Abstract
Chromatin insulators are DNA-protein complexes that regulate chromatin structure and gene expression in a wide range of organisms. These complexes also harbor enhancer blocking and barrier activities. Increasing evidence suggests that RNA molecules are integral components of insulator complexes. However, how these RNA molecules are involved in insulator function remains unclear. The Drosophila RNA-binding protein Shep associates with the gypsy insulator complex and inhibits insulator activities. By mutating key residues in the RRM domains, we generated a Shep mutant protein incapable of RNA-binding, and this mutant lost the ability to inhibit barrier activity. In addition, we found that one of many wildtype Shep isoforms but not RRM mutant Shep was sufficient to repress enhancer blocking activities. Finally, wildtype Shep rescued synthetic lethality of shep, mod(mdg4) double-mutants and developmental defects of shep mutant neurons, whereas mutant Shep failed to do so. These results indicate that the RNA-binding ability of Shep is essential for its ability to antagonize insulator activities and promote neuronal maturation. Our findings suggest that regulation of insulator function by RNA-binding proteins relies on RNA-mediated interactions.
Keywords: Chromatin insulator, Shep, RNA-binding, Enhancer blocking, Barrier activity
Chromatin insulators are DNA-protein complexes that play critical roles in shaping three-dimensional genome organization. Insulators are well known to block enhancer-promoter interactions when placed between the two elements (Bell et al. 1999). Furthermore, insulators can also function as barriers that prevent spreading of transcriptionally repressive chromatin to allow gene expression (West et al. 2002). There is increasing evidence indicating that RNA molecules are important functional components of insulator complexes in both Drosophila and mammals (Matzat et al. 2013; Kung et al. 2015). Various RNA-binding proteins have been shown to regulate insulator activities (Lei and Corces 2006; Moshkovich et al. 2011; Matzat et al. 2012; King et al. 2014), but a precise role for RNA-binding has not yet been elucidated.
We previously identified the RNA-binding protein Shep as a physical interactor of the gypsy insulator. Shep contains two RNA recognition motifs (RRMs) and binds thousands of transcripts (Dale et al. 2014; Olesnicky et al. 2018) as well as the core gypsy insulator protein components Su(Hw), Mod(mdg4)67.2, and CP190 (Matzat et al. 2012). Shep associates with the chromatin of thousands of genomic loci and overlaps extensively with core insulator proteins (Matzat et al. 2012; Dale et al. 2014). Furthermore, Shep regulates transcription of many of the genes with which Shep and insulator proteins associate (Chen et al. 2017a). Therefore, Shep may mediate a functional interaction between the gypsy insulator complex and RNA.
Shep was shown to act as an antagonist of gypsy insulator activity. Loss of Shep leads to increased enhancer blocking and barrier activities (Matzat et al. 2012). In addition, loss of Shep leads to synthetic lethality in the Mod(mdg4)67.2 null background, suggesting a critical functional interaction between the two factors to regulate development (Matzat et al. 2012). The shep gene encodes a large number of isoforms, and it remains unclear which isoform(s) participate in insulator antagonism. Previous work showed that ectopic expression of the Shep E isoform in a wildtype background can antagonize barrier activity (Matzat et al. 2012). However, existing shep mutants disrupt multiple isoforms, and specific isoform rescue experiments have not yet been performed.
In this study, we mutated key residues in the RRMs of Shep isoform E and verified loss of RNA-binding capability. We ectopically expressed wildtype and a Shep ERRM mutant isoform in vivo and found that the RNA-binding mutant lost the ability to disrupt barrier activity. Furthermore, we determined that expression of the wildtype Shep E but not the RRM mutant is capable of rescuing the defect in enhancer blocking antagonism of shep mutants. Finally, wildtype Shep E expression but not expression of the RRM mutant isoform rescues synthetic lethality of shep, mod(mdg4) double mutants and developmental defects of shep mutant neurons. Our results suggest that the RNA-binding capability of Shep is necessary to antagonize gypsy insulator activities and promote neuronal maturation.
