Abstract
Proteins secreted by skin have great potential as biomarkers for interpreting skin conditions. However, inconvenience in handling and bulky size of existing methods are existing limitations. Here, we describe a thumb-nail sized patch with the array of microdisks which captures multiple proteins from the skin surface. Microdisks with antibody on the surface enable multiplexed immunoassay. By self-assembly, microdisks are placed into 2-dimensional arrays on adhesive tape. The proposed Enzyme-Linked Immunospot array on a Patch shows sufficient sensitivity for IL-1α, IL1RA, IL-17A, IFN-g, and TNF-α, while IL-6 and IL-1β are non-detectable in some cases. As demonstrations, we quantified cytokines from different skin regions and volunteers in a high-spatial-resolution.
INTRODUCTION
The skin is the largest organ in the human body and harbors information on responses to both external stimuli and internal physiology.1–3 The skin response is orchestrated by a number of protein molecules secreted by residing cells; these secreted proteins are detected not only in the skin tissue but also on the skin surface.4–6
There have been efforts to develop non-invasive methods utilizing skin washing fluid placing open chamber and introducing protein-soluble buffer to dissolve skin residual proteins.3,7 However, the bulky features of skin washing fluid approaches generate subject discomfort and limit the spatial resolution of the measurement. Therefore, miniaturization of the overall feature size is yet to be achieved; one possible approach is to utilize a small-sized planar antibody array to make direct contact with the skin surface without the use of a buffer.
To prepare a small-sized planar antibody array, one efficient approach is to place microdisks that carry antibodies in an array.8,9 Because this microdisk preparation is a bulk process and array elements can be organized merely by selecting from each bulk and mixing, the use of microdisks is efficient, not only as regards preparation throughput but also in terms of reconfiguration flexibility for the elements of the array.10,11 Another advantage of incorporating microdisks is that this approach provides a better means of controlling the quality of each element, as a small portion of microdisks from each bulk can be sampled and tested. By placing a selected and mixed microdisk on a leveled surface by fluid-assisted self-assembly, a small-sized planar antibody array for high-spatial-resolution detection of skin residual proteins can be achieved.
Here, we propose a thumbnail-sized circular adhesive patch with a microdisk-library array, named ELIPatch (Enzyme-Linked Immunospot array on a Patch). The ELIPatch, having an array of protein-capturing microdisks, can seize target proteins directly from the human skin surface followed by fluorescent immunoassay which enables quantification of multiplexed target proteins. In this work, the ELIPatch is utilized to capture skin residual cytokines in multiplex, from different subjects or from multiple sites, with high spatial resolution.
MATERIALS AND METHODS
A microdisk is made by a microfluidic-based optofluidic maskless lithography (OFML) system that irradiates patterned ultra-violet light to photocurable polymer followed by silica-coating for surface modification. Preparation of the ELIPatch includes a self-assembly of heterogeneous encoded microdisks, called partipetting, to the microwell array on a polydimethylsiloxane (PDMS) substrate. The measurement of skin protein is peeling off ELIPatch from the substrate and placing this patch to the skin surface for an hour. After this protein capture step, ELIPatches are sonicated to release the microdisks followed by further immunoassay. Other detailed information is provided in the supplementary material.
RESULTS
Fabrication and utilization of the enzyme-linked immunospot array on the patch
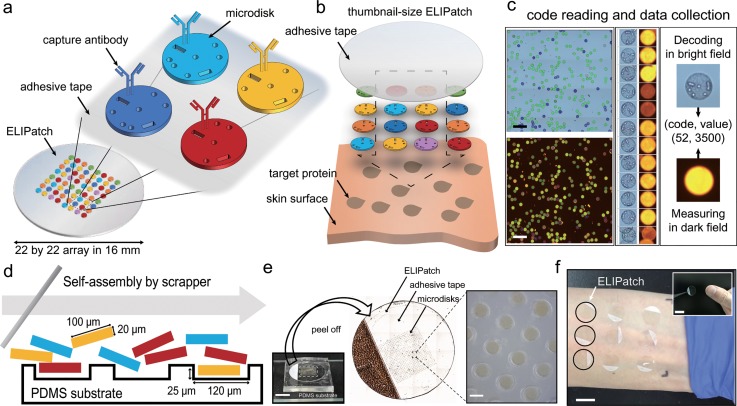
Figures 1(a) and 1(b) show the concept and schematics of the ELIPatch. One unique feature of the ELIPatch is the array of encoded microdisks that carry capture antibodies on their silica-coating. The use of microdisks allows the placement of hundreds of immunospots within a 16-mm-diameter adhesive tape. The void volume in the bar or spot shape inside every microdisk acts as a shape code, representing the target protein for antibodies. An ELIPatch is created when the microdisk array is placed on the adhesive patch, and direct contact of the ELIPatch with human skin enables simultaneous capture of target proteins [Fig. 1(b)]. Figure 1(c) shows an automated data reading and analysis process, while Fig. 1(d) shows a process of preparation of the ELIPatch by self-assembly assisted by 2.5 μl of carrier fluid.
