Abstract
Spermatogenesis in Drosophila melanogaster is characterized by a specific transcriptional program during the spermatocyte stage. Transcription of thousands of genes is regulated by the interaction of several proteins or complexes, including a tTAF-containing TFIID variant, tMAC, Mediator, and chromatin interactors, e.g., bromodomain proteins. We addressed how distinct subsets of target genes are selected. We characterized the highly similar proteins tPlus3a and tPlus3b, which contain a Plus3 domain and are enriched in the testis, mainly in spermatocytes. In tPlus3a and tplus3b deletion mutants generated using the CRISPR/Cas9 system, fertility was severely reduced and sperm showed defects during individualization. tPlus3a and tPlus3b heterodimerized with the bromodomain protein tBRD-1. To elucidate the role of the tPlus3a and tPlus3b proteins in transcriptional regulation, we determined the transcriptomes of tplus3a-tplus3b and tbrd-1 deletion mutants using next-generation sequencing (RNA-seq) and compared them to that of the wild-type. tPlus3a and tPlus3b positively or negatively regulated the expression of nearly 400 genes; tBRD-1 regulated 1,500 genes. Nearly 200 genes were regulated by both tPlus3a and tPlus3b and tBRD-1. tPlus3a and tPlus3b activated the Y-chromosomal genes kl-3 and kl-5, which indicates that tPlus3a and tPlus3b proteins are required for the function of distinct classes of genes. tPlus3a and tPlus3b and tBRD-1 repress genes relevant for seminal fluid and heat shock. We hypothesize that tPlus3a and tPlus3b proteins are required to specify the general transcriptional program in spermatocytes.
Introduction
In mammals and in Drosophila melanogaster, regulation of transcription during spermatogenesis is complex. The germ cell transcriptional program covers various processes, including meiosis, post-meiotic formation of flagella, nuclear shaping, and chromatin reorganization. In flies, the majority of transcripts needed in spermatogenesis are produced in the prolonged meiotic prophase of the spermatocyte stage. Therefore, most transcripts required for post-meiotic sperm development (spermiogenesis) are translationally repressed until they are needed in later stages [1–4]. About 50% of the predicted genes in the D. melanogaster genome are expressed in the testis [5], and a large portion of the transcripts are testis enriched or even testis-specific.
One complex that plays a pivotal role in transcriptional regulation during spermatogenesis is a germ-line-specific variant of the general transcription factor TFIID. This complex is composed of somatic TATA-binding protein (TBP)-associated factors (TAFs) and the testis-specific paralogues tTAFs [6–7]. tTAFs are recruited by the Mediator complex to chromatin, where they activate genes [8]. Mediator regulates several hundred genes, and some of its subunits are recruited to gene regions by the testis meiotic arrest complex (tMAC), which is involved in regulation of spermiogenesis-relevant genes [8–9]. Together, tTAFs, tMAC, and Mediator regulate thousands of genes [3].
It is assumed that besides these general transcription initiation factors, other factors target gene expression more specifically. These could include proteins that interact with the general transcription complex, particularly because upstream regulatory regions of genes specifically expressed in the male germ line are often very short [10]. Indeed, bromodomain-containing proteins (tBRDs), which are synthesized specifically in the testis, heterodimerize with some of the tTAFs and partly with each other [11–12]. In general, bromodomain proteins bind to acetylated lysine residues [13], which suggests that tBRDs might serve as a platform to guide parts of the transcriptional machinery to chromatin. The well-characterized tBRD-1 and tBRD-2 proteins regulate smaller sets of target genes than tTAFs and tMAC, and both depend on tTAF function. Loss of tbrd-1 or knock-down of tbrd-2 leads to post-meiotic phenotypes [11–12, 14], whereas tTAF mutants show an earlier meiotic arrest phenotype [6]. Despite heterodimerization of tBRD-1 and tBRD-2 and a similar phenotype of tbrd-1- and tbrd-2-deficient flies, microarray analyses suggest that both proteins are also part of different subcomplexes, as they regulate partially non-overlapping sets of target genes [12].
We aimed at identifying further transcriptional regulators and at clarifying whether they physically interact with one of the known tBRD molecules and share target genes. As candidates, we considered Rtf-related proteins synthesized specifically in the testis. RTF is a member of a complex that is conserved in yeast, mouse, human, and fly. Subunits of this complex—the RNA-polymerase-II-associated factor complex (Paf1C) [15]—co-purify with RNA polymerase II subunits in yeast [16] and higher eukaryotes [15, 17]. Many studies have suggested that Paf1C plays a role in transcriptional regulation, and several mechanisms have been proposed on how the effect on transcription might be achieved, including transcription elongation, RNA polymerase II pausing, or maintenance of co-transcriptional histone modifications [18]. Some subunits of Paf1C are conserved, namely Paf1, Cdc73, Leo1, Ctr9, and Rtf1 [15]. Rtf1 can also function outside of the Paf1C complex in humans and D. melanogaster [17, 19–21]. Rtf1 contains several functional domains, including a histone modification domain, a Plus3 domain, and a protein interaction domain that mediates Paf1C interaction. It has been suggested that in yeast, the histone modification domain mediates co-transcriptional histone modifications and the Plus3 domain is needed for Paf1C recruitment to chromatin [15, 22]. Strikingly, the Plus3 domain is highly conserved in yeast, human, and fly. Structural homology and in vitro experiments using recombinant Rtf1 indicate that the Plus3 domain binds nucleic acids, especially single-stranded DNA. This feature suggests a supportive role in transcription through stabilization of the transcription machinery during transcript elongation [23]. However, evidence for an interplay between the complexes involved in transcriptional regulation is fragmentary. In D. melanogaster, Rtf1 is ubiquitously transcribed [5]. In a search for proteins that might play a role in refining the transcriptional program of spermatocytes in D. melanogaster, we investigated proteins carrying the Plus3 domain that are mainly synthesized in the testes. Indeed, in a stage-specific proteome analysis, we showed that tPlus3a and tPlus3b proteins are highly enriched in stages before meiotic divisions [24]. Here, we characterized tPlus3a and tPlus3b and show that they are essential for spermiogenesis and full male fertility.
Results
Identification of three genes specifically expressed in the male germ line that encode Plus3 domain proteins
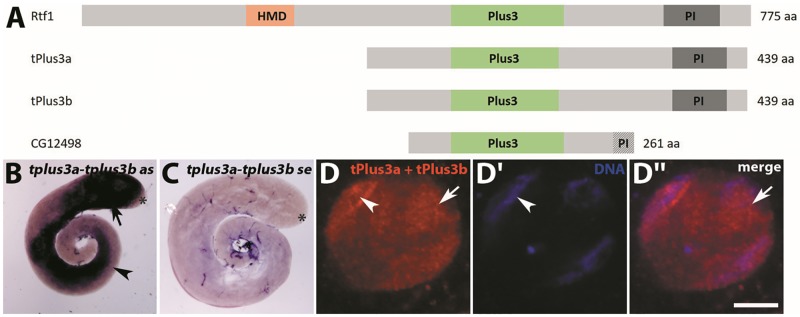
During spermatogenesis in D. melanogaster, transcription mainly takes place in spermatocytes, where distinct complexes regulate thousands of genes. We aimed at identifying testes-specific regulators responsible for distinct subsets of genes. One of the known general complexes that are involved in transcriptional regulation is Paf1C [15]. A prominent subunit of Paf1C is the Plus3 domain protein Rtf1; Rtf1 and especially its Plus3 domain are conserved in Saccharomyces cerevisiae, D. melanogaster, and Homo sapiens [23]. In D. melanogaster, the gene is ubiquitously expressed and is highly expressed in the testis. In a search for genes of other proteins with a Plus3 domain that are specifically expressed in the D. melanogaster testis, we identified three genes, namely CG12498, CG31702, and CG31703. According to RNA-seq and microarray analyses, all three genes are mostly expressed in the testis [5, 25]. The nucleotide sequences of CG31702 and CG31703 are approximately 99% identical. Based on their testis-specific transcription and their encoded Plus3 domain, we named the CG31702 and CG31703 genes tplus3a and tplus3b (testis-specifically transcribed plus3 genes), respectively. The proteins encoded by CG12498, tplus3a, and tplus3b share conserved regions (Fig 1 and S1 Fig). Due to the high similarity between tPlus3a and tPlus3b we call them tPlus3a and tPlus3b in the following. The Plus3 domains of tPlus3a and tPlus3b are identical. tPlus3a and CG12498 contain the three conserved positively charged amino acids that gave the domain its name (green in S1 Fig) and many other amino acids that are crucial for domain conformation [23]. We used Clustal Omega [26] to align the amino acid sequences of Rtf1, tPlus3a, and CG12498 (S1A Fig). The histone modification domain (HMD in Fig 1A) of Rtf1 is not present in the other Plus3 domain proteins. All four proteins have a conserved sequence near the C-terminus; in Rtf1, this sequence serves as a protein interaction domain (PI in Fig 1A). This domain is truncated in CG12498, thus it is functionality may be severely impaired (S1B Fig). The Plus3 domain sequence in tPlus3a is slightly shorter than that in Rtf1 and CG12498 (S1A Fig).
Fig 1. tPlus3a and tPlus3b proteins localize mainly to the nuclei of spermatocytes.
Schematic view of the alignment of the D. melanogaster Plus3 domain-containing proteins Rtf1, tPlus3a, tPlus3b, and CG12498. aa, length of protein in amino acids; HMD, histone modification domain; Plus3, Plus3 domain; PI, protein interaction domain. (B, C) In situ hybridization of wild-type testis with (B) tplus3a-tplus3b antisense (as) probe and (C) tplus3a-plus3b sense (se) probe. Asterisk, hub region; arrow, detection of tplus3a-tplus3b mRNA in spermatocytes; arrowhead, lack of detection in late stages. (D) Single spermatocyte nucleus from wild-type testes stained with anti-tPlus3a and tPlus3b antibody. Arrowhead, tPlus3a and tPlus3b distributed throughout nucleus, but slightly concentrated in chromosomal regions; arrow, tPlus3a and tPlus3b in distinct areas between chromosomal regions. (D’) DNA visualized with Hoechst 33258. (D”) Merged image of D and D’. Scale bar = 5 μm.
tplus3a-tplus3b transcripts are expressed and their proteins are synthesized in male germ cells
We used RNA in situ hybridization to examine the distribution of tplus3a-tplus3b, CG12498, and Rtf1 transcripts in the testis of wild-type flies (Fig 1B and S2A and S2C Fig). tplus3a-tplus3b transcripts were detected from the spermatocyte stage onward (Fig 1B), but late stages of spermatogenesis did not show any signal (Fig 1B). Thus, tplus3a-tplus3b transcripts characterize the spermatocyte and early spermatid stage. A control probe containing the sense sequence did not yield any signal (Fig 1C). CG12498 transcripts were present mainly in stages before meiotic divisions, while rtf1 transcripts were visualized in addition in post-meiotic stages (S2A and S2C Fig). We unsuccessfully attempted to generate flies that synthesized CG12498-mCherry to analyze the subcellular localization of CG12498. However, we were able to generate transgenic tplus3b-eGFP flies; tPlus3b-eGFP was expressed in the nucleoplasm of spermatocytes (S3A Fig, arrow), and the nucleolus was labeled (S3A Fig, arrowhead). In agreement with the transcript signal, we observed tPlus3b-eGFP in nuclei of round spermatids (S3B Fig, double arrow).