Materials and Methods
Drosophila strains
Stocks were raised at 25° on standard cornmeal medium. pUASt-attB shep E and pUASt-attB shep E RRM mutant constructs were transformed using PhiC31 integrase into the attP40 docking site on chromosome 2L (BestGene, Inc.) and verified by genomic PCR and Western Blotting upon driving with Gal4. The ct6 phenotype was scored in flies on the first day after eclosion. Other fly strains used in this study include luciferase transgenes (insulated and uninsulated UAS-luciferase) (Markstein et al. 2008), shepd05714 (FBal0186064), Ser::Gal4 (BSC 6791), Mef2::Gal4 (BSC 27390), arm::Gal4 (BSC 1560), UAS-Su(Hw)-RNAi (10724 GD), shepBG00836 (FBal0157046), and ct6; mod(mdg4)u1 (BSC 59960).
Cloning
RNA-binding mutations in the Shep E isoform (Y64A, F66A, F69A in the RNP1 domain of RRM1 plus V162A, F164A, R166A in the RNP1 domain of RRM2) were designed based on structural and RNA-binding studies of the Drosophila Elav and Sxl RNA-binding proteins (Lee et al. 1997; Crowder et al. 1999; Lisbin et al. 2000). The Shep E RRM mutant was generated by PCR-based site directed mutagenesis of a pENTR/D-TOPO clone (Matzat et al. 2012) and cloned into pUASt-attB (Bischof et al. 2007) for expression in flies and pET101 for production of recombinant N-terminal tagged His-Shep E RRM mutant protein. All plasmids were sequenced for verification.
Recombinant protein production
Both wildtype and RRM mutant His-tagged Shep E were produced in E. coli BL21(DE3) (Rosetta, Novagen) cells and purified with Ni-NTA-agarose (Qiagen) using manufacturer protocols.
EMSA
Single stranded RNA probes were in vitro transcribed using 32P-UTP incorporation with T7 polymerase (MegaScript, Ambion) followed by removal of unincorporated nucleotides (NucAway, Ambion). 10 pmol of labeled probe was mixed with 10 μg tRNA in binding buffer (10% glycerol, 20 mM NaCl, 60 mM KCl, 20 mM HEPES pH 7.5) and heated to 95° for 2 min, then rapidly cooled on ice. 47 pmol protein was added and incubated on ice for 30 min. Samples were run at 4° initially at 1000 V for 1.5 min then at 250 V for approximately 3 h on a pre-run 6% GTG acrylamide gel (90 mM Tris, 30 mM taurine, 0.5 mM EDTA).
Immunostaining
P14 pupae were dissected and fixed with 4% paraformaldehyde for 1 h and incubated with anti-BURS (Peabody et al. 2008) (1:5,000) overnight. Secondary antibodies (Invitrogen) were used at 1:1,000, and samples were imaged as maximum-intensity z-series projections with a Zeiss 780 confocal microscope.
Primers for in vitro transcription of EMSA probes
Data and reagent availability
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the figures. All fly strains and reagents are available upon request.
Results
Generation of Shep RNA-binding mutant
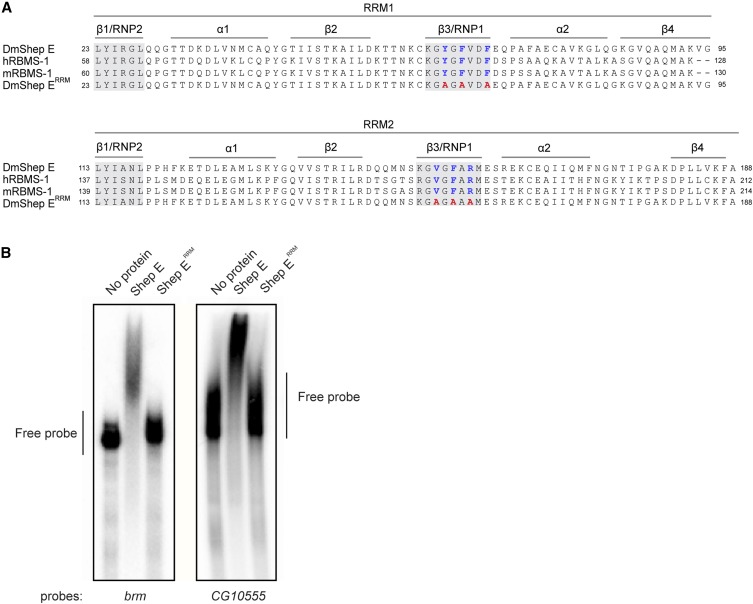
In order to test whether the RNA-binding capability of Shep is required for antagonism of gypsy insulator function, we generated point mutations in both conserved RNA-binding domains. We selected the E isoform of Shep because ectopic expression in muscle tissue was previously demonstrated to antagonize gypsy insulator barrier activity (Matzat et al. 2012). Shep harbors two RRMs, each with characteristic βαββαβ structure including RNP2 and RNP1 folds at β1 and β3, respectively (Figure 1A). Because the RNP folds are known to form the hydrophobic surface for RNA interaction, we generated three point mutations in each of the RNP1 folds in both RRM1 and RRM2 domains (Y64A, F66A, F69A in the RNP1 domain of RRM1 plus V162A, F164A, R166A in the RNP1 domain of RRM2). Mutation of each of the residues to alanine was chosen to reduce the likelihood of altered protein folding.