FIG. 1.
Schematic illustration of components and the usage of the ELIPatch. (a) ELIPatch has about 500 of encoded microdisks that have the shape code and corresponding antibody. Heterogeneous library of microdisks is placed in an array format with replicates on an adhesive tape. (b) Application of the ELIPatch to the human skin surface. (c) Automated data acquisition process. 10× images in bright field and dark field are used for decoding and fluorescence measurements. Scale bar, 500 μm. (d) Preparation of the ELIPatch by fluid-assisted self-assembly. Note that the size of the microwell fits to that of the microdisk. (e) Photographs of the ELIPatch in different scales. Microdisks in the 22 × 22 array are transferred to the 16 mm diameter tape by peeling off. Scale bar, 1 cm and 100 μm. (f) Example of application to Inner forearm. Black circles denote the array of ELIPatch. (inset) Size comparison with the ELIPatch and thumbnail. Scale bars, 1.5 cm.
Multiplexed protein detection from multiple skin sites or different volunteers
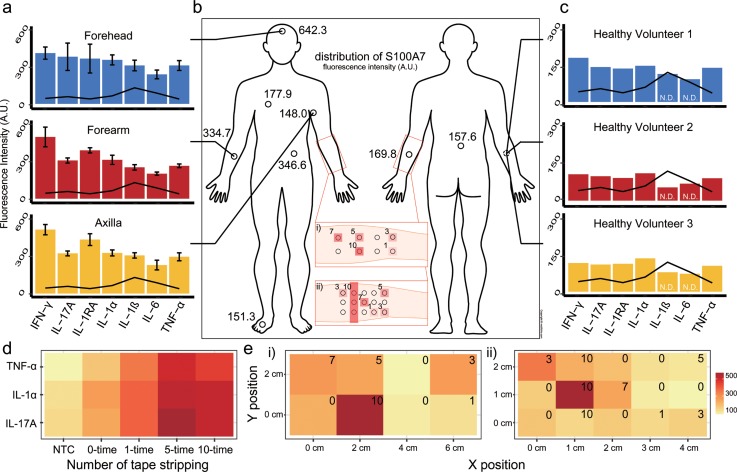
The encoded microdisk capability for multiplexed immunoassay is determined through performance of 6-plex immunoassays (Fig. S1), and preparation of the ELIPatch is conducted by self-assembly of encoded microdisks and transferring to adhesive tape (Fig. S2). To explore the ELIPatch performance for multiplexed protein detection from the human skin surface, ELIPatches with 7-plex microdisks were applied to three regions of a healthy volunteer [Fig. 2(a)], on multiple sites for singleplex assay [Fig. 2(b)], and to the same skin region on three healthy volunteers [Fig. 2(c)]. For the experiments shown in Figs. 2(a) and 2(c), ELIPatches with 7-plex microdisks were attached to each region and left in place for 1 h to minimize the effect of sweating and environment.12,13 In most cases, the measure level of cytokines is above the limit of detection, denoting that the ELIPatch has sufficient sensitivities except IL-1β and IL-6 [Fig. 2(c)].4,14,15 Figure 2(b) shows the S100A7 levels obtained from various body regions. It is known that S100A7, which is an antimicrobial protein, presents in different concentrations along the body site. In this experiment, a singleplex microdisk that targets S100A7 was used in the ELIPatch preparation and, thus, attached to the target sites for 1 h. The results show a similar profile to those previously reported, e.g., high expression at the forehead.2 Every experiment is conducted simultaneously to avoid the effect of the environment such as temperature and humidity.
FIG. 2.
Multiplexed protein detection from human skin using the ELIPatch. (a) Seven cytokines are measured from three sites of a healthy volunteer. Error bars: standard deviation. Line: limit of detection. (b) Antimicrobial protein is measured from various sites of a healthy volunteer using the ELIPatch. (c) Seven cytokines are measured from the inner forearm region of three healthy volunteers. Line: limit of detection. N.D. means non-detectable. (d) The measured values of three cytokines with the increasing stimulus and the number of tape stripping repetitions of skin. (NTC: no target control). (e) Spatial distribution of single cytokine on the inner forearm with multiple stimulus points [red boxes in (b)]. (i) IL-1α levels measured. (ii) IL-17A levels measured. All values are given in arbitrary units (A.U.).
