Abstract
The germline sex determination pathway in C. elegans determines whether germ cells develop as oocytes or sperm, with no previously known effect on viability. The mir-35 family of microRNAs are expressed in the C. elegans germline and embryo and are essential for both viability and normal hermaphroditic sex determination, preventing aberrant male gene expression in XX hermaphrodite embryos. Here we show that combining feminizing mutations with partial loss of function of the mir-35 family results in enhanced penetrance embryonic lethality that preferentially kills XO animals. This lethal phenotype is due to altered signaling through the germline sex determination pathway, and maternal germline feminization is sufficient to induce enhanced lethality. These findings reveal a surprising pleiotropy of sperm-fate promoting pathways on organismal viability. Overall, our results demonstrate an unexpectedly strong link between sex determination and embryonic viability, and suggest that in wild type animals, mir-35 family members buffer against misregulation of pathways outside the sex determination program, allowing for clean sex reversal rather than deleterious effects of perturbing sex determination genes.
Keywords: sex determination, microRNAs, embryogenesis, XO lethality, germline sex determination, Genetics of Sex
MicroRNAs are a class of endogenous 22-23-nucleotide RNAs that repress expression of complementary target mRNAs to govern diverse processes in essentially all complex eukaryotes. The seed region (nucleotides 2-7) of a microRNA is the most important portion of the sequence for determining target specificity (Bartel 2009). MicroRNAs which share the same seed sequence are classified as a “family” because they can potentially bind and redundantly regulate the same set of target mRNAs.
The mir-35-42 microRNA family is abundantly expressed in oocytes and early embryos and is essential for C. elegans embryonic development (Alvarez-Saavedra and Horvitz 2010; Wu et al. 2010). The mir-35 family consists of eight members (mir-35-42) which reside in two loci (mir-35-41 and mir-42-44). While deletion of all eight mir-35-42 microRNA genes results in completely penetrant embryonic or early larval lethality (Alvarez-Saavedra and Horvitz 2010), strains which carry a deletion affecting only the mir-35-41 cluster display a weaker temperature-sensitive lethal phenotype with low penetrance of lethality at 15° or 20° and nearly complete lethality at 25° (Alvarez-Saavedra and Horvitz 2010).
Since mir-35-41 deletion mutants can bypass embryonic lethality at permissive temperature, this genetic setting is tractable for understanding which pathways are deregulated in mir-35 family loss of function (Liu et al. 2011; Massirer et al. 2012; McJunkin and Ambros 2014). We previously demonstrated that the most strongly upregulated genes in hermaphrodite mir-35-41(nDf50) embryos are targets of the sex determination pathway that are normally repressed in hermaphrodites and highly expressed in males (McJunkin and Ambros 2017). We found that this aberrant upregulation of male-specific gene expression was driven by derepression of two mir-35-41 target genes encoding RNA binding proteins. One of these genes, SUPpressor-26 (sup-26), had previously been identified as a male-promoting modulator of the sex determination pathway (Manser et al. 2002; Mapes et al. 2010) while the other gene, NHL domain containing-2 (nhl-2), was not previously implicated in sex determination.
The fact that mir-35-41 regulate sex determination is surprising since they are maternally contributed (as well as zygotically expressed) and act partially by maternal effect. Because sex must be determined zygotically, we postulate that the mir-35 family prevents premature sex-specific gene expression. This role of the mir-35 family as a developmental timer is analogous to the roles of other essential microRNA families such as lin-4, let-7 and the let-7 sisters, each of which is temporally expressed to drive forward developmental transitions during larval development (Feinbaum and Ambros 1999; Reinhart et al. 2000; Abbott et al. 2005).
Sex determination is a rapidly evolving process that is governed by a diversity of genetic mechanisms across animal phylogeny. Because of its evolutionary plasticity, sex determination is well suited for regulation by microRNAs, whose target-specificity is defined by a minute genomic space (6-7 nucleotides), and thus is rapidly evolving.
In the C. elegans hermaphrodite, a female soma harbors a germline which produces sperm during larval development and oocytes throughout adulthood. The female somatic developmental program is initiated in animals with two X chromosomes (XX) by the dosage compensation complex (DCC), which acts to silence transcription of X-linked genes by two-fold. The DCC also silences the master regulator of sex determination, hermaphroditization of XO animals-1 (her-1), by forty-fold (Meyer 2005) (Figure 1A). Her-1 encodes a diffusible ligand that binds to and inhibits the Patched-like transmembrane receptor Transformer-2 (TRA-2) (Wolff and Zarkower 2008). Thus, low levels of HER-1 in hermaphrodites allow proteolytic activation of TRA-2. Active TRA-2 inhibits the intracellular complex of FEM proteins (FEMinization of XX and XO animals), whose activity antagonizes the transcription factor Transformer-1 (TRA-1). Thus, TRA-1 is licensed to repress its multiple target genes, ensuring proper female somatic development. Conversely, in males, only one X chromosome is present (XO), and dosage compensation is off. This permits high levels of HER-1 expression, which leads to an active FEM complex, and high levels of TRA-1 target genes, which carry out the male-specific developmental program. We previously found that the targets of mir-35-41 act at multiple levels in this pathway – both upstream and downstream of her-1 – to drive aberrant male-like gene expression in mir-35-41(nDf50) mutant hermaphrodites (McJunkin and Ambros 2017).
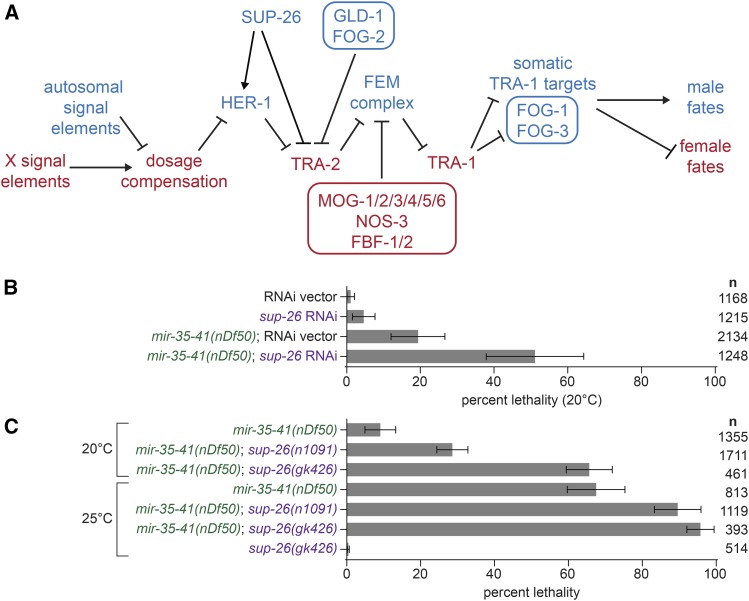
Figure 1.
