Abstract
Purpose
To assess the contribution of rare variants in the genetic background toward variability of neurodevelopmental phenotypes in individuals with rare copy-number variants (CNVs) and gene-disruptive variants.
Methods
We analyzed quantitative clinical information, exome sequencing, and microarray data from 757 probands and 233 parents and siblings who carry disease-associated variants.
Results
The number of rare likely deleterious variants in functionally intolerant genes (“other hits”) correlated with expression of neurodevelopmental phenotypes in probands with 16p12.1 deletion (n=23, p=0.004) and in autism probands carrying gene-disruptive variants (n=184, p=0.03) compared with their carrier family members. Probands with 16p12.1 deletion and a strong family history presented more severe clinical features (p=0.04) and higher burden of other hits compared with those with mild/no family history (p=0.001). The number of other hits also correlated with severity of cognitive impairment in probands carrying pathogenic CNVs (n=53) or de novo pathogenic variants in disease genes (n=290), and negatively correlated with head size among 80 probands with 16p11.2 deletion. These co-occurring hits involved known disease-associated genes such as SETD5, AUTS2, and NRXN1, and were enriched for cellular and developmental processes.
Conclusion
Accurate genetic diagnosis of complex disorders will require complete evaluation of the genetic background even after a candidate disease-associated variant is identified.
Keywords: CNV, autism, 16p11.2 deletion, phenotypic variability, modifier
Introduction
Significant advances in high-throughput genomic sequencing technologies have helped to identify hundreds of genes as risk factors for neurodevelopmental and neuropsychiatric disorders, including autism, intellectual disability, schizophrenia, and epilepsy. For example, in 2002, only 2–3% of autism cases were explained by genetic factors, whereas current studies suggest that rare disruptive variants, including copy-number variants (CNVs) and single-nucleotide variants (SNVs), account for 10–30% of autism cases.1 Despite initial claims of association with a specific disorder or syndrome, several of these pathogenic variants show incomplete penetrance and variable expressivity.2–4 For example, the 16p11.2 BP4-BP5 deletion (OMIM 611913) was first described in children with autism, but further studies on other clinical and population cohorts demonstrated that this deletion is also associated with intellectual disability and developmental delay (ID/DD), obesity, epilepsy, cardiac disease, and scoliosis, and only about 24% of cases manifest an autism phenotype.5–7 Phenotypic variability is not restricted to multigenic CNVs but has also been reported for single genes with pathogenic variants, including DISC1, PTEN, SCN1A, CHD2, NRXN1, FOXP2, and GRIN2B.3 While some of these effects could be due to allelic heterogeneity, phenotypic variability among carriers of the same molecular lesion suggests a strong role for variants in the genetic background.8,9 For example, in a large family described by St. Clair and colleagues,10 carriers of a balanced translocation disrupting DISC1 manifested a wide range of neuropsychiatric features, including schizophrenia, bipolar disorder, and depression. This phenomenon was exemplified by our delineation of a 520-kbp deletion on chromosome 16p12.1 (OMIM 136570), which is associated with developmental delay and extensive phenotypic variability.11 Interestingly, in most cases, this deletion was inherited from a parent who also manifested mild neuropsychiatric features, and the severely affected children were more likely to carry another large (>500 kbp) rare CNV. We hypothesized that while each pathogenic primary variant sensitizes the genome to varying extents, additional rare variants in the genetic background modulate the ultimate clinical manifestation.
Recent studies have identified secondary disease-associated variants that explain atypical clinical presentations of individuals carrying a primary variant.12–14 While these studies have explained phenotypic variability on a case-by-case basis, the global effect of the genetic background toward phenotypic variability among individuals sharing the same pathogenic variant has not been assessed. In this study, we evaluated 757 probands and 233 family members carrying primary variants associated with neurodevelopmental disease (17 rare CNVs or pathogenic variants in 301 genes). A comparison of the genetic background between probands and parents or siblings showed that in the presence of the same primary variant, variability and severity of neurodevelopmental disease correlates with the number of other rare variants, suggesting a global role of the genetic background toward phenotypic heterogeneity.
Materials and methods
Cohorts analyzed
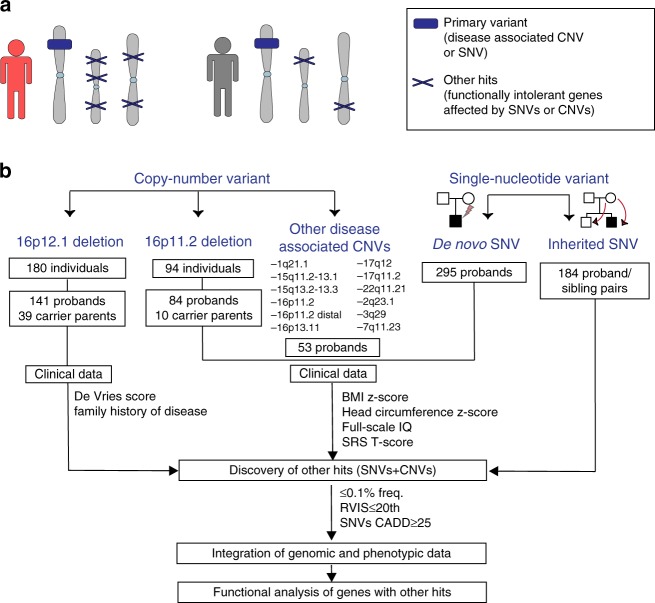
We analyzed clinical and genetic data in five subgroups of individuals carrying a disease-associated primary variant (Fig. 1): (1) 26 probands, 23 carrier parents and available family members carrying 16p12.1 deletion; (2) 53 autism probands from the Simons Simplex Collection (SSC) cohort who carry rare CNVs associated with syndromic and variably expressive genomic disorders;2 (3) 84 probands and available family members with 16p11.2 BP4-BP5 deletion from the Simons Variation in Individuals Project (SVIP) cohort; (4) 295 autism probands from the SSC cohort reported to carry severe de novo loss-of-function variants in neurodevelopmental genes;15,16 and (5) 184 probands and matched unaffected siblings from the SSC cohort who inherited the same rare (≤0.1% frequency) loss-of-function or likely damaging missense primary variants (CADD ≥25) in genes recurrently disrupted in neurodevelopmental disorders.17
Fig. 1.
