Abstract
Clathrin is a major coat protein involved in vesicle formation during endocytosis and transport in the endosomal/trans Golgi system. Clathrin is required for normal growth of yeast (Saccharomyces cerevisiae) and in some genetic backgrounds deletion of the clathrin heavy chain gene (CHC1) is lethal. Our lab defined a locus referred to as “suppressor of clathrin deficiency” (SCD1). In the presence of the scd1-v allele (“v” – viable), yeast cells lacking clathrin heavy chain survive but grow slowly, are morphologically abnormal and have many membrane trafficking defects. In the presence of scd1-i (“i”- inviable), chc1∆ causes lethality. As a strategy to identify SCD1, we used pooled linkage analysis and whole genome sequencing. Here, we report that PAL2 (YHR097C) is the SCD1 locus. pal2∆ is synthetic lethal with chc1∆; whereas a deletion of its paralog, PAL1, is not synthetic lethal with clathrin deficiency. Like Pal1, Pal2 has two NPF motifs that are potential binding sites for EH domain proteins such as the early endocytic factor Ede1, and Pal2 associates with Ede1. Also, GFP-tagged Pal2p localizes to cortical patches containing other immobile phase endocytic coat factors. Overall, our data show that PAL2 is the SCD1 locus and the Pal2 protein has characteristics of an early factor involved in clathrin-mediated endocytosis.
Keywords: Clathrin, endocytosis, membrane trafficking, pooled linkage analysis
Movement of proteins within the secretory and endocytic pathways is initiated by the binding of coat proteins to the cytosolic surface of the membrane followed by capture of cargo molecules and vesicular budding (McMahon and Mills 2004, Gomez-Navarro and Miller 2016). Clathrin and its associated proteins form a major class of vesicular transport coats (Kirchhausen et al. 2014, Robinson 2015). Clathrin-coated vesicles (CCVs) are involved in receptor mediated endocytosis, recycling of membranes, transcellular transport and transport between the TGN and endosomes (Kirchhausen et al. 1989, Pearse and Robinson 1990, Bonifacino and Glick 2004, Boettner et al. 2011, Kirchhausen et al. 2014, Robinson 2015, Elkin et al. 2016, Lu et al. 2016). Clathrin, which forms the striking polygonal surface lattice on CCVs, is a trimeric molecule, or triskelion, containing three heavy chains (HC) of ∼180 kD that radiate from a vertex and three light chains (LC) of 30 - 40 kD, which bind noncovalently near the vertex of the triskelion, one per heavy chain arm (Kirchhausen et al. 1989, Edeling et al. 2006).
CCVs have been found in virtually every eukaryotic organism that has been examined, including the yeast Saccharomyces cerevisiae (Mueller and Branton 1984, Payne and Schekman 1985, Lemmon and Jones 1987, Silveira et al. 1990). Previous studies in yeast found that deletion of the clathrin HC gene, CHC1, is lethal in some genetic backgrounds, but not in others (Payne and Schekman 1985, Lemmon and Jones 1987, Payne et al. 1987, Schekman and Payne 1988, Munn et al. 1991). The viable clathrin HC deficient cells exhibit a number of phenotypes, including slow growth, abnormal morphology and polyploidy, and defects in mating, sporulation, endocytosis and sorting in the endosomal/Trans Golgi Network (TGN) system (Payne and Schekman 1985, Lemmon and Jones 1987, Payne et al. 1987, Payne et al. 1988, Payne and Schekman 1989, Lemmon et al. 1990, Nelson and Lemmon 1993, Kaksonen et al. 2005, Newpher and Lemmon 2006). In strains studied in this laboratory, we uncovered a polymorphism in an independently segregating genetic locus that causes lethality in chc1∆ cells (Lemmon and Jones 1987). We called this locus “suppressor of clathrin deficiency” (SCD1), where in the presence of the scd1-v allele, yeast cells lacking clathrin HCs are viable but with the scd1-i allele clathrin HC deficient cells are inviable. However, the genetic basis for the lethality in scd1-i chc1∆ cells has remained a mystery for over 30 years. We initially sought to identify the SCD1 gene using a multicopy suppressor screen, and identified the genes SCD2 – SCD6 whose overexpression could supress the lethality of clathrin HC-deficient yeast cells carrying the scd1-i allele (Nelson and Lemmon 1993). However, segregation analysis showed that none of these genes were allelic to the SCD1 locus ((Nelson and Lemmon 1993, Gelperin et al. 1995, Nelson et al. 1996, Huang et al. 1997), Gelperin and S. Lemmon unpublished observations). Also, traditional complementation and other chromosomal mapping methods to identify the gene were complicated by the fact that chc1∆ cells become polyploid at high frequency and are difficult to transform (Lemmon and Jones 1987, Lemmon et al. 1990).
More recently pooled linkage analysis and whole genome sequencing has been used in yeast to identify difficult to clone mutations, polymorphisms or dominant alleles (Shendure and Ji 2008, Birkeland et al. 2010, Song et al. 2014, Lang et al. 2015, Linder et al. 2016). Taking advantage of this approach we now report that scd1-i encodes a mutation in the PAL2 (YHR097C) locus. pal2∆ is synthetic lethal with chc1∆, whereas a knockout of its paralog, PAL1, an endocytic factor (Carroll et al. 2012), is not synthetic lethal with clathrin heavy chain deficiency. Pal2-GFP localizes to cortical patches, similar to Pal1-GFP and other endocytic coat factors. Overall, our data show that PAL2 is the SCD1 locus and is likely involved in clathrin-mediated endocytosis.
Materials and Methods
Yeast strains and methods
S. cerevisiae strains used in this study are related to S288c and are listed in Tables S1 and S2. Standard methods were employed for DNA manipulations, and yeast tetrad analysis, growth and transformation. Unless otherwise indicated, the Longtine method was used for generating the fluorescently tagged reporters and deletion mutants (Longtine et al. 1998). For the triple tagged Pal1-3xGFP, pFA6a 3xGFP-KanMX6 was used as a template (Kovar et al. 2005).
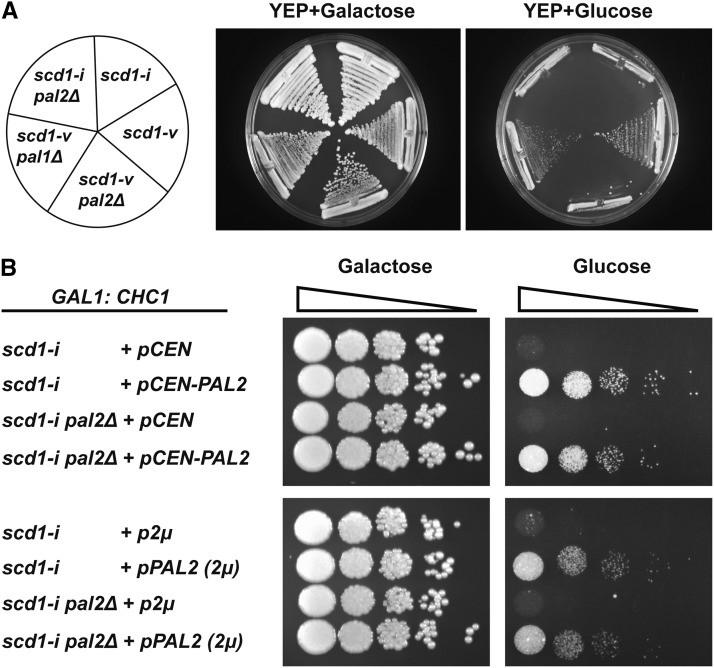
For growth tests shown in Figure 2 and Figure S2 strains were streaked from a single colony onto 1% yeast extract + 2% peptone (YEP) containing 2% galactose (YEP-Gal) and YEP + 2% Dextrose (YEPD) plates and incubated at 30° (Figure 2), 34° and 37° (Figure S2) for 4 days. For serial dilution spotting in Figure 2B, the strains were grown overnight in YEP liquid medium containing 2% galactose (YEP-Gal). Cells from these cultures were inoculated into fresh medium at 0.025 × 107 cells/ml and then grown to ∼1.0 × 107 cells/ml. Cells were pelleted, washed with dH2O and resuspended in YEPD at 0.1 × 107 cells/ml, and allowed to grow for ∼14 hr. Serial ten-fold dilutions were made in dH2O. Five µl of each of the 10−1 to 10−4 dilutions were spotted on YEP-Gal and YEPD plates and grown for 4 days.
Figure 2.
pal2∆ is synthetic lethal with loss of clathrin heavy chain. (A) GAL1:CHC1 strains with the indicated SCD1 and PAL1 genotypes were streaked on galactose and glucose medium and grown for 4 days. Strains used are: scd1-i (SL214); scd1-v (SL350); scd1-v pal2∆ (SL7249); scd1-v pal1∆ (SL7261); scd1-i pal2∆ (SL7251). (B) GAL1:CHC1 strains with the indicated SCD1 and PAL1 genotypes were transformed with vector control (pCEN or p2μ) or plasmids expressing PAL2 (pCEN-PAL2 or pPAL2 2μ). The strains were grown to log phase in YEP-galactose and then transferred to YEP-glucose for ∼14 hr. Then 10-fold serial dilutions were spotted on galactose and glucose plates and grown for 4 days. Strains used are: scd1-i +pCEN (SL7444); scd1-i +pCEN-PAL2 (SL7464); scd1-i pal2∆ +pCEN (SL7447); scd1-i pal2∆ +pCEN-PAL2 (SL7448); scd1-i +p2μ (SL7278); scd1-i +pPAL2 (2μ) (SL7279); scd1-i pal2∆ +p2μ (SL7280); scd1-i pal2∆ +pPAL2 (2μ) (SL7281).