SUP-26 promotes embryonic viability in mir-35-41(nDf50). A) A schematic of the principal genetic interactions of sex determination pathway genes in C. elegans. Genes in blue are highly active in the male soma and germline, and the spermatogenic germ cells in hermaphrodites. Genes in red are highly active specifically in the hermaphrodite soma and oogenic germ cells. Germline-specific factors are in boxes. B) Percent lethality at embryonic or early larval stages. sup-26(RNAi) has no effect on wild type, but induces synthetic lethality in mir-35-41(nDf50). C) Percent lethality at embryonic or early larval stages. sup-26(lf) alleles enhance lethality in mir-35-41(nDf50). Mean and standard error for three to four biological replicates are shown.
Sex determination of the germ cell lineage is governed by the same core pathway as the soma, but additional players act to transduce the upstream signal to produce a sperm or oocyte cell fate. Additionally, in hermaphrodites, temporal regulation of the sex determination pathway in the germline allows for the production of sperm during larval development followed by production of oocytes in the germline of adults. In both males and hermaphrodites, TRA-1 controls germ cell fate by regulating Feminization of Germline-1 and -3 (fog-1 and fog-3) (Ellis and Schedl 2007) (Figure 1A). When expressed, both FOG-1 (a cytoplasmic polyadenylation element binding protein) and FOG-3 (a Tob family protein) promote male cell fate (spermatogenesis). In hermaphrodites, additional players act to transiently repress tra-2 translation early in development to promote spermatogenesis, while other factors repress fem-3 translation later to promote the switch to oogenesis. These factors include the tra-2 repressor defective in GermLine Development 1 (GLD-1) with its partner FOG-2 (Figure 1A). Negative regulators of fem-3 translation include NanOS-related 3 (nos-3), Fem-3 mRNA Binding Factor-1 and -2 (fbf-1 and fbf-2) and Masculinization Of Germline-1-6 (mog-1 through mog-6 cyn-4).
The genes encoding the DCC are essential in XX animals because of their function in dosage compensation of X-linked genes; conversely, overexpression of the DCC in XO animals is lethal. In contrast, sex determination genes downstream of dosage compensation affect sexual dimorphism but are not essential. Generally, their mutation causes sex reversal, but not lethality. For instance, null alleles of tra-2 or tra-1 result in XX animals that undergo male development, while null alleles of her-1 or fem genes cause somatic feminization of XO animals. Mutations in germline-specific sex determination genes can result in germline-specific sex reversal, and are not known to affect viability.
In this work, we examine how the sex determination pathway genetically interacts with mir-35-41 mutant lethality phenotypes. We find that mir-35-41 prevents lethality in feminized backgrounds, surprisingly demonstrating a role for male-promoting genes (e.g., her-1) in hermaphrodite viability. Furthermore, we find that the synthetic lethality of mir-35-41 deletion with feminizing mutations preferentially affects XO animals. Finally, we observe that these synthetic lethal effects are mediated by the germline-specific module of the sex determination pathway via a maternal effect. To our knowledge this is the first description of the germline sex determination pathway modulating viability.
Materials And Methods
C. elegans culture and RNAi
C. elegans were maintained on NGM seeded with HB101. Strains were kept at 15° or 20° for 72 hr or 25° for 48 hr prior to beginning experiments conducted at the respective temperature. For quantification of lethality, single hermaphrodites were placed on individual 3cm NGM plates for approximately 24 hr. The single hermaphrodites were moved to a fresh plate each day. Approximately 24 hr after removal of the parent, larvae were counted and scored as live or dead. For him-8 strains and crosses where males and hermaphrodites were quantified, dead L1 larvae and dead embryos were counted 24 hr after removal of the parent; surviving progeny were scored as male and hermaphrodite the following day, 48 hr after removal of the parent.
For RNAi experiments, plates were supplemented with 1ug/ml IPTG, and were poured no more than two weeks prior to use, and stored at 4°. Gravid adults were placed on RNAi food either for 48 hr (at 25°) or for 72 hr (at 20°) before single L4 progeny were chosen and placed individually on RNAi plates for progeny quantification as described above. For tra-1 and tra-2 RNAi, a lag-2::GFP reporter (qIs56) was used to select L3 hermaphrodites . These L3s were placed on RNAi at 20° for 24h; after 24h, these animals were young adults and were shifted to 25°, and their progeny were scored for lethality as above. The RNAi plasmids targeting sup-26, tra-1 and tra-2 are from the Vidal ORFeome RNAi library (Rual et al. 2004).
Alleles used were mir-35-41(nDf50), sup-26(n1091), sup-26(gk426), him-8(e1489), lon-2(e678), tra-2(e2020gf), tra-2(e1095null), her-1(hv1y101null), xol-1(y9null), fem-3(e1996null), fog-2(q71null), fog-1(q325null), and fog-3(q470null). The balancer qC1 marked with qIs26 (rol-6(su1006), lag-2::GFP) was used as a balancer for sup-26(lf) alleles in the mir-35-41(nDf50) background. The mIn1 balancer marked with mIs14 (myo2::GFP, pes-10::GFP, F2B7.9::GFP) was used to balance mir-35-41(nDf50) and tra-2 alleles. Fem-3(e1996null) was balanced by nT1 with qIs51 (myo2::GFP, pes-10::GFP, F2B7.9::GFP). The X-linked mCherry transgene used to score karyotype in fem-3(null) crosses was oxTi421 [eft-3p::mCherry::tbb-2 3′UTR + Cbr-unc-119(+)].
For lon-2 crosses, one L4 hermaphrodite was mated with five males. Progeny were counted each day as described above. However, to minimize the self-progeny included in quantification, all progeny counted prior to the appearance of the first male progeny were excluded. In addition, all progeny counted on the same day as the first male progeny were excluded. For example, if males were first present on day two, then only progeny from day three and later are shown. Thus, the vast majority of animals included in quantification are cross progeny, and the rare Lon hermaphrodites likely represent self-progeny.