Strategy for understanding the role of the genetic background in phenotypic variability of neurodevelopmental disease. (a) Schematic of primary variants and other hits used in this study. Disease-associated variants shared among different individuals were considered as “primary variants” (blue box), and rare likely deleterious single-nucleotide variants (SNVs) or copy-number variants (CNVs) affecting functionally intolerant genes were defined as “other hits” (blue Xs). Individuals with a higher burden of other hits (in red) exhibit a more severe clinical manifestation compared with those carrying the same primary variant but with a lower number of other hits (in gray). (b) Combined clinical and genomic analysis of 757 probands and 233 family members carrying primary disease-associated variants (16p12.1 deletion, 16p11.2 deletion, 16 rare CNVs, de novo pathogenic variants in autism simplex cases, and inherited pathogenic variants in disease-associated genes) was performed to understand the role of rare (≤0.1%) likely deleterious variants (SNVs with CADD ≥25 and CNVs) in functionally intolerant genes (Residual Variation Intolerance Score [RVIS] ≤20th percentile) toward the variable manifestation of neurodevelopmental disease. SRS Social Responsiveness Scale
Patient recruitment and clinical data ascertainment
We obtained clinical and/or genomic data from 141 children carrying the 16p12.1 deletion, as well as 39 deletion carrier and 30 noncarrier parents. Probands and parents recruited through direct contact provided consent according to the protocol reviewed and approved by The Pennsylvania State University Institutional Review Board (IRB). When individuals were not contacted directly, de-identified phenotypic and genomic data were used; as such, these cases were exempt from IRB review and conformed to the Helsinki Declaration.
We extracted clinical information from medical records or clinical questionnaires completed by different physicians from 180 carrier individuals and available family members (Supplementary Method 1.1). We used a modified de Vries scoring system for quantifying the number and severity of phenotypic abnormalities in affected children, which allows for a uniform assessment of developmental phenotypes from clinical records (Table S1).18 Family history information was used to bin families with the deletion into strong and mild or negative family history categories based on the severity of neurodevelopmental or psychiatric features (Fig. S1, Supplementary Method 1.1). Genomic and clinical data for SSC and SVIP cohort families were obtained from the Simons Foundation Autism Research Initiative (SFARI) following appropriate approvals (see Supplementary Method 1.1).
Burden analysis of rare variants in the genetic background (“other hits”)
To identify all coding variants modulating the presentation of the 16p12.1 deletion, we performed exome sequencing and single-nucleotide polymorphism (SNP) arrays on 105 individuals from 26 families as previously reported (Supplementary Method 1.2). Variant calls (SNVs and CNVs) from 716 individuals in the SSC were obtained from exome and SNP microarray studies,15,16,19 and variant call files (VCF) and SNP array data from 84 families with 16p11.2 BP4-BP5 deletion were obtained from the Simons Foundation. We defined “rare variants” or “other hits” as additional rare likely deleterious variants (includes ≤0.1% frequency CNV or SNV with CADD ≥25) (ref. 20) affecting a functionally intolerant gene (RVIS ≤20th percentile) co-occurring in an individual who already carries a disease-associated primary variant (Fig. 1a, Supplementary Method 1.2). The Residual Variation Intolerance Score (RVIS) has been shown to be a good predictor of gene intolerance to deleterious variants and has been widely used by multiple studies for the recapitulation of known and the discovery of novel disease-associated genes.16,21 The biological function of genes with other hits was analyzed using Ingenuity Pathway Analysis (IPA, Qiagen Bioinformatics), expression data derived from the GTEx consortium,22 and Gene Ontology (GO) enrichment analysis23 (Supplementary Method 1.3).
Results
Rare variants in the genetic background and disease expressivity in 16p12.1 deletion probands
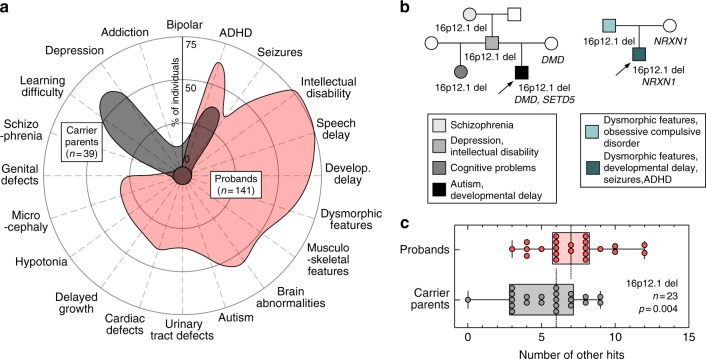
We assessed how rare likely deleterious variants in the genetic background can modulate phenotypes in concert with a primary variant by evaluating 757 affected probands and 233 family members carrying disease-associated variants (rare CNVs or pathogenic SNVs) (Fig. 1, see Methods). Using the 16p12.1 deletion as a paradigm for studying the genetic basis of variable expression of disease traits, we analyzed 180 individuals with the deletion and their noncarrier family members (Fig. 1b). The 16p12.1 deletion was inherited in 92.4% of cases, with a significant maternal bias (57.6% maternal [n=53] vs. 34.8% paternal [n=32], one-tailed binomial test p=0.01) (Table S2). In accordance with the female protective model described for neurodevelopmental disorders,2,24,25 we observed a significant gender bias among probands with the 16p12.1 deletion (67.9% males vs. 32.1% females, one-tailed binomial test p<0.0001). Detailed clinical analysis of 141 affected children with 16p12.1 deletion showed a wide heterogeneity of phenotypes, with a high prevalence of neurodevelopmental, craniofacial, and musculoskeletal features (>50%), and variable involvement of other organs and systems (Fig. 2a, Table S3). In contrast, 32 of 39 (82%) (61.5% females, 38.5% males) carrier parents showed mild cognitive, behavioral, and/or psychiatric features (Table S4), consistent with previous reports of cognitive impairment and increased risk for schizophrenia in carriers of the 16p12.1 deletion.26,27
Fig. 2.