Plasmids
Plasmids used in this study are listed in Table S3. pRS426-PAL2 was generated by PCR amplification of PAL2 (YHR097C) with ∼500 base pairs (bp) upstream and ∼300 bp downstream of the open reading frame (ORF) using clone no. 671 (plate no. 7; F12 well) from the multicopy Tiling library collection (gift of G. Prelich (Jones et al. 2008)) as a template. The PCR product was sequenced to confirm it encoded the wild type Pal2 protein (scd1-v background). The amplified PCR fragment was digested with Kpn1 (5′) and BamH1 (3′), which had been encoded in the primers, and cloned into the pRS426 vector cut with Kpn1 and BamH1. Plasmid pRS316-PAL2 was generated in the similar way as pRS426-PAL2 except that both the vector and PCR fragment were digested with Xho1 (5′) and Kpn1 (3′). Plasmid clones were verified by restriction digestion and sequencing.
Plasmid pET28c-EDE1 (EH1-3) [pBW1161] (gift of B. Wendland), contains 1261 bp of the coding sequence of the N-terminal region of EDE1 for EH domains 1, 2 and 3 inserted into the BamH1 (5′) and Xho1 (3′) sites of pET28c. This was used for bacterial expression of a N-terminal His6-tagged EH1-3 domain. The coding sequence of GFP was inserted between the Nde1 (5′) and Not1 (3′) sites of pET22b, to generate pET22b-GFP to express His6-GFP (gift of D. Patel & F. Zhang). Plasmids were verified by restriction digestion and sequencing.
Whole genome sequencing
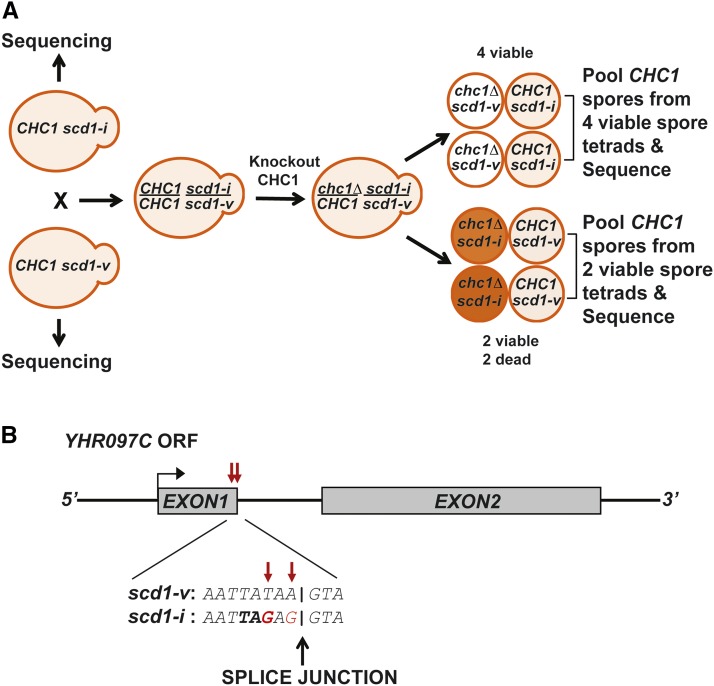
The strains used in this analysis were generated from parents and spore segregants previously described in (Lemmon and Jones 1987) (see Table S1 for the list of strains used for SCD1 identification). Parent strains BJ2700 (CHC1leu2scd1-i) and BJ2738 (CHC1leu2scd1-v) were crossed to generate the diploid BJ3068 (CHC1/CHC1leu2/leu2scd1-i/scd1-v). Then CHC1 was disrupted with LEU2 to generate chc1∆:LEU2/CHC1scd1-i/scd1-v transformants BJ3119 and BJ3120. These were subjected to tetrad analysis and wild type CHC1scd1-i or scd1-v spores were identified based on their segregation from tetrads with four viable spores or 2 viable spores, respectively. The parents of BJ3068 (BJ2700 and BJ2738) and 10 each of CHC1scd1-i or CHC1scd1-v spore segregants were analyzed.
Individual cultures of 10 segregants bearing the scd1-v allele or 10 bearing the scd1-i allele were grown in YEPD. Equal numbers of cells (1x107) from each pool member were mixed and genomic DNA extracted using the YeaStar Genomic DNA Kit (D2002; Zymo Research, Irvine CA). Cultures of parents were treated similarly except 1x108 cells were harvested for DNA isolation.
IIllumina sequencing of parents and pooled strains and alignment of the resulting reads were performed as described previously (Birkeland et al. 2010, Yau et al. 2014) by the Center for Genome Technology Sequencing Core at the Hussman Institute for Human Genomics, University of Miami, Miller School of Medicine. All samples were prepped via ’TruSeq DNA SamplePrep Guide 15026486 C’ (Illumina) with an input of 700-800ng DNA and 12 cycles of PCR. Five hundred bp long libraries were created and the two pools and parent samples were sequenced in a single multiplexed HiSeq lane with 2x101 nt paired end reads. Read alignment to the sacCer3 (R64.1.1) (Engel et al. 2014) version of the yeast genome was performed using BWA-MEM using default parameters (Li and Durbin 2010). Because of the much higher read depth in the current study, a novel approach to data analysis used samtools (samtools mpileup -DSu -d 1000 -L 1000) followed by bcftools (bcftools view -bvcg -T pair, with a ploidy of 2) to identify variants for which there was a high likelihood that the composite genotype was different for the scd1-i and scd1-v pools. Randomly segregating alleles would each appear as heterozygous in each pool while those linked to the causative mutant allele would appear as either homozygous reference or variant in different pools. The same analysis with a ploidy of 1 compared the two starting haploid strains, where candidate alleles must again have a high likelihood of having a different genotype. The two outputs were filtered for variants with a Phred-scaled log ratio of genotype likelihoods (CLR score) >225 in each of the pool and haploid strain comparisons, which resulted in two variants that proved to have only a 2 bp separation on chrVIII. Read counts (forward/reverse strand, sacCer3 coordinates) for chrVIII:298485, T > C were: wild-type pool (scd1-v), 157/90 reference and 15/12 mutant; mutant pool (scd1-i), 0/0 reference and 206/113 mutant. Read counts for chrVIII:298487, A > C were: wild-type pool (scd1-v), 157/90 reference and 20/14 mutant; mutant pool (scd1-i), 0/0 reference and 237/115 mutant. The most likely explanation for the presence of rare mutant reads in the wild-type pool was imperfect scoring of contributing spore clones.
Microscopy and image analysis
For most experiments, cells were grown to log phase at 30° in synthetic medium, concentrated, and immobilized on Concavalin-A coated coverslips. Coverslips were then mounted on slides, and imaged at 25° as indicated below.
Localization of Pal2-GFP or Pal1-(3x) GFP, and co-localization with other markers was performed on an Olympus fluorescence BX61 upright microscope equipped with Nomarski differential interference contrast (DIC) optics, a Uplan Apo 100x objective (1.35 NA), a Roper CoolSnap HQ camera, and Sutter Lambda 10-2 excitation and emission filter wheels, and a 175 watt Xenon remote source with liquid light guide. Image capture was automated using Intelligent Imaging Innovations Slidebook 6 for Windows 7. Z-stacks of 0.25 μm of fields of cells were taken and a medial plane was selected for image analysis. Image analysis was carried out using Slidebook 6 software and later exported to TIF files. The images of 300 dpi were then cropped and arranged in Adobe Photoshop CS5 and Creative Cloud. Approximately 40 – 60 cells were considered for localization of Pal2-GFP and Pal1-3xGFP. For the quantification of Pal2-GFP patches containing Ede1-mCherry, Sla2-RFP, End3-mCherry and Abp1-RFP, 10 – 18 cells with distinct Pal2-GFP patches from a single medial plane were selected and examined for the presence/overlap of mCherry/RFP signal. The percentage of Pal2-GFP patches containing Ede1-mCherry, Sla2-RFP, End3-mCherry and Abp1-RFP was calculated by the ratio between the number of GFP/RFP(mCherry) overlapping patches to the total number of Pal2-GFP patches in the cells. The percentage overlap of Pal1-3xGFP patches with Pal2-mCherry patches was determined in a similar way. Statistics were performed using the GraphPad Prism 7 software. Two-tailed t-test was carried out for each pair to measure the significance of the data.
Cortical patch to cytosol fluorescence intensity ratio analysis was carried out on the Olympus fluorescence BX61 upright fluorescence microscope as described previously (Chi et al. 2012) using Slidebook 6.0 for Windows 7 for acquisition and analysis. Strains were grown at 30° to early log phase and z-stack images (5 × 0.25 um) were captured. Analysis was performed on a medial focal plane. The fluorescence intensity of the brightest cortical patch in a cell was divided by the fluorescence from the mother cell cytosol. A representative background intensity value (outside the cell) was also subtracted from both patch and mother cell cytosol intensities before calculating the patch/cytosol ratio (n ≥ 25 cells for each strain).
To prevent actin polymerization and inhibit internalization at endocytic sites, log phase cells were treated in synthetic medium supplemented with 200 μM Latrunculin-A (LAT-A; Enzo, BML-T119) for 1 h at 30°. Control cells were treated in medium containing an equal volume of dimethyl sulfoxide (DMSO), the diluent for LAT-A.
