Summary
Therapeutic antibodies targeting ovarian cancer (OvCa)-enriched receptors have largely been disappointing due to limited tumor specific antibody-dependent cellular cytotoxicity (ADCC). Here we report a symbiotic approach that is highly selective and superior compared to investigational clinical antibodies. This Bispecific-Anchored Cytotoxicity-Activator (BaCa) antibody is rationally designed to instigate “cis” and “trans” cytotoxicity by combining specificities against folate receptor alpha-1 (FOLR1) and death receptor 5 (DR5). Whereas the in vivo agonist DR5 signaling requires FcγRIIB interaction, the FOLR1 anchor functions as a primary clustering point to retain and maintain a high-level of tumor-specific apoptosis. The presented proof of concept study strategically makes use of a tumor-cell enriched anchor receptor for agonist death-receptor targeting to potentially generate a clinically viable strategy for OvCa.
Introduction
Monoclonal antibodies (mAbs) that selectively target and eliminate cancer cells exploit multiple independent mechanisms (Tushir-Singh, 2017). Despite numerous FDA approvals for solid and blood cancers, antibodies against ovarian cancer (OvCa) enriched receptors such as folate receptor alpha-1 (FOLR1) and cancer antigen 125 (Ca125) have largely been disappointing in clinical trials (Armstrong et al., 2013; Berek et al., 2009). These antibodies have relied on IgG1 Fc dependent crosslinking of FcγRIIIA (CD16a), a widely expressed immunoglobulin superfamily receptor on natural killer (NK) cells to induce antibody directed cell cytotoxicity (ADCC) of tumor cells (Albanesi and Daeron, 2012). Their dismal clinical response is potentially due to insufficient infiltration of the NK and other immune effector cells to the hypoxic solid tumor bed (Kline et al., 2017; Sasaki et al., 2015). Interestingly, in case of farletuzumab, a humanized mAb that targets high-grade serous OvCa (HGSOC) enriched FOLR1, improvement in survival has been reported for a small subset of patients expressing low levels of Ca125 (Vergote et al., 2016). Thus it is reasonable to conclude that for the majority of patients whose OvCa highly overexpress Ca125, ADCC based strategies are not clinically feasible options. To achieve a clinically applicable response in a larger OvCa population, we hypothesized elevating the anti-tumor activity of FOLR1 targeting antibodies (such as farletuzumab) beyond the activating limit of ADCC and even independently of it.
One such approach is pro-apoptotic receptor agonists (PARA) therapy using Trail ligand (Apo2L) or epithelial cancer enriched death receptor 5 (DR5/TRAIL-R2) activating antibodies (Ashkenazi, 2008). PARA activate extrinsic apoptotic pathway by oligomerizing DR5, a hallmark of tumor necrosis factor (TNF) receptor family members (Ashkenazi and Herbst, 2008). Although several DR5 agonist antibodies as a single agent or in combination with Apo2L instigate DR5 receptor aggregation and anti-tumor response, findings from clinical studies have failed to demonstrate significant benefits in phase-2 trials (Paz-Ares et al., 2013; Soria et al., 2010). The clinical data at biochemical levels have accounted for insufficient tumor specific cell death signaling due to sub-optimal clustering of DR5 receptor (Merchant et al., 2012; Niyazi et al., 2009). As one alternative, trans-engaging (stromal cell and tumor cell) antibodies have been described to enhance DR5 clustering (Brunker et al., 2016). However, in addition to fundamental dependency on another cell type, the described fibroblast activation protein (FAP) engaging antibodies represent critical safety concerns such as severe cachexia and bone toxicity due to non-specific targeting (Tran et al., 2013). In the present study we sought to investigate whether tumor cell specific FOLR1 and DR5 targeting by a single agent Bispecific-Anchored Cytotoxicity-Activator (BaCa) antibody will result in the symbiotic gain of OvCa selectivity, safety and superior anti-tumor activity.
Results
Generation, characterization and lead BaCa antibody selection
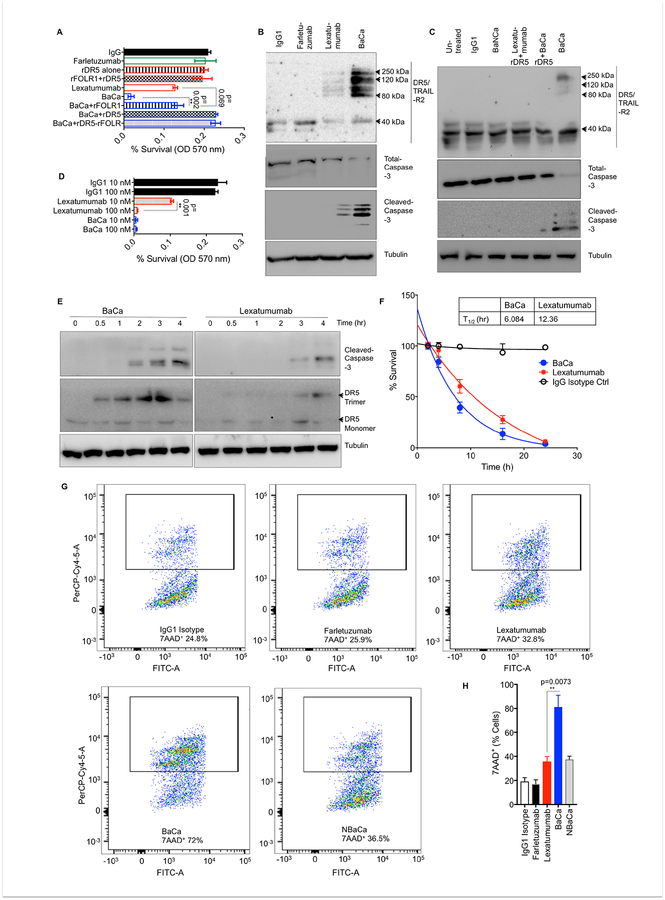
Various dual-specificity antibody configurations are in clinical trials for cancers (Brinkmann and Kontermann, 2017). To co-target FOLR1 and DR5, we engineered IgG1 Fc-based dual-specificity antibodies for the following 3 reasons: a) there is a defined requirement of FcγRIIB and IgG1 CH2 domain engagement for DR5 agonist antibodies in vivo (Li and Ravetch, 2012; Wilson et al., 2011), b) upon Apo2L ligand binding activated DR5 receptors form a tripartite structure, which is approximately ~40 Å on each side (Mongkolsapaya et al., 1999) and, c) a critical need for effective serum half-life. Hypothetically, IgG1 based antibody is best suited to provide flexible distance and longer serum half-life. Three different bispecfic antibodies were generated (Figure 1A, see STAR methods). The BaCa-1 antibody contains bivalent anti-FOLR1 (Blue) and anti-DR5 (Red) affinities at opposite ends. The BaCa-2 antibody resembles an IgG1 and is similar to CrossMab antibodies of Genentech (Ridgway et al., 1996; Schaefer et al., 2011). In BaCa-3 antibody, unlike BaCa-1, two variable domains of light and heavy chains against FOLR1 and DR5 are genetically fused next to each other via GS linkers (Gu and Ghayur, 2012). Therefore, despite being bivalent, the specificities against DR5 and FOLR1 receptors are only 10–30 Å apart. The amino acid sequences of described antibodies are provided in the STAR Methods. For BaCa-1, BaCa-2 and BaCa-3, a separating linker length of 12 GS, 45 GS, and 9 GS respectively resulted in the highest monomer recovery (Durocher and Butler, 2009) (Figure S1A). The comparison of various properties of BaCa antibodies is shown in Figure 1B. BaCa-1 antibody not only had significantly higher cytotoxicity against OvCa cells (Figure 1C), but also exhibited higher yield and stability over BaCa-2 and BaCa-3. Thus, BaCa-1 was selected as the lead antibody (BaCa or HuBaCa whenever stated) for proof of concept studies. The observed high activity of BaCa-1 antibody could be explained by geometrical flexibility of its affinities against FOLR1 and DR5 (Zhang et al., 2015). The separating distance of >170 Å between two variable domains is largest in BaCa-1 antibody to simultaneously engage FOLR1 and DR5 receptors. It is highly likely that BaCa-3 antibody once bound to FOLR1 (via inner variable domain) was not able to simultaneously engage DR5, by outer domain and vice versa, due to steric hindrance (Figure 1A). Although BaCa-2 antibody has optimal flexibility to engage FOLR1 and DR5 simultaneously, it is less effective due to being monovalent, potentially resulting in lower avidity optimized binding as described for single Fab fragments (Graves et al., 2014). As expected, when incubated in 96 wells immobilized with rFOLR1 and rDR5 receptors together, lead BaCa antibody showed the highest relative avidity index after treatment with 6 M Urea (Figure 1D) (Levett et al., 2005).
Figure 1. Engineering and characterizing BaCa antibodies with superior cytotoxicity against ovarian cancer cells.
(A) Domain organization, and SDS-PAGE analyses of BaCa-1, BaCa-2, BaCa-3, and IgG1 antibodies in native and reducing conditions. Individual gel lanes for each antibody types are cropped from the same blot.
(B) Summary of BaCa-1, BaCa-2, BaCa-3 and parental IgG1 properties
(C) NIH-OVCAR-3 cells were treated with increasing concentrations of the indicated antibodies or cisplatin. The cell death was quantified using cell viability assays (n=3).
(D) rFOLR1 and rDR5 were coated on 96 well plates in 5:1 ratio. Relative avidity index of indicated antibodies was determined in presence of 6 M urea.
(E) The binding kinetics of immobilized biotinylated rDR5 against lexatumumab and BaCa or biotinylated rFOLR1 against farletuzumab and BaCa were measured using bio-layer interferometry (BLI) optical analytical technique.
(F) Domain organization of the non-anchoring BaCa (NBaCa) antibody.
(G) Cell viabilities of NIH-OVCAR-4 cells were analyzed in the presence of BaCa, lexatumumab, or NBaCa antibodies. IC50 values are shown at the bottom (n=3).
Abbreviation used in Figure 1A and B: * = 1-step protein-A purification, ** = Total % recovery after size exclusion purification (SEC), *** = Farletuzumab data is shown (Lexatumumab was comparable), Native = Antibody run on gel with non-reducing dye, Reducing = Antibody run on gel with reducing dye, HC = Heavy chain, LC = Light chain, Fab = Fragment antigen binding, Fv = Fragment variable, scFv = Single chain fragment variable, VL = Variable domain of light chain, VH = Variable domain of heavy chain, CK = Kappa chain
Error bars in C, D and G represent SEM. Unpaired Welch’s t-test was used to determine p values.
See also Figures S1 and S2
Higher order DR5 oligomerization and activation is due to co-engagement of target receptors by BaCa antibody
To test simultaneous receptor co-engagement, we expressed and purified IgG4-Fc conjugated extracellular fragment of recombinant DR5 (r-DR5) and rFOLR1 (Figure S1B, C). Next, we analyzed the binding affinities of three BaCa and parental antibodies using ELISA (Figure S1D, E) and subsequently confirmed the binding kinetics of lead BaCa using ForteBio Octet HTX (Figure 1E, S1F, G). The individual receptor binding affinities of BaCa antibodies against FOLR1 and DR5 remained unchanged after conversion into bispecific configurations from their respective mAbs. This suggested that disparity in their cellular cytotoxic activities (IC50 values, Figure 1B) is due to their varying ability to engage, cluster and activate DR5. Thus, next we generated Non-anchoring-BaCa (NBaCa) antibodies, where anti-FOLR1 variable domain has been replaced with Praxbind, an antidote for anticoagulant medication Pradaxa (Teleb et al., 2016) (Figure 1F, S2A, B). NBaCa antibody has bivalent binding against DR5 receptor and the same structural framework of a BaCa antibody. When tested, NBaCa antibody was found to be as effective as lexatumumab (Figure 1G, S2C). Similar loss of cytotoxicity was observed when BaCa-2 antibody was engineered into NBaCa-2 antibody (Figure S2D–F).
Next, we pre-neutralized BaCa antibody (37°C, 1 hr) with rDR5, rFOLR1 or both before treating the cells. The rFOLR1 pre-neutralization reduced the cytotoxicity of BaCa to lexatumumab, while pre-blocking with rDR5 or loss of rDR5 binding domain abolished the activity (Figure 2A, S2G–I). In comparison to lexatumumab, BaCa treated lysates also showed significantly higher levels of DR5 clustering (in large molecular weight complexes) and cleaved caspase-3 levels (Figure 2B, C). Next, we tested if the lead BaCa antibody will be effective against FOLR1 anchor enriched OvCa cells independent of DR5 binding being oriented as a Fab or scFv. To this end, we engineered reverse-BaCa (R-BaCa) antibody where anti-FOLR1 affinity is scFv, while anti-DR5 affinity is a Fab. Both BaCa and R-BaCa were equally effective over lexatumumab (Figure S2J–L). Taken together, these sets of findings establish that higher order DR5 receptor clustering, signaling and activity by BaCa antibody is critically dependent on the DR5 co-engagement with the tumor-enriched anchor receptor (FOLR1).
Figure 2. BaCa antibody mediated higher order TRAIL-R2 receptor clustering requires anchor and death receptor co-engagement.
(A) Survival of OVCAR-3 cells treated with the indicated antibodies without or with pre-blocking with rDR5, rFOLR1, or rDR5+rFOLR1 (n=3).
(B) OVCAR-4 cells were treated with indicated antibodies for 24 hr, followed by lysis using RIPA buffer. DR5, total caspase-3 and cleaved caspased-3 were analyzed by immunoblotting.
(C) DR5 clustering and caspase-3 activity in OVCAR-3 cells treated with the indicated antibodies without or with pre-blocking with rDR5. Protein lysates were analyzed by immunoblotting.
(D) Survival of OVCAR-3 cells treated with 10 nM or 100 nM of lexatumumab, BaCa and IgG1 for 48 hr (n=3).
(E) Time dependent cleaved caspase-3 and DR5 trimerization (120 kDa) profile in OVCAR-3 cells treated with 100 nM lexatumumab and BaCa antibodies (n=3).
(F) Time-dependent cell killing activity of 100 nM of lexatumumab and BaCa antibodies (n=3).
(G) OVCAR-3 cells treated with indicated antibodies (100 nM), were analyzed by 7AAD+ labeling using FACS analysis (n=3).
(H) Quantitation of 7AAD+ labeling as described in G.
Error bars in (A, D F and H) represent SEM. Unpaired two-tailed Welch’s t-test was used to determine p values See also Figure S2.
Next, we asked if BaCa antibody would also positively shift the kinetics of apoptotic activation along with the overall superior cytotoxicity. Since both antibodies effectively kill >99% of cells at 100 nM in 48 hr (Figure 2D), the early time course analysis at 100 nM dose will reflect time dependent apoptotic activation function of DR5 signaling. As shown, BaCa antibody induced DR5 trimerization (120 kDa) and caspase-3 activation within 30 min and 2 hr, while lexatumumab needed 3 hr to do the same (Figure 2E). In support, BaCa antibody eliminated 50% of the OVCAR-3 cells within 6 hr, while lexatumumab needed ~12 hr (Figure 2F). Similar kinetic results were obtained in flow cytometry studies (Figure 2G, H). Importantly, the NBaCa antibody abolished the gained apoptotic kinetics as evident by equal level of 7-AAD+ staining. These findings strengthen that receptor co-engagement by BaCa antibody instigates both kinetically faster and cytotoxically superior DR5 clustering and signaling.
