Abstract
Extracellular vesicles (EVs) play an important role as active messengers in intercellular communication and distant microenvironment modeling. Increasingly, these EVs are recognized as important biomarkers for clinical diagnostics. However, current isolation methods of EVs are time-consuming and ineffective due to the high diffusive characteristics of nanoparticles coupled with fluid flow instability. Here, we develop a microfluidic CEntrifugal Nanoparticles Separation and Extraction (µCENSE) platform for the rapid and label-free isolation of microvesicles. By utilizing centrifugal microhydrodynamics, we subject the nanosuspensions between 100 nm and 1000 nm to a unique fluid flow resulting in a zonal separation into different outlets for easy post-processing. Our centrifugal platform utilizes a gentle and efficient size-based separation without the requirements of syringe pump and other accessories. Based on our results, we report a high separation efficiency of 90% and an extraction purity of 85% within a single platform. Importantly, we demonstrate high EV extraction using a table top centrifuge within a short duration of eight minutes. The simple processes and the small volume requirement further enhance the utility of the platform. With this platform, it serves as a potential for liquid biopsy extraction and point-of-care diagnostics.
I. INTRODUCTION
Extracellular vesicles (EVs), comprising microvesicles and exosomes, are recognized as key intermediary components between cell-cell communications.1–5 These phospholipid spherical vesicles, spanning between 50 nm and 1000 nm in diameter, are released from mammalian cells via exocytosis or through the fission of plasma membrane. Biomolecules are transported within the nanovesicles to distant sites without degradation. The EVs may be assimilated into recipient cells through endocytosis or fused directly with the plasma membrane, thereby releasing its exosomal content into the cytosol of the target cell. In recent years, researchers reported that the transfer of proteins and nucleic acids by EVs alters the pathways of the recipient cell at a post-transcriptional level.6 Such reconditioning facilitates an optimal environment for diseased cells to reside, and is an important criterion for cancer metastasis,7–11 cardiovascular diseases,12,13 and hematological diseases,1,14 among others. In fact, exosomes were found to be in higher quantity in patients of certain cancers,15 and could serve as an important diagnostic biomarker.16–18
Despite their diverse role in various pathologies, extraction of EVs is highly challenging due to the high diffusive characteristics of the nanoparticles. Conventionally, researchers use differential ultracentrifugation approaches to remove unwanted debris based on their size and densities. However, to isolate EVs, high centrifugal forces more than 100 000 × g are often necessary to create sufficient forces on the nanoparticles.15,19,20 These methods are not only tedious and time-consuming (>8 h), they also require large purified samples (>10 ml) which are often impractical with biofluids. Moreover, studies have shown that the high centrifugal forces resulted in exosome fusion, altering their structural properties and functions.21 Consequently, this affects the extraction yield and downstream analyses. Alternatively, precipitation kits have been commercialized in the last few years,22 but these methods require additional reagents, which may affect the yield and processing time.
In order to overcome these shortcomings, microfluidics has been proposed as a promising alternative for EV enrichment.23,24 Over the past few years, microfluidic approaches utilizing immunoaffinity capture of specific exosomes have been developed.25–28 For example, Kanwar et al. developed a microfluidic platform functionalized with antibodies against CD63 to facilitate exosome capture.25 While considerable success has been reported using this approach, the separation purity is low as CD63 is expressed in both intracellular and extracellular vesicles.29 Furthermore, sufficient time is needed to allow the nanoparticles to bind to the antibodies. More importantly, extracellular microvesicles that do not present such characteristic surface markers will inadvertently avoid capture.30,31 More recently, microfluidics utilizing label-free isolation methods have also gained significant attention owing to its ease of operation.32 Several approaches were demonstrated using microfiltration,15,33 acoustics,34,35 viscoelastic lift forces,36 and deterministic lateral displacement micropillars.37 Typically, these size-based separation methods allow the optimal capture of extracellular vesicles below 200 nm. However, while these technologies have shown early promises, separation purity is often compromised with increasing throughput.
Here, centrifugal microfluidics offers a simple and yet robust strategy to overcome these diffusive effects. Many researchers have exploited the miniaturized rotational platform for particle handling,38–40 valving,41,42 decanting,43 and sedimentation.44–46 These multiple advantages in microscale manipulation within a single platform enable sample-to-answer possibilities, and are especially useful in diagnostic applications. Despite its high utility, most of the centrifugal microfluidic applications have demonstrated success mainly towards microparticle separation.47,48 More recently, Woo et al. developed a centrifugal biodisk capable of isolating extracellular vesicles between 20 nm and 600 nm using membrane filters.49 Even so, centrifugal separation of nanoparticles is yet to be effectively established due to the complexity of forces intertwined with fluid instability.
In this study, we utilized centrifugal microfluidics to achieve nanoparticle separation. In particular, our design reduces the hydrodynamic radius and increases the external force field effects significantly. As a result, the centrifugal force requirement is significantly lower than that of conventional centrifugal methods. The processing time is also reduced by more than 100 folds compared to conventional methods. In addition, the microfluidic manifold eliminates the need for a syringe pump. Here, we demonstrate the utility of this microfluidic platform to enrich monodispersed nanosuspensions, separate mixed nanosuspensions, and extract microvesicles from the cell culture medium. Overall, our device is capable of separating bioparticles below 100 nm within minutes, and possesses obvious utility in EV extraction for disease detection and diagnosis.