Protein purification and pull-down experiment
His6-Ede1 (EH1-3) expressed from pET28c-EDE1(EH1-3) or His6-GFP expressed from pET22c-GFP were purified on Ni-NTA agarose beads (Cat No./ID: 30230, Qiagen) according to the manufacturer’s instructions. Yeast extracts were made from 8x108 cells grown in YEPD to log phase. Cell lysates were prepared by resuspending each cell pellet in 1ml of lysis buffer (10 mM Tris pH 8.0, 140 mM NaCl, 0.1% Tween-20, 1 mM β-mercaptoethanol, 1mM PMSF and 1x protease inhibitor cocktail {Protease Inhibitor Cocktail (100X); Cell Signaling, Catalog no.5871}) and subjecting it to glass bead vortexing (4 times – 1 min vortex and 2 min pause on ice), followed by centrifugation at 15000 × g in an eppendorf 5415R centrifuge for 10 min at 4°. An aliquot of supernatant was saved as input. For pull down experiments, 50 μl of Ni-NTA agarose slurry was equilibrated with lysis buffer and incubated with ∼30 μg of purified His6-Ede1 (EH1-3) or His6-GFP at 4° for 1.5 hr. Beads were washed three times with 1ml of wash buffer (lysis buffer + 20mM imidazole). Then 200μl of lysate was added to the bead alone or beads containing purified His6-Ede1(EH1-3) or His6-GFP, followed by incubation at 4° for 2 hr on a rotator. Beads were washed three times with wash buffer (lysis buffer without protease inhibitors + 10% glycerol) and resuspended in 30μl lysis buffer and 30μl of 4x SDS sample buffer. Samples were heated at 65° for 5 min and stored at -20° until used for immunoblot analysis. Twenty µl of samples from pull downs and 5μl of cell extracts for loading controls were run on Biorad precast 4–20% gradient mini gels and transferred to nitrocellulose membrane using Biorad Trans-blot turbo system (25V for 7 min). Pal2-GFP and Pal1-GFP were detected using anti-GFP antibody (1:1000, Roche; mouse monoclonal; catalog no. 11814460001), and His6-GFP and His6-Ede1 (EH1-3) were detected with 6x-His Tag monoclonal antibody (1:1000, Thermo Scientific; catalog no. 4A12E4). Goat anti-mouse conjugated with horseradish peroxidase (HRP) was used as the secondary antibody at 1:5000 dilution (Thermo Scientific; catalog no. 31430). The proteins were then detecting by Chemiluminescence using SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Scientific; catalog no 34095).
Data availability
Strains and plasmids are available upon request. Figure S1 shows the sequence alignment of the “Pal domain” of Pal2 and Pal1. Figure S2 shows the growth phenotype of GAL1:CHC1 strains with the indicated genotypes streaked on YEP+glucose at elevated temperatures. Figure S3 shows the patch to cytosol fluorescence intensity ratio for Pal2-GFP and Pal1-3xGFP in wild type and different endocytic mutants. Figure S4 shows the patch to cytosol fluorescence intensity ratio for Sla1-GFP in wild type and pal∆ mutants. Table S1 contains the yeast strains used for pooled linkage analysis and whole genome sequencing. Table S2 and Table S3 show the list of yeast strains and plasmids used in this study, respectively. Table S4 shows the tetrad data demonstrating that pal1∆ is not synthetic lethal with chc1∆. Supplemental material available at Figshare: https://doi.org/10.25387/g3.7582868.
Results
Identification of the SCD1 locus using pooled linkage analysis and next generation whole genome sequencing
To discover functional mutations in yeast among a large excess of polymorphisms and incidental mutations, a strategy was developed based on next-generation sequencing, called pooled linkage analysis (Birkeland et al. 2010, Song et al. 2014, Lang et al. 2015, Linder et al. 2016). We employed the same strategy for the identification of the SCD1 locus. In the original studies where this polymorphism was observed, two wild type strains had been crossed; one carrying the scd1-v allele (BJ2738) and the other bearing the scd1-i allele (BJ2700) generating a scd1-v/scd1-i heterozygous diploid (BJ3068) (Lemmon and Jones 1987) (see Table S1). The clathrin heavy chain gene (CHC1) was knocked out in the scd1-v/scd1-i heterozygous diploid and tetrads were dissected (Figure 1A). Tetrads from these crosses were saved enabling us to recover wild type spore segregants carrying the scd1-i or scd1-v allele (see Table S1, Figure 1A). Wild type spores of scd1-i genotype were selected from tetrads with four viable spores, since the scd1-v allele would have segregated with the two viable chc1∆ spores. Wild type spores of scd1-v genotype were selected from tetrads with 2 viable spores, as the scd1-i allele would have segregated with the two dead chc1-∆ spores. Parents of the original diploid cross and individual pools of 10 wild type spore segregants of scd1-i or scd1-v genotype were subjected to genomic DNA sequencing (Figure 1A). Incidental polymorphisms will segregate randomly and both alleles would be represented in the pools. However, causative scd1 alleles that co-segregate with the clathrin heavy chain deficient cell phenotype will be homogenous in each pool. Only one candidate allele in the scd1-i pool and the original scd1-i parent (BJ2700) emerged from this analysis, which proved to be a pair of closely spaced single-nucleotide substitutions, chrVIII:298485,T > C and chrVIII:298487,A > C in YHR097C, a spliced gene (see Materials and Methods). Careful analysis of the sequencing results showed a mutation just upstream of the 5′ splice junction, where codon 41 for tyrosine is mutated to a stop codon (Figure 1B). YHR097C has a paralog PAL1 (YDR348C) that arose from a whole genome duplication (Byrne and Wolfe 2005). Intriguingly, Pal1 has been reported to be involved in clathrin-mediated endocytosis (Carroll et al. 2012). From sequence alignment, both proteins are 43% identical. Furthermore, both share 65% identity over a conserved “Pal domain” (Figure S1). We adopted the name PAL2 for SCD1 as Kaksonen and co-workers used this gene name in a prior study (Brach et al. 2014).
Figure 1.
Identification of SCD1 locus by next generation sequencing (NGS). (A) CHC1 strains with the scd1-v (BJ2738) and scd1-i allele (BJ2700) were crossed to each other. CHC1 was deleted from the resulting heterozygous (scd1-i/scd1-v) diploid (BJ3068). Tetrads were dissected and wild type spores were selected from tetrads with 4 viable spores (CHC1 scd1-i) or 2 viable spores (CHC1 scd1-v). Parents of the original diploid and pools of 10 CHC1 spore segregants of scd1-i or scd1-v genotype were sequenced. (B) Schematic representation of YHR097C. Exon1 and Exon2 are shown as dark gray boxes. The red arrows indicate the mutation(s), where the first mutation leads to a Tyr residue to Stop codon at codon 41 of the scd1-i allele just upstream of the 5′ splice junction.
pal2∆, but not pal1∆, is synthetically lethal with clathrin heavy chain deficiency
We previously showed that CHC1 expressed via the GAL1 promoter confers regulated control of clathrin HC synthesis (Nelson and Lemmon 1993). On galactose medium GAL1:CHC1 cells express clathrin HC and grow normally, but on glucose medium CHC1 expression is repressed and cells either grow slowly (scd1-v) or are inviable (scd1-i) (Figure 2A). We then deleted PAL2 in these two strains (Figure 2A). The strains grew normally on galactose, but deletion of PAL2 in the GAL1:CHC1scd1-v background now led to inviability on glucose, similar to the GAL1:CHC1scd1-i strain. In contrast, deletion of PAL1 did not cause inviability nor did it further impair growth of the original GAL:CHC1scd1-v strain on galactose or glucose (Figure 2A), even at elevated temperature of 34° (Figure S2). All of the strains were inviable on glucose at 37°, as expected for cells lacking clathrin HC (Figure S2). These results indicate that pal2∆, but not pal1∆, is synthetic lethal with clathrin HC-deficiency.
We next tested whether PAL2 could complement GAL1:CHC1scd1-i in glucose medium, and found that PAL2 on a CEN plasmid indeed rescued the growth defect of GAL1:CHC1scd1-i strains (with or without pal2∆) (Figure 2B, upper panels). In an attempt to identify SCD1 earlier, we performed a multi-copy suppressor screen on GAL1:CHC1scd1-i yeast. SCD2-SCD6 were identified in this screen, but none were allelic to the SCD1 locus ((Nelson and Lemmon 1993, Gelperin et al. 1995, Nelson et al. 1996, Huang et al. 1997), unpublished observations). We might have expected to identify PAL2 in this screen, so we considered the possibility that overexpression of PAL2 might cause impaired growth of wild type yeast or Chc- cells. But overexpression of PAL2 (pPAL2 (2µ)) also rescued GAL1:CHC1scd1-i strains (with or without pal2∆) on glucose and had no effect on cell growth when CHC1 was expressed on galactose medium (Figure 2B, lower panels).
YHR097C (PAL2) is SCD1 locus
Our pooled linkage/sequence analysis indicates that PAL2 is SCD1. Supporting this we so far have shown that pal2∆ causes synthetic lethality in clathrin HC deficient yeast and PAL2 complements the scd1-i mutation. However to prove that PAL2 is the SCD1 locus, a classical genetic approach is required. To this end, we first deleted PAL2 in CHC1scd1-v cells and crossed this (CHC1pal2∆:NATMx6; SL7098) with a chc1∆:LEU2scd1-i strain where the spore segregant came out of tetrads protected by YCp50-CHC1 (SL98). The plasmid was dropped from the chc1∆/CHC1 heterozygous diploid and subjected to tetrad analysis. If PAL2 is SCD1, then the diploid strain would be homozygous at the SCD1 locus (pal2∆:NatMx6/scd1-i) and we expect 2 viable CHC1 and 2 dead spores (chc1∆:LEU2 with either scd1-i or pal2∆:NATMx6) in all tetrads. If PAL2 is not the SCD1 locus, then some of the chc1∆ spores would survive by receiving the scd1-v allele from the CHC1scd1-v parent. Twenty dissected tetrads segregated 2 viable:2 dead with all viable spores being CHC1 Leu- (Table 1). When we performed the same experiment in the presence of YCpCHC1 (URA3) the plasmid rescued the growth of chc1∆:LEU2 spores (Leu+Ura+), but we did not recover any Chc- spores growing in the absence of the plasmid (Leu+Ura-) (Table 1). This confirms that PAL2/YHR097C is the SCD1 locus, and scd1-i is a truncation mutant of PAL2.
