Abstract
Objectives
Candida lusitaniae is an opportunistic yeast pathogen in certain high-risk patient populations/cohorts. The species exhibits an unusual antifungal susceptibility profile with tendency to acquire rapid resistance. Here, we describe prevalence of C. lusitaniae in clinical specimens in Kuwait, its antifungal susceptibility profile and role in neonatal fungemia.
Methods
Clinical C. lusitaniae isolates recovered from diverse specimens during 2011 to 2017 were retrospectively analyzed. All isolates were identified by germ tube test, growth on CHROMagar Candida and by Vitek 2 yeast identification system. A simple species-specific PCR assay was developed and results were confirmed by PCR-sequencing of ITS region of rDNA. Antifungal susceptibility was determined by Etest. Minimum inhibitory concentrations (MICs) were recorded after 24 h incubation at 35°C.
Results
Of 7068 yeast isolates, 134 (1.89%) were identified as C. lusitaniae including 25 (2.52%) among 990 bloodstream isolates. Species-specific PCR and PCR-sequencing of rDNA confirmed identification. Of 11 cases of neonatal candidemia, 9 occurred in NICU of Hospital A and are described here. Eight of 9 neonates received liposomal amphotericin B, which was followed by fluconazole in 7 and additionally by caspofungin in 2 cases as salvage therapy. Three of 8 (37.5%) patients died. No isolate exhibited reduced susceptibility to amphotericin B, fluconazole, voriconazole, caspopfungin, micafungin and anidulafungin. The MIC ± geometric mean values for amphotericin B, fluconazole, voriconazole, and caspofungin were as follows: 0.072 ± 0.037 μg/ml, 2.32 ± 0.49 μg/ml, 0.09 ± 0.01 μg/ml and 0.16 ± 0.08 μg/ml, respectively. Only two isolates exhibited reduced susceptibility to fluconazole.
Conclusions
This study describes the prevalence and antifungal susceptibility profile of clinical C. lusitaniae isolates in Kuwait. No isolate showed reduced susceptibility to amphotericin B. The study highlights the emerging role of C. lusitaniae as a healthcare-associated pathogen capable of causing fungemia in preterm neonates and causing significant mortality.
Introduction
Candida lusitaniae (teleomorph Clavispora lusitaniae) was first described by van Uden and Carmo-Sousa as a common flora in the gastrointestinal tract of warm blooded animals [1]. It was recognized as a human pathogen in three patients with septicemia and in two of them, the isolates were reported as variants of C. tropicalis [2, 3]. The first documented case of opportunistic infection where C. lusitaniae strain developed resistance to amphotericin B during therapy was published in 1979 [4,5]. Over the years, there has been a gradual increase in the number of cases with C. lusitaniae infection, predominantly in cancer patients who received bone marrow transplantation or cytotoxic chemotherapy [6,7]. Some strains are known to exhibit intrinsic or acquired resistance to amphotericin B [5, 8–11]. This species is being increasingly isolated from cancer patients on empirical/prophylaxis antifungal therapy [12, 13]. A study from Anderson Cancer Center (Houston, Texas) during 2006–2013 revealed a high rate of occurrence of C. lusitaniae among cancer patients (1.45 episodes/100,000 inpatient days) and it was also the third most common cause of breakthrough candidemia (7/37, 19%) associated with 53% mortality [13]. In Kuwait, little information is available on the association of C. lusitaniae with human colonization and infection [14]. This study describes prevalence and susceptibility profile of clinical isolates of C. lusitaniae and its role in neonatal fungemia in Kuwait.
Materials and methods
Reference strains, clinical isolates and phenotypic identification
Reference strains or well characterized clinical isolates of C. lusitaniae (CBS4413, CBS1944, CBS6936 and ATCC38533), Candida dubliniensis (CD36), Candida albicans (ATCC56881), Candida africana (CBS9118), Candida parapsilosis (ATCC22019), Candida orthopsilosis (ATCC96139), Candida metapsilosis (ATCC96143), Candida glabrata (ATCC90030), Candida krusei (ATCC6258), Candida tropicalis (ATCC34139), Meyerozyma guilliermondii (CBS6021), Candida kefyr (ATCC28838), Candida conglobata (Kw381/16), Candida utilis (Kw3642/15), Candida haemulonii (Kw154/06), Candida duobushaemulonii (Kw3270/08) and Candida auris (Kw2611/17) were used as reference Candida species. All clinical C. lusitaniae isolates included in the study were obtained between January 2011 and December 2017. They were isolated from different clinical specimens in microbiology laboratories of various hospitals across Kuwait and were referred to Mycology Reference Laboratory (MRL) for identification and antifungal susceptibility testing as part of routine patient care. The clinical specimens were collected after obtaining informed verbal consent and this procedure was approved by the ethical Committee, Ministry of Health, Kuwait and Health Sciences Center, Kuwait University. The specimens were cultured on Sabouraud dextrose agar supplemented with chloramphenicol and growth was streaked to get isolated colonies to ensure purity of culture. All blood culture isolates were obtained using BACTEC 9240 system or BACTEC Peds Plus/F culture bottles (Becton Dickinson, Paramus, NJ, USA). All positive cultures were Gram-stained and subcultures were made on blood agar and/or Sabouraud dextrose agar. Isolated yeast colonies were processed for routine identification procedures, which included direct microscopic examination by wet mount, Germ tube test, growth characteristics on CHROMagar Candida and identification by Vitek 2 yeast identification system [15]. Laboratory details of all the isolates identified as C. lusitaniae since 2011 were retrospectively collected for source of isolation. All bloodstream isolates were also subjected to identification by matrix assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS), performed as described previously [16].
Molecular characterization
Genomic DNA was extracted from reference strains or clinical isolates from 1 ml of cell suspension in Sabouraud dextrose broth by Gentra Puregene Yeast DNA extraction kit (Qiagen, Hilden, Germany) according to kit instructions or by the rapid method using Chelex-100 as described previously [17]. A simple, low-cost PCR assay was developed for rapid molecular identification of C. lusitaniae isolates. For this purpose, one forward (CLUSITF, 5’-TTGYWTTTGCGAACAAAAAAA-3’) and one reverse (CLUSITR, 5’-TATTTCGGAGCAACGCCTA-3’) primer targeting specific sequences within ITS-1 and ITS-2 regions of rDNA of C. lusitaniae were synthesized. The unique primer sequences designed in this study were based on sequence alignment of ITS region sequences from multiple strains of all commonly encountered clinical yeast species that are available from the GenBank. The species specificity of the primers CLUSITF and CLUSITR for C. lusitaniae was indicated by BLAST searches (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi) as they showed complete sequence identity only with C. lusitaniae strains. The reaction and PCR cycling conditions were same as described previously except that primers CLUSITF and CLUSITR were used [17]. PCR amplicons were run on 2% (w/v) agarose gels, as described previously [18]. The results of species-specific identification of all C. lusitaniae isolates were confirmed by DNA sequencing of the ITS region of rDNA. The ITS region was amplified by using panfungal primers (ITS1 and ITS4) and both strands were sequenced as described previously [19, 20]. BLAST searches (http://blast.ncbi.nlm.nih.gov/Blast.cgi?) were performed and >99% sequence identity with corresponding sequence from C. lusitaniae reference strains (CBS6936 or CBS5094 or CBS4413 or ATCC38533) available in the GenBank was used for species identification [21].