Rare variants in the genetic background contribute to the phenotypic heterogeneity in 16p12.1 deletion. (a) Phenotypic spectrum of 16p12.1 deletion in probands (n=141, red) and carrier parents (n=39, gray). Probands exhibit a spectrum of severe developmental features compared with the mild cognitive and psychiatric features observed in carrier parents. Features represented were observed in ≥5% of probands or carrier parents. (b) Example of families with inherited 16p12.1 deletion. Family 1 (left) shows three generations carrying 16p12.1 deletion and multiple neurodevelopmental and psychiatric features, with the proband (P1C_01, indicated with arrow) carrying a de novo loss-of-function variant in SETD5 (p.Asp542Thrfs*3) and a stopgain variant in DMD gene (p.Trp3X) inherited from the mother without the 16p12.1 deletion (noncarrier). Family 2 (right) shows a proband (PC_11, indicated with arrow) with multiple congenital and neurodevelopmental features carrying 16p12.1 deletion and 2p16.3 deletion (encompassing NRXN1), the latter inherited from the noncarrier mother. (c) Analysis of rare (≤0.1%) likely deleterious variants (single-nucleotide variants with CADD ≥25) in genes intolerant to functional variation (RVIS ≤20th percentile) in proband–carrier parent pairs shows that probands present a higher burden of other hits compared with their carrier parents (n=23, Wilcoxon signed-rank test, p=0.004). ADHD attention deficit hyperactivity disorder
To identify variants within protein-coding regions that contribute to a more severe manifestation of the deletion in the affected children compared with their carrier parents, we performed exome sequencing and high-resolution SNP arrays in 26 families (n=105) with 16p12.1 deletion (23 inherited and 3 de novo cases, Table S5). We first evaluated whether the deletion could unmask recessive alleles, and found no rare pathogenic variants within the seven 16p12.1 genes on the nondeleted chromosome (Table S6). We next performed a case-by-case analysis of families for other hits elsewhere in the genome by focusing on rare CNVs (≤0.1%, ≥50 kbp), de novo or rare (ExAC frequency ≤0.1%) loss-of-function (LoF) variants, and rare likely damaging missense variants (Phred-like CADD ≥25) in disease-associated genes (see Methods, Tables S7–S9). For example, we identified two disease-associated variants in proband P1C_01, including a de novo LoF variant in the intellectual disability–associated gene SETD5 (OMIM 615761, c.1623_1624insAC, p.Asp542Thrfs*3) and a LoF variant in DMD (OMIM 310200, c.9G>A, p.Trp3X) transmitted from the non-16p12.1-deletion carrier mother (Fig. 2b). Similarly, a rare deletion at 2p16.3 encompassing NRXN1 (OMIM 614332), inherited from the noncarrier mother, was identified in proband PC_11 (Fig. 2b).
While private disease-associated variants may explain the variable and severe features in the affected children on a case-by-case basis, we lacked the statistical power to implicate individual genes or variants that modulate specific 16p12.1 deletion phenotypes. Therefore, to globally assess the genome-wide contribution of rare likely deleterious variants affecting functionally relevant genes, we performed an integrative analysis and quantified rare (frequency ≤0.1%), likely deleterious variants (CNVs or SNVs with CADD ≥25) within genes intolerant to functional variation (RVIS ≤20th percentile),16,20,21 hereafter referred to as the “burden of other hits.” Intrafamilial comparison showed that probands have an excess of other hits compared with their carrier parents (Wilcoxon signed-rank test, p=0.004, Fig. 2c and S2), with no change in the number of synonymous variants in all genes (p=0.29) or in RVIS ≤20th genes (p=0.36, Fig. S2E, F). Further, functional analysis of genes with other hits showed that probands presented an excess of genes that were preferentially expressed in the human brain (Wilcoxon signed-rank test, p=0.04, Fig. S3) and enriched for developmental pathways (Table S10) compared with carrier parents.
The severity and variability of neurodevelopmental features is contingent upon family history of neuropsychiatric disease.25 In fact, the cognitive and social outcomes in probands with de novo 16p11.2 BP4-BP5 deletion or 22q11.2 deletions have been reported to positively correlate with the cognitive and social skills of their parents.28,29 However, the genetic basis of such background effects has not been specifically studied. We assessed the role of other hits toward family-specific background effects and in the observed interfamilial variability of clinical features in probands with 16p12.1 deletion. We found that probands with a strong family history of neurodevelopmental and psychiatric disease presented a more severe and heterogeneous clinical presentation (Mann–Whitney one-tailed, p=0.04) and a higher burden of other hits (p=0.001) than those with mild or negative family history (Figs. 3a–c and S4A–C). Interestingly, probands with a strong family history also showed a higher difference in burden compared with their carrier parents than probands with a mild family history (p=0.003, Fig. 3d). While we did not observe a difference in burden between carrier parents based on family history (p=0.68, Fig. S4B), we found that noncarrier parents with a strong family history presented a significantly higher burden compared with those with a mild family history (p=0.01, Fig. 3e). Therefore, in families with a strong history of neurodevelopmental and psychiatric disease, a higher number of rare variants in the genetic background are more likely to be transmitted to the proband from the noncarrier parent, potentially contributing to a more severe manifestation of the disorder. These results suggest a potential role for rare variants in the genetic background in modulating intra- and interfamilial clinical variability observed in families with the 16p12.1 deletion.
Fig. 3.