Table 1. pal2∆ is synthetic lethal with chc1∆.
| Diploid genotype SL98 X SL7098 | No. of tetrads with ratio of viable to dead spores | No. of spores with phenotype | |||||
|---|---|---|---|---|---|---|---|
| 4:0 | 3:1 | 2:2 | Leu-Ura± | Leu+Ura+ | Leu+Ura- | Dead | |
| chc1∆:LEU2/CHC1 scd1-i/pal2∆:NatMX6 | 0 | 0 | 20 | 40 | 0 | 0 | 40 |
| chc1∆:LEU2/CHC1 scd1-i/pal2∆:NatMX6 (YCp50-CHC1) | 11 | 10 | 11 | 58 | 38 | 0 | 32 |
Strains SL98 and SL7098 were crossed. The heterozygous diploid was then sporulated (in the absence and presence of plasmid YCp50-CHC1), followed by tetrad dissection. Data in the table represent the number of tetrads with ratio of viable to dead spores and number of spores with different phenotypes. If pal2∆ is integrated at the SCD1 locus, we expect no viable Leu+ Ura- (chc1∆) spores in the absence of the CHC1, URA3 plasmid, YCp50-CHC1.
We also tested whether pal1∆ is synthetic lethal with chc1∆, but as shown in the GAL1:CHC1 shut down experiments (Figure 2A), pal1∆ does not cause inviability with clathrin HC deficiency. This was confirmed in tetrads from crosses of pal1∆ scd1-v and chc1∆ YCpCHC1scd1-v, where viable chc1∆:LEU2pal1∆:KanMX6 spores were recovered (Table S4).
Localization of Pal2-GFP and Pal1-GFP
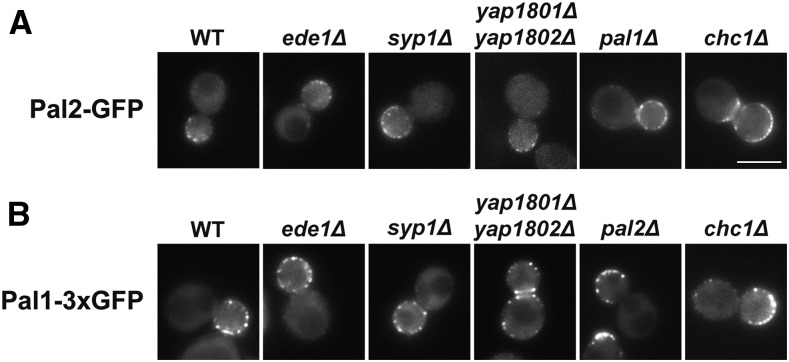
The clathrin endocytic pathway in yeast initiates with an immobile phase where the early endocytic factors, such as Syp1, Ede1 and clathrin, are first recruited to cortical patches, followed by the assembly of mid-late coat factors (e.g., Sla2, Sla1, Ent1/2). Ede1 and Syp1 leave the cortex just as the rapid mobile actin driven invagination phase commences, which is followed by scission of the coated vesicle. After the release of the nascent vesicle, it is rapidly uncoated and moves inwards and coat factors are recycled for the next round of endocytosis (Boettner et al. 2011, Weinberg and Drubin 2012, Goode et al. 2015). Previous studies have shown that Pal1-GFP forms patches at the cell cortex that are characteristic of endocytic coat proteins and it is considered an early arriving endocytic factor (Carroll et al. 2012). Global analysis of protein localization reported that GFP-tagged Pal2 localizes to the cytoplasm and nucleus (Huh et al. 2003). However, due to the homology with Pal1, we speculated that Pal2 might also localize to endocytic sites. We generated a strain expressing Pal2 with a C-terminal GFP tag. GFP-tagged Pal2 is functional as we obtained viable spores in genetic crosses with chc1∆ (not shown). Since Pal1-GFP was difficult to visualize in prior studies (Carroll et al. 2012), we generated a strain that expresses Pal1 with a C-terminal 3xGFP tag. However, pal1∆ has no phenotype even in combination with chc1∆ or pal2∆ (see below), so it was not possible to confirm Pal1-3xGFP function. But it behaved much like Pal1-GFP studied previously (Carroll et al. 2012). When we visualized the Pal proteins in live cells, we found more than 76% of cells (n = 60) had cortical patch localization (Figure 3A, 3B), a characteristic of endocytic coat proteins. Also, in addition to the cell cortex, Pal2-GFP was observed at the bud neck (32% of cells; n = 60), similar to Pal1-3xGFP (38% of cells; n = 60; Figure 3A, 3B).
Figure 3.
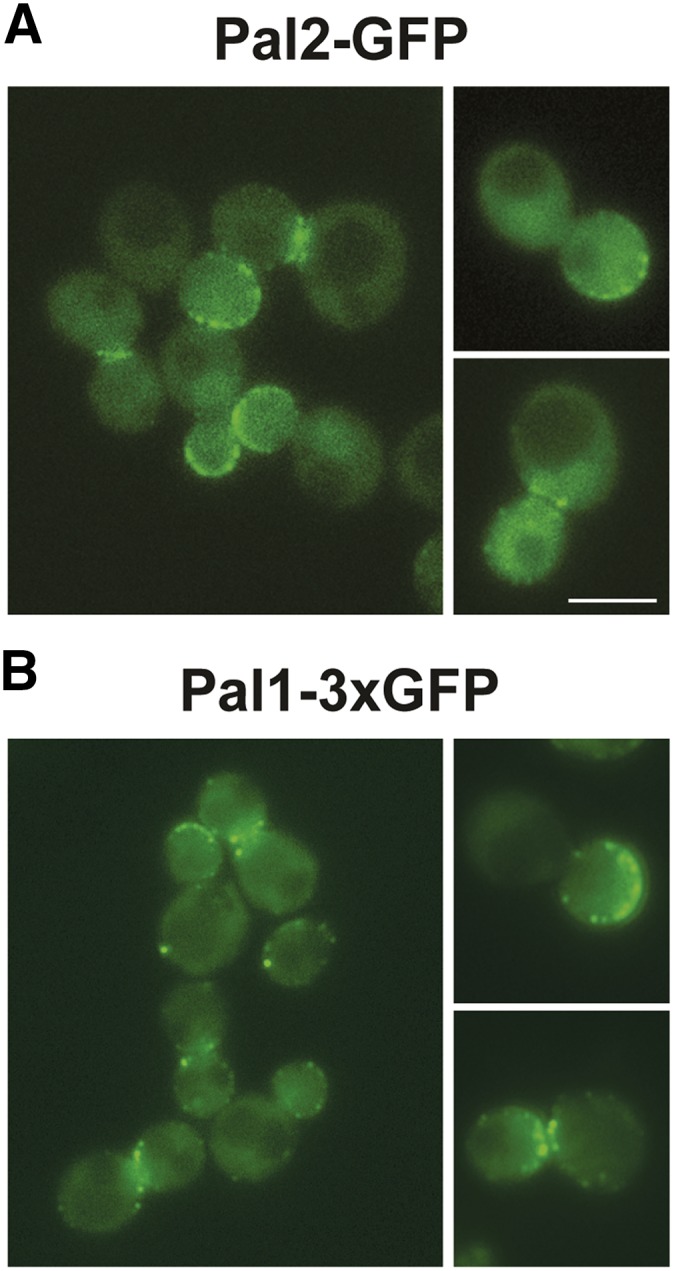
Pal2 and Pal1 localize to to cortical patches. (A) Pal2-GFP (SL7455); (B) Pal1-3xGFP (SL7335) were imaged by fluorescence microscopy. Shown are images from a single medial plane of a z-stack. Scale bar: 5μm.
Since pal2∆ shows synthetic lethality with chc1∆ and its paralog Pal1 is an early endocytic factor, we next tested growth phenotypes of pal2∆ with deletions of PAL1 and other early endocytic factor genes, EDE1, SYP1 and YAP1801/2 at normal (30°) and elevated temperatures (34° and 37°), but saw no effect either in tetrads or by direct gene deletions (data not shown). We then examined whether early endocytic proteins, Ede1, Syp1, Yap1801/2, and clathrin HC (Kaksonen et al. 2005, Newpher et al. 2005, Toshima et al. 2006, Boettner et al. 2009, Carroll et al. 2009, Reider et al. 2009, Stimpson et al. 2009, Carroll et al. 2012), are required for the recruitment of Pal2-GFP or Pal1-3xGFP to cortical sites. However, both Pal2-GFP and Pal1-3xGFP localization to cortical patches and the bud neck in null mutants of each of these genes was similar to the wildtype (Figures 4A, 4B and data not shown). We also performed cortical patch to cytosol fluorescence intensity ratio analysis to assess whether there was any defect in localization of Pal proteins in these endocytic mutants. However, there was no significant difference in patch to cytosol fluorescence intensity ratio of the Pal proteins in the mutants compared to the wildtype strain (Figure S3A & S3B). These results suggest that the early endocytic factors tested are not required cortical recruitment factors for the Pal proteins. We also verified that Pal1 and Pal2 do not affect each other’s localization (≥78% of cells had cortical patches; n = 45, upon deletion of the paralog; Figure 4A & 4B), and thus their recruitment to the cell surface is not interdependent.
Figure 4.
Localization of Pal2-GFP and Pal1-3xGFP in the absence of other endocytic factors. (A) Yeast strains expressing Pal2-GFP (WT; SL7455) and in ede1∆ (SL7505), syp1∆ (SL7405), yap1801/2∆ (SL7401), pal1∆ (SL7475), chc1∆ (SL7451). (B) Yeast strains expressing Pal1-3xGFP (WT; SL7335) and in ede1∆ (SL7502), syp1∆ (SL7579), yap1801/2∆ (SL7581), pal2∆ (SL7400), chc1∆ (SL7468). Images shown are from a single plane of a z-stack focused on the middle of the cell. Scale bar: 5μm.