II. MATERIALS AND METHODS
A. Design and fabrication of μCENSE platform
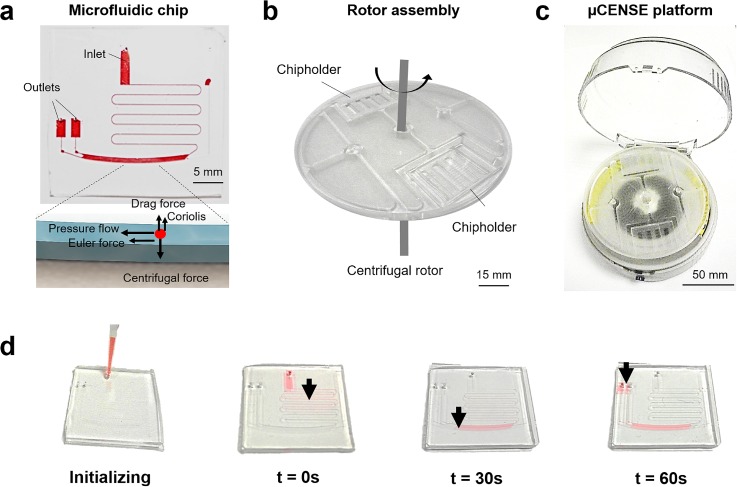
Figure 1 shows the main components of the microfluidic CEntrifugal Nanoparticles Separation and Extraction (μCENSE) platform. The μCENSE platform comprises three main parts: the microfluidic chip, the rotor assembly, and the centrifugal rotor. The microfluidic chip is detailed in Fig. 1(a). It consists of one serpentine inlet channel and two outlets. Interposed between the inlet and outlets is a separation microfluidic channel. The separation channel is 15 mm × 800 μm (L × W). Separation of particles by size occurs within this separation channel owing to the different forces acting on the nanoparticles [inset of Fig. 1(a)]. The microfluidic chip was fabricated using standard soft lithography techniques. Briefly, polydimethylsiloxane (PDMS) (Dow Corning Sylgard 184, Midland, MI) prepolymer was mixed in a 10:1 (w/w) ratio with a curing agent, poured onto the silanized wafer, and degassed for 1 h. The polymer was then cured in a drying oven at 70 °C for 2 h. The cured silicone elastomer was removed from the mold and the fluidic inlet and outlet were formed through hole-punching of diameter 1 mm. The PDMS substrate was fabricated using the same steps and covalently bonded to the PDMS microfluidic channel using oxygen plasma treatment. The device was completed after curing in the oven at 70 °C for another 3 h. Next, the rotor assembly allows multiple microfluidic chips to be placed in the platform [Fig. 1(b)]. The rotor assembly was fabricated using standard micromilling techniques. The centrifugal rotor sets the rotor assembly on a rotational speed within an enclosure of 120 mm diameter. The actual fabricated μCENSE platform is shown in Fig. 1(c). To initialize the sample extraction process, the sample fluid was loaded into the microfluidic chip using a standard micropipette [Fig. 1(d)].
FIG. 1.
Overview of the microfluidic centrifugal nanoparticles separation and extraction (μCENSE) platform. (a) Schematic of the centrifugal microfluidic chip. It consists of one inlet, two outlets, and a separation channel. The scale bar represents 5 mm. The inset describes the separation principle: centrifugal force drives fluid pressure along the separation channel and radial force pushes the particles away from the center of rotation by size. (b) Rotor assembly allowing multiple microfluidic chips to be held in place during centrifugation. The scale bar represents 15 mm. (c) Actual fabricated centrifugal microfluidic platform. The scale bar represents 50 mm. (d) Photograph depicting loading of the sample fluid via a micropipette and the movement of the red dye. The arrow indicates the position of the dye at different centrifugal intervals (t = 0 s, 30 s, and 60 s).
B. Device testing protocol
Using centrifugal hydrodynamics, particles may be separated according to their size. To test this, fluorescent nanobeads (Phosphorex, Inc.) of density 1.05 kg/m3 of various sizes were loaded separately into the microfluidic chip using a micropipette. The inlet was left open as an air vent to provide the centrifugal force in the fluid interface. The microfluidic chip was carefully placed within the chipholder and enclosed within the rotor assembly. The centrifuge was set to spin according to the stipulated duration. The microfluidic chip was then removed from the rotor assembly and observed under the fluorescence microscope. Sample liquid was extracted from the outlets using vacuum suction for further post-processing analysis. Sample was air-dried on the mica surface and scanned using atomic force microscopy (AFM).