Fingerprinting of Candida species isolates by molecular techniques is performed for epidemiological studies [22]. The genotypic relationship among bloodstream C. lusitaniae isolates collected from neonates in Hospital A was studied by comparing ITS region of rDNA sequences. The ITS region of rDNA sequences from 2 other bloodstream isolates; C. lusitaniae Kw718/11 and C. lusitaniae Kw1058/15 collected from 2 neonates from Hospital B during the period of this study and a bloodstream isolate (C. lusitaniae Kw2009/07) collected 2 years earlier from a neonate from Hospital A were also used. The rDNA sequences from C. lusitaniae CBS6936, C. lusitaniae CBS5094, C. lusitaniae CBS4413 and C. lusitaniae ATCC38533 available from GenBank were also retrieved and used for comparisons. Multiple sequence alignments were performed with Clustal omega and the phylogenetic tree was constructed with MEGA 6.1 software by using the Neighbor-joining method with Kimura-2 parameter model, as described previously [23]. The robustness of tree branches was assessed by bootstrap analysis with 1,000 replicates.
Antifungal drug susceptibility testing
The susceptibility of C. lusitaniae isolates was determined by Etest for amphotericin B, fluconazole, voriconazole, and caspofungin according to the manufacturer's instructions (bioMérieux, France) and as described previously [24]. Bloodstream isolates were also tested for micafungin and anidulafungin, by Etest and for caspofungin and micafungin by Vitek2 yeast identification system. Quality control was ensured by testing C. albicans ATCC90028, C. parapsilosis ATCC22019 and C. tropicalis ATCC750 [24]. As yet, there are no interpretive susceptibility breakpoints available for C. lusitaniae.
Statistical analysis
Statistical analysis was performed by using Fisher’s exact test or chi-square test as appropriate and probability levels <0.05 by the two-tailed test were considered as significant. Statistical analyses were performed by using WinPepi software ver. 11.65 (PEPI for Windows, Microsoft Inc., Redmond, WA, USA).
Results
Prevalence and phenotypic and molecular identification
Of 7068 yeast isolates tested during the 7-year study period, 134 isolates were identified as C. lusitaniae (Table 1). The occurrence of C. lusitaniae isolates varied from 1% in 2012 to 2.9% in 2015 while the occurrence of blood stream isolates varied from 0% in 2012 to 0.6% in 2011 and 2014 (Table 1). The overall prevalence of C. lusitaniae among yeast species isolates was 1.89%. The largest number of C. lusitaniae isolates were obtained from sputum samples (n = 54) followed by urine (n = 29) and bloodstream (n = 25). The remaining 26 C. lusitaniae isolates were obtained from wound/rectal/ear swabs (n = 11), bronchoalveolar lavage (BAL) (n = 4), peritoneal fluid (n = 3), nasopharynx swab (n = 2) and other specimens (n = 6). A total of 990 bloodstream isolates were recovered from 990 candidemia patients during the study period. Thus, the prevalence of C. lusitaniae among blood stream isolates was higher (25 of 990, 2.5%) than other specimen types (109 of 6078, 1.8%), however, the difference was not statistically significant (P = 0.119). None of the bloodstream isolates came from cancer patients. Of 25 bloodstream C. lusitaniae isolates, 11 were recovered from neonates with nine isolates originating from one hospital (Hospital A) and two isolates from another hospital (Hospital B). All (n = 134) C. lusitaniae isolates produced white to cream-colored colonies on Sabouraud dextrose agar at 30°C and shades of pink-colored colonies on CHROMagar Candida (S1 Fig). The wet mount examination showed ovoid to sub-globose budding cells with abundant pseudohyphae. All isolates were identified as C. lusitaniae by Vitek 2 yeast identification with 97–99% probability. The identity of bloodstream isolates was confirmed by MALDI-TOF MS.
Table 1. Distribution of total and bloodstream C. lusitaniae isolates among clinical yeast isolates screened during January 2011 to December 2017.
| Year of | No. of yeast | No. of C. lusitaniae | No. of bloodstream | No. of C. lusitaniae |
|---|---|---|---|---|
| isolation | isolates tested | isolates detected | C. lusitaniae isolates | isolates from neonates |
| 2011 | 926 | 26 | 6 | 3 |
| 2012 | 924 | 9 | 0 | 0 |
| 2013 | 1052 | 18 | 4 | 1 |
| 2014 | 869 | 19 | 5 | 3 |
| 2015 | 1068 | 31 | 5 | 2 |
| 2016 | 1196 | 17 | 4 | 2 |
| 2017 | 1033 | 14 | 1 | 0 |
| Total | 7068 | 134 | 25 | 11 |
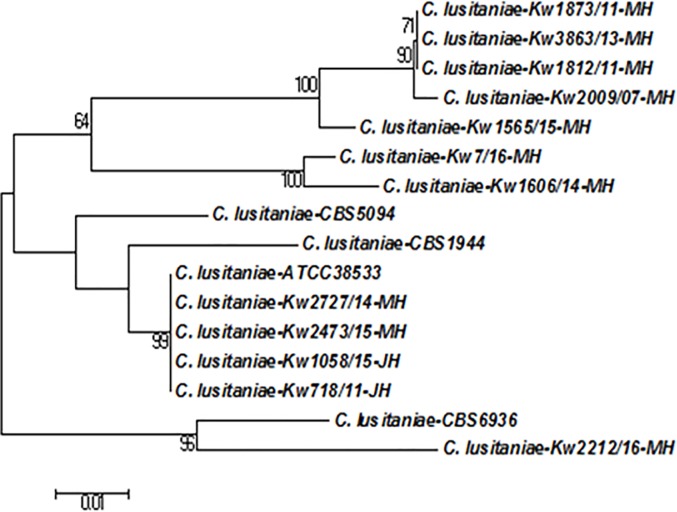
The PCR amplification performed with CLUSITF and CLUSITR primers yielded an expected size amplicon of nearly 242 bp with DNA extracted from two reference strains (CBS 1944 and CBS 4413) of C. lusitaniae only while no amplicon was obtained with genomic DNA prepared from reference strains or well characterized clinical isolates of C. dubliniensis, C. albicans, C. parapsilosis, C. orthopsilosis, C. glabrata, C. krusei, C. tropicalis, C. guilliermondii, C. kefyr, C. haemulonii, C. duobushaemulonii, and C. auris, as expected (S2 Fig). Similarly, no amplicon was also obtained with DNA from C. africana, C. metapsilosis, C. conglobata, C. utilis and with human DNA. The ITS region of rDNA sequences from 11 bloodstream isolates from neonates (Gen Bank accession numbers LS999909 to LS999920) exhibited maximum identity with corresponding sequences from reference strains of C. lusitaniae and not with other Candida species, as expected. The sequence comparisons also indicated inter-strain variations among clinical C. lusitaniae isolates from Kuwait. This prompted us to perform molecular fingerprinting studies by comparing ITS region of rDNA sequences. The data showed that 11 isolates exhibited seven different sequence types indicating that many isolates were clonally unrelated (Fig 1).
