveals increased complexity of the alternative splicing landscape in Arabidopsis. Genome Res. 22: 1184–1195. 10.1101/gr.134106.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin D. M., Toub O., Chiang A., Lo B. C., Ohse S., et al. , 2009. The bouquet of grapevine (Vitis vinifera L. cv. Cabernet Sauvignon) flowers arises from the biosynthesis of sesquiterpene volatiles in pollen grains. Proc. Natl. Acad. Sci. USA 106: 7245–7250. 10.1073/pnas.0901387106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin J. A., Wang Z., 2011. Next-generation transcriptome assembly. Nat. Rev. Genet. 12: 671–682. 10.1038/nrg3068 [DOI] [PubMed] [Google Scholar]
- Massonnet M., Fasoli M., Tornielli G. B., Altieri M., Sandri M., et al. , 2017a Ripening Transcriptomic Program in Red and White Grapevine Varieties Correlates with Berry Skin Anthocyanin Accumulation. Plant Physiol. 174: 2376–2396. 10.1104/pp.17.00311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massonnet M., Figueroa-Balderas R., Galarneau E. R. A., Miki S., Lawrence D. P., et al. , 2017b Neofusicoccum parvum Colonization of the Grapevine Woody Stem Triggers Asynchronous Host Responses at the Site of Infection and in the Leaves. Front. Plant Sci. 8: 1117 10.3389/fpls.2017.01117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael T. P., Jackson S., 2013. The first 50 plant genomes. Plant Genome 6: 1–7. 10.3835/plantgenome2013.03.0001in [DOI] [Google Scholar]
- Minio A., Lin J., Gaut B. S. B. S., Cantu D., 2017. How single molecule real-time sequencing and haplotype phasing have enabled reference-grade diploid genome assembly of wine grapes. Front. Plant Sci. 8: 826 10.3389/fpls.2017.00826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreton J., Izquierdo A., Emes R. D., 2016. Assembly, assessment, and availability of De novo generated eukaryotic transcriptomes. Front. Genet. 6: 1–9. 10.3389/fgene.2015.00361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myles S., Boyko A. R., Owens C. L., Brown P. J., Grassi F., et al. , 2011. Genetic structure and domestication history of the grape. Proc. Natl. Acad. Sci. USA 108: 3530–3535. 10.1073/pnas.1009363108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrocki E. P., Burge S. W., Bateman A., Daub J., Eberhardt R. Y., et al. , 2015. Rfam 12.0: updates to the RNA families database. Nucleic Acids Res. 43: D130–D137. 10.1093/nar/gku1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrocki E. P., Kolbe D. L., Eddy S. R., 2009. Infernal 1.0: Inference of RNA alignments. Bioinformatics 25: 1335–1337. 10.1093/bioinformatics/btp157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohmi C., Wakana A., Shiraishi S., Alexandria M., 1993. Study of the parentage of grape cultivars by genetic interpretation of GPI-2 and PGM-2 isozymes. Euphytica 65: 195–202. 10.1007/BF00023083 [DOI] [Google Scholar]
- Palumbo M. C., Zenoni S., Fasoli M., Massonnet M., Farina L., et al. , 2014. Integrated network analysis identifies fight-club nodes as a class of hubs encompassing key putative switch genes that induce major transcriptome reprogramming during grapevine development. Plant Cell Online 26: 4617–4635. 10.1105/tpc.114.133710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra G., Blanco E., Guigó R., 2000. GeneID in Drosophila. Genome Res. 10: 511–515. 10.1101/gr.10.4.511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastore C., Zenoni S., Fasoli M., Pezzotti M., Tornielli G. B., et al. , 2013. Selective defoliation affects plant growth, fruit transcriptional ripening program and flavonoid metabolism in grapevine. BMC Plant Biol. 13: 30 10.1186/1471-2229-13-30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea M., Pertea G. M., Antonescu C. M., Chang T.