Abstract
Type 2 diabetes mellitus (T2D) and obesity already represent 2 of the most prominent risk factors for cardiovascular disease, and are destined to increase in importance given the global changes in lifestyle. Ten years have passed since the first round of genome-wide association studies for T2D and obesity. During this decade, we have witnessed remarkable developments in human genetics. We have graduated from the despair of candidate gene-based studies that generated few consistently replicated genotype-phenotype associations, to the excitement of an exponential harvest of loci robustly associated with medical outcomes through ever larger genome-wide association study meta-analyses. As well as discovering hundreds of loci, genome-wide association studies have provided transformative insights into the genetic architecture of T2D and other complex traits, highlighting the extent of polygenicity and the tiny effect sizes of many common risk alleles. Genome-wide association studies have also provided a critical starting point for discovering new biology relevant to these traits. Expectations are high that these discoveries will foster development of more effective strategies for intervention, through optimization of precision medicine approaches. In this article, we review current knowledge and provide suggestions for the next steps in genetic research for T2D and obesity. We focus on four areas relevant to precision medicine: genetic architecture, pharmacogenetics and other gene-environment interactions, mechanistic inference, and drug development. As we describe, the genetic architecture of complex traits has major implications for the prospects of precision medicine, rendering some anticipated approaches decidedly unrealistic. We highlight obstacles to the translation of human genetic findings into mechanism inference but are optimistic that, as these are overcome, there is untapped potential for novel drugs and more effective strategies for treating and preventing T2D and obesity.
Keywords: diabetes mellitus, genetics, obesity, precision medicine
The pace of discovery in complex trait genetics during the past decade has been breathtaking. From the paltry returns of the candidate variant studies in a few hundred individuals which were typical until 10 to 15 years ago, we have graduated to genomewide association studies in sample sizes 1000 times larger than that. At the time of writing, >300 loci have been robustly associated with type 2 diabetes mellitus (T2D)1,2 and >500 with body mass index (BMI), waist-hip ratio, and other obesity traits.3–8 These discoveries provide the substrate for a systematic assessment of the pathophysiological basis of these conditions, one that embraces both environmental and genetic contributors to predisposition. This has led to increasing hope that these discoveries will pave the way for new approaches to diagnosing, stratifying, monitoring, preventing, and treating disease.
Such improvements in prediction, diagnosis, and prognosis have long been, and remain, one of the principal goals of genomic medicine. Once the Human Genome Project was completed9 and low-cost commodity genotyping brought the first successes from genome-wide association analyses, there was a hope that genomics would have a dramatic impact on risk stratification and therapeutic decisions regarding individual patients.10,11 This expectation was predicated on the assumption that the most important common risk variants would have at least moderate effect sizes and could, therefore, substantially move the needle in clinical decision-making for individual patients. However, 10 years after the landmark study by the Wellcome Trust Consortium,12 which represents the start of a truly remarkable period in biomedical research, there remain relatively few examples of results from genome-wide association studies (GWAS), or human genetics generally, that have impact on clinical decision-making for complex, multifactorial traits, such as obesity and T2D.
The chief reason for this is that biology has turned out to be more complicated than many believed—or perhaps hoped—15 years ago. As discussed elsewhere in this review, rather than being dominated by a small number of common variants with modest-to-large phenotypic effects, the genetic architecture of complex traits is now known to be characterized by a vast number of causal variants, most of them with only a tiny additive effect on risk.13–15 This extreme polygenicity— or, as has been recently suggested, omnigenicity16—of complex traits does not necessarily mean that GWAS findings cannot be translated into paradigm-shifting new therapies, but it substantially increases the effort required to establish and characterize causal variants, genes, and pathways.
When it comes to individual prediction of complex traits, the limited variance in disease risk attributable to the subset of common risk variants that reach genome-wide significance translates into low discriminative capacity. Although some early studies indicated a potential role for common variant risk scores as tools for predicting T2D risk,17,18 subsequent studies demonstrated limited increment in performance over clinical models that can be generated from more readily accessible risk factors (such as age, BMI, and ethnicity).19–21 The recent wave of huge GWAS has reinvigorated interest in the potential of genetic risk profiling, but the potential for clinical utility remains contested.2,22–24 In a future where genome sequence forms part of every individual’s health record, such information could prove useful for a host of clinical indications, but the benefits are not yet seen to be sufficient to motivate the dedicated collection of such data for any single specific medical purpose (eg, to obtain slightly more precise estimates of future T2D risk). Further, even though there are reports that knowledge of personal genetic risk could have some impact on individual motivation to modify behavior so as to reduce diabetes mellitus risk,25,26 randomized clinical trials have failed to show an effect of genetic risk counseling on self-reported motivation or prevention program adherence for individuals at risk of diabetes mellitus.27,28 Hence, at the moment, there is no evidence to suggest that knowledge about genetic risk for T2D can appreciably change risk assessment or improve motivation and adherence to lifestyle intervention.
The explosion of interest in the potential for precision medicine often leads to a tendency to see that potential purely in terms of genetic and genomic profiling. However, although genomics is an important and integral part of precision medicine, these terms are not equivalent. Indeed, integrative approaches using several layers of biological and clinical information are likely to be required if we are to achieve accurate diagnostic and prognostic precision for complex, multifactorial traits. For reasons discussed in detail in this review, genomics may not yet be ripe for widespread clinical implementation in enabling precision treatments for obesity and T2D. But are there other ways in which the insights from human genetics could provide a path toward precision medicine? In this review, we aim to summarize current knowledge and provide some thoughts about the next steps in this respect, with a focus on four areas that are relevant to the future implementation of precision medicine: genetic architecture, pharmacogenetics and other gene-environment interactions, mechanistic inference, and drug development.
GENETIC ARCHITECTURE AND WHY IT MATTERS
There has been a longstanding debate (dating back more than a century) as to whether the genetic differences that influence individual predisposition to common traits, such as height or T2D are mostly driven by common alleles widely shared within and across populations, or whether they are more often attributable to rare, or even unique, events, specific to an individual and their family. This question matters because the answer has profound implications for the ways in which genetic data can be used to support personalized medicine. The advent of GWAS, a decade ago, reignited this debate. The first wave of GWAS studies (for T2D, BMI, and many other traits) demonstrated the power of this approach to deliver robust discovery of novel association signals. These discoveries featured common variants (by design, of course, given the content of the early commodity genotyping arrays), but it was soon clear that most of these had, at best, modest effects on disease risk.29 The apparent disparity between the observed heritability of these conditions (derived from twin studies, for example) and the desultory performance of the GWAS signals to explain those observations (even when considered in aggregate) spawned endless discussions about the basis for this apparently missing heritability and, in particular, the extent to which this gap might be filled by rare variants of large effects that were largely invisible to GWAS studies.29–31
During the past 2 to 3 years, the ability to directly assay the contribution of lower frequency and rare alleles in complex trait predisposition has advanced to the point where this vexatious question can now be answered with confidence. Large GWAS, conducted on more informative arrays, bolstered by imputation from large, dense reference panels have extended the scope of GWAS to include variants with minor allele frequencies <1%.32 Bespoke genotyping panels (such as the exome array) and direct sequencing (of exomes predominantly) have provided access for association analysis to even rarer variants.33 In parallel, methodological advances (such as genome-wide complex trait analysis [GCTA]) have facilitated assessments of explained heritability that go beyond just the genome-wide significant hits and consider the contribution of variants genome-wide to observed variation in trait levels or disease predisposition.34 These diverse approaches have produced a clear and consistent answer: for T2D and obesity, as well as for many other complex traits, genetic risk is driven predominantly by common risk variants that are shared both within and between populations.33,35
Collectively, these variants (most of which have small effects and lie far below genome-wide significance, despite GWAS studies involving many hundreds of thousands of individuals) can explain most of the variation in disease predisposition that is attributable to genetics.2,34 It is not widely appreciated that, although there are far more sites of rare variation than there are of common, most of the genetic differences between 2 individuals reside at the latter. Thus, the allele frequency spectrum of T2D disease risk variants broadly mirrors the distribution of genetic variation at large. This should not come as a great surprise, particularly for traits of recent origin (such as T2D), which remain largely post reproductive in terms of their impact on health. These factors will have limited the extent to which T2D risk alleles were exposed to adverse selective pressure and allowed them to be maintained in populations at relatively high frequency.
