Abstract
Bumblebees (tribe Bombini, genus Bombus Latreille) play a pivotal role as pollinators in mountain regions for both native plants and for agricultural systems. In our survey of northern Thailand, four species of bumblebees (Bombus (Megabombus) montivagus Smith, B. (Alpigenobombus) breviceps Smith, B. (Orientalibombus) haemorrhoidalis Smith and B. (Melanobombus) eximius Smith), were present in 11 localities in 4 provinces (Chiang Mai, Mae Hong Son, Chiang Rai and Nan). We collected and screened 280 foraging worker bumblebees for microsporidia (Nosema spp.) and trypanosomes (Crithidia spp.). Our study is the first to demonstrate the parasite infection in bumblebees in northern Thailand. We found N. ceranae in B. montivagus (5.35%), B. haemorrhoidalis (4.76%), and B. breviceps (14.28%) and N. bombi in B. montivagus (14.28%), B. haemorrhoidalis (11.64%), and B. breviceps (28.257%).
Introduction
Bumblebees (tribe Bombini, genus Bombus Latreille) play a vitally important role as native pollinators in temperate agricultural ecosystems [1–5]. They are especially important in mountain ecosystems [6] and may be better pollinators than honey bees for many plant species in these areas [7]. Because of this, some species of bumblebees have been employed commercially, especially in greenhouses [3]. From the 1980s onwards, they have been used commercially in greenhouses to pollinate tomatoes, eggplants, and strawberries and also for fruit trees [3, 8]. Several species have been used commercially around the world, including Bombus terrestris, B. lucorum, B. occidentalis, B. ignitus and B. impatiens [3, 9, 10]. Some bumblebees species (B. terrestris, B. ruderatus, B. hortorum, and B. subterraneus) had been released in New Zealand for targeted pollination in the 19th century [11]. Among species used commercially, the most frequent are B. terrestris in Europe and B. impatiens in North America [3]. The identification of bumblebee species has been difficult because the colour patterns can be highly variable within species and convergent among species [12].
In recent years, molecular approaches have been applied for bumblebee identification using particularly a mitochondrial gene (cytochrome oxidase I (COI)) [7]. COI barcodes provide an easily obtained, dependable and cost-effective solution, especially for morphologically cryptic species [13]. Consequently, the COI gene has been used to re-evaluate species, to estimate phylogenetic relationships and to clarify species complexes in Asian bumblebees [14–18].
Similar to Apis bees, bumblebee populations are affected by a number of pathogens and parasites [19]. Crithidia bombi (Trypanosomatidae) and Nosema bombi are the most common. They are transmitted both horizontally between and vertically within colonies of their hosts [20]. Nosema bombi (Microsporidia: Nosematidae) is an obligate intracellular microsporidian parasite infecting a wide range of bumblebee species [20–24]. It is the most widespread bumblebee pathogen worldwide. Thorp (2005) suggested that N. bombi, known to infect European Bombus species [25], may have invaded North American species [25]. Imhoof et al. (1999) showed that prevalence of N. bombi was significantly higher in two declining species, B. pensylvanicus and B. occidentalis, than in other species [26]. In addition, Nosema cerana and C. bombi are associated with declining populations of bumble bees in China [27].
In this paper, we aim to study the diversity of native bumblebees in northern Thailand and to report the prevalence of microsporidians and trypanosomes parasitizing bumblebee populations in Thailand.
Materials and methods
The sample locations for which specific permission was not required and bumblebee did not involve endangered or protected species.
Collection and sample preparation
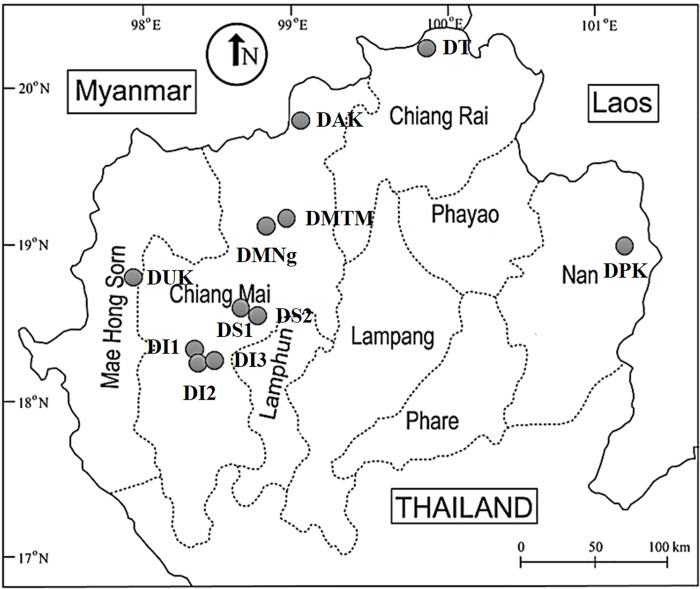
Foraging bumblebees were collected with sweep nets and as random samples from seven sites in four provinces in northern Thailand (Chiang Mai, Mae Hong Son, Chiang Rai and Nan province) in 2015 & 2016 (Table 1). After capture, they were transferred directly into RNA later Solution and stored at -20°C prior to DNA extraction. The following information was recorded for each specimen: GPS coordinates, elevation, collection-site name, and date. The samples were later analyzed in the laboratory. The exact locations are listed in Table 1 and shown in Fig 1. Bumblebee taxa were identified using an updated version of the morphological characters of Williams (2010) [28].
Table 1. Prevalence of four parasites recovered from Bombus species in northern Thailand.
| Province population | Code Name | Elevation | Latitude N | Longitude E | N Bees collected | Prevalence of parasites (%) | |||
|---|---|---|---|---|---|---|---|---|---|
| N. apis | N. ceranae | N. bombi | C. bombi | ||||||
| CHIANG MAI | |||||||||
| Doi Suthep 1 | DS1 | 1,378 | 18°48′55" | 98°55′13" | 60 | 0.00 | 3.33 | 10.00 | 0 |
| Doi Suthep 2 | DS2 | 1,378 | 18°48′55" | 98°55′13" | 20 | 0 | 5.00 | 15.00 | 0 |
| Doi Inthanon 1 | DI1 | 2,118 | 18°33′11" | 98°28′55" | 25 | 0 | 0 | 12.00 | 0 |
| Doi Inthanon 2 | DI2 | 1,297 | 18°32′41" | 98°30′58" | 40 | 0 | 7.50 | 20.00 | 0 |
| Doi Inthanon 3 | DI3 | 1,070 | 18°32′38" | 98°32′53" | 40 | 0 | 12.50 | 15.00 | 0 |
| Doi Mae Tha Man | DMTM | 1,610 | 19°31′35" | 98°83′26" | 5 | 0 | 0 | 20.00 | 0 |
| Doi Ang Khang | DAK | 1,410 | 19°54′8" | 99°2′24" | 25 | 0 | 4.00 | 8.00 | 0 |
| Doi Mon Ngao | DMNg | 930 | 19°10′60" | 99°48′35" | 20 | 0 | 0 | 15.00 | 0 |
| MAE HONG SON | |||||||||
| Doi Mae U Kho | DUK | 1,509 | 18°53′41" | 98°05′21" | 20 | 0 | 10.00 | 20.00 | 0 |
| CHIANG RAI | |||||||||
| Doi Thong | DT | 960 | 20°17′18" | 99°48′35" | 20 | 0 | 5.00 | 20.00 | 0 |
| Nan | |||||||||
| Doi Phu Kha | DPK | 1,980 | 19°12′20″ | 101°40′50" | 5 | 0 | 20.00 | 0 | 0 |
| Total | 280 | 0 | 5.71 | 13.57 | 0 | ||||
Fig 1. Map of the collection sites (grey dots) of native bumblebees in northern Thailand.