For mir-35-41(nDf50); fog-1(null) and mir-35-41(nDf50); fog-3(null) crosses and their mir-35-41(nDf50) control, all parental males were homozygous for an autosomal GFP transgene (qIs56). In these crosses, only live progeny containing qIs56 were counted (indicating that they are cross progeny). This eliminates the risk of counting self progeny, which is only a concern in the case of the control mir-35-41(nDf50) cross.
Tra-2 genetic experiments
Tra-2(e2020gf) is dominant, and prevents the development of sperm in XX animals, resulting in a male-female (rather than hermaphrodite) strain. To assess the effect of tra-2(gf) on mir-35-41(nDf50) in a pure population of XX animals, females or hermaphrodites were crossed to XX males to yield 100% XX progeny. XX males were generated by crossing tra-2(null) and xol-1(null) into to mir-35-41(nDf50). xol-1(null) was included because tra-2(null) XX pseudomales are unable to mate when xol-1 is wild type. Mir-35-41(nDf50) tra-2(null);xol-1(null) XX males were crossed to either a tra-2(gf) female or a mir-35-41(nDf50) tra-2(gf) female or a mir-35-41(nDf50) hermaphrodite. Thus, progeny are all XX, and either mir-35-41+/nDf50tra-2e2020gf/null;xol-1+/null or mir-35-41nDf50/nDf50tra-2e2020gf/null;xol-1+/null or mir-35-41nDf50/nDf50tra-2+/null;xol-1+/null. Since mir-35-41(nDf50), tra-2(null) and xol-1(null) are all recessive, and tra-2(e2020gf) is dominant, the maternal genotypes highlight the functional genotypic differences. DAPI staining was performed on unmated adults in the first 24h of gravidity by fixing in 95% ethanol for five minutes, followed by mounting in Prolong Diamond antifade mountant with DAPI (ThermoFisher).
To assess the impact of tra-2(e2020gf) on males, a tra-2(e2020gf) or mir-35-41(nDf50) tra-2(e2020gf) female was crossed to mir-35-41(nDf50) males. Here, all progeny are cross-progeny because the females are self-sterile. The control, a mir-35-41(nDf50) hermaphrodite crossed to males of the same genotype, could potentially produce hermaphrodite self-progeny if the cross is inefficient, spuriously decreasing the apparent proportion of males. However, these data do not appear to be affected by this caveat since the proportion of males observed in the mir-35-41(nDf50) strain is near 50%, and is higher than in the male-female mir-35-41(nDf50) tra-2(e2020gf) strain (due to male lethality in the latter).
Data availability
Strains are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables. Supplemental material available at Figshare: https://doi.org/10.25387/g3.7571978.
Results
mir-35-41 promotes viability in feminized genetic backgrounds
During the course of our previous work, we discovered that sup-26, a previously-characterized negative regulator of tra-2 translation (Mapes et al. 2010), is required for the aberrantly masculinized phenotypes observed in mir-35-41(nDf50) animals and that sup-26 acts both upstream and downstream of her-1 in this process (Figure 1A). Surprisingly, although sup-26(lf) suppressed most phenotypic aspects of masculinized development in mir-35-41(nDf50), knockdown or mutation of sup-26(lf) strongly enhanced the embryonic lethality phenotype, at both permissive and restrictive temperature (Figure 1B-C).
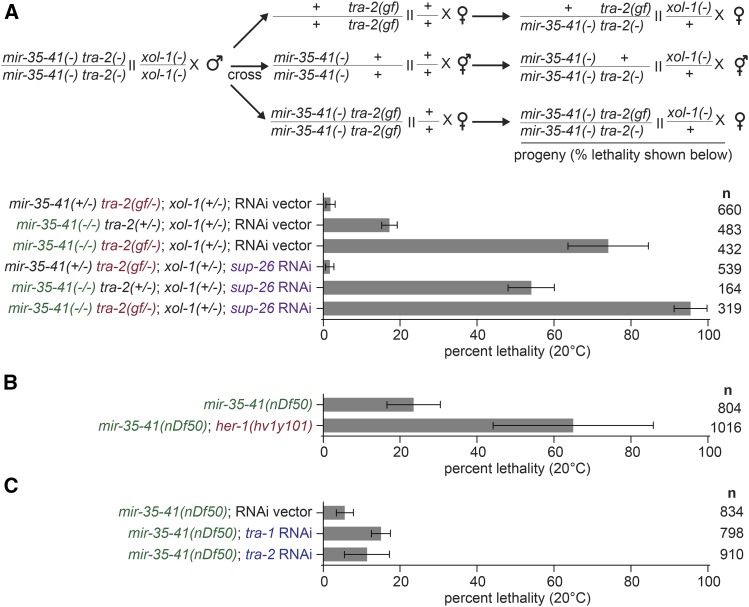
Our previous work showed that SUP-26 binds to hundreds of other RNA targets in addition to tra-2, and thus likely modulates other pathways in addition to sex determination (McJunkin and Ambros 2017). To determine whether the enhanced lethality in mir-35-41(nDf50); sup-26(lf) was due to the effects of sup-26 on sex determination, we examined the penetrance of mir-35-41(nDf50) lethality in other sex determination mutants. First we examined another feminizing mutation, tra-2(e2020gf), which should partially recapitulate the effect of sup-26(lf) on the sex determination pathway since this tra-2(e2020gf) allele deletes the primary binding site of SUP-26 (Doniach 1986; Goodwin et al. 1993; Mapes et al. 2010). The sex determination phenotype of tra-2(e2020gf) is stronger than that of sup-26(lf) because GLD-1 also binds the 3′UTR element deleted in tra-2(e2020gf) (Doniach 1986; Jan et al. 1999); thus these mutants display germline feminization, requiring a cross to XX males to examine viability of XX progeny (Figure 2A) (see Methods). Like sup-26(lf), tra-2(e2020gf) strongly enhanced mir-35-41(nDf50) lethality in XX animals (Figure 2A). The effect of tra-2(e2020gf) was further enhanced by sup-26(RNAi) (Figure 2A), possibly due to additional targets of SUP-26 in sex determination (McJunkin and Ambros 2017). Enhanced lethality is unlikely to be due to gross oocyte defects since germline development and mature oocytes appear normal in all maternal genotypes (Figure S1). Another feminizing mutation, deletion of her-1, also strongly enhanced mir-35-41(nDf50) lethality (Figure 2B).
Figure 2.