C. Biological processing protocol
Biological samples were further used to demonstrate the device utility. Human breast adenocarcinoma cell line, MCF-7, and lung adenocarcinoma cell line, H1975, were cultured in low glucose Dulbecco's modified Eagle's medium (DMEM) (Gibco®, CA) supplemented with 10% fetal bovine serum (FBS) (Invitrogen, CA) and 1% pencillin/streptomycin (Invitrogen, CA). Cell culture was maintained at 37 °C in a 5% CO2 incubator with the media replaced every 48 h until 90% confluency. The culture media was aspirated and discarded. Next, the culture flask was washed three times using phosphorus buffered saline (PBS) and the culture media was then replaced with low glucose DMEM without FBS and cultured for another 12 h following which, the cell culture solution was aspirated. The culture solution was centrifuged at 2000 × g for 30 min to remove any remaining cells and debris. The supernatant was loaded into the microfluidic chip for microvesicle extraction. The microfluidic chip was subjected to a centrifugal spin of two minutes and then removed from the centrifuge. To ensure full extraction of the separated microvesicles, 10 μl of PBS was slowly loaded into the inlet of the microfluidic chip using a syringe pump. 5 μl was extracted from each outlet and characterized using atomic force microscopy. For comparison, the culture solution was further extracted using conventional methods, as described in previous literature.50
For Western blotting, the isolated EVs were lysed in a non-reducing sodium dodecyl sulfate sample buffer (Thermo Fisher, MA), heated at 85 °C for 5 min and stored at −20 °C before analysis. Samples were loaded and resolved using polyacrylamide gel electrophoresis (SDS-PAGE) (Thermo Fisher, MA) and transferred to a nitrocellulose membrane. The membrane was blocked with a blocking buffer (5% bovine serum albumin in 0.1% Tween 20 in Tris-buffered saline solution) and incubated overnight with a primary exosomal CD63 antibody (Thermo Fisher, MA). Next, the secondary antibody was incubated for 1 h at room temperature and washed three times with 0.1% Tween 20 in Tris-buffered saline solution. Finally, enhanced chemiluminescence was used for detection.
III. RESULTS AND DISCUSSION
A. Theoretical modelling
The μCENSE platform allows pressure flow handling coupled with centrifugal hydrodynamics on a handheld centrifugal platform. At equilibrium, the resultant forces create zonal separation of nanoparticles within the microfluidic chips leading to different outlets. Upon centrifugation, a compressive pressure acts on the fluid inlet interface to initiate fluid flow, which is further assisted by the Euler acceleration. The fluid movement can be mathematically described using the Lagrangian derivative as
| (1) |
where is the Euler acceleration, is the pressure gradient, is the viscous drag term, is the centrifugal force, and is the Coriolis force. Accordingly, the serpentine inlet channel provides sufficient hydrodynamic resistance to provide fluid movement at the onset of centrifugation. To demonstrate the fluid movement within the microfluidic chip during the centrifugal process, we loaded 5 μl of red dye in the inlet and spun at various time intervals. Figure 1(d) illustrates the movement of the fluid at different centrifugal intervals (t = 0 s, 30 s, and 60 s). Notably, by the end of the centrifugation, the fluid has migrated to the outlets for subsequent retrieval.
Simultaneously, a centrifugal force field is acting perpendicular to the axis of the fluid flow direction which is counteracted by the hydrodynamic drag, buoyancy, and Coriolis force [Fig. 1(c)]. Under the steady state, the particles achieve terminal velocity up described by
| (2) |
where d is the diameter of the particle, r is the distance from the center of rotation, ω is the angular velocity, μ is the dynamic viscosity of the fluid, and ρp represents the density of the particle (refer to supplementary material). The resultant terminal velocity results in the migration of the nanoparticles towards the outer channel wall. Clearly, different terminal velocities could be achieved with nanoparticles of different sizes.
Therefore, using our centrifugal platform, we created a centrifugal speed of 5000 rpm, equivalent to approximately 1300 × g. Furthermore, by manipulating the hydrodynamic resistance, the residence time for each nanoparticle within the separation channel may be adjusted. In particular, we chose three different channel heights (100 μm, 50 μm, and 25 μm) to adjust the characteristic time for the nanoparticles (1000 nm, 500 nm, and 200 nm) within the separation channel. This further determines the vertical displacement of each particle according to its particle diameter. By doing so, a distinguishable separation between particle sizes could be established and bifurcations may be designed to separate these particles into different outlets.
B. Device performance
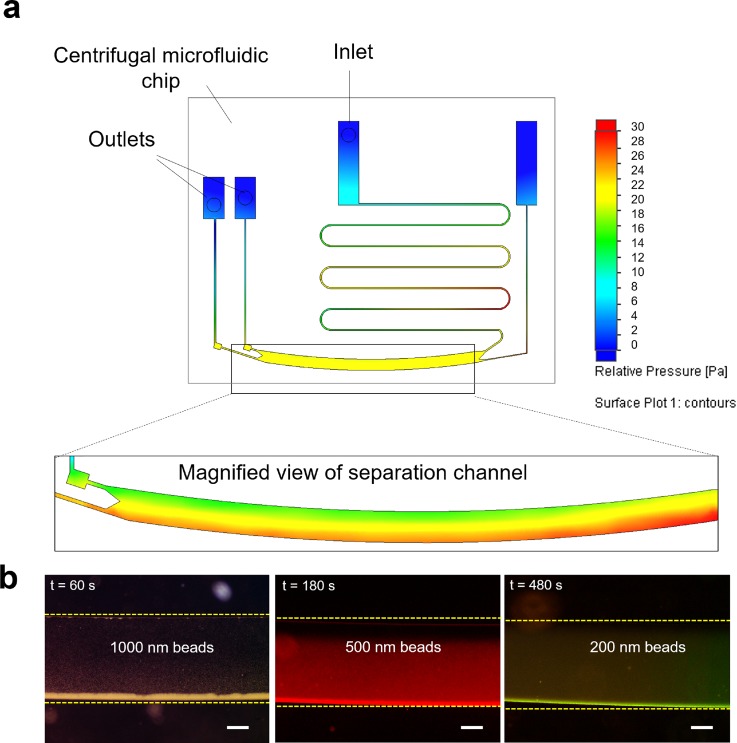
Next, we performed flow analysis within the microfluidic chip to characterize the device performance. Figure 2(a) shows the pressure difference within the microfluidic channel upon centrifugation. Notably, the difference in pressure accounts for the fluid movement from the inlet to the outlet. At the onset of centrifugation, pressurization occurs at the inlet to overcome the initial hydrodynamic resistance, resulting in fluid flow towards the outlets. When the fluid enters the separation channel, further zonal separation occurs. The inset shows the macro view of the fluid movement in the separation channel. Notably, a differential pressure exists within the separation channel due to the centrifugal force. Based on the equation, the particles migrate based on the centrifugal force which is proportional to the square of the particle diameter. Evidently, the larger particles experience larger centrifugal force leading to longer migration distances.