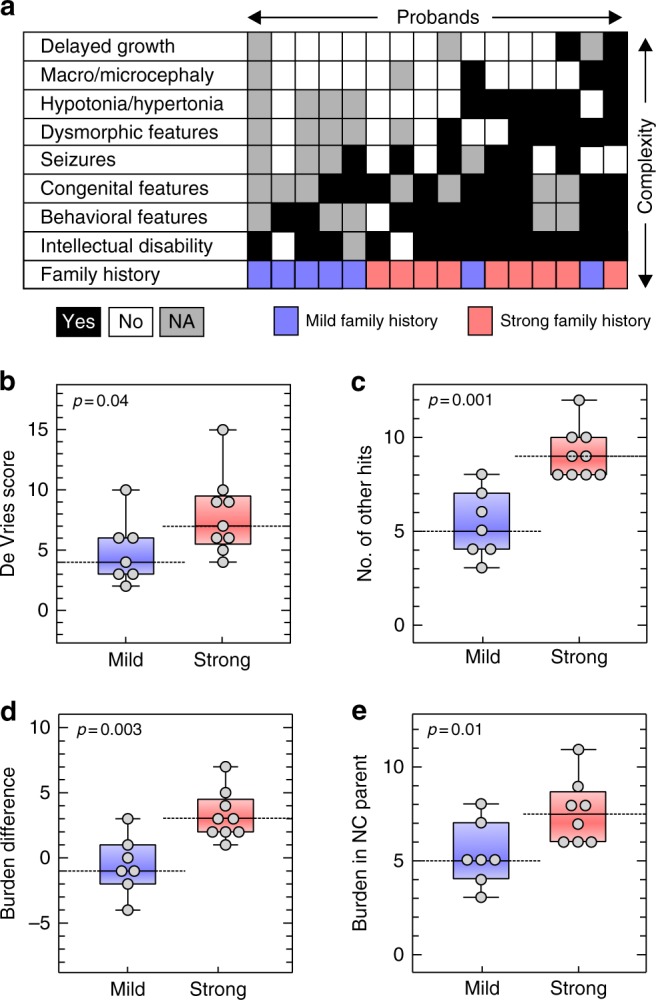
Strong family history of neurodevelopmental and psychiatric disease is associated with an excess of other hits and severe phenotypic outcome in 16p12.1 deletion probands. (a) Diagram showing phenotypic heterogeneity in 16 probands with 16p12.1 deletion (black=phenotype present, white=absent, gray=not assessed) and their family history of neurodevelopmental and psychiatric disease (red=strong, blue=mild/negative). Probands with strong family history (n=9) have (b) a more heterogeneous clinical manifestation (higher de Vries scores, one-tailed Mann–Whitney, p=0.04) and (c) a higher burden of other hits (one-tailed Mann–Whitney p=0.001) than those with mild or negative family history (n=7). (d) Probands with a strong family history exhibit a greater difference in burden of other hits compared with carrier parents (p=0.003). (e) Noncarrier parents from families with strong family history present a higher burden compared with those with mild/negative family history (one-tailed Mann–Whitney, p=0.01). NC Noncarrier
Burden of other rare variants correlates with quantitative phenotypes among individuals with 16p11.2 deletion and other rare pathogenic CNVs
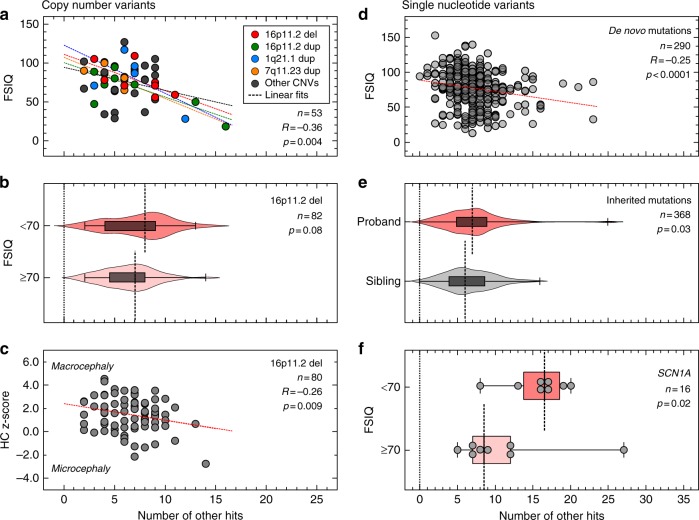
We next assessed whether the burden of other rare variants modulates quantitative phenotypes in carriers of other CNVs associated with neurodevelopmental phenotypes (Fig. 1b). In autism probands with disease-associated rare CNVs (n = 53, Table S11) from the Simons Simplex Cohort (SSC), we observed a modest but significant negative correlation (Pearson correlation, R = –0.36, p = 0.004) between the number of other hits and full-scale IQ (FSIQ) scores (Fig. 4a). This result held true when we separately analyzed individuals carrying 16p11.2 BP4-BP5 deletion (R = –0.68, p = 0.04), but did not show statistical significance for 16p11.2 BP4-BP5 duplication (R = –0.34, p = 0.17), 1q21.1 duplication (R = –0.36, p = 0.32), or 7q11.23 duplication (R = –0.74, p = 0.17), potentially due to low sample sizes (Figs. 4a and S5). Interestingly, probands with disease-associated CNVs and intellectual disability (FSIQ <70) showed a significant increase in the number of other hits compared with those without intellectual disability (FSIQ ≥70, one-tailed Mann–Whitney, p=0.02, Fig. S6).
Fig. 4.
Burden of other hits modulates quantitative phenotypes among probands with a first-hit copy-number variant (CNV) or single-nucleotide variant (SNV) associated with neurodevelopmental disease. (a) Negative correlation between the number of other hits and full-scale IQ (FSIQ) scores in individuals (n = 53) carrying 16 CNVs associated with neurodevelopmental disease (Pearson correlation, R = –0.36, p = 0.004). Probands with 16p11.2 deletion (red), 16p11.2 duplication (green), 1q21.1 duplication (blue) and 7q11.23 duplication (yellow) are highlighted, while gray circles represent probands with other rare CNVs. (b) Higher burden of other hits among probands with 16p11.2 deletion and FSIQ <70 (n = 17) compared with probands with FSIQ ≥70 (n = 65, one-tailed Mann–Whitney, p = 0.08). (c) Negative correlation between the number of other hits and head circumference z-scores (age ≥12 months, n = 80, Pearson correlation R = –0.26, p = 0.009) in probands with 16p11.2 deletion. (d) Autism probands with de novo disruptive variants and available FSIQ scores (n = 290) show a moderate negative correlation (Spearman correlation coefficient, R = –0.25, p < 0.0001) between the number of other hits and FSIQ scores. (e) Probands present an excess of other hits compared with their unaffected siblings (n = 184 pairs) carrying the same inherited pathogenic variants (loss-of-function or damaging missense CADD ≥25) in genes recurrently disrupted in neurodevelopmental disease (Wilcoxon signed-rank test, p = 0.03). (f) Enrichment of other hits among individuals with damaging variants in SCN1A (loss-of-function or missense CADD ≥25) and intellectual disability (one-tailed Mann–Whitney, p = 0.02) compared with those without intellectual disability
We further expanded our analysis by evaluating a larger set of 84 families with 16p11.2 BP4-BP5 deletion from the Simons Variation in Individuals Project (SVIP). We observed a higher median number of other hits in probands carrying the 16p11.2 deletion that had intellectual disability (FSIQ <70, median=8) compared with those with no intellectual disability (FSIQ ≥70, median=7 one-tailed Mann–Whitney, p = 0.08, Fig. 4b), without a difference in the number of synonymous variants between the two subgroups (median of 9957 synonymous changes for FSIQ <70 group versus 10,052 for the FSIQ ≥70 group, two-tailed Mann–Whitney, p = 0.51, Fig. S7A). Notably, we observed only a mild negative correlation between the burden of other hits and FSIQ, which did not attain statistical significance (Pearson correlation, R = –0.16, p = 0.08, Fig. S7B). Even though the sample size in the SSC cohort (n = 8) is small, we hypothesized that this marginal significance compared with 16p11.2 deletion probands from the SSC cohort (Figs. 4a and S5B) could be due to differences in clinical ascertainment. The SVIP cohort was selected for individuals carrying a 16p11.2 deletion who manifested a more heterogeneous set of phenotypes, while individuals from the SSC cohort were specifically ascertained for idiopathic autism.30 These differences in ascertainment were evident by different distributions of quantitative phenotypes, including body mass index (BMI), FSIQ, and SRS T-scores, in both populations (Fig. S8).