BaCa antibody is broadly effective and is superior to the described cooperativity
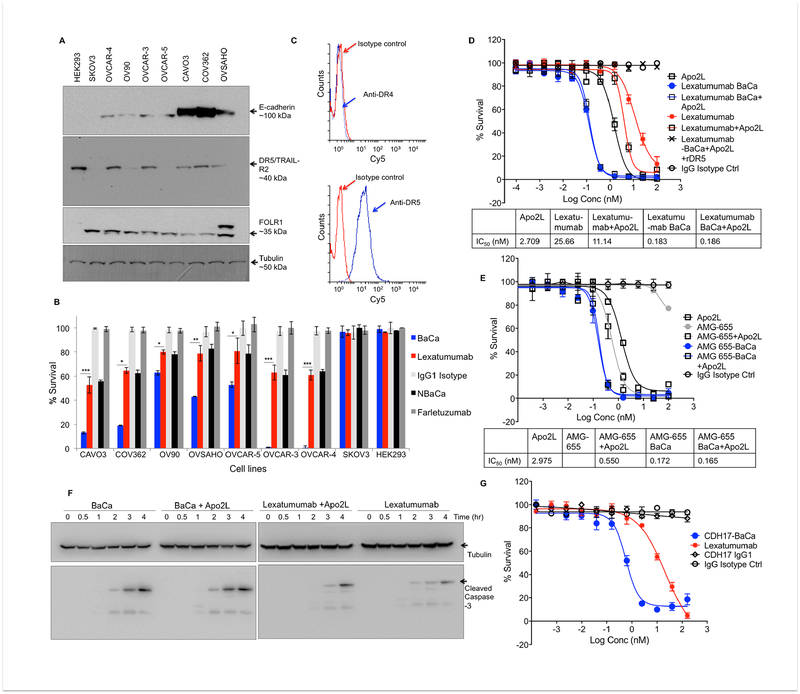
Next we extended the BaCa activity in various other likely high-grade serous ovarian carcinoma (HGSOC) cells (Domcke et al., 2013). As expected, almost all tested lines expressed high levels of FOLR1 (Figure 3A). BaCa antibody consistently instigated significantly higher cytotoxicity than lexatumumab in most of the OvCa lines and against heterogeneous patient derived OvCa cells (Figure 3B, S3A, B). The only OvCa cell line described as non-HGSOC in literature, SKOV3 (Domcke et al., 2013), did not respond to agonist DR5 therapy. Although comparable at transcript levels, the DR5 protein was significantly reduced in SKOV3 cells (Figure 3A, S3C). This promoted us to investigate if additional factors regulating DR5 stability might be differentially expressed in SKOV3 cells (Wagner et al., 2007). When tested, expression of key glycosylation regulators, N-acetylgalactosaminyltransferase-3 (GALNT3) was undetectable both at RT-PCR and qPCR levels in SKOV3 cells (Figure S3D, E). These observations indicate that loss of DR5 O-linked glycosylation in SKOV3 cells limits their sensitivity to DR5 therapy.
Figure 3. BaCa antibody is broadly effective and is highly superior over described cooperativity.
(A) E-cadherin, DR5 and FOLR1 expression profile across various OvCa cell lines. Tubulin is loading control.
(B) Survival analysis of OvCa cell lines treated with indicated antibodies for 72 hr (n=3).
(C) FACS analysis of DR4 and DR5 on the cell surface of OVCAR-3 cells.
(D) OVCAR-3 cells were treated with lexatumumab or BaCa antibody (generated with lexatumumab) with and without Apo2L as indicated (n=3). IC50 values are shown at the bottom.
(E) OVCAR-3 cells were treated with AMG-655 or BaCa antibody (generated with AMG-655) with and without Apo2L as indicated (n=3). IC50 values are shown at the bottom.
(F) Cleaved caspase-3 levels in OVCAR-3 cells treated with indicated antibodies (with and without Apo2L) for the indicated period of time. Lysates were analyzed by immunoblotting.
(G) BaCa antibody was engineered with CDH17 specific A4 antibody instead of farletuzumab and tested against Colo-205 cells (n=3).
Error bars in B, D, E, G represent SEM. Bar graphs in B were compared using Student’s t-test. * p<0.05, ** p<0.005, *** p<0.001.
See also Figure S3.
Co-treatment of Apo2L ligand and DR5 agonist antibody AMG-655 has been shown to enhance apoptotic cooperativity (Graves et al., 2014). As Apo2L ligand can induce cytotoxicity via engaging both DR4 (TRAIL-R1) and DR5 receptors, we first confirmed that OVCAR-3 cells only expressed DR5 (Figure 3C, S3F, G). Apo2L was generated in our lab and tested along with commercial Apo2L (Figure S3H). Next, we compared the cell-killing activity of BaCa antibodies (generated either with lexatumumab or AMG-655) ± Apo2L ligand. The co-treatment of Apo2L ligand was insufficient to enhance the activity of BaCa antibodies (Figure 3D, E) indicating that higher order DR5 clustering by BaCa antibody is highly superior independent of Apo2L ligand being present. In support, we observed no change in caspase-3 activation by BaCa antibody regardless of Apo2L ligand (Figure 3F).
Similar to previous reports, we also observed apoptotic cooperativity due to Apo2L and AMG-655 (Figure 3E). Interestingly, co-treatment of Apo2L and lexatumumab was not effectively cooperative (Figure 3D, F). It should also be noted that unlike lexatumumab, AMG-655 was very limitedly effective in inducing loss of OvCa cell viability, which point toward the differences in their working mechanisms. Next, we extended the BaCa targeting strategy by swapping anti-FOLR1 affinity with another cancer-enriched receptor (CDH17) targeting A4 antibody (https://patents.google.com/patent/US20160039933A1/en). CDH17 is commonly overexpressed in intestinal and colorectal cancers (Chen et al., 2012). A4-BaCa showed multiple fold higher cytotoxicity against Colo-205 cells over lexatumumab suggesting the reproducible potential of BaCa targeting to other cancers (Figure 3G).
BaCa antibody is highly selective towards FOLR1 positive OvCa cells
Selective therapeutic targeting remains a critical concern considering the minimal number of drug approvals by FDA, mostly due to non-specific accumulation and toxicity in clinical trials (Printz, 2011; Vincenzi et al., 2016). Therefore, we next compared the selective BaCa gain of function with anti-Fc crosslinking, a nonspecific way to induce DR5 receptor clustering (Wilson et al., 2011). To this end, we incubated OVCAR-3 cells with a dose titration of lexatumumab or AMG-655, either alone or together with anti-human Fc crosslinking agent (1 μg/ml). Despite non-specific crosslinking of DR5 agonists, single agent BaCa antibodies were multiple folds more effective (Figure 4A, B).
Figure 4. BaCa activity is highly selective towards FOLR1 overexpressing OvCa cancer cells.
(A) Cell viability analysis of lexatumumab and lexatumumab containing BaCa antibody ± anti-Fc crosslinking (n=3).
(B) Cell viability analysis of AMG-655 and AMG-655 containing BaCa antibody ± anti-Fc crosslinking (n=3).
(C) qRT-PCR analysis of FOLR1 and TRAIL-R2 transcripts in OVCAR-4 and Colo-205 cells (n=5).
(D) Cell viability assays using BaCa antibody in OVCAR-4 and Colo-205 cells. IC50 values are shown in right (n=3).
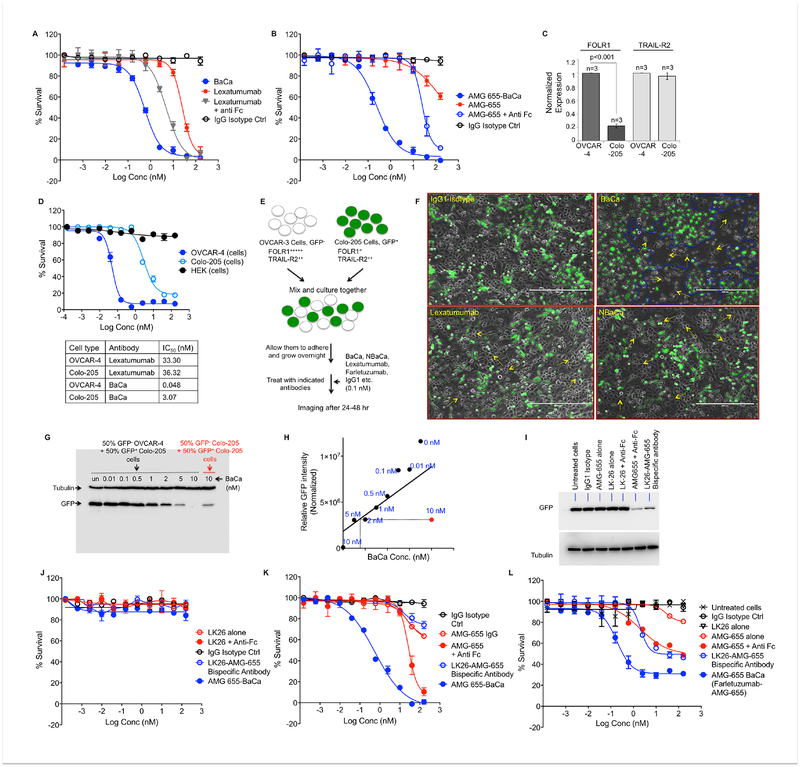
(E) Schematic of results described in F. 50% GFP− OVCAR-4 and 50% GFP+ Colo-205 cells were co-cultured. After 24 hr, cells were treated with indicated antibodies at constant 0.1 nM. After 36–48 hr, cells were analyzed using fluorescent microscope.
(F) Represented images as described in E with indicated antibody treatment (scale bar represent 400 μm).
(G) Immunoblot analysis for GFP and tubulin of GFP+ Colo-205 cells co-cultured with equal number of GFP− OVCAR-4 or GFP− Colo-205 cells and treated with the indicated concentrations of BaCa antibody
(H) The normalized relative intensities of GFP signal were plotted for the increasing BaCa dose in co-cultured conditions (filled black circles: 50% GFP− OVCAR-4 and 50% GFP+ Colo-205) against constant 10 nM dose (filled red circle: 50% GFP− Colo-205 and 50% GFP+ Colo-205).
(I) Co-cultured MC38 (GFP−) and Colo-205 (GFP+) cells were treated with 50 nM of indicated antibodies. After 48 hr, lysates were run on gel and blotted for tubulin and GFP.
(J) Cell viability of MC38 cells treated with LK26, LK26+anti-Fc, LK26-AMG-655 bispecific and BaCa antibody (n=3).
(K) Cell viability of OVCAR-3 cells treated with AMG-655, AMG-655+anti-Fc, LK26-AMG-655 bispecific and BaCa antibody (n=3).
(L) Co-cultured MC38 and OVCAR-3 cells were treated with the increasing concentration of indicated antibodies. After 48 hr, cell viability was analyzed using MTT assays.
Error bars in A, B, C, D, J, K and L represent SEM.
See also Figure S4.
An ideal anti-cancer therapeutic antibody such as BaCa should have reduced toxicity towards non or low FOLR1 expressing cells. Thus, we next compared the BaCa activity in high and low FOLR1 expressing cells. The colorectal cancer cell line Colo-205 expresses ~5 fold less FOLR1 than does OVCAR-4 ovarian cancer cells but equal levels of the DR5 and GALNT3 transcripts (Figure 4C, S3C–E). Indeed, IC50 of lexatumumab was not significantly different in Colo-205 and OVCAR-4 cells (Figure 4D). However, the BaCa antibody was ~70 fold more effective in killing OVCAR-4 cells over Colo-205 cells. This reasonably supports the dependence of gain in cytotoxicity on the increased expression of tumor specific FOLR1 anchor antigen.
Next, we asked if this cytotoxic gain would be selective to OvCa cells by co-culturing Colo-205 cells stably expressing GFP with OVCAR-4 (Figure 4E). When treated with 0.1 nM BaCa antibody for 24–36 hr, we observed selective elimination of OVCAR-4 cells (Figure 4F, S4A). Lexatumumab was completely ineffective at the same dose. Since DR5 expression was similar in both the cell types (Figure 4C), these findings indicate that at a low dose BaCa antibody is highly selective to FOLR1 anchor enriched tumor cells and prefers to engage DR5 to instigate cell death in “cis”. Similar results were obtained when BaCa antibody was generated with AMG-655 (Figure S4B).
When co-cultures were incubated at >20 fold higher concentrations (2 nM), we observed BaCa cytotoxicity towards both high anchor (OVCAR-4) and low anchor (Colo-205) expressing cells (Figure S4C). We consistently detected >95% cell death of Colo-205 cells in co-cultures and since the 2 nM concentrations of BaCa antibody were below its IC50 value (3.07 nM) against Colo-205 cells, these findings were highly suggestive of “trans” activation by BaCa antibody at higher concentrations. To reconfirm, we incubated co-cultured cells (50% GFP− OVCAR-4 and 50% GFP+ Colo-205) with the increased concentration of BaCa antibody and evaluated the loss of GFP signal as an indicator for activity in “trans”. As a control, we also co-cultured 50% GFP− Colo-205 with 50% GFP+ Colo-205 cells. At higher doses, BaCa antibody was significantly more effective (>5 fold) in killing GFP+ Colo-205 cells that were co-cultured with GFP− OVCAR-4 cells in comparison to those co-cultured with GFP− Colo-205 cell only (Figure 4G, H). Similar results were seen when co-cultured conditions had 70% GFP− OVCAR-4 and 30% GFP+ Colo-205 cells (Figure S4D, E).
Next, we made use of anchor antigen− cells to compare trans-engaging and DR5 activating bispecific antibody with the BaCa strategy. To this end, we engineered murine FOLR1 (muFOLR1) specific LK-26 antibody and huDR5 specific AMG-655 into a bispecific antibody (Figure S4F, G). Next we co-cultured GFP+ Colo-205 cells with murine MC38 cells and treated with 50 nM LK26-AMG-655 bispecific antibody. The loss of GFP in Figure 4I (most right lane) confirms bispecific antibody functioning to engage muFOLR1 to “trans” activate huDR5, similar to described for RG7386 (Brunker et al., 2016). Next, we compared the activity of trans-engaging DR5 bispecific antibody against the BaCa strategy using serial dilutions. BaCa antibody (Farletuzumab-AMG-655) induced cell killing of co-cultured OVCAR-3 at a much lower concentration than the LK26-AMG-655 bispecific antibody (Figure 4J–L). Notably, unlike BaCa, the trans-engaging bispecific antibody was totally dependent on MC38 cells (Figure 4K vs L). Similar results were obtained with OVCAR-4 cells (Figure S4H, I). The inability of cell killing assay to achieve 100% (Figure 4L, S4I) is due to presence of AMG-655 non-binding MC38 cells in the co-cultures. These findings strongly substantiate that unlike described for RG7386 that requires two different cell-types (stromal and tumor cells) to induce “trans-only” cytotoxicity, BaCa antibody has built-in function to activate both “cis” and “trans” cytotoxicity by making use of a single anchor antigen expressing cancer cell. Importantly, unlike the “trans-only” activating bispecific antibody, BaCa antibody required a significantly lower dose to achieve highly superior cytotoxicity.
Tumor specificity of BaCa antibody
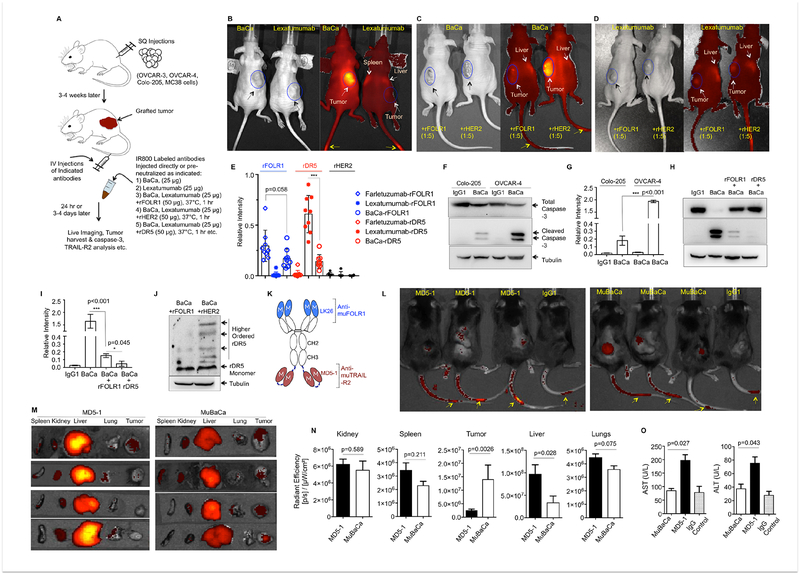
Before moving to in vivo, we tested BaCa antibody for in vitro stability (Figure S5A). For in vivo stability, we carried out serum half-life analysis as described earlier (Hutt et al., 2012). Lead BaCa antibody showed high in vivo stability with a t1/2 of ~15 days (Figure S5B, C). To test the tumor selectivity of BaCa antibody, we intravenously (IV) injected infrared dye IR800-labeled antibodies (25 μg) into the tumor bearing mice to monitor tissue localization as described (Figure 5A) (Lin et al., 2013). It was also confirmed that IR800 labeling did not change the affinities against the respective receptors (Figure S5D). BaCa antibody selectively accumulated in the grafted tumors within 24 hr, while lexatumumab showed significant more localization in the liver than tumor (Figure 5B, S5E, F). As predicted, the tumor specific enrichment of BaCa antibody was completely lost if it was neutralized with rFOLR1 before IV injection (Figure 5C). Interestingly, we observed BaCa antibody accumulation in mice liver upon FOLR1 pre-neutralization. Lexatumumab remained localized consistently in the animal liver in addition to tumors ± rFOLR1 neutralization (Figure 5D). The differential accumulation of lexatumumab was confirmed using liver specific ELISA. We observed a consistent >5 fold more accumulation of lexatumumab antibody in mice liver than BaCa antibody (Figure 5E). These results indicate that non-specific accumulation of lexatumumab in tissues (such as liver) could be responsible for its limited clinical efficacy and points to the added safety of BaCa approach due to avidity optimized retention in tumors. Next, we tested the activation of DR5 signaling in grafted OVCAR-3 tumors and also compared to Colo-205 tumors. A single IV dose of BaCa antibody produced >10 fold more cleaved caspase-3 levels in OVCAR-3 tumors and pre-blocking of BaCa antibody with rFOLR1 reduced the gain in caspase activity and DR5 oligomerization (Figure 5F–J).