Code name are abbreviated as following: DS = Doi Suthep, DI = Doi Inthanon, DMTM = Doi Mae Thaman, DAK = Doi Ang Khang, DMNg = Doi Mon Ngao, DUK = Doi Mae U Kho, DT = Doi Thong, DPK = Doi Phu Kha.
DNA extraction, mitochondrial cytochrome oxidase 1 (COI) gene sequence amplification
DNA extraction was achieved using a single crushed mid leg from each of the bumblebees. For most specimens, legs were ground in a 0.5-mL oxygen tube in liquid nitrogen using a stainless steel pestle, a Proteinase K Digestion kit was used, and the DNA was extracted following a standard phenol-chloroform protocol [29]. DNA extracts were kept at -20°C until needed as a DNA template for the PCR (polymerase chain reaction). The PCR products of the mitochondrial COI (~685 base pairs) sequence were conducted using the universal primers LCO1490 and HC02198 [30]. The PCR amplification was performed in a total volume of 25 μL containing 2 μL of DNA extract, 12.5 pM of each primer, 0.2 mM of each dNTP, 0.2 mM MgCl2, 1X reaction buffer and 2.5 units of Taq DNA polymerase (Invitrogen) under the following thermal conditions: 94°C for 1 min, 5 cycles of 94°C for 1 min, 50°C for 1.5 min, 72°C for 1 min; 35 cycles of 94°C for 1 min, 50°C for 1.5 min, 72°C for 1 min and final step 72°C for 5 min. Amplicons were checked on 1% agarose gels stained with ethidium bromide under UV light. PCR products were purified using PureLink Quick PCR Purification Kit (Invitrogen, Lithuania, USA) following the manufacturer's protocol. The purified PCR products were sequenced. Sequencing reactions were performed, and the sequences were automatically determined in a genetic analyzer (1st Base, Selangor, Malaysia) using PCR primers mentioned above.
DNA Isolation and PCR Detection for pathogen/parasite
The abdomens of 280 individual bumblebees were removed with scissors and individually homogenized in 100 μL of Krebs-Ringer solution with a sterile Eppendorf tube. Total genomic DNA was extracted from 50 μL of the homogenate of each abdomen using a DNA purification kit (PureLink Genomic DNA Mini Kit (Invitrogen)). DNA samples were stored at -20°C prior to molecular screening for parasites. Primers used for detection of N. ceranae, N. apis, N. bombi and C. bombi are listed in Table 2. The PCR amplification was performed in a total volume of 25 μL containing 2 μL DNA extract, 12.5 pM of each primer, 0.2 mM of each dNTP, 0.2 mM MgCl2, 1X reaction buffer and 2.5 unit of Taq DNA polymerase (Invitrogen). Amplification used thermal cycling profiles: initial DNA denaturation step of 4 min at 94°C followed by 40 cycles of 30s at 94°C, 30s at 56°C, and 1 min at 72°C, and terminated with a final extension step of 72°C for 10 min. For each run of the PCR reaction, negative (water) and positive (previously identified positive sample) controls were run along with DNA extracts of the samples. PCR products were electrophoresed on 1.2% agarose gels with ethidium bromide and visualized under UV light. Some of the PCR-amplified bands were purified with PureLink Quick PCR Purification Kit (Invitrogen, Lithuania, USA) following the manufacturer's protocol. After the sequencing reactions the sequences were determined automatically in a genetic analyzer (1st Base, Selangor, Malaysia) using the PCR primers mentioned above. The DNA sequences were used for estimating phylogenetic trees.
Table 2. Primers used for pathogen/parasite and mtDNA detection.
| Primer | Sequence 5′-3′ | Amplification target | Size (bp) | Reference |
|---|---|---|---|---|
| RPS5-F | AATTATTTGGTCGCTGGAATTG | Ribosomal protein S5 (reference gene) | Evans (2006)[31] | |
| RPS5-R | TAACGTCCAGCAGAATGTGGTA | |||
| LCO1490 | GGTCAACAAATCATAAAGATATTGG | mtDNA | 685 | Folmer et al. (1994)[30] |
| HCO2198 | TAAACTTCAGGGTGACCAAAAAATCA | |||
| Crith-F | GGAAACCACGGAATCACATAGACC | Crithidia (Trypanosome) | 500 | Li et al. (2012)[32] |
| Crith-R | AGGAAGCCAAGTCATCCATCGC | |||
| Napis-SSU-Jf1 | CCATGCATGTCTTTGACGTACTATG | N.apis (Microsporidium) | 325 | Klee et al. (2007)[33] |
| Napis-SSU-Jr1 | GCTCACATACGTTTAAAATG | |||
| NOS-FOR | TGCCGACGATGTGATATGAG | N.ceranae (Microsporidium) | 252 | Higes et al. (2006)[34] |
| NOS-REV | CACAGCATCCATTGAAAACG | |||
| Nbombi-SSU-Jf1 | CCATGCATGTTTTTGAAGATTATTAT | N. bombi (Microsporidium) | 323 | Klee et al. (2007)[33] |
| Nbombi-SSU-Jr1 | CATATATTTTTAAAATATGAAACAATAA |
Data analysis
Sequences were checked manually and aligned using the BioEdit (version v7.2.6; http://www.mbio.ncsu.edu/BioEdit/BioEdit.html, accessed 2017), and the primers removed from both ends (Table 2). The sequences were aligned using ClustalW and the alignments were refined by visual inspection. Sequences were used to query GenBank via the BLAST program (https://blast.ncbi.nlm.nih.gov/Blast.cgi). All covering DNA cytochrome oxidase I (COI) region and Nosema parasites sequences obtained in this study can be accessed as NCBI GenBank entries (http://www.ncbi.nlm.nih.gov; bumblebee species accession number MF582589—MF582628; Nosema parasites accession number MF776532-MF776567).
For phylogenetic analysis, multiple alignments of sequences determined in this study and reference sequences obtained from databases were taken together in the calculations of levels of sequence similarity using ClustalX2 program [35], with arithmetic averages tree-making algorithms taken from the MEGA package version 7 [36]. The topologies of the maximum likelihood phylogenetic trees were evaluated based on bootstrap analyses of 1,000 replicates.
Results
Geographical distribution
Samples were collected from Chiang Mai, Mae Hong Son, Chiang Rai and Nan province, at an elevation range of 700‒2,200 m. (sample site; Fig 1, Table 1 and Table 3).