Feminizing mutations enhance lethality in mir-35-41(nDf50) XX animals. A) Top: schematic of crosses performed to generate progeny scored in graph. All males were XX males generated via null mutations in tra-2 and xol-1. XX males were crossed to XX hermaphrodites or females of the three indicated genotypes. Bottom: percent lethality of progeny of crosses illustrated in schematic. The crosses were conducted on RNAi plates, so RNAi affects the maternal and zygotic contribution of sup-26 in the progeny. Sup-26(RNAi) and tra-2(e2020gf) enhance XX lethality in mir-35-41(nDf50). Alleles used are tra-2(e1095lf), tra-2(e2020gf), xol-1(y9). B-C) Percent lethality in mir-35-41(nDf50) combined with her-1(null) or tra-1 or tra-2 RNAi. Mean and standard error for three to four biological replicates are shown.
Masculinizing treatments, such as tra-1(RNAi) and tra-2(RNAi) had a very mild or no effect on mir-35-41(nDf50) lethality (Figure 2C), and this was not due to low potency of RNAi since surviving animals displayed transformed and intersex phenotypes (Figure S2). In summary, all feminizing genetic backgrounds examined led to a strong enhancement of lethality in the mir-35-41(nDf50) XX animals, while masculinizing mutations did not. This phenotypic effect of feminizing mutations in XX animals is surprising since XX animals are already somatically female. Thus these results demonstrate a cryptic role for her-1 in XX animals, where her-1 is generally thought to be minimally expressed and biologically inactive.
Males are preferentially affected by lethality in feminized mir-35 family mutants
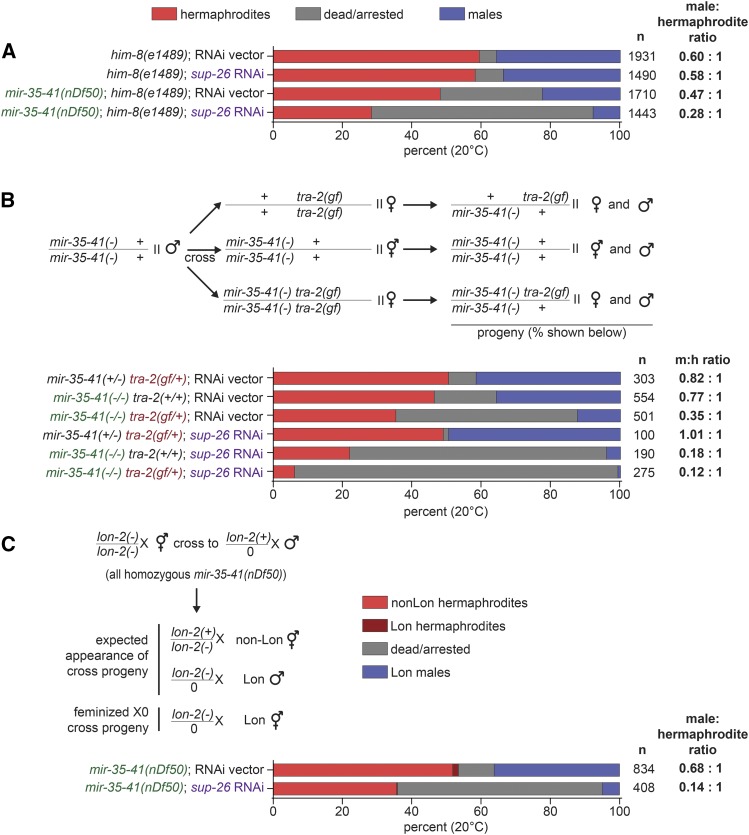
In all experiments described so far, broods of XX animals were scored for viability. We wondered whether the genetic enhancement of mir-35-41(nDf50) lethality by feminizing mutations was sex‐specific. To analyze the impact of mir-35-41(nDf50) and mir-35-41(nDf50);sup-26(lf) lethality on males, a mutation that causes a high incidence of males (him‐8(e1489)) through impaired X chromosome segregation was introduced to mir-35-41(nDf50) (Hodgkin et al. 1979). The him‐8 strain had 5.0% embryonic/early larval lethality, and segregated 37.5% males (among the surviving progeny) (Figure 3A). Both males and hermaphrodites were affected by increased lethality in the mir-35-41(nDf50); him‐8(e1489) background: the proportions of both populations were lower (in favor of dead embryos and larvae of unknown sex) (Figure 3A, compare first bar to third bar). Of the surviving animals in mir-35-41(nDf50);him‐8(e1489) populations, the proportion of males (31.8%) was slightly lower than him‐8 alone, indicating that lethality due to mir-35-41(nDf50) impacted males slightly more than hermaphrodites (Figure 3A) (McJunkin and Ambros 2017). When mir-35-41(nDf50); him‐8(e1489) was treated with sup‐26(RNAi), lethality was further increased in both sexes, and even fewer males (21.6%) were present in the surviving progeny (Figure 3A). Next, we examined the tra-2(e2020gf) genetic background. The sex determination phenotype of tra-2(e2020gf) does not affect all tissues equally, primarily affecting the XX germline (Doniach 1986). As a result, tra-2(e2020gf) produces XX females and XO males, allowing for the sex of progeny to be scored by morphology (Figure 3B). Like sup-26(RNAi), tra-2(e2020gf) enhanced overall lethality in mir-35-41(nDf50), and a smaller proportion of males were observed among the surviving progeny (Figure 3B). As observed for XX broods, these phenotypes were further enhanced when sup-26(RNAi) was performed in the tra-2(e2020gf) background, suggesting that SUP-26 acts through other targets in addition to tra-2 (Figure 3B). Together these results suggest that feminizing mutations enhance lethality of both sexes when mir‐35 family function is compromised, but preferentially enhance the lethality among males.
Figure 3.
Feminizing mutations preferentially enhance mir-35-41(nDf50) lethality in males. A) Percent dead/arrested, male or hermaphrodite progeny in a high-incidence-of-males (him) background. Sup-26(RNAi) reduces the proportion of males among surviving animals. B) Top: Schematic of crosses performed to generate progeny scored in graph. The crosses were conducted on RNAi plates, so RNAi affects the maternal and zygotic contribution of sup-26 in the progeny. Males of the same genotype were used for all crosses in order to compare equal dosage of tra-2(e2020gf) across different mir-35-41 genotypes. Colored text highlights functional genetic differences between genotypes. Sup-26(RNAi) and tra-2(e2020gf) preferentially enhance male lethality in mir-35-41(nDf50). C) Top: Schematic of cross with recessive X-linked marker (lon-2(e678)) to assess potential somatic feminization of XO cross progeny. Bottom: Percent of progeny. The rare Lon hermaphrodites likely represent self-progeny (also see methods). In addition to progeny shown, two males were scored as non-Lon on empty vector RNAi. Sup-26(RNAi) does not increase the apparent proportion of somatically feminized XO animals (Lon hermaphrodites).