After adjusting for age to allow for full manifestation of the head phenotype, we identified a negative correlation between the number of other hits in SVIP probands with 16p11.2 deletion and their head circumference (HC) z-scores (age ≥12 months, n = 80, Pearson’s R = –0.26, p = 0.009, Fig. 4c).6,7 The observation that HC z-scores decline steadily (from >2 to <–2 scores) as other hits accumulate confirms that the deletion primarily leads to macrocephaly phenotypes, and suggests that rare variants in the genetic background could explain the incomplete penetrance of this phenotype among carriers of the deletion.7 We note that the burden of other hits did not correlate with Social Responsiveness Scale (SRS) T-scores or body mass index (BMI) z-scores, measures for autism and obesity, among probands with rare CNVs from the SSC cohort (Fig. S9A, B) or those with 16p11.2 deletion from the SVIP cohort (Fig. S9C, D), suggesting other mechanisms for the variability of these phenotypes.
Rare variants in the genetic background modulate disease manifestation among individuals with disruptive variants in disease-associated genes
We next analyzed 295 autism simplex cases from the SSC cohort with previously identified de novo gene-disruptive variants within 271 genes,15,16 and observed a moderately negative correlation between the burden of other hits and FSIQ scores (Spearman’s correlation, R = –0.25, p < 0.0001, Fig. 4d). Within this cohort, individuals with intellectual disability (FSIQ <70, n = 93) presented an enrichment of other hits compared with those without intellectual disability (FSIQ ≥70, n = 197) (one-tailed Mann–Whitney, p = 0.001, Fig. S10A). We did not observe a role for the burden of other hits in modulating BMI z-scores (Spearman’s R = –0.038, p = 0.27, Fig. S10B), although we did find a mild but significant positive correlation with SRS T-scores (Spearman’s R = 0.12, p = 0.02, Fig. S10C). Moreover, when probands were separated by gender, we observed a higher burden of other hits in females compared with males (one-tailed Mann–Whitney, p=0.02, Fig. S11). This supports the hypothesis that females require a higher contribution from the genetic background to reach the genetic threshold for pathogenesis of neurodevelopmental disease than males.25
While there is a consensus on the pathogenic role of de novo gene-disruptive variants in simplex families, the interpretation of inherited pathogenic variants within the same genes is challenging. To understand the role of the genetic background in the penetrance of inherited disruptive variants in disease-associated genes, we analyzed 184 pairs of autism probands and unaffected siblings who inherited the same pathogenic variant in genes recurrently disrupted in neurodevelopmental disorders (Table S12). We found a greater enrichment of other hits in probands compared with their unaffected siblings (Wilcoxon signed-rank test p = 0.03, Fig. 4e), suggesting that rare variants likely contribute to increased penetrance of neurodevelopmental phenotypes in children with inherited pathogenic single-gene variants. When we analyzed probands carrying pathogenic variants in specific neurodevelopmental genes, we found that the severity of cognitive deficits in individuals with damaging variants in SCN1A was concordant with an excess of other hits (probands with FSIQ <70, n = 8, median=16.5, versus those with FSIQ ≥70, n = 8, median=8.5, one-tailed Mann–Whitney, p = 0.02) (Fig. 4f). This observation could also explain the diversity of other phenotypes co-occurring with the disruption of the epilepsy-associated SCN1A gene, such as motor delay and autism.31
Other hits involve disease-associated genes and affect core cellular and developmental processes
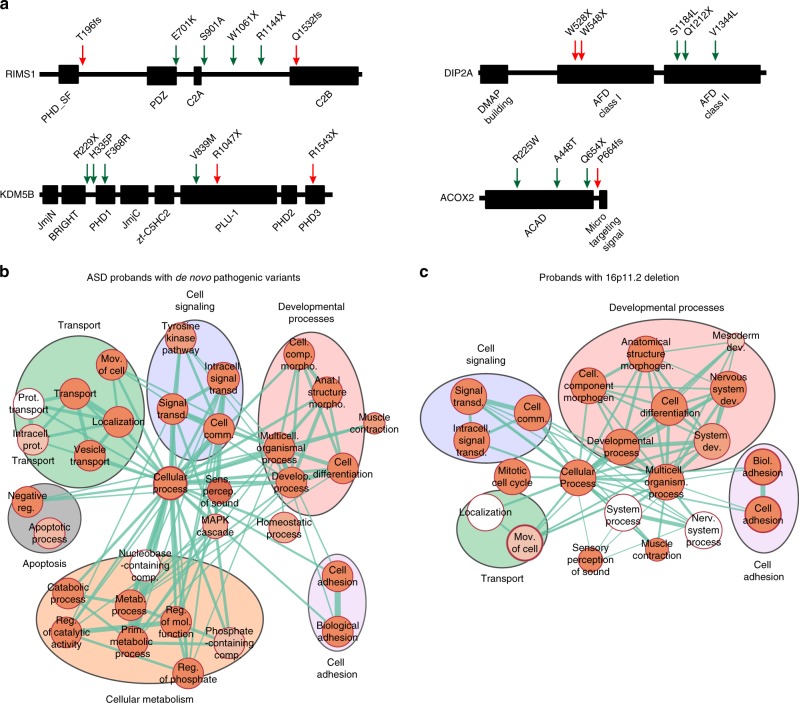
To understand how other rare variants could modulate phenotypes among probands with pathogenic first-hit variants, we explored the functionality of genes with other hits identified in all probands analyzed in our study. Overall, we identified 3197 other hits encompassing a diverse set of 1615 functionally intolerant genes. Of these, 40.9% (660/1615) were found to be extremely intolerant to loss-of-function variants (probability of loss-of-function intolerance [pLI] metric ≥0.9). These genes were also enriched for postsynaptic density genes, genes encoding FMRP targets, chromatin-associated genes, embryonically expressed genes, and essential genes compared with the whole genome (52% vs. 26%, Chi-squared test, p<0.0001).32,33 Interestingly, 44 of these genes with other hits (such as CNTNAP2, MBD5, SCN1A, CHD8, and AUTS2) have been recurrently associated with neurodevelopmental disorders17 (Fig. S12), 58 genes have been previously identified as a causative gene in simplex autism cases15,16 (Table S13), and 50 genes have been associated with skeletal, muscular, cardiovascular, or renal disorders, as classified in the human disease network (Table S14).34 We further assessed the location of other-hit variants within a subset of genes recurrently associated with disease, including RIMS1, DIP2A, KDM5B, and ACOX2, and found no specificity for the location of the other hits within the protein sequences compared with previously reported de novo pathogenic variants within these genes (Fig. 5a). In fact, in some cases, we observed stopgain variants that were more premature in the protein sequence than previously reported pathogenic variants, suggesting that the other hit can potentially exert as severe an effect as a primary variant, if these are true loss-of-function variants. The allelic diversity of other likely deleterious variants within these genes suggests that further functional analysis should be performed to understand their specific effects on modulating developmental phenotypes.
