Abstract
Morphological and molecular characteristics determine the function of biological tissues. Attempts to combine immunofluorescence and electron microscopy invariably compromise the quality of the ultrastructure of tissue sections. We developed NATIVE, a correlated light and electron microscopy approach that preserves ultrastructure while showing the locations of multiple molecular moieties even deep within tissues. This technique allowed the large-scale 3D reconstruction of a volume of mouse hippocampal CA3 tissue at nanometer resolution.
Keywords: CLEM, nanobody, ultrastructure, immunostaining, connectomics
The correlation between the morphology of a biological tissue at the nanometer scale (i.e., ultrastructure) and its molecular attributes poses major challenges, a problem acute in the study of the brain. Permeabilization of its membranes for immunolabeling will degrade the quality of any electron microscopy (EM) image of a biological specimen. Techniques that do not require permeabilization include transgenic labeling with fluorescent proteins, tags that generate a product visible in the EM, and post-embedding immunogold-EM.1–3 None of these non-permeabilization techniques however, are useful if the goal is to display many different molecular markers in the same sample.
One tissue where it would be useful to have many different markers in the same sample is the brain. The number of different cell types in the brain is vast and their organization remains poorly understood. Nevertheless, molecular markers exist for many of them. In principle correlation of immunofluorescence images with EM images of the same sample (CLEM) could provide the molecular identities of cells within an EM image. To get around the permeabilization problem antibodies can be applied after sectioning of a resin infiltrated block, as used in Array Tomography.4 However, individual staining of hundreds or thousands of sections is both labor- and cost- intensive. The resin that infiltrates the tissue must be less hydrophobic than the hardest resins, making it difficult to obtain very thin sections (i.e., below 50 nm).5 Instead of using OsO4, less than ideal heavy metal alternatives may avoid epitope destruction but with attendant loss of ultrastructural quality.5 New clearing and expansion technologies compatible with immuno-staining or fluorescent protein expression have improved the resolution of volumetric fluorescence imaging.6,7 However, rather than staining all cells and organelles, as occurs in electron microscopy, only the fluorescently labeled signals are visible. Moreover, these clearing and expansion methods eliminate the possibility of subsequently doing high resolution electron microscopy on the same samples.
To combine pre-embedding immunofluorescence and electron microscopy with preservation of ultrastructure, we developed NATIVE (Nanobody-Assisted Tissue Immunostaining for Volumetric Electron microscopy). NATIVE is based on the use of nanobodies, single-domain antibody fragments derived from camelid heavy chain-only antibodies,8 as new immunofluorescence staining agents, which allow staining for light microscopy fully decoupled from staining for EM. Therefore, each modality can operate without compromise and at its full capability, an advantage over existing CLEM strategies.
The cytoplasm of aldehyde-fixed tissues is poorly accessible to full-size antibodies, but a slight increase in permeability during paraformaldehyde fixation might make cells accessible to nanobodies instead (Fig. 1). We selected a GFP-specific nanobody (Enh), labeled it with an organic fluorophore AF647 using sortase,9,10 then tested its suitability in NATIVE. Brain slices from a YFP-16 transgenic mouse, which expresses a yellow GFP variant in its neurons under the control of the Thy-1 promoter,11 were stained with AF647-conjugated GFP-specific nanobody (Enh). The YFP signal correlated well with Enh staining (Fig. 1a), indicating that intracellular epitopes are accessible to nanobodies after chemical fixation. To see if inclusion of extracellular sucrose to the fixative, in order to prevent cell swelling and maintain extracellular space (ECS) 12, improved penetration of the nanobody, we tested Enh nanobody on 500 μm thick fixed brain sections. Enh with preservation of ECS penetrated deeply, with good staining intensity up to 100 µm on the side of application (48-hour staining at 4°C; Fig. 1b,c), beyond the imaging depth of confocal microscopes when operated without tissue clearing. An anti-GFP monoclonal antibody (~150 kDa) only stained the very surface without permeabilization. (Fig. 1d,e). Preservation of ECS for NATIVE (Fig. 1f) allows deeper penetration of the staining agent, also better represents the natural condition of brain tissue for more accurate ultrastructure and facilitates segmentation of EM data.12
Figure 1.