Because sup-26(lf) and tra-2(e2020gf) are feminizing mutations, the reduced number of males observed in mir-35-41(nDf50); him‐8(e1489) on sup-26(RNAi) and in tra-2(e2020gf) could be a result of feminization of males rather than lethality. In this case, feminized XO animals would be spuriously counted as hermaphrodites based on morphology. (Additionally, mir-35-41(nDf50); him‐8(e1489) males are abnormal, indicating that mir-35-41(nDf50) may be a genetic background that is sensitized to feminization (McJunkin and Ambros 2014).) To distinguish between feminization and preferential male lethality, mir-35-41(nDf50) hermaphrodites containing a recessive X‐linked marker (lon‐2(e678)) were crossed to non‐Lon mir-35-41(nDf50) males. If sup-26(RNAi) causes feminization of mir-35-41(nDf50); him‐8(e1489) males, then an increase in Lon hermaphrodite‐like progeny would be observed. Sup-26(RNAi) did not increase the proportion of Lon hermaphrodites, indicating that the reduced number of males observed is due to an increase in embryonic/early larval lethality among males and not feminization of XO animals (Figure 3C).
Male preferential lethality is linked to karyotype
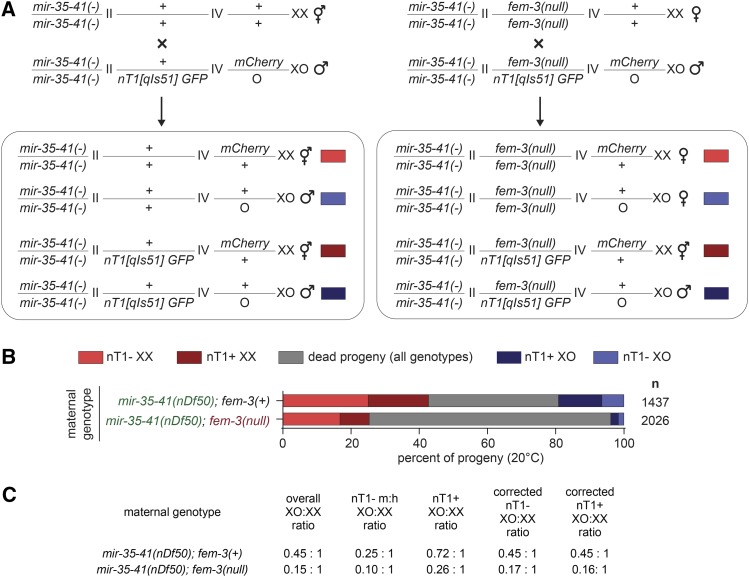
The preferential lethality of males in feminized mir-35-41(nDf50) animals examined thus far could be a result of the males’ XO karyotype or due to the male developmental program downstream of the sex determination pathway. To distinguish between these two possibilities, we decoupled XO karyotype from male development using a fem-3(e1996null) mutation. In this background, both XX and XO animals develop as females. To distinguish between morphologically similar XO and XX animals, we employed a crossing strategy in which XX progeny were marked by an mCherry transgene integrated on the X chromosome (Figure 4A). If lethality in a feminized mir-35-41(nDf50) background still preferentially affects XO animals even when they develop as females, then this preferential lethality is due to XO karyotype and not because of male development.
Figure 4.
XO karyotype underlies preferential death of males in feminized mir-35-41(nDf50) animals. A) Schematic of cross. Males contain a GFP-marked nT1 balancer and an X-linked mCherry transgene which aid in distinguishing fem-3 genotype and number of X chromosomes in progeny. B) Percent of progeny from crosses in each category. Dead/arrested embryos and larvae were not scored for fluorescent markers, and thus are likely a mixture of genotypes. C) XO:XX ratios in populations of progeny from crosses. Corrected values assume that differences in XO:XX ratio in nT1+ and nT1- population in the control mir-35-41(nDf50); fem-3(wild type) cross are due to non-Mendelian segregation of the nT1 balancer.
Overall, progeny resulting from the cross involving mir-35-41(nDf50); fem-3(null) mothers showed greater lethality than progeny from the mir-35-41(nDf50) mothers (Figure 4B). The genotypes of the dead embryos are not defined since embryos may die before expressing fluorescent markers. When examining the live mir-35-41(nDf50); fem-3(null) progeny (those that lack the nT1 balancer chromosome), the XO:XX ratio was lower than that in the comparable nT1- mir-35-41(nDf50) progeny (0.10 compared to 0.25) (Figure 4B-C). This indicates that, as in previous experiments, the feminizing mutation (fem-3(null)) preferentially enhanced lethality among XO animals. However, here, the mir-35-41(nDf50); fem-3(null) XO animals develop as females, not males. Therefore, the preferential enhancement of lethality is due to the XO karyotype since it is manifested in the absence of a male developmental outcome.
While the overall ratio of males to hermaphrodites was 0.45:1 in the control mir-35-41(nDf50) cross, this ratio was lower among animals lacking the nT1 balancer (0.25) and higher among those containing the nT1 balancer (0.72) (Figure 4C). Though this may seem like a surprising discrepancy due to the fact that nT1 is not balancing any mutant alleles in this cross, the disparity can be explained by the previously-observed tendency for nT1 to segregate preferentially with nullo-X sperm (Edgley et al. 2006). If we assume that this segregation preference accounts for the difference between the XO:XX ratio in nT1+ animals and nT1- animals in the control cross, then we can determine a correction factor for this preference (a coefficient for each value that adjusts it to the average XO:XX ratio for the cross). We can then apply the same coefficients to the XO:XX ratios of the mir-35-41(nDf50); fem-3(null) cross to correct for biases resulting from nT1 segregation alone (see below).
When examining nT1+ progeny of these crosses, mir-35-41(nDf50); fem-3(null)/nT1 progeny show a more skewed sex ratio (0.26) compared to mir-35-41(nDf50); +/nT1 progeny (0.72). In fact, after correcting the XO:XX ratios for skewed segregation of nT1, the preferential death of males occurs at the same rate in nT1+ progeny of this cross as among nT1- progeny (both 0.16-0.17). The most plausible interpretation of this finding is that the maternal fem-3 genotype has a greater impact than the zygotic fem-3 genotype on this phenotype since both nT1+ and nT1- progeny lack maternal fem-3 (while they differ in their zygotic dose of fem-3). Thus, observing the same effect size in nT1+ and nT1- progeny suggests that the enhancement of XO lethality by fem-3 loss of function occurs via a maternal effect.