Table 3. A list of Bombus subgenera with information on distribution and species number.
| Subgenus | Distribution | Species | No. sampled |
|---|---|---|---|
| Alpigenobombus | DS1. DS2 DI1, DI2, DI3 | B. breviceps | 28 |
| Megabombus | DS1, DI2, DI3, DAK, DUK | B. montivagus | 56 |
| Melanobombus | DI1 | B. eximius | 7 |
| Orientalibombus | DS1,DS2, DI2, DT, DAK, DMNg, DPK | B. haemorrhoidalis | 189 |
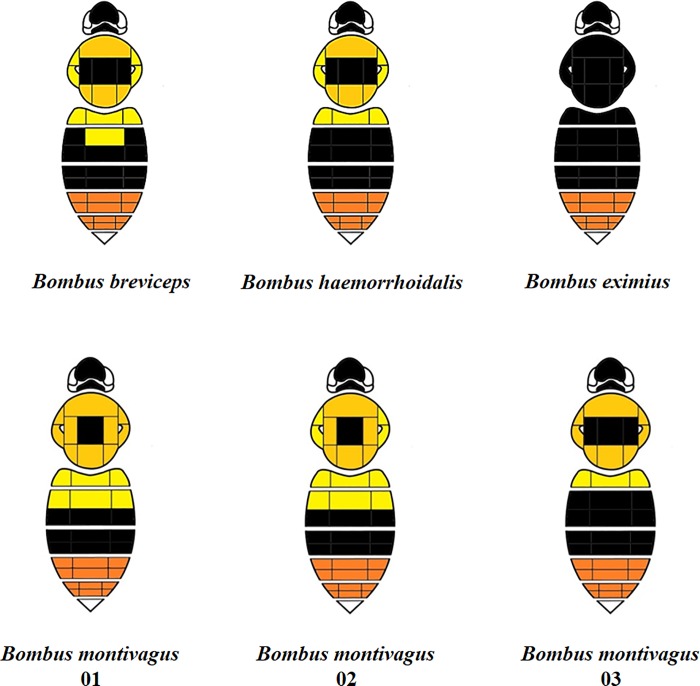
Our study of bumblebees in northern Thailand included 280 female bumblebees. Many of the bumblebees’ colour patterns were similar among species within northern Thailand. The dominant colour of the 6th abdominal segment was red in all of the specimens. Of B. montivagus, three distinct colour patterns were collected (Fig 2). In this study, similar colour patterns to those of B. montivagus were observed in co-occurring species, B. haemorrhoidalis and B. breviceps. The colour pattern of the thoracic pubescence of the workers was primarily orange. In B. breviceps, B. haemorrhoidalis, and B. montivagus, the described orange colour pattern runs anterior to posterior on the notum of the thorax. However, some species have extensive black hair on the thorax, ranging from a small patch in the center of the thorax to a transverse band between the tegulae (above the wing bases), or (in the case of B. eximius) the entire thorax. The sides of the thorax are orange or yellow in all species except B. eximius.
Fig 2. Species identification guide with simplified colour patterns of female workers.
The dorsum of the body is artificially divided into an arbitrary set of regions.
COI-sequence-based analyses
DNA was extracted and the COI gene sequence was amplified successfully from 40 individual bumblebee specimens from 11 localities. All of the sequences were 658 base pairs long after removing the primer from both ends. We found a strong A+T bias in the COI gene barcoding from mtDNA. All new sequences have been deposited in GenBank and are accessible via the sequence numbers MF582589‒MF582628 (Table 4).
Table 4. Material used in the phylogenetic analysis with the sample localities, collector, COI sequence length, depository and GenBank accession number.
| Species | Sample name | Sample locality | Collector | Latitude | Longitude | Sequence length (bp) | GenBank acc. no. |
|---|---|---|---|---|---|---|---|
| Montivagus | DS1-B01 | TH, Doi Su Thep CMP | C. Sinpoo | 18°48′55" | 98°55′13" | 658 | MF582589 |
| haemorrhoidalis | DS1-B16 | TH, Doi Su Thep CMP | C. Sinpoo | 18°48′55" | 98°55′13" | 658 | MF582590 |
| haemorrhoidalis | DS1-B21 | TH, Doi Su Thep CMP | C. Sinpoo | 18°48′55" | 98°55′13" | 658 | MF582591 |
| haemorrhoidalis | DS1-B41 | TH, Doi Su Thep CMP | C. Sinpoo | 18°48′55" | 98°55′13" | 658 | MF582592 |
| montivagus | DI2-B06 | TH, Doi Inthanon CMP | C. Sinpoo | 18°32′41" | 98°30′58" | 658 | MF582593 |
| haemorrhoidalis | DI2-B16 | TH, Doi Inthanon CMP | C. Sinpoo | 18°32′41" | 98°30′58" | 658 | MF582594 |
| haemorrhoidalis | DI2-B31 | TH, Doi Inthanon CMP | C. Sinpoo | 18°32′41" | 98°30′58" | 658 | MF582595 |
| montivagus | DI3-B11 | TH, Doi Inthanon CMP | C. Sinpoo | 18°32′38" | 98°32′53" | 658 | MF582596 |
| montivagus | DI3-B21 | TH, Doi Inthanon CMP | C. Sinpoo | 18°32′38" | 98°32′53" | 658 | MF582597 |
| breviceps | DI3-B27 | TH, Doi Inthanon CMP | C. Sinpoo | 18°32′38" | 98°32′53" | 658 | MF582598 |
| haemorrhoidalis | DMNg-B01 | TH, Doi Mon Ngao CMP | C. Sinpoo | 19°10′60" | 99°48′35" | 658 | MF582599 |
| haemorrhoidalis | DMNg-B11 | TH, Doi Mon Ngao CMP | C. Sinpoo | 19°10′60" | 99°48′35" | 658 | MF582600 |
| haemorrhoidalis | DAK-B01 | TH, Doi Ang Khang CMP | C. Sinpoo | 19°54′8" | 99°2′24" | 658 | MF582601 |
| haemorrhoidalis | DAK-B14 | TH, Doi Ang Khang CMP | C. Sinpoo | 19°54′8" | 99°2′24" | 658 | MF582602 |