Fig 1. Neighbor-joining phylogenetic tree based on ITS region of rDNA sequence data for clinical C. lusitaniae strains isolated from nine neonates from Hospital A in Kuwait together with four reference strains.
The sequence data for C. lusitaniae strains isolated from 2 neonates from Hospital B during the study period and one archived C. lusitaniae strain isolated previously from a neonate from Hospital A were also used for comparison purpose. The numbers on the node branches are bootstrap frequencies.
C. lusitaniae candidemia in neonates
During the study period, 11 cases of neonatal candidemia due to C. lusitaniae were identified. Nine of them occurred in the Neonatal Intensive Care Unit (NICU) of Hospital A and are described in detail here. The salient clinical findings and antifungal susceptibility data for eight patients (clinical details for one patient were not completely available) are presented in Table 2. Seven of eight neonates (including twins, Case 1 and Case 2) were preterm with gestational age varying between 24 to 36 weeks. The birth weight ranged between 600 to 2850 g. All neonates received prior antibiotics (ampicillin and amikacin) and had at least one catheter in place. With one exception (Case 1), all neonates developed bacterial septicemia prior to developing candidemia and received multiple antibiotics for varying periods. The intervening period between date of birth and diagnosis of candidemia ranged between 10 to 60 days (geometric mean ± standard deviation, 27.72 ± 16.12 days). Seven neonates had bacteremia 1 to 44 (5.56 ± 15.15) days before developing candidemia. All patients received amphotericin B or its lipid formulation (AmBisome) as primary therapy for varying duration. It was followed by fluconazole in all cases and additionally by caspofungin in two cases. Despite treatment, three of eight (37.5%) neonates died. Case 1 and Case 2 were twins, both developed C. lusitaniae candidemia and succumbed to infection despite treatment. The DNA isolated from the blood sample of case 2 (yielding C. lusitaniae Kw1873/11) was also used for PCR-sequencing of ITS-1 region of rDNA, as described previously [23] and the DNA sequence data of the ITS-1 region matched completely with the corresponding sequence from C. lusitaniae Kw1873/11.
Table 2. Salient features of 9 cases of C. lusitaniae fungemia in neonates and susceptibility profiles of the isolates.
| Case | DOB | GA (wks/sex) |
Birth weight (g) | Date of bacterial septicaemia | Date of Candidemia | C. lusitaniae isolate no. | Catheters | Antibacterials used | Antifungals used | Outcome | Method | Minimum Inhibitory concentration (μg/ml) by Etest | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AP | FL | VO | CS | MYC | AND | ||||||||||||
| 1 | 7/5/11 | 24/M | 600 | 1/6/11MRSE D-25 | 17/5/11 D-10 | Kw 1812/11 | CVC,UAC UVC | AMP+AK,VAN, MEM | Ambisome | Expired 14/8/11 D-89 | E-test | 0.016 | 0.5 | 0.016 | 0.19 | 0.047 | 0.032 |
| Vitek | ≤ 0.25 | ≤ 1 | ≤ 0.12 | 0.5 | 0.12 | NA | |||||||||||
| 2 | 7/5/11 | 24/M | 625 | 20/5/11 S.epidermidis D-13 | 23/5/11 D-16 | Kw 1873/11 | CVC, UAC, UVC |
AMP+AK,TEC,TZP | Ambisome, FL | Expired 23/6/11 D-31 | E-test | 0.016 | 0.5 | 0.008 | 0.125 | 0.094 | 0.023 |
| Vitek | ≤ 0.25 | ≤ 1 | ≤ 0.12 | ≤ 0.25 | 0.12 | NA | |||||||||||
| 3 | 20/10/13 | 28/F | 1200 | 14/11/13 Enterobacter D-25 | 17/11/13 D-28 | Kw 3863/13 | UVC | AMP+AK,TZP, VAN | Ambisome, FL, Cs | Discharged | E-test | 0.002 | 0.19 | 0.008 | 0.094 | 0.064 | 0.064 |
| Vitek | ≤ 0.25 | ≤ 1 | ≤ 0.12 | ≤ 0.25 | 0.12 | NA | |||||||||||
| 4 | N.A | N.A | N.A | N.A | 11/5/14 | Kw 1606/14 | N.A | N.A | N.A | N.A | E-test | 0.023 | 0.5 | 0.004 | 0.094 | 0.125 | 0.047 |
| 5 | 12/8/14 | 32/M | 1500 | 27/8/14 P.aeruginosa D-15 24/9 E.faecalis+ P.aeruginosa D-43 |
4/9/14 D-23 | Kw 2727/14 | CVC, Urinary catheter | AMP+AK,MEM | AP, FL, CS | Expired 25/9/14 D-21 | E-test | 0.016 | 0.25 | 0.006 | 0.25 | 0.032 | 0.016 |
| Vitek | ≤ 0.25 | ≤ 1 | ≤ 0.12 | ≤ 0.12 | 0.12 | NA | |||||||||||
| 6 | 11/3/15 | 25/F | 780 | 27/3/15 S.epidermidis D-16, 22/5 K.pneumoniae D-72 | 10/5/15 D-60 | Kw 1565/15 | CVC | AMP+AK MEM,VAN | Ambisome, FL | Discharged | E-test | 0.094 | 0.75 | 0.002 | 0.016 | 0.032 | 0.032 |
| Vitek | ≤ 0.25 | ≤ 1 | ≤ 0.12 | ≤ 0.25 | 0.12 | NA | |||||||||||
| 7 | 3/7/15 | 36/F | 2280 | 10/8/15 K.pneumoniae D-38 | 14/8/15 D-42 | Kw 2473/15 | ─ | AMP+AK,MEM | AP, FL | Discharged | E-test | 0.064 | 0.75 | 0.023 | 0.19 | 0.125 | 0.032 |
| Vitek | ≤ 0.25 | ≤ 1 | ≤ 0.12 | ≤ 0.25 | 0.12 | NA | |||||||||||
| 8 | 23/11/15 | 41/M | 2850 | 24/12/15 S.epidermidis D-31 | 25/12/15 D-32 | Kw 7/16 | CVC | AMP+AK TZP,MEM | Ambisome, FL | Discharged | E-test | 0.023 | 0.19 | 0.003 | 0.19 | 0.032 | 0.094 |
| Vitek | ≤ 0.25 | ≤ 1 | ≤ 0.12 | ≤ 0.25 | 0.12 | NA | |||||||||||
| 9 | 28/5/16 | 27/F | 1030 | 26/6/16 K.pneumoniae D-29 | 9/7/16 D-42 | Kw 2212/16 | CVC | AMP+AK TZP,MEM | Ambisome, FL | Discharged | E-test | 0.032 | 0.5 | 0.003 | 0.012 | 0.047 | 0.023 |
| Vitek | ≤ 0.25 | ≤ 1 | ≤ 0.12 | ≤ 0.25 | ≤ 0.06 | NA | |||||||||||
Abbreviations:-DOB-date of birth, GA-gestation age, M-male, F-female, SVD-spontaneous vaginal delivery, MRSE-methicillin resistant Staphylococcus epidermidis, CVC-central venous catheter, UAC-umbilical arterial catheter, UVC-umbilical venous catheter, AMP-ampicillin, AK-amikacin, VAN-vancomycin, MEM-meropenem, TEC-teicoplanin, TZP-piperacillin tazobactam, AmBisome- ambisome, AP-amphotericin B, FL-fluconazole, CS-caspofungin, NA- Not available in Vitek. 2 AST-YS07; N. A., not available.