Maternal germline feminization preferentially enhances lethality in males
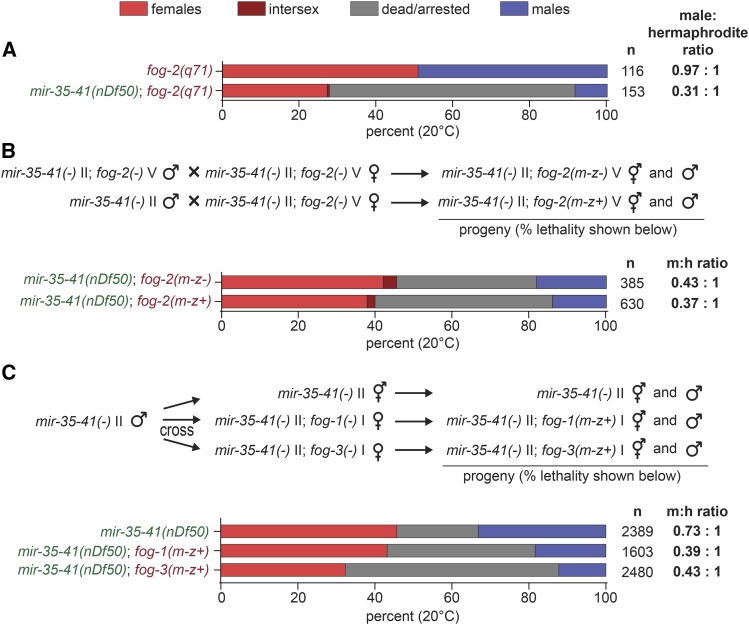
All our results thus far indicate that when mir-35-41(nDf50) is combined with feminizing genetic backgrounds (sup-26(RNAi), tra-2(e2020gf), her-1(null), fem-3(null)), higher rates of lethality occur, and that this enhanced lethality preferentially affects XO animals. Because sup-26(RNAi), her-1(null), fem-3(null) all affect both the soma and the germline (Ellis and Schedl 2007; Wolff and Zarkower 2008; McJunkin and Ambros 2017), and tra-2(e2020gf) has a much greater effect on the germline than the soma (Doniach 1986), we sought to distinguish whether these effects are mediated by the somatic or the germline sex determination pathway. To this end, we tested feminizing mutations in genes that act only in the germline sex determination pathway for their effect on mir-35-41(nDf50) lethality. First, a germline feminizing lesion, fog-2(null), was introduced to the mir-35-41(nDf50) background. Simple male-female crosses demonstrated that fog-2 loss of function enhanced preferential death of males in mir-35-41(nDf50), indicating that the germline-specific sex determination pathway is implicated in this phenotype (Figure 5A).
Figure 5.
Feminization of the germline causes preferential death of mir-35-41(nDf50) males, and this is a maternal effect. A) Percent dead/arrested, male or female progeny in a fog-2(q71) background, with wild type or deleted mir-35-41. B) Percent dead/arrested, male or female progeny. Top bar: both parents are mir-35-41(nDf50); fog-2(q71). Bottom bar: mother is mir-35-41(nDf50); fog-2(q71). Father is mir-35-41(nDf50); fog-2(wild type). C) Top: Schematic of cross. Males also contained a GFP integrated transgene to prevent the scoring of self progeny. Bottom: Percent dead/arrested, male or female progeny.
We next sought to determine whether the effect of the germline-specific sex determination pathway on this phenotype is via maternal effect. Therefore, we crossed mir-35-41(nDf50) males to mir-35-41(nDf50); fog-2(null) females and scored viability of the progeny. Like animals lacking both maternal and zygotic fog-2, mir-35-41(nDf50); fog-2(m-z+) animals showed preferential death among males, suggesting that the perturbation of germline sex determination pathway enhances this phenotype via a maternal effect (Figure 5B). These animals have normal zygotic germline sex determination. Thus, maternal germline feminization likely underlies the preferential male lethality phenotype. This is consistent with the apparent maternal effect of fem-3(null) on enhanced lethality and preferential male lethality in mir-35-41(nDf50) and the previously observed partial maternal effect of the mir-35-41 family on sex determination and other phenotypes (Alvarez-Saavedra and Horvitz 2010; McJunkin and Ambros 2014, 2017).
To determine whether other players in the canonical germline-specific sex determination pathway were also implicated in this phenotype, we tested the effect of maternal loss of fog-1 or fog-3 function in the mir-35-41(nDf50) background. Like fog-2, loss of maternal fog-1 or fog-3 enhanced lethality and male-preferential lethality of mir-35-41(nDf50), with fog-3(null) having a stronger effect than fog-1(null) (Figure 5C). Thus, the canonical germline sex determination pathway is involved in preventing the enhanced lethal phenotypes, and mir-35-41(nDf50) mutants are sensitized to maternal germline feminization, especially among XO males.
Discussion
We found that the mir-35 family, which regulates sex determination in the soma and germline, also has unexpected lethal interactions with the germline sex determination pathway. In the mir-35-41(nDf50) background, multiple germline feminizing mutations enhance lethality of both sexes, while preferentially enhancing that of XO males. To our knowledge, this is the first synthetic lethal effect of the germline sex determination pathway. In some cases, assigning significance to the enhancement of lethality in a genetic background that alone elicits lethality can be difficult. However, two factors: (1) the consistency of enhancement by feminizing mutations and failure to enhance by masculinizing mutations and (2) the preferential effect on XO males both indicate that we are observing a specific synthetic phenotype and genetic interaction of two pathways.
What is the basis of this synthetic lethality? One possibility is that mir-35-41(nDf50) mutants with feminizing mutations experience conflicting signals through the sex determination pathway due to the masculinization caused by derepression of multiple target genes downstream of mir-35-41. Such conflicting developmental signals could be deleterious. However, this is counterintuitive in light of the fact that the sex determination pathway is fairly linear, and legacy mutations show clear epistatic effects, rather than additive parallel genetic interactions. However, genetic data shows that mir-35-41 exerts a partially maternal effect on sex determination (McJunkin and Ambros 2017). Our working model is that mir-35-41 acts as a developmental timer, preventing premature sex-specific gene expression. According to this model, removal of mir-35-41 function could disrupt the clean epistatic and linear genetic relationships of the sex determination pathway, since the order of events in the pathway would be disrupted. Thus, the incoherence of premature male gene expression in mir-35-41 could conflict with feminizing mutations.