| montivagus | DAK-B05 | TH, Doi Ang Khang CMP | C. Sinpoo | 19°54′8" | 99°2′24" | 658 | MF582603 |
| haemorrhoidalis | DAK-B12 | TH, Doi Ang Khang CMP | C. Sinpoo | 19°54′8" | 99°2′24" | 658 | MF582604 |
| montivagus | DAK-B22 | TH, Doi Ang Khang CMP | C. Sinpoo | 19°54′8" | 99°2′24" | 658 | MF582605 |
| haemorrhoidalis | DAK-B06 | TH, Doi Ang Khang CMP | C. Sinpoo | 19°54′8" | 99°2′24" | 658 | MF582606 |
| haemorrhoidalis | DAK-B10 | TH, Doi Ang Khang CMP | C. Sinpoo | 19°54′8" | 99°2′24" | 658 | MF582607 |
| montivagus | DUK-B01 | TH, Doi Mae U Kho MHP | C. Sinpoo | 18°53′41" | 98°05′21" | 658 | MF582608 |
| montivagus | DUK-B08 | TH, Doi Mae U Kho MHP | C. Sinpoo | 18°53′41" | 98°05′21" | 658 | MF582609 |
| haemorrhoidalis | DT-B01 | TH, Doi Thong CRP | C. Sinpoo | 20°17′18" | 99°48′35" | 658 | MF582610 |
| haemorrhoidalis | DT-B04 | TH, Doi Thong CRP | C. Sinpoo | 20°17′18" | 99°48′35" | 658 | MF582611 |
| breviceps | DI2-B20 | TH, Doi Inthanon CMP | C. Sinpoo | 18°32′41" | 98°30′58" | 658 | MF582612 |
| breviceps | DI3-B30 | TH, Doi Inthanon CMP | C. Sinpoo | 18°32′38" | 98°32′53" | 658 | MF582613 |
| haemorrhoidalis | DS2-B01 | TH, Doi Su Thep CMP | C. Sinpoo | 18°48′55" | 98°55′13" | 658 | MF582614 |
| haemorrhoidalis | DS2-B02 | TH, Doi Su Thep CMP | C. Sinpoo | 18°48′55" | 98°55′13" | 658 | MF582615 |
| haemorrhoidalis | DS2-B03 | TH, Doi Su Thep CMP | C. Sinpoo | 18°48′55" | 98°55′13" | 658 | MF582616 |
| breviceps | DS2-B04 | TH, Doi Su Thep CMP | C. Sinpoo | 18°48′55" | 98°55′13" | 658 | MF582617 |
| breviceps | DI3-B01 | TH, Doi Inthanon CMP | C. Sinpoo | 18°32′38" | 98°32′53" | 658 | MF582618 |
| Breviceps | DI3-B03 | TH, Doi Inthanon CMP | C. Sinpoo | 18°32′38" | 98°32′53" | 658 | MF582619 |
| montivagus | DI2-B01 | TH, Doi Inthanon CMP | C. Sinpoo | 18°32′41" | 98°30′58" | 658 | MF582620 |
| haemorrhoidalis | DI2-B03 | TH, Doi Inthanon CMP | C. Sinpoo | 18°32′41" | 98°30′58" | 658 | MF582621 |
| Breviceps | DI2-B04 | TH, Doi Inthanon CMP | C. Sinpoo | 18°32′41" | 98°30′58" | 658 | MF582622 |
| haemorrhoidalis | DI2-B05 | TH, Doi Inthanon CMP | C. Sinpoo | 18°32′41" | 98°30′58" | 658 | MF582623 |
| haemorrhoidalis | DPK-B01 | TH, Doi Phu Kha NP | C. Sinpoo | 19°12′20″ | 101°40′50" | 658 | MF582624 |
| haemorrhoidalis | DPK-B02 | TH, Doi Phu Kha NP | C. Sinpoo | 19°12′20″ | 101°40′50" | 658 | MF582625 |
| eximius | DI1-B02 | TH, Doi Inthanon CMP | C. Sinpoo | 18°33′11" | 98°28′55" | 658 | MF582626 |
| eximius | DI1-B03 | TH, Doi Inthanon CMP | C. Sinpoo | 18°33′11" | 98°28′55" | 658 | MF582627 |
| Breviceps | DI1-B04 | TH, Doi Inthanon CMP | C. Sinpoo | 18°33′11" | 98°28′55" | 658 | MF582628 |
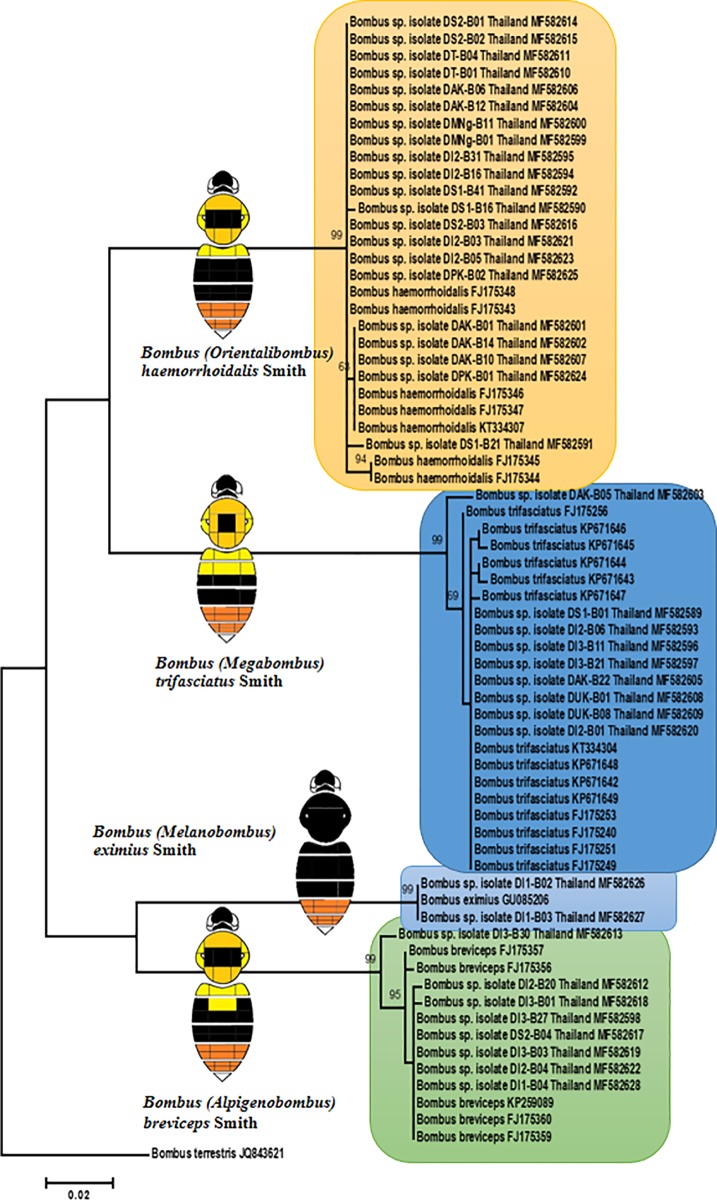
The phylogenetic analysis by maximum likelihood method (Fig 3) with COI barcode data showed strong support for all of the following four conventional Bombus subgenera: B. (Megabombus) montivagus Smith (formerly regarded as part of B. trifasciatus s. l.), B. (Alpigenobombus) breviceps Smith, B. (Orientalibombus) haemorrhoidalis Smith and B. (Melanobombus) eximius Smith (Fig 3).
Fig 3. Estimate of phylogenetic relationship of cytochrome oxidase subunit I (COI) from bumblebees (Bombus sp.) collected in northern Thailand using maximum likelihood.
The sequences of B. terrestris–JQ843621 was used as an out group. Numbers at each node represent bootstrap values as percentages and only bootstrap values greater than 70% are shown.
Microsporidian and trypanosome parasite frequencies in bumblebees
A total of 280 individual bumblebees representing four species (B. montivagus, B. haemorrhoidalis, B. breviceps, and B. eximius) were examined from samples from northern Thailand (Chiang Mai, Mae Hong Son, Chiang Rai and Nan province, sampling sites shown in Table 1). We collected and screened for the most common pathogens of foraging worker bumblebees, Nosema spp. and Crithidia spp..