FIG. 2.
Separation of monodispersed nanoparticles. (a) Simulation of the microfluidic chip upon centrifugation. Color denotes the pressure difference within the microchannels. The inset shows the magnified view of the separation channel. (b) Separation of nanoparticles of reducing sizes (i.e., d = 1000, 500, and 200 nm) observed with the microfluidic chip of reducing channel heights (i.e., h = 100, 50, and 25 μm) respectively. The dotted line indicates the position of the channel wall. The scale bar represents 200 μm.
We first demonstrate size separation efficiency by loading monodispersed fluorescent nanobeads of different sizes (d = 1000 nm, 500 nm, and 200 nm) into the inlet and subjecting them to centrifugation. We used different centrifugal intervals to assess their optimal separation duration (see supplementary material Fig. S1). We noted that migration was achieved within 6 min of centrifugation even for the 200 nm nanobeads. This is significantly shorter than most of the conventional methods of EV isolation. After centrifugation, the microfluidic chips were removed and observed using fluorescence microscopy. Figure 2(b) further illustrates the separation efficiency of the nanoparticles at the characteristic centrifugal intervals. A significantly higher fluorescence intensity was observed towards the outer channel. Clearly, this indicates the migration of nanoparticles to the outer separation channel wall. By comparing the fluorescence intensity of the outlets, we observed that monodispersed nanoparticles may be separated with 90% efficiency.
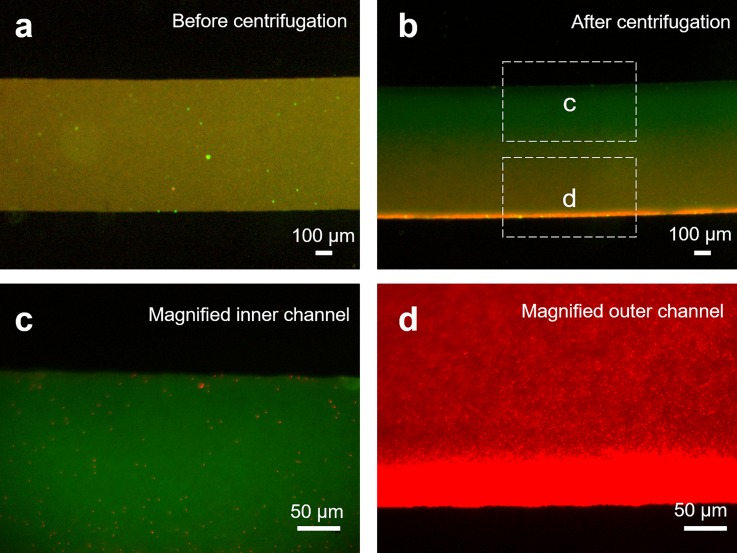
Typically, cells secrete membrane bound vesicles in two different size classifications: exosomes characterized with the size between 30 nm and 200 nm, while the size of microvesicles lies between 100 nm and 1000 nm.36,51,52 Therefore, of interest, is the ability to separate microvesicles from the exosomes. To demonstrate this, we loaded 5 μl of fluorescent mixed colloidal suspensions (200 nm and 500 nm) into the inlet of the microfluidic chip and subjected them to centrifugal spin. As the polydispersed nanobeads exhibit different excitation and emission spectra, we tracked the dispersion of the respective nanoparticles before and after centrifugation. Figure 3(a) shows the merged fluorescence image of the polydispered nanobeads within the separation channel before centrifugation, indicating a homogenous distribution of the nanoparticles of different sizes within the microfluidic channel. Upon centrifugation, we observed distinctly different spectra. In particular, we noted that the larger nanoparticles experienced a significant migration, as indicated by the change in the fluorescence emission profile near the outer channel wall in Fig. 3(b). Conversely, the centrifugal intervals are not sufficient to result in any significant migration of the smaller nanoparticles below 200 nm. By using higher optical magnification, we further observed that only the smaller nanoparticles remain dispersed towards the inner channel wall, as shown in Fig. 3(c). Similarly, we observed that the larger nanoparticles have migrated towards the outer channel wall, as shown in Fig. 3(d). By performing fluorescence intensity scattering analysis on the mixed colloidal nanoparticles, we reported separation of polydispersed nanoparticles with purity up to 85%.
FIG. 3.
Separation of polydispersed fluorescent nanoparticles. (a) Representative fluorescence micrographs of the separation channel before centrifugation filled with 0.1% v/v polydispersed fluorescent nanoparticles (500 nm in red and 200 nm in green) within the microfluidic chip. (b) Corresponding fluorescence micrographs after centrifugation for the polydispersed nanoparticles. The scale bar represents 100 μm. (c) Micrograph of the inner portion of the microfluidic channel under 20× optical magnification showed monodispersed nanoparticles, indicating that the larger nanoparticles have migrated to the outermost channel wall. (d) Fluorescence micrograph of the outer microchannel indicating a higher fluorescence intensity at the outermost channel wall. The scale bar represents 50 μm.
