Abstract
Despite numerous advancements in production protocols, manufacturing AAV to meet exceptionally high demand (1016–1017 viral genomes [VGs]) in late clinical stages and for eventual systemic delivery poses significant challenges. Here, we report an efficient, simple, scalable, robust AAV5 production process utilizing the most recent modification of the OneBac platform. An increase in volumetric yield of genomic particles by ∼6-fold and functional particles by ∼20-fold was achieved by operating a high-cell-density process in shake flasks and bioreactors that involves an Sf9-based rep/cap stable cell line grown at a density of about 10 million cells/mL infected with a single baculovirus. The overall volumetric yields of genomic (VG) and bioactive particles (enhanced transducing units [ETUs]) in representative fedbatch bioreactor runs ranged from 2.5 to 3.5 × 1014 VG/L and from 1 to 2 × 1011 ETU/L. Analytical ultracentrifugation analyses of affinity-purified AAV vector samples from side-by-side batch and fedbatch production runs showed vector preparations with a full and empty particle distribution of 20%–30% genomic and 70%–80% empty particles. Moreover, the stoichiometric analysis of capsid proteins from fedbatch production in shake flask and bioreactor run samples demonstrated the incorporation of higher VP1 subunits, resulting in better functionality.
Keywords: AAV; gene therapy vector,s insect cell baculovirus technology; OneBac; Fedbatch; process intensification; high yield production
Introduction
Recombinant adeno-associated virus (rAAV) has emerged as one of the versatile vectors for therapeutic gene delivery in both dividing and non-dividing cells for the treatment of monogenic disease conditions.1, 2 The regulatory approval of Glybera by the European Medicines Agency (EMA) in 2012 for lipoprotein lipase deficiency3 and the recent approval of Luxturna by the U.S. Food and Drug Administration (USFDA) and EMA for the treatment of hereditary retinal dystrophy are two key milestones in the AAV-based gene therapy field.
Extensive research and sustained efforts have been dedicated to the development of various production platforms to deliver large quantities of functional AAV vectors. Traditional methods of AAV production involve two-dimensional (2D) cell culture processes where adherent mammalian cell lines are transiently transfected with plasmids carrying all the necessary genes along with co-infection of a helper virus.4, 5, 6, 7, 8, 9 The latter was eventually replaced with a plasmid carrying helper functions. Restricted scalability of this system led to the development of a production method based on a suspension-adapted mammalian cell line in combination with transient transfection10 or recombinant herpes simplex virus-based infection for delivery of necessary genes. Although very promising, these systems faced challenges associated with a lower volumetric yield.10, 11 Further work involving alternative modes of cell cultivation and advancement in the processing led to the improvement of these expression systems.12, 13, 14, 15
The first report of rAAV production in Spodoptera frugiperda insect cells (Sf9) using triple Autographa californica multiple nuclear polyhedrosis baculovirus infections (ThreeBac) brought about a new excitement in the field of scalable AAV production.16 This system offered comparable per-cell yields of AAV and the possibility of enhanced volumetric yields due to the ability of Sf9 cells to grow at a high cell density in a suspension culture. This original system was further improved, addressing its key shortcomings, and TwoBac and OneBac, which were simpler systems, followed.17, 18, 19, 20, 21 The recently reported OneBac has only two components: an inducible and stable Sf9-based packaging cell line incorporating integrated copies of the rep and cap genes and a baculovirus carrying a Bac-rAAV cassette (OneBac). This system was further improved to achieve optimal VP composition and functionality in AAV5 and AAV9 vectors comparable to vectors produced on the mammalian platform. This recent improvement also demonstrated minimized encapsidation of foreign DNA in the vector particle.22, 23
Although serotype-dependent compared with TwoBac and ThreeBac, the OneBac system that we studied essentially provides an efficient packaging cell line and presents advantages for large-scale manufacturing of an AAV delivery system with serotype 5 because of the relative simplicity of operation from a process standpoint. Generating a stable cell line and establishing a master cell bank for manufacturing clinical grade material are significant undertakings. More generally, in the context of manufacturing biologic, primary work has relied on transient expression, followed thereafter by stable expression systems. In the case of viral vectors, the transient expression systems, packaging cell lines, and producing cell lines are scenarios that may be considered, depending on the viral product characteristics and end use. We believe that the stable cell line approach has the potential to be a preferable platform for well-established and clinically proven vector candidates such as AAV5 and AAV9.
Aligned with our continuing efforts to improve AAV manufacturing platforms, in this study we further explored the OneBac system from a process standpoint for AAV5 fedbatch production mode, focusing solely on the upstream process phase. The consistency of the production process was assessed in a shake flask and was further validated in a 1 L, and 3 L controlled bioreactor runs. The purified AAV was characterized for its quality attributes including in vitro functionality, capsid protein composition, and relative proportion of empty and genomic particles in affinity-purified AAV preparations.
Results
Genetic Stability of the Packaging Cell Line and Rep/Cap Copy Number Analysis
During traditional commercial-scale production, cells undergo numerous doubling cycles, and any loss of expression of integrated rep/cap genes can result in lower yields and hence it is important to assess their expression stability over the extended number of passages. The working cell bank of packaging cells was at passage number (P) 3. The cells were infected at various passages: P4 (vial thaw+1), P8 (vial thaw+5), and P35 (vial thaw+32) at an MOI of 1 PFU/cell. The clarified cell lysate containing Cap and Rep proteins was analyzed by western blot, the results of which are shown in Figure S1. The data show no significant loss of expression with either of the proteins. Furthermore, the same clarified lysate samples were analyzed for total viral genome (VG) copies via qPCR. The cell-specific yield in all three samples was around 15,000 VG/cell. This VG copy number shows no passage-dependent loss of cell-specific yield in the rep2cap5 packaging cell line, suggesting stable expression of the AAV helper genes up to 35 passages. It should be noted that this preliminary set of experiments was conducted in an early phase of the project under non-optimal conditions of MOI and the cell density at the time of infection. The Sf9 cell line (B8 clone) was found to have 9.97 copies of cap5 and 1.25 copies of rep2 integrated per cell (Table 3).
Table 3.
Determination of the Number of Integrated and Rescued Cap and Rep Genes Copies per Cell of the Sf9 B8 Stable Cell Line during AAV Production in Different Modes and Scales
| Production Method | Cap5 Copies per Cell | Rep2 Copies per Cell | Cap:Rep Ratio | Bursting Effect (Times) |
|---|---|---|---|---|
| Batch_SF, 72 hpi | 286.56 | 29.55 | 9.7 | 28.7 |
| Fedbatch SF, 72 hpi | 472.80 | 47.38 | 9.98 | 47.4 |
| Fedbatch_BR,_72 hpi | 467.64 | 48.98 | 9.55 | 46.9 |
| Non-infected B8 | 9.97 | 1.25 | 7.99 | N/A |
SF, shake flask; BR, bioreactor; hpi, hours post infection.