Figure 5. BaCa activity is highly selective towards FOLR1 overexpressing OvCa tumors in vivo.
(A) Experimental schematic of tumor formation and antibody treatments. 6–8 weeks old athymic nude or C57BL/6 mice were grafted with indicated cells via subcutaneous (SQ) injections. 3–4 weeks later (tumor ~200 mm3), mice were IV injected with indicated antibodies followed by imaging, or were harvested for biochemical analysis and ELISA as indicated.
(B) Tumor bearing mice were IV injected with IR800 labeled lexatumumab or BaCa antibody followed by live imaging.
(C, D) Tumor bearing mice were IV injected with BaCa antibody (C) or lexatumumab (D) pre-neutralized with rFOLR1 or rHER2 followed by live imaging.
(E) Relative amount of liver accumulated antibodies were detected using ELISA against coated rFOLR1, rDR5 and rHER2 from liver lysates as indicated (n=3).
(F) Harvested OVCAR-3 and Colo-205 tumors after BaCa and IgG1 treatments were analyzed by immunoblotting as indicated.
(G) Quantitation of caspase-3 activity as described in F.
(H) OVCAR-3 tumors harvested from mice injected with BaCa antibody pre-neutralized as indicated were analyzed by immunoblotting as indicated.
(I) Quantitation of caspase-3 activity as described in H.
(J) OVCAR-3 tumors harvested from mice injected with BaCa antibody pre-neutralized as indicated were analyzed for DR5 using immunoblotting.
(K) Schematic representation of muBaCa antibody consisting of LK26 and MD5–1 antibodies against muFOLR1 and muDR5, respectively.
(L) C57BL/6 mice bearing SQ tumors were IV injected with IR800 labeled MD5–1 and muBaCa antibodies followed by live imaging. Yellow arrows indicate residual signal at the site of injection.
(M) Necropsies from animals in L were analyzed by fluorescent imaging for detailed organ specific distribution of IR800 labeled antibodies (n=4).
(N) Quantitation of accumulated IR800 signal (radiant efficiency) from the indicated tissues after MD5–1 and muBaCa injections. IgG1 was used to subtract the background signal (n=4).
(O) AST, ALT assays were carried out using MC38 tumor bearing C57BL/6 mice after IV injection of MD5–1 and muBaCa antibodies (n=3). IgG was used for control.
Yellow arrows (in B, C, and D) indicate residual signal from the site of injection, white arrows mark nonspecific localization of antibody in other tissues along with tumors. Black arrows show the location of tumors.
Error bars in (E, G, I, N, and O) indicate SEM and p values were determined using unpaired t-test with Welch’s correction.
See also Figures S5 and S6.
To investigate if accumulated DR5 agonist antibody in liver would result in hepatotoxicity, we engineered a murine cross-reactive BaCa (MuBaCa) antibody consisting of LK26 and MD5–1 (Figure 5K, S5G). MD5–1 IgG1 generated in our lab was tested and confirmed along with commercial MD5–1 antibody (Figure S5H). MuBaCa, but not huBaCa, selectively eliminated murine MC38 cells without any crosslinking agent (Figure S5I). Since MC38 cells expressed >6 fold more muFOLR1 than mice ovarian ID8 cells (data not shown), we made use of MC38 cells for surrogate studies. Similar to huBaCa, C57BL/6 mice grafted with MC38 tumors showed significantly higher localization of muBaCa compared to MD5–1 antibody (Figure 5L. We also carried out detailed tissue distribution of muBaCa and MD5–1 using C57BL/6 mice necropsies. Significantly more muBaCa and MD5–1 signal was evident in tumors and livers respectively (Figure 5M, N S5J). MD5–1 also resulted in elevated serum AST and ALT levels, both of which are indicators of hepatotoxicity (Figure 5O). H&E stained liver sections from all 3 mice treated with MD5–1 showed a focal lobular hepatitis as evident with the infiltrating neutrophils near portal vein and sinusoids, while only 1 out of 4 muBaCa treated mice showed significantly minor presence of neutrophils (Figure S6A, B). These findings in surrogate animals further strengthen the selective anchor receptor (FOLR1) mediated retention, safety and activity of BaCa antibody in the grafted tumors.
Anti-tumor activity of BaCa antibody
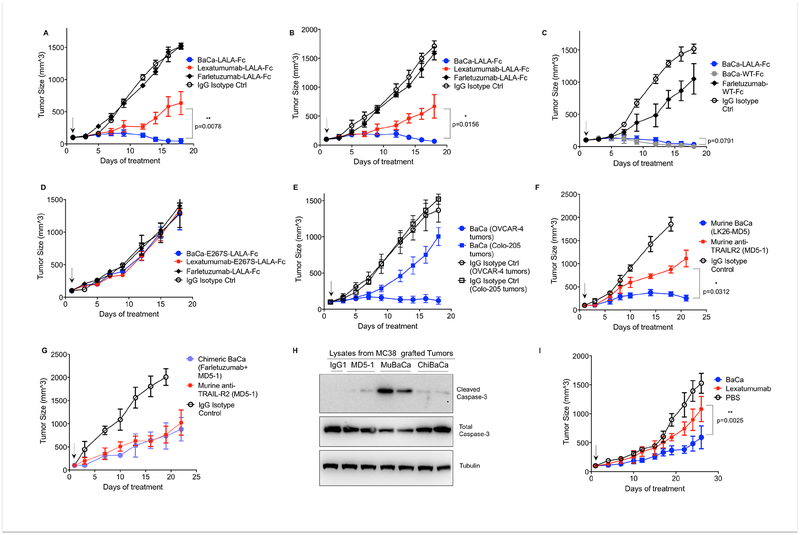
To impair FcγRIIIA binding and ADCC activity, we engineered lexatumumab, farletuzumab, and BaCa antibodies with LALA-Fc (L234A-L235A) mutations in the CH2 domain (Leabman et al., 2013). LALA mutant antibody did not exhibit measurable binding to human FcγRIIIA (Figure S7A, B). The binding affinities and activities of antibodies also remained unchanged after LALA mutations (Figure S7C–F). Next, randomly selected nude mice bearing OVCAR-3 tumors (>100 mm3) were injected intraperitoneally (IP) every third day with 25 μg dose of antibodies as indicated (Figure 6A). BaCa antibody completely regressed the tumor growth within 4 doses while lexatumumab only stabilized them. When followed for additional 4 weeks, none of the 6 BaCa injected mice showed tumor re-growth. Similar efficacy of BaCa antibody was observed in tumors generated with OVCAR-4 cells (Figure 6B). Since LALA mutant antibodies don’t engage NK cells (impaired FcγRIIIA binding), the efficacy data (Figure 6A, B) is independent of ADCC function. Next, we compared ADCC-activating farletuzumab antibody with BaCa antibody in nude animals having active NK cells and innate immunity. To this end, WT-Fc (LL234–235) containing BaCa and farletuzumab antibodies were IP injected at 25 μg dose in the mice. Farletuzumab (WT-Fc) was only limitedly effective compared to BaCa antibodies (Figure 6C). When dosed at 150 μg, farletuzumab (WT-Fc) also regressed the grafted tumors (data not shown). Both BaCa antibodies (LALA-Fc or WT-Fc) were equally effective ± ADCC activating function and the data was not statistically significant (n=6). These findings indicate that BaCa antibody potentially will be highly effective even in the immune deficient ovarian tumor microenvironment.
Figure 6. Anti-tumor activity of BaCa antibody.
(A, B) Six-eight weeks old mice bearing SQ OVCAR-3 (A) or OVCAR-4 (B) tumor were IP (A) or IV (B) injected with 25 μg of indicated antibody every third day (n=4–6). Tumor volumes were quantified at indicated days by caliper measurements.
(C) OVCAR-3 tumor volume in tumor bearing mice IP injected with BaCa antibodies (25 μg) having WT-Fc or LALA Fc.
(D) Indicated antibodies generated with E267S mutation in CH2 domain were compared for their ability to regress OVCAR-3 tumors
(E) Tumor size of OVCAR-4 and Colo-205 tumors in mice IV injected with BaCa antibodies at 25 μg dose for indicated days.
(F) C57BL/6 mice bearing SQ MC38 tumor were IP injected (25 μg) with the indicated antibodies having LALA Fc mutations. Tumor volumes were quantified at indicated days by caliper measurements.
(G) Same as (F), except chimeric BaCa (ChiBaCa) with affinity against huFOLR1 and muDR5 was compared with MD5–1 (n=5).
(H) Total and cleaved caspase-3 levels in tumors from mice after 3 doses of MD5–1, muBaCa or chiBaCa.
(I) Tumor sizes of cisplatin resistant PDX tumors in 6–8 weeks old mice IP injected with 5 mg/kg dose of indicated antibodies (Lexatumumab n=3, BaCa antibody n=4).
Error bars indicate in A, B, C, D, E, F, G and I represent SEM and p values were determined by two-tailed paired Wilcoxon Mann-Whitney test.
See also Figure S7
In support with previous reports (Li and Ravetch, 2012), antibodies engineered with E267S mutations (impaired binding to human FcγRIIB) exhibited no anti-tumor activity (Figure 6D). When tested against high vs low FOLR1 anchor expressing tumors, BaCa antibody (25 μg) completely regressed OVCAR-4 tumors while it remained limitedly effective against Colo-205 tumors at the same dose (Figure 6E). Similar caspase-3 activation results were reflected at molecular levels (Figure 5). Comparable efficacy was also observed in immunocompetent mice studies with muBaCa (Figure 6F). Since farletuzumab did not bind to muFOLR1, we made use of chiBaCa antibody (Figure S5G, H) having farletuzumab and MD5–1 domains for antigen− tumor regression analysis. When tested against MC38 tumors, chiBaCa was equally effective to MD5–1 (Figure 6G). These sets of investigations and caspase-3 activity (Figure 6H) strongly support anchor specific in vivo activity of BaCa targeting strategy. Since treatment failure in OvCa patients is mainly due to emergence of cisplatin resistance, we next compared the in vivo efficacy of BaCa antibody with lexatumumab using patent derived platinum resistant xenografts (PDX) models. As evident, in comparison to lexatumumab, BaCa antibody stabilized platinum resistant tumors (Figure 6I).
Discussion
Clinical data suggest that insufficient interactions between DR5 agonist antibodies and FcγRIIB receptor potentially limit DR5 receptor clustering, signaling, and associated anti-tumor response (Li and Ravetch, 2012; Wilson et al., 2011). A dual specificity antibody capable of engaging DR5 on tumor cells and fibroblast activating protein (FAP) receptor on stromal cells has been shown to improve DR5 activity (Brunker et al., 2016). Since FAP is also over-expressed in the disease-associated stroma of wound healing tissues and multipotent bone marrow stem cells, FAP targeting does not give specificity to the tumor. Therefore, toxicities due to non-selective activity are inevitable (Bauer et al., 2006; Tran et al., 2013). Likewise, co-administration of Apo2L and AMG-655 has been reported to enhance DR5 activity (Graves et al., 2014). Thus, reported studies require either combinatorial cell types or combinatorial agents to improve efficacy and therefore have some limitations in terms of tumor selectivity and therapeutic applicability.
To overcome these limitations, we hypothesized that a highly superior and OvCa specific death signaling could be achieved if the initial FcγRIIB crosslinking of DR5 could be supported by BaCa antibody that also co-engages FOLR1 on the same cancer cells (Figure 7). Consistent with previous reports, our findings support the importance of bivalency and flexible distance requirement for optimal DR5 activity (Jakob et al., 2013; Spiess et al., 2015). We repeatedly found that BaCa antibodies generated with either AMG-655, lexatumumab or MD5–1 were capable of inducing in vitro cytotoxicity >100 fold higher than their parental counterparts. In agreement with previous reports, BaCa antibody activity was dependent on DR5 activity regulators such as GALNT3 (Wagner et al., 2007), p53 (Ashkenazi and Herbst, 2008) and FcγRIIB (Wilson et al., 2011). However, despite high FcγRIIB affinity mutation in lexatumumab and MD5–1, FcγRIIB had its limitations to activate DR5 signaling beyond a certain threshold. On the contrary, at the same therapeutic dose, BaCa antibody was highly effective in enhancing the apoptotic threshold to significantly higher levels than the activating limit of FcγRIIB. How FOLR1 anchor co-engagement by DR5 antibodies achieves a stronger anti-tumor response could be due to multiple reasons: 1) it maintains FcγRIIB crosslinking, 2) it improves FcγRIIB affinity and stability, and 3) it is a combination of these two or other unknown events. Regardless, our findings with BaCa strategy make us believe that despite optimal expression of FcγRIIB, DR5, and other regulators in the tumor, if the DR5 agonist antibody will produce clinically applicable results will largely depend whether it has potential to induce limited (below tumor clearance threshold) or superior (above tumor clearance threshold) apoptotic signals. As reviewed elegantly, the lower DR5 activation threshold against clinical tumors by agonist antibodies accounted for the discrepancy between preclinical and clinical results (Ashkenazi, 2015). If gain in apoptotic threshold by BaCa antibody will potentially result into clinically effective outcome need to be tested?
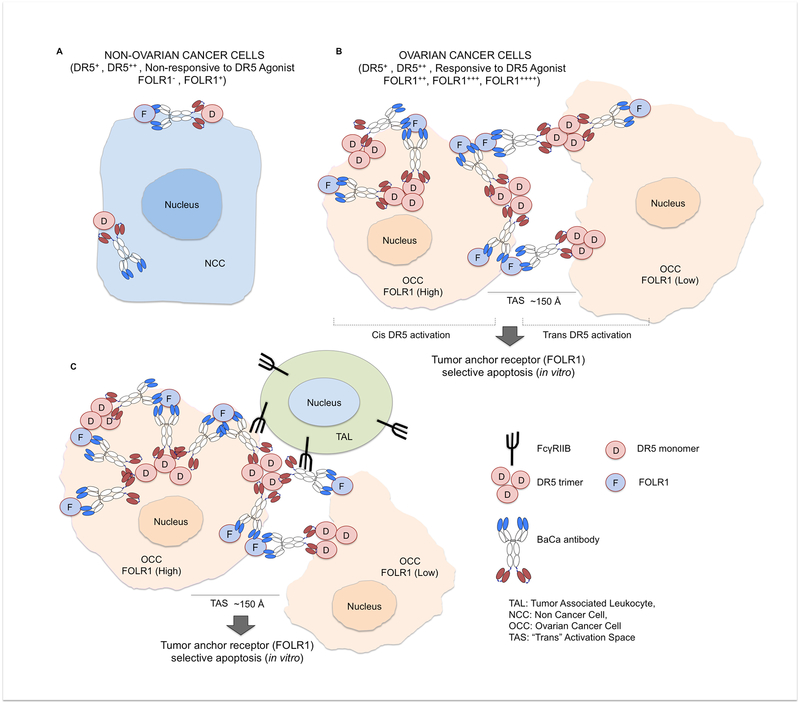
Figure 7. Working model of BaCa antibody.
(A) Healthy tissues are generally non-responsive to agonist DR5 therapy because they express no or very low level of FOLR1, thus DR5 oligomerization and activation is very minimal.
