Abstract
The Notch signaling pathway contributes to the pathogenesis of a wide spectrum of human cancers, including hematopoietic malignancies. Its functions are highly dependent on the specific cellular context. Gain-of-function NOTCH1 mutations are prevalent in human T cell leukemia, while loss of Notch signaling is reported in myeloid leukemias. Here, we report a novel oncogenic function of Notch signaling in oncogenic Kras-induced myeloproliferative neoplasm (MPN). We find that downregulation of Notch signaling in hematopoietic cells via DNMAML expression or Pofut1 deletion significantly blocks MPN development in KrasG12D mice in a cell-autonomous manner. Further mechanistic studies indicate that inhibition of Notch signaling significantly upregulates Dusp1, a dual phosphatase that inactivates p-ERK, and downregulates cytokine-evoked ERK activation in KrasG12D cells. Moreover, mitochondrial metabolism is greatly enhanced in KrasG12D cells but significantly reprogrammed by DNMAML close to that in control cells. Consequently, cell proliferation and expanded myeloid compartment in KrasG12D mice are significantly reduced. Consistent with these findings, combined inhibition of the MEK/ERK pathway and mitochondrial oxidative phosphorylation effectively inhibited the growth of human and mouse leukemia cells in vitro. Our study provides a strong rational to target both ERK signaling and aberrant metabolism in oncogenic Ras-driven myeloid leukemia.
Keywords: Notch signaling, oncogenic Kras, myeloproliferative neoplasm, ERK, oxidative phosphorylation
Introduction
The Notch receptor was first cloned in Drosophila as its genetic mutations resulted in a wing-notching phenotype (1, 2). In mammals, there are four Notch receptors, Notch1-Notch4. They are translated as a single pro-Notch precursor. After cleavage by a furin-like protease in the trans-Golgi network, Notch receptors form a noncovalently linked heterodimer with an N-terminal extracellular fragment and a C-terminal transmembrane-intracellular subunit (reviewed in (3)). The extracellular domain of Notch is modified with multiple O-fucose glycans (4). This process is catalyzed by protein O-fucosyltransferase 1 (Pofut1) (5–7) and is critical for Notch-ligand interactions and Notch signaling (8). The Notch pathway is normally activated through interactions with ligands, which are transmembrane proteins named Delta-like and Jagged. Upon ligand binding, Notch receptors undergo a series of protease cleavage events, leading to the release of their intracellular portion (termed ICN). The ICN subsequently translocates into the nucleus and forms a ternary complex with the coactivator protein mastermind-1 (MAML1) and transcription factor CSL/RBPJ to mediate target gene activation (reviewed in (3)). This canonical Notch signaling can be blocked by a dominant-negative MAML1 (DNMAML) (9).
Notch is a highly conserved signaling pathway that regulates cell-fate specification and tissue homeostasis in various contexts (reviewed in (10)). Depending on the context, Notch signaling could be oncogenic or tumor suppressive (reviewed in (3)). In the hematopoietic system, Notch1 is a master transcription factor that regulates T cell development. Gain-of-function (GOF) NOTCH1 mutations are identified in 50–70% of human T cell acute lymphoblastic leukemia/lymphoma (T-ALL) cases (11). The majority of these mutations occur in exons 26 and 27 and render ligand-independent activation of Notch or hypersensitivity to Notch ligands. Another class of NOTCH1 mutations occur in its PEST domain, which impair FBXW7-mediated proteasomal degradation and increase the cellular ICN1 concentrations. Similarly, Notch1 mutations are identified in 100% of oncogenic Ras-induced T-ALL mice during T-ALL progression (12). These mutations are predominantly Rag recombinase-mediated Type 1 deletions (13) conferring ligand-independent activation of Notch and PEST domain mutations. Functional studies demonstrate that human T-ALL-associated NOTCH1 alleles are sufficient to induce leukemia in vivo (14). They contribute to the leukemic transformation of CD8+ T cells to leukemia initiating cells in oncogenic Kras mice and thus accelerate oncogenic Kras-initiated T-ALL (12, 14). However, it remains unclear whether Notch signaling is required for the initiation of oncogenic Ras-induced T-ALL.
Compared to the oncogenic function of Notch1 GOF mutations in T-ALL, the role of Notch signaling in acute myeloid leukemia (AML) is tumor suppressive. In human AML, despite the robust expression of Notch receptors, Notch signaling is low or silenced (15, 16). Notch activation inhibits AML growth and survival, while Notch inactivation cooperates with loss of the myeloid tumor suppressor Tet2 to induce an AML-like disease in mice (16). These results indicate a tumor suppressive role of Notch signaling in AML and provide a strong rationale to use Notch receptor agonists in AML treatment.
In an independent study, Klinakis et al. reported that downregulation of Notch signaling using different genetic approaches, such as deletion of Nicastrin (an essential component for Notch processing to generate ICN) or knocking out Notch1/2 mediated by interferon-inducible Mx1-Cre, leads to a lethal myeloproliferative neoplasm (MPN), closely resembling human chronic myelomonocytic leukemia (CMML) (17). Loss-of-function mutations in Notch pathway genes were identified in a subset of CMML patients. In a similar study, ablation of FX (the homolog of human GDP-L-fucose synthase) or Pofut1 using the same Mx1-Cre line results in benign myeloid hyperplasia phenotypes in mice (18). Together, these studies suggest a tumor suppressive function of Notch signaling in MPN development as well. Here we took a genetic approach to investigate the cell-autonomous function of Notch signaling in oncogenic Kras-induced T-ALL and MPN.
Materials and Methods
Mice
All mouse lines were maintained in a pure C57BL/6 genetic background (>N10). Genotyping of KrasLSL G12D/+, Rosa26LSL DNMAML-GFP/+, Pofut1fl/fl, and Mx1-Cre was done as previously described (9, 19, 20). CD45.1-positive congenic C57BL/6 recipient mice were purchased from NCI. All animal experiments were conducted in accordance with the Guide for the Care and Use of Laboratory Animals and approved by an Animal Care and Use Committee at UW-Madison. The program is accredited by the Association for Assessment and Accreditation of Laboratory Animal Care.
Additional methods are described in Supplementary Materials and Methods.
Results
Downregulating Notch signaling inhibits both oncogenic Kras-induced T-ALL and MPN in a cell-autonomous manner
To investigate the function of Notch signaling in oncogenic Kras-induced leukemogenesis, we took two independent genetic approaches to downregulate Notch signaling. Expression of dominant-negative Mastermind-like1 (DNMAML) potently inhibits canonical Notch-mediated transcriptional activation (9), while knocking out Protein O-fucosyltransferase 1 (Pofut1) reduces interactions of Notch receptors and their ligands and downregulates Notch signaling (18). We generated KrasLSL G12D/+; Mx1-Cre (Kras), KrasLSL G12D/+; Rosa26LSL DNMAML-GFP/+; Mx1-Cre (Kras; D/+), and KrasLSL G12D/+; Pofut1fl/fl ; Mx1-Cre (Kras; P−/−) mice as previously described (21, 22). Mx1-Cre mice were used as control throughout this study.