Antifungal susceptibility
Although there are no validated EUCAST or CLSI susceptibility breakpoints for C. lusitaniae, all bloodstream isolates appeared susceptible to amphotericin B, fluconazole, voriconazole, caspofungin, micafungin and anidulafungin by Etest (Tables 2 and 3). Etest generally yielded lower MICs than Vitek 2, particularly for amphotericin B (Table 2). The geometric mean ± MIC values for amphotericin B, fluconazole, voriconazole (n = 102), and caspofungin were as follows: 0.037 ± 0.072 μg/ml, 0.49 ± 2.32 μg/ml, 0.01 ± 0.09 μg/ml and 0.16 ± 0.08 μg/ml, respectively. Only two C. lusitaniae isolates exhibited fluconazole MICs of 16 μg /ml and 24 μg /ml (Table 3).
Table 3. Antifungal susceptibility profile of clinical C. lusitaniae isolates.
| Antifungals | No. isolates tested | MIC Range (μg/ml) |
GM ± SD (μg/ml) |
|---|---|---|---|
| Amphotericin B | 129 | 0.002–0.38 | 0.037 ± 0.072 |
| Fluconazole | 130 | 0.016–24* | 0.49 ± 2.32 |
| Voriconazole | 102 | 0.002–0.5 | 0.01 ± 0.09 |
| Caspofungin | 118 | 0.008–0.5 | 0.16 ± 0.08 |
*2 isolates showed MIC values of 16 and 24 μg/ml.
Discussion
In this study, we describe the prevalence of C. lusitaniae in clinical yeast species and its role in neonatal fungemia in Kuwait. For rapid and unambiguous identification, a simple, low-cost (~1 US$ per sample excluding the cost of culture and personnel time) C. lusitaniae-specific PCR assay was developed which could be completed within 4 hours using basic PCR and gel electrophoresis equipment that are readily available in routine mycology laboratories.
C. lusitaniae has attracted world-wide attention since some strains isolated previously were found to be resistant to amphotericin B [3, 4, 5, 25]. In the present study, C. lusitaniae was isolated from diverse clinical specimens suggesting its ability to colonize different anatomical sites, and thus underscoring its potential role as a nosocomial pathogen [26, 27]. Similar to other Candida species, C. lusitaniae also enters the host via the gastrointestinal, genitourinary and respiratory tracts or through intravascular catheters and the associated risk factors for invasive infections are not very different [28, 29]. Considering the fact that C. lusitaniae forms only a minor component of human yeast flora (Table 1), its isolation from sterile sites assumes a greater clinical significance as compared to C. albicans or some other commonly encountered species. The low level of prevalence of C. lusitaniae is also reflected by the fewer number of cases of invasive infections caused by this species in the hospitalized patients [30–36]. In a comprehensive laboratory-based multicenter surveillance study determining prevalence of C. albicans and non-albicans Candida species among bloodstream isolates, the rate of occurrence of C. lusitaniae was <2% [33]. Pfaller et al [29] recently reported epidemiological data obtained from Prospective Antifungal Therapy (PATH) Registry for invasive candidiasis caused by non-albicans Candida species in North America. C. lusitaniae was associated with 1.6% (n = 41) of 2496 invasive infections.
C. lusitaniae is also an uncommon cause of fungemia in neonates with few exceptions. A review of 59 fungemic neonates during 1994–2000 in Greece showed that only two (3.3%) cases were caused by C. lusitaniae [37]. In a comprehensive prospective observational study that included infants of ≤1000g birth weight, 137 infants developed candidiasis, and in only one (0.7%) case infection was caused by C. lusitaniae [38]. Like-wise, in an international, prospective study on epidemiology of invasive candidiasis, C. lusitaniae was isolated from 4% (n = 196) of the pediatric patients but not from the any of the 25 neonates [39]. On the contrary, C. lusitaniae was found to be more frequent among 45 patients of <1 year of age causing 13.3% (n = 6) of invasive infections in PATH registry data [29]. In a retrospective analysis of 318 bloodstream yeast infections among neonates in Kuwait during 2011–2017, 11 (3.45%) were caused by C. lusitaniae including 9 described here. Other studies from the Middle East Region have also reported invasive C. lusitaniae infections. Taj-Aldeen et al. [40] reported two cases of C. lusitaniae among 17 pediatric patients from Qatar. Both the patients recovered following therapy with lipid formulation of amphotericin B and/or fluconazole (Cases 13 and 14, Table 4). In another study from Saudi Arabia, C. lusitaniae was recognized as a cause of invasive candidiasis in 5 (4 in neonates and one in an infant) of 129 pediatric patients analyzed retrospectively [41].
Table 4. Summary of cases of C. lusitaniae fungemia an deep-seated infections reported in literature in neonates and infants.