(B) In heterogeneous FOLR1 expressing OvCa cells in vitro, FOLR1 acts as an anchoring ligand to recruit BaCa antibody close to DR5 antigen at cell surface in an avidity-optimized manner. This induces a high level of DR5 clustering and activation of apoptotic pathway in both “cis” and “trans” manner selectively in FOLR1+ OvCa cells.
(C) In-vivo, tumor associated leukocytes (TAL) express inhibitory FcγRIIB receptor, which is required for the activity of DR5 agonist antibodies. Once engaged via FcγRIIB, the BaCa antibody additionally crosslinks the initial ternary complex (FcγRIIB-BaCa-DR5) via FOLR1 anchor into a high affinity stable quaternary complex (FOLR1-FcγRIIB-BaCa-DR5), which not only retains the antibody in the tumor tissue but also induces a highly superior TRAIL-R2 activation.
When tested in vitro, lexatumumab instigated superior apoptotic signals while AMG-655 and MD5–1 did not, unless cross-linked. The differential patterns of apoptotic cooperativity were also observed between Apo2L+AMG-655 and Apo2L+lexatumumab. The disparity reflects a potentially differential threshold for DR5 activation due to their independent working mechanism. One such mechanism could be distinct contact residue on DR5 receptor by these antibodies, as described for AMG-655-DR5-Apo2L ternary complex (Graves et al., 2014). However unlike AMG-655, whether lexatumumab binding to DR5 effects the conformation of DR5-Apo2L complex needs to be investigated in crystallographic studies. Regardless when engineered with FOLR1 anchor binding domain, all tested DR5 agonist antibodies pushed the apoptotic threshold multiple fold beyond the agonist antibody or ligand plus antibody. Moreover, Apo2L was inefficient to enhance the cytotoxicity when added with BaCa antibodies. These findings reveal that a ternary complex (FOLR1-BaCa-DR5) generated by a tumor anchored receptor either has already pushed the apoptotic threshold beyond the limit of Apo2L potency or has preferred cell death activation kinetics independent of Apo2L presence. The latter is also supported by the fact that in a co-culture of low and high anchor (FOLR1) expressing cells, low FOLR1 expressing Colo-205 cells survived due to lack of formation of a higher ordered anchored ternary complex compared to OVCAR-4 cells. Similar results were evident in vivo with BaCa antibody’s inability to regress Colo-205 tumors. At higher BaCa concentration, cancer cells expressing higher levels of FOLR1 helped override the apoptotic threshold in a neighboring cancer cells expressing lower FOLR1 levels potentially being engaged to form anchored complex in “trans”. These findings are in line to a hypothetical biochemical reaction where DR5 expressed in “trans” on Colo-205 cells represent a relatively lower affinity substrate for BaCa antibody while DR5 expressed in “cis” on OVCAR-4 cells represent a relatively higher affinity substrate due to avidity-optimized interactions mediated by high availability of FOLR1. Therefore, to achieve a higher enzymatic activity (Apoptosis) for a low affinity substrate, a higher enzyme (BaCa) antibody concentration is essential to increase the rate of reaction. Since stromal cell engaging antibodies such as RG7386 primarily works in “trans”, our results with LK26-AMG-655 bispecific antibody rationally indicate the higher therapeutic dose requirement for trans-engaging antibodies to achieve effective cytotoxic response as compared to BaCa antibody (Brunker et al., 2016). If a higher therapeutic dose will have a higher probability of toxicity and acquired resistance compared to a lower effective dose, need to be seen in clinical trials (Day and Read, 2016; Zuch de Zafra et al., 2016). Importantly, since intratumoural heterogeneity is one key driver of drug resistance (Saunders et al., 2012), by instigating both “cis” and “trans” signaling, BaCa antibody is ideally suited to achieve effective anti-tumor response against an OvCa having heterogeneous low and high anchor (FOLR1) expressing cancer cells. The latter is also supported by BaCa antibody’s superior ability to eliminate heterogeneous patient derived cells (in vitro) and heterogeneous cisplatin resistance PDX implants (in vitro) as compared to lexatumumab
Besides efficacy, BaCa mediated high affinity anchored ternary complex also provides critical insights for safety, tumor selectivity and therapeutic antibody retention. The liver specific ELISA and detailed tissue distribution studies in mouse models support high specificity of BaCa approach toward the grafted tumors. The observed elevated AST/ALT levels and lobular hepatitis in MD5–1 treated animal are in agreement with previous reported MD5–1 hepatotoxicity in C57BL/6 mice (Takeda et al., 2008). Although most DR5 agonist antibodies are well tolerated at a dose of 10 mg/kg, dose limiting toxicities (DLTs) have been observed with lexatumumab >12 mg/kg (Merchant et al., 2012; Wakelee et al., 2010). If anchored lexatumumab or AMG-655 (as in BaCa) will not have DLTs at a dose higher than 10 mg/kg due to their property of avidity optimized tumor retention need to be seen in clinical trials.
Disappointingly, the cellular resistance due to Bcl-2 up-regulation, Bax mutations (LeBlanc et al., 2002), NF-kB activation (Godwin et al., 2013), and loss of surface DR5 (Jin et al., 2004) has been reported against many DR5 agonists (Wang et al., 2014). If BaCa antibody will encounter same degree of resistance and discrepancy between preclinical and clinical results, it is difficult to predict (Ashkenazi, 2015). However, because of its anchored binding properties, BaCa antibody exhibited superior activity, a higher ordered DR5 activation function to induce “cis” and “trans” signaling, differential tissue distribution in animals, and significantly faster apoptotic kinetics. If the described gain of constructive functions would potentially limit the required time for cellular resistance compared to antibodies having slower apoptotic kinetics and random tissue distribution need to be seen in clinical trials. The in vivo efficacy differences in nude and surrogate animals, between anchor antigen positive and negative tumors supports a favorable cytotoxicity index of BaCa strategy. In addition, the selective >10 fold activation of cleaved caspase-3 levels in anchor (FOLR1) expressing tumors without focal hepatitis underscores the clinical safety of BaCa therapy. This also supports the idea that along with increased efficacy by BaCa antibody in clinics, a therapeutic safety window is highly achievable in patients experiencing potential toxicity by administration of an extracellular fragment of anchor antigen similar to idarucizumab, a selective reversal agent against Pradaxa (Glund et al., 2016).
In summary, we have identified a tumor cell specific anchor based DR5 activation mechanism that is highly superior over clinically tested DR5 agonist antibodies and other described strategies. The central role of anchor in retaining and maintaining tumor-restricted activity of BaCa antibody provides insights with implications to improve clinical safety that can be broadly applied. Our findings are highly relevant to clinical investigations and offer a promising path to revive the death receptor agonsim field beyond phase-II trials in ovarian and other solid cancers.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to, and will be fulfilled by Lead contact: Jogender Tushir-Singh (jogi@virginia.edu)
EXPERIMENTAL MODELS AND SUBJECT DETAILS
Patient derived cell lines and human subjects
Patient derived V565, patient derived V584, patient derived 135R, and patient derived 111 cells were isolated from ovarian cancer patients with respective age’s of 65.9, 69.4, 64.5, and 54.4 years at diagnosis. Following are the tissue sources of patient derived cells: V565 – metastatic high-grade serous carcinoma, V584 – high-grade endometrioid adenocarcinoma, 135R – stage 3 serous ovarian cancer, 111 – stage 3C serous ovarian cancer. Remnant surgical resections of omental metastatic ovarian cancer tissues (as indicated above) used for cell culture and patient-derived xenograft experiments were collected into a tissue bank by waiver of consent and approved by the University of Virginia Institutional Review Board for Health Sciences Research. The UVa Biorepository and Tissue Research Facility procured remnant samples under this protocol from UVa Pathology. De-identified tissues were pulled from this bank and used in experiments approved by UVa IRB-HSR.
Mouse tumor animal models:
6–8 weeks old (Age), 20–25 gram (Weight) female (Sex) mice were used for in vivo efficacy, imaging and safety studies. 4–6 weeks old (Age), 20–24 gram (Weight), randomized male/female (Sex) mice were used for serum half-life assays. Tumor xenografts live animal imaging, and liver ELISA studies with human cancer cells were carried out using immunodeficient Balb/c derived athymic Nude Foxn1nu/Foxn1+ (Envigo) mice model carrying functional B cell and NK (innate immunity) cells. Randomly selected and weight matched male and female Crl:CD1(ICR) mice (Charles River), a well establish strain for pharmacokinetics studies, were used for serum half-life studies. For surrogate animal studies female (Sex) 6–8 weeks-old (Age) C57BL/6J mice, 22–26 gram (Weight) were used for investigating liver toxicity, detailed tissue distribution, H&E staining, AST/ALT assays, and surrogate in vivo efficacy using MD5–1, muBaCa and chiBaCa antibodies as indicated in Figure legends. For assessing plat resistant patient derived xenografts, female (Sex) SCID C.B-17/IcrHsd-Prkdc (Envigo, Dublin, VA) mice that were 6–8 weeks old (Age) with 20–25 grams were used. Procedures involving animals handling, tumor xenografts and serum half-life studies were reviewed and approved by the Institutional Animal Care and Use Committee here at the University of Virginia and conform to the relevant regulatory standards.
Cell lines
The following cell lines were used in the study: OVCAR-3, OVCAR-4, OVCAR-5, OV90, OVSAHO, COV362, CAVO362, SKOV3, Colo-205, MC38, ID8 and patient derived cell lines (next section). All the cell lines were maintained in RPMI-1640 medium supplemented with 10% heat-inactivated fetal bovine serum (FBS), 2 mM glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin (complete medium) unless otherwise specified. MC38 cells (provided by S. Ostrand-Rosenberg, University of Maryland) were cultured in DMEM supplemented with 10% (vol/vol) FCS and 1 mM penicillin/streptomycin. Patient derived cells lines were maintained in 20% FBS and 100 mM sodium pyruvate in RPMI 1640 media supplemented with glutamax (Gibco) and 1% penicillin/streptomycin (Gibco). Various cell lines were trypsinized and expanded as follow: After digestion, the cell suspension was neutralized with complete media and centrifuged 5 min at 1500 rpm. The cell pellets were suspended in relevant DMEM/RPMI media and either expanded or seeded after counting using countess II (Life technologies). Passaged cell lines were routinely tested for mycoplasma using MycoAlert Detection Kit (Lonza).
METHOD DETAILS
Recombinant antibody Cloning
Various BaCa antibodies were engineered by genetically linking variable regions of farletuzumab (Anti-FOLR1 antibody) and lexatumumab (Anti-TRAIL-R2/DR5 antibody) into human IgG1 framework as shown in Figure 1. The DNA sequences were retrieved from the open sources (IMGT.ORG or publically available patents) and synthesized as gene string using Invitrogen GeneArt. After PCR amplification, DNA was gel purified and inserted into pcDNA 3.1+ vector (CMV promoter) by making use of In-Fusion HD Cloning Kits (Takara Bio). EcoR1 and HindIII digested vector was incubated with overlapping PCR fragments (of various different recombinant DNAs, see list of clones in Key Resource Table) with infusion enzyme (1:2, vector: insert ratio) at 55°C for 30 min, followed by additional 30 min incubation on ice after adding E. coli Stellar™ cells (Clontech). Transformation and bacterial screening was carried out using standard cloning methods. Positive clones were sequence confirmed in a 3-tier method. Confirmed bacterial colonies were Sanger sequenced upon PCR followed by re-sequencing of mini-prep DNA extracted from the positive colonies. Finally, maxiprep were re-sequenced prior to each transfection. Recombinant antibodies were also re-confirmed by ELISA and flow cytometry surface binding studies. MuBaCa, chiBaCa, LK26, MD5–1, AMG-655, NBaCa and other indicated bispecific antibodies were similarly engineered.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-DR5 (Human) | Cell Signaling Technology | Cat#3696 |
| Anti-DR4 (Human) | Cell Signaling Technology | Cat#42533 |
| Anti-FOLR1 | Thermo Fisher | Cat#PA5–24186 |
| α-Tubulin Antibody | Cell Signaling Technology | Cat#2144 |
| Cleaved Caspase-3 Antibody | Cell Signaling Technology | Cat#9661 |
| Capase-3 Antibody | Cell Signaling Technology | Cat#9668 |
| GAPDH Antibody | Cell Signaling Technology | Cat#5174 |
| Caspase-8 Antibody | Santa Cruz Biotechnology | Cat#8CSP03 |
| E-Cadherin Antibody | Cell Signaling Technology | Cat#14472 |
| Anti-Rabbit-HRP antibody | Cell Signaling Technology | Cat#7074 |
| Anti-Mouse-HRP antibody | Cell Signaling Technology | Cat#7076 |
| Anti-DR5 (Mouse) antibody | Abcam | Cat#Ab8416 |
| Cleaved PARP antibody | Cell Signaling Technology | Cat#9953 |
| Commercial LK26 (anti-murine FOLR1) antibody | Abcam | Cat#Ab3361 |
| Commercial MD5–1 (anti-Murine DR5) antibody | Abcam | Cat#ab171248 |
| Anti-Human IgG1 HRP | Life Technologies | Ref#A10648 |
| Anti-GFP antibody | Santa-Cruz Biotechnology | Cat#sc-9996 |
| Mouse anti-human IgG1 Fc | Thermo Fisher | A10648 |
| Bacterial and Virus Strains | ||
| E. coli Stellar™ | F′, endA1, supE44, thi1, recA1, relA1, gyrA96, phoA, Φ80d lacZΔM15, Δ(lacZYA-argF) U169, Δ(mrr-hsdRMS-mcrBC), ΔmcrA, λ- | Clontech |
| BL21 Competent E. coli | NE Biolabs | C2530H |
| DH5α | Thermo Fisher | 18258012 |
| Biological Samples | ||
| PDX cell line: V584 | Dr. Chip Landen, | https://cancer.uvahealth.com/research/shared-resources/maps |
| PDX cell line: V565 | Dr. Chip Landen, | https://cancer.uvahealth.com/research/shared-resources/maps |
| PDX cell line: 111 | Dr. Chip Landen, | https://cancer.uvahealth.com/research/shared-resources/maps |
| PDX cell line: 135R | Dr. Chip Landen, | https://cancer.uvahealth.com/research/shared-resources/maps |
| Patient-derived xenografts (PDX) | UVA MAPS core/Tumor bank | https://cancer.uvahealth.com/research/shared-resources/maps |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Recombinant Apo2L | R&D systems | Cat#375-TL |
| Recombinant HER2 | R&D systems | 1129-ER-050 |
| Recombinant IgG4-Fc-DR5 (rDR5) | This paper | |
| Recombinant IgG4-Fc-FOLR1 (rFOLR1) | This paper | |
| Cisplatin | Sigma | NC0837572 |
| EZ-Link Sulfo-NHS-SS-Biotin | Thermo Fisher | 21331 |
| Critical Commercial Assays | ||
| Endpoint Chromogenic LAL endotoxin assay kit | Lonza | 50–648U |
| AlamarBlue Cell viability reagent | Thermo Fisher | DAL1100 |
| MTT reagent | Thermo Fisher | |
| AST reagent | Pointe Scientific | 23-666-1221 |
| EnzyChrom ALT Assay Kit | Bioassay Systems | EASTR-100 |
| Deposited Data | ||
| doi:10.17632/tgnzd6m8y5.1 | This Paper | Shivange et al., |
| Experimental Models: Cell Lines | ||
| Human: OVCAR-3 | ATCC | HTB-161 |
| Human: OVCAR-4 | ExPASy | CVCL_1627 |
| Human: OVCAR-5 | ExPASy | CVCL_1628 |
| Human: CAVO3 | ATCC | HTB-75 |
| Human: COV362 | ExPASy | CVCL_2420 |
| Human: OV90 | Kind gift from Dr. Chip Landen, UVA | N/A |
| Human: OVSAHO | Kind gift from Dr. Chip Landen, UVA | N/A |