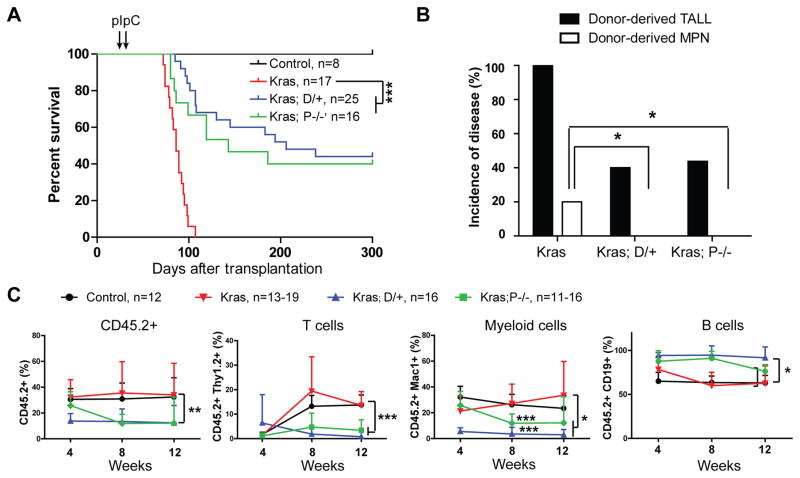
We first took a bone marrow transplantation approach to study how downregulating Notch signaling affects oncogenic Kras-induced leukemogenesis in a cell-autonomous manner. The same number of control, Kras, Kras;D/+, or Kras;P−/− bone marrow cells (CD45.2+) were transplanted along with congeneic competitor cells (CD45.1+) into lethally irradiated mice (CD45.1+). Three weeks after transplantation, recipients were injected with polyinosinic-polycytidylic acid (pI-pC) to induce expression of oncogenic Kras and DNMAML-GFP and deletion of Pofut1. Consistent with previous reports (21, 22), all of the recipients transplanted with Kras cells died of T-ALL quickly (Figure 1A and 1B). As expected, inhibition of Notch signaling significantly inhibited T-cell development (Figure 1C), reduced the penetrance, and delayed onset of T-ALL in recipients with Kras;D/+ or Kras;P−/− cells (Figure 1A and 1B). Importantly, the T-ALL that did develop in these recipients, while having undergone Cre-mediated activation of the mutant Kras allele (Figure S1A), had not expressed DNMAML (as evidenced by the lack of GFP expression – Figure S1B) nor deleted Pofut1 (Figure S1C), arguing that Notch signaling is absolutely required for the initiation of Kras mediated T-ALL. Consistent with our previous finding (12), all T-ALL specimens contained a Notch1 Type 1 deletion (Figure S1D), which renders ligand-independent activation of Notch1 signaling (13). In addition, we found that ~20% of the recipient mice transplanted with Kras cells developed a donor-derived MPN, whereas none of Kras; D/+ and Kras; P−/− recipients developed this disease (P=0.02) (Figure 1B). Donor-derived MPN is defined as previously described (21): donor-derived CD45.2+ cells constitute >50% in the peripheral blood of recipients and >20% of donor-derived cells are Mac1+ Gr1− monocytes (Figure S2A). This disease often associates with splenomegaly (Figure S2B) and extramedullary hematopoiesis in spleen (Figure S2C) (19, 21). Together, both genetic approaches yielded essentially identical results (Figure 1 and our unpublished observations in primary mice).
Figure 1. Downregulating Notch signaling inhibits oncogenic Kras-induced T-ALL in a cell-autonomous manner.
Lethally irradiated mice (CD45.1+) were transplanted with 2.5×105 bone marrow cells (CD45.2+) from control (Mx1-Cre), KrasLSL G12D/+;Mx1-Cre (Kras), KrasLSL G12D/+;Rosa26LSL DNMAML-GFP/+;Mx1-Cre (Kras; D/+) or KrasLSL G12D/+; Pofutfl/fl ;Mx1-Cre (Kras; P−/−) mice along with 2.5×105 competitor cells (CD45.1+). Four weeks after transplantation, Cre expression was induced using pI-pC injections as described in Methods. Moribund recipients transplanted with Kras, Kras; D/+ or Kras; P−/− cells and age-matched recipients transplanted with control cells were sacrificed for analysis. (A) Kaplan-Meier survival curves of different groups of recipient mice were plotted against days after transplantation. P values were determined using the Log-rank test. (B) Disease incidence in different groups of recipients. Chi-square analysis was performed. (C) Total donor-derived cells and donor-derived myeloid cells, B cells or T cells (CD45.2+) in different groups of recipients were evaluated regularly after transplantation. Of note, 4-week data were collected right before pI-pC injections. The results are presented as mean ± SD. * P<0.05; ** P<0.01; *** P<0.001.
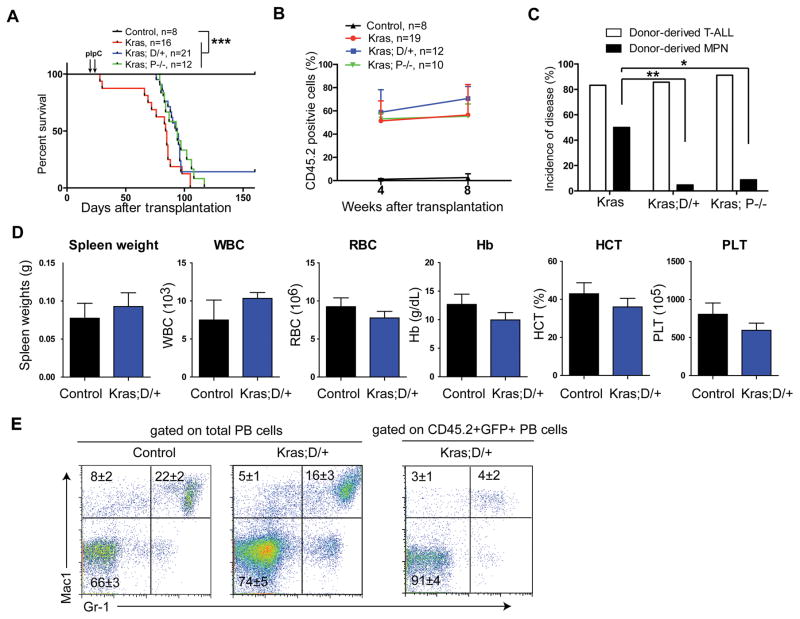
The absence of donor-derived MPN in Kras; D/+ and Kras; P−/− recipients associated with significantly lower percentages of total donor-derived cells and donor-derived myeloid cells in the peripheral blood (Figure 1C), which could result from significantly lower frequencies of hematopoietic stem cell (HSC) in Kras;D/+ and Kras; P−/− bone marrows than those in Kras bone marrows (Figure S3). Therefore, it was unclear whether the absence of donor-derived MPN in Kras;D/+ and Kras; P−/− recipients was due to the reduced HSC reconstitution in these animals or the inhibitory functions of downregulating Notch signaling. To distinguish between these two possibilities, we next took a splenocyte transplant approach because Kras, Kras;D/+, and Kras;P−/− splenocytes contained comparable numbers of HSCs mobilized from bone marrow (Figure S3) and because Kras splenocyte transplant yielded a robust MPN with a much higher penetrance (~50%) (23). As expected, Kras, Kras;D/+, and Kras;P−/− splenocytes reconstituted the recipients at a comparable level (Figure 2B). The majority of the recipients died with a donor-derived T-ALL; T-ALL developed in recipient mice transplanted with Kras;D/+ or Kras;P−/− splenocytes were derived from donor cells that expressed oncogenic Kras, preserved intact Notch signaling, and carried Notch1 Type 1 deletion (Figure 2C, S4, S5A, and S5B).
Figure 2. Downregulating Notch signaling inhibits oncogenic Kras-induced acute MPN in a cell-autonomous manner.
Lethally irradiated mice (CD45.1+) were transplanted with 2 ×106 splenocytes (CD45.2+) from control (Mx1-Cre), KrasLSL G12D/+;Mx1-Cre (Kras), KrasLSL G12D/+;Rosa26LSL DNMAML-GFP/+;Mx1-Cre (Kras; D/+), or KrasLSL G12D/+; Pofutfl/fl ;Mx1-Cre (Kras; P−/−) mice along with 2.5×105 competitor cells (CD45.1+). Four weeks after transplantation, Cre expression was induced using pI-pC injections as described in Methods. Moribund recipients transplanted with Kras, Kras; D/+ or Kras; P−/− cells and age-matched recipients transplanted with control cells were sacrificed for analysis. (A) Kaplan-Meier survival curves of different groups of recipient mice were plotted against days after transplantation. P values were determined using the Log-rank test. (B) Quantification of donor-derived cells (CD45.2+) in the peripheral blood of recipients. Of note, 4-week data were collected right before pI-pC injections. The results are presented as mean ± SD. (C) Disease incidence in different groups of recipient mice. Chi-square analysis was performed. (D, E) Three recipients transplanted with Kras; D/+ cells did not develop T-ALL and were sacrificed for analysis 160 days after transplantation with age-matched control recipients. (D) Quantification of spleen weight and CBC analysis results. Numbers of WBC (white blood cell), RBC (red blood cell), Hb (hemoglobin), HCT (hematocrit), and PLT (platelet) are shown. (E) Flow cytometric analysis of peripheral blood (PB) cells using myeloid lineage markers. Total (left) or donor-derived (right) live nucleated cells are gated for analysis. The results are presented as mean ± SD. * P<0.05; ** P<0.01; *** P<0.001.