AAV Production Culture Characteristics
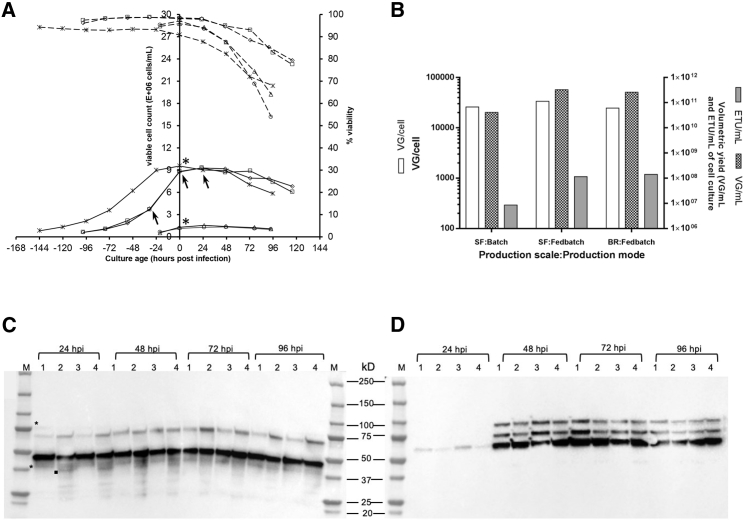
In the present study, we investigated AAV production using regular- and high-cell-density cultures. For ease of understanding throughout the paper, the regular-cell-density and high-cell-density production processes are referred to as batch and fedbatch processes (modes), respectively. Also, in the batch and fedbatch processes, cells were infected at low and high cell densities, respectively. In the latter case, the cultures in the pre- and post-infection phases were supplemented with an additional nutrient feed, as has been reported on several occasions by research groups.24, 25, 26 The cell density and viability profiles during AAV production in batch (triplicate shake flask runs) and fedbatch (triplicate shake flask and bioreactor runs) modes are shown in Figure 1A. In both modes, the cells were infected in their mid-exponential growth phase which is followed by a characteristic baculovirus-induced cell growth arrest and protein expression phase. Preliminary experiments were conducted to screen the suitable cell density at the time of infection and the multiplicity of infection in the low-cell-density process. The findings suggest that 1.2–1.4 million cells/mL was a suitable cell density for infection and an MOI of 3 PFU/cell would give higher cell-specific yield (data not shown). Any combination of cell density and MOI that resulted in an overall cell density above 2–2.5 million cells/mL during AAV production resulted in reduced cell-specific and volumetric yield. This could be due to culture medium limitation and its inability to support protein expression in the post-infection phase. Similar work was conducted for AAV production in fedbatch mode for preliminary screening (Figure S2). In batch mode, when cells were infected at a cell density of around 1.2–1.4 million cells/mL, cells density increased to ∼1.5–1.7 million cells/mL within 24 hpi, followed by progressive declines in cell density and viability. Similarly, in the high-cell-density process (fedbatch mode), the cells were infected at 9.5–10 million cells/mL, and cell density increased to 10.5–11 million cells/mL within 24 hpi, followed by a sequential decline in both viable cell density and viability. In fedbatch mode, the addition of nutrient feed and baculovirus stock resulted in partial dilution of the culture, the effect of which is reflected in the final cell density. The viability of the cells at the time of harvest (96–120 hpi) was found to be between 70% and 80% in fedbatch mode and 50% and 60% in batch mode. After infection at MOI 3, the majority of the cells were infected within 24 h (Figure S3). The peak expression of Rep and Cap proteins was obtained at 48 h (Figures 1C and 1D). A graphical representation of AAV yield on the different scales (shake flask and bioreactor run) and in the different production modes (batch and fedbatch) is shown in Figure 1B. The cell-specific (VG/cell) and volumetric (VG/mL) yields of genomic particles were found to be 25,000–27,000 VG/cell and 4–5 × 1010 VG/mL of culture in batch mode, respectively. In shake flask and bioreactor runs under fedbatch mode, around 25,000–35,000 VG/cell and 2–3 × 1011 VG/mL titers were obtained. The volumetric yields of enhanced transducing units (ETUs), otherwise reported as infective virus particles,27 were around 1 × 107 and 1–2 × 108 ETU/mL in the batch and fedbatch processes, respectively. The inline profile of various bioreactor sensors and process parameters during AAV production in a fedbatch bioreactor run is also shown in Figure 2.
Figure 1.
AAV Production Characteristics
(A) Insect cell culture characteristics during AAV5 production runs. The data shown are for AAV batch production (shake flasks 1 and 2) and fedbatch production (shake flasks 1 and 2 and bioreactor). Asterisks represent the time point of infection in the batch and fedbatch processes. The three arrows represent the time points of nutrient feed supplementation. The cell density data are shown by solid lines, and the percentage of cell viability is shown by dotted lines. ○, SF batch F1; ☐, SF fedbatch F1; Δ, SF batch F2; ◊, SF fedbatch F2; ✻, bioreactor. (B) Bar graph representation of overall cell-specific and volumetric yield of AAV5 production in batch and fedbatch mode in shake flask (SF) and bioreactor (BR). Note the logarithmic scale on the left y axis. (C) Kinetic of AAV Rep expression in the post-infection phase. Lane 1: SF batch flask 1; lane 2: SF fedbatch flask 1; lane 3: SF batch flask 2; and lane 4: SF fedbatch flask 2. Rep 78 and 40 are indicated by asterisks, and the low-molecular-weight fragment of Rep 52 is indicated by a dot. (D) Kinetics of AAV5 Cap expression in the post-infection phase. Lane 1, SF batch flask 1; lane 2, SF fedbatch flask 1; lane 3, SF batch flask 2; lane 4, SF fedbatch flask 2. BR, bioreactor; ETU, enhanced transducing units; hpi, hours post infection, M, molecular weight marker; SF, shake flask; VG, viral genome.
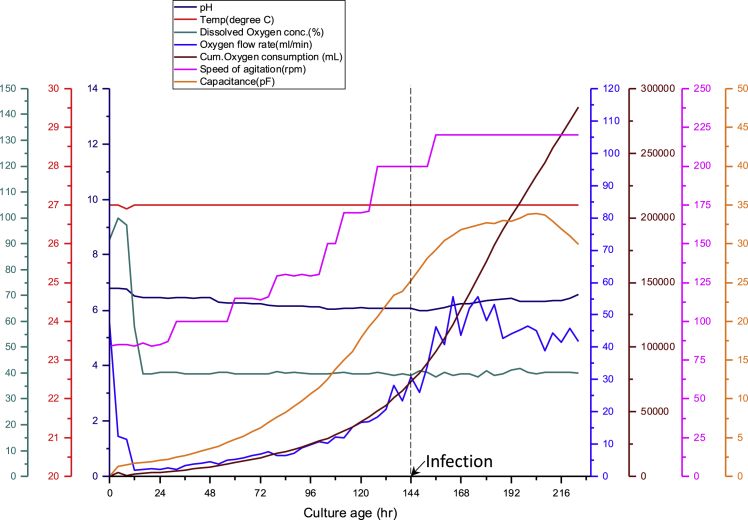
Figure 2.
In-Line Sensors Profile during AAV Production in a Bioreactor
Profile of various bioreactor sensors and process parameters during AAV production in a bioreactor. The speed of agitation was increased gradually as the cells grew. The pH, dissolved oxygen concentration, and temperature were maintained at their respective set points throughout the run. Note the change in capacitance and oxygen consumption after infection, an indication of cells undergoing the productive infection phase. pF, picofarads.
AAV5 Characterization
Analysis of AAV5 Vector Heterogeneity via Analytical Ultracentrifugation
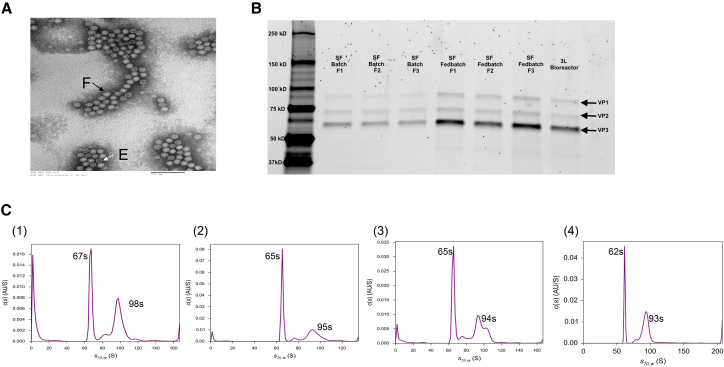
Analytical ultracentrifugation (AUC) is increasingly becoming a valuable tool for routine analysis of structural variants in AAV vector preparations where the real-time sedimentation analysis data provide a qualitative and quantitative measurement of vector heterogeneity, i.e., empty particle, full particle, and continuum of the population in between.28 To obtain the samples with a purity necessary for AUC analysis, the AAV5 produced in multiple runs in shake flasks and bioreactors were prepared by affinity chromatography. The clarified cell lysate was subjected to single-step affinity chromatography, and the eluted AAV was collected and neutralized and the buffer exchanged against a suitable buffer. The characteristic chromatogram is shown in Figure S4 with a magnified image of the eluate peak containing the virus.
