The domain of unknown function protein CgnJ from the crocagin-biosynthetic gene cluster has no sequence homologues in the Protein Data Bank, but is a structural homologue of ComJ. CgnJ appears to bind the natural product crocagin with weak affinity. The structural homology may imply a function for CgnJ.
Keywords: crocagin, RiPPs, CgnJ, domains of unknown function
Abstract
Natural products often contain interesting new chemical entities that are introduced into the structure of a compound by the enzymatic machinery of the producing organism. The recently described crocagins are novel polycyclic peptides which belong to the class of ribosomally synthesized and post-translationally modified peptide natural products. They have been shown to bind to the conserved prokaryotic carbon-storage regulator A in vitro. In efforts to understand crocagin biosynthesis, the putative biosynthetic genes were expressed and purified. Here, the first crystal structure of a protein from the crocagin-biosynthetic gene cluster, CgnJ, a domain of unknown function protein, is reported. Possible functions of this protein were explored by structural and sequence homology analyses. Even though the sequence homology to proteins in the Protein Data Bank is low, the protein shows significant structural homology to a protein with known function within the competency system of Bacillus subtilis, ComJ, leading to the hypothesis of a similar role of the protein within the producing organism.
1. Introduction
Natural products have been a source of bioactive substances with often interesting and novel chemical scaffolds since the discovery of penicillin in 1929 (Lobanovska & Pilla, 2017 ▸). Ribosomally synthesized and post-translationally modified peptide (RiPP) natural products are a fast-growing family of chemically and structurally diverse compounds with interesting and potent bioactivities (Dunbar & Mitchell, 2013 ▸). RiPPs are produced from a ribosomal precursor peptide, which undergoes one or more maturation steps to yield the final, often heavily modified, natural product (Hetrick & van der Donk, 2017 ▸). As is common for prokaryotes, the proteins involved in RiPP biosynthesis are usually organized as gene clusters (Doroghazi & Metcalf, 2013 ▸). Modern advances in the field of systems biology, such as secondary metabolome mining, allow an efficient search for novel natural products and allow the biosynthetic potential of natural producers to be uncovered (Maansson et al., 2016 ▸). The use of these techniques led to the recent identification and isolation of the crocagins, a novel group of polycyclic RiPPs from Chondromyces crocatus Cm c5 containing a tetrahydropyrrolo-[2,3-b]-indole core (Fig. 1 ▸ a) (Viehrig et al., 2017 ▸). While searching for potential activities of these natural products, they were discovered to bind carbon-storage regulator A (CsrA; Viehrig et al., 2017 ▸). CsrA is a highly conserved key player in the Csr-type regulatory system, an important post-transcriptional control mechanism involving small regulatory RNAs (sRNAs), which coordinates the expression of a variety of proteins, including specific virulence factors, as well as biofilm formation (Maurer et al., 2016 ▸). CsrA-type proteins have been found in over 1200 bacterial genomes, including those of important human Gram-negative pathogens such as Legionella pneumophila, Pseudomonas aeruginosa, Escherichia coli, Acinetobacter baumannii and Salmonella typhimurium (Heroven et al., 2012 ▸). CsrA is currently not the target of any known antibiotic, and the potential ability of crocagins to interfere with CsrA-controlled processes, especially in Gram-negative bacteria, makes them highly attractive compounds for further research.
Figure 1.
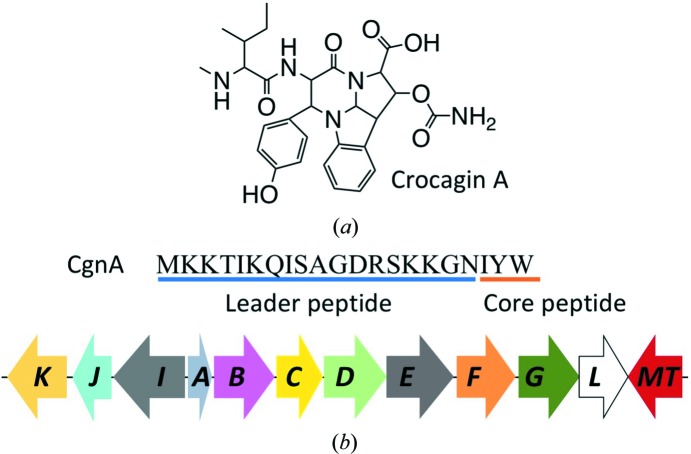
(a) Chemical structure of crocagin A. (b) Sequence of the crocagin precursor peptide CgnA and the crocagin-biosynthetic gene cluster. The leader peptide is underlined in blue and the core peptide in orange. All genes encoding proteins thought to be involved in crocagin biosynthesis are shown (cgnB–cgnL; cgnMT, methyltransferase).