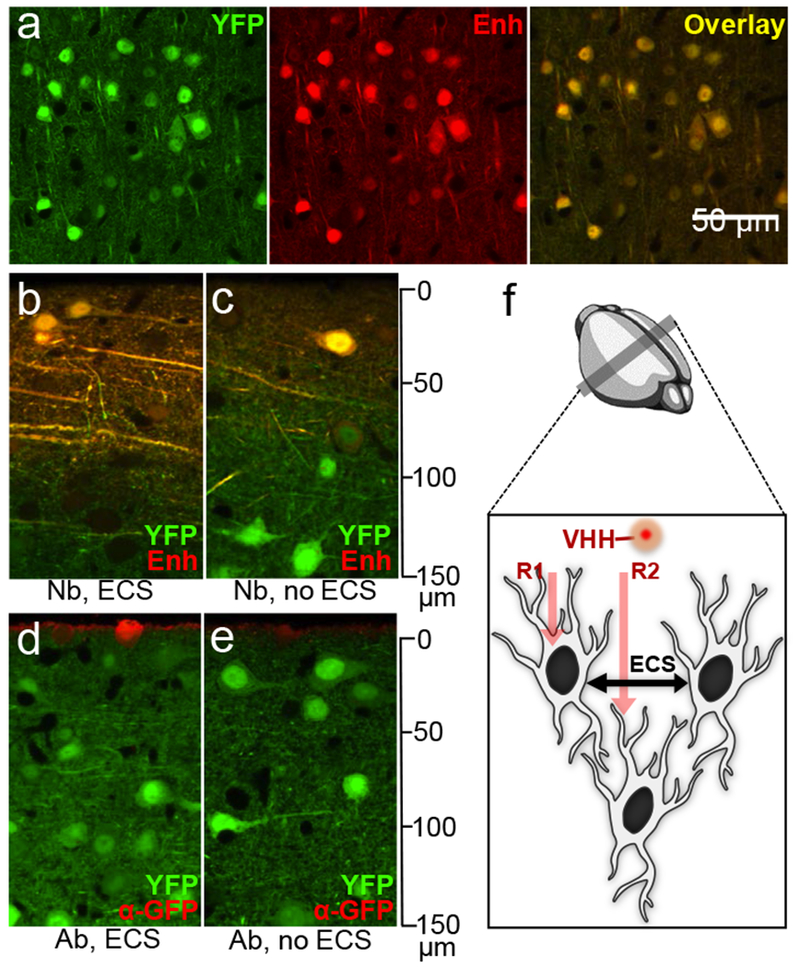
Optimization of NATIVE. (a) Brain tissue from YFP-16 mice was stained by a GFP-specific nanobody (Enh) to show the extent of co-registration. This experiment was performed seven times independently, with same results obtained each time. (b-e) Penetration comparison: tissues were stained with nanobodies. This experiment was performed twice independently, with same results obtained each time. (b, c) and commercial full-size antibodies (d, e) against GFP. Images were taken at cross-sections to show penetration with (b, d) or without (c, e) ECS. [Nb = nanobody; Ab = full-size antibody; ECS = extracellular space]. (f) Proposed dual-mechanism for enhanced penetration of nanobodies. Route 1 (R1): direct penetration of chemically fixed cell membrane; Route 2 (R2): penetration through preserved extracellular space.
We expanded the selection of nanobodies to VHH-DC1313, VHH-2114, and VHH-E915, which recognize cell surface markers integrin-αM (CD11b; microglia) and lymphocyte antigen Ly-6C/6G (endothelial cells), and an intracellular marker glial fibrillary acidic protein (GFAP), respectively. VHH-DC13, 21 and E9 were each sortagged to a different oligoglycine-modified fluorophore (AF488, AF647, and TexasRed respectively). We subsequently performed confocal fluorescence imaging on nanobody-stained intact thick tissue sections retrieved from perfusion-fixed (4% paraformaldehyde) and ECS-preserved (3% sucrose) animals. Laser scanning confocal microscopy with a 40× 1.3 NA oil immersion lens in z-stack mode was used to reconstruct a 400 µm × 400 µm × 42 µm volume of mouse hippocampal CA3 (xyz pixel size: X-Y 0.208 μm, Z 1 μm; Fig. 2a, Supplementary Figure 1 & 2, Supplementary Video 1 & 2). Confocal imaging was immediately followed by secondary chemical fixation (2.5 % glutaraldehyde + 2% paraformaldehyde) in preparation for EM.