The AUC analysis of affinity eluate samples from multiple AAV production runs showed two major peaks at around 62–67 and 93–98 s. The sharp peak at 62–67 s corresponded to empty particles, whereas the intermediate population and the peak around 93–98 s corresponded to capsids with a genomic content and a prevalence of capsid incorporating complete rAAV genome (93–98 s). The AUC profile at the 260 nm scan of each representative sample from batch and fedbatch runs at various scales is shown in Figure 3C. The relative proportion of empty and genomic capsids was found to be ∼70%–80% and ∼20%–30%, respectively. The AUC histogram for each of the AAV production runs (n = 3) in batch and fedbatch mode in shake flasks is provided in Figure S5, along with detailed data in Table S1. A detailed description of the calculation method is provided in the Supplemental Materials and Methods.
Figure 3.
AAV Purification and Characterization
(A) TEM image of affinity-purified AAV5 showing a mixed population of full particles (F) and empty capsids (E) with a characteristic particle diameter of 20–23 nm. (B) VP stoichiometry analysis via a Flamingo-stained SDS-PAGE gel of an affinity-purified sample of three production runs of AAV in batch mode (shake flask [SF]) and fedbatch mode (shake flask: SF and 3 L bioreactor). The total VG loading per well was in the range of 5 × 108–7 × 108. The VP band intensities in all samples are similar and comparable to that of the 1 L bioreactor sample. (C) AUC histogram representing a 260 nm absorbance profile of representative samples of an affinity-purified AAV5 from production in (1) batch mode in shake flask, (2) fedbatch mode in shake flask, (3) fedbatch mode in 3 L bioreactor, and (4) fedbatch mode in 1 L bioreactor. Different components in an affinity-purified AAV5 sample as a function of sedimentation coefficient are shown. A very sharp peak around 62–67 s represents the presence of an empty capsid, and a major peak around 93–98 s and the intermediate population between these two major peaks represent genomic capsids. The peaks corresponding to the intermediate population may represent the form of a particle with encapsidation of partial AAV genome or collaterally packaged contaminating DNA. (See also Figures S4–S6, and Tables S1 and S2.) SF, shake flask.
Transmission Electron Microscopy of AAV
The sample used for AUC was recovered from the sample chamber after analysis and used for transmission electron microscopy (TEM) analysis. The uranyl acetate negatively stained AAV sample was analyzed at ×180,000 magnification. A representative image is shown in Figure 3A. The TEM image shows a mixed population of empty and genomic particles. The diameter of the particles was in the range of 20–23 nm, which is consistent with the reported values.
AAV5 VP Composition Analysis via SDS-PAGE and Densitometry
The best possible linearity between VP band intensity and VG was found to be in the concentration range of 5.5 × 108–7.3 × 108 VGs loaded (Figure S6). The stoichiometric composition of VP subunits was somewhat different compared with that in the previous report.23 The incorporation of more VP1 is reported to be associated with the enhanced functionality of the vector preparation.23, 29, 30, 31 The results of VP composition from analysis of AAV samples of multiple production runs are shown in Figure 3B. Please see Table S2 for details of VP composition data for each sample.
Assessment of In Vitro Functionality via Gene Transfer Assay
The titer values obtained from gene transfer assay (GTA) for the different samples analyzed are summarized in Tables 1 (clarified lysate) and 2 (AVB-affinity eluate samples). For batch and fedbatch production processes, the functional titers were found to be of 1 × 107 and 1 × 108 ETU/mL, respectively, wherein the latter reflected a 1 log increase in titer in the fedbatch process, corresponding to high cell density.
Table 1.
Summary of AAV Production Yield in a Clarified Lysate
| Sample | Culture Volume (mL) | ETU/mLa | VG/mLb | ETUs/Cell | VG:ETU |
|---|---|---|---|---|---|
| Shake flask-batchc | 25 | 1.1 × 107 ± 2.53 × 106 | 4.0 × 1010 ± 0.56 × 1010 | 6 ± 1.4 | ∼3,600:1 |
| Shake flask-fedbatchd | 25 | 1.9 × 108 ± 0.43 × 108 | 2.7 × 1011 ± 0.37 × 1011 | 20 ± 4.6 | ∼1,400:1 |
| Bioreactor 1-fedbatch | 740 | 2.1 × 108 ± 0.48 × 108 | 3.8 × 1011 ± 0.53 × 1011 | 22 ± 5 | ∼1,700:1 |
| Bioreactor 2-fedbatch | 2,250 | 1.4 × 108 ± 0.32 × 108 | 2.6 × 1011 ± 0.27 × 1011 | 15 ± 3.5 | ∼1,700:1 |
ETUs, enhanced transducing units; GTA, gene transfer assay; VG, viral genome copies or genomic particles.
GTA variability: relative SD 23%.
qPCR assay variability: relative SD 14%.
Values shown for multiple runs (n = 6); relative SD 19%.
Values shown for multiple runs (n = 6); relative SD 18%.
Table 2.
Summary of AAV5 Characterization in AVB Affinity-Purified Samples
| Sample | Eluate Volume (mL) | ETU/mLa | VG/mLb | VG: ETU | % Empty Capsid | % Genomic Capsids |
|---|---|---|---|---|---|---|
| Shake flask-batchc | 3.5 | 8.2 × 107 ± 1.9 × 107 | 3.2 × 1011 ± 0.41 × 1011 | ∼3,800:1 | 74.1 ± 0.6d | 25.9 ± 0.6d |
| Shake flask-fedbatchc | 3.5 | 5.3 × 108 ± 1.23 × 108 | 1.3 × 1012 ± 0.29 × 1012 | ∼2,500:1 | 80.6 ± 1.0d | 19.3 ± 1.1d |
| Bioreactor 1-fedbatch | 3.5 | 4.0 × 108 ± 0.92 × 108 | 1.0 × 1012 ± 0.14 × 1012 | ∼2,500:1 | 68 | 32 |
| Bioreactor 2-fedbatch | 3.5 | 3.57 × 108 ± 0.82 × 108 | 8.48 × 1011 ± 1.35 × 1011 | ∼2,400:1 | 78.8 | 21.2 |
ETU, enhanced transducing units; VG: viral genome copies or genomic particles.
GTA variability: relative SD 23%.
qPCR assay variability: relative SD 14%.
Values shown for triplicate runs (n = 3).
Average value for three runs.