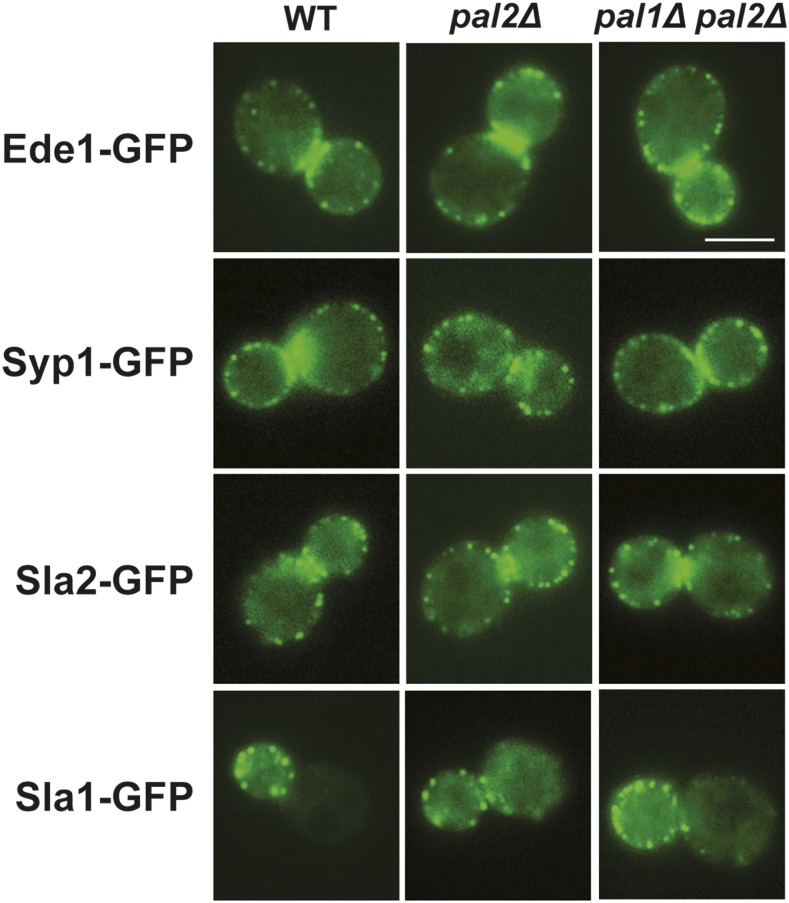
To determine whether Pal2 and Pal1 have a role in the recruitment of other endocytic factors, we examined the localization of GFP tagged Ede1, Syp1, Sla2 and Sla1 in pal2∆ or pal1∆ pal2∆ cells. However, no significant difference in the localization of these endocytic factors was observed compared to wild type (Figure 5). We also noted that there was more cytosolic Sla1 in pal2∆ or pal1∆ pal2∆ cells (Figure 5 and see cortical patch to cytosol fluorescence intensity ratio in Figure S4). Altogether, our data suggest that Pal2 and Pal1 localization at endocytic sites is independent of early endocytic factors and both proteins are not required for recruitment of Ede1, Syp1 and Sla2, but they affect recruitment of Sla1 to the cortex during the immobile phase of internalization.
Figure 5.
Recruitment of endocytic factors does not depend on Pal2 and Pal1. Localization of Ede1-GFP (WT; SL5755), Syp1-GFP (WT; SL5806), Sla2-GFP (WT; SL5927), Sla1-GFP (WT; SL5311); in pal2∆ (SL 7330; 7298; 7295 and 7301 respectively) and in pal1∆ pal2∆ (SL 7500; 7473; 7497 and 7476 respectively). Scale bar: 5μm.
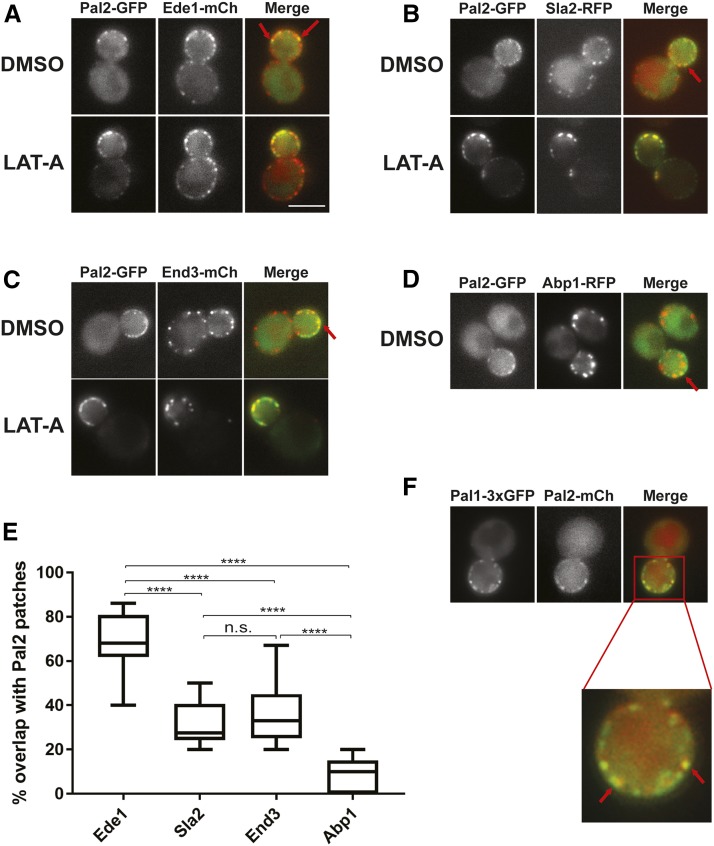
Pal2 localizes to cortical patches containing other endocytic factors
To determine whether Pal2 is actually in endocytic sites at the cortex, we tested for co-localization of Pal2-GFP with endocytic factors that mark different phases of clathrin-mediated endocytosis. These include Ede1 – early immobile phase, Sla2 and End3 – mid/late immobile phase, and Abp1 – actin/mobile phase (Boettner et al. 2011). To enhance any co-localization signal, we treated cells with Latrunculin-A (LAT-A), a drug that inhibits actin polymerization by sequestering monomers of actin and thereby stalling the endocytosis process (Ayscough 1998). The Pal2-GFP signal overlapped significantly with Ede1-mCherry, Sla2-mCherry and End3-mCherry, even in the absence of LAT-A (DMSO control cells) (Figure 6A – 6C). However, Pal2-GFP appeared to be largely at the surface more like Ede1 and not in invaginating structures. Consistent with this Pal2-GFP did not show significant overlap with Abp1-RFP (Figure 6D), which marks the mobile invagination phase of internalization and arrives at or just after Syp1 and Ede1 leave the cell surface (Boettner et al. 2009, Stimpson et al. 2009).
Figure 6.
Localization of Pal2-GFP with other endocytic factors. Representative micrographs of yeast cells co-expressing Pal2-GFP with Ede1-mCherry (A; SL7483), Sla2-RFP (B; SL7479), End3-mCherry (C; SL7487), Abp1-RFP (D; SL7411) grown to log phase and treated for 1 h with DMSO vehicle control or 200 μM Latrunculin A (LAT-A). Red arrows indicate examples of co-localization. (E) Whisker plot showing the percentage overlap of Ede1, Sla2, End3 and Abp1 patches with Pal2 patches. Two-tailed t-test was performed for all the pairs. Except for the Sla2-End3 pair which is not significant (n.s.), all other pairs were significant at P < 0.0001 indicated by “****”. (F). Co-localization of Pal2-mCherry and Pal1-3xGFP (SL7459). Images shown are from a single medial plane of a z-axis collection. Scale bar: 5μm.
We also quantified the number of Pal2 patches that contained the early (Ede1) and middle/late endocytic coat factors (Sla2/End3), as well as actin (Abp1). Consistent with Pal2 arriving early at endocytic sites, 68% of Pal2 patches contained Ede1 (n = 111 from 18 cells), 31% contained Sla2 (n = 67 from 12 cells), 35% contained End3 (n = 56 from 13 cells), and only 8% contained Abp1 (n = 77 from 11 cells) (Figure 6E). As predicted, Pal1-3xGFP and Pal2 tagged with mCherry also colocalized at cortical sites (67%; n = 78 from 11 cells; Figure 6F). This supports the idea that Pal2 arrives before Sla2, End3 and Abp1. Overall these results suggest that Pal2 is an early endocytic factor that may leave the cortex without internalization, possibly similar to Ede1 and Syp1(Boettner et al. 2009, Reider et al. 2009, Stimpson et al. 2009).