Consistent with our previous observation, only 1/21 Kras;D/+ recipient mice and 1/12 Kras;P−/− recipients developed a donor-derived MPN disease, while 8/16 Kras recipients died with donor-derived MPN (Figure 2C). In particular, 3 Kras;D/+ recipients did not develop T-ALL. We sacrificed them 160 days after transplantation for analysis (Figure 2A). Although their hematopoietic system was infiltrated with Kras;D/+ cells (Figure S5C), no sign of an MPN was evident as measured by spleen weights, CBC results, and myeloid compartment in peripheral blood (Figure 2D and 2E).
To better assess the role of canonical Notch signaling in oncogenic Kras-induced MPN, we took two independent approaches to minimize the prevalence of T-ALL in the Kras model. First, we used a more myeloid-restricted Cre, LysM-Cre (24), to drive oncogenic Kras expression. Due to the embryonic expression of LysM-Cre, KrasLSL G12D/+; LysM-Cre mice died significantly earlier than KrasLSL G12D/+; Mx1-Cre mice without pI-pC injections (Figure S6A). Approximately 30% of KrasLSL G12D/+; LysM-Cre mice died without a hematopoietic disease (perhaps due to oncogenic Kras expression in non-hematopoietic tissues), while the remaining 70% mice died with a significant MPN. Consistent with the previous report that LysM-Cre labels ~8% HSCs and subsequently ~8% of T- and B-cells (25), ~10% of KrasLSL G12D/+; LysM-Cre mice died with a significant T-ALL, which was comparable to that in KrasLSL G12D/+; Mx1-Cre mice without pI-pC injections. We further explored the possibility of transplanting KrasLSL G12D/+; LysM-Cre fetal liver cells into lethally irradiated recipients (Figure S6B). These recipients survived moderately but significantly longer than those with KrasLSL G12D/+; Mx1-Cre splenocytes, but they developed donor-derived MPN (~50%) and T-ALL (~90%) at similar frequencies as the latter. Our results demonstrated that compared to Mx1-Cre, LysM-Cre-driven Kras expression does not significantly reduce T-ALL incidence.
Second, we transplanted GFP+ LSK cells isolated from Kras; D/+ mice into lethally irradiated recipients and pI-pC injections were performed to further ensure DNMAML expression in these cells (Figure S7). To our surprise, all recipients died of a severe T-ALL, similarly to recipients transplanted with same number of Kras mutant LSK cells. Upon immunophenotypic examination, we found that despite predominant DNMAML expressing, GFP+ donor cells in the recipient bone marrow and spleen, T-ALL cells shut down DNMAML expression and became GFP−. Consistent with our previous result, none of these recipients developed a donor-derived MPN (Figure S7B). Together, our results indicate that downregualtion of Notch signaling inhibits both T-ALL and MPN development in Kras mice in a cell-autonomous manner.
Downregulating Notch signaling reduces Kras myeloid compartment
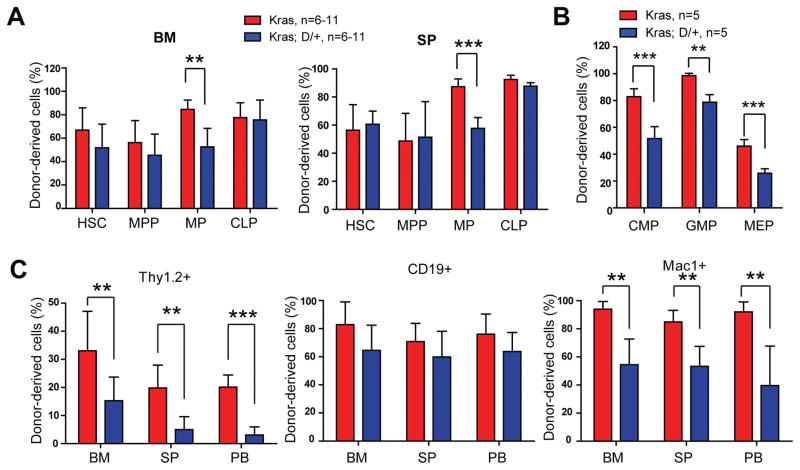
To investigate the mechanism(s) underlying MPN inhibition induced by downregulation of Notch signaling, we analyzed donor derived hematopoiesis in recipients with Kras or Kras;D/+ splenocytes 6 weeks after transplantation. In the remaining study, we define Kras derived cells as CD45.2+ cells and Kras;D/+ derived cells as CD45.2+ GFP+ cells. At 6 weeks after transplantation, the recipients had not displayed overt T-ALL or MPN phenotypes. We found that donor-derived HSC, multi-potential progenitor (MPP), and common lymphoid progenitor (CLP) compartments were comparable in two groups of recipient animals, while the myeloid progenitor (MP) compartment in Kras;D/+ recipients was significantly reduced compared to that in Kras recipients (Figure 3A). Consistent with the prior study of mapping Notch pathway activity in vivo (26), we found that the reduction of MP compartment mainly occurred in the common myeloid progenitor (CMP) and megakaryocyte-erythroid progenitor (MEP) compartments (Figure 3B). Consequently, in various hematopoietic tissues, percentages of donor-derived T cells and myeloid lineage cells in Kras;D/+ recipients were significantly decreased, while donor-derived B cells were comparable between these two groups of recipients (Figure 3C). We observed similar reduction of donor-derived T cell and myeloid compartments in Kras;P−/− recipients as well (Figure S8). In the subsequent mechanistic studies, we primarily focused on D/+ mice due to its higher breeding efficiency.
Figure 3. DNMAML expression in Kras hematopoietic system reduces myeloid compartment.
Lethally irradiated mice (CD45.1+) were transplanted with 2 ×106 splenocytes (CD45.2+) from KrasLSL G12D/+;Mx1-Cre (Kras) or KrasLSL G12D/+;Rosa26LSL DNMAML-GFP/+;Mx1-Cre (Kras; D/+) mice along with 2.5×105 competitor cells (CD45.1+). Three weeks after transplantation, Cre expression was induced using pI-pC injections as described in Methods. Recipients transplanted with Kras or Kras; D/+ cells were sacrificed 3 weeks after pI-pC injections. Donor-derived cells are defined as CD45.2+ cells in Kras recipients and CD45.2+ GFP+ cells in Kras; D/+ recipients. (A) Quantification of donor-derived hematopoietic stem cells (HSCs), multi-potential progenitors (MPPs), myeloid progenitors (MPs), and common lymphoid progenitors (CLPs) in the bone marrow (BM) and spleen (SP) of recipients. (B) Quantification of donor-derived common myeloid progenitors (CMPs), granulocyte-macrophage progenitors (GMPs), and megakaryocyte-erythroid progenitors (MEPs) in the bone marrow of recipients. (C) Quantification of donor-derived differentiated cells in BM, SP, and peripheral blood (PB). Data are presented as mean ± SD. * P<0.05, ** P<0.01; *** P<0.001.