The Relative Ratio of Genomic and Functional Virus Particles
To quantify the respective ratio (VG:ETU), the data were compiled from AAV analyses via qPCR and GTA for the clarified cell lysate and the affinity-purified sample. The genomic-to-functional particle ratio (VG:ETU) for the cell lysate sample of the batch production was around 3,600:1, and that of the fedbatch production was 1,400:1 (shake flask) and about 1,700:1 (bioreactors). In the case of the purified sample, this ratio was somewhat higher, eventually resulting from possible loss of functional particles during the low-pH elution necessary in affinity chromatography. Additionally, the variability of the assay may contribute to the variability of this ratio. The cell-specific yield of the functional particles was found to be around 6 ETU/cell (shake flask: batch), 20 ETU/cell (shake flask: fedbatch), and around 15–20 ETU/cell (bioreactors: fedbatch). It is important to note that the values of ETU obtained from the GTA were highly dependent on the detection method and the cell line used in the bioassay. In particular, the transduction susceptibility of the cell line used in the GTA was AAV serotype dependent. Our group has previously reported that HEK293- and HeLa-cell-based GTAs display significant differences in the AAV serotype titer values. Specifically, when AAV2 and AAV5 serotypes were produced side by side via a triple transfection process in suspension-adapted HEK293 cells, the genomic titer (VG/mL) of both serotypes was identical, whereas the functional titer via GTA assay resulted in approximately 2 log lower quantification values for AAV5 (∼1 × 107 ETU/mL) in the HEK293-based GTA and 1 log lower quantification value (∼1 × 108 ETU/mL) in the HeLa-based GTA, compared with AAV2 (∼1 × 109 ETU/mL). For of these reasons, caution should be exercised when comparing the functional titers of different serotypes with that of AAV2.10
Characterization of the Fedbatch Production Process
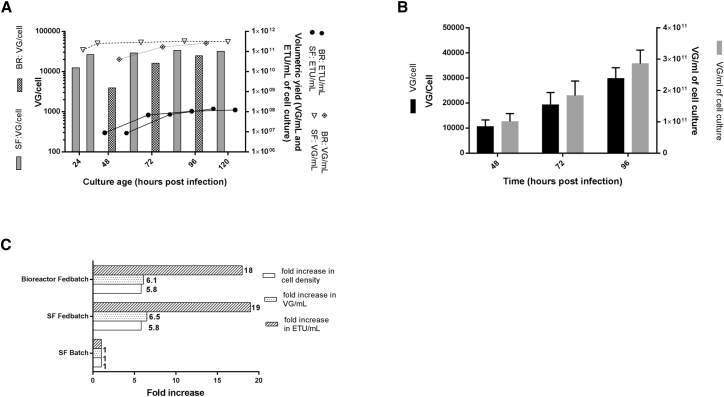
It is expected that AAV5 production at high cell density in fedbatch mode would add a dimension to the complexity and possible process variability; therefore, it is important to assess the robustness and consistency of the AAV production process under this mode of operation. Three AAV5 fedbatch productions in shake flasks were run in parallel and analyzed for consistency of production, as well as the final titer of genomic and functional virus particles. Moreover, side-by-side AAV productions in the fedbatch mode in shake flask and bioreactor runs were conducted to validate the process. The kinetics data of AAV VG productions in shake flasks are provided in Figure 4B. The variability in cell-specific and volumetric yields of three replicates at different time points after infection is shown, wherein the error bar represents the relative SD. As the data suggest, there was significant consistency in AAV5 VG kinetics at different time points and in the final titer of the three replicates. There was a consistent increase of approximately 7,000–10,000 VG/(cell · day) between 48 and 96 hpi. The overall cell-specific and volumetric yield at the time of harvest was in the range of 27,000–30,000 VG/cell and 2–3 × 1011 VG/mL, respectively. In the case of the side-by-side comparison of AAV production kinetics in a shake flask and a bioreactor (Figure 4A), a similar trend in AAV production kinetics was observed for both scales. The genomic particles increased up to 96 hpi followed by a plateau between 96 and 120 hpi. The ETU/mL data followed a similar trend but with a late onset, where the functional particles were only detected at 48 hpi. This trend represents a typical AAV production kinetics where capsid protein expression commences immediately after the baculovirus infection followed by transgene rescue, replication, and packaging into preformed capsids. It is postulated that the preformed capsid with encapsidated genome undergoes conformational changes and capsid maturation in later stage and hence the functional titer values appear typically after 24 hpi. The final volumetric yield of AAV5 genomic titer in shake flask, bioreactor1, and bioreactor2 runs are 2.7 × 1014 VG/L, 3.8 × 1014 VG/L, and 2.6 × 1014 VG/L, respectively, and that of the functional titer is 1.9 × 1011 ETU/L, 2.1 × 1011 ETU/L, and 1.4 × 1011 ETU/L, respectively. The overall summary of these results expressed as a factorial increase in volumetric yield is given in Figure 4C. To further extend the process characterization, representative samples from AAV production in batch mode and fedbatch mode at 72 hpi were analyzed for rep/cap copy number to determine the degree of rep/cap amplification after baculovirus infection along with non-infected Sf9 B8 as a control (Table 3). The data suggest around 47× and 28× amplification in fedbatch and batch mode of production, respectively.
Figure 4.
Fedbatch Process Characterization
(A) AAV production kinetics data of parallel runs of AAV productions in a shake flask and bioreactor in fedbatch mode. At different post-infection time points, the VG/cell, VG/mL, and ETU/mL values are shown via a bar graph, dotted trend line, and solid trend line, respectively. The maximum cell-specific and volumetric yields are achieved by 96 hpi with no further noticeable increase afterward, and hence the harvest time was set at 96 hpi. (B) Consistency analysis of production kinetics and overall yield of AAV in fedbatch mode in shake flasks in triplicate parallel runs. The error bar in the bar graph represents the relative SD of titers. (C) Comparative analysis of batch and fedbatch processes on the shake flask and bioreactor scales. In the fedbatch process, the overall cell density was increased by a factor of ∼6 compared with the batch process. This resulted in an ∼6-fold increase in volumetric yield of genomic particles and an ∼20-fold increase in that of transducing particles during AAV productions in shake flask and bioreactor runs. This exponential increase in volumetric yield is believed to be due to a confounding positive effect of nutrient supplementation in a fedbatch mode of production. (See also Table S3.) BR, bioreactor; SF, shake flask; ETU, enhanced transducing units; VG, viral genome.
Discussion
Progression of AAV-based gene delivery vector candidates into late clinical stages necessitates a higher amount of clinical grade vector material. This poses a critical challenge and confers a further burden on vector production processes because of the low volumetric yield of current AAV production platforms. The therapeutic dosage of AAV for systemic administration in humans ranges from 1012 to 1014 VG/kg, depending on indication.32, 33 This translates into an overall requirement of 1016–1018 VG in a typical phase 3 clinical trial. Using traditional plasmid transfection protocols with adherent HEK293 cells to generate that quantity of virus makes it impractical for commercial-scale production.16 Recently, various bioreactors based on alternative modes of adherent cell cultivation, such as CellFactory, CellSTACKs, and iCELLis, have been used to generate AAV material for clinical studies.14, 34, 35 The most straightforward approach is a three-dimensional suspension cell culture that offers linear scalability and eventually a higher volumetric yield. Production platforms based on suspension-adapted HEK293 cell and insect cell cultures represent a more viable option, where the typical volumetric yield of AAV at regular cell density (∼1 million cells/mL) cultures is about 1 × 1010 VG/mL and requires 100–10,000-L scale bioreactor production for late-stage clinical studies.10, 36, 37, 38 Among the insect cell culture-based systems for AAV production, the OneBac platform is the simplest from a process standpoint, as it consists of only two components. It reportedly provides cell-specific yields comparable to that of the other two systems.22 It was further fine tuned for the VP1:VP2:VP3 capsid composition and minimum encapsidation of foreign DNA.23 In the current study, we examined this system from a process standpoint. The volumetric yield was enhanced by around 6-fold (Figure 4C), using the fedbatch mode of production in shake flask and bioreactor formats.
The OneBac system generates functional AAV particles upon infection with a single baculovirus. The detailed description of this system has been reported previously.20, 21, 22 The expression level of Rep and Cap proteins is reported to have a direct impact on AAV yield.17 The passage stability of the expression of the Rep protein helper suggests a potential shift to higher molecular weight proteins Rep78 and -52; however, there is no noticeable effect on the per cell yield of VGs. It can be inferred that the necessary expression levels of these two Rep proteins for maximum replication and encapsidation of vector DNA are maintained, irrespective of the number of passages up to 35 (the maximum passage tested). For high-cell-density AAV production in fedbatch mode, the timely addition of a nutrient feed is crucial for two key attributes: (1) in the pre-infection phase, it supports and helps to maintain high-cell-density culture in an exponential growth phase prior to infection, and (2) in the post-infection phase, it supports elevated nutrient demand to sustain high protein and genomic material expression. A high-MOI infection that results in a cell growth arrest within 24 hpi metabolically shifts the overall culture from the cell growth phase to the protein expression phase, unlike a low-MOI (e.g., MOI of 0.1) infection, where this shift is often delayed by an additional cell growth cycle.26, 39 As shown here, the peak expression of Rep and Cap proteins is attained within 48 hpi and the nutrient supplementation addition at the time of infection and at 24 hpi sufficiently fulfills the metabolic need of the culture. Similar findings have been reported with a three baculovirus system where the high cell density was achieved via infection of the Sf9 cell culture at 5 million cells/mL with a low MOI of 0.1 PFU/cell.26 Comparative analysis of the volumetric yields in batch and fedbatch modes shows that, even at high cell density, the cell-specific yield was maintained. There was a proportionate increase in volumetric yield of genomic and transducing virus particle titers with an increase in cell density. This further suggests that the feeding formulation and regimen applied at this high cell density successfully leads to process intensification. The data from the bioreactor production runs provide further validation of the process. Also, improved cell-specific yield of functional virus particles (ETU/cell) in fedbatch compared with batch processes (Table 1) suggests that the requirement of essential nutrients to support cellular processing in the late infection phase was fulfilled with the fedbatch feeding regimen. The overall cellular processing seems to be impacted by the cell culture and extracellular environmental parameters (cell viability, pH, osmolarity, accumulation of toxic metabolites, and depletion of essential nutrient). The VP1 capsid protein subunit can be directly correlated with virus infectivity due to the presence of an integrated phospholipase A2-like enzymatic domain and its postulated role in virion escape from a late endosomal stage. It has been reported on several occasions that insect cell-produced AAV vectors exert inferior infectivity compared with that produced in mammalian cells due to an altered capsid composition, resulting in low incorporation of VP1.17, 23 The analysis of capsid composition of the bioreactor-produced AAV5 showed a higher ratio of VP1 compared with that in a previous report,23 and it is believed to be responsible for the improved infective virus particle titer.