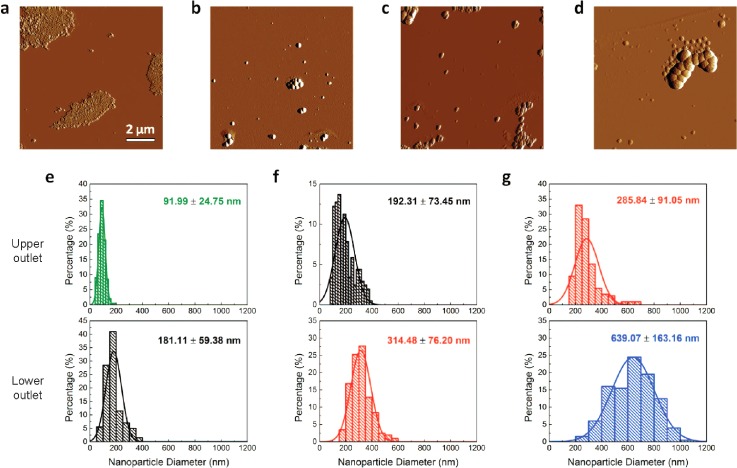
As further confirmation, we retrieved the nanosuspensions from the outlets and scanned the fluid sample by atomic force microscopy. Figure 4(a) through Fig. 4(c) shows the representative images of the nanobeads retrieved from the upper outlets of the microfluidic chips of increasing channel heights (h = 25 μm, 50 μm, and 100 μm), respectively. The images show the distinct size difference in particles, indicating its extraction efficiency. Figure 4(d) further shows the representative image of the larger nanoparticles that are retrieved from the lower outlet of the microfluidic chip. To acquire statistical information about the size distribution of the nanobeads in each outlet, we evaluated more than 200 nanobeads for each outlet of the microfluidic chip. Figure 4(e) shows the size distribution obtained from the upper outlet (labeled in green) and the lower outlet (labeled in black) for a microfluidic chip of height 25 μm. Expectedly, the average size of the nanoparticles from the lower outlet is distinctly larger than the average size of the nanoparticles from the upper outlet. Similarly, Fig. 4(f) shows the size distribution for a microfluidic chip of 50 μm. Notably, the nanoparticles extracted from the lower outlet (labeled in red) are distinctly larger. Finally, Fig. 4(g) shows the size distribution of the extracted nanoparticles from a microfluidic chip of height 100 μm. Again, as expected, the nanoparticles obtained from the upper outlet are significantly larger. We further observed that the average size of the extracted nanoparticles from each outlet represents a close approximate of the sizes of mixed colloidal particles mixed into the microfluidic chip. Overall, we noted that the size distribution of the nanosuspensions is distinctly different from each outlet, indicating effective nanobead separation and extraction by size.
FIG. 4.
Characterization of extracted fluorescent nanoparticles separated in the μCENSE platform. (a)–(d) Representative topological profile of nanoparticles as characterized by tapping mode atomic force microscopy. Nanoparticles are extracted from the upper outlets of microfluidic chips of: (a) height 25 μm, (b) height 50 μm, and (c) height 100 μm, as well as (d) from the lower outlet of the microfluidic chip of height 100 μm. The scale bar represents 2 μm. (e)–(g) Size distribution of the nanoparticles (n = 200) retrieved from the outlets from the microfluidic chip of: (e) height 25 μm, (f) height 50 μm, and (g) height 100 μm. The top and bottom figures represent the size distribution from the upper outlets and lower outlets, respectively.
C. Extraction of EVs
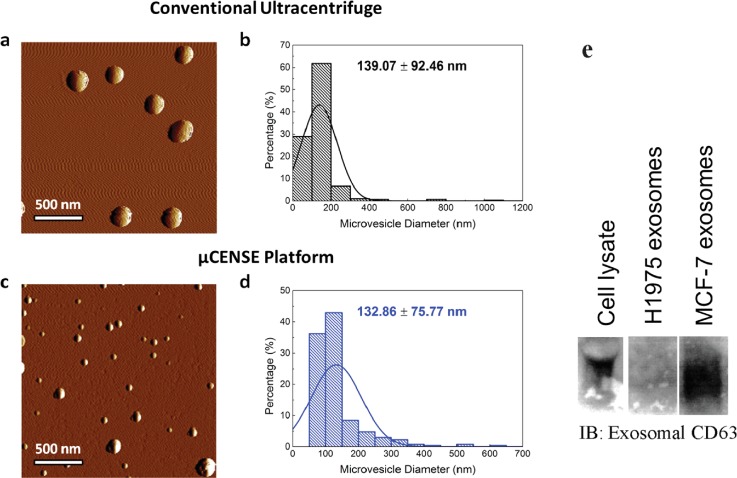
Finally, to demonstrate device utility, cell culture mediums of cancer cell lines, MCF-7 and H1975, were aspirated and loaded into the microfluidic chip using a micropipette. The chip was then mounted on the platform and centrifuged for eight minutes. Microvesicles were then extracted from the upper outlet and scanned using AFM. This approach allows the characterization of extracellular vesicles in their native environment without any fixation, staining or labeling requirements, which is ideal for biological samples.53 As a comparison, we extracted microvesicles using conventional ultracentrifugal methods (at 10 000 × g for 1 h) and scanned the sample using AFM, as shown in Fig. 5(a). The size distribution of the extracted particles was represented in Fig. 5(b). We compared the size of the nanoparticles extracted using our centrifugal microfluidic platform, in Figs. 5(c) and 5(d), respectively. Figure 5(c) shows the representative images of the scanned microvesicles, which correlate well with previously reported literature.54–56 Furthermore, we noted that the microvesicles extracted from the platform were similar in size to those extracted with ultracentrifugation, further highlighting their separation utility [Fig. 5(d)]. With this, we demonstrated that even with centrifugation speeds 10 folds lower than traditional methods, microvesicles may be extracted from the μCENSE platform. Furthermore, we performed immunoblotting on the extracted samples from the outlets to determine the presence of exosomal CD 63 markers in microvesicles of different cell lines [Fig. 5(e)]. Western blot analysis confirmed the presence of CD63 exosmal markers on microvesicles extracted from the MCF-7 cell line. Interestingly, microvesicles extracted from the cancer cell line H1975 showed the absence of CD63 exosmal markers. This is consistent with other exosomal studies on lung cancer patients of low CD63 expression,26,57 further indicating the heterogeneity of surface markers on the microvesicles.