Another model is that feminizing mutations in the sex determination pathway ameliorate aberrant masculinization of mir-35-41(nDf50) mutants, while having a synthetic lethal effect with another pathway misregulated in mir-35-41(nDf50). This is consistent with the concept that the mir-35-41(nDf50) family controls multiple targets, pathways, and phenotypes (Alvarez-Saavedra and Horvitz 2010; Liu et al. 2011; Massirer et al. 2012; McJunkin and Ambros 2014, 2017; Kagias and Pocock 2015). This latter model would imply that the germline sex determination pathway controls other aspects of development outside of sex determination. FOG-1 and FOG-3 bind to the mRNA of 81 and ∼1000 target genes, respectively (Noble et al. 2016; Aoki et al. 2018). While regulation of many of these targets must mediate the sex determination function of FOG-1 and FOG-3, regulation of some targets may also contribute to other biological pathways we have yet to understand. Mutation of a fog gene may require homeostatic changes in downstream non-sex determination genes to result in a clean germline feminization phenotype without deleterious pleiotropic effects. The mir-35 family may be important in buffering these homeostatic changes, thus revealing potentially deleterious consequences in the microRNA family mutant.
The above models provide frameworks for conceptualizing the generalized enhanced lethality caused by feminizing mutations in mir-35-41(nDf50), but what could be the basis of the preferential effect on XO animals? Because the enhanced lethality preferentially affects animals with an XO karyotype more than those with an XX karyotype, they may result from perturbation of chromatin modifying complexes that bind and modulate gene expression on the X chromosome. One of these complexes is the DCC. In wild type animals, the DCC is stable and loaded onto X chromosomes only in XX animals (Meyer 2005). Aberrant activation of the DCC in XO animals causes XO-specific lethality. A feedback loop whereby TRA-1 reinforces repression of xol-1 transcription could explain some of the genetic effects seen here, though not those of fog-1 and fog-3 (Hargitai et al. 2009). While an effect on the DCC is one possible model for the phenotype we observe here, two observations oppose this model. First, the mir-35-41(nDf50) mutation causes masculinization of gene expression, which would be expected if the DCC were inactivated, not aberrantly activated. Second, the enhancement of lethality by feminizing mutations observed here importantly affects both XX and XO (with a stronger effect on XO) animals; this would be somewhat unusual for a DCC-activating mutation since DCC activation in XX animals is tolerated and essential.
A second system that modifies X-linked chromatin differently than autosomes is the maternal effect sterile (MES) proteins. MES-2/3/6 make up the polycomb repressive complex 2 (PRC2) in C. elegans and are critical in silencing the X chromosome in the germline via H3K27 methylation (Bender et al. 2004). Their activity is opposed by MES-4, which modifies autosomes with activating H3K36 methylation and excludes autosomal binding by MES-2/3/6 (Bender et al. 2006). Many of the synthetic multivulva class B (synMuv B) genes are required to contain the germline activity of the MES proteins to their proper compartment. In certain synMuv B mutants, including inactivation of lin-35/Rb, somatic cells take on a germline-like gene expression signature which leads to early larval arrest at high temperature (Petrella et al. 2011; Wu et al. 2012). In these contexts, loss of function of either MES-2/3/6 or MES-4 suppresses larval arrest and germline-like gene expression in the soma. Whether this larval arrest could preferentially affect XO animals is not known, but this would be a reasonable prediction since the arrest is due to a germline-like chromatin environment in the soma (and thus somatic X chromosome silencing).
In mir-35-41(nDf50) mutant embryos, lin-35 does not accumulate to wild type levels, leading to a lin-35(lf)-like enhanced RNAi phenotype (Massirer et al. 2012). Since lin-35 also promotes female development, the low lin-35 level in mir-35-41(nDf50) could also underlie the observed masculine gene expression pattern (Grote and Conradt 2006). If low lin-35 in mir-35-41(nDf50) also causes a germline-like chromatin state in the soma, this could possibly account for the lethality phenotype observed here, including the deleterious effect on both sexes as well as the XO-preferential effect.
Future studies should attempt to distinguish between these models. Is the male-like gene expression of mir-35-41(nDf50) required for the enhanced lethality and preferential death among XO? Is a germline-like chromatin state present in the mir-35-41(nDf50) soma? Is the low level of lin-35 responsible for these phenotypes? Delineating the pathways responsible for these phenotypes downstream of mir-35-41 will be facilitated by CRISPR/Cas9-mediated genome editing. By relieving single target genes from mir-35-41-mediated repression, we can begin to understand which phenotypes are downstream of each axis of the pathway, and thus eventually sort out which gene expression changes and phenotypes are causative of each other or genetically separable.
Acknowledgments
Thank you to Victor Ambros in whose lab this work was initiated. Thank you to Rosalind Lee for strains and ongoing support. We acknowledge Eric Haag for critical reading of the manuscript. The McJunkin lab is funded by the NIDDK Intramural Research Program (1ZII DK075147-01). Some strains were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440).
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25387/g3.7571978.