Multiplexed protein detection from the skin surface under stimulus in a high spatial resolution
We then explored the ELIPatch capability to detect the human skin response to the stimulus. The inner forearm region was stripped with adhesive tape various times, and ELIPatches were applied. Among the seven cytokines, the concentrations of TNF-α, IL-1α, and IL-17A varied in response to an increased number of tape stripping repetitions, as shown in Fig. 2(d).
Figure 2(e) is a demonstration of skin protein detection with high spatial resolution. The inner forearm region was divided into a grid, and each section was stripped with adhesive tape a different number of times [insets of Fig. 2(b)]. The spatial densities of the ELIPatch differed, as 8 patches were applied in a 6 cm × 2 cm area in Figs. 2(e) and 2(i) while the second case involved 15 patches in a 4 cm × 2 cm area, having higher spatial resolution. The numbers in the insets indicate the number of tape stripping repetitions, and the circles in the insets of Fig. 2(b) represent the applied ELIPatch. As expected, the results from each section yield intensities corresponding to the repeated number of tape stripping. These results indicate that the ELIPatch has the potential to detect skin residual proteins with centimeter resolution and thereby to compare the cytokine profile from multi-regions of skin, such as diseased region and normal region, at the same time.15,16
DISCUSSION
In this paper, we introduced a method to quantify multiple skin residual proteins with high spatial resolution. In this approach, a thumbnail-sized circular adhesive patch with an array of microdisks acting as immunospot is fabricated and demonstrated. The small size of the proposed patch enables high-spatial-density application to the skin surface. We also utilized this platform to detect the skin response under the stimulus by quantifying the level of cytokines of the stimulated skin sites. The results indicate that our platform can not only detect changes in the cytokine concentration in accordance with the degree of stimulus but also resolve the spatial distribution of the stimulus. We believe that this method will provide a simple and comfortable method of detecting skin residual proteins for both clinicians and subjects. Although the improvement on sensitivities is required to have a diagnostic value,17–20 there is potential to be advanced in antibody quality and design of the microdisk and patch itself. Therefore, we expect that this proposed patch contributes to not only accumulation of information on the profiles of skin residual proteins but also the objective complementation based on the biomolecules to diagnosis and prescription of skin inflammatory diseases such as psoriasis and atopic dermatitis.21–23
SUPPLEMENTARY MATERIAL
See supplementary material for figures of the result of multiplexed immunoassay using the microdisk [Fig. S1]; preparation of the ELIPatch by self-assembly of the microdisk [Fig. S2]; optofluidic maskless lithography system for microdisk fabrication [Fig. S3]; additional tables of limit of detection [Table S1] and coefficient of variance [Table S2]; comparison of limit of detection and baseline of cytokine levels from previous research [Tables S3 and S4]; complete methods and experimental data.
ACKNOWLEDGMENTS
This research was supported by Nano Material Technology Development Program (2012M3A7A9671610) and Global Research Development Center Program (2015K1A4A3047345) through the National Research Foundation of Korea (NRF) funded by the Ministry of Science and ICT (MSIT).
References
- 1. Keum H., Mccormick M., Liu P., Zhang Y., and Omenetto F. G., Science 333, 838 (2011). 10.1126/science.1206157 [DOI] [PubMed] [Google Scholar]
- 2. Gläser R., Harder J., Lange H., Bartels J., Christophers E., and Schröder J.-M., Nat. Immunol. 6, 57 (2005). 10.1038/ni1142 [DOI] [PubMed] [Google Scholar]
- 3. Orro K., Smirnova O., Arshavskaja J., Salk K., Meikas A., Pihelgas S., Rumvolt R., Kingo K., Kazarjan A., Neuman T., and Spee P., Biomarker Res. 2, 20 (2014). 10.1186/2050-7771-2-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kinn P. M., Holdren G. O., Westermeyer B. A., Abuissa M., Fischer C. L., Fairley J. A., Brogden K. A., and Brogden N. K., Sci. Rep. 5, 10472 (2015). 10.1038/srep10472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dutkiewicz E. P., Der Lin J., Tseng T. W., Wang Y. S., and Urban P. L., Anal. Chem. 86, 2337 (2014). 10.1021/ac4039338 [DOI] [PubMed] [Google Scholar]