FIG. 5.
Comparison of extracted microvesicle separation using a commercial kit and the μCENSE platform. (a) Representative topological profile of microvesicles extracted from the conventional extraction kit using tapping mode atomic force microscopy. (b) Size distribution of the microvesicles (n = 287). (c) Representative topological profile of microvesicles extracted from the μCENSE platform using tapping mode atomic force microscopy. (d) Size distribution of the microvesicles (n = 270). The scale bar represents 500 nm. (e) Immunoblotting indicates the presence of exosomal CD63 markers in the microfluidic outlets for the MCF-7 cell line but absence in the H1975 cell line.
IV. CONCLUSIONS
Effective capture of EVs will aid tremendously in early disease detection and diagnosis. Despite the heightened interest in exosomes for diagnostic or prognostic utility, extraction methods are still labor intensive, time-consuming, and require expensive reagents or equipment. To this end, we developed a centrifugal microfluidic system (μCENSE) for label-free fractionation of EVs. By implementing the novel combination of fluid pressure driven flow and centrifugal sedimentation in a single platform, we optimized a distinguishable separation of mixed colloidal nanosuspensions into different outlets within minutes. By altering the microchannel dimensions, we significantly reduced the hydrodynamic instability that is characteristic of nanosuspension. Analytical analyses further confirmed the capability of the centrifugal platform in extracting microvesicles. The size selection separation microfluidic chip is highly versatile as different microfluidic chip designs can be easily implemented in the system. Furthermore, the implementation of a low centrifugal speed platform and the minimal use of accessories not only increase its cost effectiveness, but also its viability as a point-of-care diagnostic kit. Remarkably, the entire process takes less than ten minutes, enabling a rapid and cost-effective nanoparticle separation technology.
V. SUPPLEMENTARY MATERIAL
See supplementary material for detailed derivation of centrifugal microhydrodynamics, channel height calculation, and separation characteristics at different centrifugal intervals.
ACKNOWLEDGMENTS
This research was supported by the use of the laboratory facilities in the Mechanobiology Institute (MBI) and the MechanoBioEngineering Laboratory at the Department of Biomedical Engineering of the National University of Singapore.
References
- 1. Braicu C., Tomuleasa C., Monroig P., Cucuianu A., Berindan-Neagoe I., and Calin G. A., Cell Death Differ. 22(1), 34–45 (2015). 10.1038/cdd.2014.130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Taylor D. D. and Gercel-Taylor C., Front. Genet. 4, 142 (2013). 10.3389/fgene.2013.00142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cocucci E., Racchetti G., and Meldolesi J., Trends Cell Biol. 19(2), 43–51 (2009). 10.1016/j.tcb.2008.11.003 [DOI] [PubMed] [Google Scholar]
- 4. Raposo G. and Stoorvogel W., J. Cell Biol. 200(4), 373 (2013). 10.1083/jcb.201211138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Valadi H., Ekström K., Bossios A., Sjöstrand M., Lee J. J., and Lötvall J. O., Nat. Cell Biol. 9(6), 654–659 (2007). 10.1038/ncb1596 [DOI] [PubMed] [Google Scholar]
- 6. Lötvall J., Hill A. F., Hochberg F., Buzás E. I., Di Vizio D., Gardiner C., Gho Y. S., Kurochkin I. V., Mathivanan S., Quesenberry P., Sahoo S., Tahara H., Wauben M. H., Witwer K. W., and Théry C., J. Extracell. Vesicles 3(1), 26913 (2014). 10.3402/jev.v3.26913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kosaka N., Iguchi H., and Ochiya T., Cancer Sci. 101(10), 2087–2092 (2010). 10.1111/j.1349-7006.2010.01650.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Thakur B. K., Zhang H., Becker A., Matei I., Huang Y., Costa-Silva B., Zheng Y., Hoshino A., Brazier H., Xiang J., Williams C., Rodriguez-Barrueco R., Silva J. M., Zhang W., Hearn S., Elemento O., Paknejad N., Manova-Todorova K., Welte K., Bromberg J., Peinado H., and Lyden D., Cell Res. 24(6), 766–769 (2014). 10.1038/cr.2014.44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Shao H., Chung J., Balaj L., Charest A., Bigner D. D., Carter B. S., Hochberg F. H., Breakefield X. O., Weissleder R., and Lee H., Nat. Med. 18(12), 1835–1840 (2012). 10.1038/nm.2994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Xu R., Greening D. W., Rai A., Ji H., and Simpson R. J., Methods 87, 11–25 (2015). 10.1016/j.ymeth.2015.04.008 [DOI] [PubMed] [Google Scholar]