The results showed that 16 out of 280 individual bumblebees (5.71%) were infected with N. ceranae. This parasite was found in specimens of B. montivagus (5.35%), B. breviceps (14.28%), and B. haemorrhoidalis (4.76%). Nosema bombi was found in 38 individuals (13.57%) from the three species of Bombus as shown in Table 5. Infection rates of N. ceranae and N. bombi were higher in B. breviceps than in other bumblebee species. Nosema bombi was also more prevalent than N. ceranae in the three species of bumblebees. When considering the geographical areas, the highest prevalence values of N. ceranae (20% and 12.5% respectively) were found at the locations Doi Phu Kha (Nan) and Doi Inthanon 3 (Chiang Mai). Prevalence of N. bombi of 20% was found at Doi Inthanon 2, Doi Mae Tha Man (Chiang Mai) and Doi Mae U Kho (Mae Hong Son).
Table 5. Overall occurrence of four parasites in host species (Bombus spp.) (Identities confirmed from barcodes).
| Species | N Bees collected | N. apis a | N. ceranaea | N. bombia | C. bombia |
|---|---|---|---|---|---|
| B. montivagus | 56 | 0.00 | 5.35 | 14.28 | 0.00 |
| B. haemorrhoidalis | 189 | 0.00 | 4.76 | 11.64 | 0.00 |
| B. breviceps | 28 | 0.00 | 14.28 | 28.57 | 0.00 |
| B. eximius | 7 | 0.00 | 0.00 | 0.00 | 0.00 |
| Total | 280 | 0.00 | 5.71 | 13.57 | 0.00 |
N = Total number of individual each Bombus species collected.
a = Prevalence (%)
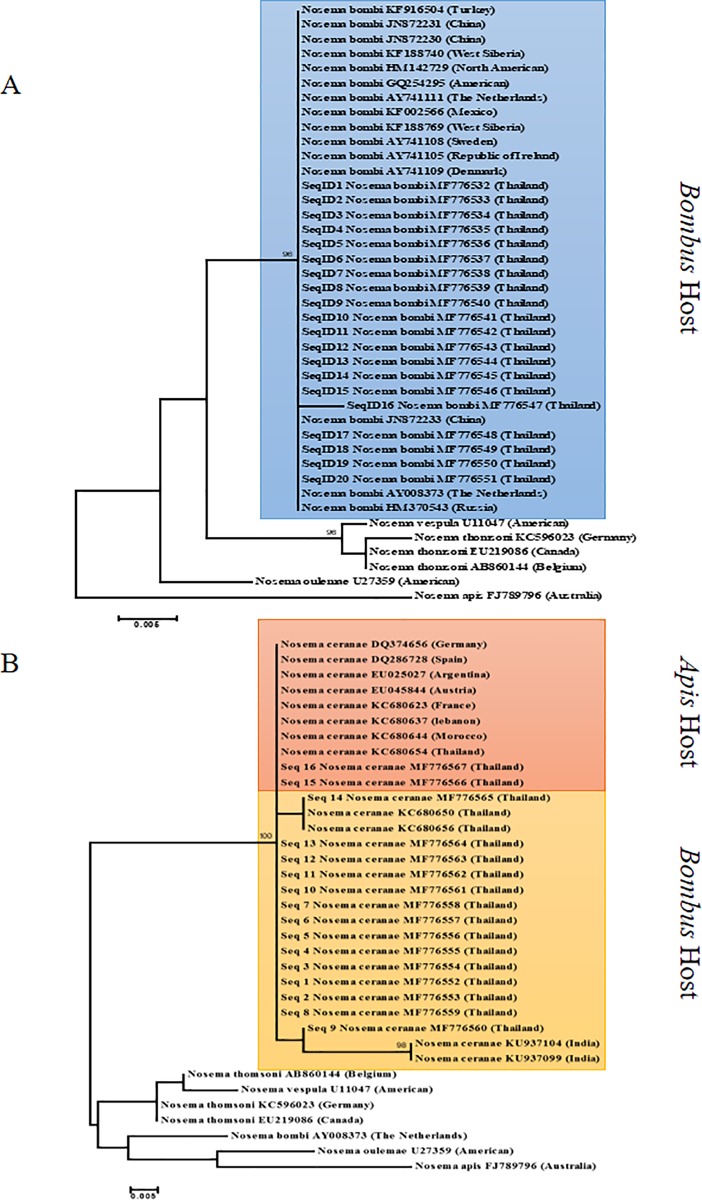
Phylogenetic trees were estimated to assess relationships between the samples of Nosema as shown in Fig 4A and 4B. This included a total of 36 sequences from infected Bombus with a length of 269 bp for 20 sequences of N. bombi and 212 bp for 16 sequences of N. ceranae, after removing the primers from both ends. New sequences of Nosema have been deposited in GenBank and are accessible with the numbers MF776532–MF776567 (Table 6).
Fig 4. The phylogenetic tree showing the relationship of Nosema.
Unrooted consensus of phylogenetic tree showing the relationship of Nosema isolate the partial sequences of 16S ribosomal RNA of Nosema (4-A; N bombi, 4-B; N. ceranae) from Bombus spp. collected in northern Thailand. The tree was estimated using Maximum Likelihood. Numbers at each node represent bootstrap values as percentages and only bootstrap values greater than 70% are shown.
Table 6. Material used in the phylogenetic analysis with the sample locality, collector, sequence length, depository and GenBank accession number.