DNMAML expression upregulates Dusp1 expression and blocks GM-CSF-stimulated ERK hyperactivation in Kras myeloid progenitors
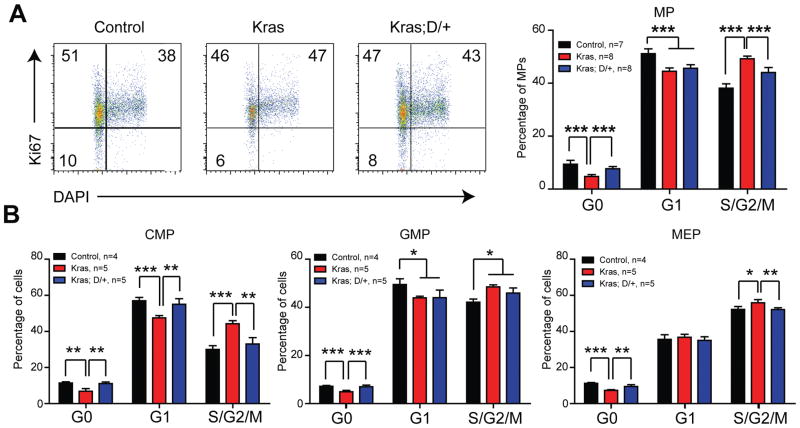
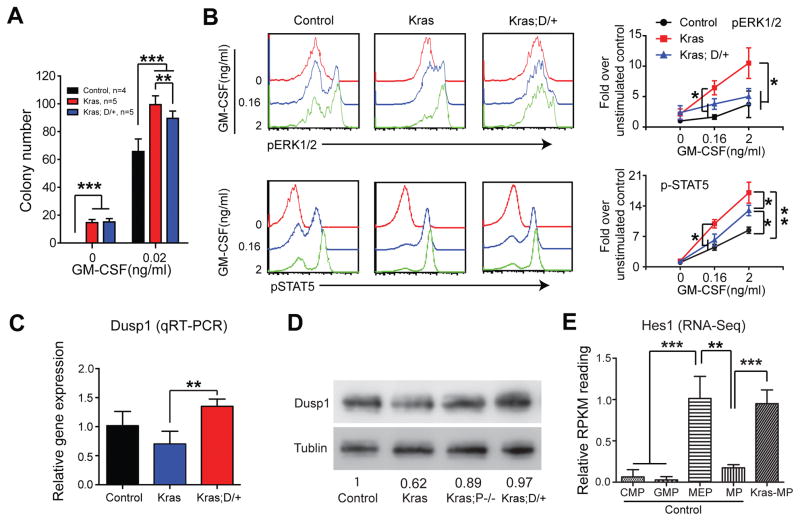
Because expansion of Kras myeloid compartment is driven by hyperproliferation and hyperactivation of GM-CSF-evoked ERK1/2 in MP cells (21), we subsequently examined whether reduced myeloid compartment in Kras;D/+ recipients associates with reduced cell proliferation and/or reduced ERK1/2 activation. Consistent with our previous report (23), cell cycle analysis showed that Kras MPs were significantly hyperpoliferative than control MPs, while this hyperprolferation phenotype was significantly reduced in Kras;D/+ MPs (Figure 4A). Further fractionation of MP compartment revealed that DNMAML expression mainly reduced cell proliferation in CMPs and MEPs (Figure 4B). Consistent with our cell cycle analysis, Kras;D/+ cells formed significantly less colonies than Kras cells in the presence of 0.02 ng/ml of GM-CSF (Figure 5A).
Figure 4. DNMAML expression reduces hyperproliferation of Kras myeloid progenitors.
Lethally irradiated mice (CD45.1+) were transplanted with 2 ×106 splenocytes (CD45.2+) from KrasLSL G12D/+;Mx1-Cre (Kras) or KrasLSL G12D/+;Rosa26LSL DNMAML-GFP/+;Mx1-Cre (Kras; D/+) mice along with 2.5×105 competitor cells (CD45.1+). The control group was transplanted with 1×106 bone marrow cells (CD45.2+) along with 2.5×105 competitor cells (CD45.1+). Three weeks after transplantation, Cre expression was induced using pI-pC injections as described in Methods. Recipients transplanted with control, Kras or Kras; D/+ cells were sacrificed 4–5 weeks after pI-pC injections. Donor-derived cells are defined as CD45.2+ cells in control and Kras recipients or CD45.2+ GFP+ cells in Kras; D/+ recipients. Cell cycle analysis of bone marrow donor-derived myeloid progenitors (MPs) (A), common myeloid progenitors (CMPs), granulocyte-macrophage progenitors (GMPs), and megakaryocyte-erythroid progenitors (MEPs) (B) using Ki67 and DAPI.
Figure 5. DNMAML expression leads to Dusp1 upregulation and downregulation of GM-CSF-stimulated ERK activation in Kras myeloid progenitors.
Lethally irradiated mice (CD45.1+) were transplanted with 2 ×106 splenocytes (CD45.2+) from KrasLSL G12D/+;Mx1-Cre (Kras) or KrasLSL G12D/+;Rosa26LSL DNMAML-GFP/+;Mx1-Cre (Kras; D/+) mice along with 2.5×105 competitor cells (CD45.1+). The control group was transplanted with 1×106 bone marrow cells (CD45.2+) along with 2.5×105 competitor cells (CD45.1+). Three weeks after transplantation, Cre expression was induced using pI-pC injections as described in Methods. Recipients transplanted with control, Kras or Kras; D/+ cells were sacrificed 4–5 weeks after pI-pC injections. Donor-derived cells are defined as CD45.2+ cells in control and Kras recipients or CD45.2+ GFP+ cells in Kras; D/+ recipients. (A) 5X104 donor-derived bone marrow cells from recipients were plated in duplicate in semi-solid medium with or without GM-CSF. (B) Donor-derived whole bone marrow cells were sorted using flow cytometry and serum- and cytokine-starved for 2 hours at 37°C. Cells were then stimulated with different concentrations of mGM-CSF for 10 minutes at 37°C. Levels of p-ERK1/2 and pSTAT5 were measured using phospho-flow cytometry. Lin−/low c-Kit+ cells, which are enriched for myeloid progenitors, were gated for analysis. (C, D) Dusp1 expression was quantified in donor-derived Lin− bone marrow cells using qRT-PCR (C) or Western blot (D). (E) Quantification of Hes1 expression in different populations of progenitor cells using RNA-Seq. Data are presented as mean ± SD. * P<0.05, ** P<0.01; *** P<0.001.
We also investigated GM-CSF-stimulated ERK1/2 activation in Lin−/low c-Kit+ cells (enriched for MPs). Consistent with previous reports (21, 27), the ERK1/2 pathway was significantly hyperactivated in Kras cells upon GM-CSF stimulation but was restored to the level comparable to control cells in Kras;D/+ cells (Figure 5B). Concomitantly, STAT5 activation was significantly reduced in Kras;D/+ cells compared to that in Kras cells (Figure 5B). We further validated that GM-CSF-stimulated ERK activation was restored in Kras;P−/− cells (Figure S9). These results suggest that reduced myeloid compartment in recipients of Kras;D/+ cells could result from decreased cell proliferation and inhibition of ERK1/2 hyperactivation.
To further understand the molecular mechanism(s) underlying the inhibition of ERK hyperactivation in Kras;D/+ and Kras;P−/− cells, we performed RNA-Seq analysis using donor-derived MPs isolated from Kras and Kras;D/+ recipients as well as MPs from control animals. We reasoned that this signaling phenotype may be caused by upregulation of negative regulator(s) of the Ras/MEK/ERK pathway. Indeed, we found that Dusp1, a dual phosphatase that inactivates ERK1/2, was one of the top genes upregulated in Kras;D/+ vs Kras MPs. This result was further validated in donor-derived Lin− cells using qRT-PCR (Figure 5C) and Western blot (Figure 5D).
Hes1 has been reported to mediate the transcriptional repression of Dusp1 in a Kras-driven lung cancer model (28). Therefore, we examined Hes1 expression in our multiple data sets. Consistent with the previous study of in vivo Notch activity (26), our RNA-Seq data detected low level of Hes1 expression in control MEPs but not in control CMPs and GMPs (Figure 5E). Hes1 expression in control MEPs was further validated using qRT-PCR (Figure S10). Not surprisingly, Hes1 expression was marginally detectable in control MPs (Figure 5E). In contrast, Hes1 was expressed in Kras MPs at a level significantly higher than that in control MPs (Figure 5E) but downregulated in Kras;D/+ MPs (Figure S11A), indicating that canonical Notch signaling is activated in Kras cells but downregulated by DNMAML expression. Consistent with this idea, we found that Hes1 expression is significantly upregulated in NrasG12D; p53−/− AML-MPs (29). Our data suggest that DNMAML expression results in upregulation of Dusp1 and inhibition of ERK signaling, likely through a Hes1-dependent mechanism.
DNMAML expression alters the transcriptional levels of Notch target genes
To determine whether DNMAML perturbs the expression levels of Notch target genes in Kras cells, we first examined the expression of “Notch-Targets” in Kras and Kras;D/+ MPs. “Notch-Targets” are comprised of genes previously reported to be transcriptional targets of NOTCH1 (30) and are enriched in acute promyelocytic leukemia (APL) cells compared to normal promyelocytes (31). This gene signature demonstrated distinct expression patterns in Kras vs Kras;D/+ cells (Figure S11A). Next, by using previously published Rbpj chromatin immunoprecipitation sequencing data (32) that were collected from Notch-dependent mouse T–ALL cells, we identified 1,594 genes with significant Rbpj binding peaks in their promoter regions. This gene signature is also enriched in Kras MPs compared to Kras;D/+ MPs (Figure S11B). These results provide strong bioinformatics evidence that DNMAML regulates Notch target gene expression in Kras cells.