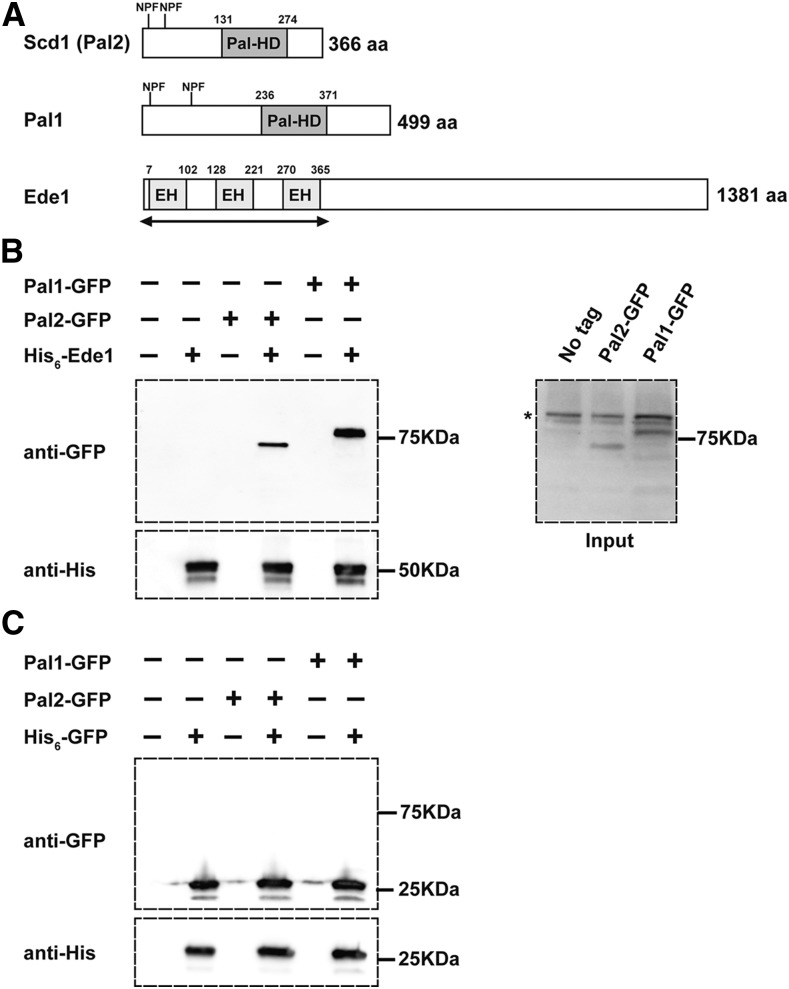
Eps15 homology (EH)-domain containing protein, Ede1, interacts with Pal2 and Pal1
Ede1, the homolog of mammalian Eps15 (Gagny et al. 2000), contains 3 N-terminal EPS15 homology (EH) domains (EH1: 7-102; EH2: 128-221; EH3: 270-365) which recognize the Asn-Pro-Phe (NPF) motifs (Figure 7A) in target ligands (de Beer et al. 2000, Gagny et al. 2000, Morgan et al. 2003, Suzuki et al. 2012). A previous study used immunoprecipitation to show Pal1 interacts with Ede1 in whole cell extracts (Carroll et al. 2012). To investigate whether Pal2 interacts with Ede1, we generated a recombinant protein expressing only the 3 EH domains (EH1-3) of Ede1 fused to a His6-tag (His6-Ede1 (EH1-3)) for pull downs of Pal1-1xGFP or Pal2-1xGFP from yeast cell extracts. Both Pal2 and Pal1 associated with His6-Ede1 (EH1-3) (Figure 7B), but not a His6-tagged-GFP control (Figure 7C). Our results indicate that both Pal1 and Pal2 associate with the N-terminal EH domain region of Ede1.
Figure 7.
Interaction between Pal2-GFP and Pal1-GFP with His6-Ede1(EH1-3). (A) Schematic representation of Pal2, Pal1 and Ede1 proteins showing the Pal homology domain and NPF motifs in Pal2 and Pal1; and 3 EH-domains in Ede1. The double arrow under the Ede1 EH domain is the region used in the pulldown experiments in panel B. (B) Protein lysates from yeast cells expressing Pal2-GFP (SL7455) or Pal1-GFP (SL7442) were incubated with Ni-NTA agarose beads containing recombinant His6-Ede1(EH1-3). Both Pal2 and Pal1 co-purify with Ede1 and are detected by anti-GFP antibody. Inputs containing Pal2-GFP and Pal1-GFP are shown at the right; ‘*’ shows non-specific bands. (C) Protein lysates as in (B) from yeast cells expressing Pal2-GFP or Pal1-GFP were incubated with Ni-NTA agarose beads containing recombinant His6-GFP (His6-GFP) as a control. Neither Pal2 nor Pal1 co-purifies with His6-GFP.
Discussion
It has been over 30 years since the identification of a polymorphism in the gene referred to as SCD1, suppressor of clathrin deficiency, where one allele (scd1-i) resulted in inviability of clathrin HC deficient yeast, but the other (scd1-v) allowed survival of cells lacking clathrin HC (Lemmon and Jones 1987). At the time this was surprising because the concept of synthetic lethality in yeast was relatively novel and there were limited examples in the literature (Huffaker et al. 1987, Kaiser and Schekman 1990). It was also unanticipated to discover this random polymorphism in what were considered to be relatively isogenic yeast strains (Mortimer and Johnston 1986). Controversy over this finding ensued because it was unexpected that yeast cells should even survive without clathrin (Payne and Schekman 1985, Lemmon and Jones 1987, Payne et al. 1987, Schekman and Payne 1988, Munn et al. 1991, Robinson 2015). Indeed, viable chc1∆ yeast exhibit very severe growth defects and a number of trafficking and other cell phenotypes, so their survival was considered tenuous at best. In fact, it was argued that mutations in many additional genes might cause a very sick strain to be inviable, and thus the possibility that the SCD1 locus would have anything to do with clathrin function seemed remote (Payne et al. 1987, Schekman and Payne 1988, Munn et al. 1991). Moreover, use of the term “suppressor” for this polymorphism, though genetically valid, may have implied to some that the “mutant allele” was a viability conferring “suppressing” allele (scd1-v) (Lemmon and Jones 1987, Schekman and Payne 1988) (see response by Lemmon & Jones to Schekman & Payne 1988). Also, some studies showed that knockouts of CHC1 in a number of different yeast strains usually led to viability of Chc-deficient yeast (Payne et al. 1987, Schekman and Payne 1988, Munn et al. 1991). This was used as an argument that scd1-i was the less common allele and likely an inactivating mutation in SCD1. However, a number of mutations are propagated in our commonly used yeast strains to make them more amenable to genetic manipulation, so this still left the nature of the SCD1 locus and the scd1-i allele unclear (see response of Lemmon & Jones to (Schekman and Payne 1988)).
The search for the SCD1 locus proved to be highly challenging. We initially used a multicopy suppression strategy, seeking genes whose overexpression could suppress the lethality of scd1-i clathrin HC-deficient cells in which CHC1 was under control of the repressible GAL1 promoter (Nelson and Lemmon 1993). We reasoned that overexpression might overcome the need to know which allele of SCD1 was dominant or recessive. In the process we identified SCD2-SCD6 and uncovered very interesting biology, including genes encoding proteins that linked directly to membrane trafficking and endocytosis (Nelson and Lemmon 1993, Gelperin et al. 1995, Nelson et al. 1996, Huang et al. 1997, Chang et al. 2002, Henry et al. 2002, Henry et al. 2003, Newpher et al. 2006, Chi et al. 2012). However, genetic analysis demonstrated that none of these multicopy suppressors are allelic to the SCD1 locus ((Nelson and Lemmon 1993, Gelperin et al. 1995, Nelson et al. 1996, Huang et al. 1997); Gelperin and S. Lemmon unpublished). A caveat to this approach was we did not know the nature of the SCD1 locus in existing plasmid libraries. In addition, other genetic mapping strategies were thwarted by the propensity of clathrin-deficient yeast to become polyploid and their poor transformation efficiency (Lemmon and Jones 1987, Lemmon et al. 1990).
The advent of powerful pooled linkage analysis and whole genome sequencing (Birkeland et al. 2010, Song et al. 2014, Lang et al. 2015, Linder et al. 2016) allowed us to finally identify the SCD1 locus, solving this long unresolved question. We found that scd1-i is a mutation in YHR097c, also referred to as PAL2 due to the existence of a paralogue, PAL1 (Carroll et al. 2012, Brach et al. 2014). The scd1-i allele has a stop codon that results in a truncated protein, and the scd1-v allele is the wild type SCD1/PAL2 gene. This confirms that it is a loss of function allele that is synthetic lethal with clathrin HC deficiency. Of particular interest, though, our data indicate that the Pal2/Scd1 protein plays a role in clathrin-mediated endocytosis, arguing against the concept that this mutation was not likely relevant to clathrin.
The first Pal1 protein was characterized in S. pombe cells, where it localizes to the cell tips during interphase and at the cell division plane during mitosis and cytokinesis (Ge et al. 2005). A null mutation causes morphological and polarity phenotypes, including pear shaped and spherical cells, thus the name Pal1 for pears and lemons. A possible role in endocytosis was suggested by an association with Sla2, related to HIP1/R in mammalian cells (Ge et al. 2005). More recently Pal1 was studied in S. cerevisiae where it was found to localize to sites of clathrin-mediated endocytosis and behave like an early endocytic coat factor (Carroll et al. 2012). Our studies here indicate its paralog, Pal2, also may play an endocytic role. It localizes to cortical patches containing other endocytic coat factors, including Pal1, and it interacts with the early coat factor Ede1, like Pal1. For technical reasons, we have not been able to study Pal2’s dynamics by live cell imaging. However, it appears to localize more like early endocytic factors that do not internalize, such as Ede1 and Syp1. There was no obvious displacement from the cortex, and limited colocalization with actin as seen in factors found in invaginating vesicles. This would be a distinction from Pal1 which seems to internalize with endocytic vesicles (Carroll et al. 2012).
A few questions still need to be answered about the Pal proteins in yeast. First, are Pal1 and Pal2 redundant? Arguing against this idea is pal2∆ is synthetic lethal with chc1∆, while pal1∆ is not. However, it could be that Pal2 is just more abundant than Pal1, so depletion of Pal2 has more of an effect in the absence of clathrin HC. However, proteomic quantification of the number of Pal proteins per cell showed little difference in abundance (Saccharomyces Genome Database, (Kulak et al. 2014)).
Furthermore, we still don’t know what are the exact function(s) of the Pal proteins. There is no obvious phenotype of the null mutations alone or in combination, or combined with other early endocytic factors tested so far, except for the genetic interaction of PAL2 with CHC1. Although, further supporting an endocytic role for the Pal proteins, pal2∆ and pal1∆pal2∆ did lead to an increase in the cytoplasmic level of Sla1 compared to the wild type strain. This is similar to the effects seen on Sla1 when genes for other early endocytic factors are deleted (Kaksonen et al. 2005, Brach et al. 2014). Also, both proteins have NPF motifs and bind to the EH region of Ede1. Prior studies suggested that Ede1 is needed to recruit Pal1 to the cell cortex (Carroll et al. 2012), but our results did not confirm these findings, as both Pal1 and Pal2 were in cortical patches without Ede1.