Figure 2.
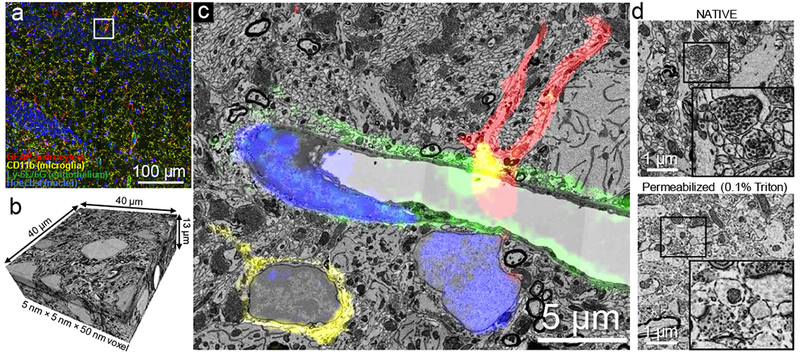
NATIVE enables tissue-scale, multiplex CLEM. (a) Fluorescence confocal image stack is used to identify a region of hippocampus with abundant astrocytes, microglial cells and capillaries, by virtue of nanobody-labeled GFAP (astrocyte marker) in red, CD11b (microglia marker) in yellow, Ly-6C/6G (endothelial cell marker for capillaries) in green and cell nuclei stained by Hoechst in blue. This experiment was performed five times independently, with same results obtained each time. (b) High resolution serial sectioning EM dataset acquired in the region marked by the white box in a. (c) A section in the acquired EM dataset shows that NATIVE-stained tissue is suitable for high quality CLEM. The aligned confocal image layer confirms molecular identities of the different cell types (a fuller demonstration of the alignment between fluorescence and electron microscopy can be seen in Supplementary Video 2. (d) Tissue ultrastructure was compared between NATIVE (top) and full-size antibody staining conditions with 0.1% Triton X100 (bottom). Full resolution pictures are provided in Supplementary Figure 5, Supplementary Figure 6. This experiment was performed twice independently, with same results obtained each time.
We used an automated tape-collecting ultramicrotome (ATUM) to collect 800 serial sections of 50 nm thickness on carbon-coated (i.e., conductive) Kapton tape, covering 40 μm of depth. Strips of tape containing the sections were affixed to silicon wafers and imaged using a scanning EM.16,17 We acquired a low-resolution EM volume of 400 μm × 400 μm × 13 μm and a high-resolution EM volume of 40 μm × 40 μm × 13 μm that coincided with part of the confocal volume (Fig. 2b, Supplementary Figure 3, Supplementary Video 3).
The low resolution EM data was used to map the position of the sections on the wafers for automated EM imaging and also to locate the region imaged by fluorescence.17 However, the confocal and EM stacks were rotated, tilted and stretched differently. To obtain registration of the two data sets, we performed a 3D affine transformation based on 16 corresponding-point pairs placed on nucleoli, blood vessel branches and lysosomes in both the fluorescence microscopy and the EM datasets. In any small region of the aligned confocal and EM image stacks we could identify fluorescently labeled molecular markers and their ultrastructural correlates. In Fig. 2c, the fluorescence images of the CA3 region of hippocampus identify microglial cells (CD11b staining, yellow) and astrocytes (GFAP staining, red) near the endothelial cells of a blood vessel (Ly6C/6G staining, green), presumably representing the blood-brain barrier (BBB) and nuclei (Hoechst, blue).
The preservation of ultrastructure in NATIVE-stained samples was excellent and identical to staining of EM samples without immunostaining or fluorescence imaging. Synaptic vesicles, synapse contacts, mitochondria, intracellular microtubule bundles, axons and dendritic processes were well preserved with excellent membrane contrast throughout the entire CLEM dataset (i.e. 40 μm; Supplementary Figure 4). In contrast, the use of 0.1% Triton as a mild permeabilization reagent showed extensive deterioration of membrane structure with complete loss of fine processes (Fig. 2d).