-C., Mendell J. T., et al. , 2015. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33: 290–295. 10.1038/nbt.3122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price A., Gibas C., 2017. The quantitative impact of read mapping to non-native reference genomes in comparative RNA-Seq studies. PLoS One 12: e0180904 10.1371/journal.pone.0180904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryszcz L. P., Gabaldón T., 2016. Redundans: An assembly pipeline for highly heterozygous genomes. Nucleic Acids Res. 44: e113 10.1093/nar/gkw294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy A. S. N., Marquez Y., Kalyna M., Barta A., 2013. Complexity of the alternative splicing landscape in plants. Plant Cell 25: 3657–3683. 10.1105/tpc.113.117523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricker N., Shen S. Y., Goordial J., Jin S., Fulthorpe R. R., 2016. PacBio SMRT assembly of a complex multi-replicon genome reveals chlorocatechol degradative operon in a region of genome plasticity. Gene 586: 239–247. 10.1016/j.gene.2016.04.018 [DOI] [PubMed] [Google Scholar]
- Safonova Y., Bankevich A., Pevzner P. A., 2015. dipSPAdes: Assembler for Highly Polymorphic Diploid Genomes. J. Comput. Biol. A J. Comput. Mol. Cell Biol. 22: 528–545. 10.1089/cmb.2014.0153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmela L., Rivals E., 2014. LoRDEC: Accurate and efficient long read error correction. Bioinformatics 30: 3506–3514. 10.1093/bioinformatics/btu538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savoi S., Wong D. C. J., Arapitsas P., Miculan M., Bucchetti B., et al. , 2016. Transcriptome and metabolite profiling reveals that prolonged drought modulates the phenylpropanoid and terpenoid pathway in white grapes (Vitis vinifera L.). BMC Plant Biol. 16: 67 10.1186/s12870-016-0760-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savoi S., Wong D. C. J., Degu A., Herrera J. C., Bucchetti B., et al. , 2017. Multi-Omics and Integrated Network Analyses Reveal New Insights into the Systems Relationships between Metabolites, Structural Genes, and Transcriptional Regulators in Developing Grape Berries (Vitis vinifera L.) Exposed to Water Deficit. Front. Plant Sci. 8: 1124 10.3389/fpls.2017.01124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sefc K. M., Steinkellner H., Glössl J., Kampfer S., Regner F., 1998. Reconstruction of a grapevine pedigree by microsatellite analysis. Theor. Appl. Genet. 97: 227–231. 10.1007/s001220050889 [DOI] [Google Scholar]
- Seo J., Rhie A., Kim J., Lee S., Sohn M., et al. , 2016. De novo assembly and phasing of a Korean human genome. Nature 538: 243–247. 10.1038/nature20098 [DOI] [PubMed] [Google Scholar]
- Serrano A., Espinoza C., Armijo G., Inostroza-Blancheteau C., Poblete E., et al. , 2017. Omics Approaches for Understanding Grapevine Berry Development: Regulatory Networks Associated with Endogenous Processes and Environmental Responses. Front. Plant Sci. 8: 1486 10.3389/fpls.2017.01486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simão F. A., Waterhouse R. M., Ioannidis P., Kriventseva E. V., Zdobnov E. M., 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31: 3210–3212. 10.1093/bioinformatics/btv351 [DOI] [PubMed] [Google Scholar]
- Simon S., Zhai J., Nandety R. S., McCormick K. P., Zeng J., et al. , 2009. Short-read sequencing technologies for transcriptional analyses. Annu. Rev. Plant Biol. 60: 305–333. 10.1146/annurev.arplant.043008.092032 [DOI] [PubMed] [Google Scholar]
- Smit, A. F. A., and R. Hubley, 2008 RepeatModeler Open-1.0. http://www.repeatmasker.org.
- Smit, A. F. A., R. Hubley, and P. Green, 2013 RepeatMasker Open-4.0. http://www.repeatmasker.org 2013–2015.