In addition to the natural products, the biosynthetic gene cluster of crocagins was identified by a combination of genome analysis, targeted gene inactivation of the producer and in vitro experiments (Fig. 1 ▸ b; Viehrig et al., 2017 ▸). Within the gene cluster, nine potentially biosynthetic genes (cgnB–cgnE, cgnI–cgnL and cgnMT) have been identified and putative functions have been assigned by sequence homology (Viehrig et al., 2017 ▸). cgnA encodes the precursor peptide containing the core peptide sequence IYW (Fig. 1 ▸ b), which is modified to yield crocagin. By utilizing a targeted gene-inactivation approach and in vivo metabolome studies, the functions of two biosynthetic genes, cgnB and cgnI, could be explored, leaving the actual biosynthetic functions of all of the other genes in the cluster as hypothetical (Viehrig et al., 2017 ▸). Of these, CgnJ was the only protein determined to be a domain of unknown function (DUF) protein, leaving questions as to its potential role and biosynthetic function. Even though functions for DUF proteins cannot be routinely determined using sequence homology to known proteins, they can be found in all species, leading to the hypothesis that they fulfil an essential role owing to being evolutionarily conserved (Goodacre et al., 2013 ▸). In the post-genomic era, the relevance of DUF proteins became visible, with well over 3700 different families in the Pfam database (Mudgal et al., 2015 ▸). Combining these results with a structural study of 240 DUF proteins conducted by the Protein Structure Initiative, it was shown that only as few as 20% contain completely novel folds, suggesting the use of X-ray crystallography as an important tool in the elaboration of potential functions of DUF proteins (Jaroszewski et al., 2009 ▸). Furthermore, this sometimes allows the co-crystallization of a ligand in the process, providing clues about the putative role of the protein (Bateman et al., 2010 ▸).
Here, we present the first structural and biophysical study of a protein from the crocagin-biosynthetic gene cluster: the domain of unknown function protein CgnJ.
2. Materials and methods
2.1. Macromolecule production and affinity measurements
2.1.1. Expression of native and l-selenomethionine-labelled CgnJ
The open reading frame for CgnJ was cloned from C. crocatus genomic DNA into the pHIS-SUMO-TEV plasmid with an N-terminal His6 tag and a Tobacco etch virus (TEV) protease site (a kind gift from Dr David Owen). The protein was expressed in E. coli BL21 (DE3) cells grown in Luria–Bertani (LB) medium supplemented with 50 µg ml−1 kanamycin. Cultures were grown at 37°C and 200 rev min−1 until the OD600 reached 0.6, and the cells were then induced by the addition of isopropyl β-d-1-thiogalactopyranoside (IPTG; final concentration 0.1 mM) and further grown at 16°C and 180 rev min−1 overnight.
l-Selenomethionine-labelled (SeMet) CgnJ was expressed from E. coli BL21 (DE3) cells grown in minimal medium supplemented with glucose-free nutrient mix, 50 µg ml−1 kanamycin and 5% glycerol. This medium was inoculated with an overnight culture grown in LB, which was washed three times in minimal medium. After 20 min of growth at 37°C, 40 mg l−1 l-selenomethionine was added. The cultures were returned to 37°C and 200 rev min−1 and grown until an OD600 of 0.6 was reached, whereupon 100 mg l−1 lysine, phenylalanine and threonine and 50 mg l−1 isoleucine and valine were added. After incubation for a further 20 min, expression was induced with IPTG (final concentration 1 mM) and the cells were grown at 20°C for 24 h. For both the native and SeMet variants, the cells were harvested by centrifugation (4000g, 4°C, 15 min).