In addition, we raised a peptide antibody that could detect tPlus3a and tPlus3b. In immunofluorescence stainings of squash preparations of wild-type testes, tPlus3a and tPlus3b proteins were detected only in the nuclei of spermatocytes. In contrast to visualization with tPlus3b-eGFP, the nucleolus was not labeled. This lack of labeling might be due to inaccessibility of the antigenic region of tPlus3a and tPlus3b for the antibody or to artificial localization of eGFP in the nucleolus. The signal was fairly homogenously distributed in the nucleus and slightly concentrated in chromosomal regions (Fig 1D–1D”) and in distinct areas between the chromosomal regions in the nucleus (Fig 1D and 1D”). This region between the Hoechst-positive chromosomal regions contains large lampbrush loops, which correspond to three of the Y-chromosomal fertility loci [27]. We conclude that tPlus3a and tPlus3b expression is characteristic for the highly transcriptionally active spermatocyte phase.
tPlus3a and tPlus3b proteins are required for full male fertility
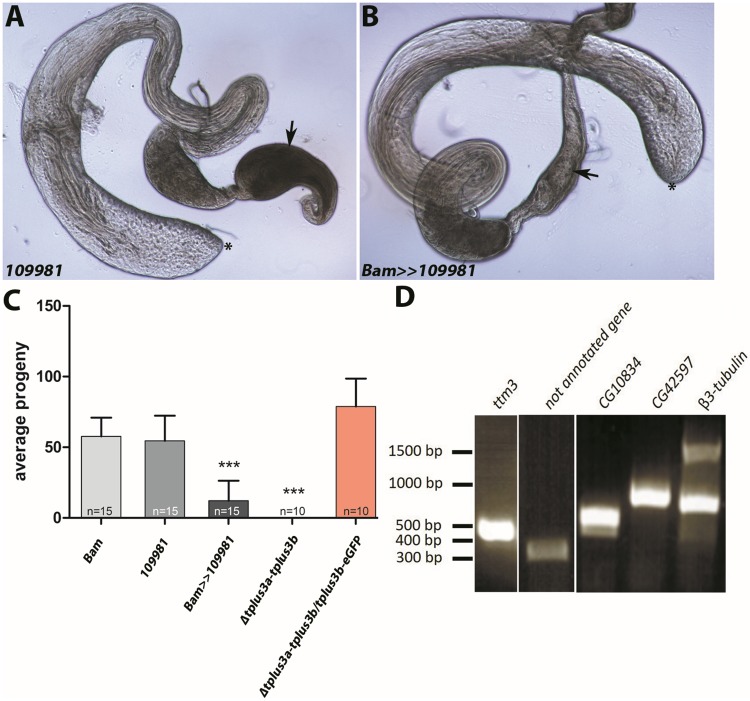
To determine whether tPlus3a and tPlus3b proteins are essential for male fertility, we first used an RNAi approach to reduce the transcript levels of tplus3a-tplus3b (Fig 2). The analysis of testes and seminal vesicles of male flies revealed that the seminal vesicles of knock-down males (Fig 2B, arrow) contained much less sperm than seminal vesicles of control males (Fig 2A, arrow). This is in agreement with strongly reduced fertility of tplus3a-tplus3b knock-down males (Fig 2C).
Fig 2. Knock-down of tplus3a-tplus3b leads to severely reduced male fertility.
(A) Seminal vesicle (arrow) of a male of the undriven tplus3a-tplus3b RNAi line filled with sperm. (B) Seminal vesicle (arrow) of a male from the bam GAL4 driven RNAi line v109981 contains few sperm. (C) Fertility tests of tplus3a-tplus3b knock-down males and sterility rescue of homozygous Δtplus3a-tplus3b mutants by tPlus3b-eGFP. (D) Analysis of genomic DNA of Δtplus3a-tplus3b mutants showed the presence of ttm3, a not-annotated gene, CG10834, CG42597, and the β3-tubulin gene as control. Dashes indicate the position of marker ladder.
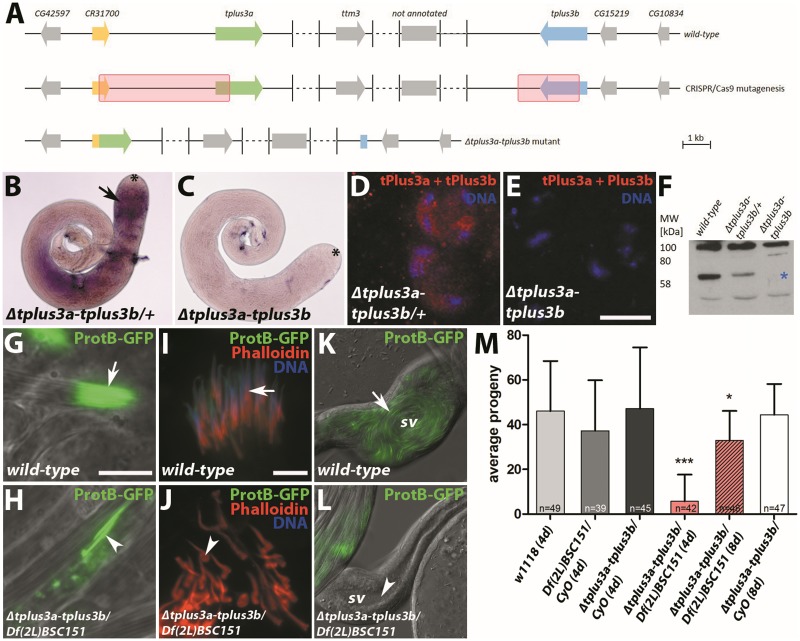
As these data suggested a crucial role of tplus3a-tplus3b in male fertility, we generated deletion mutants using the CRISPR/Cas9 system. We established transgenic flies carrying a construct coding for a single guide RNA able to detect both tplus3a and tplus3b and crossed these with flies expressing Cas9 in the germ line [28]. We screened 50 independent single crossings of putative mutants via PCR and identified the deletion mutant Δtplus3a-tplus3b, in which large parts of the open reading frame (ORF) of both tplus3a and tplus3b were deleted. We did not obtain any mutants with solely a tplus3a or tplus3b deletion. The deletions led to a fusion of tplus3a within its ORF to CR31700, an inactive pseudogene approximately 4,000 base pairs (bp) upstream of tplus3a [5, 25] and yielded a null allele of tplus3a. In addition, it was possible to PCR amplify regions flanking tplus3b but not its ORF, which indicated that most of the tplus3b ORF was deleted. Thus, this mutant likely carries loss-of-function mutations for both genes; the genes neighboring tplus3a and tplus3b, other than CR31700, remained intact (Fig 2D), this also held true for the non-annotated gene with expression in accessory glands (modENCODE). The tplus3b-eGFP construct was able to strongly reduce sterility of homozygous Δtplus3a-tplus3b mutants (Fig 2C). This rescue indicated, that mainly the loss of tplus3a and tplus3b was responsible for the observed sterility defects and that tplus3b-eGFP can rescue sterility, which might argue for functional redundancy between tPlus3a and tPlus3b.
In situ hybridization detected transcripts in testis of heterozygous Δtplus3a-tplus3b mutants (Fig 3B, arrow) but not in homozygous mutants (Fig 3C). Analogous results were obtained in anti-tPlus3a and tPlus3b immunofluorescence stainings. tPlus3a and tPlus3b proteins were detected in spermatocytes of heterozygous Δtplus3-tplus3b mutant flies (Fig 3D) but not in homozygous ΔtPlus3a-tplus3b mutants (Fig 3E).
Fig 3. Deletion of tplus3a-tplus3b leads to severely reduced male fertility and defects during sperm individualization.
(A) Schematic view of the tplus3a and tplus3b genomic region and the CRISPR/Cas9-mediated generation of deletions (red boxes), which resulted in the fusion of tplus3a and pseudogene CR31700 and a deletion of most of the tplus3b ORF. Dashed lines, regions not fully depicted. The region between ttm3 and tplus3b contains a not-annotated gene which is expressed in the accessory glands of the male reproductive track (modENCODE). (B, C) In situ hybridization of testis of (B) heterozygous ΔpPlus3a-tplus3b mutant (arrow, tplus3a-tplus3b signal detected) and (C) homozygous Δtplus3a-tplus3b mutant. Asterisk, hub region. (D, E) Spermatocytes of (D) heterozygous and (E) homozygous Δtplus3a-tplus3b mutant testes stained with anti-tPlus3a and tPlus3b. Scale bar = 10 μm. (F) Western blot of protein extracts from wild-type and homozygous and heterozygous Δtplus3a-tplus3b mutant testes probed with anti-tPlus3a and tPlus3b. Asterisk, lack of signal at ca. 60 kDa. (G, H) Whole-mount preparations of (G) wild-type and (H) Δtplus3a-tplus3b/Df(2L)BSC151 testes. Sperm nuclei visualized with ProtB-eGFP. Arrow, sperm nuclei aligned in tight bundles; arrowhead, sperm nuclei distributed across the testis. Scale bar = 10 μm. (I, J) Squash preparations of (I) wild-type and (J) Δtplus3a-tplus3b/Df(2L)BSC151 testes. Sperm nuclei visualized with ProtB-eGFP, DNA stained with Hoechst 33258, and individualization complex stained with TRITC-Phalloidin. Arrow, sperm nuclei aligned in tight bundles; arrowhead, disturbed individualization complex formation with one end thickened. Scale bar = 10 μm. (K, L) Whole-mount preparations of (K) wild-type and (L) Δtplus3a-tplus3b/Df(2L)BSC151 seminal vesicles (sv) of 1-day-old males. Sperm nuclei visualized with ProtB-eGFP. Arrow, seminal vesicles filled with sperm; arrowhead, empty seminal vesicles. (M) Fertility of 4-day-old wild-type, 4-day-old Df(2L)BSC151/CyO, 4- and 8-day-old Δtplus3a-tplus3b/CyO, and 4- and 8-day-old Δtplus3a-tplus3b/Df(2L)BSC151 flies. *, p < 0.05; ***, p < 0.005.