| Case Nos. |
Reference | Country | Age, Sex | Source of isolation | Underlying disease or condition | Chemotherapy/ steroids /neutropenia | Intravascular catheter | Prior antibiotics | Antifungal therapy | Outcome |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Christenson et al.; 1987 [44] | USA | 2 m, M | Bloo d, RT | Congenital heart disease | Yes | Yes | Yes | AP | Recovery |
| 2 | Sanchez & Cooper; 1987 [57] | USA | 7 d, M | Blood, Urine, CSF | Prematurity | No | Yes | Yes | AP | Recovery |
| 3 | Yinnon et al; 1992 [58] | USA | 14 d, M | Blood, Urine, Catheter | Prematurity, CVL | No | Yes | Yes | AP, 5FC, KE | Recovery |
| 4 | Oleinik et al; 1993 [45] | USA | 8 d, M | blood | Prematurity | - | - | - | AP, FL, 5FC | Recovery |
| 5 | Nguyen et al; 1996 [30] | USA | 4 m, M | Blood, Catheter | CVL, cardiac surgery | - | Yes | - | AP | Recovery |
| 6 | Fowler et al; 1998 [27] | USA | 21 d, NA | Urine | Prematurity | No | Catheter | Yes | AP, FL, 5FC | Recovery |
| 7 | Fowler et al; 1998 [27] | USA | 35 d, NA | blood, Urine, CSF | Prematurity | No | Catheter | Yes | AP, 5FC, FL | Death |
| 8 | Fowler et al; 1998 [27] | USA | 35 d, NA | blood | Prematurity | No | Yes | Yes | AP, FL, 5FC | Recovery |
| 9 | Levy et al; 2002 [59] | USA | 3 m, M | Blood, Urine | Chronic granulomatous disease | No | - | Yes | IT, AP, FL | Death |
| 10 | Viudes et al; 2002 [31] | Spain | 46 d, F | Blood, urine | Prematurity, Congenital nephrotic syndrome | Yes | - | Yes | L-AmB | Recovery |
| 11 | Favel et al; 2003 [49] | France | 12 d, M | Blood, Urine, Nephrostomy catheter | Prematurity | No | Yes | Yes | L-AmB, AP, FL | Death |
| 12 | Estrada et al; 2006 [60] | USA | 3 m, M | Lymph node | Chronic granulomatous disease | - | - | Yes | FL | Recovery |
| 13 | Taj-Aldeen et al; 2014 [40] | Qatar | 7 m, F | Blood | Malabsorption | - | - | - | L-AmB, FL | Recovery |
| 14 | Taj-Aldeen et al; 2014 [40] | Qatar | 20 d, M | Blood | Motor development delay | - | - | - | L-AmB | Recovery |
| 15 | Gautam et al; 2014 [61] | Nepal | 7 d twins, F & M | blood | Prematurity | Yes | Yes | Yes | FL | Recovery |
| 16 | Chorro-Mari et al; 2015 [43] | UK | 189 d | NA | Prematurity, Pyelonephritis | L-AmB, MYC | NA | |||
| 17 | Sariguzel et al; 2017 [62] | Turkey | 8 m, M | CSF | Meningitis,Ventricular drainage | Yes | - | Yes | FL | Death |
Abbreviations:- M-male, F- female, d- day, w-week, m-month, y- year, NA- not available, CVL- central venous line, ST-solid tumor, VPS- ventriculoperitoneal shunt, RT- respiratory tract, CSF- cerebrospinal fluid, NA- not available,DT-digestive tract AP- amphotericine B, FL- fluconazole, 5FC- 5 fluorocytosine, L-AmB- liposomal Amphotericin B, IT- itraconazole, VO- voriconazole, CS- caspofungin, MYC-micafungin
While echinocandins are the first-line drugs for the treatment of candidemia in adult patients, amphotericin B or its lipid formulations are the recommended options for neonates [42]. In the present study, all neonates with C. lusitaniae fungemia received amphotericin B or its lipid formulation. Interestingly, two of the neonates were twins (Case 1 and Case 2), developing C. lusitaniae fungemia on day 10 and day 16 and died despite AmBisome therapy for 89 and 31 days, respectively (Table 2). None of the isolates appeared resistant to amphotericin B. Our experience for treating C. lusitaniae fungemia is limited. Of the two neonates where caspofungin was used as a salvage therapy, one expired apparently due to other complications. The crude mortality of C. lusitaniae fungemia in our study may be taken as 33.3% as the three expired neonates also had bacterial septicemia. Recently Chorri-Mari & Christyiansen [43] used micafungin (15 mg/kg/day) in a preterm neonate for treating C. lusitaniae pyelonephritis for 24 days with no adverse effects. This patient was previously given liposomal amphotericin B without any clinical response.
Contrary to some early reports [44, 45], the occurrence of intrinsic or acquired resistance to amphotericin B is not as common as generally perceived [9, 46]. It is also supported by amphotericin B MIC values of 129 C. lusitaniae obtained in the present study (0.037 ± 0.072 μg/ml) (Table 3). In an early report, Blinkhorn et al. [28] described two adult cases of C. lusitaniae fungemia caused by amphotericin B-susceptible strains, who responded to amphotericin B therapy and recovered. The authors emphasized that isolates from patients responding poorly to amphotericin B therapy should be tested for susceptibility. The rarity of amphotericin B resistance among C. lusitaniae isolates was also supported by the study of Diekema et al. [32]. Of 171 C. lusitaniae isolates collected globally, only 3 (1.75%) isolates showed MIC values of ≥2 μg/ml against amphotericin B [27]. It is possible that despite in vitro susceptibility of C. lusitaniae isolates to amphotericin B, there are other factors which may impact clinical efficacy. It has been demonstrated by time-kill curve analysis that susceptible C. lusitaniae strains may be less prone to fungicidal activity of amphotericin B as compared to C. glabrata or C. albicans and thus may be less amenable to treatment particularly in neutropenic cancer patients [12].
It has also been suggested that C. lusitaniae in its haploid state may be prone to developing drug resistance [47]. The molecular mechanisms of amphotericin B resistance are being elucidated in Candida species [48]. There are also reports of acquisition of multidrug resistance in C. lusitaniae in association with phenotypic switching [49–51]. Favel et al. [49] described a case of fatal kidney infection in a preterm neonate due to C. lusitaniae. The isolates developed multidrug resistance to amphotericin B and fluconazole during therapy, which was accompanied with switching in colonial morphology. Asner et al. [11] recently reported a case of rapid development of multidrug resistance in C. lusitaniae to amphotericin B, caspofungin, flucytosine and azoles during treatment of persistent candidemia in an immunosuppressed child. The authors hypothesized that selection pressure created due to use of multiple antifungal drugs might have facilitated successive emergence of resistant strains during the course of therapy. C. lusitaniae is a heterothallic ascomycetous yeast and mating is only possible between two haploid cells of opposite mating types [47]. Little is known about the mating type that is predominantly associated with human infection or prone to developing resistance or undergo phenotypic switching [51].
As stated above, there are no CLSI/EUCAST antifungal susceptibility breakpoints available for this species. However, several studies have determined epidemiologic cut-off values for C. lusitaniae to discriminate wild-type and non-wild-type strains [52–54]. Our C. lusitaniae isolates appeared susceptible to amphotericin B, voriconazole, caspofungin and resistance to fluconazole (≥8 μg/ml) was observed in only two strains (Table 3). Pfaller et al. [55] determined caspofungin susceptibility of 105 C. lusitaniae isolates obtained over a 4-year period (2001 and 2004) from 91 institutions under global surveillance program. All the isolates were inhibited at a concentration of ≤4 μg/ml. None of our isolates showed MIC value for caspofungin above >0.5 μg/ml (Table 3). We have no comparative susceptibility data for micafungin and anidulafungin, which is a limitation of our study. Here attention may be drawn to a recent study demonstrating loss of fungicidal or fungistatic activity of micafungin in the presence of serum proteins, which is not predicted by MICs in case of C. lusitaniae [56]. The authors suggested that micafungin and probably other echinocandins should be used with caution against rare Candida species including C. lusitaniae.