| Species | Sample name | Sample locality | Collector | Sequence length (bp) | GenBank |
|---|---|---|---|---|---|
| 1 N. bombi | BomDS2-B06 | TH, Doi Su Thep CMP | C. Sinpoo | 269 | MF776532 |
| 2 N. bombi | BomDS2-B12 | TH, Doi Su Thep CMP | C. Sinpoo | 269 | MF776533 |
| 3 N. bombi | BomDS2-B20 | TH, Doi Su Thep CMP | C. Sinpoo | 269 | MF776534 |
| 4 N. bombi | BomDS1-B04 | TH, Doi Su Thep CMP | C Sinpoo | 269 | MF776535 |
| 5 N. bombi | BomDS1-B10 | TH, Doi Su Thep CMP | C. Sinpoo | 269 | MF776536 |
| 6 N. bombi | BomDS1-B37 | TH, Doi Su Thep CMP | C Sinpoo | 269 | MF776537 |
| 7 N. bombi | BomDS1-B45 | TH, Doi Su Thep CMP | C. Sinpoo | 269 | MF776538 |
| 8 N. bombi | BomDS1-B55 | TH, Doi Su Thep CMP | C. Sinpoo | 269 | MF776539 |
| 9 N. bombi | BomDI1-B04 | TH, Doi Inthanon CMP | C. Sinpoo | 269 | MF776540 |
| 10 N. bombi | BomDI1-B07 | TH, Doi Inthanon CMP | C. Sinpoo | 269 | MF776541 |
| 11 N. bombi | BomDI1-B11 | TH, Doi Inthanon CMP | C. Sinpoo | 269 | MF776542 |
| 12 N. bombi | BomDI2-B17 | TH, Doi Inthanon CMP | C. Sinpoo | 269 | MF776543 |
| 13 N. bombi | BomDI2-B24 | TH, Doi Inthanon CMP | C. Sinpoo | 269 | MF776544 |
| 14 N. bombi | BomDI3-B07 | TH, Doi Inthanon CMP | C. Sinpoo | 269 | MF776545 |
| 15 N. bombi | BomDMNg-B05 | TH, Doi Mon Ngao CMP | C. Sinpoo | 269 | MF776546 |
| 16 N. bombi | BomDMNg-B11 | TH, Doi Mon Ngao CMP | C. Sinpoo | 269 | MF776547 |
| 17 N. bombi | BomDMNg-B15 | TH, Doi Mon Ngao CMP | C. Sinpoo | 269 | MF776548 |
| 18 N. bombi | BomDMTM-B03 | TH, Doi Mae Tha Man CMP | C. Sinpoo | 269 | MF776549 |
| 19 N. bombi | BomDAK-B10 | TH, Doi Ang Khang CMP | C. Sinpoo | 269 | MF776550 |
| 20 N. bombi | BomDAK-B12 | TH, Doi Ang Khang CMP | C. Sinpoo | 269 | MF776551 |
| 1 N. ceranae | BomDS2-B16 | TH, Doi Su Thep CMP | C. Sinpoo | 212 | MF776552 |
| 2 N. ceranae | BomDS1-B10 | TH, Doi Su Thep CMP | C. Sinpoo | 212 | MF776553 |
| 3 N. ceranae | BomDS1-B37 | TH, Doi Su Thep CMP | C. Sinpoo | 212 | MF776554 |
| 4 N. ceranae | BomDI2-B02 | TH, Doi Inthanon CMP | C. Sinpoo | 212 | MF776555 |
| 5 N. ceranae | BomDI3-B02 | TH, Doi Inthanon CMP | C. Sinpoo | 212 | MF776556 |
| 6 N. ceranae | BomDAK-B10 | TH, Doi Ang Khang CMP | C. Sinpoo | 212 | MF776557 |
| 7 N. ceranae | BomDUK-B10 | TH, Doi Mae U Kho CMP | C. Sinpoo | 212 | MF776558 |
| 8 N. ceranae | BomDT-B16 | TH, Doi Thong CRP | C. Sinpoo | 212 | MF776559 |
| 9 N. ceranae | BomDT-B16 | TH, Doi Thong CRP | C. Sinpoo | 212 | MF776560 |
| 10 N. ceranae | BomDI2-B04 | TH, Doi Inthanon CMP | C. Sinpoo | 212 | MF776561 |
| 11 N. ceranae | BomDI2-B38 | TH, Doi Inthanon CMP | C. Sinpoo | 212 | MF776562 |
| 12 N. ceranae | BomDI3-B05 | TH, Doi Inthanon CMP | C. Sinpoo | 212 | MF776563 |
| 13 N. ceranae | BomDI3-B23 | TH, Doi Inthanon CMP | C. Sinpoo | 212 | MF776564 |
| 14 N. ceranae | BomDI3-B27 | TH, Doi Inthanon CMP | C. Sinpoo | 212 | MF776565 |
| 15 N. ceranae | BomDI3-B39 | TH, Doi Inthanon CMP | C. Sinpoo | 212 | MF776566 |
| 16 N. ceranae | BomDUK-B19 | TH, Doi Inthanon MHS | C. Sinpoo | 212 | MF776567 |
Discussion
In this study we aimed to identify native bumblebees from multiple sites in northern Thailand (Chiang Mai, Mae Hong Son, Chiang Rai and Nan province). Three bumblebee species (B. montivagus Smith, B. haemorrhoidalis Smith, and B. breviceps Smith) show similar colour patterns. These colour patterns are similar to others in Southeast Asia and may have evolved though mutually protective Mullerian mimicry [37]. We have identified similar colour patterns for bumblebee workers (Fig 2) (three of them for B montivagus in northern Thailand). Hines and Williams (2012) examined colour-pattern evolution in bumblebees in this Southeast Asian mimicry group, which includes B. (Megabombus) montivagus Smith, B. (Alpigenobombus) breviceps Smith, and B. (Orientalibombus) haemorrhoidalis Smith [37]. Moreover, they reported that because these bumblebees also have high variability of colour patterns within species it is sometimes difficult to make reliable species identifications. Considerable colour variation within bumblebee species has been known for more than a century [38]. Our work reaffirms that only some morphological data can be used to accurately distinguish species.
When possible, additional molecular data should therefore be used to confirm species identification [15, 37, 39, 40]. According to our results, the bumblebee species are supported by groups identified from the (COI) gene. This confirms the value of evidence from barcodes for examining the more closely related bumblebee species despite the variation within species [15, 40, 41].
This study is the first survey of the prevalence of major bumblebee pathogens in native bumblebees in northern Thailand, showing the detection and infection rates of N. cerana and N. bombi among 280 female bumblebee specimens. In this sample, N. bombi was present in three species of Bombus (i.e. B. montivagus, B. haemorrhoidalis, and B. breviceps). The complete gene encoding ssrRNA sequences of Nosema isolates were identical to those reported previously from the bumblebee species B. terrestris, B. hortorum, and B. lucorum [21]. Cameron et al. (2011) and Kissinger et al. (2011) could only analyze N. bombi in samples of various Bombus spp. from the southern states of the USA, which were genetically similar to the European isolates screened by these authors [5, 42]. In our results, the gene sequences showed small variations. In the past it was believed that among all Nosema taxa identified to date, only N. bombi was an established parasite of Bombus spp. [21] in which it may be present at varying levels [19, 43]. Thorp (2005) and Tay et al. (2005) suggested that N. bombi was the only microsporidian known to infect European Bombus species [20, 25].
Our study found that N. ceranae was also present in three Bombus spp. (B. montivagus, B. haemorrhoidalis, and B. breviceps). Normally, N. ceranae infects honey bees (originally isolated from A. cerana [44] now infecting A. mellifera as well [33, 45]), but Plischuk et al (2009) found N. ceranae in bumblebees in South America [46]. Our work also is similar to the findings of researchers who have reported the presence of N. ceranae in native bumblebees of Argentina (B. atratus, B. bellicosus, and B. morio) [46]. Mean prevalence values of N. ceranae found in B. breviceps (14.28%) are lower than those reported in B. atratus (72%) and B. bellicosus (63%) from Argentina [47] as well as from these same species in other countries [32, 48]. On the other hand, the lower infection intensity found in native bumblebees of northern Thailand may prevent infection from increasing further as natural reservoirs with high prevalence of the pathogen have not yet been found.
We collected and screened the most common pathogens for total of 280 native foraging worker bumblebees. The trypanosome C. bombi was not observed in this study. Kissinger et al. (2011) also reported few C. bombi in his extensive survey [42]. Similarly, prevalence of Crithidia was less than 10% of all Bombus species examined in United States [49].
Previous studies have proposed that N. ceranae is closer phylogenetically to N. bombi than to N. apis [21, 50, 51], although there is a report to the contrary [52]. Shafer et al. (2009) suggest that N. apis is a basal member of the clade and, therefore, N. bombi is closer to N. ceranae [53]. In our study, N. ceranae strains present in three species of Bombus (B. montivagus, B. haemorrhoidalis, and B. breviceps) from northern Thailand were closely related to the N. ceranae strains reported from A. mellifera. This reaffirms that N. ceranae has a broad host range and may cross between host genera. Nosema ceranae was first discovered in A. cerana, however although it is now spreading to A. mellifera. This pathogen has potential as an emerging threat to bumblebees among the indigenous pollinators [54].