In western blots using protein extracts from testes and probed with anti-tPlus3a and tPlus3b, a prominent band at approximately 60 kDa was detected in the wild-type (predicted molecular mass of tPlus3a and tPlus3b ca. 50 kDa). The signal of protein extracts of heterozygous Δtplus3a-tplus3b testes was lower, and no signal was detected when protein extracts of homozygous Δtplus3a-tplus3b testes were probed (Fig 3F).
To exclude off-target effects arising during CRISPR/Cas9 mutagenesis, we crossed Δtplus3-tplus3b/CyO with Df(2L)BSC151/CyO, which is a fly line deficient in tplus3a and tplus3b. All further phenotypic analyses were carried out with trans-heterozygous Δtplus3a-tplus3b/Df(2L)BSC151 flies. These flies were mildly defective in sperm individualization compared to wild-type flies; in wild-type flies, the nuclei of spermatids in an individual cyst with synchronously developing spermatids are arranged strictly parallel, as visualized with protB-eGFP transgene expression (Fig 3G). In Δtplus3a-tplus3b/Df(2L)BSC151 flies, many sperm bundles were arranged in parallel, but some sperm were dispersed in the testis and lost their cyst organization (Fig 3H). In wild-type testis squash preparations, many cysts appeared and the individualization complex formed normally, as visualized with TRITC-phalloidin (Fig 3I). In Δtplus3a-tplus3b/Df(2L)BSC151 testis squash preparations, a number of sperm bundles were disorganized. In some cases, the individualization complex formed was abnormal in that one end was thickened (Fig 3J). The seminal vesicles of wild-type flies were already filled with sperm 1 day after hatching (Fig 3K, arrow), whereas the seminal vesicles of Δtplus3a-tplus3b/Df(2L)BSC151 males of the same age were empty (Fig 3L, arrowhead). Two days after hatching, also seminal vesicles of Δtplus3a-tplus3b/Df(2L)BSC151 males contained sperm, the number of sperm in the seminal vesicles increased with age as exemplarily shown in S4C and S4D Fig in comparison to wild-type (S4A and S4B Fig). For fertility assays, we mated one day old males and analyzed the number of progeny after further 3 days (4 day old males), alternatively, we mated 5 day old males with virgins and analyzed the number of progeny after 3 days (8 day old males). In these sterility tests comparing 4-day-old wild-type, Df(2L)BSC151/CyO, Δtplus3a-tplus3b/CyO, and Δtplus3a-tplus3b/Df(2L)BSC151 males, the fertility of Δtplus3a-tplus3b/Df(2L)BSC151 males was reduced to ca. 12% of that of the wild-type control (Fig 3M). As 2-day-old Δtplus3a-tplus3b/Df(2L)BSC151 males started to have sperm in the seminal vesicles, the reduced fertility after 4 days suggests that sperm found in the seminal vesicles were less than in wild-type males or have reduced in motility. Sterility tests with 8-day-old Δtplus3a-tplus3b/Df(2L)BSC151 males revealed that fertility is partially restored after several days (Fig 3M). We conclude that defects in late sperm development result in delayed fertility of Δtplus3a-tplus3b/Df(2L)BSC151 males and that the proportion of sperm in Δtplus3a-tplus3b/Df(2L)BSC151 males increases with age. At first glance, this was surprising, but lack of tplus3a-tplus3b was possibly compensated by the increase in synthesis of the CG12498-encoded tPlus3 domain protein or by dRtf1, which in contrast to CG12498 contains a complete PI domain (Fig 1A).
tPlus3a and tPlus3b heterodimerize with the bromodomain protein tBRD-1
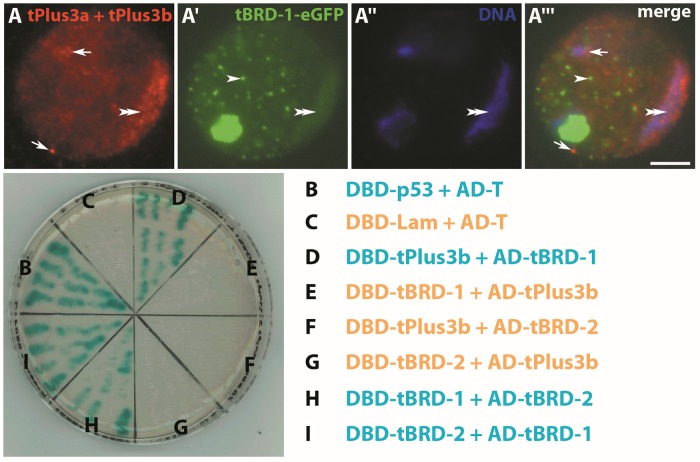
We then addressed whether tPlus3a and tPlus3b proteins interact with other known proteins of transcription complexes in spermatocytes. We previously observed a late arrest in spermatogenesis and infertility of tbrd-1 mutants and after tbrd-2 knock-down in male germ cells [12, 14]. We analyzed tPlus3a and tPlus3b immunofluorescence stainings at high magnification in flies synthesizing tBRD-1-eGFP to determine whether tPlus3a and tPlus3b co-localize with tBRD-1 in nuclei of spermatocytes (Fig 4A–4A”‘). The tBRD-1-eGFP signal was concentrated in the chromosomal regions (Fig 4A’) and overlapped with the tPlus3a and tPlus3b staining in this region (Fig 4A and 4A”‘). The tBRD-1-eGFP signal also concentrated in speckles widely distributed in the nucleus (Fig 4A’), but these areas did not overlap significantly with the prominent tPlus3a and tPlus3b staining between the chromosomal regions (Fig 4A and 4A”‘).
Fig 4. tPlus3a and tPlus3b partially co-localize with tBRD-1 and interact with tBRD-1 but not with tBRD-2 in yeast two-hybrid experiments.
(A–A”‘) Anti-tPlus3a and tPlus3b immunofluorescence staining of a single spermatocyte nucleus of flies synthesizing tBRD-1-eGFP. (A) tPlus3a and tPlus3b signal present throughout the nucleus but concentrated in chromosomal regions (double arrowhead) and in distinct spots (arrows). (A’) tBRD-1 found mainly in the nucleolus, in chromosomal regions (double arrowhead), and in a speckled pattern (arrowhead). (A”) DNA stained with Hoechst. Double arrowhead, chromosomal regions. (A”‘) Merged image of A, A’, and A” showing co-localization of tPlus3a and tPlus3b and tBRD-1-eGFP in the chromosomal regions (double arrowhead). Scale bar = 5 μm. (B–I) Yeast two-hybrid interaction assay. (B) Positive control. (C) Negative control. (D) Heterodimerization of tPlus3b bait and tBRD-1 prey. (E) Lack of heterodimerization of tPlus3b prey and tBRD-1 bait. (F, G) Lack of heterodimerization of tPlus3b as bait or prey and tBRD-2 as prey or bait. (H, I) Heterodimerization of tBRD-1 and tBRD-2 as prey or bait.
The partial co-localization of tPlus3a and tPlus3b and tBRD-1 in the chromosomal regions prompted us to test whether tBRD-1 and also tBRD-2, whose synthesis overlaps with that of tBRD-1, interact with tPlus3a and tPlus3b. We analyzed whether tPlus3a and tPlus3b heterodimerize with tBRD-1 or tBRD-2 using yeast two-hybrid assays. The coding regions of tPlus3b, tBRD-1, and tBRD-2 were fused to the GAL4 DNA binding domain (DBD), which represented the bait protein, and to the GAL4 activation domain (AD), which represented the prey protein. Interaction of bait and prey fusion proteins leads to activation of a MEL1 reporter gene, which stains yeast colonies blue. The positive control, with DBD-p53 as bait protein and the large T antigen (AD-T) as prey protein, activated the reporter gene (Fig 4B), whereas the negative control, with Lamin C as bait protein and the large T antigen as prey protein, did not (Fig 4C).
In the assay, the bait fusion DBD-tPlus3b heterodimerized with the prey fusion AD-tBRD-1 (Fig 4D). To exclude auto-activation of reporter genes that might be caused by bait or prey fusion proteins, we tested DBD-tPlus3b with an unfused activation domain; the reporter gene was not activated (not shown). We did not detect heterodimerization of DBD-tBRD-1 with AD-tPlus3b (Fig 4E), possibly because of conformational changes in the tPlus3b structure caused by the fusion to the activation domain or insufficient synthesis of the AD-tPlus3b fusion. DBD-tPlus3b did not heterodimerize with AD-tBRD-2 (Fig 4F), and DBD-tBRD-2 did not heterodimerize with AD-tPlus3b (Fig 4G). As a control for the functionality of the yeast two-hybrid constructs, we tested the previously described heterodimerization of tBRD-1 and tBRD-2 [11]; the proteins formed a heterodimer (Fig 4H and 4I).
tPlus3a and tPlus3b and tBRD-1 activate and repress transcription individually and together
Next, we addressed whether tPlus3a and tPlus3b proteins are important for gene activation and/or repression. As our yeast two-hybrid results showed that tPlus3b and tBRD-1 form a heterodimer, we asked whether tPlus3a and tPlus3b and tBRD-1 share target genes. We sequenced testis RNA from Δtplus3a-tplus3b/Df(2L)BSC151 flies, tbrd-11 mutants, and wild-type flies in parallel and identified differentially expressed genes (log2 FC ≥ 1 and ≤ −1). In Δtplus3a-tplus3b/Df(2L)BSC151 testes, 398 transcripts were differentially expressed, of which 209 were down-regulated and 189 were up-regulated compared to the wild-type (Fig 5A). In tbrd-11 mutant flies, 1,499 genes were differentially expressed, of which 1,196 were down-regulated and 303 were up-regulated compared to the wild-type.