We succeeded in segmenting a most difficult target in brain tissue reconstruction, which are the glia. Glia show great variability in the shape of their processes and have many small membranous extrusions that are below the diffraction limit of confocal fluorescence microscopes. Using a computer-assisted manual segmentation approach (VAST),16 we segmented both a nanobody-stained astrocyte and a nanobody stained microglial cell (Fig. 3a and full 3D view Supplementary Figure 7, Supplementary Video 4). The complicated spatial orientation and cell-cell contacts were well-preserved by NATIVE in 3D reconstruction. The reconstructed microglial cell and astrocyte in an overlay with the fluorescence confocal stack (3D-CLEM) show the advantage of EM for ascertaining fine structure (Fig. 3b, c). We found a complicated process of a microglial cell that envelopes parts of other cells and some debris (Fig. 3b arrow). An enveloped synapse is consistent with the role of microglia in synapse elimination.19 A magnified section from this region of the cell (Fig. 3d, Supplementary Figure 8) shows the many finger-like protrusions near debris and other cellular material that was not immunolabeled. We identified an expanded perivascular terminal structure (endfoot) of the astrocyte on the surface of the blood vessel, along with smaller contacts from the same cell (Fig. 3c, e). We also identified a tripartite synapse (Fig. 3c, arrows), where the astrocyte forms a sheath surrounding a pair of presynaptic and postsynaptic elements (Fig. 3f). Correlation of the GFAP signal in fluorescence with GFAP fiber bundles in the electron micrographs (Supplementary Figure 9) shows a continuous distribution of this fibrillary protein around the nucleus, forming a belt-like structure around the astrocyte’s dumbbell-shaped nucleus (Fig. 3g-h, Supplementary Figure 10 & 11, Supplementary Video 5 & 6).
Figure 3.
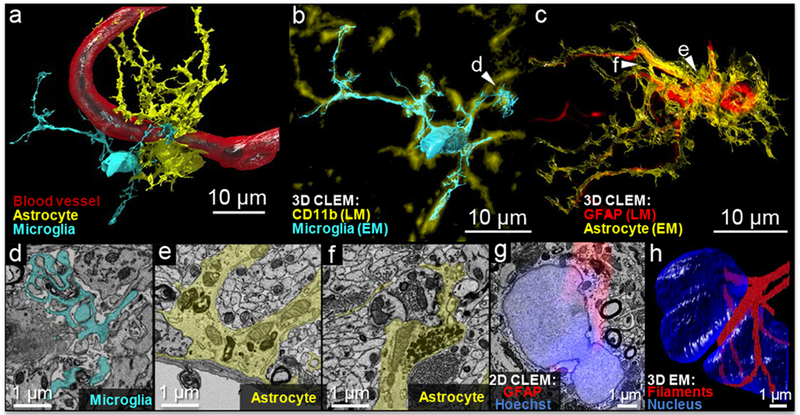
NATIVE-enabled correlative 3D volumetric reconstruction. (a) Volumetric 3D reconstruction based on the segmentation of data from scanning electron microscopy and guided by correlative LM imaging with unambiguous molecular marker identification. (b) and (c) 3D-CLEM overlay of high-resolution EM reconstructed cell contours with fluorescence-based reconstructed molecular markers. The fluorescence and EM reconstructions were displayed in different colors, as indicated by the legends, to show the correlations more clearly. The slight shifts in the fluorescence and EM cell boundaries derive from the different lateral and depth resolution of the light and electron-based datasets and small deformations that were not corrected with the global 3D affine transformation (<2 μm). (d) Phagocytosis by a microglial cell (pseudo-color: cyan) with finger-like protrusions enveloping debris. (e) A cross-section of blood-brain-barrier with the astrocyte (pseudo-color: yellow) around the blood endothelium cells. (f) A tripartite synapse formed by an astrocyte (pseudo-color: yellow) around a pair of pre/post synapses. (g) In 2D-CLEM, fluorescence layer reveals a continuous distribution of the fibrillary protein around the nucleus; (f) 3D-EM reconstruction confirms belt-like GFAP filaments around the astrocyte’s dumbbell-shaped nucleus.
NATIVE-enabled 3D-CLEM also provides a reference for robust proofreading during serial section electron microscopy reconstruction. EM reconstruction guided by fluorescence provides a useful check for manual tracing and maybe even automated segmentation (Fig. 3b, c).