- Smith-Unna R. D., Boursnell C., Patro R., Hibberd J. M., Kelly S., et al. , 2016. TransRate: reference-free quality assessment of de novo transcriptome assemblies. Genome Res. 26: 1134–1144. 10.1101/gr.196469.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparvoli F., Martin C., Scienza A., Gavazzi G., Tonelli C., 1994. Cloning and molecular analysis of structural genes involved in flavonoid and stilbene biosynthesis in grape (Vitis vinifera L.). Plant Mol. Biol. 24: 743–755. 10.1007/BF00029856 [DOI] [PubMed] [Google Scholar]
- Stanke M., Keller O., Gunduz I., Hayes A., Waack S., et al. , 2006. AUGUSTUS: ab initio prediction of alternative transcripts. Nucleic Acids Res. 34: W435–W439. 10.1093/nar/gkl200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbiss S., Willhoeft U., Gremme G., Kurtz S., 2009. Fine-grained annotation and classification of de novo predicted LTR retrotransposons. Nucleic Acids Res. 37: 7002–7013. 10.1093/nar/gkp759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strefeler M. S., Weeden N. F., Reisch B. I., 1992. Inheritance of chloroplast DNA in two full-sib Vitis populations. Vitis 31: 183–187. [Google Scholar]
- Swarup R., Crespi M., Bennett M. J., 2016. One gene, many proteins: mapping cell-specific alternative splicing in plants. Dev. Cell 39: 383–385. 10.1016/j.devcel.2016.11.002 [DOI] [PubMed] [Google Scholar]
- Tapia A. M., Cabezas J. A., Cabello F., Lacombe T., Martínez-Zapater J. M., et al. , 2007. Determining the Spanish origin of representative ancient American grapevine varieties. Am. J. Enol. Vitic. 58: 242–251. [Google Scholar]
- Thatcher S. R., Danilevskaya O. N., Meng X., Beatty M., Zastrow-Hayes G., et al. , 2016. Genome-wide analysis of alternative splicing during development and drought stress in maize. Plant Physiol. 170: 586–599. 10.1104/pp.15.01267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tombácz D., Csabai Z., Oláh P., Balázs Z., Likó I., et al. , 2016. Full-length isoform sequencing reveals novel transcripts and substantial transcriptional overlaps in a herpesvirus. PLoS One 11: e0162868 10.1371/journal.pone.0162868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungaro A., Pech N., Martin J. F., McCairns R. J. S., Mévy J. P., et al. , 2017. Challenges and advances for transcriptome assembly in non-model species. PLoS One 12: e0185020 10.1371/journal.pone.0185020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasco R., Zharkikh A., Troggio M., Cartwright D. A., Cestaro A., et al. , 2007. A high quality draft consensus sequence of the genome of a heterozygous grapevine variety. PLoS One 2: e1326 10.1371/journal.pone.0001326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venturini L., Ferrarini A., Zenoni S., Tornielli G. B. G. B., Fasoli M., et al. , 2013. De novo transcriptome characterization of Vitis vinifera cv. Corvina unveils varietal diversity. BMC Genomics 14: 41 10.1186/1471-2164-14-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vij S., Kuhl H., Kuznetsova I. S., Komissarov A., Yurchenko A. A., et al. , 2016. Chromosomal-level assembly of the asian seabass genome using long sequence reads and multi-layered scaffolding. PLOS Genet. 12: e1005954 10.1371/journal.pgen.1005954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitulo N., Forcato C., Carpinelli E. C., Telatin A., Campagna D., et al. , 2014. A deep survey of alternative splicing in grape reveals changes in the splicing machinery related to tissue, stress condition and genotype. BMC Plant Biol. 14: 99 10.1186/1471-2229-14-99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B., Tseng E., Regulski M., Clark T. A., Hon T., et al. , 2016. Unveiling the complexity of the maize transcriptome by single-molecule long-read sequencing. Nat. Commun. 7: 11708 10.1038/ncomms11708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnes, G. R., B. Bolker, L. Bonebakker, R. Gentleman, W. Huber et al., 2016 Package “gplots”: Various R programming tools for plotting data. R Packag. version 2.17.0 https://CRAN.R-project.org/package=gplots.