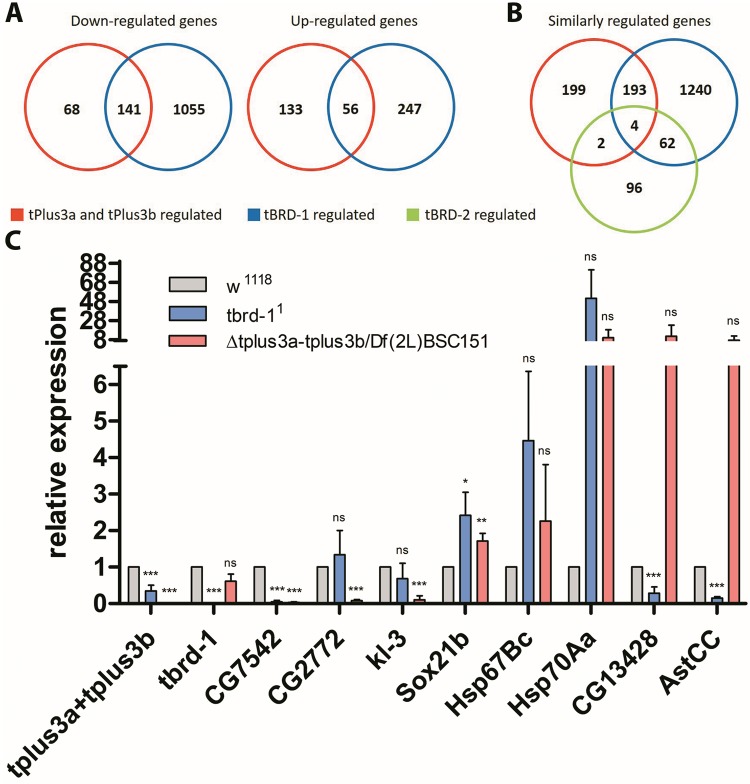
Fig 5. tPlus3a and tPlus3b and tBRD-1 regulate a common set of genes.
Transcript expression was measured with RNA-seq in three replicates each of testes RNA of the wild-type, Δtplus3a-tplus3b/Df(2L)BSC151, and tbrd-11 mutants. Expression levels in mutants are relative to those in the wild-type, which were set to 1. Differentially expressed genes were identified using a log2 FC ≥ 1 and ≤ −1. (A-C) Numbers indicate the number of genes that were shown to be regulated (A) Venn diagrams showing the overlap between down-regulated and up-regulated genes in Δtplus3a-tplus3b/Df(2L)BSC151 (tPlus3a and tPlus3b regulated) and tbrd-11 (tBRD-1 regulated) mutants relative to the wild-type. (B) Venn diagram showing the overlap of similarly regulated genes in Δtplus3a-tplus3b/Df(2L)BSC151, tbrd-11, and tbrd-2 knock-down testes (tBRD-2 regulated). (C) Validation of RNA-seq results using qPCR, normalized to Rpl32. As controls, no tplus3a-tplus3b and tbrd-1 transcripts were expressed in the respective mutants. P-values for significance Δtplus3a-tplus3b/Df(2L)BSC151 or tbrd-11 compared to wild-type: * p < 0.025, ** p < 0.005 and *** p < 0.0025.
The number of genes down-regulated in both Δtplus3a-tplus3b/Df(2L)BSC151 and tbrd-11 mutants, namely 141, represents about two-thirds of the genes that are activated by tPlus3a and tPlus3b (Fig 5A). On the other hand, only 12% of the differentially expressed genes in tbrd-11 mutants overlap with genes expressed in Δtplus3a-tplus3b/Df(2L)BSC151, which suggests that most genes regulated by tPlus3a and tPlus3b are also targeted by tBRD-1, but that tBRD-1 itself activates a much larger set of genes. Of the up-regulated genes in Δtplus3a-tplus3b/Df(2L)BSC151 and tbrd-11 mutants, 56 overlap, which corresponds to about 30% of the tPlus3a and tPlus3b up-regulated genes and 17% of the genes up-regulated in tbrd-11 mutants. Our results suggested that bromodomain proteins specifically synthesized in the testis serve as regulators of different sets of target genes in the germ line transcription machinery. Therefore, we also compared our RNA-seq data of Δtplus3a-tplus3b/Df(2L)BSC151 and tbrd-11 mutants to RNA microarray data from tbrd-2 knock-down testes [12]. In contrast to the large number of target genes common to tPlus3a and tPlus3b and tBRD-1, the expression of only six genes changed in both the absence of tPlus3a and tPlus3b and upon knock-down of tBRD-2. Four of these genes are also regulated by tBRD-1; 66 genes are regulated by both tBRD-1 and tBRD-2 (Fig 5B). These findings suggest a role for tPlus3a and tPlus3b in defining a subset of genes regulated by tBRD-1.
To validate our RNA-seq results, we performed real-time quantitative PCR (qPCR) with cDNA from wild-type and Δtplus3a-tplus3b/Df(2L)BSC151 and tbrd-11 mutants. We first validated down-regulation of tbrd-1 and tplus3a-tplus3b in the respective mutants and observed a clear decrease in the number of the transcripts expressed. tplus3a-tplus3b transcripts in tbrd-11 mutants were also visibly down-regulated. According to our RNA-seq analysis, tplus3a-tplus3b transcripts were reduced by 40–45% in tbrd-11 mutants; these transcripts do not appear in the results owing to a less than twofold decrease in the log2-fold change (Fig 5C). We determined the transcript levels of the down-regulated target genes CG7542, CG2772, and kl-3. Our results confirmed that CG7542 is down-regulated in both mutants, and CG2772 and kl-3 are down-regulated only in Δtplus3a-tplus3b/Df(2L)BSC151 testes (Fig 5C). We then validated the results for the up-regulated genes Sox21b, Hsp67Bc, Hsp70Aa, CG13428, and AstCC (Fig 5C). Hsp67Bc was slightly up-regulated only in Δtplus3a-tplus3b/Df(2L)BSC151, Sox21b was up-regulated, and Hsp70Aa was strongly up-regulated in both Δtplus3a-tplus3b/Df(2L)BSC151 and tbrd-11 mutants.
The top 40 most down- or up-regulated genes in both Δtplus3a-tplus3b/Df(2L)BSC151 and tbrd-11 mutant testes or in only Δtplus3a-tplus3b/Df(2L)BSC151 testes are given in Table 1 (highest logFC as top).
Table 1. Genes down-regulated by tPlus3a and tPlus3b (from -7,1 to -1,3 log2FC) and by both tPlus3a and tPlus3b and tBRD-1 (from -8,1 to -2,7 log2FC).
Genes up-regulated by tPlus3a and tPlus3b (from 6,2 to 1,7 log2FC) and by both tPlus3a and tPlus3b and tBRD-1 (from 4,0 to 1,3).
| Down-regulated genes | Up-regulated genes | ||
|---|---|---|---|
| Δtplus3a-tplus3b/ Df(2L)BSC151 only | Δtplus3a-tplus3b/ Df(2L)BSC151 and tbrd-11 | Δtplus3a-tplus3b/ Df(2L)BSC151 only | Δtplus3a-tplus3b/ Df(2L)BSC151 and tbrd-11 |
| CG31703 | Salt | CG34436 | Sfp87B |
| CG31702 | CG14245 | CG13428 | Peb |
| Muc30E | Muc11A | AstCC | Sfp24Bb |
| CG2772 | CG7542 | CG12990 | Sox21b |
| CG43401 | Mur18B | Elo68alpha | PebII |
| Dh31-R | CG16762 | CG10764 | Obp56g |
| CG9259 | CG8028 | CG43320 | Sfp60F |
| CG42370 | CG3285 | Uro | CG17572 |
| CG6293 | CG18095 | CG18180 | CG14304 |
| CG12017 | CG34426 | CG12268 | CG3868 |
| CG12009 | CG32023 | Acp24A4 | ppk19 |
| CG43707 | CG45072 | CG34113 | CG30427 |
| dpr7 | CG14292 | Eip63F-1 | Hsp70Aa |
| CG41434 | CG31198 | GstZ2 | Hsp70Ab |
| IM23 | CG3290 | CG42313 | AttB |
| kl-3 | Irk3 | CG11951 | Hsp70Ba |
| CG34393 | CG42235 | kek3 | Hsp70Bb |
| CG8641 | Smvt | ninaC | Hsp70Bc |
| Ory | unc80 | CG4267 | Hsp70Bbb |
| CG8093 | CG2187 | CG14535 | CG11893 |
| mthl8 | Scp2 | rho-6 | CG2065 |
| IM1 | CG34284 | CG13038 | Hsp67Bc |
| CG31897 | CG11626 | CG2663 | CG18278 |
| CG31600 | CG13309 | Snoo | CG1441 |
| l(2)34Fc | CG2736 | CG5697 | CG30059 |
| CG6967 | CG14219 | CG11598 | CG10550 |
| CG14982 | Oatp58Da | CG31710 | CG10096 |
| CG8301 | CG7882 | asparagine-synthetase | CG10097 |
| CG8145 | NaPi-T | CG10657 | fd102C |
| CG12869 | CG10560 | CG7214 | CG5337 |
| CG42368 | CG34043 | NimC2 | CG31157 |
| CG17162 | CG9512 | TwdlT | CG13285 |
| CG16898 | CG3690 | CG16820 | GstD5 |
| CG11634 | CG31202 | SA-2 | sisA |
| CG13748 | CG31373 | CG12780 | Cyp4e3 |
| stum | CG1809 | CG45546 | Arr1 |
| CG34166 | CG31106 | Adh | Adgf-D |
| CG17159 | CG5791 | Adhr | CG6488 |
| H15 | SA | CG13516 | CG31300 |
| CG11629 | CG7084 | Rdl | CG31777 |
1: indicates the tbrd-1 allele used for this studies
tPlus3a and tPlus3b activate the Y-chromosomal fertility factors kl-3 and kl-5
The set of genes down-regulated in Δtplus3a-tplus3b/Df(2L)BSC151, i.e., regulated by tPlus3a and tPlus3b, includes kl-3 and kl-5, which encode Y-chromosomal fertility factors. These genes were not co-regulated by tBRD-1. As kl-3 and kl-5 encode dynein heavy chains of the axoneme, their down-regulation could contribute to the severely reduced fertility in Δtplus3a-tplus3b/Df(2L)BSC151 flies that we observed.