Fig. 5.
Rare variants in the genetic background affect core biological processes and disease-associated genes. (a) Examples of nonspecificity in the location of other hits in protein domains compared with first-hits. Location of variants in the protein sequences of RIMS1, DIP2A, KDM5B and ACOX2, genes with other hits (green arrows) and previously reported de novo disruptive variants in simplex autism cases (red arrows). Genes with other hits found in (b) autism spectrum disorder (ASD) probands carrying de novo disruptive variants (Simons Simplex Cohort; SSC) and (c) probands with the 16p11.2 deletion (Simons Variation in Individuals Project; SVIP) are enriched in core biological processes (FDR <0.05 with Bonferroni correction). Clusters of enriched Gene Ontology (GO) terms for “developmental processes,” “cell signaling,” “cell adhesion,” and “transport” functions are present among other hits found in each cohort. The size of each circle represents the number of genes annotated for each GO term; red shading of each circle represents the FDR for enrichment of each GO term among genes with other hits, with darker shades indicating a lower FDR. Line thickness represents the number of shared genes between pairs of GO terms. FDR values of the enriched GO terms are detailed in Tables S15-S16. FDR False discovery rate
To further understand the functional role of genes carrying rare variants, we performed Gene Ontology enrichment analysis of genes with other hits in probands with de novo pathogenic variants from the SSC and 16p11.2 deletion probands from the SVIP cohort. We found that genes carrying other hits in probands from both cohorts were enriched for core processes, including cell signaling, cell adhesion, and developmental processes (Fig. 5b, c, Tables S15, S16). Although some of these genes have been individually associated with a disease phenotype, further functional analyses are required to understand potential interactions between genes affected by both primary variants and other hits, their cumulative burden, and ultimately their potential contribution toward phenotypic variability.
Discussion
In this study, we have explored the contribution of rare variants in the genetic background toward the phenotypic heterogeneity of disease-associated variants. Recently, exome and genome sequencing studies have reported an increased burden of rare deleterious variants toward risk for neurodevelopmental disorders16,35,36 and combined parent-of-origin inherited risk effects for autism.16,32,37 Our analysis supports a complex model for neurodevelopmental disorders, and further postulates how rare variants in the genetic background modulate specific phenotypes in the presence of the same disease-causing variant.2,11–14 We propose that a higher burden of rare variants increases the likelihood of involving a modifier gene within disease-related pathways, as well as allows for a higher number of oligogenic combinations potentially modulating the phenotype associated with the first hit. Some primary variants more tolerant to changes in the genetic background, such as the 16p12.1 deletion, are transmitted through generations and only surpass the threshold for severe disease with the accumulation of several additional rare pathogenic variants. Other primary variants that are often de novo, such as the 16p11.2 deletion, push the genetic background closer to the threshold for severe manifestation and therefore require a lesser contribution from other hits. Similarly, highly penetrant syndromic CNVs such as Smith–Magenis syndrome and Sotos syndrome, which are mostly de novo and encompass genes more intolerant to functional variation compared with variably expressive CNVs (p=0.03, one-tailed Mann–Whitney, Table S17, Fig. S13), would push the genetic liability beyond the threshold for severe disease on their own.2,11 While additional variants may not be necessary for complete penetrance of these disorders, these variants can modify specific phenotype traits when present. For example, deleterious variants in histone modifier genes have been reported to contribute to heart defects in 22q11.2 deletion syndrome.38 This model would also apply for single-gene disorders, where other hits potentially explain discordant clinical features reported among affected carriers of the same molecular alteration, as described for Rett syndrome and individuals with pathogenic variants in the intellectual disability gene PACS1.39,40
Our observations that probands with a strong family history exhibit severe clinical manifestation and a higher burden of other hits also provide insights into the role of rare variants in the genetic background toward the reported correlation between parental profiles and clinical outcome in probands carrying rare CNVs.28,29 Moreover, the observed higher burden of other hits in the noncarrier parents in families with strong family history suggests assortative mating, and transmission of these hits to the proband potentially explains the increased clinical severity. Similarly, in 16p11.2 deletion, we observed that children who inherited the CNV presented lower FSIQ scores (n=8, median FSIQ=75) than probands with a de novo deletion (n=57, median FSIQ=85, one-tailed Mann–Whitney, p=0.006, Fig. S14A), in agreement with previous reports.6 This was also consistent with a nonsignificant excess of other hits among probands with an inherited 16p11.2 deletion (one-tailed Mann–Whitney, p=0.06, Fig. S14B) compared with probands with a de novo deletion. These results highlight the importance of eliciting family history of psychiatric and neurodevelopmental disease for more accurate diagnostic assessment of the affected children.