| Human: SKOV-3 | ATCC | HTB-77 |
| Human: Colo-205 | ATCC | CCL-222 |
| CHO-S cells | Thermo Scientific | R80007 |
| Mouse: ID8 | Kind gift from Dr. Chip Landen, UVA | N/A |
| Mouse: MC38 | Kind gift from Dr. Suzanne Ostrand-Rosenberg, UMBC | N/A |
| PDX cell line: V584 | Dr. Chip Landen, UVA | UVA MAPS core/Tumor bank |
| PDX cell line: V565 | Dr. Chip Landen, UVA | UVA MAPS core/Tumor bank |
| PDX cell line: 111 | Dr. Chip Landen, UVA | UVA MAPS core/Tumor bank |
| PDX cell line: 135R | Dr. Chip Landen, UVA | UVA MAPS core/Tumor bank |
| K562 cells | Kind gift from Dr. Golam Mohi, UVA | N/A |
| HEK293 | Kind gift from Dr. Sanchita Bhatnagar, UVA | N/A |
| Human: Colo-205-GFP stable | This Paper | Generated in laboratory of novel biologics, UVA |
| Mouse: MC38-Luc stable | This Paper | Generated in laboratory of novel biologics, UVA |
| Mouse: ID8-Luc stable | This Paper | Generated in laboratory of novel biologics, UVA |
| HEK | ATCC | CRL-1533 |
| Experimental Models: Organisms/Strains | ||
| Mouse: athymic Nude Foxn1nu/Foxn1+ | Envigo | RRID:MGI:5652489 |
| Mouse: C56BL/6 | Bhatnagar lab (UVA animal facility) | NA |
| Mouse: CD1(Crl:CD1(ICR) | Charles River | RRID:IMSR_CRL:22 |
| Mouse: C.B-17/IcrHsd-Prkdcscid | UVA MAPS core, Envigo, Dublin, VA | RRID:MGI:2160375 |
| Oligonucleotides | ||
| Primer FOLR1 Forward: GTCGACCCTGGAGGAAGAAT | This Paper | N/A |
| Primer FOLR1 Reverse: AGTCCAGTTCCAGCCCTTGT |
This Paper | N/A |
| Primer TRAIL-R2/DR5 Forward: GATGGTCAAGGTCGGTGATT |
This Paper | N/A |
| Primer TRAIL-R2/DR5 Reverse: TGGACTTCCATTTCCTGCTC |
This Paper | N/A |
| Primer GALNT3 Reverse: ACAGAGGTTCTAGCCAACCAT |
This Paper | N/A |
| Primer FUT3 Forward: CTGTCCCGCTGTTCAGAGATG |
This Paper | N/A |
| Primer FUT3 Reverse: AGGCGTGACTTAGGGTTGGA |
This Paper | N/A |
| Recombinant DNA | ||
| Full Length Lexatumumab IgG and scFv | This Paper | GeneArt, Thermo Fisher |
| Full Length Farletuzumab IgG and scFv | This Paper | GeneArt, Thermo Fisher |
| Full Length AMG-655 IgG and scFv | This Paper | GeneArt, Thermo Fisher |
| Full Length LK26 IgG and scFv | This Paper | GeneArt, Thermo Fisher |
| Full Length MD5–1 IgG and scFv | This Paper | GeneArt, Thermo Fisher |
| Full Length Recombinant DNA, huFOLR1-IgG4-Fc | This Paper | GeneArt, Thermo Fisher |
| Full Length Recombinant DNA, huDR5-IgG4-Fc | This Paper | GeneArt, Thermo Fisher |
| Full Length Idarucizumab IgG and scFv | GeneArt, Thermo Fisher | GeneArt, Thermo Fisher |
| pCDNA3.1+ | Thermo Fisher | V79020 |
| pET-28a | Addgene | 69864–3 |
| pTT5 | (Durocher and Butler, 2009) | Addgene (52326) |
| Software and Algorithms | ||
| Vector NTI | Thermo scientific | N/A |
| GraphPad Prism | GraphPad Software | www.graphpad.com |
| FlowJo | FlowJo, LLC | www.flowjo.com |
| FCS Express | De Novo Software | www.denovosoftware.com |
| Other | ||
| PowerUp SYBR Green Master mix | Thermo Fisher | A25742 |
| Superscript II | Invitrogen | 18064014 |
| Halt protease inhibitor | Thermo Fisher | 78430 |
| CHO CD efficient Feed B | Thermo Fisher | A1024001 |
| Tryptone Feed | Thermo Fisher | BP9726–2 |
| PEI transfection reagent | Thermo Fisher | BMS1003A |
| Matrigel | Corning | 354234 |
| Infusion | Takara BioScience | 638989 |
| IRDye 800CW | LI-COR | Cat#929–70020 |
| TMB Substrate Reagent Set | BD OptEIA | Cat # 555214 |
| 7 Aminoactinomycin-D (7-AAD) | Thermo Fisher | A1310 |
| CHO free style Media | Thermo Fisher | 12651014 |
| HiTrap MabSelect Sure column | GE | 11003493 |
| Protein-A resin | Thermo Fisher | P153142 |
| HisPur Ni-NTA resin | Thermo Fisher | 88221 |
| HiPure Plasmid Maxiprep kit | Invitrogen | K21007 |
Recombinant antibody expression
Free style CHO-S cells (Invitrogen, Key Resource Table) were cultured and maintained according to supplier’s recommendations (Life technologies) biologics using free style CHO expression system (life technologies) and as previously described (Durocher and Butler, 2009). A ratio of 2:1 (light chain, VL: heavy chain, VH) DNA was transfected using 1 μg/ml polyethylenimine (PEI). After transfection, cells were kept at 37°C for 24 hr. After 24 hr, transfected cells were shifted to 32°C to slow down the growth for 9 additional days. Cells were routinely fed (every 2nd day) with 1:1 ratio of Tryptone feed and CHO Feed B. After 10 days, supernatant from cultures was harvested and antibodies were purified using protein-A affinity columns. The detailed amino acid sequences of recombinant BaCa antibodies are provided below. Various recombinant antibodies used in this study (Parental antibodies: farletuzumab, lexatumumab, AMG-655, LK26, MD5–1 and BaCa, NBaCa, R-BaCa, BaNCa, muBaCa, chiBaCa etc.) and recombinant target antigens were engineered, expressed and purified in Singh Laboratory of Novel Biologics as described above. Recombinant human Apo2L/TRAIL was obtained from R&D systems. His-tag Apo2L was also expressed and purified using nickel NTA columns using standard BL21 bacterial expression system. His-Apo2L generated in our laboratory was confirmed (along side commercial Apo2L) using multiple cancer lines (Figure S3S). Similarly the activity of commercial MD5–1 antibody was compared next to recombinant MD5–1 generated in our laboratory using two different cell lines (Figure S4I and data not shown).
Antibody purification
Various transfected monospecific and bispecific antibodies (as indicated in text and Figure legends) were affinity purified using HiTrap MabSelect SuRe (GE, 11003493) protein-A columns. Transfected cultures were harvested after 10 days and filtered through 0.2 micron PES membrane filters (Milipore Express Plus). Cleaning-in-place (CIP) was performed for each column using 0.2 M NaOH wash (20 min). Following cleaning, columns were washed 3 times with Binding buffer (20 mM sodium phosphate, 0.15 M NaCl, pH 7.2). Filtered supernatant containing recombinant antibodies or antigens were passed through the columns at 4°C. Prior to elution in 0.1 M sodium citrate, pH 3.0–3.6, the columns were washed 3 times with binding buffer (pH 7.0). The pH of eluted antibodies was immediately neutralized using sodium acetate (3 M, pH 9.0). After protein measurements at 280 nm, antibodies were dialyzed in PBS using Slide-A-Lyzer 3.5K (Thermo Scientific, 66330). Antibodies were run on gel filtration columns (next section) to analyze the percent monomers. Whenever necessary a second step size exclusion chromatography (SEC) was performed. Recombinants IgG4-Fc tagged extracellular domain antigens such as FOLR1, DR5, and HER2 were also similarly harvested and purified using protein-A columns.
Size Exclusion chromatography
The percent monomer of purified antibodies was determined by size exclusion chromatography. 0.1 mg of purified antibody was injected into the AKTA protein purification system (GE Healthcare Life Sciences) and protein fractions were separated using a Superdex 200 10/300 column (GE Healthcare Life Sciences) with 50 mM Tris (pH 7.5) and 150 mM NaCl. The elution profile was exported as Excel file and chromatogram was developed. The protein sizes were determined by comparing the elution profile with the gel filtration standard (BioRad 151–1901). Any protein peak observed in void fraction was considered as antibody aggregate. The area under the curve was calculated for each peak and a relative percent monomer fraction was determined. The percent monomers of various BaCa-1, BaCa-2 and BaCa-3 antibodies generated with various linker lengths were determined as above (See Figure S1A).
BaCa antibodies details and structural integrity confirmation on SDS-PAGE
Schematic of genetic construction and domain organization of BaCa antibodies are shown in Figure 1A. The BaCa-1 antibody configuration contains bivalent anti-FOLR1 and anti-TRAIL-R2 affinities. The average distance of a N-terminal of variable (Fv) domain to the C-terminal of CH3 domain in an IgG1 is 150 Å, the genetic ligation of anti-FOLR1-IgG1-CH3 domain to TRAIL-R2 single-chain-Fv (scFv) with 12 GS linkers add an extra linear and flexible distance of ~20 Å and ~35–50 Å respectively in BaCa-1 antibody (Zhang et al., 2015). Therefore, because of total separating distance of >170 Å (150 Å + ~20 Å) BaCa-1 antibody affinities against FOLR1 and TRAIL-R2 receptors are at the opposite ends (Blue and Red). BaCa-1 antibody when run on SDS-PAGE has ~75 kDa heavy (FOLR1-VH chain joined with TRAIL-R2 scFv) and ~25 kDa light chain (FOLR1-VL) in reducing conditions. The BaCa-2 antibody configuration resembles an IgG1 and is similar to CrossMab antibodies of Genentech. In this configuration, the affinities against TRAIL-R2 and FOLR1 are monovalent (Blue and Red). BaCa-2 was engineered by making use of: a) knob/hole mutations to allow heterodimerization of two IgG chains that only differ in Fv domain (Ridgway et al., 1996), b) H435R and Y436F mutations in the CH3 domain of the hole chain in order to prevent protein-A binding to the hole-hole homodimers, and c) Glycine-serine linkers (45 GS) that are genetically linked between 3’ end of c-kappa and 5’ end of VH for proper light chain pairing. Therefore, when run in reducing conditions, BaCa-2 antibody showed a single band of ~75 kDa as light chain and heavy chain are genetically linked by GS linkers (Figure 1A). In BaCa-3 antibody (similar to Dual-Variable-Domain Ig platform of Abbvie Inc.), two different FOLR1 and TRAIL-R2 light and heavy chains are genetically linked via 12 GS linkers next to each other. Thus, despite being bivalent, the specificities against TRAIL-R2 and FOLR1 receptors are only 10–30 Å apart (Blue and Red). Thus, ~67 kDa (TRAIL-R2-VH joined with FOLR1-IgG1) and ~36 kDa (TRAIL-R2-VL joined with FOLR1-VL-Ck) bands are evident upon reduction, which are of different size than heavy and light chains of BaCa-1 antibody. Multiple bands in intact BaCa-3 (native conditions lane) indicate aggregated forms. IgG1 isotype antibody produced a heavy chain (~50 kDa) and a light chain (~25 kDa) after reduction. Fab= Fragment antigen binding, scFv= Single-chain-Fv, Fv= Variable fragment, VL= variable domain light chain, VH= variable domain heavy chain, GS= Glycine-Serine linkers, IA=Intact Antibody, HC= Heavy Chain, LC= Light Chain, NR= Antibody run on gel with non-reducing dye, R= Antibody run on gel with reducing dye, K, H= Knob-hole chains.
BaCa antibody in vitro stability assay:
Freshly purified antibodies were dialyzed in PBS using Slide-A-Lyzer 3.5K (Thermo Scientific, 66330). From the same lot, equal amount of antibodies (in PBS) were distributed in various 1.5 ml tubes. One tube was left at 4°C and others were stored at 25°C, 37°C, or −80°C (followed by multiple freeze thaw cycles) as indicated in Figure S5A. At the end of various incubation periods, all antibodies were quantified and tested for antigen binding and cytotoxicity activity together. As a positive control, farletuzumab and FDA approved adalimumab antibody (standard in our lab) were incubated together and were analyzed for percent monomer.
Binding studies by ELISA
Binding specificity and affinity of various described IgG1 subclasses were determined by ELISA using the recombinant extracellular domain of FOLR1 and/or DR5/TRAIL-R2. For coating 96-well ELISA plates (Olympus), the protein solutions (2 μg/ml) were prepared in coating buffer (100 mM Sodium Bicarbonate pH 9.2) and 100 μl was distributed in each well. The plates were then incubated overnight at 4°C. Next day, the unbound areas were blocked by cell culture media containing 10% FBS, 1% BSA and 0.5% sodium azide for 2 hr at room temperature. The serial dilutions of antibodies (2-fold dilution from 50 nM to 0.048 nM) were prepared in blocking solution and incubated in target protein coated plates for 1 hr at 37°C. After washing with PBS solution containing 0.1% Tween20, the plates were incubated for 1 hr with horseradish peroxidase (HRP) conjugated anti-human IgG1 (Thermo Scientific, A10648). Detection was performed using a two-component peroxidase substrate kit (BD biosciences) and the reaction was stopped with the addition of 2 N Sulfuric acid. Absorbance at 450 nm was immediately recorded using a Synergy Spectrophotometer (BioTech), and background absorbance from negative control samples was subtracted. The antibody affinities (Kd) were calculated by non-linear regression analysis using GraphPad Prism software.
Binding studies by BioLayer Interferometry (BLI)
Binding kinetics measurements were performed using Bio-Layer Interferometry on FortéBio Red Octet 96 instrument (Pall). Biotin-Streptavidin based sensors were employed for the studies. Recombinant Fc linked antigens, DR5-Fc and FOLR1-Fc were biotinylated using EZ-Link Sulfo-NHS-SS-Biotin (Thermo Scientific 21331) following the manufacturer’s instructions. Unbound Sulfo-NHS-SS-Biotin was removed via dialysis in PBS. For kinetic analysis biotinylated antigens (1 μg/mL) were immobilized on streptavidin (SA) biosensors (Pall) for 300 sec to ensure saturation. The 96-well microplates used in the Octet were filled with 200 μL of test antibody dilutions or buffer per well. All interaction analyses were conducted at 35°C in PBS buffer containing 2 mg/ml BSA. Following a washing step, association and dissociation measurements were carried out using serial dilutions of antibodies (4 to 160 nM). Kinetic parameters (Kon and Koff) and affinities (KD) were analyzed using Octet data analysis software, version 9.0 (Pall).
In Vitro Cell Viability Assays
Cell viability following lexatumumab, AMG-655, MD5–1, BaCa, NBaCa, muBaCa etc treatments (as indicated in various figures) either alone or in combination with human Apo2L/TRAIL ligand or in combination with an antihuman (Fab’)2 reagent were determined using the AlamarBlue cell viability assays and MTT cell proliferation assays as per manufactured protocols. Briefly, cells (indicated cells in main text or Figure legends) were treated with increasing concentration of various antibodies (as indicated) along with relevant positive and negative control antibodies for 6 hr, 24 hr or 48 hr (as indicated according to the experiment). For each cell killing assay, the Figures show the representative profiles from n= 2–4 with different cultured confluency. Whenever used for immunoblotting, following antibodies treatment, caspase-3 processing in tumor cells was monitored using selective antibodies that recognize cleaved human caspase-3 or total caspase-3 (Cell signaling, 9661 and 9668). TRAIL-R2 receptor in oligomerization was determined using immunoblotting assays (cell signaling Rabbit mAb, 8074). Cell viability was additionally examined by flow cytometry based apoptotic detection methods using 7-aminoactinomycin D (7-ADD) exclusion from live cells. Statistical significance for 7AAD FACS studies was calculated using unpaired two-tailed parametric Welch’s t-test. Figure 2H: Lexatumumab vs BaCa p=0.0073 (**). Error bars show ± SEM.
IC50 Determination
IC50 values were calculated using MTT assays. Cells were seeded in 96 well plates. Next day, when cultures became adherent, cells were incubated for 48 hr at 37°C (5% CO2) with the increasing concentrations of the antibodies or drug (such as cisplatin) as indicated in experiments. Before treatments, various antibodies were dialyzed into PBS and typically had a pH of 7.5. Values obtained after reading the 96 well plates were normalized to IgG control antibody control and IC50 values were calculated using nonlinear dose-response regression curve fits using GraphPad Prism software. The final results shown in the histograms were obtained from three independent experiments. Whenever provided in the curves, the error bars show ± SEM.