A PubMed-based literature search revealed 17 case reports of C. lusitaniae invasive infections in neonates/children since 1984 (Table 4) [27, 30, 31, 40, 43–45, 49, 57–62]. All had underlying conditions, which included congenital defects (n = 4), prematurity (n = 10) and chronic granulomatous diseases (n = 2) and meningitis/extraventricular shunt (n = 1). Their age ranged between 7 days to 8 months. All patients received antifungal therapy, mainly with amphotericin B (n = 10) or liposomal amphotericin B (n = 5). Three patients were treated with fluconazole alone and one died. Mortality in these 17 cases was about 24% (Table 4).
In conclusion, we have described prevalence and antifungal susceptibility of C. lusitaniae isolates obtained from diverse clinical specimens over a seven-year period in Kuwait. Additionally, 9 cases of C. lusitaniae fungemia in neonates are also described. All isolates appeared susceptible to amphotericin B.
Supporting information
Pink-colored colonies of five Candida species on CHROMagar Candida: (a) C. auris, (b) C. famata, (c) C. guilliermondii, (d) C. lusitaniae, strain No. Kw 1812/11-MH, (e) C. lusitaniae strain No. Kw 2212/16-MH and (f) C. glabrata.
(DOCX)
Lane M is 100 bp DNA ladder and the positions of migration of 100 bp, 300 bp and 600 bp fragments are marked.
(DOCX)
Acknowledgments
The study was supported by Kuwait University Research Sector grant No. MI01/15. Authors are thankful to Sandhya Vayalil, Soumya Varghese and Omar Al-Musallam for technical support.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
The work was supported by Kuwait University Research Sector grant No. MI01/15 (granted to Ziauddin Khan). The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.van Uden N, Buckley H. Candida Berkhout p. 893–1087. In Lodder J. (ed.). The yeasts. A taxonomic study. North-Holland Publishing Co; Amsterdam: 1970. [Google Scholar]
- 2.Merz WG, Sandford GR. Isolation and characterization of a polyene-resistant variant of Candidt tropicalis. J Clin. Microbiol. 1979; 9:677–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Merz WG. Candida lusitaniae: frequency of recovery, colonization, infection, and amphotericin B resistance. J Clin Microbiol.1984; 20:1194–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Holzschu DL, Presley HL, Miranda M, Phaff HJ. Identification of Candida lusitaniae as an opportunistic yeast in humans. J Clin Microbiol. 1979; 10: 202–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pappagianis D, Collins MS, Hector R, Remington J. Development of resistance to amphotericin B in Candida lusitaniae infecting a human. Antimicrob Agents Chemother. 1979; 16:123–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wingard JR. Importance of Candida species other than C. albicans as pathogens in oncology patients. Clin Infect Dis. 1995; 20: 115–125. [DOI] [PubMed] [Google Scholar]
- 7.Minari A, Hachem R, Raad I. Candida lusitaniae: A cause of breakthrough fungemia in cancer patients. Clin Infect Dis. 2001; 32:186–190. 10.1086/318473 [DOI] [PubMed] [Google Scholar]
- 8.Guinet R., Chanas J, Goullier A, Bonnefoy G, Ambroise-Thomas P. Fatal septicemia due to amphotericin B-resistant Candidia lusitaniae. J. Clin. Microbiol. 1983; 18:443–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clancy C, Nguyen M, Yu V. Fungemia caused by Candida lusitaniae Infections in medicine, 1996; 13: 940, 948–951. [Google Scholar]
- 10.Hawkins JL, Baddour LM. Candida lusitaniae infections in the era of fluconazole availability. Clin Infect Dis. 2003; 36: e14–8. 10.1086/344651 [DOI] [PubMed] [Google Scholar]
- 11.Asner SA, Giulieri S, Diezi M, Marchetti O, Sanglard D. Acquired Multidrug Antifungal Resistance in Candida lusitaniae during Therapy. Antimicrob Agents Chemother. 2015; 59: 7715–7722. 10.1128/AAC.02204-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Atkinson BJ, Lewis RE, Kontoyinnis DP. Candida lusitaniae fungemia in cancer patients: risk factors for amphotericin B failure and outcome. Med Mycol. 2008, 46: 541–546. 10.1080/13693780801968571 [DOI] [PubMed] [Google Scholar]
- 13.Jung DS, Farmakiotis D, Jiang Y, Tarrand JJ, Kontoyiannis DP. Uncommon Candida species fungemia among cancer patients, Houston, Texas, USA. Emerg Infect Dis. 2015; 21:1942–1950. 10.3201/eid2111.150404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jeragh A, Ahmad S, Naseem J, Khan ZU. Candida lusitaniae arthritis in an intravenous drug user. Mycoses. 2007; 50: 430–432. 10.1111/j.1439-0507.2007.01394.x [DOI] [PubMed] [Google Scholar]
- 15.Ahmad S, Khan Z. Invasive candidiasis: a review of nonculture-based laboratory diagnostic methods. Indian J Med Microbiol 2012; 30: 264–269. 10.4103/0255-0857.99482 [DOI] [PubMed] [Google Scholar]
- 16.Jamal WY, Ahmad S, Khan ZU, Rotimi VO. Comparative evaluation of two matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) systems for the identification of clinically significant yeasts. Int J Infect Dis. 2014; 26:167–170. 10.1016/j.ijid.2014.05.031 [DOI] [PubMed] [Google Scholar]
- 17.Asadzadeh M, Ahmad S, Hagen F, Meis JF, Al-Sweih N, Khan Z. Simple, low-cost detection of Candida parapsilosis complex isolates and molecular fingerprinting of Candida orthopsilosis strains in Kuwait by ITS region sequencing and amplified fragment length polymorphism analysis. PLoS One 2015; 10: e0142880 10.1371/journal.pone.0142880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ahmad S, Mustafa AS, Khan Z, Al-Rifaiy AI, Khan ZU. PCR-enzyme immunoassay of rDNA in the diagnosis of candidemia and comparison with amplicon detection by agarose gel electrophoresis. Int J Med Microbiol 2004; 294: 45–51. 10.1016/j.ijmm.2004.01.002 [DOI] [PubMed] [Google Scholar]
- 19.Khan ZU, Ahmad S, Mokaddas E, Chandy R, Cano J, Guarro J. Actinomucor elegans var. kuwaitiensis isolated from the wound of a diabetic patient. Antonie van Leeuwoenhoek 2008; 94: 343–352. [DOI] [PubMed] [Google Scholar]