Crucially, this is not to deny any role for rare risk alleles in predisposition to these traits. Rare alleles that contribute to T2D risk and variation in BMI are being increasingly revealed as the sample sizes exposed to dense imputed genotyping, exome, or whole-genome sequencing grow.2,7,8,33 Because detectable rare variant association signals will necessarily be dominated by rare alleles with relatively large effects, their identification can provide valuable and rapid biological insights, particularly when they map to well-annotated sequence (such as coding regions).36–38 However, the evidence from T2D indicates that, with the possible exception of isolated or endogamous populations, rare variant signals are neither numerous enough nor of sufficient collective effective size to explain much of the variation in phenotype.
These discoveries about the genetic architecture of traits such as T2D are hugely relevant to the perspectives we should be adopting with regard to patient stratification. It is increasingly evident that personal risk of T2D or obesity is driven by an individual’s pattern of genotypes across many hundreds (or, more likely, thousands) of, mostly common, variant sites. These act in concert with environmental exposures that, judging by the global nature of these conditions, are likely to be pervasive. Thus, although each individual’s predisposition barcode (across variants and exposures) is likely to be unique, at the same time, these profiles are substantially overlapping across individuals.
In contrast to many Mendelian diseases, this makes it inherently unlikely that it will be possible to define robust phenotypic clusters that map onto distinct etiological pathways, in a manner consistent with a traditional view of precision medicine. One recent study indicating the potential to stratify individuals with newly-diagnosed diabetes mellitus on the basis of clinical and biochemical characteristics suggests that this assessment may be overly pessimistic, but it remains unclear how the group-level structure uncovered in this study maps onto clinical response and the extent to which this provides a platform for individual decision-making.39
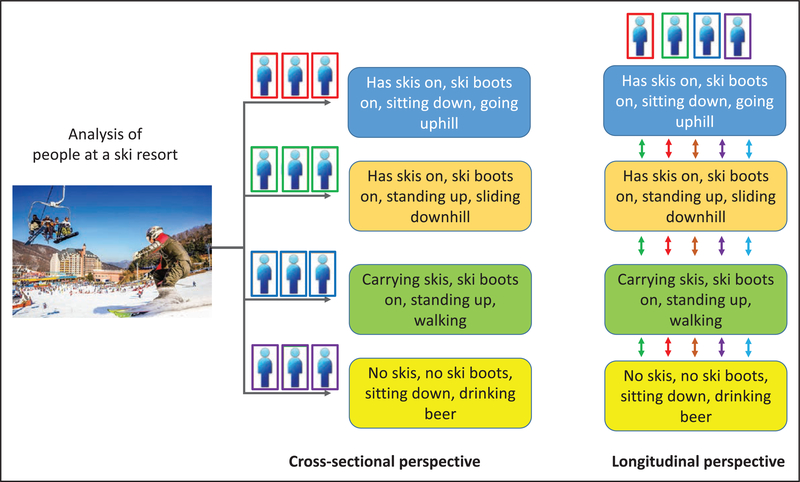
In addition, longitudinal studies are required to ensure that any clustering detected in cross-sectional studies is truly reflective of etiological heterogeneity and does not simply reflect different stages of disease evolution or other temporal variation (Figure). This is particularly true for biological readouts, such as novel proteomic or metabolomic biomarkers, as well as traditional risk factors for T2D, which, in contrast to DNA variation, change over time and may provide valuable opportunities for dynamic tracking of disease progression.
Figure. Drawing the wrong conclusions from clustering of cross-sectional data.
Your alien spacecraft crashes near a ski resort. In an effort to understand the diversity of human life, you apply your favorite clustering algorithm to the life forms visible through the window. You conclude that there are 14 subtypes of human life with distinctive and nonoverlapping appearances and behaviors. However, as you await rescue, you realize that this clustering was only transient and an artifact arising from the restriction to a single cross-sectional view. Over time, you observe that almost all life forms progress through all 4 stages at various points and that membership of a given cluster at a single point of time provides little insight into long-term behavior.
The above argues for a shift to a more nuanced, probabilistic, molecular taxonomy of disease, which recognizes that what we consider disease is, for complex traits at least, usually the consequence of a confluence of risk across multiple processes (in the case of T2D, these would include insulin secretion, insulin sensitivity, islet autoimmunity, fat distribution, etc).40 This model allows for a redefined perspective on precision medicine, one focused on understanding the mix of abnormalities contributing to disease risk and progression for a given individual at a given point of time, and in developing the tools (eg, biomarkers for each component process) and interventions (capable of reversing as many of those defects as possible) that allow for a more targeted response. We discuss some of the clinical implications of this model in the next section.
One last question needs to be posed. What can we expect to learn about disease mechanisms when genetics points to the involvement of thousands of regulatory elements and genes influencing many disparate pathways? This is an important and thought-provoking question to which we only have partial answers.16 First, it is worth emphasizing that thousands of genes and many pathways are not the same as all genes and all pathways, and as we define more of the processes through which disease risk is mediated, we will better understand whether the mechanisms involved in disease-specific biology continue to proliferate or whether they start to converge on a limited set of key pathways. Second, it is important to consider the importance of temporal and tissue specificity, which after all is what defines many diseases in the first place: the dimensionality of disease mechanisms is not merely measured in genes and pathways but in their impact across cell and tissue types and across the life course.
PHARMACOGENETICS AND OTHER GENE-ENVIRONMENT INTERACTIONS
Pharmacogenomic testing to personalize diabetes mellitus treatment has been an area of intense research in the past 10 years.41 Given the large interindividual variations in efficacy and side effects of different treatment regimens, more precise ways to predict individual treatment response would be of huge importance to clinical care. The hope is that genetic information can be used to define subgroups that share more similar responses to the various preventative and therapeutic treatments available. Such developments would epitomize precision medicine, which seeks to deliver the right treatment to the right person at the right time.
Needless to say, there is also a fair bit of hype surrounding these concepts. One frequent expectation arising from the evident heterogeneity in the presentation, course, and treatment response in T2D is that it will become possible to decompose the T2D monolith into some number of etiologically distinct and more phenotypically homogeneous subtypes, each of which will be exquisitely responsive to interventions targeted to reversing the specific lesion responsible. However, as we described above, such a model is at variance with what we know of the genetic architecture of T2D.
In the palette model described earlier,40 there may well be opportunities to match phenotypic presentation and disease course to specific genetic profiles, but this is likely to be the most effective in those (possibly exceptional) individuals in whom the phenotype is dominated by a single phenotypic defect (ie, those with monogenic disease or something approaching it). In such circumstances, it is likely that genetic information will allow us to stratify individuals who may optimally respond to different treatments. However, we have some way to go before that such strategies are clinically validated, and the effects are likely to be more nuanced than the more extreme expectations referred to above. It is possible that genetic information may be more useful for target engagement (ie, selecting a specific diabetes mellitus therapy) than for prediction of disease risk because the genetic effects on drug response are likely to be stronger than those seen for associations with disease. This is, in part, because of the closer biological connection between a genetic variant in or near a gene encoding a drug target and the response to the corresponding drug and, in part, because there may be less selective pressure against variants of large effect for novel drug exposures.
Many of those with typical T2D will have several processes contributing in parallel to their metabolic state (elevated BMI, unfavorable fat distribution, suboptimal β-cell function, etc), and neither their genetic profiles nor their clinical features will necessarily provide a distinctive pattern that can be matched to available therapies. Much will depend on the extent to which it becomes possible, through the integration of information on genetic predisposition, measures of external and internal environmental exposures, clinical phenotype, and biomarker readouts of relevant pathophysiological processes, to characterize the dominant mechanisms contributing to ill health in a given individual at a given point of time and to define specific interventions and treatments that demonstrably have differential merit for returning that individual toward health.