It is possible that the Pal proteins have an endocytic function that is only critical under certain conditions, such as cell stress or during morphogenesis events like mating. Alternatively, they are selective cargo adaptors for membrane proteins that have yet to be identified. Deletions of AP1801/2, yeast AP2, and Syp1 adaptors cause no major general growth and limited endocytic defects (Huang et al. 1999, Kaksonen et al. 2005, Boettner et al. 2009, Stimpson et al. 2009), but they participate in internalization of specific cargos. AP180’s are adaptors for Snc1, a vesicle SNARE (Burston et al. 2009), while AP2 is important for Killer Toxin toxicity (Carroll et al. 2009) and for internalization of the stress sensor Mid2 (Chapa-y-Lazo et al. 2014). Mid2 is also a cargo of Syp1 (Reider et al. 2009), a F-Bar -µHomology domain protein. Syp1’s uHomology region was recently shown to interact with cargo with DxY motifs, including those in Mid2, Snc1, Ptr2 and Mep3 (Apel et al. 2017). Perhaps the Pal1 homology domain also binds specific cargo motifs. Further studies will be needed to investigate these possibilities and to understand the roles of the Pal proteins.
Acknolwedgements
We thank Dr. Beverly Wendland and Dr. Derek Prosser for the pET28c-EDE1(EH1-3) construct. We are grateful to Dr. Patel and Dr. Zhang for the gift of HIS6-GFP. This work was supported by National Institutes of Health grants R01 GM120767 to T.E.W. and R01-GM055796 to S. K. L.
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25387/g3.7582868.
Communicating editor: N. Rhind
Literature Cited
- Apel A. R., Hoban K., Chuartzman S., Tonikian R., Sidhu S., et al. , 2017. Syp1 regulates the clathrin-mediated and clathrin-independent endocytosis of multiple cargo proteins through a novel sorting motif. Mol. Biol. Cell 28: 2434–2448. 10.1091/mbc.e15-10-0731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayscough K., 1998. Use of latrunculin-A, an actin monomer-binding drug. Methods Enzymol. 298: 18–25. 10.1016/S0076-6879(98)98004-1 [DOI] [PubMed] [Google Scholar]
- Birkeland S. R., Jin N., Ozdemir A. C., Lyons R. H., Jr., Weisman L. S., et al. , 2010. Discovery of mutations in Saccharomyces cerevisiae by pooled linkage analysis and whole-genome sequencing. Genetics 186: 1127–1137. 10.1534/genetics.110.123232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boettner D. R., Chi R. J., Lemmon S. K., 2011. Lessons from yeast for clathrin-mediated endocytosis. Nat. Cell Biol. 14: 2–10. 10.1038/ncb2403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boettner D. R., D’Agostino J. L., Torres O. T., Daugherty-Clarke K., Uygur A., et al. , 2009. The F-BAR protein Syp1 negatively regulates WASp-Arp2/3 complex activity during endocytic patch formation. Curr. Biol. 19: 1979–1987. 10.1016/j.cub.2009.10.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifacino J. S., Glick B. S., 2004. The mechanisms of vesicle budding and fusion. Cell 116: 153–166. 10.1016/S0092-8674(03)01079-1 [DOI] [PubMed] [Google Scholar]
- Brach T., Godlee C., Moeller-Hansen I., Boeke D., Kaksonen M., 2014. The initiation of clathrin-mediated endocytosis is mechanistically highly flexible. Curr. Biol. 24: 548–554. 10.1016/j.cub.2014.01.048 [DOI] [PubMed] [Google Scholar]
- Burston H. E., Maldonado-Baez L., Davey M., Montpetit B., Schluter C., et al. , 2009. Regulators of yeast endocytosis identified by systematic quantitative analysis. J. Cell Biol. 185: 1097–1110. 10.1083/jcb.200811116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne K. P., Wolfe K. H., 2005. The Yeast Gene Order Browser: combining curated homology and syntenic context reveals gene fate in polyploid species. Genome Res. 15: 1456–1461. 10.1101/gr.3672305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll S. Y., Stimpson H. E., Weinberg J., Toret C. P., Sun Y., et al. , 2012. Analysis of yeast endocytic site formation and maturation through a regulatory transition point. Mol. Biol. Cell 23: 657–668. 10.1091/mbc.e11-02-0108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll S. Y., Stirling P. C., Stimpson H. E., Giesselmann E., Schmitt M. J., et al. , 2009. A yeast killer toxin screen provides insights into a/b toxin entry, trafficking, and killing mechanisms. Dev. Cell 17: 552–560. 10.1016/j.devcel.2009.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J. S., Henry K., Wolf B. L., Geli M., Lemmon S. K., 2002. Protein phosphatase-1 binding to scd5p is important for regulation of actin organization and endocytosis in yeast. J. Biol. Chem. 277: 48002–48008. 10.1074/jbc.M208471200 [DOI] [PubMed] [Google Scholar]
- Chapa-y-Lazo B., Allwood E. G., Smaczynska-de R., II, Snape M. L., Ayscough K. R., 2014. Yeast endocytic adaptor AP-2 binds the stress sensor Mid2 and functions in polarized cell responses. Traffic 15: 546–557. 10.1111/tra.12155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi R. J., Torres O. T., Segarra V. A., Lansley T., Chang J. S., et al. , 2012. Role of Scd5, a protein phosphatase-1 targeting protein, in phosphoregulation of Sla1 during endocytosis. J. Cell Sci. 125: 4728–4739. 10.1242/jcs.098871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Beer T., Hoofnagle A. N., Enmon J. L., Bowers R. C., Yamabhai M., et al. , 2000. Molecular mechanism of NPF recognition by EH domains. Nat. Struct. Biol. 7: 1018–1022. 10.1038/80924 [DOI] [PubMed] [Google Scholar]
- Edeling M. A., Smith C., Owen D., 2006. Life of a clathrin coat: insights from clathrin and AP structures. Nat. Rev. Mol. Cell Biol. 7: 32–44. 10.1038/nrm1786 [DOI] [PubMed] [Google Scholar]
- Elkin S. R., Lakoduk A. M., Schmid S. L., 2016. Endocytic pathways and endosomal trafficking: a primer. Wien. Med. Wochenschr. 166: 196–204. 10.1007/s10354-016-0432-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel S. R., Dietrich F. S., Fisk D. G., Binkley G., Balakrishnan R., et al. , 2014. The reference genome sequence of Saccharomyces cerevisiae: then and now. G3 (Bethesda) 4: 389–398. 10.1534/g3.113.008995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagny B., Wiederkehr A., Dumoulin P., Winsor B., Riezman H., et al. , 2000. A novel EH domain protein of Saccharomyces cerevisiae, Ede1p, involved in endocytosis. J. Cell Sci. 113: 3309–3319. [DOI] [PubMed] [Google Scholar]
- Ge W., Chew T. G., Wachtler V., Naqvi S. N., Balasubramanian M. K., 2005. The novel fission yeast protein Pal1p interacts with Hip1-related Sla2p/End4p and is involved in cellular morphogenesis. Mol. Biol. Cell 16: 4124–4138. 10.1091/mbc.e04-11-0976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelperin D., Weigle J., Nelson K., Roseboom P., Irie K., et al. , 1995. 14–3-3 proteins: potential roles in vesicular transport and Ras signaling in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 92: 11539–11543. 10.1073/pnas.92.25.11539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Navarro N., Miller E. A., 2016. COP-coated vesicles. Curr. Biol. 26: R54–R57. 10.1016/j.cub.2015.12.017 [DOI] [PubMed] [Google Scholar]
- Goode B. L., Eskin J. A., Wendland B., 2015. Actin and endocytosis in budding yeast. Genetics 199: 315–358. 10.1534/genetics.112.145540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry K. R., D’Hondt K., Chang J., Newpher T., Huang K., et al. , 2002. Scd5p and clathrin function are important for cortical actin organization, endocytosis, and localization of sla2p in yeast. Mol. Biol. Cell 13: 2607–2625. 10.1091/mbc.e02-01-0012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry K. R., D’Hondt K., Chang J. S., Nix D. A., Cope M. J., et al. , 2003. The actin-regulating kinase Prk1p negatively regulates Scd5p, a suppressor of clathrin deficiency, in actin organization and endocytosis. Curr. Biol. 13: 1564–1569. 10.1016/S0960-9822(03)00579-7 [DOI] [PubMed] [Google Scholar]
- Huang K. M., D’Hondt K., Riezman H., Lemmon S. K., 1999. Clathrin functions in the absence of heterotetrameric adaptors and AP180-related proteins in yeast. EMBO J. 18: 3897–3908. 10.1093/emboj/18.14.3897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K. M., Gullberg L., Nelson K. K., Stefan C. J., Blumer K., et al. , 1997. Novel functions of clathrin light chains: clathrin heavy chain trimerization is defective in light chain-deficient yeast. J. Cell Sci. 110: 899–910. [DOI] [PubMed] [Google Scholar]
- Huffaker T. C., Hoyt M. A., Botstein D., 1987. Genetic analysis of the yeast cytoskeleton. Annu. Rev. Genet. 21: 259–284. 10.1146/annurev.ge.21.120187.001355 [DOI] [PubMed] [Google Scholar]
- Huh W. K., Falvo J. V., Gerke L. C., Carroll A. S., Howson R. W., et al. , 2003. Global analysis of protein localization in budding yeast. Nature 425: 686–691. 10.1038/nature02026 [DOI] [PubMed] [Google Scholar]
- Jones G. M., Stalker J., Humphray S., West A., Cox T., et al. , 2008. A systematic library for comprehensive overexpression screens in Saccharomyces cerevisiae. Nat. Methods 5: 239–241. 10.1038/nmeth.1181 [DOI] [PubMed] [Google Scholar]
- Kaiser C. A., Schekman R., 1990. Distinct sets of SEC genes govern transport vesicle formation and fusion early in the secretory pathway. Cell 61: 723–733. 10.1016/0092-8674(90)90483-U [DOI] [PubMed] [Google Scholar]
- Kaksonen M., Toret C. P., Drubin D. G., 2005. A modular design for the clathrin- and actin-mediated endocytosis machinery. Cell 123: 305–320. 10.1016/j.cell.2005.09.024 [DOI] [PubMed] [Google Scholar]