- 6. Dutkiewicz E. P., Chiu H. Y., and Urban P. L., Anal. Chem. 89, 2664 (2017). 10.1021/acs.analchem.6b04276 [DOI] [PubMed] [Google Scholar]
- 7. Portugal-Cohen M., Oron M., Ma'or Z., Boaz M., Shtendik L., Biro A., Cernes R., Barnea Z., Kazir Z., and Kohen R., Biomed. Pharmacother. 65, 280 (2011). 10.1016/j.biopha.2011.02.001 [DOI] [PubMed] [Google Scholar]
- 8. Chung S. E., Kim J., Oh D. Y., Song Y., Lee S. H., Min S., and Kwon S., Nat. Commun. 5, 3468 (2014). 10.1038/ncomms4468 [DOI] [PubMed] [Google Scholar]
- 9. Kim J. J., Bong K. W., Reátegui E., Irimia D., and Doyle P. S., Nat. Mater. 16, 139 (2017). 10.1038/nmat4747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nolan J. P. and Sklar L. A., Trends Biotechnol. 20, 9 (2002). 10.1016/S0167-7799(01)01844-3 [DOI] [PubMed] [Google Scholar]
- 11. Kim L. N., Kim M., Jung K., Bae H. J., Jang J., Jung Y., Kim J., and Kwon S., Chem. Commun. 51, 12130 (2015). 10.1039/C5CC02048D [DOI] [PubMed] [Google Scholar]
- 12. Munje R. D., Muthukumar S., Selvam A. P., and Prasad S., Sci. Rep. 5, 14586 (2015). 10.1038/srep14586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Munje R. D., Muthukumar S., Jagannath B., and Prasad S., Sci. Rep. 7, 1950 (2017). 10.1038/s41598-017-02133-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Perkins M. A., Cardin C. W., Osterhues M. A., and Robinson M. K., Skin Res. Technol. 8, 187 (2002). 10.1034/j.1600-0846.2002.20337.x [DOI] [PubMed] [Google Scholar]
- 15. Gläser R., Meyer-Hoffert U., Harder J., Cordes J., Wittersheim M., Kobliakova J., Fölster-Holst R., Proksch E., Schröder J.-M., and Schwarz T., J. Invest. Dermatol. 129, 641 (2009). 10.1038/jid.2008.268 [DOI] [PubMed] [Google Scholar]
- 16. Falcone D., Spee P., Salk K., Peppelman M., van de Kerkhof P. C. M., and van Erp P. E. J., Skin Res. Technol. 23, 336–345 (2017). 10.1111/srt.12340 [DOI] [PubMed] [Google Scholar]
- 17. Sonner Z., Wilder E., Heikenfeld J., Kasting G., Beyette F., Swaile D., Sherman F., Joyce J., Hagen J., Kelley-Loughnane N., and Naik R., Biomicrofluidics 9, 031301 (2015). 10.1063/1.4921039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gao W., Emaminejad S., Nyein H. Y. Y., Challa S., Chen K., Peck A., Fahad H. M., Ota H., Shiraki H., Kiriya D., Lien D. H., Brooks G. A., Davis R. W., and Javey A., Nature 529, 509 (2016). 10.1038/nature16521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Marques-Deak A., Cizza G., Eskandari F., Torvik S., Christie I. C., Sternberg E. M., and Phillips T. M., J. Immunol. Methods 315, 99 (2006). 10.1016/j.jim.2006.07.011 [DOI] [PubMed] [Google Scholar]
- 20. Cizza G., Marques A. H., Eskandari F., Christie I. C., Torvik S., Silverman M. N., Phillips T. M., and Sternberg E. M., Biol. Psychiatry 64, 907 (2008). 10.1016/j.biopsych.2008.05.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nomura I., Goleva E., Howell M. D., Hamid Q. A., Ong P. Y., Hall C. F., Darst M. A., Gao B., Boguniewicz M., Travers J. B., and Leung D. Y. M., J. Immunol. 171, 3262 (2003). 10.4049/jimmunol.171.6.3262 [DOI] [PubMed] [Google Scholar]
- 22. Deeva I., Mariani S., De Luca C., Pacifico V., Leoni L., Raskovic D., Kharaeva Z., Korkina L., and Pastore S., Cytokine 49, 163 (2010). 10.1016/j.cyto.2009.09.014 [DOI] [PubMed] [Google Scholar]
- 23. Szegedi K., Lutter R., Res P. C., Bos J. D., Luiten R. M., Kezic S., and Middelkamp-Hup M. A., J. Euro. Acad. Derm. Venerology 29, 2136 (2015). 10.1111/jdv.13160 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
See supplementary material for figures of the result of multiplexed immunoassay using the microdisk [Fig. S1]; preparation of the ELIPatch by self-assembly of the microdisk [Fig. S2]; optofluidic maskless lithography system for microdisk fabrication [Fig. S3]; additional tables of limit of detection [Table S1] and coefficient of variance [Table S2]; comparison of limit of detection and baseline of cytokine levels from previous research [Tables S3 and S4]; complete methods and experimental data.