Genetic testing in maturity onset diabetes of the young and neonatal diabetes mellitus has already provided the diabetes mellitus field with proof-of-principle examples, where genetic knowledge guides treatment.42 However, it will be more challenging to develop robust pharmacogenomic tests to aid therapy decisions in T2D than for these monogenic disorders because of the complex web of genetic and environmental factors influencing T2D risk. Indeed, there has been a multitude of small, post hoc studies addressing the pharmacogenomics of different diabetes mellitus drugs, such as metformin43 and sulfonylureas.41 In a systematic review of 34 studies published in 2014, there was some evidence of pharmacogenetic interactions for metformin, sulfonylureas, repaglinide, thiazolidinediones, and acarbose consistent with their pharmacokinetics and pharmacodynamics but a lack of large, prospective, high-quality studies.44 An example of the kind of large, high-quality study that will be crucial for clinical implementation was published by the Metformin Genetics Consortium last year.45 In this study of 10 577 participants of European ancestry, an allele in the intron of SLC2A2, which encodes GLUT2 (Glucose transporter 2), was associated with greater metformin response. Among obese individuals, homozygote carriers had a 0.33% (3.6 mmol/mol) greater absolute Hemoglobin A1c (HbA1c) reduction than reference allele carriers. Although this effect size is likely too modest to justify genotyping of this variant for this indication alone, this study provides some evidence of pharmacogenomic markers with potential to inform precision medicine in T2D. Crucially, this is another example of the kind of association that could have clinical utility in a world where individual genome sequence (or GWAS) data were widely available. It also illustrates the need to extend large, international collaborations—the single most important success factor for GWAS of complex traits—into the pharmacogenomic arena.
There have been parallel efforts to find genetic variants that modify the response to lifestyle interventions that might prevent obesity and T2D and which might, therefore, support optimization of individually tailored behavioral modifications. Most such gene-environment studies have suffered from the same problems as the candidate gene studies of the 1990s. They have been based on questionable hypotheses: candidates from known biology, or even GWAS loci associated with main effects on the trait of interest, are not the best candidates for gene-environment interaction effects because of the nature of their discovery (which would generally be biased against discovery of signals that are contingent on environmental exposures). Further, most studies have been way too small and underpowered, lacking replication in independent study samples.46 A recent systematic review reported that none of the eight gene-macronutrient interactions for T2D risk that had been reported in prior literature could be replicated.47 The most promising advance in this area has come from analyses that suggest the combination of clinical information and assays of gut microbiome content can predict individual excursions in glucose profile in response to specific dietary components.48
Although there are some successful studies of gene-environment interactions of obesity, they have typically combined risk alleles into genetic risk scores49–51—an approach that increases power but does not provide insights on specific loci. Also, these studies too have typically been performed in small samples with limited replication, so their value is still uncertain. For gene-environment effects of physical activity on obesity, there is one locus, FTO, which shows a well-replicated interaction. The effect of this variant on BMI has been reported, in a study of ≈200 000 individuals, to be attenuated by ≈30% in physically active individuals compared with inactive individuals.52 As in the case of the Metformin Genetics Consortium, this highlights the importance for investigators interested in gene-environment interactions to follow the successes of the GWAS era and understand the importance of forming alliances to maximize sample sizes and replicate findings. Indeed, there are strong reasons to argue that this is going to be even more important for gene-environment studies than for analyses of main effects.
Unfortunately, this paucity of robust findings for gene-environment interactions in T2D and obesity has not prevented commercial companies from attempts to monetize the field of consumer genetics. Although there are some serious businesses out there, there are all too many examples of genetic tests being oversold without much, if any, science to support them. There are companies that claim that their genetic tests can provide personalized diet or exercise advice53–57; predicting what wine you like,58 whether you are likely to like Marmite,59 or indicating that your child is likely to be a successful soccer player.60
LOST IN TRANSLATION: MECHANISTIC CHALLENGES
One of the key goals of human genetic enquiry for complex traits, such as diabetes mellitus and obesity, is to reveal the fundamental mechanisms, which lead to disease predisposition and development, as well as those that influence the risk of subsequent complications. In contrast to the findings from traditional approaches to physiological and epidemiological investigation from which it can be difficult to disentangle causal effects from those which reflect reverse causation or confounding, human genetics offers the opportunity to define mechanistic relationships that link inherited variants to consequent changes at the molecular, cellular, tissue, and physiological level.61,62
Mechanistic insights can provide the foundation for uncovering novel interventions that better prevent and treat disease and for the detection of biomarkers that monitor disease progression. Those same insights may also presage a more rational mechanistic taxonomy of disease, which enables a more personalized approach to disease prevention and management.63 The value of this approach has been reinforced time and again from studies of monogenic and syndromic conditions, including many early-onset familial forms of diabetes mellitus and obesity. The demonstration that many cases of neonatal diabetes mellitus result from mutations in the KCNJ11 gene, encoding a component of the β-cell KATP channel, suggested that patients with this rare condition might be amenable to treatment with sulphonylureas, rather than insulin—a hypothesis that received rapid empirical confirmation.64,65 In similar fashion, the identification, in some rare instances of extreme early obesity and hyperphagia, of causal mutations in the leptin gene enabled successful treatment with replacement leptin.66
The discoveries just described led to rapid clinical advances and some of the clearest examples of personalized medicine in current medical practice. This was, in part, because the disease-causing mutations had dramatic and unequivocal impacts on the protein sequence of genes that could be easily positioned within existing understanding of disease pathogenesis. At the same time, the specific phenotypic presentation in these cases provided confidence that the remedial treatments concerned were likely to be both effective and safe. The situation for a multifactorial disease like T2D is a deal more complex. As described above, the genetic contribution to T2D predisposition is made up from many hundreds (likely thousands) of mostly common variants of individually small effects.33 Each of us (diabetic or not) carries our own particular configuration of risk and protective alleles across these variants. Because they are common, causal variants at these loci are typically hidden among a forest of highly correlated alleles (because of linkage disequilibrium), frustrating efforts to identify the specific variants involved. And, because most of the signals map to noncoding sequence, the identity of the genes (protein coding or otherwise) through which they operate is often obscure.67 All of these aspects represent challenges to proceeding from genetic discovery (for example, of a signal detected by GWAS) to enumeration of the mechanisms involved. Indeed, many thousands of GWAS loci have been identified in the past decade, but a compelling mechanistic description has been forthcoming for only a small proportion. Having said that, recent advances are propelling far more complex trait loci toward mechanistic resolution.
One illustrative example from the T2D arena concerns a GWAS signal mapping near MTNR1B, encoding one of the receptors for the hormone melatonin.68–70 The progressive accretion of GWAS data through consortium efforts provided replication of this signal and the capacity to disentangle the correlations between adjacent single nucleotide polymorphisms (SNPs) through fine mapping, such that the association signal could be resolved to a single variant (rs10830963).71 Analysis of intermediate metabolic traits in nondiabetic individuals indicated that the T2D-predisposing action of the risk allele was, at least in part, mediated through a deficit in insulin secretion, highlighting the pancreatic islet as one key tissue of interest.72 In genome-wide analyses, it had been demonstrated that T2D GWAS signals were enriched for overlap with binding sites (in islet and liver) for the Forkhead Box A2 (FOXA2) transcription factor, and the MTNR1B risk variant was shown to map into an islet enhancer region containing such a site (although rs10830963 appeared to be modifying a binding site in that enhancer for Neurogenic differentiation factor 1 (NEUROD1) rather than FOXA2 itself). The T2D risk allele was subsequently shown to increase enhancer activity and to preferentially bind NEUROD1 in islet-derived cells.71 These findings were consistent with evidence from human islets that the T2D risk allele was associated with increased MTNR1B RNA expression. This, in turn, chimed with the hypothesis that one of the desirable physiological consequences of the circadian (nocturnal) release of melatonin from the pineal gland is suppression of insulin secretion during a period of restricted energy intake.73,74
However, there are reasons to consider this reductionist narrative—focused around identifying a single causal variant, a clearly defined effector gene, and a single plausible physiological mechanism—as overly simplistic. At some loci, at least, the truth may be a deal more complex.