2.1.2. Purification of native and SeMet CgnJ
The cell pellets were resuspended in lysis buffer [50 mM Tris pH 7.8, 200 mM NaCl, 20 mM imidazole, 10% glycerol, 3 mM β-mercaptoethanol (BME)] supplemented with 0.4 mg g−1 DNAse and cOmplete EDTA-free protease-inhibitor tablets (Roche; one tablet per 50 ml of resuspension). The cells were lysed via passage through a cell disruptor at 207 MPa (Microfluidics) and the cell debris was removed by centrifugation (30 000g, 4°C, 20 min). The supernatant was loaded onto a pre-equilibrated Ni–NTA column at 4°C and the column was washed with 20 column volumes of lysis buffer. The protein was eluted from the column in elution buffer (lysis buffer supplemented with 250 mM imidazole) and fractions containing protein were pooled and passed over a desalting column pre-equilibrated in lysis buffer (16/10 Desalting, GE Healthcare) to remove excess imidazole. TEV protease was added at a mass:mass ratio of 1:10 and the sample was incubated for 1 h at 20°C to remove the N-terminal His6-SUMO tag. The digested sample was passed over a second Ni–NTA column pre-equilibrated in lysis buffer and the flowthrough was collected, concentrated and loaded onto a Superdex 200 gel-filtration column (GE Healthcare) equilibrated in GF buffer (10 mM HEPES pH 7.4, 150 mM NaCl, 1 mM TCEP; Supplementary Fig. S1). Protein yields were 24 and 16 mg l−1 for native and SeMet CgnJ, respectively. The purity of the protein was determined by SDS–PAGE (Supplementary Fig. S1). Pure CgnJ protein was concentrated to 3.75 mg ml−1.
2.1.3. Expression and purification of His6-CgnJ
To allow His6-tag labelling of CgnJ for microscale thermophoresis experiments (see below), the open reading frame for CgnJ was subcloned into the pHIS-TEV plasmid with an N-terminal His6 tag and a Tobacco etch virus (TEV) protease site (Liu & Naismith, 2009 ▸). The protein was expressed as described for native pHIS-SUMO-TEV CgnJ. The cell pellets were resuspended in lysis buffer (20 mM Tris pH 8.0, 500 mM NaCl, 20 mM imidazole, 3 mM BME) supplemented with 0.4 mg g−1 DNAse and cOmplete EDTA-free protease-inhibitor tablets (Roche; one tablet per 50 ml of resuspension). The cells were lysed via passage through a cell disruptor at 207 MPa (Microfluidics) and the cell debris was removed by centrifugation (30 000g, 4°C, 20 min). The supernatant was loaded onto a pre-equilibrated Ni–NTA column at 4°C and the column was washed with 20 column volumes of lysis buffer. The protein was eluted from the column in elution buffer (lysis buffer supplemented with 250 mM imidazole) and the fractions containing the largest amounts of protein were pooled and loaded onto a Superdex 200 gel-filtration column (GE Healthcare) equilibrated in GF buffer (10 mM HEPES pH 7.4, 150 mM NaCl, 1 mM TCEP). The purity of the His6-tagged protein was determined by SDS–PAGE.
2.1.4. Microscale thermophoresis (MST)
MST experiments were performed using a Monolith NT.115 instrument (NanoTemper) in MST buffer (50 mM Tris–HCl pH 7.4, 150 mM NaCl, 10 mM MgCl2, 0.05% Tween 20) at 20°C. His6-CgnJ was dialysed into freshly prepared MST buffer, while a 100 mM stock of crocagin in 100% DMSO was diluted to a starting concentration of 5 mM in MST buffer. The proteins were fluorescently labelled using the Monolith His-tag labelling kit RED-tris-NTA (NanoTemper) according to the manufacturer’s instructions. For MST measurements, the manufacturer’s instructions were followed using a starting concentration of 2.5 mM crocagin A, 20% excitation power and 40% MST power. The K d value was estimated using the MO.Affinity software suite (NanoTemper). Macromolecule-production information is summarized in Table 1 ▸.
Table 1. Macromolecule-production information.