Acknowledgments
This research project is supported by Chiang Mai university.
Data Availability
All relevant data are within the paper.
Funding Statement
This research project is supported by Chiang Mai University.
References
- 1.Bingham RA, Orthner AR. Efficient pollination of alpine plants. Nature. 1998;391(6664):238. [Google Scholar]
- 2.Kremen C, Williams NM, Thorp RW. Crop pollination from native bees at risk from agricultural intensification. Proceedings of the National Academy of Sciences. 2002;99(26):16812–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Velthuis HH, Van Doorn A. A century of advances in bumblebee domestication and the economic and environmental aspects of its commercialization for pollination. Apidologie. 2006;37(4):421–51. [Google Scholar]
- 4.Williams PH, Osborne JL. Bumblebee vulnerability and conservation world-wide. Apidologie. 2009;40(3):367–87. [Google Scholar]
- 5.Cameron AC, Gelbach JB, Miller DL. Robust inference with multiway clustering. Journal of Business & Economic Statistics. 2011;29(2):238–49. [Google Scholar]
- 6.Macior LW, Tang Y. A preliminary study of the pollination ecology of Pedicularis in the Chinese Himalaya. Plant Species Biology. 1997;12(1):1–7. [Google Scholar]
- 7.Winter K, Adams L, Thorp R, Inouye D, Day L, Ascher J, et al. Importation of non-native bumble bees into North America: potential consequences of using Bombus terrestris and other non-native bumble bees for greenhouse crop pollination in Canada, Mexico, and the United States. San Francisco. 2006;33. [Google Scholar]
- 8.Dias B, Raw A, Imperatriz-Fonseca V, editors. International pollinators initiative: The São Paulo declaration on pollinators. Report on the recommendations of the workshop on the conservation and sustainable use of pollinators in agriculture with emphasis on bees; 1999.
- 9.Ruz L. Bee pollinators introduced to Chile: a review. Pollinating bees. 2002:155–67. [Google Scholar]
- 10.Li J, Wu J, Cai W, Peng W, An J, Huang J. Comparison of the colony development of two native bumblebee species Bombus ignitus and Bombus lucorum as candidates for commercial pollination in China. Journal of apicultural research. 2008;47(1):22–6. [Google Scholar]
- 11.Macfarlane R, Gurr L. Distribution of bumble bees in New Zealand. New Zealand Entomologist. 1995;18(1):29–36. [Google Scholar]
- 12.Williams P. The distribution of bumblebee colour patterns worldwide: possible significance for thermoregulation, crypsis, and warning mimicry. Biological Journal of the Linnean Society. 2007;92(1):97–118. [Google Scholar]
- 13.Hebert PD, Ratnasingham S, de Waard JR. Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species. Proceedings of the Royal Society of London B: Biological Sciences. 2003;270(Suppl 1):S96–S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Williams PH, An J, Huang J. The bumblebees of the subgenus Subterraneobombus: integrating evidence from morphology and DNA barcodes (Hymenoptera, Apidae, Bombus). Zoological Journal of the Linnean Society. 2011;163(3):813–62. [Google Scholar]
- 15.Williams PH, An J, Brown MJ, Carolan JC, Goulson D, Huang J, et al. Cryptic bumblebee species: consequences for conservation and the trade in greenhouse pollinators. PloS one. 2012;7(3):e32992 10.1371/journal.pone.0032992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Williams PH, Byvaltsev A, Sheffield C, Rasmont P. Bombus cullumanus—an extinct European bumblebee species? Apidologie. 2013;44(2):121–32. [Google Scholar]
- 17.Huang J, Jie W, Jiandong A, Williams PH. Newly discovered colour-pattern polymorphism of Bombus koreanus females (Hymenoptera: Apidae) demonstrated by DNA barcoding. Apidologie. 2015;46(2):250–61. [Google Scholar]
- 18.Williams PH, Byvaltsev AM, Cederberg B, Berezin MV, Ødegaard F, Rasmussen C, et al. Genes suggest ancestral colour polymorphisms are shared across morphologically cryptic species in arctic bumblebees. PLoS One. 2015;10(12):e0144544 10.1371/journal.pone.0144544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schmid-Hempel P. On the evolutionary ecology of host-parasite interactions: addressing the question with regard to bumblebees and their parasites. Naturwissenschaften. 2001;88(4):147–58. [DOI] [PubMed] [Google Scholar]
- 20.Tay WT, O'MAHONY EM, Paxton RJ. Complete rRNA gene sequences reveal that the microsporidium Nosema bombi infects diverse bumblebee (Bombus spp.) hosts and contains multiple polymorphic sites. Journal of Eukaryotic Microbiology. 2005;52(6):505–13. 10.1111/j.1550-7408.2005.00057.x [DOI] [PubMed] [Google Scholar]
- 21.Fries I, De Ruijter A, Paxton RJ, da Silva AJ, Slemenda SB, Pieniazek NJ. Molecular characterization of Nosema bombi (Microsporidia: Nosematidae) and a note on its sites of infection in Bombus terrestris (Hymenoptera: Apoidea). Journal of Apicultural research. 2001;40(3–4):91–6. [Google Scholar]
- 22.Larsson JR. Cytological variation and pathogenicity of the bumble bee parasite Nosema bombi (Microspora, Nosematidae). Journal of invertebrate pathology. 2007;94(1):1–11. 10.1016/j.jip.2006.07.006 [DOI] [PubMed] [Google Scholar]
- 23.Otti O, Schmid‐Hempel P. A field experiment on the effect of Nosema bombi in colonies of the bumblebee Bombus terrestris. Ecological Entomology. 2008;33(5):577–82. [Google Scholar]
- 24.Rutrecht ST, Brown MJ. Differential virulence in a multiple‐host parasite of bumble bees: resolving the paradox of parasite survival? Oikos. 2009;118(6):941–9. [Google Scholar]
- 25.Thorp R, Shepherd M, Vaughan D. Red list of pollinator insects of North America. The Xerces Society for Invertebrate Conservation. 2005. [Google Scholar]
- 26.Imhoof B, Schmid-Hempel P. Colony success of the bumble bee, Bombus terrestris, in relation to infections by two protozoan parasites, Crithidia bombi and Nosema bombi. Insectes Sociaux. 1999;46(3):233–8. [Google Scholar]
- 27.Li J, Chen J, Wang S. Introduction Risk Management of Supply and Cash Flows in Supply Chains: Springer; 2011. p. 1–48. [Google Scholar]
- 28.Williams P.H., Ito M., Matsumura T. & Kudo I. The bumblebees of the Nepal Himalaya (Hymenoptera: Apidae). Insecta Matsumurana. 2010;66:115–151. [Google Scholar]
- 29.Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual: Cold spring harbor laboratory press; 1989. [Google Scholar]
- 30.Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Molecular marine biology and biotechnology. 1994;3(5):294–9. [PubMed] [Google Scholar]
- 31.Evans JD. Beepath: An ordered quantitative-PCR array for exploring honey bee immunity and disease. Journal of Invertebrate Pathology. 2006;93(2):135–9. 10.1016/j.jip.2006.04.004 [DOI] [PubMed] [Google Scholar]
- 32.Li J, Chen W, Wu J, Peng W, An J, Schmid-Hempel P, et al. Diversity of Nosema associated with bumblebees (Bombus spp.) from China. International journal for parasitology. 2012;42(1):49–61. 10.1016/j.ijpara.2011.10.005 [DOI] [PubMed] [Google Scholar]
- 33.Klee J, Besana AM, Genersch E, Gisder S, Nanetti A, Tam DQ, et al. Widespread dispersal of the microsporidian Nosema ceranae, an emergent pathogen of the western honey bee, Apis mellifera. Journal of Invertebrate Pathology. 2007;96(1):1–10. 10.1016/j.jip.2007.02.014 [DOI] [PubMed] [Google Scholar]
- 34.Higes M, Martín R, Meana A. Nosema ceranae, a new microsporidian parasite in honeybees in Europe. Journal of Invertebrate Pathology. 2006;92(2):93–5. 10.1016/j.jip.2006.02.005 [DOI] [PubMed] [Google Scholar]
- 35.Larkin MA, Blackshields G Fau—Brown NP, Brown Np Fau—Chenna R, Chenna R Fau—McGettigan PA, McGettigan Pa Fau—McWilliam H, McWilliam H Fau—Valentin F, et al. Clustal W and Clustal X version 2.0. (1367–4811 (Electronic)). [DOI] [PubMed]
- 36.Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Molecular biology and evolution. 2016;33(7):1870–4. 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hines HM, Williams PH. Mimetic colour pattern evolution in the highly polymorphic Bombus trifasciatus (Hymenoptera: Apidae) species complex and its comimics. Zoological Journal of the Linnean Society. 2012;166(4):805–26. [Google Scholar]
- 38.Vogt O. Studien über das Artproblem. Über das Variieren der Hummeln. Mitt. 1 u. 2. Sitzgsber Ges naturforsch Freunde Berl. 1909;1911.