For a start, the concept of a single causal variant may, for some GWAS signals, be illusory. Not only may there be multiple independent signals in a given region (revealed by conditional analysis for example)75 but each independent signal may not be resolvable to a single variant. This could arise where multiple highly correlated alleles on a single haplotype are causal, each contributing to disease predisposition, for example through combinatorial effects on enhancer activity. An alternative mechanism, relevant to alleles that create or destroy CpG sites, could involve a joint haplotype-specific impact on local methylation status. For obvious reasons, examples of this kind of behavior have been hard to demonstrate, although the haplotype-specific methylation signal at the FTO locus may be one.76 At some loci, potentially including PPARG, predisposition may reflect the joint effects of both regulatory and coding alleles.32,77 The potential for such scenarios to confound efforts to obtain functional evidence for the causal variant is obvious and speaks to the need to consider potential risk variants in relation to their genomic context.
A subsequent challenge lies in connecting the causal variant(s) identified through fine mapping and genomic integration with the effector transcripts through which they operate. In the absence of a compelling biological candidate in the region (for example, a gene known to harbor rare severe mutations that result in a broadly similar, albeit more extreme, phenotype), these connections are often made on the basis of the detection of cis-expression signals or evidence of physical interaction with target gene promoters (typically derived from conformational capture methods).78,79 There are issues here with ensuring that the GWAS and cis expression quantitative trait loci (eQTL) signals are truly colocalized (rather than merely overlapping)80 and in establishing that the tissues or cells analyzed are relevant to both the disease of interest and the specific locus under consideration. As studies become larger, increasing numbers of GWAS and cis eQTL signals are seen to be composed of multiple independent signals, posing challenges to interpretation when only a subset of these colocalize. We still lack unbiased information on the sensitivity and specificity of these approaches, although the current evidence would suggest that neither is sufficiently high to command robust inference from any single quantum of experimental data.
A third consideration concerns the phenotypic consequences of the variants of interest. It is interesting to note that the first-discovered landmark GWAS loci for BMI, T2D, and coronary artery disease (FTO, TCF7L2, and CDKN2A, respectively) have proven particularly refractory to mechanistic resolution, despite large effect sizes and the most extended—and most intensive— research efforts.81–84 Might it be that the reason that these loci have the largest common variant impacts for their respective traits (prompting their early discovery) is because they influence multiple processes? Recent work has shown that common variants near the FTO gene that influence BMI and obesity compromise the thermogenic potential of adipose tissue via altered regulation of IRX3 and IRX5,85 but that does not necessarily prove that this is the only mechanism through which the variants operate. Other genes in the region (RPGRIP1L and FTO itself) are reasonable candidates with strong experimental support, and central, as well as peripheral effects on BMI at this locus are plausible.86,87
Also, going beyond cis effects, it is likely that some GWAS loci tag genes or other elements acting as master regulators of disease. A well-known example of this is KLF14, where an imprinted trans eQTL has multiple downstream effects on gene expression88 and metabolic traits, including T2D and high-density lipoprotein (HDL) cholesterol.62 Distinguishing between such true master regulators of gene expression acting in trans and correlations resulting from the downstream consequences of variation in one phenotype impacting others will require careful attention and thoughtful experiments because the pathways and mechanisms underlying obesity and T2D are complex and involve many negative and positive feedback loops.
Given the difficulties in assigning function to weak regulatory variants in humans, it is natural for researchers to seek to fold in additional ways of confirming the causal role of proposed effector transcripts. The most obvious route is to observe the consequences of direct perturbation of gene function in humans, rodent models, or cellular systems. Here again, there are limits to the facile interpretation of the data arising from such studies. Most obviously, the cellular system used in such studies may lack authenticity (a poorly chosen cell line or a tissue inappropriate for the disease of interest or the specific locus under consideration) or the rodent model fail to recapitulate the human phenotype.
Less widely appreciated is the likely divergence in the phenotypic expression of coding and regulatory variants even when both impinge on the function or expression of the same gene. Most GWAS signals map to tissue-specific enhancers, and their phenotypic consequences will be filtered by that tissue specificity. In contrast, the impact of a coding variant will be felt in any tissue that expresses that gene (or, more precisely, any tissue that expresses the isoform in which that variant is transcribed), leading in many cases to a broader or more divergent phenotype.36,89 This is analogous to the difference between a tissue-specific and a global knockout mouse. This distinction is highly relevant from the perspective of therapeutic intervention, given that most pharmaceuticals will be active across all tissues in which the target is expressed, and the profile of action (and side effects) may be better captured by coding than regulatory variants. This may explain some of the apparent discrepancies between regulatory and coding variants at the MTNR1B locus. Whereas the T2D risk allele discovered by GWAS is, as described earlier, associated with increased MTNR1B expression in islets,90 a study of rare functionally deficient coding alleles in the same gene demonstrated an aggregate effect, which was also supportive of increased T2D risk.91 One explanation for this directional divergence could involve the impact of coding variants on melatonin receptor function at sites outside the islet (such as the brain).74,92
In the face of these challenges, it may be tempting to despair of our capacity to make mechanistic headway. Our view is that, although some GWAS loci may feature daunting mechanistic complexity (along the lines described), there is no reason to think that this is universal. The ability to build a mechanistic narrative from diverse lines of functional evidence and to place findings in their genome-wide, tissue-specific context is allowing researchers to reduce reliance on single data points of uncertain significance. Emerging high-throughput genome-wide techniques, such as HiChIP93,94 and Assay for Transposase-Accessible Chromatin using sequencing (ATAC-seq),95,96 that can map chromatin interactions and accessibility with higher resolution than previous methods will improve our ability to disentangle GWAS loci; while single-cell RNA sequencing97,98 and CRISPR-based pooled gene perturbation methods99–103 provide unprecedented opportunities for studies of how RNA expression patterns differ between cells within tissues and how those tissues and cells react to perturbation of multiple genes in parallel.
In this way, more and more GWAS loci will start to reveal their secrets: the hope is that solving the biological puzzle at a subset of loci will provide clues that allow other, less immediately tractable, loci to gather mechanistic momentum. Although there is no guarantee that the puzzle can ever be fully solved or even that solving the puzzle would lead to important clinical insights, we think that combining genomic information with characterization of relevant disease pathways in a temporal and tissue-specific manner is more likely to be successful than sequencing of static DNA variation alone.
CAN GENETICS INCREASE EFFICIENCY OF THE T2D DRUG DEVELOPMENT PIPELINE?
Although there have been indisputable scientific and technological advances in biomedicine during the past 50 years, this has not translated to an improved efficiency of pharmaceutical drug research and development (R&D). On the contrary, we have seen a remarkable decline in R&D efficiency because the number of drugs approved per amount spent on R&D has halved every 9 years since 1950.104 The consequence of this disturbing and clearly unsustainable trend—described by some as Eroom’s law, to highlight the contrast with Moore’s law regarding computing costs—is that every new drug reaching market costs at least $1 billion104 (and according to some estimates up to $3–5 billion USD) to develop.105–107 The reasons for this failure of pharmaceutical R&D have been discussed widely. Even when considering other reasons for drugs not reaching market, such as strategic, commercial, or operational aspects, there seems general agreement that the main obstacle lies in lack of efficacy rather than safety issues.108–111 This may be particularly true for drugs failing late in development (phase III trials), especially problematic from an economical and ethical standpoint.109
Many have argued that the main reason for the lack of efficacy is the limited predictive value of the preclinical models of disease that have dominated the discovery and evaluation of therapeutic targets.112 The dramatic advances in human genetics during the past decade offer the prospect for improved assessment of drug development efficacy in early pharmaceutical R&D through provision of a stronger evidence base for linking perturbation of a given target or pathway to a desirable phenotypic outcome in humans. In addition, human genetics can offer an opportunity to detect less welcome consequences of perturbation that may presage safety or toxicity concerns. There are already several examples in the public domain demonstrating these concepts, and more are likely to follow given the current reallocation of resources to human genetics at a growing number of pharmaceutical companies.