| Source organism | C. crocatus Cm c5 |
| DNA source | Genomic DNA |
| Forward primer | CTTCCATGGCAGAGGAAGTCGCCGAG |
| Reverse primer | CTTAAGCTTTCATCTCTCGGGCCACAACGT |
| Cloning vector | pHis-SUMO-TEV |
| Expression vector | pHis-SUMO-TEV |
| Expression host | E. coli |
| Complete amino-acid sequence of the construct produced | MGSSHHHHHHGSDSEVNQEAKPEVKPEVKPETHINLKVSDGSSEIFFKIKKTTPLRRLMEAFAKRQGKEMDSLRFLYDGIRIQADQTPEDLDMEDNDIIEAHREQIGGDIPTTENLYFQGAMAEEVAEIILPASTWILFFDASCSINSPAFWSTNDAVDRIWRLKIAHELVLLQVVLEGYFKVRCILRSSAPAFEMVNADVSELVSIVLPSGRLVACTTDEPTLNRHVLTVPPGRYRVLREWSVHEESKHYDVESAEAYPADEGPDGIITLWPER |
2.2. Crystallization
Crystal screens for SeMet CgnJ were set up by the sitting-drop vapour-diffusion method using a Gryphon robot (Art Robbins). The protein was screened against The Classics Suite (Qiagen) at 20°C. Initial crystals formed in condition C2 (1 M ammonium phosphate, 0.1 M trisodium citrate pH 5.6) after 48 h. Diffraction-quality crystals (tetragonal bipyramids) were grown in a condition consisting of 0.75 M ammonium phosphate, 0.1 M trisodium citrate pH 5.8. Crystals of native CgnJ grew in the same condition as the SeMet CgnJ protein. A single SeMet CgnJ crystal was cryoprotected in mother liquor supplemented with 34% glycerol and flash-cooled in liquid nitrogen. Crystallization information is summarized in Table 2 ▸.
Table 2. Crystallization.
| Method | Vapour diffusion |
| Plate type | Swissci |
| Temperature (K) | 291 |
| Protein concentration (mg ml−1) | 3.75 |
| Buffer composition of protein solution | 150 mM NaCl, 10 mM HEPES, 1 mM TCEP pH 7.4 |
| Composition of reservoir solution | 0.75 M ammonium phosphate, 0.1 M trisodium citrate pH 5.8 |
| Volume and ratio of drop | 1:1 µl, 1:2 µl (reservoir:protein) |
| Volume of reservoir (µl) | 500 |
2.3. Data collection and processing
A single-wavelength anomalous dispersion (SAD) data set was collected at the Se K absorption edge at 100 K on beamline X06DA at the Swiss Light Source (SLS). Data were reduced and scaled with maximum redundancy for optimal SAD phasing using xia2 (Winter, 2010 ▸). For the highest possible resolution, the native data set was reduced and scaled at an outer shell 〈I/σ(I)〉 value of 2.9 using xia2. Table 3 ▸ shows the data-collection statistics for the high-resolution native and the SAD data sets.
Table 3. Data collection and processing.
Values in parentheses are for the outer shell.
| Native | SAD | |
|---|---|---|
| Diffraction source | BL14.1, BESSY II | X06DA, SLS |
| Wavelength (Å) | 0.91841 | 0.97941 |
| Temperature (K) | 100 | 100 |
| Detector | PILATUS 6M | PILATUS 2M-F |
| Crystal-to-detector distance (mm) | 349.9 | 152.25 |
| Rotation range per image (°) | 0.10 | 0.20 |
| Total rotation range (°) | 100 | 360 |
| Exposure time per image (s) | 0.4 | 0.1 |
| Space group | P41212 | P41212 |
| a, b, c (Å) | 132.56, 132.56, 112.98 | 132.90, 132.90, 112.85 |
| α, β, γ (°) | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 |
| Mosaicity (°) | 0.063 | 0.060 |
| Resolution range (Å) | 57.17–2.00 (2.05–2.00) | 42.06–2.19 (2.25–2.19) |
| Total No. of reflections | 504776 (38046) | 1416110 (97301) |
| No. of unique reflections | 68294 (4964) | 52596 (3814) |
| Completeness (%) | 99.97 (99.99) | 100.0 (100.0) |
| Multiplicity | 7.4 (7.7) | 26.9 (25.5) |
| 〈I/σ(I)〉 | 18.5 (2.9) | 23.9 (5.1) |
| R r.i.m. (%) | 4.8 (38.7) | 3.8 (22.2) |
| Overall B factor from Wilson plot (Å2) | 24.006 | 23.291 |
2.4. Structure solution and refinement
The structure was solved using the AutoSol package (Terwilliger et al., 2009 ▸) and the chains were built into electron density using the AutoBuild package (Terwilliger et al., 2008 ▸) from the PHENIX crystallographic software suite (Adams et al., 2010 ▸). The initial model was used for molecular replacement (MR) of a higher resolution native data set. MR was performed using Phaser (McCoy et al., 2007 ▸) and the structure was manually rebuilt in Coot (Emsley et al., 2010 ▸) and refined using phenix.refine (Afonine et al., 2012 ▸). TLS restraints were generated in PHENIX (Adams et al., 2010 ▸) and the structure was validated using MolProbity (Chen et al., 2010 ▸). Refinement statistics are summarized in Table 4 ▸.