- 11. Syn N., Wang L., Sethi G., Thiery J.-P., and Goh B.-C., Trends Pharmacol. Sci. 37(7), 606–617 (2016). 10.1016/j.tips.2016.04.006 [DOI] [PubMed] [Google Scholar]
- 12. Lai R. C., Arslan F., Lee M. M., Sze N. S., Choo A., Chen T. S., Salto-Tellez M., Timmers L., Lee C. N., El Oakley R. M., Pasterkamp G., de Kleijn D. P., and Lim S. K., Stem Cell Res. 4(3), 214–222 (2010). 10.1016/j.scr.2009.12.003 [DOI] [PubMed] [Google Scholar]
- 13. Sluijter J. P., Verhage V., Deddens J. C., van den Akker F., and Doevendans P. A., Cardiovasc. Res. 102(2), 302–311 (2014). 10.1093/cvr/cvu022 [DOI] [PubMed] [Google Scholar]
- 14. Aharon A., Rebibo-Sabbah A., Tzoran I., and Levin C., Rambam Maimonides Med. J. 5(4), e0032 (2014). 10.5041/RMMJ.10166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Muller L., Hong C. S., Stolz D. B., Watkins S. C., and Whiteside T. L., J. Immunol. Methods 411, 55–65 (2014). 10.1016/j.jim.2014.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kalluri R., J. Clin. Invest. 126(4), 1208–1215 (2016). 10.1172/JCI81135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cappello F., Logozzi M., Campanella C., Bavisotto C. C., Marcilla A., Properzi F., and Fais S., Eur. J. Pharm. Sci. 96, 93–98 (2017). 10.1016/j.ejps.2016.09.010 [DOI] [PubMed] [Google Scholar]
- 18. Panagiotara A., Markou A., Lianidou E. S., Patrinos G. P., and Katsila T., Public Health Genomics 20(2), 116–125 (2017). 10.1159/000478253 [DOI] [PubMed] [Google Scholar]
- 19. Kalra H., Adda C. G., Liem M., Ang C. S., Mechler A., Simpson R. J., Hulett M. D., and Mathivanan S., Proteomics 13(22), 3354–3364 (2013). 10.1002/pmic.201300282 [DOI] [PubMed] [Google Scholar]
- 20. Tauro B. J., Greening D. W., Mathias R. A., Ji H., Mathivanan S., Scott A. M., and Simpson R. J., Methods 56(2), 293–304 (2012). 10.1016/j.ymeth.2012.01.002 [DOI] [PubMed] [Google Scholar]
- 21. Linares R., Tan S., Gounou C., Arraud N., and Brisson A. R., J. Extracell. Vesicles 4(1), 29509 (2015). 10.3402/jev.v4.29509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Van Deun J., Mestdagh P., Sormunen R., Cocquyt V., Vermaelen K., Vandesompele J., Bracke M., De Wever O., and Hendrix A., J. Extracell. Vesicles 3(1), 24858 (2014). 10.3402/jev.v3.24858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yang F., Liao X., Tian Y., and Li G., Biotechnol. J. 12(4), 1600699 (2017). 10.1002/biot.201600699 [DOI] [PubMed] [Google Scholar]
- 24. Contreras-Naranjo J. C., Wu H.-J., and Ugaz V. M., Lab Chip 17(21), 3558–3577 (2017). 10.1039/C7LC00592J [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kanwar S. S., Dunlay C. J., Simeone D. M., and Nagrath S., Lab Chip 14(11), 1891–1900 (2014). 10.1039/C4LC00136B [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. He M., Crow J., Roth M., Zeng Y., and Godwin A. K., Lab Chip 14(19), 3773–3780 (2014). 10.1039/C4LC00662C [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fang S., Tian H., Li X., Jin D., Li X., Kong J., Yang C., Yang X., Lu Y., Luo Y., Lin B., Niu W., and Liu T., PLoS One 12(4), e0175050 (2017). 10.1371/journal.pone.0175050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhang P., He M., and Zeng Y., Lab Chip 16(16), 3033–3042 (2016). 10.1039/C6LC00279J [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pols M. S. and Klumperman J., Exp. Cell Res. 315(9), 1584–1592 (2009). 10.1016/j.yexcr.2008.09.020 [DOI] [PubMed] [Google Scholar]
- 30. Li P., Kaslan M., Lee S. H., Yao J., and Gao Z., Theranostics 7(3), 789–804 (2017). 10.7150/thno.18133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Furi I., Momen-Heravi F., and Szabo G., Ann. Transl. Med. 5(12), 263 (2017). 10.21037/atm.2017.03.95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Im H., Shao H., Park Y. I., Peterson V. M., Castro C. M., Weissleder R., and Lee H., Nat. Biotechnol. 32(5), 490–495 (2014). 10.1038/nbt.2886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Davies R. T., Kim J., Jang S. C., Choi E.-J., Gho Y. S., and Park J., Lab Chip 12(24), 5202–5210 (2012). 10.1039/c2lc41006k [DOI] [PubMed] [Google Scholar]