Figure 1.
Shep ERRM mutant is defective for RNA binding in vitro. A. Conservation of Shep E RRM domains compared to human and mouse orthologs. Consensus resides of RNP1 and RNP2 in RRM domains are shaded for Shep human and mouse orthologs. Residues mutated to alanine of the RNP1 domains are indicated as blue mutated to red. B. EMSA of wildtype Shep E vs. Shep ERRM mutant. 455nt ssRNA fragment of brm or 470nt ssRNA fragment of CG10555 was mixed with buffer, recombinant wildtype Shep E, or Shep ERRM mutant and run on a 6% GTG polyacrylamide gel.
In order to verify that the Shep ERRM mutant is disrupted for RNA-binding, we expressed recombinant His-tagged versions of wildtype and mutant Shep in E. coli and tested their RNA binding capability in electrophoretic mobility shift assays (EMSA). We generated single stranded 32P labeled RNA probes corresponding to fragments of two transcripts, brahma (455 nt) and CG10555 (470 nt), which are full length transcripts shown to stably associate with Shep in nuclear extracts of CNS-derived BG3 cells (Dale et al. 2014). Both RNA fragments contain preferred consensus binding sequences for Shep E previously identified by RNAcompete (Ray et al. 2013). As expected, we observed retarded mobility of both probes when wildtype Shep but not RRM mutant was added (Figure 1B).
Shep RNA-binding is required to antagonize barrier activity
We next generated transgenes of either wildtype Shep or RRM mutant under UAS control in order to achieve tissue-specific expression in flies. These constructs were integrated into the attP40 docking site on chromosome 2L using PhiC31 integrase. We verified by Western blotting that both transgenes express at equivalent levels when driven with arm::Gal4 (Figure 2A).
Figure 2.
Ectopic expression of wildtype Shep E but not Shep ERRM mutant decreased gypsy barrier activity in muscle. A. Anterior thirds of wildtype larvae expressing no transgene, UAS-shep E, or UAS-shep ERRM driven by arm::Gal4 were used for western blotting for Shep E and a loading control Protein on ecdysone puffs (Pep). Normalized band intensity of Shep E relative to Pep was quantified by CCD imaging of chemiluminescence followed by Photoshop analysis. B. Schematic diagram of UAS-luciferase system shows flanking gypsy insulator sequences act as a barrier to allow luciferase expression. C. Relative luciferase activity of non-insulated (left) or insulated (right) reporters in individual larvae expressing UAS-su(Hw) RNAi, UAS-shep E, or UAS-shep ERRM driven by Mef2::Gal4. Luciferase activities are reported as boxplots with boxes representing the first and third quartiles. Luciferase activities across genotypes were compared by One-way ANOVA followed by Tukey HSD post hoc tests, with statistical threshold at 0.05. For each genotype, luciferase signals were read for 12 individuals, each normalized to input protein level. Samples showing signal variation more than 100-fold from median of all replicates were discarded as outliers. At least 10 valid replicate samples of each genotype were used for statistics.
It was previously shown that ectopic expression of Shep E can antagonize gypsy insulator barrier activity in muscle tissue; therefore, we tested whether the RRM mutant retains this activity. We used the muscle-specific Mef2::Gal4 driver to activate expression of a UAS-luciferase reporter flanked by gypsy insulator sites inserted at the attP3 docking site (Matzat et al. 2012) (Figure 2B). High expression is achieved only if the reporter is insulated, and luciferase expression was highly reduced when Su(Hw) was depleted by RNAi (Figure 2C). When wildtype Shep E was ectopically expressed in this tissue, luciferase expression of the insulated reporter was significantly decreased, indicating reduced insulator activity. However, expression of Shep ERRM had no effect on luciferase activity. These results suggest that RNA-binding of Shep E is required to antagonize gypsy insulator barrier activity.