Table 4. Structure refinement.
Values in parentheses are for the outer shell.
| Resolution range (Å) | 57.17–2.00 (2.071–2.000) |
| Completeness (%) | 99.97 (99.99) |
| σ Cutoff | 2.9 |
| No. of reflections, working set | 64810 |
| No. of reflections, test set | 3484 |
| Final R cryst (%) | 17.5 |
| Final R free (%) | 19.9 |
| No. of non-H atoms | |
| Total | 4157 |
| Protein | 3695 |
| Ligand | 18 |
| Water | 444 |
| R.m.s. deviations | |
| Bonds (Å) | 0.003 |
| Angles (°) | 0.66 |
| Average B factors (Å2) | |
| Overall | 33.2 |
| Protein | 31.8 |
| Ligand | 76.0 |
| Water | 43.2 |
| Ramachandran plot | |
| Favoured regions (%) | 98.04 |
| Additionally allowed (%) | 1.96 |
| Outliers (%) | 0.00 |
All structural images were presented using PyMOL (https://pymol.org/2/support.html). All sequence alignments were created using Clustal Omega (Sievers & Higgins, 2014 ▸).
3. Results and discussion
3.1. Crystal structure of CgnJ
The open reading frame annotated as CgnJ encoding a 154-amino-acid protein was amplified from genomic DNA isolated from C. crocatus Cm c5 (Accession No. WP_050430638). No homologues of this protein can be found using standard BLAST parameters, except for a hypothetical protein from Pelomonas sp. Root1217 (accession No. WP_057297704), which possesses 30% sequence identity over 110 amino acids. CgnJ was expressed and purified as described in Section 2, and the protein purity and integrity were determined by SDS–PAGE (Supplementary Fig. S1).
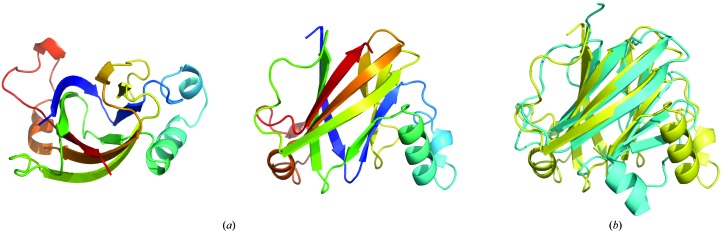
Native and SeMet CgnJ crystallized in space group P41212, and high-redundancy data were collected to 2.19 Å resolution from a SeMet CgnJ crystal at the selenium edge. The data-collection and refinement statistics can be found in Tables 3 ▸ and 4 ▸. The structure was determined using Se-SAD, and the initial model was used for molecular replacement of a 2.0 Å resolution native data set. The native crystal contained three CgnJ molecules in the asymmetric unit, and in the final model each protomer contains residues 1–154 (the full length). The structure has a fold that is comprised of four α-helices and nine β-strands (Fig. 2 ▸ a). The β-strands are arranged as four- and five-stranded sheets that form a β-barrel-like core decorated with loops and helices. Chains B and C appeared to form a dimer, but analysis with PISA (Krissinel & Henrick, 2007 ▸) gives a complex-formation significance score of 0, indicating that this is a crystal contact rather than a biological dimer. When analysed using dynamic light scattering, CgnJ appears to be monomeric in solution (Supplementary Fig. S2).
Figure 2.
(a) X-ray crystal structure of CgnJ represented as a cartoon using a rainbow colour scheme: top and side views (90° rotation). (b) Superposition of the X-ray crystal structures of CgnJ (yellow) and ComJ (cyan).