Retrospective proof-of-principle examples of this concept (where the drug was already in use before the genetic finding was made) include PPARG113 and thiazolidinediones,114 as well as HMGCR115 and statins.116 There are also several recent examples where genetic studies have predicted the outcome of an ongoing randomized controlled trial, even though the genetic finding may not have been the basis for initiating drug development. These include the reports of inactivating variants in NPC1L1117 that correctly predicted the clinical effect of ezetimibe on recurrent cardiovascular events118 and of loss-of-function variants in apolipoprotein C3 (APOC3)119 that predicted a clinical effect of ISIS 304801—an antisense inhibitor of APOC3 synthesis—on triglyceride levels.120
However, there are also several emerging prospective examples where the genetic discovery preceded and underpinned the development of the drug. These include variation in Cystic fibrosis transmembrane conductance regulator (CFTR; causing cystic fibrosis)121 and ivacaftor (a CFTR potentiator)122 and from the cardiovascular area, the well-known example of PCSK9 inhibitors.123–127 The CFTR-ivacaftor gene-drug pair illustrates a time frame typical for pharmaceutical R&D, with >20 years from initial scientific discovery to commercial availability of a drug. In contrast, the PCSK9 story highlights how the new era in human genetics can accelerate this process. The first study describing a gain-of-function variant in PCSK9 causing hypercholesterolemia was published in 2003.125 It was rapidly followed by landmark studies that described loss-of-function alleles, particularly in African Americans, that were associated with decreased low-density lipoprotein (LDL) cholesterol124 and, importantly, protection against coronary heart disease.123 After a tight head-to-head competition between several pharmaceutical companies, two antibody-based PCSK9 inhibitors—evolocumab and alirocumab—were approved by the Food and Drug Administration and European Medicines Agency in 2015, barely a decade after the initial genetic discovery. It would be misleading to claim that the PCSK9 target was discovered using GWAS—on the contrary, it was mapped using a traditional family-based design—but we argue that it is likely that there will be many other examples arising from GWAS that can be developed into successful drugs.
Currently, many hope that large-scale exome sequencing will provide new leads for development of drugs treating T2D and other diseases. Certainly, an advantage of exome sequencing is the focus on exonic variants, which makes the translation easier than with lead variants from GWAS, as discussed in detail in the previous section. An example lies in the study of loss-of-function alleles in the SLC30A8 gene (encoding an islet-specific zinc transporter): these have been associated with decreased risk of T2D and highlight zinc transporter 8 (ZnT8) inhibition as a potential novel strategy for T2D therapy.128 However, most exome sequencing studies so far have been too small to provide much additional value beyond previous GWAS. As recently demonstrated for T2D, most variants discovered on exome sequencing were common or located in regions already identified by GWAS.33
It is important, therefore, not to overlook the enormous potential in already existing knowledge about loci robustly associated with T2D and other diseases that has been generated through the past decade of GWAS. Presumably, there are many pointers to novel therapeutic regimens among these loci: the challenge is to systematically work through this treasure trove of associations to find them, starting with establishing causal genes in the loci, followed by careful characterization of the downstream mechanisms to understand underlying biology and to identify druggable mechanisms.89,129 There are strong indications that GWAS loci are enriched with genes that are successfully targeted with existing drugs130,131 and that a drug development pipeline built on support from human genetics will be more efficient and lead to improvements in clinical development success.131,132 For example, the proportion of drug targets with genetic support from GWAS or the Online Mendelian Inheritance in Man database increases significantly across stages in the drug development pipeline; from ≈2% in the preclinical stage to ≈8% for approved drugs—a proportion that is even higher for drugs treating diabetes mellitus and metabolic diseases.131
Even if the often-cited PCSK9 example may represent particularly low-hanging fruit, there is no good reason to think that human genetics as a drug discovery vehicle should be less efficient for obesity and T2D than for lipid-lowering treatment. Indeed, there are several examples of GWAS loci with genes encoding targets of already established drugs against T2D.71,133 In addition to the already mentioned PPARG-thiazolidinediones113,114 pair, other examples of GWAS hit drug combinations include ABCC8-sulfonylurea134,135 and GLP1R–GLP-1 agonists.136,137 Although these genes and corresponding drugs were established before the GWAS era – a necessity given the lead time in drug development – such reidentification of druggable targets illustrates the promise of GWAS for discovery of new targets that can be entered into the drug development pipeline.
CONCLUSIONS
Ten years have passed since the first round of GWAS for T2D and obesity. This decade has been transformative for human genetics and has already had a major impact on the understanding of these diseases. We have learned much about their genetic architecture, including the high degree of polygenicity and tiny effect sizes of most risk variants, which have major implications for precision medicine. Several mechanistic challenges complicate the translation of novel loci, but overcoming these will expose untapped potential for the development of novel drugs to treat T2D and obesity. In parallel, there is the expectation that we will move to a more informed molecular taxonomy of disease that provides a more precise framework for the delivery of preventative and therapeutic interventions. There is a clear need for increased focus on translational and mechanistic research to make sense of the hundreds of loci associated with these diseases, but this work can be conducted with the knowledge that, provided we can find ways to better capitalize on them, the rewards will be momentous.
Acknowledgments
Sources of Funding
Dr Ingelsson is supported by grants from the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK; 1R01DK106236–01A1) and the National Heart, Lung, and Blood Institute (1R01HL135313–01) when writing this article. Dr McCarthy is a Wellcome Trust Senior Investigator and a National Institute for Health Research (NIHR) Senior Investigator, and work relevant to this review was supported by the Wellcome Trust (090367, 090532, 098381, 106130, and 203141) and NIDDK (U01DK085545, R01DK098032, and U01DK105535). The work was partly supported by the Oxford NIHR Biomedical Research Centre based at Oxford University Hospitals National Health Service (NHS) Trust and University of Oxford. The views expressed are those of the authors and not necessarily those of the National Institutes of Health, NHS, or NIHR.
Disclosures
Dr Ingelsson is a scientific advisor for Precision Wellness and Olink Proteomics for work unrelated to the present project. Dr McCarthy is a scientific advisor for Pfizer and Novo Nordisk and in receipt of research funding from Pfizer, Novo Nordisk, Eli Lilly, Janssen, Roche, Merck, Takeda, Sanofi-Aventis, Servier, Boehringer Ingelheim, and AstraZeneca.
Contributor Information
Erik Ingelsson, Division of Cardiovascular Medicine, Department of Medicine, Stanford University School of Medicine, CA; Stanford Cardiovascular Institute, Stanford University, CA.
Mark I. McCarthy, Wellcome Centre for Human Genetics, University of Oxford, United Kingdom; Oxford Centre for Diabetes, Endocrinology and Metabolism, Churchill Hospital, University of Oxford, United Kingdom; Oxford NIHR Biomedical Research Centre, Oxford University Hospitals Trust, United Kingdom.