Communicating editor: K. Gunsalus
Literature Cited
- Abbott A. L., Alvarez-Saavedra E., Miska E. A., Lau N. C., Bartel D. P., et al. , 2005. The let-7 MicroRNA family members mir-48, mir-84, and mir-241 function together to regulate developmental timing in Caenorhabditis elegans. Dev. Cell 9: 403–414. 10.1016/j.devcel.2005.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Saavedra E., Horvitz H. R., 2010. Many families of C. elegans microRNAs are not essential for development or viability. Curr. Biol. 20: 367–373. 10.1016/j.cub.2009.12.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki S. T., Porter D. F., Prasad A., Wickens M., Bingman C. A., et al. , 2018. An RNA-Binding Multimer Specifies Nematode Sperm Fate. Cell Reports 23: 3769–3775. 10.1016/j.celrep.2018.05.095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D. P., 2009. MicroRNAs: target recognition and regulatory functions. Cell 136: 215–233. 10.1016/j.cell.2009.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender L. B., Cao R., Zhang Y., Strome S., 2004. The MES-2/MES-3/MES-6 Complex and Regulation of Histone H3 Methylation in C. elegans. Curr. Biol. 14: 1639–1643. 10.1016/j.cub.2004.08.062 [DOI] [PubMed] [Google Scholar]
- Bender L. B., Suh J., Carroll C. R., Fong Y., Fingerman I. M., et al. , 2006. MES-4: an autosome-associated histone methyltransferase that participates in silencing the X chromosomes in the C. elegans germ line. Development 133: 3907–3917. 10.1242/dev.02584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doniach T., 1986. Activity of the sex-determining gene tra-2 is modulated to allow spermatogenesis in the C. elegans hermaphrodite. Genetics 114: 53–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgley M. L., Baillie D. L., Riddle D. L., Rose A. M., 2006. Genetic balancers. WormBook, ed. The C. elegans Research Community WormBook, 10.1895/wormbook.1.89.1, http://www.wormbook.org. 1–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis R., Schedl T., 2007. Sex determination in the germ line. WormBook, ed. The C. elegans Research Community WormBook, 10.1895/wormbook.1.82.2, http://www.wormbook.org. 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinbaum R., Ambros V., 1999. The timing of lin-4 RNA accumulation controls the timing of postembryonic developmental events in Caenorhabditis elegans. Dev. Biol. 210: 87–95. 10.1006/dbio.1999.9272 [DOI] [PubMed] [Google Scholar]
- Goodwin E. B., Okkema P. G., Evans T. C., Kimble J., 1993. Translational regulation of tra-2 by its 3′ untranslated region controls sexual identity in C. elegans. Cell 75: 329–339. 10.1016/0092-8674(93)80074-O [DOI] [PubMed] [Google Scholar]
- Grote P., Conradt B., 2006. The PLZF-like protein TRA-4 cooperates with the Gli-like transcription factor TRA-1 to promote female development in C. elegans. Dev. Cell 11: 561–573. 10.1016/j.devcel.2006.07.015 [DOI] [PubMed] [Google Scholar]
- Hargitai B., Kutnyánszky V., Blauwkamp T. A., Steták A., Csankovszki G., et al. , 2009. xol-1, the master sex-switch gene in C. elegans, is a transcriptional target of the terminal sex-determining factor TRA-1. Development 136: 3881–3887. 10.1242/dev.034637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin J., Horvitz H. R., Brenner S., 1979. Nondisjunction Mutants of the Nematode CAENORHABDITIS ELEGANS. Genetics 91: 67–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan E., Motzny C. K., Graves L. E., Goodwin E. B., 1999. The STAR protein, GLD-1, is a translational regulator of sexual identity in Caenorhabditis elegans. EMBO J. 18: 258–269. 10.1093/emboj/18.1.258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagias K., Pocock R., 2015. microRNA regulation of the embryonic hypoxic response in Caenorhabditis elegans. Sci. Rep. 5: 11284 10.1038/srep11284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M., Liu P., Zhang L., Cai Q., Gao G., et al. , 2011. mir-35 is involved in intestine cell G1/S transition and germ cell proliferation in C. elegans. Cell Res. 21: 1605–1618. 10.1038/cr.2011.102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manser J., Wood W. B., Perry M. D., 2002. Extragenic suppressors of a dominant masculinizing her-1 mutation in C. elegans identify two new genes that affect sex determination in different ways. Genesis 34: 184–195. 10.1002/gene.10118 [DOI] [PubMed] [Google Scholar]
- Mapes J., Chen J.-T., Yu J.-S., Xue D., 2010. Somatic sex determination in Caenorhabditis elegans is modulated by SUP-26 repression of tra-2 translation. Proc. Natl. Acad. Sci. USA 107: 18022–18027. 10.1073/pnas.1004513107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massirer K. B., Perez S. G., Mondol V., Pasquinelli A. E., 2012. The miR-35–41 family of microRNAs regulates RNAi sensitivity in Caenorhabditis elegans. PLoS Genet. 8: e1002536 10.1371/journal.pgen.1002536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McJunkin K., Ambros V., 2017. A microRNA family exerts maternal control on sex determination in C. elegans. Genes Dev. 31: 422–437. 10.1101/gad.290155.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McJunkin K., Ambros V., 2014. The embryonic mir-35 family of microRNAs promotes multiple aspects of fecundity in Caenorhabditis elegans. G3 (Bethesda) 4: 1747–1754. 10.1534/g3.114.011973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer B. J., 2005. X–Chromosome dosage compensation. WormBook, ed. The C. elegans Research Community WormBook, 10.1895/wormbook.1.8.1, http://www.wormbook.org. 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble D. C., Aoki S. T., Ortiz M. A., Kim K. W., Verheyden J. M., et al. , 2016. Genomic Analyses of Sperm Fate Regulator Targets Reveal a Common Set of Oogenic mRNAs in Caenorhabditis elegans. Genetics 202: 221–234. 10.1534/genetics.115.182592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrella L. N., Wang W., Spike C. A., Rechtsteiner A., Reinke V., et al. , 2011. synMuv B proteins antagonize germline fate in the intestine and ensure C. elegans survival. Development 138: 1069–1079. 10.1242/dev.059501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart B. J., Slack F. J., Basson M., Pasquinelli A. E., Bettinger J. C., et al. , 2000. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403: 901–906. 10.1038/35002607 [DOI] [PubMed] [Google Scholar]
- Rual J.-F., Ceron J., Koreth J., Hao T., Nicot A.-S., et al. , 2004. Toward improving Caenorhabditis elegans phenome mapping with an ORFeome-based RNAi library. Genome Res. 14: 2162–2168. 10.1101/gr.2505604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff J. R., Zarkower D., 2008. Somatic sexual differentiation in Caenorhabditis elegans. Curr. Top. Dev. Biol. 83: 1–39. 10.1016/S0070-2153(08)00401-8 [DOI] [PubMed] [Google Scholar]
- Wu X., Shi Z., Cui M., Han M., Ruvkun G., 2012. Repression of germline RNAi pathways in somatic cells by retinoblastoma pathway chromatin complexes. PLoS Genet. 8: e1002542 10.1371/journal.pgen.1002542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu E., Thivierge C., Flamand M., Mathonnet G., Vashisht A. A., et al. , 2010. Pervasive and cooperative deadenylation of 3′UTRs by embryonic microRNA families. Mol. Cell 40: 558–570. 10.1016/j.molcel.2010.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Strains are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables. Supplemental material available at Figshare: https://doi.org/10.25387/g3.7571978.