In conclusion, the NATIVE correlated light and EM technique, by use of fluorescently tagged nanobodies, allows, in one sample, complete methodological separation of immunolabelling and fluorescence imaging from electron microscopy staining and imaging without compromising the quality of images in either regime. Light sheet or super-resolution microscopy therefore can also be coupled to NATIVE. For the EM module, automated serial section EM enables routine and rapid volumetric tissue volumes at nanometer-scale resolution. NATIVE should allow exploration of biological samples other than brain. Data analysis will be limited primarily by computational capacity 20 but a larger selection of nanobodies will be critical as well. 8
METHODS
Step-by-step instructions for the NATIVE workflow are available as a Supplementary Protocol {LINK?}. Methods, including statements of data availability and analysis codes and references are available in the online version of the paper.
Note: Supplementary Information and source Data Files are available in the online version of the paper
Further information on experimental design is available in the Nature Research Reporting Summary linked to this article.
Online Methods
Step-by-step instructions for the NATIVE workflow are available as a Supplementary Protocol {LINK?}.
Sortase reaction.
Detailed protocols have been described before.21 Briefly, in a reaction buffer containing Tris·HCl (50 mM, pH 7.5), CaCl2 (10 mM), NaCl (150 mM), N-terminal triglycine-modified peptides/organic fluorophores (500 μM) and LPETG-containing VHHs (100 μM) were added, followed by the addition of sortase A pentamutant (2.5 μM, Addgene: 51140). After incubation at 12 °C with gentle agitation for 2~4 h, Ni-NTA beads were added to the reaction mixture with gentle shaking for another 30 min at 4 °C. At the end of incubation, the total mixture with beads was directly loaded onto a PD-10 column (GE Healthcare). Fractions corresponding to the desired product fail to stick to Ni-NTA beads and were retrieved in the void volume to yield pure VHH-fluorophore conjugate. The labeled VHH was adjusted to final concentration of 1mg/mL and stored at −80 °C with 10% (v/v) glycerol.
Perfusion and fixation.
Adult C57BL/6 mice, except where noted, were used in developing NATIVE (All animal procedures were performed according to US National Institutes of Health guidelines and approved by the Committee on Animal Care at Harvard University). Mice were anesthetized with isoflurane inhalation and perfused transcardially with a Masterflex Peristaltic pump at a flow rate of 10 mL/min. 15 mL carbonated aCSF buffer was perfused to clear the blood, followed by 30 mL ice-cold fixative mixture of 3% sucrose, 4% paraformaldehyde in 0.1M phosphate buffered saline (PBS, pH 7.4). Brains were carefully removed from the skull to avoid damage to the tissue. On a Leica VT1000 S vibrating blade microtome, 120 µm slices were cut at 4 °C in the fixative mixture. Slices were transferred into fresh fixative solution and post-fixed for 24 hrs with gentle agitation at 4 °C.
Nanobody staining.
Fixed brain slices were trimmed into smaller pieces containing the target area and transferred to a clean 24-well plate. After rinsing 3 × 5 min in 0.1M PBS, a solution containing 0.1M PBS, 0.1M glycine and 0.05% sodium azide (glycine blocking solution) was added. Nanobody mixture staining solution was made by diluting fluorophore-conjugated nanobody stock solutions (~1 mg/ml) 1:250 in the glycine blocking solution. After 20 min of blocking, 1 mL of the nanobody staining mixture was added to each well. Nanobody staining was performed for 48 hrs at 4 °C with gentle agitation. Samples were then rinsed 3 × 5 min in 0.1M PBS. Nuclei were stained with a Hoechst solution (1:5000 dilution in 0.1M PBS) for 5 hrs at 4 °C and rinsed 3 × 5 min in 0.1M PBS. Tissues were protected from light exposure during the entire staining process.
Fluorescence microscopy.
For imaging, samples were mounted between two glass coverslips separated by a 120 µm secure-seal spacer (Thermo Fisher) to protect samples from compression. Fluorescence imaging was performed on a Zeiss LSM 880 confocal laser scanning microscope equipped with a 40x/1.3 NA oil-objective lens. After imaging, a low resolution image of the entire sample was taken and fluorescently imaged regions were labeled as a reference for future sample trimming. The sample was carefully removed from the glass slides and transferred to a fixative mixture containing 2% paraformaldehyde, 2.5% glutaraldehyde, 0.15M sodium cacodylate buffer (pH 7.4) for post-fixation overnight.