Shep RNA-binding is required to antagonize enhancer blocking activity and rescue synthetic lethality with loss of Mod(mdg4)67.2
Furthermore, we examined whether RNA-binding ability is required for Shep antagonism of insulator enhancer blocking activity. It was previously shown that the shepd05714 loss-of-function mutation restores enhancer blocking activity of the mod(mdg4)u1 null mutant at the ct6 locus (Matzat et al. 2012). This allele harbors an insertion of the gypsy retrotransposon between the wing margin enhancer and promoter of ct, resulting in reduced expression of ct and disruption of the wing margin (Figure 3A). Enhancer blocking activity of the mod(mdg4)u1 null mutant is low, but additional mutation of shepd05714 results in improvement of enhancer blocking activity. We found that expression of wildtype UAS-shep E in the shepd05714, mod(mdg4)u1 mutant background using the Ser::Gal4 driver substantially decreased enhancer blocking activity, represented as reduced wing notching (Figure 3A). However, similar expression of the UAS-shep ERRM mutant did not result in prominent rescue of wing morphology. Moreover, we found that expression of wildtype UAS-shep E rescued synthetic lethality observed in the shepd05714, mod(mdg4)u1 double mutant (Matzat et al. 2012), but expression of the RRM mutant had no effect on viability (Figure 3B).
Figure 3.
Wildtype Shep E but not Shep ERRM mutant rescues enhancer blocking antagonism of shep mutant and its synthetic lethality with mod(mdg4)u1. A. Schematic diagram and scoring of ct6 reporter activities. A gypsy retrotransposon situated between the ct promoter and enhancer disrupts ct expression and wing development. Insulator activity for ct6 was scored in female wings of indicated genotypes on a scale of 0-3. “0”, no notching; “1”, slight notching in distal tip of wing; “2”, moderate notching throughout distal proportion of wing; “3”, extensive notching in both distal and proximal wing. N represents the total number of flies scored. UAS-shep E or UAS-shep ERRM was driven with Ser::Gal4 driver. Kruskal-Wallis Test was performed (χ2df = 3 = 166.61, p-value < 2.2e-16) followed by post hoc pairwise Wilcox-Mann-Whitney U-tests. Statistical significance cutoff of Bonferroni-corrected p value was 0.05. B. Viability was scored in female flies of indicated genotypes. N represents the examined sample size for each genotype. UAS-shep E or UAS-shep ERRM was driven with Ser::Gal4 driver in homozygous mod(mdg4)u1 background. Pearson’s Chi Squared Test was performed (χ2df = 3 = 469.52, p-value < 2.2e-16) followed by post hoc pairwise Fisher’s Exact Tests. Statistical significance cutoff of Bonferroni-corrected p value was 0.05.
Shep regulation of neuronal development requires its RNA-binding capacity
Since Shep is required for proper neuronal maturation, we examined whether the RRM mutant was sufficient to rescue developmental defects of shep mutant neurons. Consistent with previous studies (Chen et al. 2014; Chen et al. 2017b), we observed smaller cell bodies and fewer projections of homozygous shepBG00836 bursicon neurons that are rescued by expression of wildtype Shep E (Figure 4A-C, E-G, and I-J). However, the RRM mutant Shep failed to rescue these morphological defects (Figure 4D and H-J). These results suggest that Shep RNA-binding capacity is essential for its regulation of neuronal development.
Figure 4.
Wildtype Shep E but not Shep ERRM mutant rescues developmental defects of shep mutant bursicon neurons. A-H. Homozygous but not heterozygous loss of shep (shepBG00836) leads to smaller cell bodies and fewer projections. Expression of wildtype Shep but not the RRM mutant using a Gal4 driver inserted into the shep locus rescues these morphological defects. Bursicon neurons and their projections are visualized by immunostaining with anti-BURS. I. Soma sizes are quantified by Photoshop and compared by One-way ANOVA followed by Tukey HSD post hoc tests, with statistical threshold at 0.05, ** P < 0.01. Sample sizes are indicated in parentheses. J. Projections of bursicon neurons are quantified by Sholl analysis. Sample sizes are indicated in parentheses.