tPlus3a and tPlus3b and tBRD-1 negatively regulate heat shock genes and seminal fluid protein genes in the germ line
Strikingly, many heat-shock-related genes, e.g., Hsp70Aa, are among the genes up-regulated in both mutants (Table 1). Therefore, tPlus3a and tPlus3b and tBRD-1 might fulfill a repressive function for this gene class by limiting the activation of heat shock genes during the spermatocyte phase. Another class that appears to be regulated by both factors is represented by genes such as Sfp87B and Sfp24Bb, which encode seminal fluid proteins. These are synthesized by somatic parts of the reproductive tract and are first needed after sperm development [29]. Acp24A4, which encodes a male accessory gland protein [30], was only regulated by tPlus3a and tPlus3b. We found these transcripts up-regulated in all three biological samples (S5A Fig) and hardly any variation was found between the independent samples (S5A Fig). However, we asked if these transcripts could be up-regulated due to contaminations of testes preparations by accessory gland material. First, we searched for corresponding proteins in proteome data (containing 5500 proteins and recently generated in our group) from adult testes of wild-type males [24]. We did not detect accessory gland proteins in our proteome data which indicates clean testes preparation. As RNA-seq might be more sensitive we aimed at clarifying whether our testes RNA preparations were contaminated with transcripts from accessory glands. We searched for transcripts of other abundant seminal-fluid-relevant transcripts in our RNA-seq data. We searched for Mst57Da, Mst57Db, Mst57Dc [31], Sfp96F (FlyBase), and GGT-1 [32]. We found that these genes were not regulated by tPlus3a and tPlus3b and/or tBRD-1 (S5B Fig). Transcripts for the ejaculatory bulb proteins Ebp and EbpII (FlyBase), however, were found to be repressed directly or indirectly by tPlus3a and tPlus3b and tBRD-1 (S5A Fig). The data for transcripts of somatic parts of the male reproductive system are summarized in Table 2.
Table 2. Up-regulation of transcripts in Δtplus3a-tplus3b and tbrd-1 mutant testes which are synthesized in somatic parts of the male reproductive track.
| Transcript | Up-regulated in Δtplus3a-tplus3b mutants | Up-regulated in tbrd-1 mutants |
|---|---|---|
| Acp24A4 | + | - |
| Ggt-1 | - | - |
| Mst57Da | - | - |
| Mst57Db | - | - |
| Mst57Dc | - | - |
| Ebp | ++ | + |
| EbpII | ++ | + |
| Sfp24Bb | ++ | + |
| Sfp60F | ++ | + |
| Sfb87B | ++ | + |
| Sfp96F | + | - |
- no up-regulation of transcripts, + clear up-regulation, ++ high level of up-regulation
These findings suggested that transcriptional repression of Ebp, EbpII, Sfp24Bb, Sfp60F, and Sfp87B, depended directly or indirectly on tPlus3a and tPlus3b and tBRD-1 in spermatocytes, while Sfp96F and Acp24A appeared solely regulated by tPlus3a and tPlus3b.
Discussion
D. melanogaster spermatogenesis is characterized by a unique transcriptional program in spermatocytes. Thousands of transcripts are synthesized in this germ cell stage, but most are only needed much later in post-meiotic development [1–3]. Jiang et al. [33] reported that one-third of the tTAF-dependent genes is regulated by Modulo or Acj6. In our previous studies, we described a possible interplay between tBRDs and the testis transcription machinery and a role of tBRDs in guiding the complexes Mediator, tMAC, and TFIID to specific subsets of target genes [11–12]. We hypothesize that in addition to tBRD-1 and tBRD-2, other factors are required for fine-tuning gene expression during spermatogenesis. In the current study, we characterized synthesis and function of tPlus3a and tPlus3b. We show that tPlus3a and tPlus3b and the bromodomain protein tBRD-1 co-regulate distinct groups of target genes, which underscores their potential for fine-tuning the transcriptional regulatory network in spermatocytes. tplus3a-tplus3b are target genes of Aly and MED22 [8, 34], thus probably of the tMAC and Mediator complexes and are also slightly down-regulated in tbrd-11 mutants, which indicates that regulators of transcription are interdependent.
Plus3 domain proteins are synthesized in the testis and might be functionally redundant
The genome of D. melanogaster has the potential to synthesize only a few proteins containing a Plus3 domain. Here, we showed that the Plus3 domain proteins tPlus3a and tPlus3b are essential for full male fertility. In male germ cells, tPlus3a and tPlus3b proteins were limited to nuclei of spermatocytes and localize in the chromosomal regions. tPlus3a and tPlus3b proteins also localize to distinct spots in some regions of the nucleus, which might correspond to areas of Y-chromosomal lampbrush loops. The lampbrush loops harbor genes encoding male fertility factors, e.g., kl-3 and kl-5. In contrast to the Plus3 domain protein Rtf1, tPlus3a and tPlus3b lack the histone modification domain, which suggests that tPlus3a and tPlus3b are not able to directly modify histones by themselves. The Plus3 domain might be needed for DNA binding, as described for human Rtf1 [23]. Therefore, tPlus3a and tPlus3b could be required as a platform to recruit other histone-modifying enzymes. Based on a C-terminal consensus sequence in Rft1, tPlus3a, and tPlus3b, we conclude that a Paf1C interaction domain like that in Rtf1 is also present in tPlus3a and tPlus3b, but interaction between tPlus3a and tPlus3b and members of Paf1C remains to be examined. Interestingly, all homologues of Paf1C subunits in D. melanogaster are also synthesized in the testis [5], which might point to an interaction of at least one of the Paf1C components with tPlus3a and/or tPlus3b. In addition, a role in spermatogenesis for the remaining Plus3 domain protein-coding gene, CG12498, should be considered. Based on structural homologies between tPlus3a, tPlus3b, and CG12498, it seems likely that these proteins would be able to functionally replace each other to a certain degree. Functional redundancy between tPlus3a and tPlus3b and CG12498 might be the reason for the increasing fertility of 8-day-old Δtplus3a-tplus3b/Df(2L)BSC151 males and could also explain why younger Δtplus3a-tplus3b/Df(2L)BSC151 males are not completely sterile. We consider the possibility that the highly similar Plus3 domain protein CG12498 can replace the loss of tPlus3a and tPlus3b by increasing the CG12498 transcript level or translational activity. The truncation of the PI domain of CG12498 might lead to an inefficient replacement of tPlus3a and tPlus3b. This is in agreement with our observation that it takes days to rescue fertility defects, albeit not to the wild-type level. Furthermore, RTF1 with an intact PI domain might compensate.
tPlus3a and tPlus3b proteins regulate Y-chromosomal genes essential for full male fertility
Generation of Δtplus3a-tplus3b deletion mutants resulted in cysts with abnormal individualization during sperm individualization. The inefficient individualization of sperm likely contributes to the reduced fertility of males. In our RNA-seq data of Δtplus3a-tplus3b/Df(2L)BSC151, the kl-3 and kl-5 genes were down-regulated. Both of these targets encode axonemal outer arm dynein heavy chains, which suggests a role in assembly of the axoneme and motility of sperm [35–37]. As flies bearing kl-3 or kl-5 deletions lose the outer dynein arm of the flagellar axoneme [38], down-regulation of kl-3 and kl-5 might explain our finding that Δtplus3a-tplus3b/Df(2L)BSC151 mutants have a reduced fertility [39–40]. kl-3 and kl-5 are both transcribed in the spermatocyte Y-loops [41]. Three of these loop structures can be observed in spermatocytes during spermatocyte development [27]. According to our RNA-seq data, kl-3 and kl-5 are regulated by tPlus3a and tPlus3b but not by tBRD-1. This might point to a specific function of tPlus3a and tPlus3b in the regulation of these male fertility factors. Along these lines, co-localization of tBRD-1 and tPlus3a and tPlus3b was only observed in chromosomal regions, whereas speckles of tBRD-1 and distinct spots of tPlus3a and tPlus3b did not overlap.
tPlus3a and tPlus3b and tBRD-1 co-repress heat shock genes in spermatocytes
Our RNA-seq data also suggest a repressor function of tPlus3a and tPlus3b, as many genes were up-regulated in Δtplus3a-tplus3b/Df(2L)BSC151. Strikingly, several genes seemed to be up-regulated also in tbrd-11 mutant testes. One group of genes (Sfp87B, Sfp24Bb, and Sfp60F) encode several seminal fluid proteins (Sfp). These are needed for fertilization and can found in the ejaculate of male flies [42]. Peb (Ebp, Peb-me) and PebII (EbpII), which encode proteins of the ejaculatory bulb, are likewise not expressed in germ cells of wild-type flies. Their proteins are transferred together with sperm in the ejaculate of males to female flies during mating to prevent early remating [43–44]. These protein classes are first needed during fertilization; therefore, it is not surprising that a repression mechanism controls these genes in germ cells. As the general transcription rate is extremely high in the spermatocyte phase, tPlus3a and tPlus3b and tBRD-1 might prevent untimely transcription of sperm fluid genes.
Interestingly, AstCC and CG13428 were upregulated in Δtplus3a-tplus3b/Df(2L)BSC151 but strongly down-regulated in tbrd-11 mutants. This might indicate that these genes are regulated oppositely by tPlus3a and tPlus3b and tBRD-1.
Another gene group repressed by both tPlus3a and tPlus3b and tBRD-1 are heat shock genes. Many Hsp70 genes as well as Hsp67Bc were up-regulated in Δtplus3a-tplus3b/Df(2L)BSC151 and tbrd-11. This finding might explain why primary spermatocytes were reported to poorly respond to heat shock in spermatocytes [45].
tPlus3a and tPlus3b share a large subset of target genes with tBRD-1 but not with tBRD-2
As previously reported, the transcription machinery in spermatocytes is supported by several interactions between various complexes, such as TFIID, tMAC, and Mediator [8]. Recent findings indicate how the specificity for different sets of genes might be achieved by cooperation of general transcription factors with bromodomain proteins [11–12]. Our RNA-seq results for tplus3a-tplus3b-deficient fly testes and tbrd-11 mutant testes suggest a role for Plus3 domain proteins in transcription regulation. Comparative analyses of our Δtplus3a-tplus3b/Df(2L)BSC151 and tbrd-11 RNA-seq datasets with microarray data from tbrd-2 knock-down testis revealed that many genes are co-regulated by tPlus3a and tPlus3b and tBRD-1, whereas only a few genes are commonly regulated by tPlus3a and tPlus3b and tBRD-2. However, a direct comparison of microarray data and RNA-seq data should be handled with caution. A potential cooperation between tPlus3b and tBRD-1 is supported by their heterodimerization in the yeast two-hybrid system. We hypothesize that tPlus3a and tPlus3b proteins either activate or repress sets of target genes. In RNA-seq experiments, we cannot distinguish between direct binding to regulated genes or indirect action by regulation of other activation or repression relevant components. During gene activation tPlus3a and tPlus3b probably also interact with some factors of the general or testis-specific transcription machinery. Indeed, tBRD-1 heterodimerizes with the tTAF Sa [11].