Loss of canonical Notch signaling targets oxidative phosphorylation in Kras myeloid progenitor and precursor cells
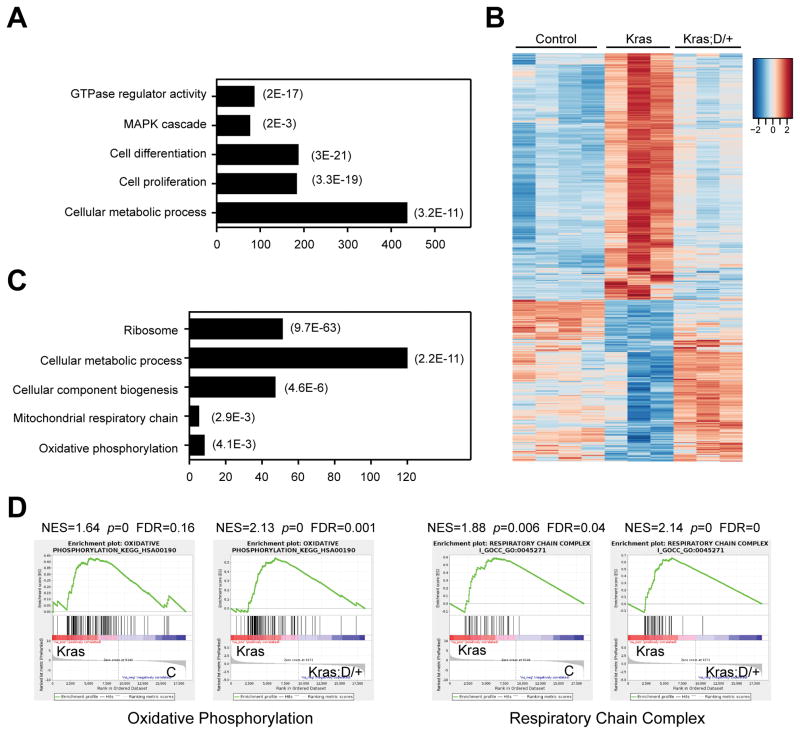
We further analyzed the RNA-Seq data obtained from control, Kras, and Kras;D/+ MPs to investigate molecular mechanisms underlying MPN inhibition in Kras; D/+ mice. Compared to control MPs, 1,820 genes were significantly up- or down-regulated in Kras MPs (fold change > 2 and FDR<0.05). Not surprisingly, this aberrant transcriptome was enriched for genes involved in small GTPase activity, ERK1/2 signaling cascade, and cell proliferation and differentiation (Figure 6A). In addition, RNA-Seq analysis identified a significant enrichment of genes regulating cell metabolism. Interestingly, the aberrant expression levels of ~430 genes in Kras MPs were restored or partially restored to control levels in Kras;D/+ MPs (Figure 6B). These genes are predominantly involved in cell metabolism and mitochondria biogenesis/functions (Figure 6C). Consistent with our observation, genome-wide gene set enrichment analysis revealed that genes involved in oxidative phosphorylation (OXPHO) and mitochondria respiration were significantly upregulated in Kras MPs, while their aberrant expression was restored to control levels upon expression of DNMAML (Figure 6D).
Figure 6. DNMAML expression restores transcription levels of metabolic genes in Kras myeloid progenitors.
Lethally irradiated mice (CD45.1+) were transplanted with 2 ×106 splenocytes (CD45.2+) from KrasLSL G12D/+;Mx1-Cre (Kras) or KrasLSL G12D/+;Rosa26LSL DNMAML-GFP/+;Mx1-Cre (Kras; D/+) mice along with 2.5×105 competitor cells (CD45.1+). Three weeks after transplantation, Cre expression was induced using pI-pC injections as described in Methods. Recipients transplanted with Kras or Kras; D/+ cells were sacrificed 3 weeks after pI-pC injections. Donor-derived myeloid progenitors (MPs) and MPs from control mice (Ctrl) were sorted for RNA-Seq analysis. Donor-derived cells are defined as CD45.2+ cells in control and Kras recipients or CD45.2+ GFP+ cells in Kras; D/+ recipients. (A) Gene Ontology (GO) analysis of differentially expressed genes in Kras MPs using DAVID bioinformatics program. The representative biological processes are shown with numbers of genes in each category (represented by the bar lengths) and corresponding P values. (B) Heatmap analysis of gene expression signature perturbed in Kras MPs but restored in Kras;D/+ MPs. (C) Go analysis of genes that were perturbed in Kras MPs but restored in Kras;D/+ MPs. The representative biological processes are shown with corresponding P values. (D) Gene Set Enrichment Analysis (GSEA) identified that oxidative phosphorylation and respiratory chain complex were upregulated in Kras MPs but restored to control levels in Kras;D/+ MPs. Data are presented as mean ± SD. * P<0.05, ** P<0.01; *** P<0.001.
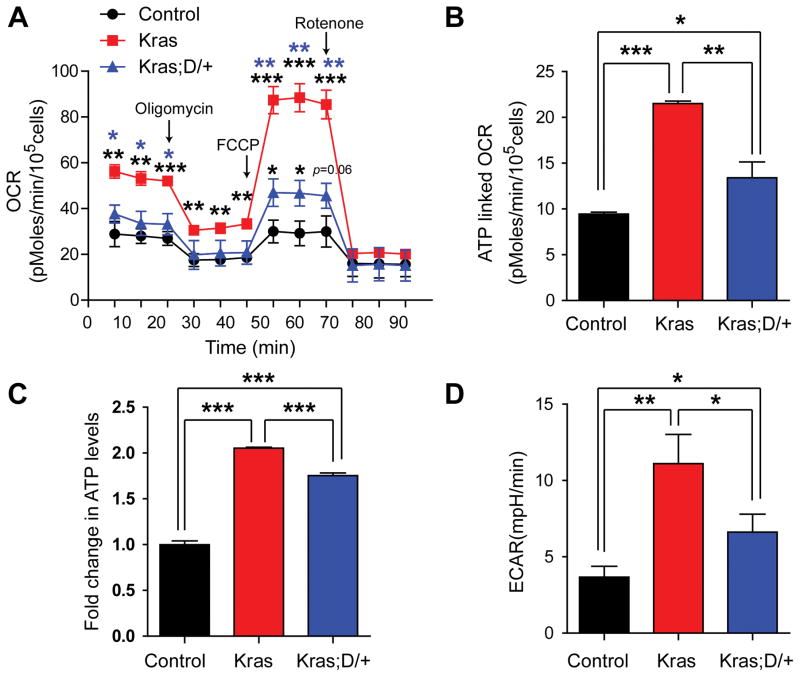
We next sought to functionally validate the impact of DNMAML expression on the mitochondrial metabolism. We analyzed mitochondrial aerobic metabolism in intact, viable CD45.2+ Lin− cells (donor-derived myeloid progenitors and precursors) using real-time measurement of oxygen consumption (Figure 6). Kras cells displayed much higher basal oxygen consumption and maximal oxidative capacity as compared with control cells. These elevated metabolic parameters were profoundly restored to approximate control levels in Kras;D/+ cells (Figure 7A). Using the ATP synthase inhibitor oligomycin, we determined that ATP-linked respiration was significantly increased in Kras cells and inhibition of canonical Notch signaling significantly lowered it in Kras;D/+ cells (Figure 7B). Consistent with this result, the steady state total cellular ATP concentration in Kras;D/+ cells was also significantly decreased compared with that of Kras cells (Figure 7C). Measurement of extracellular proton flux revealed that Kras cells had significantly increased extracellular acidification rates (ECAR) relative to control cells (Figure 7D), while the ECAR in Kras;D/+ cells was significantly reduced compared with Kras cells, suggesting that downregulation of Notch signaling alleviates enhanced glycolysis in Kras cells. Together, our data demonstrated that downregulation of Notch signaling inhibits both oxidative phosphorylation and glycolysis in Kras myeloid progenitor and precursor cells in a cell-autonomous manner.
Figure 7. Downregulating Notch signaling targets mitochondrial metabolism in Kras myeloid progenitor and precursor cells.