Western Blotting
Cells were cultured overnight in tissue culture-treated 6-well plates prior to treatment. After antibody treatment for 48 hr (or indicated time), cells were rinsed with PBS and then lysed with RIPA buffer supplemented with protease inhibitor cocktail (Thermo Scientific). After spinning at 14000 rpm for 30 min cleared protein lysates were quantified by Pierce BCA protein assay kit. Western blotting was performed using the Bio-Rad SDS-PAGE Gel system. Briefly, 30 μg of protein was resolved on 10% Bis-Tris gels and then transferred onto PVDF membrane. Membranes were blocked for one hour at room temperature in TBS + 0.1% Tween (TBST) with 5% non-fat dry milk. Membranes were probed overnight at 4°C with primary antibodies. Membranes were washed three times in TBST and then incubated with anti-rabbit or anti-mouse secondary antibodies (1/10,000 dilution, coupled to peroxidase) for 1 hr at room temperature. Membranes were then washed three times with TBST and Immunocomplexes were detected with SuperSignal West Pico Chemiluminescent Substrate (Thermo Fisher Scientific). Images were taken using a Bio-Rad Gel Doc Imager system. Primary antibodies are listed in the Key Resource Table.
Pre-neutralization assays
Whenever indicated throughout the manuscript or in Figure legends, variable domain pre-neutralization of BaCa antibody (or lexatumumab, or farletuzumab) was carried out to confirm the function of FOLR1 anchor in gain in cytotoxicity. For in vitro and in vivo studies, indicated antibodies and recombinant antigens (rFOLR1, rDR5 etc.) were incubated together (either 1:1 or 1:5 ratio, as indicated) at 37°C for 1 hr shaking on a platform. As a control, indicated non-preneutralized antibodies were also incubated at 37°C for 1 hr shaking on a platform either with PBS alone or with recombinant non-specific proteins such as rHER2 or rGFP. Following pre-neutralization, antibodies were either used in vitro for cell killing assays, for cellular/tumor lysates generation (immunoblotting), or for live in vivo live imaging etc. as indicated. Statistical significance was calculated using unpaired two-tailed parametric Welch’s t-test. The following are the values for Figure 2A: Lexatumumab vs BaCa+rFOLR1 p=0.6669 (ns), BaCa vs BaCa+rFOLR1 p=0.0022 (**) and Figure 2D: Lexatumumab 10 nM vs Lexatumumab 100 nM p=0.0015 (**). Error bars show ± SEM.
Liver accumulation of antibodies
To examine the liver accumulation of BaCa (HuBaCa) and lexatumumab, 6–8 weeks old weight matched female athymic Nude Foxn1nu/Foxn1+ mice (envigo) were allowed to develop tumor. When tumor reached ~200 mm3, a single dose (50 μg) of lexatumumab, farletuzumab and BaCa antibodies were injected intravenously (n=6). All injected therapeutic antibodies had LALA mutations in Fc to avoid any interference with FcR binding (Li and Ravetch, 2012). Roughly after 4 days of treatments, mice were euthanized for liver study. Following animal necropsies, liver lysates were prepared in RIPA buffer supplemented with protease inhibitor cocktail (Thermo Scientific). rFOLR1, rDR5 and rHER2 antigens were coated on the 96-well ELISA plate and relative quantity of the antibody in liver lysate was determined by binding experiment as described above (n=6). Quantification of liver accumulation (by ELISA) for each antibody treatment (n = 6) was performed by unpaired two-tailed Welch’s t-test. The following were the p values: BaCa-rDR5 vs Lexatumumab-rDR5, p=<0.0001 (***), BaCa-rFOLR1 vs Farletuzumab-rFOLR1, p=0.0584 (ns), as shown in Figure 5E.
AST/ALT assays and Hemanoxylin/Eosin Staining
To study the hepatotoxic effect of BaCa antibody, female C57BL/6 mice (n=4–5) were treated intraperitoneally with 50 μg of MD5–1, murine BaCa, or IgG1 control (in PBS) at two-day interval for 10 days. At the end of the experiment, serum was isolated from blood samples and assessed for aspartate aminotransferase (AST) and alanine aminotransferase (ALT) levels using Liquid AST (SGOT) reagent set (Pointe Scientific A7561450) and EnzyChrom Alanine Transaminase Assay Kit (Bioassay Systems EALT-100) respectively, as per manufacturer instructions and as described earlier (Takeda et al., 2008). Following blood collection, the mice were perfused with 10% neutral buffered formalin and isolated liver sections were fixed in 10% neutral buffered formalin overnight at 4°C. The paraffin embedding and H&E staining was performed by Research Histology Core here at University of Virginia School of Medicine (National Cancer Institute P30 UVA Center Grant). For AST/ALT assays, p values were determined by unpaired t-test with Welch’s correction. ALT: p=0.043 (*), AST: p=0.0274 (*) as shown in Figure 5Q.
Flow cytometry
The cell surface expression of DR4/DR5 was analyzed by flow cytometry. Overnight grown OVCAR-3 cells were trypsinized and suspended in FACS buffer (PBS containing 2% FBS). The single cell suspension was then incubated with primary DR4/DR5 antibodies for 1 hr at 4°C with gentle mixing. Following the wash with FACS buffer, the cells were then incubated with fluorescently labeled anti-Rabbit antibody for 1 hr. Cells were washed and flow cytometry was performed using FACSCalibur. The data was analyzed by FCS Express (De Novo Software) and FlowJo. Similar FACS studies were performed for farletuzumab, lexatumumab, AMG-655, LK26, MD5–1 and BaCa antibodies whenever necessary (as indicated in text and Figure legends).
Quantitative RT-PCR (qRT-PCR)
For qRT-PCR assays, RNA was extracted using the Trizol Reagent (Invitrogen). cDNA was prepared by amplifying 500 ng of RNA by the SuperScript-II cDNA Synthesis Kit (Life Technologies). Quantitative PCR was performed using PowerUp SYBR Green Master mix (Applied Biosystems) following manufacturer’s instructions. Data was analyzed using StepOneV2.0 software (Applied Biosystems). The relative expression levels were normalized to GAPDH. Statistical significance was determined by an unpaired t-test with Welch’s correction using Graph Pad Prism software (n=4). Error bar show ± SEM. Primers used are as follows: huFOLR1 FOR: GTCGACCCTGGAGGAAGAAT, huFOLR1 REV: AGTCCAGTTCCAGCCCTTGT, huTRAIL-R2 FOR: GATGGTCAAGGTCGGTGATT, huTRAIL-R2 REV: TGGACTTCCATTTCCTGCTC, GALNT3 FOR: CACCTGCAATACTGCTGAAGG, GALNT3 REV: ACAGAGGTTCTAGCCAACCAT, FUT3 FOR: CTGTCCCGCTGTTCAGAGATG, FUT3 REV: AGGCGTGACTTAGGGTTGGA
Live Imaging and Tissue distribution studies
Indicated antibodies (Lexatumumab, HuBaCa, MD5–1, MuBaCa and IgG1 control) were tagged with IRDye® 800CW NHS Ester (Li-Cor) fluorochrome. Briefly, antibody solutions were prepared in 100 mM phosphate buffer pH 8.5 and mixed with IRdye 800-NHS (0.04 mg dye per 1 mg of antibodies). The conjugation was carried out at 20°C for 2 hr and unconjugated dye was separated by dialysis in PBS. It was confirmed that IR800 dye labeling did not affect antibody binding to respective antigens for all the antibodies (Figure S5D and data not shown). Subcutaneous tumors were generated by injecting either 1×106 OVCAR-3 cells, 2×106 OVCAR-4, 2×106 Colo-205, or 5×105 MC38 cells (in matrigel) respectively as described in earlier section. OVCAR-3 or MC38 tumors were grown in athymic nude or WT C57BL/6 mice respectively. 25 μg of fluorescent antibody was injected intravenously (IV) as indicated and the mice were imaged after 24 hr using Xenogen IVIS spectrum In Vivo Imaging System (PerkinElmer Inc.). For tissue distribution studies, various organs (Spleen, Kidney, Liver, lungs and stomach) were isolated along with tumor and exposed directly to the excitation wavelength (772 nm) to monitor the tissue specific fluorescent signal of each antibody. Radiant efficiency (fluorescent intensity) was calculated after subtracting the fluorescent signal from IgG1 (IRDye® 800 labeled) injected animals in the exactly similar conditions. Statistical significance of differential distribution was determined by an unpaired t-test with Welch’s correction using Graph Pad Prism software (n=4). Error bar show ± SEM. Following are the p values in Figure 5P, Kidney: p=0.5893 (ns), Liver: p=0.0283 (*), Lung: p=0.0750 (ns), Spleen: p=0.2118 (ns), Tumor: p=0.0026 (**).
Serum Half-life
Animal care and all experiments performed were in accordance with IACUC guidelines and have been approved by university ACUC authorities. Male and female CD1 mice (4–6 weeks, 20–25 grams) were randomized in groups (See Figure S5B, C) and injected intravenous with 25 μg of antibodies in a total volume of 100 μl. The blood samples (50–100 μl) were collected by pricking tail vein at indicated time interval and allowed to clot at room temperature for 30 min as described earlier (Hutt et al., 2012). Clotted blood was centrifuged at 13000 × g for 20 min at 4°C and serum sample was stored in −80°C in small aliquots. As described above, serum concentration of antibodies were determined using ELISA. Sets of 4–5 mice were used for each antibody study. The serum half-lives of antibodies were determined using one phase exponential decay equation model fitted by non-linear regression of % concentration of leftover antibody in serum vs. time using Prism version 5.01 software (Graph Pad Software, Inc.). For comparison, the antibody concentration of first collected serum sample (30 min) was set as 100% and relative % concentrations of each antibody were determined for different time point earlier(Hutt et al., 2012). Next, we transformed the data in semi-log plot (shown in Figure S5B, C). For that, we recalculated the serum antibody concentrations in μg/ml for each time point and analyzed by two-phase exponential decay model fitted by log antibody concentrations vs. time using Prism 5.01 software. The serum half-life calculations of the elimination phase were determined using the formula t1/2 = ln2/β, where β is the negative slope of the line. Very similar values were obtained with semi-log plot and two-phase exponential analyses.
Avidity assays
To assess the collective binding affinity of the lexatumumab, farletuzumab and various BaCa antibodies, both of the target antigens rFOLR1 and rTRAILR2 were coated in 96 well plates in 5:1 ratio. 20 nM of parental and BaCa antibodies (BaCa-1, BaCa-2, BaCa-3) were allowed to bind the target protein for 60 min at 37°C. Wells were then washed with PBS and exposed to 6 M urea for 10 min as described earlier (Levett et al., 2005). After washing, the concentrations of remaining antibodies were determined as described above. A relative avidity index was calculated for each antibody by representing the percentage of reactivity remained in urea treated wells compared to PBS treated wells. Statistical significance was determined by an unpaired t-test with Welch’s correction using Graph Pad Prism software (n=4), p=0.0286(*). Error bar show ± SEM.
Nude Tumor Xenograft studies
All animal procedures were conducted under the accordance of University of Virginia Institutional Animal Care and Use Committee (IACUC) with approved protocol (#4112). Following different cell lines were used for tumor nude xenograft studies: 1) OVCAR-3 cells, 2) OVCAR-4 cells, and 3) Colo-205 cells. Since OvCa is a female pathology, female animals were given a 2 weeks acclimation period after arrival to the vivarium and all animal procedures were conducted under institutional policies. Weight and age (6–8 weeks old) matched female athymic Nude Foxn1nu/Foxn1+ (Envigo) mice were injected subcutaneously (SC) in their right flank with indicated cell lines in matrigel. 1 × 106 OVCAR-3 cells, 1 × 106 COLO-205 cells or 2 × 106 OVCAR-4 cells were injected in 100 μl volume. Colo-205 cells formed tumors between 2–3 weeks, while both OVCAR-3 and OVCAR-4 produced tumors after ~3–4 weeks. For antitumor efficacy studies, mice bearing ~100 mm3 tumors weight matched animals were randomly assigned into groups and injected (either 25 μg or indicated different dose) either intraperitoneally or intravenously (as indicated in Figure legends) three times per week with lexatumumab IgG1 (WT-Fc or KO-Fc or E267 mutation as indicated), farletuzumab IgG1 (WT-Fc or KO-Fc or E267S mutation as indicated), BaCa antibody (WT-Fc or KO-Fc or E267 mutation as indicated) or IgG1 isotype control (WT-Fc or KO-Fc or E267 mutation as indicated). Tumors were measured in two dimensions using a caliper as described previously (Graves et al., 2014; Wilson et al., 2011). Tumor volume was calculated using the formula: V = 0.5axb2, where a and b are the long and the short diameters of the tumor respectively. (n=4–6 animals were used for each therapeutic antibody injection). The p values are determined by two-tailed paired Wilcoxon Mann-Whitney test. Figure 6A: BaCa-LALA-Fc vs Lexatumumab-LALA-Fc, p=0.0078 (**), Figure 6B: BaCa-LALA-Fc vs Lexatumumab-LALA-Fc, p=0.0156 (*), Figure 6C: BaCa-S267E-WTFc vs BaCa-S267E-LALA-Fc, p=0.0781 (ns).
Surrogate Tumor Xenograft studies
All animal procedures were conducted under the accordance of University of Virginia Institutional Animal Care and Use Committee (IACUC) with approved protocol (#4112). Since OvCa is a female pathology, female mice (C57BL/6J) were used for surrogate xenograft studies. MC38 cells were used for surrogate tumor grafts. 6–8 weeks old female littermate of matched size and weight C57BL/6J mice were injected subcutaneously (SC) in their right flank with 0.5 × 106 MC38 cells lines in matrigel. MC38 cells consistently formed tumors within 2–3 weeks as described (Takeda et al., 2008). For tumor regression studies, mice bearing ~100 mm3 tumors were (after matching tumor size, n=4–6) randomly assigned into groups and injected with therapeutic antibodies (25 μg dose) intraperitoneally three times per week. For surrogate efficacy studies, MD5–1, muBaCa or chiBaCa and IgG1 control were engineered with KO-Fc and S267E mutations. Tumors were measured three times a week and volumes were calculated as the product of three orthogonal diameters similar to nude animal studies as described in previous section. The p values are determined by two-tailed paired Wilcoxon Mann-Whitney test. Figure 6F: MD5–1 vs MuBaCa, p=0.0312 (*), Figure 6G: ChiBaCa vs MD5–1, p=0.745, ns). For Biochemical analysis of tumors, mice were euthanized when tumor diameter reached >100 mm. For in vivo caspase-3 activity and comparison, tissues were harvested and processed as described earlier (n=2) (Li and Ravetch, 2012; Wilson et al., 2011).
Cisplatin Resistant Patient Derived Xenografts (PDXs) efficacy studies
Mice and surgical procedures: All animal procedures were conducted under the approval of the Institutional Animal Care and Use Committee (IACUC) of the University of Virginia (#4111). Procedures with mice were conducted in collaboration with the University of Virginia Molecular Assessments and Preclinical Studies (MAPS) Core Facility. The grant funding CCSG grant number 2P30CA044579-26 supports MAPS core facility. Adult (6–8 weeks of age) female SCID C.B-17/IcrHsd-Prkdc (Envigo, Dublin, VA) mice were given a 2 week acclimation period after arrival to the facility where they were maintained on a 10:14 light:dark schedule (lights on at 6am) in a dedicated immune compromised housing room for mice with filter top cages, distilled H2O and a diet optimized for immune compromised mice consisting of every other week feeding of Teklad LM-485 irradiated standard rodent diet (Envigo, 7912) and Uniprim (Envigo, TD.06596). For surgical implantation, mice were administered a cocktail of Ketamine (60–80 mg/kg) and Xylazine (5–10 mg/kg) intraperitoneally and prepped for sterile surgery using aseptic technique. A small dorsal incision was made and the skin undermined along the flanks of each mouse to prep the site for subcutaneous implantation of tumor. Previously frozen cisplatin resistant patient-derived xenograft (PDX) tumors were implanted bilaterally into the flanks and the incision site was closed with wound clips or skin adhesive. PDX tumor from this model was confirmed to be human by RNA-Seq alignment to both mouse and human genomes, and by comparing original human tumor sequencing to PDX (data not shown). Mice were monitored after surgery until recovery, and administered analgesic for several days at the end of which wound clips were removed. Mice were monitored closely for tumor growth and overall health throughout the study. Tumor take rate was >90%, with 13 out of 14 mice implanted successfully growing tumor and treated. Mice were treated with PBS, lexatumumab (LALA-Fc) and BaCa (LALA-Fc) antibodies at 5 mg/kg dose, as indicated, and tumor measurements were carried out as described in earlier sections. The p values are determined by two-tailed paired Wilcoxon Mann-Whitney test. Figure 6I: BaCa vs Lexatumumab, p=0.0020 (**).