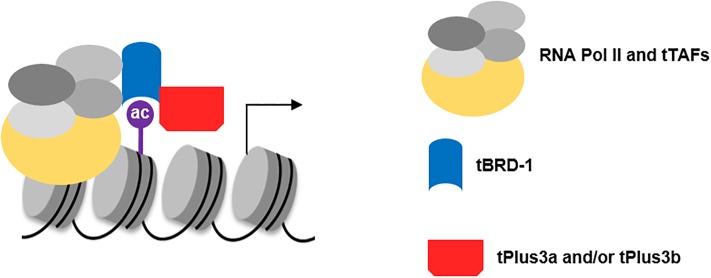
We previously showed that tBRD-1 binds to H3 peptides acetylated at lysines 9 and 14 and to H4 peptides acetylated at lysines 5, 8, and 12, which suggested a direct role of tBRD-1 in gene activation [12]. A small number of genes regulated by tPlus3a and tPlus3b and tBRD-1 and are also regulated by the tTAF Sa [46]. The genes regulated by all three proteins comprise ca. 12% of tPlus3a and tPlus3b-regulated genes, ca. 3% of tBRD-1-regulated genes, and ca. 1% of Sa-regulated genes. During transcription of these genes we propose a direct molecular interaction of Sa with tBRD-1, tBRD-1 with tPlus3a and tPlus3b, and tBRD1 binding to acetylated histones in an RNA Polymerase B containing complex (Fig 6).
Fig 6. Model of the complex formation of tPlus3a and tPlus3b, tBRD-1, and other factors in gene activation.
tPlus3a and tPlus3b molecularly interact with tBRD-1, which binds to acetylated histones. tBRD-1 interacts with the tTAF Sa to activate RNA Polymerase mediated transcription.
Material and methods
Fly strains and cultures
Flies were maintained at 25 °C on standard medium. w1118 or w1118; CyO/Sp; ProtB-eGFP [47] flies were used as the wild-type control. tbrd-11 mutants and flies synthesizing tBRD-1-eGFP [14] were used for RNA-seq and analysis of co-localization with tTPlus3a and tPlus3b, respectively. w1118; Df(2L)BSC151/CyO (Bloomington stock number: 9510) was used as deficiency fly line for tplus3a and tplus3b. Δtplus3a and Δtplus3b (Δtplus3a-tplus3b) mutants were maintained in a y1cho2v1 background, and Δtplus3a-tplus3b/Df(2L)BSC151 trans-heterozygotes were made in a w1118 background. w1118; Δtplus3a-tplus3b/Df(2L)BSC151; ProtB-eGFP was used for visualization of nuclei phenotypic analyses.
Generation of tplus3b-eGFP flies
The tplus3b-eGFP fusion construct was generated by PCR amplification of the tplus3b putative promoter region, 5’-UTR, and open reading frame (ORF) on w1118 genomic DNA with primers 5’-GGTACCTCGTGTAGCTCTTTTTAGC-3’ and 5’-ACTAGTGCCTTTCATTATCTCGTCCC-3’. The PCR product was subcloned into the pCRII-TOPO vector (Invitrogen). The recombinant plasmid was digested with KpnI and SpeI and ligated into pChabΔsalΔlacZ (modified after [48]), which carries the gene encoding eGFP, yielding a construct with eGFP at the C-terminus. Transgenic flies were established by injection into the w1118 strain.
In situ hybridization
In situ hybridization of w1118 (wild-type), y1cho2v1; Δtplus3a-tplus3b and y1cho2v1; Δtplus3a-tplus3b/CyO was performed as described in Morris et al. [49]. tplus3a-tplus3b RNA probes were synthesized by PCR amplification of 610 bp of the tplus3b ORF with primers 5’-CAAATAAACTGGAGTCTGGTGG-3’ and 5’-TTGGCGATGTCATTAAGCGTTG-3’. The amplified product was cloned into the pCRII-TOPO vector (Invitrogen). Genes were transcribed in vitro for probe synthesis using SP6 and T7 polymerase (Roche) after linearization of the vector with KpnI or EcoRI, respectively. Probes were then precipitated with ethanol and dissolved in hybridization solution [49]. CG12498 and rtf1 In situ hybridization was performed using probes, which are based on constructs synthesized with the respective primers given in S1 Table.
Immunofluorescence staining and antibody generation
Immunofluorescence stainings were performed according to Hime et al. [50] with modifications [51] DNA was stained with Hoechst 33258, and actin components, e.g., the individualization complex, were stained with TRITC-phalloidin. A tPlus3a and tPlus3b peptide antibody was raised against peptide KRTAKPEQHELEKYMRRKY (aa 329 to aa 347 of tPlus3a and tPlus3b) in rabbit, affinity purified (Pineda Antibody Service, http://www.pineda-abservice.de), and diluted 1:500. The secondary antibody anti-rabbit-Cy3 (Dianova) was diluted 1:100. Stainings were analyzed with an AxioPlan2 microscope (Zeiss). Images were processed using Photoshop (Adobe).
RNA-interference and CRISPR/Cas9 mutant generation and analysis
For initial tplus3a-tplus3b functional studies the RNAi line 109981 (P{KK115200}VIE-260B, Vienna Drosophila Resource Center) was applied. For the tplus3 and tplus3b knock down, v109981 males were crossed against virgin females of bam-Gal4/bam-Gal4; Sp/CyO; bam-Gal4-VP16/MKRS (Bam). Crossings for knock down (Bam>>109981) experiments were maintained at 30 °C in parallel with 109981 and Bam controls.
y1cho2v1; Δtplus3a-tplus3b mutants were generated using a single target construct in the CRISPR/Cas9 system as described by Kondo and Ueda [28]. For cloning of the target construct, primers 5’-CTTCGCAGAATGGATGAGCGCTTA-3’ and 5’-AAACTAAGCGCTCATCCATTCTGC-3’ were used, and the product was ligated to pBFv-U6.2. The target construct was injected into y2cho2v1P{nos-phiC31\int.NLS}X;attP2(III), and established transgenic flies were crossed with y2cho2v1;attP40{nos-Cas9}/CyO flies for mutagenesis. Fifty founder flies were crossed with y2cho2v1; Sco/CyO flies to establish potential mutants. Genomic regions next to the target sequence of homozygous offspring were analyzed by PCR and screened for deletions. Flies with tplus3b-eGFP in homozygous Δtplus3a-tplus3b situation were analyzed in fertility assays.
Western blotting
Protein extracts for western blots were prepared from 20 testes of w1118, y1cho2v1; Δtplus3a -tplus3b/CyO and y1cho2v1; Δtplus3a-tplus3b flies. Testes were homogenized with 20 μl 2×SDS sample buffer by sonication for 20 min at 4 °C. Protein extracts were incubated for 5 min at 95 °C and separated on SDS-10% polyacrylamide gels. Western blotting followed standard procedures. Anti-tTPlus3a and tPlus3b was diluted 1:1,000 in 5% dry milk powder/1×TBS. HRP-conjugated anti-rabbit (Jackson Immunology) was diluted 1:1,000 in 5% dry milk powder/1×TBS. Enhanced chemiluminescence (ECL, Invitrogen) was detected on an Odyssey Fc Imaging System (Licor) following the manufacturer’s instructions.
Sterility tests
For sterility tests, individual 1-day (after hatching)-old males (w1118 (wild-type), w1118;Df(2L)BSC151/CyO, y1cho2v1; Δtplus3a-tplus3b/CyO and w1118; Δtplus3a-tplus3b/Df(2L)BSC151) and 5-day (after hatching)-old w1118; Δtplus3a-tplus3b/Df(2L)BSC151 males were placed with two virgin w1118 females in separate vials at 25 °C and were allowed to mate for three days. The final age of the flies was 4 days and 8 days, respectively. Sterility test for tplus3a and tplus3b knock down was performed at 25 °C using males which were grown at 30 °C (Bam, 109981 controls and knock down Bam>>109981). The number of offspring per vial was counted after 2 weeks and the mean is depicted with standard deviation (SD). Data were statistically analyzed using one-way ANOVA with Tukey’s multiple comparison test and Graph Pad Prism Version 5.
Yeast two-hybrid experiments
Yeast two-hybrid experiments were carried out using the Matchmaker GAL4 Two-Hybrid System 3 (Takara Clontech) according to the manufacturer’s instructions. For construct generation, the tplus3b ORF of genomic w1118 DNA was PCR amplified with primers 5’-CCATGGCGATGGATGAGCGCTTAC-3’ and 5’-GTCGACCTAGCCTTTCATTATCTCGTC-3’; the products were cloned into the pCRII-TOPO vector (Invitrogen). The prey vector pGBKT7 was digested with NcoI and SalI, and the bait vector pGADT7 was digested with NcoI and XhoI. The tplus3b ORF was excised from pCRII-TOPO with NcoI and SalI and ligated into pGBKT7 for synthesis of the DBD-tPlus3b fusion protein (fusion with GAL4-DNA-binding domain) and into pGADT7 for synthesis of the AD-tPlus3b fusion protein (fusion with GAL4-activation domain). Yeast two-hybrid constructs for tbrd-1 and tbrd-2 are described in Theofel et al. [11].