- 20.Khan ZU, Ahmad S, Hagen F, Fell JW, Kowshik T, Chandy R, Boekhout T. Cryptococcus randhawai sp. nov., a novel anamorphic basidiomycetous yeast isolated from tree trunk hollow of Ficus religiosa (peepal tree) from New Delhi, India. Antonie Van Leeuwenhoek 2010; 97: 253–259. 10.1007/s10482-009-9406-8 [DOI] [PubMed] [Google Scholar]
- 21.Schoch CL, Seifert KA, Huhndorf S, Robert V, Spouge JL, Levesque CA, et al. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for fungi. Proc Natl Acad Sci USA 2012; 109, 6241–6246. 10.1073/pnas.1117018109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ahmad S, Khan Z, Mustafa AS, Khan ZU. Epidemiology of Candida colonization in an intensive care unit of a teaching hospital in Kuwait. Med Mycol 2003; 41: 487–493. [DOI] [PubMed] [Google Scholar]
- 23.Al-Haqqan A, Al-Sweih N, Ahmad S, Khan S, Joseph L, et al. Azole-resistant Candida blankii as a newly recognized cause of bloodstream infection. New Microbes New Infect. 2018; 26: 25–29. 10.1016/j.nmni.2018.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Asadzadeh M, Al-Sweih NA, Ahmad S, Khan ZU. Antifungal susceptibility of clinical Candida parapsilosis isolates in Kuwait. Mycoses 2008; 51: 318–323. 10.1111/j.1439-0507.2008.01492.x [DOI] [PubMed] [Google Scholar]
- 25.Hadfield TL, Smith MB, Winn RE, Rinaldi MG, Guerra C. Mycoses caused by Candida lusitaniae. Rev Infect Dis. 1987; 9:1006–12. [DOI] [PubMed] [Google Scholar]
- 26.Sanchez V, Vazquez JA, Barth-Jones D, Dembry L, Sobel JD, Zervos MJ. Epidemiology of nosocomial acquisition of Candida lusitaniae. J Clin Microbiol. 1992; 30:3005–3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fowler SL, Rhoton B, Springer SC, Messer SA, Hollis RJ, Pfaller MA. Evidence for person-to-person transmission of Candida lusitaniae in a neonatal intensive-care unit. Infect Control Hosp Epidemiol. 1998; 19:343–5. [DOI] [PubMed] [Google Scholar]
- 28.Blinkhorn RJ, Adelstein D, Spagnuolo PJ. Emergence of a new opportunistic pathogen, Candida lusitaniae. J Clin Microbiol. 1989; 27: 236–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pfaller MA, Andes DR, Diekema DJ, Horn DL, Reboli AC, Rotstein C., et al. Epidemiology and outcomes of invasive candidiasis due to non-albicans species of Candida in 2,496 patients: data from the Prospective Antifungal Therapy (PATH) registry 2004–2008. PLoS One. 2014; 9: e101510 10.1371/journal.pone.0101510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nguyen MH, Morris AJ, Dobson ME, Yu VL. Candida lusitaniae: An important emerging cause of candidemia. Infect Dis Clini Pract. 1996,5:273–278. [Google Scholar]
- 31.Viudes A, Pemán J, Cantón E, Salavert M, Ubeda P, López-Ribot JL, et al. Two cases of fungemia due to Candida lusitaniae and a literature review. Eur J Clin Microbiol Infect Dis. 2002; 21:294–299. 10.1007/s10096-002-0713-5 [DOI] [PubMed] [Google Scholar]
- 32.Diekema DJ, Messer SA, Boyken LB, Hollis RJ, Kroeger J, Tendolkar S, et al. In vitro activity of seven systemically active antifungal agents against a large global collection of rare Candida species as determined by CLSI broth microdilution methods. J Clin Microbiol. 2009; 47: 3170–3177. 10.1128/JCM.00942-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Falagas ME, Roussos N, Vardakas KZ. Relative frequency of albicans and the various non-albicans Candida spp among candidemia isolates from inpatients in various parts of the world: a systematic review. Int J Infect Dis. 2010; 14: e954–e966. 10.1016/j.ijid.2010.04.006 [DOI] [PubMed] [Google Scholar]
- 34.Faria-Ramos I, Neves-Maia J, Ricardo E, Santos-Antunes J, Silva AT, Costa-de-Oliveira S, et al. Species distribution and in vitro antifungal susceptibility profiles of yeast isolates from invasive infections during a Portuguese multicenter survey. Eur J Clin Microbiol Infect Dis. 2014; 33: 2241–2247. 10.1007/s10096-014-2194-8 [DOI] [PubMed] [Google Scholar]
- 35.Montagna MT, Lovero G, Borghi E, Amato G, Andreoni S, Campion L, et al. Candidemia in intensive care unit: a nationwide prospective observational survey (GISIA-3 study) and review of the European literature from 2000 through 2013. Eur Rev Med Pharmacol Sci. 2014; 18: 661–674. [PubMed] [Google Scholar]
- 36.Orasch C, Marchetti O., Garbino J., Schrenzel J., Zimmerli S., Muhlethaler K., et al. Candida species distribution and antifungal susceptibility testing according to European Committee on Antimicrobial Susceptibility Testing and new vs. old Clinical and Laboratory Standards Institute clinical breakpoints: a 6-year prospective candidaemia survey from the fungal infection network of Switzerland. Clin Microbiol Infect. 2014; 20: 698–705 10.1111/1469-0691.12440 [DOI] [PubMed] [Google Scholar]
- 37.Roilides E, Farmaki E, Evdoridou J, Dotis J, Hatziioannidis E, Tsivitanidou M, et al. Neonatal candidiasis: analysis of epidemiology, drug susceptibility, and molecular typing of causative isolates. Eur J Clin Microbiol Infect Dis. 2004; 23:745–50. 10.1007/s10096-004-1210-9 [DOI] [PubMed] [Google Scholar]
- 38.Benjamin DK Jr, Stoll BJ, Gantz MG, Walsh MC, Sánchez PJ, Das A, et al. Neonatal candidiasis: epidemiology, risk factors, and clinical judgment. Pediatrics. 2010; 126: e865–73. 10.1542/peds.2009-3412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Steinbach WJ, Roilides E, Berman D, Hoffman JA, Groll AH, Bin-Hussain I, et al. Results from a prospective, international, epidemiologic study of invasive candidiasis in children and neonates. Pediatr Infect Dis J 2012; 31: 1252–1257. 10.1097/INF.0b013e3182737427 [DOI] [PubMed] [Google Scholar]
- 40.Taj-Aldeen SJ, AbdulWahab A, Kolecka A, Deshmukh A, Meis JF, Boekhout T. Uncommon opportunistic yeast bloodstream infections from Qatar. Med Mycol. 2014; 52:552–556. 10.1093/mmycol/myu016 [DOI] [PubMed] [Google Scholar]