EM staining.
A modified ROTO (reduced osmium-thiocarbohydrazide-osmium) protocol was applied to stain the sample for EM imaging. After rinsing 3 × 5 min in 0.15M sodium cacodylate buffer, the sample was first stained in a solution containing 1% OsO4, 1.5% potassium ferrocyanide and 0.15M sodium cacodylate at room temperature for 1 hr. After washing 3 × 5 min in deionized water, the sample was incubated in filtered 1% w/v thiocarbohydrazide at 40 °C for 30 min. The sample was washed 3 × 5 min with deionized water and stained again with 2% OsO4 aqueous solution at room temperature for 1 hr. The sample was then washed 3 × 5 min and transferred to 1% uranyl acetate solution. The sample stayed in uranyl acetate solution at 4 °C overnight, protected from light. On the next day, the sample was warmed up to 50 °C in the same uranyl acetate solution for 1hr. The sample was washed 3 × 5 min and ready for embedding.
Resin embedding.
The sample was dehydrated through a graded (25, 50, 75, 100%, 5 min each) acetonitrile series and then placed in anhydrous acetonitrile for 10 min. After infiltration in 25% EPON:acetonitrile for 1 hr, 50% EPON:acetonitrile for 2 hr, 75% EPON:acetonitrile for 2 hr, the sample was placed in 100% EPON overnight on a rotator. The infiltrated sample was then transferred to the embedding capsule and incubated in fresh EPON in a 60 °C oven for two days.
Serial ultrathin sections collection with ATUM.
The block face of the sample cylinder was trimmed into a 1.5 × 1.5 mm square containing the ROI imaged by light microscopy. The low resolution image was used as reference for trimming. 50 nm thin sections were cut using a Leica ultramicrotome and collected on a home-built automated tape collecting system16 on Kapton tape that was carbon-coated and plasma-treated. Upon collection of the sections, the tape was cut into appropriate lengths and mounted on 100 mm silicon wafers.
EM imaging.
The sections were imaged with a Zeiss Sigma SEM using a backscattered electron detector. A few sections were mapped at low resolution and compared with the light microscopy (LM) images. Blood vessels and nuclei were employed as fiducial markers in correlating the LM and EM images to locate the ROI. High resolution images of the ROI (12k × 12k pixel image, 5 nm per pixel) were acquired with the working distance of ~7.3 mm, 8 keV incident electron energy and dwell time of 2 µs. The EM image stack was aligned by Fiji’s plugin “Linear Stack Alignment with SIFT” to form a coherent 3D EM volume.
LM-EM coregistration and data processing.
LM images and aligned EM images were loaded into VAST separately (https://software.rc.fas.harvard.edu/lichtman/vast/). Characteristic features visible in both image stacks were identified, including nucleoli, branching of blood vessels, and lysosomes, and corresponding point pairs were placed in each feature in the two stacks. The coordinates of these points in both LM and EM were recorded, and a three-dimensional affine transformation was calculated for translating the LM coordinates to the EM coordinates using the pseudo-inverse method. We then translated the LM image stack to the EM coordinate system and loaded both stacks in VAST together. The microglia, the astrocyte and the blood vessel endothelial cell were traced in VAST. Traced objects and isosurfaces of the fluorescence were extracted as 3D surfaces and rendered in 3ds Max (Autodesk Inc.).
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Shu-Hsien Sheu for helpful discussions of the tissue staining protocols, Dr. Kim Swee, Dr. Mohammad Rashidian and Vincent Verschoor for providing the Ly-6C/G-specific nanobody. This research is supported by grant D16PC00002, GG008784, P50 MH094271, U19 NS104653-01 to J.L.; R01AI087879 to H.P; F32CA220990 to T. F.
Footnotes
Competing Interests
A provisional patent application has been filed.
Data Availability
The authors declare that the main data supporting the findings of this study within the article and its Supplementary Information files are available.
The high-resolution EM, low resolution EM and aligned LM image stacks can be downloaded from https://software.rc.fas.harvard.edu/lichtman/temp/NATIVE.