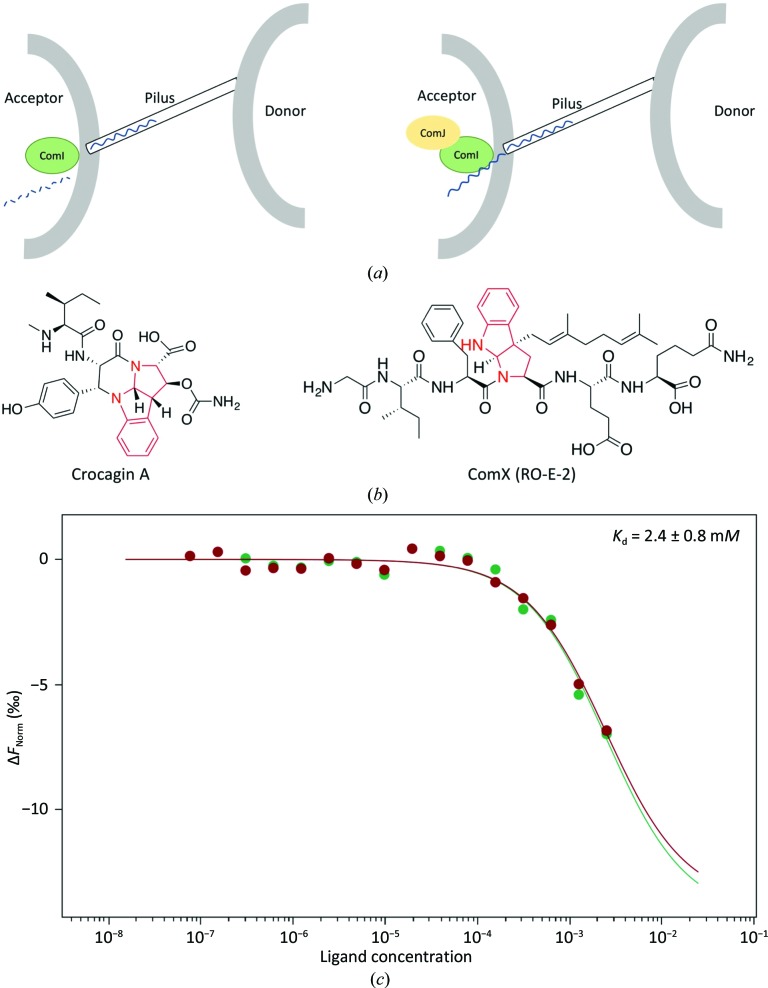
To search for structural homologues that may point to the function of CgnJ, we used the DALI server (Holm & Laakso, 2016 ▸) for a heuristic PDB search with a CgnJ monomer as the search model. When using a cutoff of 50% sequence coverage, the search returns only one protein, the nuclease inhibitor ComJ from Bacillus subtilis (PDB entry 4mqd; Midwest Center for Structural Genomics, unpublished work), with a Z-score of 12.2 and a Cα r.m.s.d. of 2.6 Å over 117 residues. The β-strands are very well conserved between the two proteins, and the main differences are found in the helices and loops at the periphery of the two proteins (Fig. 2 ▸ b). The good overall structural conservation was surprising, since the sequence identity between the two proteins is only 15.6% (Supplementary Fig. S3). The function of ComJ is known (Fig. 3 ▸ a). It is part of the natural competence machinery in B. subtilis and plays a pivotal role during the bacterial conjugation process (Zhang et al., 2012 ▸). As donor DNA tries to enter the recipient’s cell, it has to pass the nuclease ComI (also known as NucA). This nuclease is active and prevents DNA from entering until it is bound by ComJ, which renders the nuclease inactive and thus facilitates horizontal gene transfer (Zhang et al., 2012 ▸). Interestingly, ComJ and ComI are part of the Gram-negative quorum-sensing system involving the competence-stimulating peptide (CSP) ComX (Fig. 3 ▸ b). This bacterial pheromone also belongs to the RiPP family of natural products and does not share sequence homology with crocagin (Magnuson et al., 1994 ▸), yet both natural products contain a tryptophan residue that is cyclized via its side-chain C2 atom to its amide nitrogen. While ComX is polyprenylated at the Cγ atom, crocagins are hydroxylated (and subsequently carbamoylated) at the Cβ position. Since no biological activity has yet been reported for crocagins, it is possible that they play a role in quorum sensing.
Figure 3.
(a) Cartoon representing the role of ComJ. The nuclease ComI degrades incoming DNA unless it is bound by ComJ, which inactivates ComI and allows foreign DNA to enter the cell. (b) Chemical structures of crocagin and ComX (RO-E-2). The similar tryptophan modifications are highlighted in red. (c) Two independent MST measurements of CgnJ with crocagin A (green and red). Since saturation could not be reached (owing to compound solubility), the K d is estimated to be 2.4 ± 0.8 mM.