Discussion
In this manuscript, we tested whether the RNA-binding capacity of Shep is essential for antagonism of gypsy insulator activities. We generated six point-mutations in the RNP1 domains of Shep E that caused Shep to become incapable of RNA binding in vitro. Subsequent in vivo insulator activity assays indicated that expression of wildtype Shep E was sufficient to inhibit gypsy enhancer blocking activity in a shep mutant genetic background. However, RRM mutant Shep failed to inhibit either enhancer blocking or barrier activities of the gypsy insulator. Moreover, RRM mutant Shep did not resuce the synthetic lethality of shep, mod(mdg4) double mutants or developmental defects of shep mutant neurons as did wildtype Shep E. Our results suggest that Shep antagonism of the gypsy insulator requires its RNA-binding capacity.
Shep E expression is sufficient to rescue phenotypes of a shep loss-of-function mutant
We showed that expression of Shep E was sufficient to restore the inhibition of gypsy enhancer blocking activities and the viability of the shep, mod(mdg4) double mutant. The observation further extends previous findings that Shep E is sufficient to inhibit the gypsy barrier activities (Matzat et al. 2012) and are consistent with the finding that expression of Shep E can rescue developmental defects of shep mutants (Chen et al. 2014). One of the shortest isoforms, Shep E contains both RRM domains, which are present in all isoforms, and therefore may represent a minimal isoform that may be partially functionally redundant with others. Since expression and function of Shep isoforms vary across tissues (Matzat et al. 2012), the diverse N termini may contain residues that are needed for tissue-specific functions. To date, FlyBase has reported eight different protein isoforms of Shep. However, we have cloned cDNAs that contain various alternatively spliced microexons that appear to further increase the complexity of protein isoforms and possibly their function (data not shown). Finally, extension of the 3′ UTR of shep has been shown to be regulated in a tissue and stage-specific manner (Hilgers et al. 2011; Smibert et al. 2012), suggesting an additional layer of isoform diversity. Future work will be required to address the function and regulation of these various Shep isoforms.
Shep may be recruited by RNA molecules to inhibit insulators
Our findings are consistent with a recently proposed model that Shep is co-transcriptionally recruited to regulate insulator activities. Previous studies observed that stably associated RNA immunoprecipitation targets of Shep are often derived from genes to which Shep binds in chromatin, suggesting Shep recruitment in cis (Dale et al. 2014; Chen et al. 2017a). Moreover, Shep-dependent genes revealed by RNA-seq of shep mutants are highly enriched as genomic and transcript binding targets of Shep, implying co-transcriptional regulation of gene expression by Shep (Chen et al. 2017a). Notably, a significant proportion of these genomic and transcript targets of Shep are also associated with Su(Hw) and Mod(mdg4)67.2 (Chen et al. 2017a), and Shep shares many other genomic and transcript targets with Su(Hw) (Matzat et al. 2012; Dale et al. 2014). Our findings further support that the ability of Shep to antagonize the gypsy insulator and regulate neuronal maturation relies on its RNA-binding capacity, raising the possibility that RNA-mediated interactions underlie these functions. These RNA molecules may be co-transcriptionally captured as insulator components that nucleate complexes or further recruit other regulatory factors. Future studies will address these intriguing possibilities.
Acknowledgments
We thank I. Bag for comments on the manuscript. This work was funded by the Intramural Program of the National Institute of Diabetes and Digestive and Kidney Diseases of the National Institutes of Health.
Footnotes
Communicating Editor: H. Salz
Literature Cited
- Bell A. C., West A. G., Felsenfeld G., 1999. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell 98: 387–396. 10.1016/S0092-8674(00)81967-4 [DOI] [PubMed] [Google Scholar]