- Weirather J. L., Afshar P. T., Clark T. A., Tseng E., Powers L. S., et al. , 2015. Characterization of fusion genes and the significantly expressed fusion isoforms in breast cancer by hybrid sequencing. Nucleic Acids Res. 43: e116 10.1093/nar/gkv562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Workman R. E., Myrka A. M., Tseng E., Wong G. W., Welch K. C., et al. , 2017. Single molecule, full-length transcript sequencing provides insight into the extreme metabolism of ruby-throated hummingbird Archilochus colubris. bioRxiv 117218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T. D., Watanabe C. K., 2005. GMAP: a genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics 21: 1859–1875. 10.1093/bioinformatics/bti310 [DOI] [PubMed] [Google Scholar]
- Xi H., Ma L., Liu G., Wang N., Wang J., et al. , 2014. Transcriptomic Analysis of Grape (Vitis vinifera L.) Leaves after Exposure to Ultraviolet C Irradiation. PLoS One 9: e113772 10.1371/journal.pone.0113772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin H., Zhang J., Zhu W., Wang N., Fang P., et al. , 2013. The effects of artificial selection on sugar metabolism and transporter genes in grape. Tree Genet. Genomes 9: 1343–1349. 10.1007/s11295-013-0643-7 [DOI] [Google Scholar]
- Yan K., Liu P., Wu C. A., Yang G. D., Xu R., et al. , 2012. Stress-induced alternative splicing provides a mechanism for the regulation of microRNA processing in Arabidopsis thaliana. Mol. Cell 48: 521–531. 10.1016/j.molcel.2012.08.032 [DOI] [PubMed] [Google Scholar]
- Yang I. S., Kim S., 2015. Analysis of whole transcriptome sequencing data: Workflow and software. Genomics Inform. 13: 119–125. 10.5808/GI.2015.13.4.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J., Fang L., Zheng H., Zhang Y., Chen J., et al. , 2006. WEGO: a web tool for plotting GO annotations. Nucleic Acids Res. 34: W293–W297. 10.1093/nar/gkl031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenoni S., Dal Santo S., Tornielli G. B., D’Incà E., Filippetti I., et al. , 2017. Transcriptional responses to pre-flowering leaf defoliation in grapevine berry from different growing sites, years, and genotypes. Front. Plant Sci. 8: 630 10.3389/fpls.2017.00630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Yang H., Yang H., 2015. Evolutionary character of alternative splicing in plants. Bioinform. Biol. Insights 9: 47–52. 10.4137/BBI.S33716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Massonnet M., Sanjak J. S., Cantu D., Gaut B. S., et al. , 2017. Evolutionary genomics of grape (Vitis vinifera ssp. vinifera) domestication. Proc. Natl. Acad. Sci. USA 114: 11715–11720. 10.1073/pnas.1709257114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimin A. V., Puiu D., Luo M. C., Zhu T., Koren S., et al. , 2017. Hybrid assembly of the large and highly repetitive genome of Aegilops tauschii, a progenitor of bread wheat, with the MaSuRCA mega-reads algorithm. Genome Res. 27: 787–792. 10.1101/gr.213405.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zulkapli M. M., Rosli M. A. F., Salleh F. I. M., Mohd Noor N., Aizat W. M., et al. , 2017. Iso-Seq analysis of Nepenthes ampullaria, Nepenthes rafflesiana and Nepenthes × hookeriana for hybridisation study in pitcher plants. Genom. Data 12: 130–131. 10.1016/j.gdata.2017.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Sequencing data are accessible through NCBI (PRJNA433195) and other relevant datasets, such as protein-coding gene and repeat coordinates, can be retrieved from the Cantu lab github repository (http://cantulab.github.io/data.html). Supplemental material available at Figshare: https://doi.org/10.25387/g3.7291943.