Overall, our results suggest a multidimensional effect of rare variants in the genetic background toward clinical features, and their contribution to specific phenotypic domains depends on the extent to which the primary variant sensitizes an individual toward a specific phenotypic trajectory. An important observation from our study is that a large number of disease-associated variants deemed to be solely causative for the disorder are in fact accompanied by a substantial amount of rare genetic variation. Longitudinal and quantitative phenotyping across multiple developmental domains in all family members, along with genome sequencing studies in affected and seemingly asymptomatic individuals with a primary variant, are necessary for a more accurate understanding of these complex disorders. Therefore, it is critical that even after identifying a likely diagnostic pathogenic variant, further analysis of the genetic background must be performed to provide appropriate counseling and management.
Electronic supplementary material
Acknowledgements
This work was supported by NIH R01-GM121907, Brain and Behavior Foundation (NARSAD 22535), SFARI Pilot Grant (SFARI 399894, S.G.) and resources from the Huck Institutes of the Life Sciences to S.G. L.P. was supported by Fulbright Commission Uruguay–ANII and the Huck Institutes of the Life Sciences. M.J. was supported by NIH T32-GM102057. C.R., L.C., O.G., and E.A. were supported by the Italian Ministry of Health and “5 per mille” funding. K.M. is a Jacobs Foundation Research Fellow. A.R. is supported by the Swiss National Science Foundation (31003A_160203). Dedicated to the memory of Ethan Francis Schwartz, 1996–1998. We are grateful to all of the families in each cohort (16p12.1 deletion, SVIP and SSC) who participated in the study. We thank the SSC principal investigators (A. Beaudet, R. Bernier, J. Constantino, E. Cook, E. Fombonne, D. Geschwind, R. Goin-Kochel, E. Hanson, D. Grice, A. Klin, D. Ledbetter, C. Lord, C. Martin, D. Martin, R. Maxim, J. Miles, O. Ousley, K. Pelphrey, B. Peterson, J. Piggot, C. Saulnier, M. State, W. Stone, J. Sutcliffe, C. Walsh, Z. Warren, E. Wijsman) as well as the Simons VIP Consortium. We appreciate obtaining access to genomic and phenotypic data on SFARI Base. Approved researchers can obtain the SSC and SVIP population data sets described in this study by applying at https://www.base.sfari.org.
Disclosure
The authors declare no conflicts of interest.
Electronic supplementary material
The online version of this article (10.1038/s41436-018-0266-3) contains supplementary material, which is available to authorized users.
References
- 1.Vorstman JAS, Parr JR, Moreno-De-Luca D, Anney RJL, Nurnberger JI, Jr, Hallmayer JF. Autism genetics: opportunities and challenges for clinical translation. Nat Rev Genet. 2017;18:362–376. doi: 10.1038/nrg.2017.4. [DOI] [PubMed] [Google Scholar]
- 2.Girirajan S, Rosenfeld JA, Coe BP, et al. Phenotypic heterogeneity of genomic disorders and rare copy-number variants. New Engl J Med. 2012;367:1321–1331. doi: 10.1056/NEJMoa1200395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moreno-De-Luca A, Myers SM, Challman TD, Moreno-De-Luca D, Evans DW, Ledbetter DH. Developmental brain dysfunction: revival and expansion of old concepts based on new genetic evidence. Lancet Neurol. 2013;12:406–414. doi: 10.1016/S1474-4422(13)70011-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mannik K, Magi R, Mace A, et al. Copy number variations and cognitive phenotypes in unselected populations. JAMA. 2015;313:2044–2054. doi: 10.1001/jama.2015.4845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bijlsma E, Gijsbers A, Schuurs-Hoeijmakers J, et al. Extending the phenotype of recurrent rearrangements of 16p11. 2: deletions in mentally retarded patients without autism and in normal individuals. Eur J Med Genet. 2009;52:77–87. doi: 10.1016/j.ejmg.2009.03.006. [DOI] [PubMed] [Google Scholar]
- 6.D'Angelo D, Lebon S, Chen Q, et al. Defining the effect of the 16p11.2 duplication on cognition, behavior, and medical comorbidities. JAMA Psychiatry. 2016;73:20–30. doi: 10.1001/jamapsychiatry.2015.2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jacquemont S, Reymond A, Zufferey F, et al. Mirror extreme BMI phenotypes associated with gene dosage at the chromosome 16p11.2 locus. Nature. 2011;478:97–102. doi: 10.1038/nature10406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hammer MF, Ishii A, Johnstone L, et al. Rare variants of small effect size in neuronal excitability genes influence clinical outcome in Japanese cases of SCN1A truncation-positive Dravet syndrome. PLoS One. 2017;12:e0180485. doi: 10.1371/journal.pone.0180485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Todarello G, Feng N, Kolachana BS, et al. Incomplete penetrance of NRXN1 deletions in families with schizophrenia. Schizophr Res. 2014;155:1–7. doi: 10.1016/j.schres.2014.02.023. [DOI] [PubMed] [Google Scholar]
- 10.St Clair D, Blackwood D, Muir W, et al. Association within a family of a balanced autosomal translocation with major mental illness. Lancet. 1990;336:13–16. doi: 10.1016/0140-6736(90)91520-K. [DOI] [PubMed] [Google Scholar]