- Kirchhausen T., Nathanson K. L., Matsui W., Vaisberg A., Chow E. P., et al. , 1989. Structural and functional division into two domains of the large (100- to 115-kDa) chains of the clathrin-associated protein complex AP-2. Proc. Natl. Acad. Sci. USA 86: 2612–2616. 10.1073/pnas.86.8.2612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchhausen T., Owen D., Harrison S. C., 2014. Molecular structure, function, and dynamics of clathrin-mediated membrane traffic. Cold Spring Harb. Perspect. Biol. 6: a016725 10.1101/cshperspect.a016725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovar D. R., Wu J. Q., Pollard T. D., 2005. Profilin-mediated competition between capping protein and formin Cdc12p during cytokinesis in fission yeast. Mol. Biol. Cell 16: 2313–2324. 10.1091/mbc.e04-09-0781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulak N. A., Pichler G., Paron I., Nagaraj N., Mann M., 2014. Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nat. Methods 11: 319–324. 10.1038/nmeth.2834 [DOI] [PubMed] [Google Scholar]
- Lang A., John Peter A. T., Kornmann B., 2015. ER-mitochondria contact sites in yeast: beyond the myths of ERMES. Curr. Opin. Cell Biol. 35: 7–12. 10.1016/j.ceb.2015.03.002 [DOI] [PubMed] [Google Scholar]
- Lemmon S. K., Freund C., Conley K., Jones E. W., 1990. Genetic instability of clathrin-deficient strains of Saccharomyces cerevisiae. Genetics 124: 27–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon S. K., Jones E. W., 1987. Clathrin requirement for normal growth of yeast. Science 238: 504–509. 10.1126/science.3116672 [DOI] [PubMed] [Google Scholar]
- Li H., Durbin R., 2010. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26: 589–595. 10.1093/bioinformatics/btp698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder R. A., Seidl F., Ha K., Ehrenreich I. M., 2016. The complex genetic and molecular basis of a model quantitative trait. Mol. Biol. Cell 27: 209–218. 10.1091/mbc.E15-06-0408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M. S., McKenzie A., 3rd, Demarini D. J., Shah N. G., Wach A., et al. , 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14: 953–961. [DOI] [PubMed] [Google Scholar]
- Lu R., Drubin D. G., Sun Y., 2016. Clathrin-mediated endocytosis in budding yeast at a glance. J. Cell Sci. 129: 1531–1536. 10.1242/jcs.182303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon H. T., Mills I. G., 2004. COP and clathrin-coated vesicle budding: different pathways, common approaches. Curr. Opin. Cell Biol. 16: 379–391. 10.1016/j.ceb.2004.06.009 [DOI] [PubMed] [Google Scholar]
- Morgan J. R., Prasad K., Jin S., Augustine G. J., Lafer E. M., 2003. Eps15 homology domain-NPF motif interactions regulate clathrin coat assembly during synaptic vesicle recycling. J. Biol. Chem. 278: 33583–33592. 10.1074/jbc.M304346200 [DOI] [PubMed] [Google Scholar]
- Mortimer R. K., Johnston J. R., 1986. Genealogy of principal strains of the yeast genetic stock center. Genetics 113: 35–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller S. C., Branton D., 1984. Identification of coated vesicles in Saccharomyces cerevisiae. J. Cell Biol. 98: 341–346. 10.1083/jcb.98.1.341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munn A. L., Silveira L., Elgort M., Payne G. S., 1991. Viability of clathrin heavy-chain-deficient Saccharomyces cerevisiae is compromised by mutations at numerous loci: implications for the suppression hypothesis. Mol. Cell. Biol. 11: 3868–3878. 10.1128/MCB.11.8.3868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson K. K., Holmer M., Lemmon S. K., 1996. SCD5, a suppressor of clathrin deficiency, encodes a novel protein with a late secretory function in yeast. Mol. Biol. Cell 7: 245–260. 10.1091/mbc.7.2.245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson K. K., Lemmon S. K., 1993. Suppressors of clathrin deficiency: overexpression of ubiquitin rescues lethal strains of clathrin-deficient Saccharomyces cerevisiae. Mol. Cell. Biol. 13: 521–532. 10.1128/MCB.13.1.521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newpher T. M., Idrissi F. Z., Geli M. I., Lemmon S. K., 2006. Novel function of clathrin light chain in promoting endocytic vesicle formation. Mol. Biol. Cell 17: 4343–4352. 10.1091/mbc.e06-07-0606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newpher T. M., Lemmon S. K., 2006. Clathrin is important for normal actin dynamics and progression of Sla2p-containing patches during endocytosis in yeast. Traffic 7: 574–588. 10.1111/j.1600-0854.2006.00410.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newpher T. M., Smith R. P., Lemmon V., Lemmon S. K., 2005. In vivo dynamics of clathrin and its adaptor-dependent recruitment to the actin-based endocytic machinery in yeast. Dev. Cell 9: 87–98. 10.1016/j.devcel.2005.04.014 [DOI] [PubMed] [Google Scholar]
- Payne G. S., Baker D., van Tuinen E., Schekman R., 1988. Protein transport to the vacuole and receptor-mediated endocytosis by clathrin heavy chain-deficient yeast. J. Cell Biol. 106: 1453–1461. 10.1083/jcb.106.5.1453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne G. S., Hasson T. B., Hasson M. S., Schekman R., 1987. Genetic and biochemical characterization of clathrin-deficient Saccharomyces cerevisiae. Mol. Cell. Biol. 7: 3888–3898. 10.1128/MCB.7.11.3888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne G. S., Schekman R., 1985. A test of clathrin function in protein secretion and cell growth. Science 230: 1009–1014. 10.1126/science.2865811 [DOI] [PubMed] [Google Scholar]
- Payne G. S., Schekman R., 1989. Clathrin: a role in the intracellular retention of a Golgi membrane protein. Science 245: 1358–1365. 10.1126/science.2675311 [DOI] [PubMed] [Google Scholar]
- Pearse B. M., Robinson M. S., 1990. Clathrin, adaptors, and sorting. Annu. Rev. Cell Biol. 6: 151–171. 10.1146/annurev.cb.06.110190.001055 [DOI] [PubMed] [Google Scholar]
- Reider A., Barker S. L., Mishra S. K., Im Y. J., Maldonado-Baez L., et al. , 2009. Syp1 is a conserved endocytic adaptor that contains domains involved in cargo selection and membrane tubulation. EMBO J. 28: 3103–3116. 10.1038/emboj.2009.248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M. S., 2015. Forty Years of Clathrin-coated Vesicles. Traffic 16: 1210–1238. 10.1111/tra.12335 [DOI] [PubMed] [Google Scholar]
- Schekman R., Payne G., 1988. Clathrin: a matter of life or death? Science 239: 919 10.1126/science.3277285 [DOI] [PubMed] [Google Scholar]
- Shendure J., Ji H., 2008. Next-generation DNA sequencing. Nat. Biotechnol. 26: 1135–1145. 10.1038/nbt1486 [DOI] [PubMed] [Google Scholar]
- Silveira L. A., Wong D. H., Masiarz F. R., Schekman R., 1990. Yeast clathrin has a distinctive light chain that is important for cell growth. J. Cell Biol. 111: 1437–1449. 10.1083/jcb.111.4.1437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Q., Johnson C., Wilson T. E., Kumar A., 2014. Pooled segregant sequencing reveals genetic determinants of yeast pseudohyphal growth. PLoS Genet. 10: e1004570 10.1371/journal.pgen.1004570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stimpson H. E., Toret C. P., Cheng A. T., Pauly B. S., Drubin D. G., 2009. Early-arriving Syp1p and Ede1p function in endocytic site placement and formation in budding yeast. Mol. Biol. Cell 20: 4640–4651. 10.1091/mbc.e09-05-0429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki R., Toshima J. Y., Toshima J., 2012. Regulation of clathrin coat assembly by Eps15 homology domain-mediated interactions during endocytosis. Mol. Biol. Cell 23: 687–700. 10.1091/mbc.e11-04-0380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toshima J. Y., Toshima J., Kaksonen M., Martin A. C., King D. S., et al. , 2006. Spatial dynamics of receptor-mediated endocytic trafficking in budding yeast revealed by using fluorescent alpha-factor derivatives. Proc. Natl. Acad. Sci. USA 103: 5793–5798. 10.1073/pnas.0601042103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg J., Drubin D. G., 2012. Clathrin-mediated endocytosis in budding yeast. Trends Cell Biol. 22: 1–13. 10.1016/j.tcb.2011.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yau R. G., Peng Y., Valiathan R. R., Birkeland S. R., Wilson T. E., et al. , 2014. Release from myosin V via regulated recruitment of an E3 ubiquitin ligase controls organelle localization. Dev. Cell 28: 520–533. 10.1016/j.devcel.2014.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Strains and plasmids are available upon request. Figure S1 shows the sequence alignment of the “Pal domain” of Pal2 and Pal1. Figure S2 shows the growth phenotype of GAL1:CHC1 strains with the indicated genotypes streaked on YEP+glucose at elevated temperatures. Figure S3 shows the patch to cytosol fluorescence intensity ratio for Pal2-GFP and Pal1-3xGFP in wild type and different endocytic mutants. Figure S4 shows the patch to cytosol fluorescence intensity ratio for Sla1-GFP in wild type and pal∆ mutants. Table S1 contains the yeast strains used for pooled linkage analysis and whole genome sequencing. Table S2 and Table S3 show the list of yeast strains and plasmids used in this study, respectively. Table S4 shows the tetrad data demonstrating that pal1∆ is not synthetic lethal with chc1∆. Supplemental material available at Figshare: https://doi.org/10.25387/g3.7582868.