Given that the same GTA protocol was used for the analysis of both samples, it provides an opportunity for direct comparison of two different platforms (OneBac and HEK293). The functional virus particle titer of the fedbatch bioreactor-produced AAV5 is comparable to that of AAV5 produced by the suspension-adapted, HEK293-based, triple-plasmid transfection method, as reported previously by our research group.10 The data confirm that the OneBac system used for AAV5 production in the present study, in conjunction with the proposed production process, generates AAV5 with identical functionality.
Materials and Methods
Cells and Medium
The stably transformed rep2cap5 packaging Sf9 cell line B8 was designed, essentially as described previously.23 These cells were used for the production of AAV. rAAV-encoding baculovirus expression vector (BEV) stocks used to infect B8 cells were grown in naive Sf9 cells in serum-free insect cell medium: sf900-II or sf900-III (Thermo Fisher Scientific, Waltham, Massachusetts). For high-cell-density AAV productions in fedbatch mode, B8 cells were supplemented with a single proprietary nutrient feed formulation. Both insect cell cultures were maintained in sterile PETG disposable flasks (Fisher Scientific, Hampton, New Hampshire) in serum-free medium in a 27°C incubator at 110 rpm agitation speed. The suspension-adapted HEK293 cells used for the gene transfer assay are described in our previous studies and are maintained in CD HEK293 medium (Irvine Scientific, Santa Ana, California) supplemented with L-glutamine at a final concentration of 4 mM. The suspension cell culture was maintained in a shaker incubator set at 37°C, 5% CO2, and 85% relative humidity. The cell density analyses from routine maintenance flasks and virus production runs were performed with the Vi-Cell XR cell counter (Beckman Coulter, Brea, California). This instrument works on the principle of Trypan blue dye exclusion and captures 50 images and provides an average value for viable cell density, percentage of viable cells, and cell diameter. The accuracy of the instrument is ±10% in the range of 0.2–10 × 106 cells/mL.
Baculovirus
The recombinant baculovirus carrying the AAV transgene expression cassette (Bac-GFP) consisted of AAV-ITR flanking eGFP under the control of chicken β-actin-CMV hybrid promoter. The baculovirus stock was plaque purified, and five baculovirus isolates expressing a high level of transgene cassette were collected. A highly potent Bac-GFP isolate was further selected based on AAV5 DNase-resistant particle titer analyzed from cell lysates of B8 cells infected with these individual isolates. The selected working isolate was volumetrically expanded using naive Sf9 cells where the clarified cell culture supernatant from 72 to 96 hpi was stored in the dark at 4°C (for short-term use) or at −80°C (long-term use). This Bac-GFP working stock, quantified via a standard baculovirus plaque assay protocol, was found to have a functional titer of 3–5 ×108 PFU/mL. This working baculovirus stock was at P3 with respect to the primary stock.
Analysis of the Genetic Stability of the Packaging B8 Cell Line
The rep2cap5-expressing insect cells at an extended number of passages (vial thaw+1: P4, vial thaw+5: P8 and vial thaw+32: P35) were infected with Bac-GFP at an MOI of 1 PFU/cell, and cells were harvested 96 hpi and lysed via a triple freeze-thaw cycle. Clarified lysate was used for the analysis of Rep and Cap expression by western blotting. A detailed description of the culture harvest and cell lysis protocols is given in a separate section that follows later.
Analysis of Rep2/Cap5 Copy Number
To characterize the AAV5-producing B8 Sf9 stable cell line for the number of rep and cap copies, the real-time qPCR using Cap5-, and Rep2-specific primers, as well as the Sf9 housekeeping gene Innexin2 (Inx2)20 was used. Briefly, 0.3 μg of total DNA isolated by Quick DNA kit (Zymo Research, Irvine, California) was digested by 20 U BamH HF enzyme (New England Biolabs, Ipswich, Massachusetts) for 1 h. Then, 30 ng of digested DNA was amplified using PowerUp SYBR Green Master Mix kit (Applied Biosystems, Foster City, California). Linearized plasmid DNA spiked by Sf9 wild-type gDNA was used as a reference for calculation of Cap5 and Rep2 copies per cell. For calculation of input genomes per qPCR reaction, the Spodoptera frugiperda Sf9 insect cell line genome size was assumed to be 451 Mbp.40 The sequences of primers used in this study are provided in Table S3.
AAV5 Production in Low- and High-Cell-Density Cultures at Shake Flask Scale
For all AAV production runs at shake flask scale, 125 mL sterile PETG flasks were used with 25 mL working volume, and the cells were maintained in a 27°C incubator at 110 rpm agitation speed. The insect cells at mid-exponential phase (2.5–3.5 × 106 cells/mL) were used to seed the subsequent culture and the seeding density at the beginning of the culture was always kept at around 0.65–0.75 million cells/mL. In the low-cell-density production process, the B8 cells at a density of 1.2–1.4 million cells/mL were infected with Bac-GFP at an MOI of 3 PFU/cell. At 96 hpi, the culture was harvested, cells were lysed, and the clarified lysate was collected for subsequent analysis. In the high-cell-density culture process, the insect cells were infected when the cell density was around 9.5–10 million cells/mL. Cell culture was harvested at 96 hpi and processed for AAV recovery and analysis. In the high-cell-density process, the pre- and post-infection phases were supported by fedbatch mode where cells were supplemented with a single nutrient cocktail at specific time points: at 24 h before infection (4% v/v), at the time of infection (0 hpi, 8% v/v) and at 24 hpi (8% v/v). Cell culture samples were collected every 24 h for analysis throughout the run. The addition of nutrient cocktail in two aliquots at 24 h intervals during the post-infection phase provided a nutrient-rich environment to support maximum protein expression for up to 48 hpi—a time phase where the cell density reaches a plateau and AAV Rep/Cap expression is at its peak.
Fedbatch Process Characterization
For fedbatch process characterization, process consistency and AAV production kinetics analysis were studied. AAV production runs were performed in triplicate in 125 mL sterile shake flasks with a 25 mL working volume. After infection, samples were collected every 24 h for analysis.
AAV5 Production at High Cell Density in Bioreactors
High cell density in 1 L (bioreactor 1) and 3 L (bioreactor 2) bioreactors was achieved by growing cells in fedbatch mode, applying the same nutrient feeding strategy as described previously for the shake flask. The bioreactor (Applikon Biotechnology, Delft, the Netherlands) was equipped with a single marine impeller, a pH sensor, a temperature sensor, a dissolved oxygen (DO) concentration sensor, and a micro sparger with 100 μm pore size. The pH, temperature, and DO parameters were set at 6.3 ± 0.3, 27°C ± 1°C, and 40% air saturation ± 2% at their corresponding units, respectively. In addition, the 3 L bioreactor was equipped with a capacitance probe for in-line monitoring of the culture characteristics, such as cell growth and progress of infection. The bioreactor control unit, equipped with a proportional-integral-derivative (PID) controller suite, ensured controlled conditions of process parameters within the given operational range throughout the run. The B8 cells from shake flask at a mid-exponential growth phase were used to inoculate the bioreactor at 0.65–0.75 million cells/mL density. When cells were at 9.5–10 million cells/mL, they were infected with a Bac-GFP at an MOI of 3 (PFU/cell) and harvested at 96 hpi for further processing.