3.2. Investigating the potential binding of CgnJ to crocagin
Since the structural homology of CgnJ to ComJ hinted at a possible function, we searched for additional homologues of the competency system in C. crocatus using standard BLAST parameters. We were unable to find additional components based on sequence homology, perhaps owing to the low degree of sequence conservation. Attempts to identify a homologue of the nuclease ComI as a potential binding partner of CgnJ were also unsuccessful. We wondered whether CgnJ may be able to bind the precursor peptide CgnA or modify it biochemically. After 24 h at 37°C, no modification of CgnA was observed (Supplementary Fig. S4) and no binding between CgnJ and CgnA was detected using microscale thermophoresis (MST; Supplementary Fig. S4). Given the small size of the protein and its lack of homology to known RiPP recognition proteins, we decided to investigate whether CgnJ was able to bind the natural product crocagin itself. To this end, we used purified crocagin A to determine the affinity of these two components by MST. We were able to demonstrate that crocagin A binds to CgnJ with low affinity (Fig. 3 ▸ c). While the binding was robust and reproducible, compound solubility prevented us from using sufficiently high concentrations of crocagin to reach saturation. The K d was estimated to be 2.8 ± 0.8 mM. The yield of crocagin was reported to be 0.2 mg l−1 of pure compound after isolation and purification (Viehrig et al., 2017 ▸). This equates to roughly 0.4 µM and would indicate that even if the yield had been 0.1% of the compound found in the actual culture, an interaction would be unlikely to occur in vivo. However, whether crocagin is produced at a steady rate or in a burst remains unclear, as does the concentration of crocagin that can be reached inside the producer during production. Further experiments will thus be required to investigate the biological relevance of this in vitro interaction.
4. Conclusion
The DUF protein CgnJ has only one sequence homologue in publicly available sequence data. X-ray crystallography revealed the structure of CgnJ to be related to that of the nuclease inhibitor ComJ from B. subtilis, prompting us to search for homologous nucleases in C. crocatus. Owing to the very low sequence homology, we are unsure whether our inability to find ComI homologues rules out the presence of a structural homologue. In a search for a possible role of CgnJ, we tested whether this protein is able to bind the natural product itself. Despite a measurable affinity, the interaction is unlikely to be biologically relevant. Molecules which are structurally related to crocagin, such as kawaguchipeptin (Ishida et al., 1997 ▸), ComX (Okada et al., 2005 ▸) and kapakahine (Nakao et al., 1995 ▸) (Supplementary Fig. S5), have been assigned biological functions. It will require further study to determine whether the interaction of crocagin with CsrA is biologically relevant.
Supplementary Material
PDB reference: CgnJ, 6i86
Materials and methods and supplementary figures.. DOI: 10.1107/S2053230X19000712/ek5012sup1.pdf
Acknowledgments
We thank the staff of beamline X06DA at the Swiss Light Source for their support with data collection. SA would like to thank Rebekka Christmann for help with DLS experiments.
Funding Statement
This work was funded by Deutsche Forschungsgemeinschaft grant 4116/3-1 to Jesko Koehnke. Boehringer Ingelheim Stiftung grant .
References
- Adams, P. D., Afonine, P. V., Bunkóczi, G., Chen, V. B., Davis, I. W., Echols, N., Headd, J. J., Hung, L.-W., Kapral, G. J., Grosse-Kunstleve, R. W., McCoy, A. J., Moriarty, N. W., Oeffner, R., Read, R. J., Richardson, D. C., Richardson, J. S., Terwilliger, T. C. & Zwart, P. H. (2010). Acta Cryst. D66, 213–221. [DOI] [PMC free article] [PubMed]