- 11.Girirajan S, Rosenfeld JA, Cooper GM, et al. A recurrent 16p12. 1 microdeletion supports a two-hit model for severe developmental delay. Nat Genet. 2010;42:203–209. doi: 10.1038/ng.534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karaca Ender, Posey Jennifer E, Coban Akdemir Zeynep, Pehlivan Davut, Harel Tamar, Jhangiani Shalini N, Bayram Yavuz, Song Xiaofei, Bahrambeigi Vahid, Yuregir Ozge Ozalp, Bozdogan Sevcan, Yesil Gozde, Isikay Sedat, Muzny Donna, Gibbs Richard A, Lupski James R. Phenotypic expansion illuminates multilocus pathogenic variation. Genetics in Medicine. 2018;20(12):1528–1537. doi: 10.1038/gim.2018.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Posey JE, Harel T, Liu P, et al. Resolution of disease phenotypes resulting from multilocus genomic variation. N Engl J Med. 2017;376:21–31. doi: 10.1056/NEJMoa1516767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schaaf CP, Sabo A, Sakai Y, et al. Oligogenic heterozygosity in individuals with high-functioning autism spectrum disorders. Hum Mol Genet. 2011;20:3366–3375. doi: 10.1093/hmg/ddr243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iossifov I, O’Roak BJ, Sanders SJ, et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature. 2014;515:216–221. doi: 10.1038/nature13908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krumm N, Turner TN, Baker C, et al. Excess of rare, inherited truncating mutations in autism. Nat Genet. 2015;47:582–588. doi: 10.1038/ng.3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gonzalez-Mantilla AJ, Moreno-De-Luca A, Ledbetter DH, Martin CL. A cross-disorder method to identify novel candidate genes for developmental brain disorders. JAMA Psychiatry. 2016;73:275–283. doi: 10.1001/jamapsychiatry.2015.2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De Vries B, White S, Knight S, et al. Clinical studies on submicroscopic subtelomeric rearrangements: a checklist. J Med Genet. 2001;38:145–150. doi: 10.1136/jmg.38.3.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sanders SJ, He X, Willsey AJ, et al. Insights into autism spectrum disorder genomic architecture and biology from 71 risk loci. Neuron. 2015;87:1215–1233. doi: 10.1016/j.neuron.2015.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kircher M, Witten DM, Jain P, O'Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Petrovski S, Wang Q, Heinzen EL, Allen AS, Goldstein DB. Genic intolerance to functional variation and the interpretation of personal genomes. PLoS Genet. 2013;9:e1003709. doi: 10.1371/journal.pgen.1003709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Consortium GT. The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mi H, Muruganujan A, Casagrande JT, Thomas PD. Large-scale gene function analysis with the PANTHER classification system. Nat Protoc. 2013;8:1551–1566. doi: 10.1038/nprot.2013.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jacquemont S, Coe BP, Hersch M, et al. A higher mutational burden in females supports a “female protective model” in neurodevelopmental disorders. Am J Human Genet. 2014;94:415–425. doi: 10.1016/j.ajhg.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Polyak A, Rosenfeld JA, Girirajan S. An assessment of sex bias in neurodevelopmental disorders. Genome Med. 2015;7:94. doi: 10.1186/s13073-015-0216-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rees E, Walters JT, Chambert KD, et al. CNV analysis in a large schizophrenia sample implicates deletions at 16p12. 1 and SLC1A1 and duplications at 1p36. 33 and CGNL1. Hum Mol Genet. 2014;23:1669–1676. doi: 10.1093/hmg/ddt540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stefansson H, Meyer-Lindenberg A, Steinberg S, et al. CNVs conferring risk of autism or schizophrenia affect cognition in controls. Nature. 2014;505:361–366. doi: 10.1038/nature12818. [DOI] [PubMed] [Google Scholar]
- 28.Moreno-De-Luca A, Evans DW, Boomer KB, et al. The role of parental cognitive, behavioral, and motor profiles in clinical variability in individuals with chromosome 16p11.2 deletions. JAMA Psychiatry. 2015;72:119–126. doi: 10.1001/jamapsychiatry.2014.2147. [DOI] [PubMed] [Google Scholar]
- 29.Klaassen P, Duijff S, Swanenburg de Veye H, et al. Explaining the variable penetrance of CNVs: parental intelligence modulates expression of intellectual impairment caused by the 22q11.2 deletion. Am J Med Genet B. 2016;171:790–796. doi: 10.1002/ajmg.b.32441. [DOI] [PubMed] [Google Scholar]
- 30.Simons Vip C. Simons Variation in Individuals Project (Simons VIP): a genetics-first approach to studying autism spectrum and related neurodevelopmental disorders. Neuron. 2012;73:1063–1067. doi: 10.1016/j.neuron.2012.02.014. [DOI] [PubMed] [Google Scholar]
- 31.Guerrini R, Cellini E, Mei D, et al. Variable epilepsy phenotypes associated with a familial intragenic deletion of the SCN1A gene. Epilepsia. 2010;51:2474–2477. doi: 10.1111/j.1528-1167.2010.02790.x. [DOI] [PubMed] [Google Scholar]
- 32.Iossifov I, Levy D, Allen J, et al. Low load for disruptive mutations in autism genes and their biased transmission. Proc Natl Acad Sci U S A. 2015;112:E5600–5607. doi: 10.1073/pnas.1516376112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lek M, Karczewski KJ, Minikel EV, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goh KI, Cusick ME, Valle D, Childs B, Vidal M, Barabási AL. The human disease network. Proc Natl Acad Sci U S A. 2007;104:8685–8690. doi: 10.1073/pnas.0701361104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Singh T, Walters JTR, Johnstone M, et al. The contribution of rare variants to risk of schizophrenia in individuals with and without intellectual disability. Nat Genet. 2017;49:1167–1173. doi: 10.1038/ng.3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Turner TN, Coe BP, Dickel DE, et al. Genomic patterns of de novo mutation in simplex autism. Cell. 2017;171:710–22 e712. doi: 10.1016/j.cell.2017.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brandler WM, Antaki D, Gujral M, et al. Paternally inherited cis-regulatory structural variants are associated with autism. Science. 2018;360:327–331. doi: 10.1126/science.aan2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guo T, Chung JH, Wang T, et al. Histone modifier genes alter conotruncal heart phenotypes in 22q11.2 deletion syndrome. Am J Hum Genet. 2015;97:869–877. doi: 10.1016/j.ajhg.2015.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grillo E, Rizzo CL, Bianciardi L, et al. Revealing the complexity of a monogenic disease: Rett syndrome exome sequencing. PLoS One. 2013;8:e56599. doi: 10.1371/journal.pone.0056599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schuurs-Hoeijmakers JH, Oh EC, Vissers LE, et al. Recurrent de novo mutations in PACS1 cause defective cranial-neural-crest migration and define a recognizable intellectual-disability syndrome. Am J Hum Genet. 2012;91:1122–1127. doi: 10.1016/j.ajhg.2012.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.