Culture Harvesting and AAV Recovery
The whole-cell culture at 96 hpi was lysed in situ in a shake flask and a bioreactor via addition of 10% v/v lysis buffer (20 mM MgCl2, 500 mM Tris-HCl [pH 7.5], and 1% Triton X-100) supplemented at final concentration of 50 U/mL Benzonase DNase (Millipore Sigma, Burlington, Massachusetts). The cell culture was incubated with this lysis buffer for 1.5–2 h at 27°C with agitation, followed by addition of MgSO4 to achieve a final concentration of 37.5 mM. This step was followed by an additional incubation for 30 min at the same temperature. The latter step resulted in an ionic strength of more than 200 mM, where the presence of a high salt concentration assists in AAV release and prevents or minimizes interparticle AAV aggregation (Mg+2 being more effective) and AAV cell debris and AAV-DNA complexes.36 In the case of samples for virus titration, the crude lysate underwent a further three freeze-thaw cycles to ensure efficient cell lysis of less fragile insect cells and complete AAV release in the high ionic strength lysate. Following this, the lysate was clarified via centrifugation at 3,000 g for 30 min to eliminate cellular debris, and the supernatant containing released AAV5 particles was collected for further processing.
AAV Characterization
Clarified cell lysate and affinity-purified material containing AAV5 was used for quantitative and qualitative analytical assays.
The purified samples for various characterization assays were generated via a single-step immune-affinity chromatography method. AAV5 purification was performed by single-step affinity chromatography using AVB Sepharose resin and the ÄKTA Avant25 (GE Healthcare, Chicago, Illinois) chromatographic system. AVB resin, harboring a single-domain monoclonal antibody fragment, is reported to selectively bind a majority of AAV serotypes.21 Based on SDS-PAGE and western blot analysis, this single-step column purification yields a high-purity vector free of major cellular and baculovirus components (data not shown). Specifically, a 1 mL Hi-Trap prepacked AVB Sepharose column was equilibrated with 10 column volumes of loading buffer (20 mM Tris and 500 mM NaCl [pH 8]) followed by the loading of 100 mL lysate filtered via a 0.45 μm modified polyethersulfone membrane filter. The column was washed with loading buffer until the 260/280 nm baseline returned to zero. The elution was done by the introduction of a low-pH elution buffer (0.1 M glycine [pH 2.5]) which resulted in a sharp pH gradient and the low-pH environment inside the column induced detachment of bound AAV. The acidic eluate containing AAV5 was immediately neutralized via dropwise addition of neutralizing buffer (1M Tris [pH 8.8]) until the pH shifted to neutral or slightly alkaline values. This neutralized AAV material was further buffer exchanged in a suitable buffer using PD-10 desalting columns (GE Healthcare, Chicago, Illinois). The chromatographic process flowrate throughout the operation was kept at 75 cm/h. The process was monitored with built-in UV-VIS, conductivity, and pH. The affinity-purified material was used for subsequent quantitative and qualitative characterization.
Quantitative Analysis
Quantification of DNase-Resistant Genomic Particles by qPCR
Clarified lysate or affinity-purified material was incubated with 5 U/mL Benzonase for 30 min at 37°C before viral DNA extraction. Benzonase-treated undiluted and 1:10 diluted samples were used for viral DNA extraction using the High Pure Viral DNA Extraction kit (Roche Diagnostics, Risch-Rotkreuz, Switzerland). In the end, 200 μL lysate sample resulted in 50 μL purified and concentrated viral DNA. This viral DNA, serially diluted to 1:10, 1:100, and 1:1,000, was used as a template for subsequent qPCR analysis via probe-based reaction. The forward primer (5′-ATAGGGACTTTCCATTGACGTC-3′), the reverse primer (5′-TGATACACTTGATGTACTGCCAAG-3′), and the probe (FAM 5′-TGGGTGGACTATTTACGGTAAACTGCC-3′ BHQ) targeting the cytomegalovirus (CMV) enhancer sequence used in the qPCR reaction were purchased from Integrated DNA Technologies (Coralville, Iowa). The 20 μL qPCR reaction mixture consisted of 10 μL SsoAdvanced Universal Probes Supermix (Bio-Rad Laboratories, Hercules, California), 1.2 μL of 10 μM forward primer, 1.2 μL of 10 μM reverse primer, 1 μL of 5 μM probe, 1.6 μL of qPCR-grade water, and 5 μL of DNA template. The temperature programming for qPCR reaction used was preincubation at 95°C/3 min for denaturation and 40 cycles of 95°C/15 s and 54.5°C /30 s. Vector genome standard was prepared using an AAV-GFP plasmid (Addgene, Cambridge, Massachusetts). Purified and linearized plasmid in the concentration range of 1.7 × 102–1.7 × 107 copies/μL was used for the qPCR standard curve. The qPCR reaction was performed using the CFX-96 Touch real-time PCR system (Bio-Rad Laboratories, Hercules, California).
GTA for Quantification of ETU or Functional Particles
Over time, various protocols have been developed for in vitro assessment of AAV vector functionality. The suspension-adapted HEK293 cells were infected with serial dilutions of AAV5 (clarified lysate or affinity purified) and co-infected with an E1, E3 deleted adenovirus serotype 5-(ΔE1E3Ad5) at an MOI of 5 infectious particles/cell. In the case of cell lysates, the samples were heated at 60°C for 15 min before infection to deactivate existing baculovirus and to prevent false-positive results. At 22–24 hpi, the cells were harvested via centrifugation at 300 g for 5 min, and the cell pellet was resuspended and fixed in freshly prepared cold fixation buffer (final concentration, 2% paraformaldehyde in PBS) at 4°C for a minimum of 30 min. Later, the cell suspension was analyzed for GFP-positive cells via flow cytometry. Here, only ΔE1E3Ad5-infected negative control samples were used to set the gate, and a total of 10,000 events were recorded for data analysis. The linear range of quantification was established between 2%–20% GFP-positive cells.
Qualitative Analysis
AAV2 Rep and AAV5 Cap Western Blot
Clarified cell lysate samples were filtered through a 0.45 μm filter and used for western blot analysis. Lysate samples, equivalent of 6 × 104 cells, were mixed with preformulated 4× Laemmli sample buffer, to which was added a reducing agent per the manufacturer’s instructions (Bio-Rad Laboratories, Hercules, California), heated to 95°C for 15 min, and loaded onto a TGX stain-free gel, 4%–15% acrylamide (Bio-Rad Laboratories, Hercules, California). The gel was run at 120 V for 90 min. Once the run was complete, the protein was transferred to a nitrocellulose membrane using the Transblot Turbo Transfer System (Bio-Rad Laboratories, Hercules, California), per the manufacturer’s instructions. Next, the nitrocellulose membrane was placed in a blocking buffer-PBS containing 0.1% v/v Tween 20 (PBST) and 5% w/v skimmed milk and incubated for 1 h at room temperature. Afterward, the membrane was incubated overnight at 2°C–8°C with mouse IgG anti-AAV5 VP primary antibody (American Research Products, Waltham, Massachusetts) at a 1:2,000 dilution prepared in PBST (PBS containing 0.1% v/v Tween-20), and anti-AAV2 Rep primary antibody (American Research Products, Waltham, Massachusetts) at a 1:100 dilution prepared in PBST. Following this, the membrane was washed three times with PBST and incubated at room temperature with a horseradish peroxidase-conjugated rabbit polyclonal antibody against mouse IgG secondary antibody (Abcam, Cambridge, United Kingdom) at a 1:10,000 dilution prepared in PBST. Finally, the membrane was washed three times with PBST, incubated with enhanced chemiluminescence (ECL) reagent (Bio-Rad Laboratories, Hercules, California), and visualized using Bio-Rad’s ChemiDoc imager.