- Afonine, P. V., Grosse-Kunstleve, R. W., Echols, N., Headd, J. J., Moriarty, N. W., Mustyakimov, M., Terwilliger, T. C., Urzhumtsev, A., Zwart, P. H. & Adams, P. D. (2012). Acta Cryst. D68, 352–367. [DOI] [PMC free article] [PubMed]
- Bateman, A., Coggill, P. & Finn, R. D. (2010). Acta Cryst. F66, 1148–1152. [DOI] [PMC free article] [PubMed]
- Chen, V. B., Arendall, W. B., Headd, J. J., Keedy, D. A., Immormino, R. M., Kapral, G. J., Murray, L. W., Richardson, J. S. & Richardson, D. C. (2010). Acta Cryst. D66, 12–21. [DOI] [PMC free article] [PubMed]
- Doroghazi, J. R. & Metcalf, W. W. (2013). BMC Genomics, 14, 611. [DOI] [PMC free article] [PubMed]
- Dunbar, K. L. & Mitchell, D. A. (2013). ACS Chem. Biol. 8, 473–487. [DOI] [PMC free article] [PubMed]
- Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. (2010). Acta Cryst. D66, 486–501. [DOI] [PMC free article] [PubMed]
- Goodacre, N. F., Gerloff, D. L. & Uetz, P. (2013). MBio, 5, e00744-13. [DOI] [PMC free article] [PubMed]
- Heroven, A. K., Böhme, K. & Dersch, P. (2012). RNA Biol. 9, 379–391. [DOI] [PubMed]
- Hetrick, K. J. & van der Donk, W. A. (2017). Curr. Opin. Chem. Biol. 38, 36–44. [DOI] [PMC free article] [PubMed]
- Holm, L. & Laakso, L. M. (2016). Nucleic Acids Res. 44, W351–W355. [DOI] [PMC free article] [PubMed]
- Ishida, K., Matsuda, H., Murakami, M. & Yamaguchi, K. (1997). J. Nat. Prod. 60, 724–726. [DOI] [PubMed]
- Jaroszewski, L., Li, Z., Krishna, S. S., Bakolitsa, C., Wooley, J., Deacon, A. M., Wilson, I. A. & Godzik, A. (2009). PLoS Biol. 7, e1000205. [DOI] [PMC free article] [PubMed]
- Krissinel, E. & Henrick, K. (2007). J. Mol. Biol. 372, 774–797. [DOI] [PubMed]
- Liu, H. & Naismith, J. H. (2009). Protein Expr. Purif. 63, 102–111. [DOI] [PMC free article] [PubMed]
- Lobanovska, M. & Pilla, G. (2017). Yale J. Biol. Med. 90, 135–145. [PMC free article] [PubMed]
- Maansson, M., Vynne, N. G., Klitgaard, A., Nybo, J. L., Melchiorsen, J., Nguyen, D. D., Sanchez, L. M., Ziemert, N., Dorrestein, P. C., Andersen, M. R. & Gram, L. (2016). mSystems, 1, e00028-15. [DOI] [PMC free article] [PubMed]
- Magnuson, R., Solomon, J. & Grossman, A. D. (1994). Cell, 77, 207–216 [DOI] [PubMed]
- Maurer, C. K., Fruth, M., Empting, M., Avrutina, O., Hossmann, J., Nadmid, S., Gorges, J., Herrmann, J., Kazmaier, U., Dersch, P., Müller, R. & Hartmann, R. W. (2016). Future Med. Chem. 8, 931–947. [DOI] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst. 40, 658–674. [DOI] [PMC free article] [PubMed]
- Mudgal, R., Sandhya, S., Chandra, N. & Srinivasan, N. (2015). Biol. Direct, 10, 38. [DOI] [PMC free article] [PubMed]
- Nakao, Y., Yeung, B. K. S., Yoshida, W. Y., Scheuer, P. J. & Kelly-Borges, M. (1995). J. Am. Chem. Soc. 117, 8271–8272.
- Okada, M., Sato, I., Cho, S. J., Iwata, H., Nishio, T., Dubnau, D. & Sakagami, Y. (2005). Nature Chem. Biol. 1, 23–24. [DOI] [PubMed]
- Sievers, F. & Higgins, D. G. (2014). Curr. Protoc. Bioinformatics, 48, 3.13.1–3.13.16. [DOI] [PubMed]
- Terwilliger, T. C., Adams, P. D., Read, R. J., McCoy, A. J., Moriarty, N. W., Grosse-Kunstleve, R. W., Afonine, P. V., Zwart, P. H. & Hung, L.-W. (2009). Acta Cryst. D65, 582–601. [DOI] [PMC free article] [PubMed]
- Terwilliger, T. C., Grosse-Kunstleve, R. W., Afonine, P. V., Moriarty, N. W., Zwart, P. H., Hung, L.-W., Read, R. J. & Adams, P. D. (2008). Acta Cryst. D64, 61–69. [DOI] [PMC free article] [PubMed]
- Viehrig, K., Surup, F., Volz, C., Herrmann, J., Abou Fayad, A., Adam, S., Köhnke, J., Trauner, D. & Müller, R. (2017). Angew. Chem. Int. Ed. 56, 7407–7410. [DOI] [PubMed]
- Winter, G. (2010). J. Appl. Cryst. 43, 186–190.
- Zhang, D., de Souza, R. F., Anantharaman, V., Iyer, L. M. & Aravind, L. (2012). Biol. Direct, 7, 18. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: CgnJ, 6i86
Materials and methods and supplementary figures.. DOI: 10.1107/S2053230X19000712/ek5012sup1.pdf