- 34. Lee K., Shao H., Weissleder R., and Lee H., ACS Nano 9(3), 2321–2327 (2015). 10.1021/nn506538f [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wu M., Ouyang Y., Wang Z., Zhang R., Huang P.-H., Chen C., Li H., Li P., Quinn D., Dao M., Suresh S., Sadovsky Y., and Huang T. J., Proc. Natl. Acad. Sci. U.S.A. 114(40), 10584–10589 (2017). 10.1073/pnas.1709210114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Liu C., Guo J., Tian F., Yang N., Yan F., Ding Y., Wei J., Hu G., Nie G., and Sun J., ACS Nano 11(7), 6968–6976 (2017). 10.1021/acsnano.7b02277 [DOI] [PubMed] [Google Scholar]
- 37. Santana S. M., Antonyak M. A., Cerione R. A., and Kirby B. J., Biomed. Microdevices 16(6), 869–877 (2014). 10.1007/s10544-014-9891-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Burger R., Kirby D., Glynn M., Nwankire C., O'Sullivan M., Siegrist J., Kinahan D., Aguirre G., Kijanka G., Gorkin R. A. 3rd, and Ducree J., Curr. Opin. Chem. Biol. 16(3–4), 409–414 (2012). 10.1016/j.cbpa.2012.06.002 [DOI] [PubMed] [Google Scholar]
- 39. Burger R. and Ducrée J., Expert Rev. Mol. Diagn. 12(4), 407–421 (2012). 10.1586/erm.12.28 [DOI] [PubMed] [Google Scholar]
- 40. Yu Z. T. F., Joseph J. G., Liu S. X., Cheung M. K., Haffey P. J., Kurabayashi K., and Fu J., Sens Actuators B Chem 245, 1050–1061 (2017). 10.1016/j.snb.2017.01.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Clime L., Brassard D., Geissler M., and Veres T., Lab on a Chip 15(11), 2400–2411 (2015). 10.1039/C4LC01490A [DOI] [PubMed] [Google Scholar]
- 42. Al-Faqheri W., Ibrahim F., Thio T. H., Bahari N., Arof H., Rothan H. A., Yusof R., and Madou M., Sensors 15(3), 4658–4676 (2015). 10.3390/s150304658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Haeberle S., Brenner T., Zengerle R., and Ducree J., Lab Chip 6(6), 776–781 (2006). 10.1039/B604145K [DOI] [PubMed] [Google Scholar]
- 44. Strohmeier O., Keller M., Schwemmer F., Zehnle S., Mark D., von Stetten F., Zengerle R., and Paust N., Chem. Soc. Rev. 44(17), 6187–6229 (2015). 10.1039/C4CS00371C [DOI] [PubMed] [Google Scholar]
- 45. Ducrée J., Haeberle S., Lutz S., Pausch S., Stetten F. v., and Zengerle R., J. Micromech. Microeng. 17(7), S103–S115 (2007). 10.1088/0960-1317/17/7/S07 [DOI] [Google Scholar]
- 46. Son J. H., Lee S. H., Hong S., Park S. M., Lee J., Dickey A. M., and Lee L. P., Lab Chip 14(13), 2287–2292 (2014). 10.1039/C4LC00149D [DOI] [PubMed] [Google Scholar]
- 47. Yeo J. C., Wang Z., and Lim C. T., Biomicrofluidics 9(5), 054114 (2015). 10.1063/1.4931953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lee A., Park J., Lim M., Sunkara V., Kim S. Y., Kim G. H., Kim M. H., and Cho Y. K., Anal. Chem. 86(22), 11349–11356 (2014). 10.1021/ac5035049 [DOI] [PubMed] [Google Scholar]
- 49. Woo H.-K., Sunkara V., Park J., Kim T.-H., Han J.-R., Kim C.-J., Choi H.-I., Kim Y.-K., and Cho Y.-K., ACS Nano 11(2), 1360–1370 (2017). 10.1021/acsnano.6b06131 [DOI] [PubMed] [Google Scholar]
- 50. Schageman J., Zeringer E., Li M., Barta T., Lea K., Gu J., Magdaleno S., Setterquist R., and Vlassov A. V., BioMed Res. Int. 2013, 253957. 10.1155/2013/253957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Gould S. J. and Raposo G., J. Extracell. Vesicles 2(1), 20389 (2013). 10.3402/jev.v2i0.20389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Budnik V., Ruiz-Cañada C., and Wendler F., Nat. Rev. Neurosci. 17(3), 160–172 (2016). 10.1038/nrn.2015.29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Palanisamy V., Sharma S., Deshpande A., Zhou H., Gimzewski J., and Wong D. T., PLoS One 5(1), e8577 (2010). 10.1371/journal.pone.0008577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Buzas E. I., Gyorgy B., Nagy G., Falus A., and Gay S., Nat. Rev. Rheumatol. 10(6), 356–364 (2014). 10.1038/nrrheum.2014.19 [DOI] [PubMed] [Google Scholar]
- 55. Tian T., Wang Y., Wang H., Zhu Z., and Xiao Z., J. Cell. Biochem. 111(2), 488–496 (2010). 10.1002/jcb.22733 [DOI] [PubMed] [Google Scholar]
- 56. Sharma S. and Gimzewski J. K., J. Nanomed. Nanotechnol. 3(7), 1000e115 (2012). 10.4172/2157-7439.1000e115 [DOI] [Google Scholar]
- 57. Kwon M. S., Shin S.-H., Yim S.-H., Lee K. Y., Kang H.-M., Kim T.-M., and Chung Y.-J., Lung Cancer 57(1), 46–53 (2007). 10.1016/j.lungcan.2007.01.032 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
See supplementary material for detailed derivation of centrifugal microhydrodynamics, channel height calculation, and separation characteristics at different centrifugal intervals.