Recombinant antibody sequences
BaCa-1 (HuBaCa, BaCa, Lexatumumab BaCa) amino acid sequence
Heavy Chain (Farletuzumab and Lexatumumab):
EVQLVESGGGVVQPGRSLRLSCSASGFTFSGYGLSWVRQAPGKGLEWVAMISSGGSYTYYADSVKGRFAISRDNAKNTLFLQMDSLRPEDTGVYFCARHGDDPAWFAYWGQGTPVTVSSASTKGPSVFPLAPSSKSTSGGTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQSSGLYSLSSVVTVPSSSLGTQTYICNVNHKPSNTKVDKKVEPKSCDKTHTCPPCPAPELLGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVEHEDPEVKFNWYVDGVEVHNAKTKPREEQYNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKALPAPIEKTISKAKGQPREPQVYTLPPSREEMTKNQVSLTCLVKGFYPSDIAVEWESNGQPENNYKTTPPVLDSDGSFFLYSKLTVDKSRWQQGNVFSCSVMHEALHNHYTQKSLSLSLGKGGGSGGGSGGGSSSELTQDPAVSVALGQTVRITCQGDSLRSYYASWYQQKPGQAPVLVIYGKNNRPSGIPDRFSGSSSGNTASLTITGAQAEDEADYYCNSRDSSGNHVVFGGGTKLTVLGGGGSGGGDSGGGGSGGGGSEVQLVQSGGGVERPGGSLRLSCAASGFTFDDYGMSWVRQAPGKGLEWVSGINWNGGSTGYADSVKGRVTISRDNAKNSLYLQMNSLRAEDTAVYYCAKILGAGRGWYFDLWGKGTTVTVSS
Light chain (Farletuzumab):
DIQLTQSPSSLSASVGDRVTITCSVSSSISSNNLHWYQQKPGKAPKPWIYGTSNLASGVPSRFSGSGSGTDYTFTISSLQPEDIATYYCQQWSSYPYMYTFGQGTKVEIKRTVAAPSVFIFPPSDEQLKSGTASVVCLLNNFYPREAKVQWKVDNALQSGNSQESVTEQDSKDSTYSLSSTLTLSKADYEKHKVYACEVTHQGLSSPVTKSFNRGEC
BaCa-2 amino acid sequence
Farletuzumab Knob single chain variable Fragment:
DIQLTQSPSSLSASVGDRVTITCSVSSSISSNNLHWYQQKPGKAPKPWIYGTSNLASGVPSRFSGSGSGTDYTFTISSLQPEDIATYYCQQWSSYPYMYTFGQGTKVEIKRTVAAPSVFIFPPSDEQLKSGTASVVCLLNNFYPREAKVQWKVDNALQSGNSQESVTEQDSKDSTYSLSSTLTLSKADYEKHKVYACEVTHQGLSSPVTKSFNRGECGGGGSGGGGSGGGGSGGGGSGGGGSGGGGSGGGGSGGGGSGGGGS
GGGGSEVQLVESGGGVVQPGRSLRLSCSASGFTFSGYGLSWVRQAPGKGLEWVAMISSGGSYTYYADSVKGRFAISRDNAKNTLFLQMDSLRPEDTGVYFCARHGDDPAWFAYWGQGTPVTVSSASTKGPSVFPLAPSSKSTSGGTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQSSGLYSLSSVVTVPSSSLGTQTYICNVNHKPSNTKVDKRVEPKSCDKTHTCPPCPAPELLGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSHEDPEVKFNWYVDGVEVHNAKTKPREEQYNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKALPAPIEKTISKAKGQPREPQVYTLPPSREEMTKNQVSLYCLVKGFYPSDIAVEWESNGQPENNYKTTPPVLDSDGSFFLYSKLTVDKSRWQQGNVFSCSVMHEALHNHYTQKSLSLSPG
Lexatumumab Hole single chain variable Fragment:
SSELTQDPAVSVALGQTVRITCQGDSLRSYYASWYQQKPGQAPVLVIYGKNNRPSGIPDRFSGSSSGNTASLTITGAQAEDEADYYCNSRDSSGNHVVFGGGTKLTVLRTVAAPSVFIFPPSDEQLKSGTASVVCLLNNFYPREAKVQWKVDNALQSGNSQESVTEQDSKDSTYSLSSTLTLSKADYEKHKVYACEVTHQGLSSPVTKSFNRGECGGGGSGGGGSGGGGSGGGGSGGGGSGGGGSGGGGSGGGGSGGGGS
GGGGSEVQLVQSGGGVERPGGSLRLSCAASGFTFDDYGMSWVRQAPGKGLEWVSGINWNGGSTGYADSVKGRVTISRDNAKNSLYLQMNSLRAEDTAVYYCAKILGAGRGWYFDLWGKGTTVTVSSASTKGPSVFPLAPSSKSTSGGTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQSSGLYSLSSVVTVPSSSLGTQTYICNVNHKPSNTKVDKRVEPKSCDKTHTCPPCPAPELLGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSHEDPEVKFNWYVDGVEVHNAKTKPREEQYNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKALPAPIEKTISKAKGQPREPQVYTLPPSREEMTKNQVSLTCLVKGFYPSDIAVEWESNGQPENNYKTTPPVLDSDGSFFLTSKLTVDKSRWQQGNVFSCSVMHEALHNRFTQKSLSLSPG
BaCa-3 amino acid sequence
Heavy Chain (Farletuzumab and Lexatumumab):
EVQLVQSGGGVERPGGSLRLSCAASGFTFDDYGMSWVRQAPGKGLEWVSGINWNGGSTGYADSVKGRVTISRDNAKNSLYLQMNSLRAEDTAVYYCAKILGAGRGWYFDLWGKGTTVTVSSGGSGGSGGSGGSEVQLVESGGGVVQPGRSLRLSCSASGFTFSGYGLSWVRQAPGKGLEWVAMISSGGSYTYYADSVKGRFAISRDNAKNTLFLQMDSLRPEDTGVYFCARHGDDPAWFAYWGQGTPVTVSSASTKGPSVFPLAPSSKSTSGGTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQSSGLYSLSSVVTVPSSSLGTQTYICNVNHKPSNTKVDKKVEPKSCDKTHTCPPCPAPELLGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVEHEDPEVKFNWYVDGVEVHNAKTKPREEQYNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKALPAPIEKTISKAKGQPREPQVYTLPPSREEMTKNQVSLTCLVKGFYPSDIAVEWESNGQPENNYKTTPPVLDSDGSFFLYSKLTVDKSRWQQGNVFSCSVMHEALHNHYTQKSLSLSLG
Light Chain (Farletuzumab and Lexatumumab):
SSELTQDPAVSVALGQTVRITCQGDSLRSYYASWYQQKPGQAPVLVIYGKNNRPSGIPDRFSGSSSGNTASLTITGAQAEDEADYYCNSRDSSGNHVVFGGGTKLTVLGGSGGSGGSGGSDIQLTQSPSSLSASVGDRVTITCSVSSSISSNNLHWYQQKPGKAPKPWIYGTSNLASGVPSRFSGSGSGTDYTFTISSLQPEDIATYYCQQWSSYPYMYTFGQGTKVEIKRTVAAPSVFIFPPSDEQLKSGTASVVCLLNNFYPREAKVQWKVDNALQSGNSQESVTEQDSKDSTYSLSSTLTLSKADYEKHKVYACEVTHQGLSSPVTKSFNRGEC
Recombinant DR5 (rDR5) amino acid sequence
ITQQDLAPQQRAAPQQKRSSPSEGLCPPGHHISEDGRDCISCKYGQDYSTHWNDLLFCLRCTRCDSGEVELSPCTTTRNTVCQCEEGTFREEDSPEMCRKCRTGCPRGMVKVGDCTPWSDIECVHKESGGGSGGSESKYGPPCPPCPAPEFLGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSQEDPEVQFNWYVDGVEVHNAKTKPREEQFNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKGLPSSIEKTISKAKGQPREPQVYTLPPSQEEMTKNQVSLTCLVKGFYPSDIAVEWESNGQPENNYKTTPPVLDSDGSFFLYSKLTVDKSRWQEGNVFSCSVMHEALHNHYTQKSLSLSLG
Recombinant FOLR1 (rFOLR1) amino acid sequence
AQRMTTQLLLLLVWVAVVGEAQTRIAWARTELLNVCMNAKHHKEKPGPEDKLHEQCRPWRKNACCSTNTSQEAHKDVSYLYRFNWNHCGEMAPACKRHFIQDTCLYECSPNLGPWIQQVDQSWRKERVLNVPLCKEDCEQWWEDCRTSYTCKSNWHKGWNWTSGFNKCAVGAACQPFHFYFPTPTVLCNEIWTHSYKVSNYSRGSGRCIQMWFDPAQGNPNEEVARFYAAAGGSGGSESKYGPPCPPCPAPEFLGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSQEDPEVQFNWYVDGVEVHNAKTKPREEQFNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKGLPSSIEKTISKAKGQPREPQVYTLPPSQEEMTKNQVSLTCLVKGFYPSDIAVEWESNGQPENNYKTTPPVLDSDGSFFLYSKLTVDKSRWQEGNVFSCSVMHEALHNHYTQKSLSLSLG
Murine BaCa (muBaCa) amino acid sequence
Heavy chain (LK26-MD5–1):
QVQLQESGGDLVKPGGSLKLSCAASGFTFSGYGLSWVRQTPDKRLEWVAMISSGGSYTYYADSVKGRFAISRDNAKNSLFLQMSSLKSDDTAIYICARHGDDPAWFAYWGQGTLVTVSAASTKGPSVFPLAPSSKSTSGGTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQSSGLYSLSSVVTVPSSSLGTQTYICNVNHKPSNTKVDKKVEPKSCDKTHTCPPCPAPEAAGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVEHEDPEVKFNWYVDGVEVHNAKTKPREEQYNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKALPAPIEKTISKAKGQPREPQVYTLPPSREEMTKNQVSLTCLVKGFYPSDIAVEWESNGQPENNYKTTPPVLDSDGSFFLYSKLTVDKSRWQQGNVFSCSVMHEALHNHYTQKSLSLSLGKGGSGGSGGSGGSDIQVTQSPSLLSASFGDKVTINCLVTQDITYYLSWYQQKSGQPPTLLIYNGNSLQSGVPSRFSGQYSGRTFTLSLSSLEPEDAGTYYCLQHYSVPFTFGGGTRLEIKGGGGSGGGDSGGGGSGGGGSQIQLQESGPGLVKPAQSLSLTCSITGFPITAGGYWWTWIRQFPGQKLEWMGYIYSSGSTNYNPSIKSRISITRDTAKNQFFLQLNSVTTEEDTAIYYCARAGTSYSGFFDSWGQGTLVTVSS
Light chain (LK26):
DIELTQSPALNAASPGEKVTITCSVSSSISSNNLHWYQQKSETSPKPWIYGTSNLASGVPLRFRGFGSGTSYSLTISSNEAEDAATYYCQQWSSYPYMYTFGGGTKLEIKRTVAAPSVFIFPPSDEQLKSGTASVVCLLNNFYPREAKVQWKVDNALQSGNSQESVTEQDSKDSTYSLSSTLTLSKADYEKHKVYACEVTHQGLSSPVTKSFNRGEC
Chimeric BaCa (ChiBaCa) amino acid sequence
Heavy chain (Farletuzumab-MD5–1):
EVQLVESGGGVVQPGRSLRLSCSASGFTFSGYGLSWVRQAPGKGLEWVAMISSGGSYTYYADSVKGRFAISRDNAKNTLFLQMDSLRPEDTGVYFCARHGDDPAWFAYWGQGTPVTVSSASTKGPSVFPLAPSSKSTSGGTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQSSGLYSLSSVVTVPSSSLGTQTYICNVNHKPSNTKVDKKVEPKSCDKTHTCPPCPAPEAAGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVEHEDPEVKFNWYVDGVEVHNAKTKPREEQYNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKALPAPIEKTISKAKGQPREPQVYTLPPSREEMTKNQVSLTCLVKGFYPSDIAVEWESNGQPENNYKTTPPVLDSDGSFFLYSKLTVDKSRWQQGNVFSCSVMHEALHNHYTQKSLSLSLGKGGSGGSGGSGGSDIQVTQSPSLLSASFGDKVTINCLVTQDITYYLSWYQQKSGQPPTLLIYNGNSLQSGVPSRFSGQYSGRTFTLSLSSLEPEDAGTYYCLQHYSVPFTFGGGTRLEIKGGGGSGGGDSGGGGSGGGGSQIQLQESGPGLVKPAQSLSLTCSITGFPITAGGYWWTWIRQFPGQKLEWMGYIYSSGSTNYNPSIKSRISITRDTAKNQFFLQLNSVTTEEDTAIYYCARAGTSYSGFFDSWGQGTLVTVSS
Light chain (Farletuzumab):
DIQLTQSPSSLSASVGDRVTITCSVSSSISSNNLHWYQQKPGKAPKPWIYGTSNLASGVPSRFSGSGSGTDYTFTISSLQPEDIATYYCQQWSSYPYMYTFGQGTKVEIKRTVAAPSVFIFPPSDEQLKSGTASVVCLLNNFYPREAKVQWKVDNALQSGNSQESVTEQDSKDSTYSLSSTLTLSKADYEKHKVYACEVTHQGLSSPVTKSFNRGEC
AMG-655 BaCa amino acid sequence
Heavy Chain (Farletuzumab and AMG-655):
EVQLVESGGGVVQPGRSLRLSCSASGFTFSGYGLSWVRQAPGKGLEWVAMISSGGSYTYYADSVKGRFAISRDNAKNTLFLQMDSLRPEDTGVYFCARHGDDPAWFAYWGQGTPVTVSSASTKGPSVFPLAPSSKSTSGGTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQSSGLYSLSSVVTVPSSSLGTQTYICNVNHKPSNTKVDKKVEPKSCDKTHTCPPCPAPELLGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVEHEDPEVKFNWYVDGVEVHNAKTKPREEQYNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKALPAPIEKTISKAKGQPREPQVYTLPPSREEMTKNQVSLTCLVKGFYPSDIAVEWESNGQPENNYKTTPPVLDSDGSFFLYSKLTVDKSRWQQGNVFSCSVMHEALHNHYTQKSLSLSLGKGGSGGSEIVLTQSPGTLSLSPGERATLSCRASQGISRSYLAWYQQKPGQAPSLLIYGASSRATGIPDRFSGSGSGTDFTLTISRLEPEDFAVYYCQQFGSSPWTFGQGTKVEIKGGGGSGGGDSGGGGSGGGGSQVQLQESGPGLVKPSQTLSLTCTVSGGSISSGDYFWSWIRQLPGKGLEWIGHIHNSGTTYYNPSLKSRVTISVDTSKKQFSLRLSSVTAADTAVYYCARDRGGDYYYGMDVWGQGTTVTVSS
Light chain (Farletuzumab):
DIQLTQSPSSLSASVGDRVTITCSVSSSISSNNLHWYQQKPGKAPKPWIYGTSNLASGVPSRFSGSGSGTDYTFTISSLQPEDIATYYCQQWSSYPYMYTFGQGTKVEIKRTVAAPSVFIFPPSDEQLKSGTASVVCLLNNFYPREAKVQWKVDNALQSGNSQESVTEQDSKDSTYSLSSTLTLSKADYEKHKVYACEVTHQGLSSPVTKSFNRGEC
QUANTITATION AND STATISTICAL ANALYSIS
Data unless indicated otherwise are presented as mean ± SEM. In general, when technical replicates were shown for in vitro experiments (Figure 3B, 4C, S3P), student t-test was used for statistical analysis and the same experiment was at least repeated once with similar trend observed. When data from multiple experiments was merged into one Figure (Figure 2A, 2D, 2H, 3B, 5G, 5I, 5Q, 5P, S4A), statistical significance was determined by an unpaired t-test with Welch’s correction using Graph Pad Prism 5.0 software. Quantification of tumor burden in described experiments performed with mice samples were analyzed using Wilcoxon Mann-Whitney test. For comparative therapeutic antibody in vivo efficacy analysis (Figures 6A, 6B, 6C, 6E, 6F, 6G), on average, tumor bearing mice (n= 4–6) were quantified and group comparison from animals injected with indicated antibody (such as BaCa vs Lexatumumab, muBaCa vs MD5–1) was performed and calculated with 95% confidence with the two-tailed paired nonparametric t-test. Tumor growth curves are displayed as mean ± SEM. For all the statistical experiments p values, p<0.05 (*), p<0.01 (**) and p<0.001 (***) were considered statistically different whereas specific p values indicated otherwise or “ns” indicates non-significant.