RNA isolation and RNA-seq
Total RNA from 200 testes of w1118; tbrd-11 and w1118; Δtplus3a-tplus3b/Df(2L)BSC151 flies was isolated using TRIzol (Invitrogen). DNA was digested with TURBO DNase (Invitrogen) and purified with the RNeasy Mini Kit (Qiagen) according to the manufacturer’s instructions. Transcriptomes were analyzed using next-generation sequencing (RNA-seq) in three replicates. Prior to library preparation, RNA quality was assessed using the Experion RNA StdSens Analysis Kit (BioRad). Libraries were prepared using the TruSeq Stranded mRNA LT Kit (Illumina) according to the manufacturer’s instructions. The quality of libraries was controlled using a Bioanalyzer 2100 and the Agilent High Sensitivity DNA Kit (Agilent). Pooled libraries were quantified with digital PCR (QuantStudio 3D, Thermo Fisher) and sequenced on the HiSeq 1500 platform (Illumina) in rapid-run mode with 50-base single reads. Reads were aligned to the D. melanogaster genome retrieved from Ensembl revision 89 (BDGP6) with STAR 2.4.1a [52]. Default parameters were used, except for outFilterScoreMin and outFilterMatchNmin, which were set to 60% of the read length. For analysis of RNA-seq data, tag counts were calculated and normalized to one million mapped exonic reads and gene length (FPKM). To generate the set of expressed genes, only genes with a minimum read count of 50 and a minimum FPKM of 0.3 were kept. Fold change was calculated between the technical replicates of two conditions. Differential expression was assessed using the log2 of the median fold change. Only genes with an increase or decrease of at least twofold were considered to be differentially expressed.
PCR and qPCR
RNA from 200 testes of w1118; tbrd-11 and w1118; Δtplus3a-tplus3b/Df(2L)BSC151 was isolated using TRIzol (Invitrogen). DNA was digested with TURBO DNase (Invitrogen) and purified with RNeasy Mini Kit (Qiagen) according to the manufacturer’s instructions. RNA (1 μg) was used for reverse transcription with random hexamer primers using the Transcriptor First Strand cDNA Synthesis Kit (Roche). The qPCR reaction (20 μl) contained 10 μl iTaq Universal SYBR Green Supermix (Bio-Rad), 12.5 ng cDNA, and 10 μM gene-specific forward and reverse primers (S1 Table), and was run on an Agilent Stratagene Mx3000P cycler with an annealing temperature of 60 °C. Ct-values for three technical replicates were normalized to the expression level of Rpl32. Depicted is the mean value and standard error (SE) of three replicates using GraphPad PRISM version 5.03. Statistical analysis was calculated with Student’s t-test and Bonferroni correction.
PCR for amplification of flanking genes was performed using primers for the respective genes listed in S1 Table. DNA was isolated from homozygous y1cho2v1; Δtplus3a-tplus3b males via S-Fly PCR.
ArrayExpress accession data
RNA-seq data were deposited in the ArrayExpress [53] database at EMBL-EBI (www.ebi.ac.uk/arrayexpress) under accession number E-MTAB-7013.
Supporting information
(A) Shown are the Plus3 domain of Drosophila Rtf1 (dRtf1), tPlus3a and tPlus3b and CG12498. The three conserved positively charged amino acids that gave the Plus3 domain its name (Plus3) are marked in green and with *, other conserved amino acids are marked in grey. (B) PI domains of human RTF (hRTF), see [21]), Drosophila Rtf1 (dRTF1), and tPlus3a and tPlus3b. The region corresponding to the putative protein interaction domain (PI) is truncated in CG12498. Conserved amino acids in all shown proteins are marked in yellow, those conserved between the Drosophila Plus3 domains are marked in grey, those conserved only between dRtf1 and CG12498 are marked in blue.
(TIFF)
(A) CG12498 transcripts were present mainly in stages before meiotic divisions. (B) Sense control for CG12498. (C) rtf1 transcripts were visualized in spermatocytes and in post-meiotic stages. (D) Sense control for rtf1.
(TIF)
Whole-mount preparations of testis from flies expressed tPlus3b-eGFP (A) in the nucleoplasm of spermatocytes (arrow) and in the nucleolus (arrowhead). (B) tPlus3b-eGFP is expressed in the nucleus of round spermatids. Asterisk marks the hub region Scale bar = 20 μm.
(TIF)
(A B) ProtB-eGFP marked sperm in seminal vesicles of wild-type males. (D, E) ProtB-eGFP marked sperm accumulate in seminal vesicles of Δtplus3a-tplus3b/Df(2L)BSC151 males.
(TIF)
Three biological samples were prepared and analyzed by RNA-seq. Only little variation within an individual genotype was observed. The data for genes, which are transcribed in somatic parts of the reproductive tract are analyzed for loss of repression in the male germ line (A). Transcripts levels increased in mutants; FPKM values between 0 and 50, the w1 data were set to 0. (B) Transcript levels do not increase in mutants; FPKM values between 100 and 4000.
(TIF)
(PDF)
Acknowledgments
We thank Rainer Renkawitz and Christian Bökel for helpful discussions and support, and Igor Macinkovic, Stephan Awe, and Julia Seiz for help with qPCR experiments and evaluation. We thank Christine Dottermusch-Heidel for help with in situ hybridizations for S2 Fig, Ruth Hyland, Ljubinka Cigoja, and Sabina Huhn for excellent technical assistance, and Katja Gessner for support with graphic programs. We thank Karen A. Brune for linguistic revision of the manuscript. We are grateful to Ryu Ueda and the NIG Japan for sending us the CRISPR/Cas9 flies.
Data Availability
All RNAseq data were deposited in the ArrayExpress database at EMBL-EBI (www.ebi.ac.uk/arrayexpress) under accession number E-MTAB-7013.
Funding Statement
This work was supported by the Deutsche Forschungsgemeinschaft (SFB TRR81 "Chromatin Changes in Differentiation and Malignancies" to AB, CR, and RR-P).
References
- 1.Olivieri G, Olivieri A. Autoradiographic study of nucleic acid synthesis during spermatogenesis in Drosophila melanogaster. Mutat Res. 1965. August;2(4):366–80. [DOI] [PubMed] [Google Scholar]
- 2.White-Cooper H, Schäfer MA, Alphey LS, Fuller MT. Transcriptional and post-transcriptional control mechanisms coordinate the onset of spermatid differentiation with meiosis I in Drosophila. Development. 1998. January;125(1):125–34. [DOI] [PubMed] [Google Scholar]
- 3.White-Cooper H, Davidson I. Unique aspects of transcription regulation in male germ cells. Cold Spring Harb Perspect Biol. 2011. July 1;3(7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rathke C, Baarends WM, Awe S, Renkawitz-Pohl R. Chromatin dynamics during spermiogenesis. Biochim Biophys Acta. 2014. March;1839(3):155–68. 10.1016/j.bbagrm.2013.08.004 [DOI] [PubMed] [Google Scholar]
- 5.Chintapalli VR, Wang J, Dow JAT. Using FlyAtlas to identify better Drosophila models of human disease. Nat Genet. 2007. June;39(6):715–20. 10.1038/ng2049 [DOI] [PubMed] [Google Scholar]
- 6.Hiller MA, Lin TY, Wood C, Fuller MT. Developmental regulation of transcription by a tissue-specific TAF homolog. Genes Dev. 2001. April 15;15(8):1021–30. 10.1101/gad.869101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hiller MA, Chen X, Pringle MJ, Suchorolski M, Sancak Y, Viswanathan S, et al. Testis-specific TAF homologs collaborate to control a tissue-specific transcription program. Development. 2004. November;131(21):5297–308. 10.1242/dev.01314 [DOI] [PubMed] [Google Scholar]
- 8.Lu C, Fuller MT. Recruitment of mediator complex by cell type and stage-specific factors required for tissue-specific TAF dependent gene activation in an adult stem cell lineage. PLoS Genet. 2015. December 1;11(12):e1005701 10.1371/journal.pgen.1005701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beall EL, Lewis PW, Bell M, Rocha M, Jones DL, Botchan MR. Discovery of tMAC: a Drosophila testis-specific meiotic arrest complex paralogous to Myb-Muv B. Genes Dev. 2007. April 15;21(8):904–19. 10.1101/gad.1516607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barckmann B, Chen X, Kaiser S, Jayaramaiah-Raja S, Rathke C, Dottermusch-Heidel C et al. Three levels of regulation lead to protamine and Mst77F expression in Drosophila. Dev Biol. 2013. May 1;377(1):33–45. 10.1016/j.ydbio.2013.02.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Theofel I, Bartkuhn M, Hundertmark T, Boettger T, Gärtner SM, Leser K, et al. tBRD-1 selectively controls gene activity in the Drosophila testis and interacts with two new members of the bromodomain and extra-terminal (BET) family. PLoS One. 2014. September 24;9(9):e108267 10.1371/journal.pone.0108267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Theofel I, Bartkuhn M, Boettger T, Gärtner SMK, Kreher J, Brehm A et al. tBRD-1 and tBRD-2 regulate expression of genes necessary for spermatid differentiation. Biol Open. 2017. April 15;6(4):439–448. 10.1242/bio.022467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dhalluin C, Carlson JE, Zeng L, He C, Aggarwal AK, Zhou M-M. Structure and ligand of a histone acetyltransferase bromodomain. Nature. 1999. June 3;399(6735):491–6. 10.1038/20974 [DOI] [PubMed] [Google Scholar]
- 14.Leser K, Awe S, Barckmann B, Renkawitz-Pohl R, Rathke C. The bromodomain-containing protein tBRD-1 is specifically expressed in spermatocytes and is essential for male fertility. Biol Open. 2012. June 15;1(6):597–606. 10.1242/bio.20121255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tomson BN, Arndt KM. The many roles of the conserved eukaryotic Paf1 complex in regulating transcription, histone modifications, and disease states. Biochim Biophys Acta. 2013. January;1829(1):116–26. 10.1016/j.bbagrm.2012.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wade PA, Werel W, Fentzke RC, Thompson NE, Leykam JF, Burgess RR, et al. A novel collection of accessory factors associated with yeast RNA polymerase II. Protein Expr Purif. 1996. August;8(1):85–90. 10.1006/prep.1996.0077 [DOI] [PubMed] [Google Scholar]