REFERENCES
- 1.Scott RA, et al. An expanded genome-wide association study of type 2 diabetes in Europeans. Diabetes. 2017;66:2888–2902. doi: 10.2337/db16-1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mahajan A, et al. Fine-mapping of an expanded set of type 2 diabetes loci to single-variant resolution using high-density imputation and islet-specific epigenome maps. bioRxiv. 2018. doi: 10.1101/245506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lu Y, et al. New loci for body fat percentage reveal link between adiposity and cardiometabolic disease risk. Nat Commun. 2016;7:10495. doi: 10.1038/ncomms10495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Locke AE, et al. Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518:197–206. doi: 10.1038/nature14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shungin D, et al. New genetic loci link adipose and insulin biology to body fat distribution. Nature. 2015;518:187–196. doi: 10.1038/nature14132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Winkler TW, et al. The influence of age and sex on genetic associations with adult body size and shape: a large-scale genome-wide interaction study. PLoS Genet. 2015;11:e1005378. doi: 10.1371/journal.pgen.1005378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Turcot V, et al. Protein-altering variants associated with body mass index implicate pathways that control energy intake and expenditure in obesity. Nat Genet. 2018;50:26–41. doi: 10.1038/s41588-017-0011-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pulit SL, et al. Meta-analysis of genome-wide association studies for body fat distribution in 694,649 individuals of European ancestry. bioRxiv. 2018. doi: 10.1101/304030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lander ES, et al. ; International Human Genome Sequencing Consortium. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 10.O’Donnell CJ, et al. Cardiovascular genomics, personalized medicine, and the National Heart, Lung, and Blood Institute: part I: the beginning of an era. Circ Cardiovasc Genet. 2008;1:51–57. doi: 10.1161/CIRCGENETICS.108.813337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Collins FS, et al. Implications of the human genome project for medical science. JAMA. 2001;285:540–544. [DOI] [PubMed] [Google Scholar]
- 12.Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu Z, et al. Dominance genetic variation contributes little to the missing heritability for human complex traits. Am J Hum Genet. 2015;96:377–385. doi: 10.1016/j.ajhg.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wood AR, et al. Defining the role of common variation in the genomic and biological architecture of adult human height. Nat Genet. 2014;46:1173–1186. doi: 10.1038/ng.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hemani G, et al. Inference of the genetic architecture underlying BMI and height with the use of 20,240 sibling pairs. Am J Hum Genet. 2013;93:865–875. doi: 10.1016/j.ajhg.2013.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boyle EA, et al. An expanded view of complex traits: from polygenic to omnigenic. Cell. 2017;169:1177–1186. doi: 10.1016/j.cell.2017.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lyssenko V, et al. Clinical risk factors, DNA variants, and the development of type 2 diabetes. N Engl J Med. 2008;359:2220–2232. doi: 10.1056/NEJMoa0801869. [DOI] [PubMed] [Google Scholar]
- 18.Meigs JB, et al. Genotype score in addition to common risk factors for prediction of type 2 diabetes. N Engl J Med. 2008;359:2208–2219. doi: 10.1056/NEJMoa0804742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vassy JL, et al. Polygenic type 2 diabetes prediction at the limit of common variant detection. Diabetes. 2014;63:2172–2182. doi: 10.2337/db13-1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vassy JL, et al. A genotype risk score predicts type 2 diabetes from young adulthood: the CARDIA study. Diabetologia. 2012;55:2604–2612. doi: 10.1007/s00125-012-2637-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vassy JL, et al. Is genetic testing useful to predict type 2 diabetes? Best Pract Res Clin Endocrinol Metab. 2012;26:189–201. doi: 10.1016/j.beem.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khera AV, et al. Genome-wide polygenic score to identify a monogenic risk-equivalent for coronary disease. bioRxiv. 2017. doi: 10.1101/218388. [DOI] [Google Scholar]
- 23.Knowles JW, et al. Cardiovascular disease: the rise of the genetic risk score. PLoS Med. 2018;15:e1002546. doi: 10.1371/journal.pmed.1002546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Natarajan P, et al. Polygenic risk score identifies subgroup with higher burden of atherosclerosis and greater relative benefit from statin therapy in the primary prevention setting. Circulation. 2017;135:2091–2101. doi: 10.1161/CIRCULATIONAHA.116.024436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Markowitz SM, et al. Perceived impact of diabetes genetic risk testing among patients at high phenotypic risk for type 2 diabetes. Diabetes Care. 2011;34:568–573. doi: 10.2337/dc10-1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tarini BA, et al. Effects of hypothetical type 2 diabetes genetic testing on parents’ efforts to prevent diabetes in children. Clin Pediatr (Phila). 2013;52:821–828. doi: 10.1177/0009922813488644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li SX, et al. The effect of communicating the genetic risk of cardiometabolic disorders on motivation and actual engagement in preventative lifestyle modification and clinical outcome: a systematic review and meta-analysis of randomised controlled trials. Br J Nutr. 2016;116:924–934. doi: 10.1017/S0007114516002488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grant RW, et al. Personalized genetic risk counseling to motivate diabetes prevention: a randomized trial. Diabetes Care. 2013;36:13–19. doi: 10.2337/dc12-0884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Manolio TA, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lupski JR, et al. Clan genomics and the complex architecture of human disease. Cell. 2011;147:32–43. doi: 10.1016/j.cell.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dickson SP, et al. Rare variants create synthetic genome-wide associations. PLoS Biol. 2010;8:e1000294. doi: 10.1371/journal.pbio.1000294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mahajan A, et al. Refining the accuracy of validated target identification through coding variant fine-mapping in type 2 diabetes. Nat Genet. 2018;50:559–571. doi: 10.1038/s41588-018-0084-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fuchsberger C, et al. The genetic architecture of type 2 diabetes. Nature. 2016;536:41–47. doi: 10.1038/nature18642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang J, et al. Genetic variance estimation with imputed variants finds negligible missing heritability for human height and body mass index. Nat Genet. 2015;47:1114–1120. doi: 10.1038/ng.3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Agarwala V, et al. Evaluating empirical bounds on complex disease genetic architecture. Nat Genet. 2013;45:1418–1427. doi: 10.1038/ng.2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pal A, et al. Loss-of-function mutations in the cell-cycle control gene CDKN2A impact on glucose homeostasis in humans. Diabetes. 2016;65:527–533. doi: 10.2337/db15-0602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moltke I, et al. A common Greenlandic TBC1D4 variant confers muscle insulin resistance and type 2 diabetes. Nature. 2014;512:190–193. doi: 10.1038/nature13425. [DOI] [PubMed] [Google Scholar]
- 38.Steinthorsdottir V, et al. Identification of low-frequency and rare sequence variants associated with elevated or reduced risk of type 2 diabetes. Nat Genet. 2014;46:294–298. doi: 10.1038/ng.2882. [DOI] [PubMed] [Google Scholar]
- 39.Ahlqvist E, et al. Novel subgroups of adult-onset diabetes and their association with outcomes: a data-driven cluster analysis of six variables. Lancet Diabetes Endocrinol. 2018;6:361–369. doi: 10.1016/S2213-8587(18)30051-2. [DOI] [PubMed] [Google Scholar]
- 40.McCarthy MI. Painting a new picture of personalised medicine for diabetes. Diabetologia. 2017;60:793–799. doi: 10.1007/s00125-017-4210-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Florez JC. Pharmacogenetics in type 2 diabetes: precision medicine or discovery tool? Diabetologia. 2017;60:800–807. doi: 10.1007/s00125-017-4227-1. [DOI] [PubMed] [Google Scholar]
- 42.Hattersley AT, et al. Precision diabetes: learning from monogenic diabetes. Diabetologia. 2017;60:769–777. doi: 10.1007/s00125-017-4226-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Florez JC. The pharmacogenetics of metformin. Diabetologia. 2017;60:1648–1655. doi: 10.1007/s00125-017-4335-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maruthur NM, et al. The pharmacogenetics of type 2 diabetes: a systematic review. Diabetes Care. 2014;37:876–886. doi: 10.2337/dc13-1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou K, et al. Variation in the glucose transporter gene SLC2A2 is associated with glycemic response to metformin. Nat Genet. 2016;48:1055–1059. doi: 10.1038/ng.3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Franks PW, et al. Gene-environment and gene-treatment interactions in type 2 diabetes: progress, pitfalls, and prospects. Diabetes Care. 2013;36:1413–1421. doi: 10.2337/dc12-2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li SX, et al. Interaction between genes and macronutrient intake on the risk of developing type 2 diabetes: systematic review and findings from European Prospective Investigation into Cancer (EPIC)-InterAct. Am J Clin Nutr. 2017;106:263–275. doi: 10.3945/ajcn.116.150094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zeevi D, et al. Personalized nutrition by prediction of glycemic responses. Cell. 2015;163:1079–1094. doi: 10.1016/j.cell.2015.11.001. [DOI] [PubMed] [Google Scholar]
- 49.Wang T, et al. Improving adherence to healthy dietary patterns, genetic risk, and long term weight gain: gene-diet interaction analysis in two prospective cohort studies. BMJ. 2018;360:j5644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Qi Q, et al. Fried food consumption, genetic risk, and body mass in-dex: gene-diet interaction analysis in three US cohort studies. BMJ. 2014;348:g1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Qi Q, et al. Sugar-sweetened beverages and genetic risk of obesity. N Engl J Med. 2012;367:1387–1396. doi: 10.1056/NEJMoa1203039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Graff M, et al. Genome-wide physical activity interactions in adiposity - a meta-analysis of 200,452 adults. PLoS Genet. 2017;13:e1006528. doi: 10.1371/journal.pgen.1006528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.DNA Fit. https://www.dnafit.com/us/.
- 54.DNActiv8. https://www.dnactiv8.co.uk/.
- 55.Vitagene. https://vitagene.com/.
- 56.Helix. https://www.helix.com/.
- 57.Orig3n. https://orig3n.com/. Accessed August 24, 2017.
- 58.Vinome. https://www.vinome.com/.
- 59.Marmite Gene Project. https://www.marmite.co.uk/geneproject. Accessed September 14, 2017.