Supplementary Material
Significance.
Ovarian Cancer (OvCa) is the most lethal gynecological disease with no effective treatments. We discovered and characterized that FOLR1 and DR5 co-targeting by a single-agent antibody symbiotically compensates each other limitations to promote OvCa specific anti-tumor activity. The described BaCa strategy is also highly superior to combinatorial Apo2L/TRAIL ligand and DR5 agonist antibodies. Three clinical bispecific configurations highlight the critical need of domain flexibility in choosing the optimal geometry for enhanced death receptor clustering and support biological insights for safe and selective tumor localization. In summary, BaCa antibody not only provides rational argument into limited preclinical efficacy of DR5 agonist antibodies but also offers a paradigm to clinically revive the ADCC activating antibodies using death receptor targeting approach.
Acknowledgements
We thank Drs. Patrick Grant and Larry Lum for helpful discussions. We are thankful for core imaging, core FACS and core vivariam facility for assistance. Special Thanks to Marya Dunlap-Brown (MAPS core: CCSG: 2P30CA044579-26) for assisting with platinum resistant studies. IRG-ACS (88-001-30-IRG) grant to J. T-S supported this work. DoD Research Breakthrough Award to S.B (BC170197P1) and J. T-S (BC170197) supported part of this work.
Footnotes
Declaration of Interests
The authors declare no competing interests. BaCa antibody mediated targeting of FOLR1 and DR5, and “Cis”-”Trans” mechanism of apoptotic activation is part of a provisional patent here at University of Virginia Licensing and Venture Group.
References
- Albanesi M, and Daeron M (2012). The interactions of therapeutic antibodies with Fc receptors. Immunol Lett 143, 20–27. [DOI] [PubMed] [Google Scholar]
- Armstrong DK, White AJ, Weil SC, Phillips M, and Coleman RL (2013). Farletuzumab (a monoclonal antibody against folate receptor alpha) in relapsed platinum-sensitive ovarian cancer. Gynecol Oncol 129, 452–458. [DOI] [PubMed] [Google Scholar]
- Ashkenazi A (2008). Directing cancer cells to self-destruct with pro-apoptotic receptor agonists. Nat Rev Drug Discov 7, 1001–1012. [DOI] [PubMed] [Google Scholar]
- Ashkenazi A (2015). Targeting the extrinsic apoptotic pathway in cancer: lessons learned and future directions. J Clin Invest 125, 487–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashkenazi A, and Herbst RS (2008). To kill a tumor cell: the potential of proapoptotic receptor agonists. J Clin Invest 118, 1979–1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer S, Jendro MC, Wadle A, Kleber S, Stenner F, Dinser R, Reich A, Faccin E, Godde S, Dinges H, et al. (2006). Fibroblast activation protein is expressed by rheumatoid myofibroblast-like synoviocytes. Arthritis Res Ther 8, R171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berek J, Taylor P, McGuire W, Smith LM, Schultes B, and Nicodemus CF (2009). Oregovomab maintenance monoimmunotherapy does not improve outcomes in advanced ovarian cancer. J Clin Oncol 27, 418–425. [DOI] [PubMed] [Google Scholar]
- Brinkmann U, and Kontermann RE (2017). The making of bispecific antibodies. MAbs 9, 182–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunker P, Wartha K, Friess T, Grau-Richards S, Waldhauer I, Koller CF, Weiser B, Majety M, Runza V, Niu H, et al. (2016). RG7386, a Novel Tetravalent FAP-DR5 Antibody, Effectively Triggers FAP-Dependent, Avidity-Driven DR5 Hyperclustering and Tumor Cell Apoptosis. Mol Cancer Ther 15, 946–957. [DOI] [PubMed] [Google Scholar]
- Chen RY, Cao JJ, Chen J, Yang JP, Liu XB, Zhao GQ, and Zhang YF (2012). Single nucleotide polymorphisms in the CDH17 gene of colorectal carcinoma. World J Gastroenterol 18, 7251–7261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day T, and Read AF (2016). Does High-Dose Antimicrobial Chemotherapy Prevent the Evolution of Resistance? PLoS Comput Biol 12, e1004689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domcke S, Sinha R, Levine DA, Sander C, and Schultz N (2013). Evaluating cell lines as tumour models by comparison of genomic profiles. Nat Commun 4, 2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durocher Y, and Butler M (2009). Expression systems for therapeutic glycoprotein production. Curr Opin Biotechnol 20, 700–707. [DOI] [PubMed] [Google Scholar]
- Glund S, Stangier J, van Ryn J, Schmohl M, Moschetti V, Haazen W, De Smet M, Gansser D, Norris S, Lang B, et al. (2016). Restarting Dabigatran Etexilate 24 h After Reversal With Idarucizumab and Redosing Idarucizumab in Healthy Volunteers. J Am Coll Cardiol 67, 1654–1656. [DOI] [PubMed] [Google Scholar]
- Godwin P, Baird AM, Heavey S, Barr MP, O’Byrne KJ, and Gately K (2013). Targeting nuclear factor-kappa B to overcome resistance to chemotherapy. Front Oncol 3, 120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves JD, Kordich JJ, Huang TH, Piasecki J, Bush TL, Sullivan T, Foltz IN, Chang W, Douangpanya H, Dang T, et al. (2014). Apo2L/TRAIL and the death receptor 5 agonist antibody AMG 655 cooperate to promote receptor clustering and antitumor activity. Cancer Cell 26, 177–189. [DOI] [PubMed] [Google Scholar]
- Gu J, and Ghayur T (2012). Generation of dual-variable-domain immunoglobulin molecules for dual-specific targeting. Methods Enzymol 502, 25–41. [DOI] [PubMed] [Google Scholar]
- Hutt M, Farber-Schwarz A, Unverdorben F, Richter F, and Kontermann RE (2012). Plasma half-life extension of small recombinant antibodies by fusion to immunoglobulin-binding domains. J Biol Chem 287, 4462–4469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakob CG, Edalji R, Judge RA, DiGiammarino E, Li Y, Gu J, and Ghayur T (2013). Structure reveals function of the dual variable domain immunoglobulin (DVD-Ig) molecule. MAbs 5, 358–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Z, McDonald ER 3rd, Dicker DT, and El-Deiry WS (2004). Deficient tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) death receptor transport to the cell surface in human colon cancer cells selected for resistance to TRAIL-induced apoptosis. J Biol Chem 279, 35829–35839. [DOI] [PubMed] [Google Scholar]
- Kline JB, Kennedy RP, Albone E, Chao Q, Fernando S, McDonough JM, Rybinski K, Wang W, Somers EB, Schweizer C, et al. (2017). Tumor antigen CA125 suppresses antibody-dependent cellular cytotoxicity (ADCC) via direct antibody binding and suppressed Fc-gamma receptor engagement. Oncotarget 8, 52045–52060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leabman MK, Meng YG, Kelley RF, DeForge LE, Cowan KJ, and Iyer S (2013). Effects of altered FcgammaR binding on antibody pharmacokinetics in cynomolgus monkeys. MAbs 5, 896–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBlanc H, Lawrence D, Varfolomeev E, Totpal K, Morlan J, Schow P, Fong S, Schwall R, Sinicropi D, and Ashkenazi A (2002). Tumor-cell resistance to death receptor--induced apoptosis through mutational inactivation of the proapoptotic Bcl-2 homolog Bax. Nat Med 8, 274–281. [DOI] [PubMed] [Google Scholar]
- Levett PN, Sonnenberg K, Sidaway F, Shead S, Niedrig M, Steinhagen K, Horsman GB, and Drebot MA (2005). Use of immunoglobulin G avidity assays for differentiation of primary from previous infections with West Nile virus. J Clin Microbiol 43, 5873–5875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, and Ravetch JV (2012). Apoptotic and antitumor activity of death receptor antibodies require inhibitory Fcgamma receptor engagement. Proc Natl Acad Sci U S A 109, 10966–10971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Spidel JL, Maddage CJ, Rybinski KA, Kennedy RP, Krauthauser CL, Park YC, Albone EF, Jacob S, Goserud MT, et al. (2013). The antitumor activity of the human FOLR1-specific monoclonal antibody, farletuzumab, in an ovarian cancer mouse model is mediated by antibody-dependent cellular cytotoxicity. Cancer Biol Ther 14, 1032–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant MS, Geller JI, Baird K, Chou AJ, Galli S, Charles A, Amaoko M, Rhee EH, Price A, Wexler LH, et al. (2012). Phase I trial and pharmacokinetic study of lexatumumab in pediatric patients with solid tumors. J Clin Oncol 30, 4141–4147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mongkolsapaya J, Grimes JM, Chen N, Xu XN, Stuart DI, Jones EY, and Screaton GR (1999). Structure of the TRAIL-DR5 complex reveals mechanisms conferring specificity in apoptotic initiation. Nat Struct Biol 6, 1048–1053. [DOI] [PubMed] [Google Scholar]
- Niyazi M, Marini P, Daniel PT, Humphreys R, Jendrossek V, and Belka C (2009). Efficacy of triple therapies including ionising radiation, agonistic TRAIL antibodies and cisplatin. Oncol Rep 21, 1455–1460. [DOI] [PubMed] [Google Scholar]
- Paz-Ares L, Balint B, de Boer RH, van Meerbeeck JP, Wierzbicki R, De Souza P, Galimi F, Haddad V, Sabin T, Hei YJ, et al. (2013). A randomized phase 2 study of paclitaxel and carboplatin with or without conatumumab for first-line treatment of advanced non-small-cell lung cancer. J Thorac Oncol 8, 329–337. [DOI] [PubMed] [Google Scholar]
- Printz C (2011). Cancer drug safety presents challenges: oncologists, researchers, and government balance risks with benefits. Cancer 117, 5023–5025. [DOI] [PubMed] [Google Scholar]
- Ridgway JB, Presta LG, and Carter P (1996). ‘Knobs-into-holes’ engineering of antibody CH3 domains for heavy chain heterodimerization. Protein Eng 9, 617–621. [DOI] [PubMed] [Google Scholar]
- Sasaki Y, Miwa K, Yamashita K, Sunakawa Y, Shimada K, Ishida H, Hasegawa K, Fujiwara K, Kodaira M, Fujiwara Y, et al. (2015). A phase I study of farletuzumab, a humanized anti-folate receptor alpha monoclonal antibody, in patients with solid tumors. Invest New Drugs 33, 332–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders NA, Simpson F, Thompson EW, Hill MM, Endo-Munoz L, Leggatt G, Minchin RF, and Guminski A (2012). Role of intratumoural heterogeneity in cancer drug resistance: molecular and clinical perspectives. EMBO Mol Med 4, 675–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer G, Haber L, Crocker LM, Shia S, Shao L, Dowbenko D, Totpal K, Wong A, Lee CV, Stawicki S, et al. (2011). A two-in-one antibody against HER3 and EGFR has superior inhibitory activity compared with monospecific antibodies. Cancer Cell 20, 472–486. [DOI] [PubMed] [Google Scholar]
- Soria JC, Smit E, Khayat D, Besse B, Yang X, Hsu CP, Reese D, Wiezorek J, and Blackhall F (2010). Phase 1b study of dulanermin (recombinant human Apo2L/TRAIL) in combination with paclitaxel, carboplatin, and bevacizumab in patients with advanced non-squamous non-small-cell lung cancer. J Clin Oncol 28, 1527–1533. [DOI] [PubMed] [Google Scholar]
- Spiess C, Zhai Q, and Carter PJ (2015). Alternative molecular formats and therapeutic applications for bispecific antibodies. Mol Immunol 67, 95–106. [DOI] [PubMed] [Google Scholar]
- Takeda K, Kojima Y, Ikejima K, Harada K, Yamashina S, Okumura K, Aoyama T, Frese S, Ikeda H, Haynes NM, et al. (2008). Death receptor 5 mediated-apoptosis contributes to cholestatic liver disease. Proc Natl Acad Sci U S A 105, 10895–10900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teleb M, Salire K, Wardi M, Alkhateeb H, Said S, and Mukherjee D (2016). Idarucizumab: Clinical Role of a Novel Reversal Agent for Dabigatran. Cardiovasc Hematol Disord Drug Targets 16, 25–29. [DOI] [PubMed] [Google Scholar]
- Tran E, Chinnasamy D, Yu Z, Morgan RA, Lee CC, Restifo NP, and Rosenberg SA (2013). Immune targeting of fibroblast activation protein triggers recognition of multipotent bone marrow stromal cells and cachexia. J Exp Med 210, 1125–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tushir-Singh J (2017). Antibody-siRNA conjugates: drugging the undruggable for anti-leukemic therapy. Expert Opin Biol Ther 17, 325–338. [DOI] [PubMed] [Google Scholar]
- Vergote I, Armstrong D, Scambia G, Teneriello M, Sehouli J, Schweizer C, Weil SC, Bamias A, Fujiwara K, Ochiai K, et al. (2016). A Randomized, Double-Blind, Placebo-Controlled, Phase III Study to Assess Efficacy and Safety of Weekly Farletuzumab in Combination With Carboplatin and Taxane in Patients With Ovarian Cancer in First Platinum-Sensitive Relapse. J Clin Oncol 34, 2271–2278. [DOI] [PubMed] [Google Scholar]
- Vincenzi B, Armento G, Spalato Ceruso M, Catania G, Leakos M, Santini D, Minotti G, and Tonini G (2016). Drug-induced hepatotoxicity in cancer patients - implication for treatment. Expert Opin Drug Saf 15, 1219–1238. [DOI] [PubMed] [Google Scholar]
- Wagner KW, Punnoose EA, Januario T, Lawrence DA, Pitti RM, Lancaster K, Lee D, von Goetz M, Yee SF, Totpal K, et al. (2007). Death-receptor O-glycosylation controls tumor-cell sensitivity to the proapoptotic ligand Apo2L/TRAIL. Nat Med 13, 1070–1077. [DOI] [PubMed] [Google Scholar]
- Wakelee HA, Patnaik A, Sikic BI, Mita M, Fox NL, Miceli R, Ullrich SJ, Fisher GA, and Tolcher AW (2010). Phase I and pharmacokinetic study of lexatumumab (HGS-ETR2) given every 2 weeks in patients with advanced solid tumors. Ann Oncol 21, 376–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Lin J, and Xu R (2014). The molecular mechanisms of TRAIL resistance in cancer cells: help in designing new drugs. Curr Pharm Des 20, 6714–6722. [DOI] [PubMed] [Google Scholar]
- Wilson NS, Yang B, Yang A, Loeser S, Marsters S, Lawrence D, Li Y, Pitti R, Totpal K, Yee S, et al. (2011). An Fcgamma receptor-dependent mechanism drives antibody-mediated target-receptor signaling in cancer cells. Cancer Cell 19, 101–113. [DOI] [PubMed] [Google Scholar]
- Zhang X, Zhang L, Tong H, Peng B, Rames MJ, Zhang S, and Ren G (2015). 3D Structural Fluctuation of IgG1 Antibody Revealed by Individual Particle Electron Tomography. Sci Rep 5, 9803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuch de Zafra CL, Ashkenazi A, Darbonne WC, Cheu M, Totpal K, Ortega S, Flores H, Walker MD, Kabakoff B, Lum BL, et al. (2016). Antitherapeutic antibody-mediated hepatotoxicity of recombinant human Apo2L/TRAIL in the cynomolgus monkey. Cell Death Dis 7, e2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.