- 39.Duennes MA, Lozier JD, Hines HM, Cameron SA. Geographical patterns of genetic divergence in the widespread Mesoamerican bumble bee Bombus ephippiatus (Hymenoptera: Apidae). Molecular Phylogenetics and Evolution. 2012;64(1):219–31. 10.1016/j.ympev.2012.03.018 [DOI] [PubMed] [Google Scholar]
- 40.Williams PH, Brown MJ, Carolan JC, An J, Goulson D, Aytekin AM, et al. Unveiling cryptic species of the bumblebee subgenus Bombus s. str. worldwide with COI barcodes (Hymenoptera: Apidae). Systematics and Biodiversity. 2012;10(1):21–56. [Google Scholar]
- 41.Carolan JC, Murray TE, Fitzpatrick Ú, Crossley J, Schmidt H, Cederberg B, et al. Colour patterns do not diagnose species: quantitative evaluation of a DNA barcoded cryptic bumblebee complex. PloS one. 2012;7(1):e29251 10.1371/journal.pone.0029251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kissinger CN, Cameron SA, Thorp RW, White B, Solter LF. Survey of bumble bee (Bombus) pathogens and parasites in Illinois and selected areas of northern California and southern Oregon. Journal of invertebrate pathology. 2011;107(3):220–4. 10.1016/j.jip.2011.04.008 [DOI] [PubMed] [Google Scholar]
- 43.Shykoff J, Schmid-Hempel P. Incidence and effects of four parasites in natural populations of bumble bees in Switzerland. Apidologie. 1991;22(2):117–25. [Google Scholar]
- 44.Fries I, Feng F, Silva A, Slemenda SB, Pieniazek NJ. Nosema ceranae (Microspora, Nosematidae), morphological and molecular characterization of a microsporidian parasite of the Asian honeybee Apis cerana (Hymenoptera, Apidae). Eur J Protistol. 1996;32. [Google Scholar]
- 45.Chen Y, Evans JD, Smith IB, Pettis JS. Nosema ceranae is a long-present and wide-spread microsporidian infection of the European honey bee (Apis mellifera) in the United States. Journal of Invertebrate Pathology. 2008;97(2):186–8. 10.1016/j.jip.2007.07.010 [DOI] [PubMed] [Google Scholar]
- 46.Plischuk S, Martín‐Hernández R, Prieto L, Lucía M, Botías C, Meana A, et al. South American native bumblebees (Hymenoptera: Apidae) infected by Nosema ceranae (Microsporidia), an emerging pathogen of honeybees (Apis mellifera). Environmental Microbiology Reports. 2009;1(2):131–5. 10.1111/j.1758-2229.2009.00018.x [DOI] [PubMed] [Google Scholar]
- 47.Arbulo N, Antúnez K, Salvarrey S, Santos E, Branchiccela B, Martín-Hernández R, et al. High prevalence and infection levels of Nosema ceranae in bumblebees Bombus atratus and Bombus bellicosus from Uruguay. Journal of invertebrate pathology. 2015;130:165–8. 10.1016/j.jip.2015.07.018 [DOI] [PubMed] [Google Scholar]
- 48.Graystock P, Yates K, Darvill B, Goulson D, Hughes WO. Emerging dangers: deadly effects of an emergent parasite in a new pollinator host. Journal of invertebrate pathology. 2013;114(2):114–9. 10.1016/j.jip.2013.06.005 [DOI] [PubMed] [Google Scholar]
- 49.Cordes N, Huang W-F, Strange JP, Cameron SA, Griswold TL, Lozier JD, et al. Interspecific geographic distribution and variation of the pathogens Nosema bombi and Crithidia species in United States bumble bee populations. Journal of invertebrate pathology. 2012;109(2):209–16. 10.1016/j.jip.2011.11.005 [DOI] [PubMed] [Google Scholar]
- 50.Wang LL, Chen KP, Zhang Z, Yao Q, Gao GT, Zhao Y. Phylogenetic analysis of Nosema antheraeae (Microsporidia) isolated from Chinese oak silkworm, Antheraea pernyi. Journal of Eukaryotic Microbiology. 2006;53(4):310–3. 10.1111/j.1550-7408.2006.00106.x [DOI] [PubMed] [Google Scholar]
- 51.Chen Y, Evans JD, Zhou L, Boncristiani H, Kimura K, Xiao T, et al. Asymmetrical coexistence of Nosema ceranae and Nosema apis in honey bees. Journal of invertebrate pathology. 2009;101(3):204–9. 10.1016/j.jip.2009.05.012 [DOI] [PubMed] [Google Scholar]
- 52.Slamovits CH, Fast NM, Law JS, Keeling PJ. Genome compaction and stability in microsporidian intracellular parasites. Current Biology. 2004;14(10):891–6. 10.1016/j.cub.2004.04.041 [DOI] [PubMed] [Google Scholar]
- 53.Shafer AB, Williams GR, Shutler D, Rogers RE, Stewart DT. Cophylogeny of Nosema (Microsporidia: Nosematidae) and bees (Hymenoptera: Apidae) suggests both cospeciation and a host-switch. Journal of Parasitology. 2009;95(1):198–203. 10.1645/GE-1724.1 [DOI] [PubMed] [Google Scholar]
- 54.Fürst MA, McMahon DP, Osborne JL, Paxton RJ, Brown MJ. Disease associations between honeybees and bumblebees as a threat to wild pollinators. Nature. 2014. February;506(7488):364 10.1038/nature12977 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.