- 60.Soccer Genomics. https://www.soccergenomics.com/home/.
- 61.Evans DM, et al. Mining the human phenome using allelic scores that index biological intermediates. PLoS Genet. 2013;9:e1003919. doi: 10.1371/journal.pgen.1003919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Small KS, et al. Regulatory variants at KLF14 influence type 2 diabetes risk via a female-specific effect on adipocyte size and body composition. Nat Genet. 2018;50:572–580. doi: 10.1038/s41588-018-0088-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ashley EA. Towards precision medicine. Nat Rev Genet. 2016;17:507–522. doi: 10.1038/nrg.2016.86. [DOI] [PubMed] [Google Scholar]
- 64.Sagen JV, et al. Permanent neonatal diabetes due to mutations in KCNJ11 encoding Kir6.2: patient characteristics and initial response to sulfonylurea therapy. Diabetes. 2004;53:2713–2718. [DOI] [PubMed] [Google Scholar]
- 65.Gloyn AL, et al. Activating mutations in the gene encoding the ATP-sensitive potassium-channel subunit Kir6.2 and permanent neonatal diabetes. N Engl J Med. 2004;350:1838–1849. doi: 10.1056/NEJMoa032922. [DOI] [PubMed] [Google Scholar]
- 66.Montague CT, et al. Congenital leptin deficiency is associated with severe early-onset obesity in humans. Nature. 1997;387:903–908. doi: 10.1038/43185. [DOI] [PubMed] [Google Scholar]
- 67.McCarthy MI, et al. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet. 2008;9:356–369. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- 68.Bouatia-Naji N, et al. A variant near MTNR1B is associated with in-creased fasting plasma glucose levels and type 2 diabetes risk. Nat Genet. 2009;41:89–94. doi: 10.1038/ng.277. [DOI] [PubMed] [Google Scholar]
- 69.Prokopenko I, et al. Variants in MTNR1B influence fasting glucose levels. Nat Genet. 2009;41:77–81. doi: 10.1038/ng.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lyssenko V, et al. Common variant in MTNR1B associated with increased risk of type 2 diabetes and impaired early insulin secretion. Nat Genet. 2009;41:82–88. doi: 10.1038/ng.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gaulton KJ, et al. Genetic fine mapping and genomic annotation de-fines causal mechanisms at type 2 diabetes susceptibility loci. Nat Genet. 2015;47:1415–1425. doi: 10.1038/ng.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dimas AS, et al. Impact of type 2 diabetes susceptibility variants on quantitative glycemic traits reveals mechanistic heterogeneity. Diabetes. 2014;63:2158–2171. doi: 10.2337/db13-0949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tuomi T, et al. Increased melatonin signaling is a risk factor for type 2 diabetes. Cell Metab. 2016;23:1067–1077. doi: 10.1016/j.cmet.2016.04.009. [DOI] [PubMed] [Google Scholar]
- 74.Mulder H Melatonin signalling and type 2 diabetes risk: too lit-tle, too much or just right? Diabetologia. 2017;60:826–829. doi: 10.1007/s00125-017-4249-8. [DOI] [PubMed] [Google Scholar]
- 75.Mahajan A, et al. Genome-wide trans-ancestry meta-analysis provides insight into the genetic architecture of type 2 diabetes susceptibility. Nat Genet. 2014;46:234–244. doi: 10.1038/ng.2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bell CG, et al. Integrated genetic and epigenetic analysis identifies haplotype-specific methylation in the FTO type 2 diabetes and obesity susceptibility locus. PLoS One. 2010;5:e14040. doi: 10.1371/journal.pone.0014040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lappalainen T, et al. Epistatic selection between coding and regulatory variation in human evolution and disease. Am J Hum Genet. 2011;89:459–463. doi: 10.1016/j.ajhg.2011.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schmitt AD, et al. Genome-wide mapping and analysis of chromo-some architecture. Nat Rev Mol Cell Biol. 2016;17:743–755. doi: 10.1038/nrm.2016.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.GTEx Consortium. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nica AC, et al. Candidate causal regulatory effects by integration of expression QTLs with complex trait genetic associations. PLoS Genet. 2010;6:e1000895. doi: 10.1371/journal.pgen.1000895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Frayling TM, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316:889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Grant SF, et al. Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nat Genet. 2006;38:320–323. doi: 10.1038/ng1732. [DOI] [PubMed] [Google Scholar]
- 83.Helgadottir A, et al. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science. 2007;316:1491–1493. doi: 10.1126/science.1142842. [DOI] [PubMed] [Google Scholar]
- 84.McPherson R, et al. A common allele on chromosome 9 associated with coronary heart disease. Science. 2007;316:1488–1491. doi: 10.1126/science.1142447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Claussnitzer M, et al. FTO obesity variant circuitry and adipocyte browning in humans. N Engl J Med. 2015;373:895–907. doi: 10.1056/NEJMoa1502214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Stratigopoulos G, et al. Hypomorphism of Fto and Rpgrip1l causes obesity in mice. J Clin Invest. 2016;126:1897–1910. doi: 10.1172/JCI85526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Church C, et al. Overexpression of Fto leads to increased food in-take and results in obesity. Nat Genet. 2010;42:1086–1092. doi: 10.1038/ng.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Civelek M, et al. Genetic regulation of adipose gene expression and cardio-metabolic traits. Am J Hum Genet. 2017;100:428–443. doi: 10.1016/j.ajhg.2017.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Thomsen SK, et al. Human genetics as a model for target validation: finding new therapies for diabetes. Diabetologia. 2017;60:960–970. doi: 10.1007/s00125-017-4270-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Fadista J, et al. Global genomic and transcriptomic analysis of human pancreatic islets reveals novel genes influencing glucose metabolism. Proc Natl Acad Sci USA. 2014;111:13924–13929. doi: 10.1073/pnas.1402665111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bonnefond A, et al. Rare MTNR1B variants impairing melatonin receptor 1B function contribute to type 2 diabetes. Nat Genet. 2012;44:297–301. doi: 10.1038/ng.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bonnefond A, et al. The case for too little melatonin signalling in increased diabetes risk. Diabetologia. 2017;60:823–825. doi: 10.1007/s00125-017-4255-x. [DOI] [PubMed] [Google Scholar]
- 93.Mumbach MR, et al. Enhancer connectome in primary human cells identifies target genes of disease-associated DNA elements. Nat Genet. 2017;49:1602–1612. doi: 10.1038/ng.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mumbach MR, et al. HiChIP: efficient and sensitive analysis of protein-directed genome architecture. Nat Methods. 2016;13:919–922. doi: 10.1038/nmeth.3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kumasaka N, et al. Fine-mapping cellular QTLs with RASQUAL and ATAC-seq. Nat Genet. 2016;48:206–213. doi: 10.1038/ng.3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Buenrostro JD, et al. ATAC-seq: a method for assaying chromatin accessibility genome-wide. Curr Protoc Mol Biol. 2015;109:21.29.1–21.29.9. doi: 10.1002/0471142727.mb2129s109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Macosko EZ, et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Jaitin DA, et al. Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science. 2014;343:776–779. doi: 10.1126/science.1247651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Adamson B, et al. A multiplexed single-cell CRISPR screening platform enables systematic dissection of the unfolded protein response. Cell. 2016;167:1867–1882.e21. doi: 10.1016/j.cell.2016.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Dixit A, et al. Perturb-seq: dissecting molecular circuits with scalable single-cell RNA profiling of pooled genetic screens. Cell. 2016;167:1853–1866.e17. doi: 10.1016/j.cell.2016.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Datlinger P, et al. Pooled CRISPR screening with single-cell transcriptome readout. Nat Methods. 2017;14:297–301. doi: 10.1038/nmeth.4177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Jaitin DA, et al. Dissecting immune circuits by linking CRISPR-pooled screens with single-cell RNA-seq. Cell. 2016;167:1883–1896.e15. doi: 10.1016/j.cell.2016.11.039. [DOI] [PubMed] [Google Scholar]
- 103.Xie S, et al. Multiplexed engineering and analysis of combinatorial enhancer activity in single cells. Mol Cell. 2017;66:285–299.e5. doi: 10.1016/j.molcel.2017.03.007. [DOI] [PubMed] [Google Scholar]
- 104.Scannell JW, et al. Diagnosing the decline in pharmaceutical R&D efficiency. Nat Rev Drug Discov. 2012;11:191–200. doi: 10.1038/nrd3681. [DOI] [PubMed] [Google Scholar]
- 105.Herper M How much does pharmaceutical innovation cost? A look at 100 companies. Forbes. 2013. http://www.forbes.com/sites/matthewherper/2013/08/11/the-cost-of-inventing-a-new-drug-98-companies-ranked/.