Code Availability
The Matlab codes to compute a 3D transformation are available in the Supplementary Information.
References
- 1.Paez-Segala MG et al. Fixation-resistant photoactivatable fluorescent proteins for CLEM. Nat. Methods 12, 215–218 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lam SS et al. Directed evolution of APEX2 for electron microscopy and proximity labeling. Nat. Methods 12, 51–54 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baude A et al. The metabotropic glutamate receptor (mGluRlα) is concentrated at perisynaptic membrane of neuronal subpopulations as detected by immunogold reaction. Neuron 71, 771–787 (1993).. [DOI] [PubMed] [Google Scholar]
- 4.Micheva KD & Smith SJ Array Tomography: A New Tool for Imaging the Molecular Architecture and Ultrastructure of Neural Circuits. Neuron 55, 25–36 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collman F et al. Mapping Synapses by Conjugate Light-Electron Array Tomography. J. Neurosci. 35, 5792–5807 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chung K & Deisseroth K CLARITY for mapping the nervous system. Nat. Methods 10, 508–513 (2013). [DOI] [PubMed] [Google Scholar]
- 7.Ku T et al. Multiplexed and scalable super-resolution imaging of three-dimensional protein localization in size-adjustable tissues. Nat. Biotechnol. 34, 973–981 (2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ingram JR, Schmidt FI & Ploegh HL Exploiting Nanobodies’ Singular Traits. Annu. Rev. Immunol. 36, annurev-immunol-042617–053327 (2018). [DOI] [PubMed] [Google Scholar]
- 9.Fang T et al. Targeted antigen delivery by an anti-class II MHC VHH elicits focused αMUC1(Tn) immunity. Chem. Sci 8, 5591–5597 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fang T et al. Structurally Defined αMHC-II Nanobody-Drug Conjugates: A Therapeutic and Imaging System for B-Cell Lymphoma. Angew. Chemie Int. Ed 55, 2416–2420 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feng G et al. Imaging Neuronal Subsets in Transgenic Mice Expressing Multiple Spectral Variants of GFP. Neuron 28, 41–51 (2000). [DOI] [PubMed] [Google Scholar]
- 12.Pallotto M, Watkins PV, Fubara B, Singer JH & Briggman KL Extracellular space preservation aids the connectomic analysis of neural circuits. Elife 4, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duarte JN et al. Generation of Immunity against Pathogens via Single-Domain Antibody–Antigen Constructs. J. Immunol. 197, 4838–4847 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bachran C et al. The activity of myeloid cell-specific VHH immunotoxins is target-, epitope-, subset- and organ dependent. Sci. Rep 7, 17916 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li T et al. Cell-penetrating anti-GFAP VHH and corresponding fluorescent fusion protein VHH-GFP spontaneously cross the blood-brain barrier and specifically recognize astrocytes: application to brain imaging. FASEB J. 26, 3969–3979 (2012). [DOI] [PubMed] [Google Scholar]
- 16.Kasthuri N et al. Saturated Reconstruction of a Volume of Neocortex. Cell 162, 648–661 (2015). [DOI] [PubMed] [Google Scholar]
- 17.Hayworth KJ et al. Imaging ATUM ultrathin section libraries with WaferMapper: a multi-scale approach to EM reconstruction of neural circuits. Front. Neural Circuits 8, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lana D, Iovino L, Nosi D, Wenk GL & Giovannini MG The neuron-astrocyte-microglia triad involvement in neuroinflammaging mechanisms in the CA3 hippocampus of memory-impaired aged rats. Exp. Gerontol. 83, 71–88 (2016). [DOI] [PubMed] [Google Scholar]
- 19.Hong S & Stevens B Microglia: Phagocytosing to Clear, Sculpt, and Eliminate. Dev. Cell 38, 126–128 (2016). [DOI] [PubMed] [Google Scholar]
- 20.Lichtman JW, Pfister H & Shavit N The big data challenges of connectomics. Nat. Neurosci. 17, 1448–1454 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods-only References
- 21.Antos JM, Ingram J, Fang T, Pishesha N, Truttmann MC, Ploegh HL Site-specific protein labeling via sortase-mediated transpeptidation. Current Protocols in Protein Science, 89, 15.3.1–15.3.19 (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