- Bischof J., Maeda R. K., Hediger M., Karch F., Basler K., 2007. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc. Natl. Acad. Sci. USA 104: 3312–3317. 10.1073/pnas.0611511104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D., Dale R. K., Lei E. P., 2017a Shep regulates Drosophila neuronal remodeling by controlling transcription of its chromatin targets. Development 145: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D., Gu T., Pham T. N., Zachary M. J., Hewes R. S., 2017b Regulatory Mechanisms of Metamorphic Neuronal Remodeling Revealed Through a Genome-Wide Modifier Screen in Drosophila melanogaster. Genetics 206: 1429–1443. 10.1534/genetics.117.200378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D., Qu C., Bjorum S. M., Beckingham K. M., Hewes R. S., 2014. Neuronal remodeling during metamorphosis is regulated by the alan shepard (shep) gene in Drosophila melanogaster. Genetics 197: 1267–1283. 10.1534/genetics.114.166181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowder S. M., Kanaar R., Rio D. C., Alber T., 1999. Absence of interdomain contacts in the crystal structure of the RNA recognition motifs of Sex-lethal. Proc. Natl. Acad. Sci. USA 96: 4892–4897. 10.1073/pnas.96.9.4892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale R. K., Matzat L. H., Lei E. P., 2014. metaseq: a Python package for integrative genome-wide analysis reveals relationships between chromatin insulators and associated nuclear mRNA. Nucleic Acids Res. 42: 9158–9170. 10.1093/nar/gku644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilgers V., Perry M. W., Hendrix D., Stark A., Levine M., et al. , 2011. Neural-specific elongation of 3′ UTRs during Drosophila development. Proc. Natl. Acad. Sci. USA 108: 15864–15869. 10.1073/pnas.1112672108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King M. R., Matzat L. H., Dale R. K., Lim S. J., Lei E. P., 2014. The RNA-binding protein Rumpelstiltskin antagonizes gypsy chromatin insulator function in a tissue-specific manner. J. Cell Sci. 127: 2956–2966. 10.1242/jcs.151126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kung J. T., Kesner B., An J. Y., Ahn J. Y., Cifuentes-Rojas C., et al. , 2015. Locus-specific targeting to the X chromosome revealed by the RNA interactome of CTCF. Mol. Cell 57: 361–375. 10.1016/j.molcel.2014.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A. L., Volkman B. F., Robertson S. A., Rudner D. Z., Barbash D. A., et al. , 1997. Chemical shift mapping of the RNA-binding interface of the multiple-RBD protein sex-lethal. Biochemistry 36: 14306–14317. 10.1021/bi970830y [DOI] [PubMed] [Google Scholar]
- Lei E. P., Corces V. G., 2006. RNA interference machinery influences the nuclear organization of a chromatin insulator. Nat. Genet. 38: 936–941. 10.1038/ng1850 [DOI] [PubMed] [Google Scholar]
- Lisbin M. J., Gordon M., Yannoni Y. M., White K., 2000. Function of RRM domains of Drosophila melanogaster ELAV: Rnp1 mutations and rrm domain replacements with ELAV family proteins and SXL. Genetics 155: 1789–1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markstein M., Pitsouli C., Villalta C., Celniker S. E., Perrimon N., 2008. Exploiting position effects and the gypsy retrovirus insulator to engineer precisely expressed transgenes. Nat. Genet. 40: 476–483. 10.1038/ng.101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzat L. H., Dale R. K., Lei E. P., 2013. Messenger RNA is a functional component of a chromatin insulator complex. EMBO Rep. 14: 916–922. 10.1038/embor.2013.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzat L. H., Dale R. K., Moshkovich N., Lei E. P., 2012. Tissue-specific regulation of chromatin insulator function. PLoS Genet. 8: e1003069 10.1371/journal.pgen.1003069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moshkovich N., Nisha P., Boyle P. J., Thompson B. A., Dale R. K., et al. , 2011. RNAi-independent role for Argonaute2 in CTCF/CP190 chromatin insulator function. Genes Dev. 25: 1686–1701. 10.1101/gad.16651211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olesnicky E. C., Antonacci S., Popitsch N., Lybecker M. C., Titus M. B., et al. , 2018. Shep interacts with posttranscriptional regulators to control dendrite morphogenesis in sensory neurons. Dev. Biol. 444: 116–128. 10.1016/j.ydbio.2018.09.022 [DOI] [PubMed] [Google Scholar]
- Peabody N. C., Diao F. Q., Luan H. J., Wang H., Dewey E. M., et al. , 2008. Bursicon Functions within the Drosophila CNS to Modulate Wing Expansion Behavior, Hormone Secretion, and Cell Death. J. Neurosci. 28: 14379–14391. 10.1523/JNEUROSCI.2842-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray D., Kazan H., Cook K. B., Weirauch M. T., Najafabadi H. S., et al. , 2013. A compendium of RNA-binding motifs for decoding gene regulation. Nature 499: 172–177. 10.1038/nature12311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smibert P., Miura P., Westholm J. O., Shenker S., May G., et al. , 2012. Global patterns of tissue-specific alternative polyadenylation in Drosophila. Cell Reports 1: 277–289. 10.1016/j.celrep.2012.01.001; Erratum, Cell Reports 3: 969, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West A. G., Gaszner M., Felsenfeld G., 2002. Insulators: many functions, many mechanisms. Genes Dev. 16: 271–288. 10.1101/gad.954702 [DOI] [PubMed] [Google Scholar]