Lethally irradiated mice (CD45.1+) were transplanted with 2 ×106 splenocytes (CD45.2+) from KrasLSL G12D/+;Mx1-Cre (Kras) or KrasLSL G12D/+;Rosa26LSL DNMAML-GFP/+;Mx1-Cre (Kras; D/+) mice along with 2.5×105 competitor cells (CD45.1+). The control group was transplanted with with 1×106 bone marrow cells (CD45.2+) along with 2.5×105 competitor cells (CD45.1+). Three weeks after transplantation, Cre expression was induced using pI-pC injections as described in Methods. Recipients transplanted with control, Kras or Kras; D/+ cells were sacrificed 5 weeks after pI-pC injections. Donor-derived Lin− bone marrow cells were sorted using flow cytometry. Donor-derived cells are defined as CD45.2+ cells in control and Kras recipients or CD45.2+ GFP+ cells in Kras; D/+ recipients. (A) Oxygen consumption rates (OCR) were measured in the presence of the mitochondrial inhibitor (oligomycin, 1μM), the uncoupling agent (FCCP, 2.5 μM), and the respiratory chain inhibitor (rotenone, 1 μM). (B) Quantification of ATP-linked OCR, which is the calculated difference between the basal OCR level and the OCR level after oligomycin treatment. (C) Total cellular ATP concentrations were measured using the CellTiter Glo assay. (D) Quantification of extracellular acidification rates (ECAR). Data are presented as mean ± SD. * P<0.05, ** P<0.01; *** P<0.001.
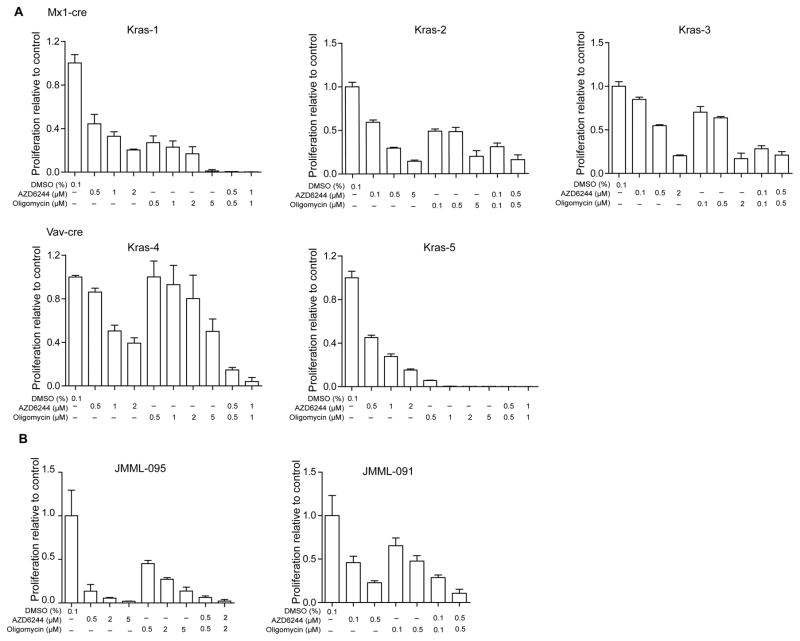
Combined AZD6244 and oligomycin treatment effectively inhibits the growth of human and mouse leukemia cells in vitro
To determine whether inhibition of ERK and/or mitochondria metabolism effectively controls Kras cell growth, we isolated bone marrow cells from moribund Kras mice (3 with Mx1-Cre and 2 with Vav-Cre) and cultured them in the absence or presence of AZD6244 (a MEK inhibitor (33, 34)) and/or oligomycin (a specific inhibitor of mitochondrial ATP synthase (35)) (Figure 8A). Although Kras cells demonstrated variable sensitivity to AZD6244 alone and oligomycin alone, they were consistently more sensitive to the combinatorial treatment. We also tested AZD6244 and oligomycin on human JMML samples, one with unknown mutation (JMML-095) and the other with a germline PTPN11 mutation (JMML-091) (Figure 8B). In both cases, combinatorial treatment of AZD6244 and oligomycin inhibited JMML cell growth more effectively than single drug alone.
Figure 8. Combined AZD6244 and oligomycin treatment effectively inhibits the growth of human and mouse leukemia cells in vitro.
Leukemia cells from moribund Kras G12D/+ mice (carrying Mx1-Cre or Vav-Cre) with advanced JMML-like phenotypes (n=5) (A) or from human JMML patients (n=2) (B) were cultured in triplicate in 96-well plates in the presence of vehicle or various concentrations of AZD6244 and/or oligomycin for 5 days (A) or 14 days (B). Cell number was quantified using the CellTiter-Glo assay. Data are presented as mean ± s.d.
Discussion
In this study, we show that Notch signaling is necessary for the initiation of oncogenic Kras-induced T-ALL; downregulating this pathway in Kras mice completely blocks T-ALL formation. Surprisingly, downregulation of Notch signaling in Kras hematopoietic cells also inhibits MPN in a cell autonomous manner. Our data suggest that inhibition of Notch signaling lowers cytokine-stimulated ERK1/2 hyperactivation and shifts abnormal metabolic state of Kras MPs close to that of control MPs, leading to reduced cell proliferation and consequently a reduction of myeloid compartment (Figure S12).
It was previously shown that GOF NOTCH1 mutations associated with human T-ALL patients are sufficient to drive T-ALL formation (14). In addition, they cooperate with oncogenic Ras to transform normal CD8+ T cells into leukemia initiating cells and accelerate oncogenic Ras-initiated T-ALL (12, 14). Therefore, 100% of oncogenic Ras-induced T-ALL carried GOF Notch1 mutations, including Type 1 deletions (100%) and PEST domain mutations (~70%) (12, 36). In this study, we further demonstrated that Notch signaling is essential for oncogenic Kras-initiated T-ALL. Downregulating Notch signaling completely blocked T-ALL development in Kras cells. In the recipients that eventually developed T-ALL, the leukemia cells were all derived from rare donor cells that expressed oncogenic Kras and did not downregulate Notch (Figure S1 and S5). Moreover, these leukemia cells contained Notch1 Type 1 deletions (Figure S1 and S5). Therefore, GOF Notch1 mutations are both necessary and sufficient for oncogenic Ras-induced T-ALL.
Our study identified an oncogenic function of Notch signaling in KrasG12D-induced MPN. This is consistent with an activated Notch gene signature in human APL (31) but in sharp contrast to previous studies reporting a tumor suppressor function of Notch signaling in CMML and other types of AML (15, 16), suggesting a highly dynamic, perhaps genetic context-dependent role of the Notch pathway in myeloid leukemogenesis. In support of this hypothesis, we and others did not detect loss of function mutations in Notch pathway genes in CMML patients with mutations in Ras pathway genes (37, 38). Nevertheless, our results are highly consistent with a recent study finding that γ–secreatase and RBPJ, two critical components of Notch signaling, are essential for the formation of KrasG12V-driven non-small cell lung carcinomas (28). Moreover, therapeutic effects of inhibiting Notch signaling were reported in treating Kras-driven lung cancers (28, 39).
Our results suggest multiple mechanisms underlying how inhibition of Notch signaling blocks KrasG12D-induced MPN. First, loss of canonical Notch pathway significantly downregulates cytokine-evoked ERK1/2 signaling in Kras MPs (Figure 5B and S9). As in KrasG12V-driven lung cancer (28), downregulating Notch-mediated inhibition of ERK activation might be attributed to derepression of Dusp1 (Figure 5C and 5D). Second, Kras enhances oxidative phosphorylation and glycolysis, while inhibition of canonical Notch signaling profoundly restores these aberrant metabolic changes (Figure 7). The involvement of Notch signaling in mitochondrial metabolism is quite surprising to us but indeed reflects a novel biological function of this pathway, which was just recently revealed in literature. Xu et al. reported that Notch reprograms mitochondrial metabolism for M1 macrophage activation (40), while Kishton et al. found that oncogenic Notch signaling in T-ALL activates AMPK to balance mitochondrial metabolism and glycolysis (41). Similarly, we demonstrated that downregulating Notch signaling reprograms the aberrant mitochondrial metabolism and glycolysis in Kras cells, which could contribute to reduced cell proliferation and MPN inhibition. Our observation is consistent with previous reports that myeloid leukemia cells often stay in an abnormal metabolic state and genetic/pharmacologic inhibition of oxidative phosphorylation leads to antileukemic activity (42, 43). In agreement with this conclusion, recent studies in Kras-driven solid tumors identified metabolic susceptibilities as potential therapeutic targets (44, 45). We believe that the two mechanisms proposed above are likely to cooperate and contribute to the in vivo phenotypes we observed in Kras;D/+ mice.