- 17.Kim J, Guermah M, Roeder RG. The human PAF1 complex acts in chromatin transcription elongation both independently and cooperatively with SII/TFIIS. Cell. 2010. February 19;140(4):491–503. 10.1016/j.cell.2009.12.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van Oss SB, Cucinotta CE, Arndt KM. Emerging insights into the roles of the Paf1 Complex in gene regulation. Trends Biochem Sci. 2017. October;42(10):788–798. 10.1016/j.tibs.2017.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rozenblatt-Rosen O, Hughes CM, Nannepaga SJ, Shanmugam KS, Copeland TD, Guszczynski T, et al. The parafibromin tumor suppressor protein is part of a human Paf1 complex. Mol Cell Biol. 2005. January;25(2):612–20. 10.1128/MCB.25.2.612-620.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Adelman K, Wei W, Ardehali MB, Werner J, Zhu B, Reinberg D, et al. Drosophila Paf1 modulates chromatin structure at actively transcribed genes. Mol Cell Biol. 2006. January;26(1):250–60. 10.1128/MCB.26.1.250-260.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cao QF, Yamamoto J, Isobe T, Tateno S, Murase Y, Chen Y, et al. Characterization of the Human Transcription Elongation Factor Rtf1: Evidence for Nonoverlapping Functions of Rtf1 and the Paf1 Complex. Mol Cell Biol. 2015. October;35(20):3459–70. 10.1128/MCB.00601-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Warner MH, Roinick KL, Arndt KM. Rtf1 is a multifunctional component of the Paf1 complex that regulates gene expression by directing cotranscriptional histone modification. Mol Cell Biol. 2007. September;27(17):6103–15. 10.1128/MCB.00772-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.De Jong RN, Truffault V, Diercks T, Eiso AB, Daniels MA, Kaptein R, et al. Structure and DNA Binding of the Human Rtf1 Plus3 Domain. Structure. 2008. January;16(1):149–59. 10.1016/j.str.2007.10.018 [DOI] [PubMed] [Google Scholar]
- 24.Gärtner S, Hundertmark T, Nolte H, Theofel I, Eren-Ghiani Z, Tetzner C et al. Stage-specific testes proteomics of Drosophila melanogaster identifies essential proteins for male fertility. EJCB 10.1016/j.ejcb.2019.01.001 [DOI] [PubMed] [Google Scholar]
- 25.Brown JB, Boley N, Eisman R, May GE, Stoiber MH, Duff MO, et al. Diversity and dynamics of the Drosophila transcriptome. Nature. 2014. August 28;512(7515):393–9. 10.1038/nature12962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011. October 11;7:539 10.1038/msb.2011.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bonaccorsi S, Pisano C, Puoti F, Gatti M. Y chromosome loops in Drosophila melanogaster. Genetics. 1988. December;120(4):1015–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kondo S, Ueda R. Highly Improved Gene Targeting by Germline-Specific Cas9 Expression in Drosophila. Genetics. 2013. November;195(3):715–21. 10.1534/genetics.113.156737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Adams EM, Wolfner MF. Seminal proteins but not sperm induce morphological changes in the Drosophila melanogaster female reproductive tract during sperm storage. J Insect Physiol. 2007. April;53(4):319–31. 10.1016/j.jinsphys.2006.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kubli E. Sex-peptides: seminal peptides of the Drosophila male. Cell Mol Life Sci. 2003. August;60(8):1689–704. 10.1007/s00018-003-3052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Simmerl E, Schäfer M, Schäfer U. Structure and regulation of a gene cluster for male accessory gland transcripts in Drosophila melanogaster. Insect Biochem Mol Biol. 1995. January;25(1):127–37. [DOI] [PubMed] [Google Scholar]
- 32.Walker MJ, Rylett CM, Keen JN, Audsley N, Sajid M, Shirras AD et al. Proteomic identification of Drosophila melanogaster male accessory gland proteins, including a pro-cathepsin and a soluble gamma-glutamyl transpeptidase. Proteome Sci. 2006. May 2;4:9 10.1186/1477-5956-4-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jiang M, Gao Z, Wang J, Nurminsky DI. Evidence for a hierarchical transcriptional circuit in Drosophila male germline involving testis-specific TAF and two gene-specific transcription factors, Mod and Acj6. FEBS Lett. 2018. January;592(1):46–59. 10.1002/1873-3468.12937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Doggett K, Jiang J, Aleti G, White-Cooper H. Wake-up-call, a lin-52 paralogue, and Always early, a lin-9 homologue physically interact, but have opposing functions in regulating testis-specific gene expression. Dev Biol. 2011. July 15;355(2):381–93. 10.1016/j.ydbio.2011.04.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goldstein LS, Hardy RW, Lindsley DL. Structural genes on the Y chromosome of Drosophila melanogaster. Proc Natl Acad Sci U S A. 1982. December;79(23):7405–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gepner J, Hays TS. A fertility region on the Y chromosome of Drosophila melanogaster encodes a dynein microtubule motor. Proc Natl Acad Sci U S A. 1993. December 1;90(23):11132–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Carvalho AB, Dobo BA, Vibranovski MD, Clark AG. Identification of five new genes on the Y chromosome of Drosophila melanogaster. Proc Natl Acad Sci U S A. 2001. November 6;98(23):13225–30. 10.1073/pnas.231484998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hardy RW, Tokuyasu KT, Lindsley DL. Analysis of spermatogenesis in Drosophila melanogaster bearing deletions for Y-chromosome fertility genes. Chromosoma. 1981;83(5):593–617. [DOI] [PubMed] [Google Scholar]
- 39.Kennison JA. The Genetic and Cytological Organization of the Y Chromosome of Drosophila melanogaster. Genetics. 1981. July;98(3):529–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang P, Stankiewicz RL. Y-linked male sterile mutations induced by P-Element in Drosophila melanogaster. Genetics. 1998. October;150(2):735–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ceprani F, Raffa GD, Petrucci R, Piergentili R. Autosomal mutations affecting Y chromosome loops in Drosophila melanogaster. BMC Genet. 2008. April 11;9:32 10.1186/1471-2156-9-32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Findlay GD, Yi X, MacCoss MJ, Swanson WJ. Proteomics reveals novel Drosophila seminal fluid proteins transferred at mating. PLoS Biol. 2008. July 29;6(7):e178 10.1371/journal.pbio.0060178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bretman A, Lawniczak MK, Boone J, Chapman T. A mating plug protein reduces early female remating in Drosophila melanogaster. J Insect Physiol. 2010. January;56(1):107–13. 10.1016/j.jinsphys.2009.09.010 [DOI] [PubMed] [Google Scholar]
- 44.Avila FW, Cohen AB, Ameerudeen FS, Duneau D, Suresh S, Mattei AL, et al. Retention of ejaculate by Drosophila melanogaster females requires the male-derived mating plug protein PEBme. Genetics. 2015. August;200(4):1171–9. 10.1534/genetics.115.176669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bendena WG, Ayme-Southgate A, Garbe JC, Pardue ML. Expression of heat-shock locus hsr-omega in nonstressed cells during development in Drosophila melanogaster. Dev Biol. 1991. March;144(1):65–77. [DOI] [PubMed] [Google Scholar]
- 46.Lu C, Kim J, Fuller MT. The polyubiquitin gene Ubi-p63E is essential for male meiotic cell cycle progression and germ cell differentiation in Drosophila. Development. 2013. September;140(17):3522–31. 10.1242/dev.098947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jayaramaiah Raja S, Renkawitz-Pohl R. Replacement by Drosophila melanogaster protamines and Mst77F of histones during chromatin condensation in late spermatids and role of sesame in the removal of these proteins from the male pronucleus. Mol Cell Biol. 2005. July;25(14):6165–77. 10.1128/MCB.25.14.6165-6177.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thummel CS, Boulet AM, Lipshitz HD. Vectors for Drosophila P-element-mediated transformation and tissue culture transfection. Gene. 1988. December 30;74(2):445–56. [DOI] [PubMed] [Google Scholar]
- 49.Morris CA, Benson E, White-Cooper H. Determination of gene expression patterns using in situ hybridization to Drosophila testes. Nat Protoc. 2009;4(12):1807–19. 10.1038/nprot.2009.192 [DOI] [PubMed] [Google Scholar]
- 50.Hime GR, Brill JA, Fuller MT. Assembly of ring canals in the male germ line from structural components of the contractile ring. J Cell Sci. 1996. December;109 (Pt 12):2779–88. [DOI] [PubMed] [Google Scholar]
- 51.Hundertmark T, Theofel I, Eren-Ghiani Z, Miller D, Rathke C. Analysis of chromatin dynamics during Drosophila spermatogenesis. Methods Mol Biol. 2017;1471:289–303. 10.1007/978-1-4939-6340-9_17 [DOI] [PubMed] [Google Scholar]
- 52.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013. January 1;29(1):15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kolesnikov N, Hastings E, Keays M, Melnichuk OY, Tang A, Williams E, et al. ArrayExpress update—simplifying data submissions. Nucleic Acids Res. 2015. January;43(Database issue):D1113–6. 10.1093/nar/gku1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Shown are the Plus3 domain of Drosophila Rtf1 (dRtf1), tPlus3a and tPlus3b and CG12498. The three conserved positively charged amino acids that gave the Plus3 domain its name (Plus3) are marked in green and with *, other conserved amino acids are marked in grey. (B) PI domains of human RTF (hRTF), see [21]), Drosophila Rtf1 (dRTF1), and tPlus3a and tPlus3b. The region corresponding to the putative protein interaction domain (PI) is truncated in CG12498. Conserved amino acids in all shown proteins are marked in yellow, those conserved between the Drosophila Plus3 domains are marked in grey, those conserved only between dRtf1 and CG12498 are marked in blue.
(TIFF)
(A) CG12498 transcripts were present mainly in stages before meiotic divisions. (B) Sense control for CG12498. (C) rtf1 transcripts were visualized in spermatocytes and in post-meiotic stages. (D) Sense control for rtf1.
(TIF)
Whole-mount preparations of testis from flies expressed tPlus3b-eGFP (A) in the nucleoplasm of spermatocytes (arrow) and in the nucleolus (arrowhead). (B) tPlus3b-eGFP is expressed in the nucleus of round spermatids. Asterisk marks the hub region Scale bar = 20 μm.
(TIF)
(A B) ProtB-eGFP marked sperm in seminal vesicles of wild-type males. (D, E) ProtB-eGFP marked sperm accumulate in seminal vesicles of Δtplus3a-tplus3b/Df(2L)BSC151 males.
(TIF)
Three biological samples were prepared and analyzed by RNA-seq. Only little variation within an individual genotype was observed. The data for genes, which are transcribed in somatic parts of the reproductive tract are analyzed for loss of repression in the male germ line (A). Transcripts levels increased in mutants; FPKM values between 0 and 50, the w1 data were set to 0. (B) Transcript levels do not increase in mutants; FPKM values between 100 and 4000.
(TIF)
(PDF)
Data Availability Statement
All RNAseq data were deposited in the ArrayExpress database at EMBL-EBI (www.ebi.ac.uk/arrayexpress) under accession number E-MTAB-7013.