AAV5 VP Proteins Stoichiometry Analysis
Stoichiometric analysis of AAV5 VP subunits was performed via SDS-PAGE of the affinity-purified sample. The purified vector was serially diluted within 1 log range of the VGs, with the resulting amount loaded per well in the range of 2.2 × 108–2.2 × 109 VGs. After the run, the gel was stained with Flamingo fluorescent stain, per the manufacturer’s protocol (Bio-Rad Laboratories, Hercules, California). This stain was selected because it offers similar sensitivity and better linear range of quantification in comparison to silver stain. The stained gel was visualized via the ChemiDoc Imager, and the densitometric analysis of the resolved bands was performed using the Image Lab software suite. The linear range of quantification was established by plotting the VP band intensity as a function of VG amount. The best possible linearity between VP band intensity and VG was found to be in the concentration range 5.5 × 108–7.3 × 108 VG loaded (Figure S5). Also within this range, the VP3 band intensity was normalized—arbitrarily set to 10—and the relative band intensities of the three VP subunits were determined.
AUC of the Affinity-Purified AAV5
For AUC analysis, the affinity-purified AAV5 material (2 × 1012 VG/mL) was buffer exchanged into PBS and concentrated using a 30 kDa molecular weight cutoff centrifugal filter device, such that the 260 nm absorbance value for the concentrate was between 0.4 and 0.8. The concentrated sample was then analyzed using the Proteomelab XL-1 (Beckman Coulter, Brea, California). The sample chamber was charcoal-filled Epon with two-sector centerpieces with a 1.2 cm pathlength. The reference chamber was filled with 420 μL PBS (blank) and the sample chamber with concentrated AAV5. After the sample chamber was placed in the rotor, it was allowed to equilibrate at 20°C with full vacuum applied for 1 h. The sedimentation analysis run was performed at 20,000 rpm for 2 h at 20°C. The real-time data were captured with 260 nm optics. The Sedfit software was used for post-run data analysis. This free-access software was developed by a research group from NIH.
AAV Analysis via TEM
Of the virus dispersion, 10 μL, fixed in 2% paraformaldehyde, was dropped on a carbon-formvar-coated nickel grid for 10 min. Grids containing fixed viruses were washed with water prior to staining, and 10 μL 1% uranyl acetate was applied to the grid for 30 s. The excess of sample and negative stain solution was blotted with Whatman filter paper (Fisher Scientific, Hampton, New Hampshire), and the grid was air-dried prior to observation in TEM. The examination was performed with a Philips CM100 transmission electron microscope (Phillips, Eindhoven, the Netherlands). Electron micrographs were captured with an AMT XR80 digital camera (Advanced Microscopy Techniques, Woburn, Massachusetts).
Author Contributions
Conceptualization, P.R.H.J., P.S.C., and A.A.K.; Methodology, P.R.H.J., L.C., I.A., J.S., O.K., S.Z., P.S.C., and A.A.K.; Execution P.R.H.J., Writing – Original Draft, P.R.H.J.; Writing – Review & Editing, P.S.C., S.Z., and A.A.K.; Funding Acquisition, A.A.K.; Resources, P.S.C. and A.K.; and Supervision, P.S.C. and A.A.K.
Conflicts of Interest
S.Z. is a co-founder of Lacerta Therapeutics, a licensee of the insect cell technology used in this research.
Acknowledgments
P.R.H.J. is financially supported by a fellowship from the Faculty of Engineering, McGill University and the project by a grant from the Natural Sciences and Engineering Research Council (NSERC RGPIN-2015-05132) of the government of Canada. The authors thank Diane Gingras for performing the AAV TEM analysis, Marie-Hélène Venne for initial setup of the GTA, and Alice Bernier for assistance in chromatographic processing and a critical reading of the manuscript.
Footnotes
Supplemental Information includes six figures, three tables, and Supplemental Materials and Methods and can be found with this article online at https://doi.org/10.1016/j.omtm.2019.02.003.
Supplemental Information
References
- 1.Huang S., Kamihira M. Development of hybrid viral vectors for gene therapy. Biotechnol. Adv. 2013;31:208–223. doi: 10.1016/j.biotechadv.2012.10.001. [DOI] [PubMed] [Google Scholar]
- 2.Waehler R., Russell S.J., Curiel D.T. Engineering targeted viral vectors for gene therapy. Nat. Rev. Genet. 2007;8:573–587. doi: 10.1038/nrg2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ylä-Herttuala S. Endgame: glybera finally recommended for approval as the first gene therapy drug in the European union. Mol. Ther. 2012;20:1831–1832. doi: 10.1038/mt.2012.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clark K.R., Liu X., McGrath J.P., Johnson P.R. Highly purified recombinant adeno-associated virus vectors are biologically active and free of detectable helper and wild-type viruses. Hum. Gene Ther. 1999;10:1031–1039. doi: 10.1089/10430349950018427. [DOI] [PubMed] [Google Scholar]
- 5.Collaco R.F., Cao X., Trempe J.P. A helper virus-free packaging system for recombinant adeno-associated virus vectors. Gene. 1999;238:397–405. doi: 10.1016/s0378-1119(99)00347-9. [DOI] [PubMed] [Google Scholar]
- 6.Collaco R.F., Trempe J.P. A method for helper virus-free production of adeno-associated virus vectors. Methods Mol. Med. 2003;76:237–254. doi: 10.1385/1-59259-304-6:237. [DOI] [PubMed] [Google Scholar]
- 7.Gao G.P., Qu G., Faust L.Z., Engdahl R.K., Xiao W., Hughes J.V., Zoltick P.W., Wilson J.M. High-titer adeno-associated viral vectors from a Rep/Cap cell line and hybrid shuttle virus. Hum. Gene Ther. 1998;9:2353–2362. doi: 10.1089/hum.1998.9.16-2353. [DOI] [PubMed] [Google Scholar]
- 8.Harris J.D., Beattie S.G., Dickson J.G. Novel tools for production and purification of recombinant adeno-associated viral vectors. Methods Mol. Med. 2003;76:255–267. doi: 10.1385/1-59259-304-6:255. [DOI] [PubMed] [Google Scholar]
- 9.Xiao X., Li J., Samulski R.J. Production of high-titer recombinant adeno-associated virus vectors in the absence of helper adenovirus. J. Virol. 1998;72:2224–2232. doi: 10.1128/jvi.72.3.2224-2232.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chahal P.S., Schulze E., Tran R., Montes J., Kamen A.A. Production of adeno-associated virus (AAV) serotypes by transient transfection of HEK293 cell suspension cultures for gene delivery. J. Virol. Methods. 2014;196:163–173. doi: 10.1016/j.jviromet.2013.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clément N., Knop D.R., Byrne B.J. Large-scale adeno-associated viral vector production using a herpesvirus-based system enables manufacturing for clinical studies. Hum. Gene Ther. 2009;20:796–806. doi: 10.1089/hum.2009.094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Adamson-Small L., Potter M., Byrne B.J., Clément N. Sodium Chloride Enhances Recombinant Adeno-Associated Virus Production in a Serum-Free Suspension Manufacturing Platform Using the Herpes Simplex Virus System. Hum. Gene Ther. Methods. 2017;28:1–14. doi: 10.1089/hgtb.2016.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clément N., Grieger J.C. Manufacturing of recombinant adeno-associated viral vectors for clinical trials. Mol. Ther. Methods Clin. Dev. 2016;3:16002. doi: 10.1038/mtm.2016.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Emmerling V.V., Pegel A., Milian E.G., Venereo-Sanchez A., Kunz M., Wegele J., Kamen A.A., Kochanek S., Hoerer M. Rational plasmid design and bioprocess optimization to enhance recombinant adeno-associated virus (AAV) productivity in mammalian cells. Biotechnol. J. 2016;11:290–297. doi: 10.1002/biot.201500176. [DOI] [PubMed] [Google Scholar]