- 41.Almoosa Z, Ahmed GY, Omran A, AlSarheed A, Alturki A, Alaqeel A, et al. Invasive candidiasis in pediatric patients at King Fahad Medical City in Central Saudi Arabia. A 5-year retrospective study. Saudi Med J. 2017; 38:1118–1124. 10.15537/smj.2017.11.21116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pappas PG, Kauffman CA, Andes DR, Clancy CJ, Marr KA, Ostrosky-Zeichner L, et al. Clinical Practice Guideline for the Management of Candidiasis: 2016 Update by the Infectious Diseases Society of America. Clin Infect Dis. 2016; 62: e1–50. 10.1093/cid/civ933 Epub 2015 Dec 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chorro-Mari V, Christiansen N. Safety of high dose micafungin for the treatrment of fungal infections in neonates. Abstracts from the Neonatal and Paediatric Pharmacists Group (NPPG) 20th Annual Meeting (P. 13). Arch Dis Child. 2015; 100: e1. [Google Scholar]
- 44.Christenson JC, Guruswamy A, Mukwaya G, Rettig PJ. Candida lusitaniae: an emerging human pathogen. Pediatr Infect Dis J. 1987; 6:755–757. [PubMed] [Google Scholar]
- 45.Oleinik EM, Della-Latta P, Rinaldi MG, Saiman L. Candida lusitaniae osteomyelitis in a premature infant. Am J Perinatol. 1993; 10:313–315. 10.1055/s-2007-994749 [DOI] [PubMed] [Google Scholar]
- 46.Favel A, Michel-Nguyen A, Datry A, Challier S, Leclerc F, Chastin C, et al. Susceptibility of clinical isolates of Candida lusitaniae to five systemic antifungal agents. J Antimicrob Chemother. 2004; 53:526–529. 10.1093/jac/dkh106 [DOI] [PubMed] [Google Scholar]
- 47.François F, Noël T, Pépin R, Brulfert A, Chastin C, Favel A, et al. Alternative identification test relying upon sexual reproductive abilities of Candida lusitaniae strains isolated from hospitalized patients. J Clin Microbiol. 2001; 39: 3906–3914. 10.1128/JCM.39.11.3906-3914.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ahmad S, Joseph L, Parker JE, Asadzadeh M, Kelly SL, Meis JF, et al. ERG6 and ERG2 are major targets conferring reduced susceptibility to amphotericin B in clinical Candida glabrata isolates in Kuwait. Antimicrob Agents Chemother. 2019; 63: e01900–18. 10.1128/AAC.01900-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Favel A, Michel-Nguyen A, Peyron F, Martin C, Thomachot L, Datry A, et al. Colony morphology switching of Candida lusitaniae and acquisition of multidrug resistance during treatment of a renal infection in a newborn: case report and review of the literature. Diagn Microbiol Infect Dis. 2003; 47:331–339. [DOI] [PubMed] [Google Scholar]
- 50.McClenny NB, Fei H, Baron EJ, Gales AC, Houston A, Hollis RJ, et al. Change in colony morphology of Candida lusitaniae in association with development of amphotericin B resistance. Antimicrob Agents Chemother. 2002; 46:1325–1328. 10.1128/AAC.46.5.1325-1328.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Favel A, Michel-Nguyen A, Datry A, Challier S, Leclerc F, Chastin C, et al. Occurrence of Clavispora lusitaniae, the teleomorph of Candida lusitaniae, among clinical isolates. J Clin Microbiol. 1990; 28: 2224–2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Castanheira M, Messer SA, Rhomberg PR, Pfaller MA. Antifungal susceptibility patterns of a global collection of fungal isolates: results of the SENTRY Antifungal Surveillance Program (2013). Diagn Microbiol Infect Dis. 2016; 85:200–204. 10.1016/j.diagmicrobio.2016.02.009 [DOI] [PubMed] [Google Scholar]
- 53.Pfaller MA, Espinel-Ingroff A, Bustamante B, Canton E, Diekema DJ, Fothergill A, et al. Multicenter study of anidulafungin and micafungin MIC distributions and epidemiological cutoff values for eight Candida species and the CLSI M27-A3 broth microdilution method. Antimicrob Agents Chemother. 2014; 58:916–922. 10.1128/AAC.02020-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lockhart SR, Pham CD, Kuykendall RJ, Bolden CB, Cleveland AA. Candida lusitaniae MICs to the echinocandins are elevated but FKS-mediated resistance is rare.Diagn Microbiol Infect Dis. 2016; 84: 52–54. 10.1016/j.diagmicrobio.2015.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pfaller MA, Boyken L, Hollis RJ, Messer SA, Tendolkar S, Diekema DJ. In vitro susceptibilities of Candida spp. to caspofungin: four years of global surveillance. J Clin Microbiol. 2006; 44: 760–763. 10.1128/JCM.44.3.760-763.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Qasem S, Renátó K, Gábor K, Rudolf G, Tamás K, Aliz B, László M. Decreased killing activity of micafungin against Candida guilliermondii, Candida lusitaniae, and Candida kefyr in the presence of human serum. Microb Drug Resist.2017; 23: 764–770. 10.1089/mdr.2016.0241 [DOI] [PubMed] [Google Scholar]
- 57.Sanchez PJ, Cooper BH. Candida lusitaniae: sepsis and meningitis in a neonate. Pediatr Infect Dis J. 1987; 6:758–759. [PubMed] [Google Scholar]
- 58.Yinnon AM, Woodin KA, Powell KR. Candida lusitaniae infection in the newborn: case report and review of the literature. Pediatr Infect Dis J. 1992; 11: 878–880. [DOI] [PubMed] [Google Scholar]
- 59.Levy O, Bourquin JP, McQueen A, Cantor AB, Lachenauer C, Malley R. Fatal disseminated Candida lusitaniae infection in an infant with chronic granulomatous disease. Pediatr Infect Dis J. 2002; 21:262–264. [DOI] [PubMed] [Google Scholar]
- 60.Estrada B, Mancao MY, Polski JM, Figarola MS. Candida lusitaniae and chronic granulomatous disease. Pediatr Infect Dis J. 2006; 25: 758–759. 10.1097/01.inf.0000227802.32163.a4 [DOI] [PubMed] [Google Scholar]
- 61.Gautam MK, Li J. Neonatal Candida lusitaniae Septicemia. J Nepal Paediatr Soc 2014; 34: 160–162. [Google Scholar]
- 62.Sariguzel FM, Koc AN, Ozturk M. Candida Utilis and Candida lusitaniae Meningitis in an infant with extraventricular drainage. Van Tıp Derg 2017; 24: 40–43. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Pink-colored colonies of five Candida species on CHROMagar Candida: (a) C. auris, (b) C. famata, (c) C. guilliermondii, (d) C. lusitaniae, strain No. Kw 1812/11-MH, (e) C. lusitaniae strain No. Kw 2212/16-MH and (f) C. glabrata.
(DOCX)
Lane M is 100 bp DNA ladder and the positions of migration of 100 bp, 300 bp and 600 bp fragments are marked.
(DOCX)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.