In summary, our results show that inhibition of Notch signaling effectively blocks oncogenic Kras-induced MPN in vivo in a cell-autonomous manner. We attribute this MPN inhibitory effect to inhibition of ERK signaling and reprogramming of mitochondrial metabolism. Combined inhibition of the MEK/ERK pathway and mitochondrial oxidative phosphorylation effectively inhibited the growth of human and mouse leukemia cells in vitro, providing a strong rationale to target both pathways in treating oncogenic Kras-driven malignancies.
Supplementary Material
Acknowledgments
We are grateful to Patrick Nyman and Dr. Paul Lambert for providing the Rosa26LSL DNMAML-GFP/+ mice and to Dr. Pamela Stanley for sharing the Pofut1fl/fl mice. We appreciate the critical comments from Drs. Emery Bresnick and Paul Lambert on the manuscript. We would like to thank the University of Wisconsin Carbone Comprehensive Cancer Center (UWCCC) for use of its Shared Services (Flow Cytometry Laboratory and Experimental Pathology Laboratory) to complete this research. This work was supported by the National Natural Science Foundation of China (NO.81600100) to G.K., Alexander von Humboldt Foundation (Alfred Toepfer Faculty Fellow) and NIH-MIRA grant R35GM124806 to X.Z., grants from American Cancer Society LIB-125064 and NIH HL103827 to L.Z., and R01 grants CA152108 and HL113066, and a Scholar Award from the Leukemia & Lymphoma Society to J.Z.. This work was also supported in part by NIH/NCI P30 CA014520--UW Comprehensive Cancer Center Support.
Footnotes
Mouse genotype abbreviations:
Control (Mx1-Cre)
Kras, spontaneously recombined KrasG12D/+ heterozygous (KrasLSL G12D/+; Mx1-Cre)
Kras; D/+, spontaneously recombined KrasG12D/+ heterozygous expressing DNMAML (KrasLSL G12D/+; Rosa26LSL DNMAML-GFP/+; Mx1-Cre)
Kras; P−/−, spontaneously recombined KrasG12D/+ heterozygous deficient for Pofut1 (KrasLSL G12D/+; Pofut1fl/fl; Mx1-Cre)
Conflict of Interest Disclosures:
We declare that no conflict of interest exists.
Authorship Contributions
Conception and design: G. Kong, X. You, and J. Zhang
Acquisition of data: G. Kong, X. You, Z. Wen, Y.-I Chang, C. Letson, J. F. Zhang, Y. Zhou, Y. Liu, A. Rajagopalan
Analysis and interpretation of data: G. Kong, J. F. Zhang, X. Zhang, E. A. Ranheim, A. Rajagopalan, E. Padron, W. S. Pear, L. Zhou, and J. Zhang
Writing, review, and/or revision of the manuscript: G. Kong, E. A. Ranheim, W. S. Pear, L. Zhou, and J. Zhang
Technical or material support: S. Qian, I. Hofmann-Zhang, M. Fleming, X. Zhong, W. S. Pear and L. Zhou
Study supervision: J. Zhang
References
- 1.Wharton KA, Johansen KM, Xu T, Artavanis-Tsakonas S. Nucleotide sequence from the neurogenic locus notch implies a gene product that shares homology with proteins containing EGF-like repeats. Cell. 1985 Dec;43(3 Pt 2):567–581. doi: 10.1016/0092-8674(85)90229-6. [DOI] [PubMed] [Google Scholar]
- 2.Kidd S, Kelley MR, Young MW. Sequence of the notch locus of Drosophila melanogaster: relationship of the encoded protein to mammalian clotting and growth factors. Mol Cell Biol. 1986 Sep;6(9):3094–3108. doi: 10.1128/mcb.6.9.3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ntziachristos P, Lim JS, Sage J, Aifantis I. From fly wings to targeted cancer therapies: a centennial for notch signaling. Cancer Cell. 2014 Mar 17;25(3):318–334. doi: 10.1016/j.ccr.2014.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moloney DJ, Panin VM, Johnston SH, Chen J, Shao L, Wilson R, et al. Fringe is a glycosyltransferase that modifies Notch. Nature. 2000 Jul 27;406(6794):369–375. doi: 10.1038/35019000. [DOI] [PubMed] [Google Scholar]
- 5.Wang Y, Shao L, Shi S, Harris RJ, Spellman MW, Stanley P, et al. Modification of epidermal growth factor-like repeats with O-fucose. Molecular cloning and expression of a novel GDP-fucose protein O-fucosyltransferase. J Biol Chem. 2001 Oct 26;276(43):40338–40345. doi: 10.1074/jbc.M107849200. [DOI] [PubMed] [Google Scholar]
- 6.Luo Y, Haltiwanger RS. O-fucosylation of notch occurs in the endoplasmic reticulum. J Biol Chem. 2005 Mar 25;280(12):11289–11294. doi: 10.1074/jbc.M414574200. [DOI] [PubMed] [Google Scholar]
- 7.Okajima T, Xu A, Lei L, Irvine KD. Chaperone activity of protein O-fucosyltransferase 1 promotes notch receptor folding. Science. 2005 Mar 11;307(5715):1599–1603. doi: 10.1126/science.1108995. [DOI] [PubMed] [Google Scholar]
- 8.Okajima T, Irvine KD. Regulation of notch signaling by o-linked fucose. Cell. 2002 Dec 13;111(6):893–904. doi: 10.1016/s0092-8674(02)01114-5. [DOI] [PubMed] [Google Scholar]
- 9.Maillard I, Koch U, Dumortier A, Shestova O, Xu L, Sai H, et al. Canonical notch signaling is dispensable for the maintenance of adult hematopoietic stem cells. Cell Stem Cell. 2008 Apr 10;2(4):356–366. doi: 10.1016/j.stem.2008.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999 Apr 30;284(5415):770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 11.Weng AP, Ferrando AA, Lee W, Morris JPt, Silverman LB, Sanchez-Irizarry C, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004 Oct 8;306(5694):269–271. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- 12.Kong G, Du J, Liu Y, Meline B, Chang YI, Ranheim EA, et al. Notch1 gene mutations target KRAS G12D-expressing CD8+ cells and contribute to their leukemogenic transformation. J Biol Chem. 2013 Jun 21;288(25):18219–18227. doi: 10.1074/jbc.M113.475376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ashworth TD, Pear WS, Chiang MY, Blacklow SC, Mastio J, Xu L, et al. Deletion-based mechanisms of Notch1 activation in T-ALL: key roles for RAG recombinase and a conserved internal translational start site in Notch1. Blood. 2010 Dec 16;116(25):5455–5464. doi: 10.1182/blood-2010-05-286328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chiang MY, Xu L, Shestova O, Histen G, L’Heureux S, Romany C, et al. Leukemia-associated NOTCH1 alleles are weak tumor initiators but accelerate K-ras-initiated leukemia. J Clin Invest. 2008 Sep;118(9):3181–3194. doi: 10.1172/JCI35090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kannan S, Sutphin RM, Hall MG, Golfman LS, Fang W, Nolo RM, et al. Notch activation inhibits AML growth and survival: a potential therapeutic approach. J Exp Med. 2013 Feb 11;210(2):321–337. doi: 10.1084/jem.20121527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lobry C, Ntziachristos P, Ndiaye-Lobry D, Oh P, Cimmino L, Zhu N, et al. Notch pathway activation targets AML-initiating cell homeostasis and differentiation. J Exp Med. 2013 Feb 11;210(2):301–319. doi: 10.1084/jem.20121484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klinakis A, Lobry C, Abdel-Wahab O, Oh P, Haeno H, Buonamici S, et al. A novel tumour-suppressor function for the Notch pathway in myeloid leukaemia. Nature. 2011 May 12;473(7346):230–233. doi: 10.1038/nature09999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yao D, Huang Y, Huang X, Wang W, Yan Q, Wei L, et al. Protein O-fucosyltransferase 1 (Pofut1) regulates lymphoid and myeloid homeostasis through modulation of Notch receptor ligand interactions. Blood. 2011 May 26;117(21):5652–5662. doi: 10.1182/blood-2010-12-326074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang J, Wang J, Liu Y, Sidik H, Young KH, Lodish HF, et al. Oncogenic Kras-induced leukemogeneis: hematopoietic stem cells as the initial target and lineage-specific progenitors as the potential targets for final leukemic transformation. Blood. 