- 15.Durocher Y., Pham P.L., St-Laurent G., Jacob D., Cass B., Chahal P., Lau C.J., Nalbantoglu J., Kamen A. Scalable serum-free production of recombinant adeno-associated virus type 2 by transfection of 293 suspension cells. J. Virol. Methods. 2007;144:32–40. doi: 10.1016/j.jviromet.2007.03.014. [DOI] [PubMed] [Google Scholar]
- 16.Urabe M., Ding C., Kotin R.M. Insect cells as a factory to produce adeno-associated virus type 2 vectors. Hum. Gene Ther. 2002;13:1935–1943. doi: 10.1089/10430340260355347. [DOI] [PubMed] [Google Scholar]
- 17.Kohlbrenner E., Aslanidi G., Nash K., Shklyaev S., Campbell-Thompson M., Byrne B.J., Snyder R.O., Muzyczka N., Warrington K.H., Jr., Zolotukhin S. Successful production of pseudotyped rAAV vectors using a modified baculovirus expression system. Mol. Ther. 2005;12:1217–1225. doi: 10.1016/j.ymthe.2005.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen H. Intron splicing-mediated expression of AAV Rep and Cap genes and production of AAV vectors in insect cells. Mol. Ther. 2008;16:924–930. doi: 10.1038/mt.2008.35. [DOI] [PubMed] [Google Scholar]
- 19.Smith R.H., Levy J.R., Kotin R.M. A simplified baculovirus-AAV expression vector system coupled with one-step affinity purification yields high-titer rAAV stocks from insect cells. Mol. Ther. 2009;17:1888–1896. doi: 10.1038/mt.2009.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aslanidi G., Lamb K., Zolotukhin S. An inducible system for highly efficient production of recombinant adeno-associated virus (rAAV) vectors in insect Sf9 cells. Proc. Natl. Acad. Sci. USA. 2009;106:5059–5064. doi: 10.1073/pnas.0810614106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mietzsch M., Grasse S., Zurawski C., Weger S., Bennett A., Agbandje-McKenna M., Muzyczka N., Zolotukhin S., Heilbronn R. OneBac: platform for scalable and high-titer production of adeno-associated virus serotype 1-12 vectors for gene therapy. Hum. Gene Ther. 2014;25:212–222. doi: 10.1089/hum.2013.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mietzsch M., Casteleyn V., Weger S., Zolotukhin S., Heilbronn R. OneBac 2.0: sf9 cell lines for production of AAV5 vectors with enhanced infectivity and minimal encapsidation of foreign DNA. Hum. Gene Ther. 2015;26:688–697. doi: 10.1089/hum.2015.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kondratov O., Marsic D., Crosson S.M., Mendez-Gomez H.R., Moskalenko O., Mietzsch M., Heilbronn R., Allison J.R., Green K.B., Agbandje-McKenna M. Direct head-to head evaluation of recombinant adeno-associated viral vectors manufactured in human versus insect cells. Mol. Ther. 2017;25:2661–2675. doi: 10.1016/j.ymthe.2017.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bédard C., Kamen A., Tom R., Massie B. Maximization of recombinant protein yield in the insect cell/baculovirus system by one-time addition of nutrients to high-density batch cultures. Cytotechnology. 1994;15:129–138. doi: 10.1007/BF00762387. [DOI] [PubMed] [Google Scholar]
- 25.Elias C.B., Zeiser A., Bédard C., Kamen A.A. Enhanced growth of Sf-9 cells to a maximum density of 5.2 x 10(7) cells per mL and production of beta-galactosidase at high cell density by fed batch culture. Biotechnol. Bioeng. 2000;68:381–388. doi: 10.1002/(sici)1097-0290(20000520)68:4<381::aid-bit3>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 26.Mena J.A., Aucoin M.G., Montes J., Chahal P.S., Kamen A.A. Improving adeno-associated vector yield in high density insect cell cultures. J. Gene Med. 2010;12:157–167. doi: 10.1002/jgm.1420. [DOI] [PubMed] [Google Scholar]
- 27.Aucoin M.G., Perrier M., Kamen A.A. Critical assessment of current adeno-associated viral vector production and quantification methods. Biotechnol. Adv. 2008;26:73–88. doi: 10.1016/j.biotechadv.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 28.Burnham B., Nass S., Kong E., Mattingly M., Woodcock D., Song A., Wadsworth S., Cheng S.H., Scaria A., O’Riordan C.R. Analytical ultracentrifugation as an approach to characterize recombinant adeno-associated viral vectors. Hum. Gene Ther. Methods. 2015;26:228–242. doi: 10.1089/hgtb.2015.048. [DOI] [PubMed] [Google Scholar]
- 29.Girod A., Wobus C.E., Zádori Z., Ried M., Leike K., Tijssen P., Kleinschmidt J.A., Hallek M. The VP1 capsid protein of adeno-associated virus type 2 is carrying a phospholipase A2 domain required for virus infectivity. J. Gen. Virol. 2002;83:973–978. doi: 10.1099/0022-1317-83-5-973. [DOI] [PubMed] [Google Scholar]
- 30.Dorsch S., Liebisch G., Kaufmann B., von Landenberg P., Hoffmann J.H., Drobnik W., Modrow S. The VP1 unique region of parvovirus B19 and its constituent phospholipase A2-like activity. J. Virol. 2002;76:2014–2018. doi: 10.1128/JVI.76.4.2014-2018.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vihinen-Ranta M., Wang D., Weichert W.S., Parrish C.R. The VP1 N-terminal sequence of canine parvovirus affects nuclear transport of capsids and efficient cell infection. J. Virol. 2002;76:1884–1891. doi: 10.1128/JVI.76.4.1884-1891.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wells D.J. Systemic AAV gene therapy close to clinical trials for several neuromuscular diseases. Mol. Ther. 2017;25:834–835. doi: 10.1016/j.ymthe.2017.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Galibert L., Merten O.-W. Latest developments in the large-scale production of adeno-associated virus vectors in insect cells toward the treatment of neuromuscular diseases. J. Invertebr. Pathol. 2011;107(Suppl):S80–S93. doi: 10.1016/j.jip.2011.05.008. [DOI] [PubMed] [Google Scholar]
- 34.Allay J.A., Sleep S., Long S., Tillman D.M., Clark R., Carney G., Fagone P., McIntosh J.H., Nienhuis A.W., Davidoff A.M. Good manufacturing practice production of self-complementary serotype 8 adeno-associated viral vector for a hemophilia B clinical trial. Hum. Gene Ther. 2011;22:595–604. doi: 10.1089/hum.2010.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Powers A.D., Piras B.A., Clark R.K., Lockey T.D., Meagher M.M. Development and optimization of AAV hFIX particles by transient transfection in an iCELLis® fixed-bed bioreactor. Hum. Gene Ther. Methods. 2016;27:112–121. doi: 10.1089/hgtb.2016.021. [DOI] [PubMed] [Google Scholar]
- 36.Chahal P.S., Aucoin M.G., Kamen A. Primary recovery and chromatographic purification of adeno-associated virus type 2 produced by baculovirus/insect cell system. J. Virol. Methods. 2007;139:61–70. doi: 10.1016/j.jviromet.2006.09.011. [DOI] [PubMed] [Google Scholar]
- 37.Meghrous J., Aucoin M.G., Jacob D., Chahal P.S., Arcand N., Kamen A.A. Production of recombinant adeno-associated viral vectors using a baculovirus/insect cell suspension culture system: from shake flasks to a 20-L bioreactor. Biotechnol. Prog. 2005;21:154–160. doi: 10.1021/bp049802e. [DOI] [PubMed] [Google Scholar]
- 38.Nagrete A., Kotin R.M. Production of recombinant adeno-associated vectors using two bioreactors configurations at different scales/ J. Virol Methods. 2007;145:155–161. doi: 10.1016/j.jviromet.2007.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mena J.A., Ramírez O.T., Palomares L.A. Population kinetics during simultaneous infection of insect cells with two different recombinant baculoviruses for the production of rotavirus-like particles. BMC Biotechnol. 2007;7:39. doi: 10.1186/1472-6750-7-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nandakumar S., Ma H., Khan A.S. Whole-genome sequence of the Spodoptera frugiperda Sf9 Insect Cell Line. Genome Announc. 2017;5:e00829-17. doi: 10.1128/genomeA.00829-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.