- 106.DiMasi JA, et al. Innovation in the pharmaceutical industry: New estimates of R&D costs. J Health Econ. 2016;47:20–33. doi: 10.1016/j.jhealeco.2016.01.012. [DOI] [PubMed] [Google Scholar]
- 107.DiMasi JA, et al. The cost of drug development. N Engl J Med. 2015;372:1972. doi: 10.1056/NEJMc1504317. [DOI] [PubMed] [Google Scholar]
- 108.Cook D, et al. Lessons learned from the fate of AstraZeneca’s drug pipe-line: a five-dimensional framework. Nat Rev Drug Discov. 2014;13:419–431. doi: 10.1038/nrd4309. [DOI] [PubMed] [Google Scholar]
- 109.Arrowsmith J, et al. Trial watch: phase II and phase III attrition rates 2011–2012. Nat Rev Drug Discov. 2013;12:569. doi: 10.1038/nrd4090. [DOI] [PubMed] [Google Scholar]
- 110.DiMasi JA, et al. Trends in risks associated with new drug develop-ment: success rates for investigational drugs. Clin Pharmacol Ther. 2010;87:272–277. doi: 10.1038/clpt.2009.295. [DOI] [PubMed] [Google Scholar]
- 111.Morgan P, et al. Can the flow of medicines be improved? Fundamental pharmacokinetic and pharmacological principles toward improving phase II survival. Drug Discov Today. 2012;17:419–424. doi: 10.1016/j.drudis.2011.12.020. [DOI] [PubMed] [Google Scholar]
- 112.Plenge RM, et al. Validating therapeutic targets through human genetics. Nat Rev Drug Discov. 2013;12:581–594. doi: 10.1038/nrd4051. [DOI] [PubMed] [Google Scholar]
- 113.Altshuler D, et al. The common PPARgamma Pro12Ala polymorphism is associated with decreased risk of type 2 diabetes. Nat Genet. 2000;26:76–80. doi: 10.1038/79216. [DOI] [PubMed] [Google Scholar]
- 114.Lehmann JM, et al. An antidiabetic thiazolidinedione is a high affinity ligand for peroxisome proliferator-activated receptor gamma (PPAR gamma). J Biol Chem. 1995;270:12953–12956. [DOI] [PubMed] [Google Scholar]
- 115.Kathiresan S, et al. Six new loci associated with blood low-density lipo-protein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. Nat Genet. 2008;40:189–197. doi: 10.1038/ng.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Endo A The discovery and development of HMG-CoA reductase inhibi-tors. J Lipid Res. 1992;33:1569–1582. [PubMed] [Google Scholar]
- 117.Stitziel NO, et al. Inactivating mutations in NPC1L1 and protection from coronary heart disease. N Engl J Med. 2014;371:2072–2082. doi: 10.1056/NEJMoa1405386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Cannon CP, et al. Ezetimibe added to statin therapy after acute coronary syndromes. N Engl J Med. 2015;372:2387–2397. doi: 10.1056/NEJMoa1410489. [DOI] [PubMed] [Google Scholar]
- 119.Crosby J, et al. Loss-of-function mutations in APOC3, triglycer-ides, and coronary disease. N Engl J Med. 2014;371:22–31. doi: 10.1056/NEJMoa1307095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Gaudet D, et al. Antisense inhibition of apolipoprotein C-III in patients with hypertriglyceridemia. N Engl J Med. 2015;373:438–447. doi: 10.1056/NEJMoa1400283. [DOI] [PubMed] [Google Scholar]
- 121.Riordan JR, et al. Identification of the cystic fibrosis gene: cloning and characterization of complementary DNA. Science. 1989;245:1066–1073. [DOI] [PubMed] [Google Scholar]
- 122.Wainwright CE, et al. Lumacaftor-ivacaftor in patients with cystic fibrosis homozygous for Phe508del CFTR. N Engl J Med. 2015;373:1783–1784. doi: 10.1056/NEJMc1510466. [DOI] [PubMed] [Google Scholar]
- 123.Cohen JC, et al. Sequence variations in PCSK9, low LDL, and protection against coronary heart disease. N Engl J Med. 2006;354:1264–1272. doi: 10.1056/NEJMoa054013. [DOI] [PubMed] [Google Scholar]
- 124.Cohen J, et al. Low LDL cholesterol in individuals of African descent resulting from frequent nonsense mutations in PCSK9. Nat Genet. 2005;37:161–165. doi: 10.1038/ng1509. [DOI] [PubMed] [Google Scholar]
- 125.Abifadel M, et al. Mutations in PCSK9 cause autosomal dominant hyper-cholesterolemia. Nat Genet. 2003;34:154–156. doi: 10.1038/ng1161. [DOI] [PubMed] [Google Scholar]
- 126.Sabatine MS, et al. Efficacy and safety of evolocumab in reducing lip-ids and cardiovascular events. N Engl J Med. 2015;372:1500–1509. doi: 10.1056/NEJMoa1500858. [DOI] [PubMed] [Google Scholar]
- 127.Robinson JG, et al. Efficacy and safety of alirocumab in reducing lipids and cardiovascular events. N Engl J Med. 2015;372:1489–1499. doi: 10.1056/NEJMoa1501031. [DOI] [PubMed] [Google Scholar]
- 128.Flannick J, et al. Loss-of-function mutations in SLC30A8 protect against type 2 diabetes. Nat Genet. 2014;46:357–363. doi: 10.1038/ng.2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Grotz AK, et al. Prioritising causal genes at type 2 diabetes risk loci. Curr Diab Rep. 2017;17:76. doi: 10.1007/s11892-017-0907-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Finan C, et al. The druggable genome and support for target identification and validation in drug development. Sci Transl Med. 2017;9. doi: 10.1126/scitranslmed.aag1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Nelson MR, et al. The support of human genetic evidence for approved drug indications. Nat Genet. 2015;47:856–860. doi: 10.1038/ng.3314. [DOI] [PubMed] [Google Scholar]
- 132.Hurle MR, et al. Trial watch: Impact of genetically supported target selec-tion on R&D productivity. Nat Rev Drug Discov. 2016;15:595–597. doi: 10.1038/nrd.2016.164. [DOI] [PubMed] [Google Scholar]
- 133.Morris AP, et al. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat Genet. 2012;44:981–990. doi: 10.1038/ng.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Flanagan SE, et al. Update of mutations in the genes encoding the pan-creatic beta-cell K(ATP) channel subunits Kir6.2 (KCNJ11) and sulfonylurea receptor 1 (ABCC8) in diabetes mellitus and hyperinsulinism. Hum Mutat. 2009;30:170–180. doi: 10.1002/humu.20838. [DOI] [PubMed] [Google Scholar]
- 135.Nagashima K, et al. Sulfonylurea and non-sulfonylurea hypoglycemic agents: pharmachological properties and tissue selectivity. Diabetes Res Clin Pract. 2004;66(suppl 1):S75–S78. doi: 10.1016/j.diabres.2003.12.011. [DOI] [PubMed] [Google Scholar]
- 136.Maiorino MI, et al. Insulin and glucagon-like peptide 1 receptor agonist combination therapy in type 2 diabetes: a systematic review and metaanalysis of randomized controlled trials. Diabetes Care. 2017;40:614–624. doi: 10.2337/dc16-1957. [DOI] [PubMed] [Google Scholar]
- 137.Potts JE, et al. The effect of glucagon-like peptide 1 receptor agonists on weight loss in type 2 diabetes: a systematic review and mixed treatment comparison meta-analysis. PLoS One. 2015;10:e0126769. doi: 10.1371/journal.pone.0126769. [DOI] [PMC free article] [PubMed] [Google Scholar]