2009 Feb 5;113(6):1304–1314. doi: 10.1182/blood-2008-01-134262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shi S, Stanley P. Protein O-fucosyltransferase 1 is an essential component of Notch signaling pathways. Proc Natl Acad Sci U S A. 2003 Apr 29;100(9):5234–5239. doi: 10.1073/pnas.0831126100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Du J, Liu Y, Meline B, Kong G, Tan LX, Lo JC, et al. Loss of CD44 attenuates aberrant GM-CSF signaling in Kras G12D hematopoietic progenitor/precursor cells and prolongs the survival of diseased animals. Leukemia. 2013;27(3):754–757. doi: 10.1038/leu.2012.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chang YI, You X, Kong G, Ranheim EA, Wang J, Du J, et al. Loss of Dnmt3a and endogenous Kras cooperate to regulate hematopoietic stem and progenitor cell functions in leukemogenesis. Leukemia. 2015 Mar 24; doi: 10.1038/leu.2015.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kong G, Chang Y-I, Damnernsawad A, You X, Du J, Ranheim RA, et al. Loss of wild-type Kras promotes activation of all Ras isoforms in oncogenic Kras-induced leukemogenesis. Leukemia. 2016;30(7):1542–1551. doi: 10.1038/leu.2016.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clausen BE, Burkhardt C, Reith W, Renkawitz R, Forster I. Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Res. 1999 Aug;8(4):265–277. doi: 10.1023/a:1008942828960. [DOI] [PubMed] [Google Scholar]
- 25.Ye M, Iwasaki H, Laiosa CV, Stadtfeld M, Xie H, Heck S, et al. Hematopoietic stem cells expressing the myeloid lysozyme gene retain long-term, multilineage repopulation potential. Immunity. 2003 Nov;19(5):689–699. doi: 10.1016/s1074-7613(03)00299-1. [DOI] [PubMed] [Google Scholar]
- 26.Oh P, Lobry C, Gao J, Tikhonova A, Loizou E, Manent J, et al. In vivo mapping of notch pathway activity in normal and stress hematopoiesis. Cell Stem Cell. 2013 Aug 1;13(2):190–204. doi: 10.1016/j.stem.2013.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van Meter ME, Diaz-Flores E, Archard JA, Passegue E, Irish JM, Kotecha N, et al. K-RasG12D expression induces hyperproliferation and aberrant signaling in primary hematopoietic stem/progenitor cells. Blood. 2007 May 1;109(9):3945–3952. doi: 10.1182/blood-2006-09-047530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maraver A, Fernandez-Marcos PJ, Herranz D, Canamero M, Munoz-Martin M, Gomez-Lopez G, et al. Therapeutic effect of gamma-secretase inhibition in KrasG12V-driven non-small cell lung carcinoma by derepression of DUSP1 and inhibition of ERK. Cancer Cell. 2012 Aug 14;22(2):222–234. doi: 10.1016/j.ccr.2012.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang J, Kong G, Rajagopalan A, Lu L, Song J, Hussaini M, et al. p53−/− synergizes with enhanced NrasG12D signaling to transform megakaryocyte-erythroid progenitors in acute myeloid leukemia. Blood. 2017 Jan 19;129(3):358–370. doi: 10.1182/blood-2016-06-719237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Palomero T, Lim WK, Odom DT, Sulis ML, Real PJ, Margolin A, et al. NOTCH1 directly regulates c-MYC and activates a feed-forward-loop transcriptional network promoting leukemic cell growth. Proc Natl Acad Sci U S A. 2006 Nov 28;103(48):18261–18266. doi: 10.1073/pnas.0606108103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grieselhuber NR, Klco JM, Verdoni AM, Lamprecht T, Sarkaria SM, Wartman LD, et al. Notch signaling in acute promyelocytic leukemia. Leukemia. 2013 Jul;27(7):1548–1557. doi: 10.1038/leu.2013.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang H, Zou J, Zhao B, Johannsen E, Ashworth T, Wong H, et al. Genome-wide analysis reveals conserved and divergent features of Notch1/RBPJ binding in human and murine T-lymphoblastic leukemia cells. Proc Natl Acad Sci U S A. 2011 Sep 06;108(36):14908–14913. doi: 10.1073/pnas.1109023108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yeh TC, Marsh V, Bernat BA, Ballard J, Colwell H, Evans RJ, et al. Biological characterization of ARRY-142886 (AZD6244), a potent, highly selective mitogen-activated protein kinase kinase 1/2 inhibitor. Clin Cancer Res. 2007 Mar 1;13(5):1576–1583. doi: 10.1158/1078-0432.CCR-06-1150. [DOI] [PubMed] [Google Scholar]
- 34.Kong G, Wunderlich M, Yang D, Ranheim EA, Young KH, Wang J, et al. Combined MEK and JAK inhibition abrogates murine myeloproliferative neoplasm. J Clin Invest. 2014 Jun 2;124(6):2762–2773. doi: 10.1172/JCI74182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hitosugi T, Fan J, Chung TW, Lythgoe K, Wang X, Xie J, et al. Tyrosine phosphorylation of mitochondrial pyruvate dehydrogenase kinase 1 is important for cancer metabolism. Mol Cell. 2011 Dec 23;44(6):864–877. doi: 10.1016/j.molcel.2011.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kindler T, Cornejo MG, Scholl C, Liu J, Leeman DS, Haydu JE, et al. K-RasG12D-induced T-cell lymphoblastic lymphoma/leukemias harbor Notch1 mutations and are sensitive to gamma-secretase inhibitors. Blood. 2008 Oct 15;112(8):3373–3382. doi: 10.1182/blood-2008-03-147587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chang YI, Damnernsawad A, Allen LK, Yang D, Ranheim EA, Young KH, et al. Evaluation of allelic strength of human TET2 mutations and cooperation between Tet2 knockdown and oncogenic Nras mutation. Br J Haematol. 2014 Apr 3; doi: 10.1111/bjh.12871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Merlevede J, Droin N, Qin T, Meldi K, Yoshida K, Morabito M, et al. Mutation allele burden remains unchanged in chronic myelomonocytic leukaemia responding to hypomethylating agents. Nature communications. 2016;7:10767. doi: 10.1038/ncomms10767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ambrogio C, Gomez-Lopez G, Falcone M, Vidal A, Nadal E, Crosetto N, et al. Combined inhibition of DDR1 and Notch signaling is a therapeutic strategy for KRAS-driven lung adenocarcinoma. Nat Med. 2016 Mar;22(3):270–277. doi: 10.1038/nm.4041. [DOI] [PubMed] [Google Scholar]
- 40.Xu J, Chi F, Guo T, Punj V, Lee WN, French SW, et al. NOTCH reprograms mitochondrial metabolism for proinflammatory macrophage activation. J Clin Invest. 2015 Apr;125(4):1579–1590. doi: 10.1172/JCI76468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kishton RJ, Barnes CE, Nichols AG, Cohen S, Gerriets VA, Siska PJ, et al. AMPK Is Essential to Balance Glycolysis and Mitochondrial Metabolism to Control T-ALL Cell Stress and Survival. Cell metabolism. 2016 Apr 12;23(4):649–662. doi: 10.1016/j.cmet.2016.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lagadinou ED, Sach A, Callahan K, Rossi RM, Neering SJ, Minhajuddin M, et al. BCL-2 inhibition targets oxidative phosphorylation and selectively eradicates quiescent human leukemia stem cells. Cell Stem Cell. 2013 Mar 7;12(3):329–341. doi: 10.1016/j.stem.2012.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jacque N, Ronchetti AM, Larrue C, Meunier G, Birsen R, Willems L, et al. Targeting glutaminolysis has antileukemic activity in acute myeloid leukemia and synergizes with BCL-2 inhibition. Blood. 2015 Sep 10;126(11):1346–1356. doi: 10.1182/blood-2015-01-621870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu Y, Marks K, Cowley GS, Carretero J, Liu Q, Nieland TJ, et al. Metabolic and functional genomic studies identify deoxythymidylate kinase as a target in LKB1-mutant lung cancer. Cancer discovery. 2013 Aug;3(8):870–879. doi: 10.1158/2159-8290.CD-13-0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kerr EM, Gaude E, Turrell FK, Frezza C, Martins CP. Mutant Kras copy number defines metabolic reprogramming and therapeutic susceptibilities. Nature. 2016 Mar 3;531(7592):110–113. doi: 10.1038/nature16967. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.