Abstract
Venom is a complex molecular phenotype that shows high levels of variation in expressed proteins between individuals within and between populations. However, the functional significance of this variation in terms of toxicity towards prey is largely unknown. Here, we assessed the relative toxicity of venom from individual pygmy rattlesnakes (Sistrurus miliarius) on brown anoles (Anolis sagrei) using a novel assay involving tests of fixed doses of venom from individual snakes on individual lizards. We found high levels of functional variation between individual venoms within populations with individual differences (nested within population) explaining 3.6 times more variation in toxicity than population differences. Our results suggest a previously unappreciated adaptive significance to within-population variation in venom. They argue that selective mechanisms that maintain variation within populations may be of equal or greater importance to divergent selection leading to local adaption between populations as evolutionary explanations of venom variation within species.
Keywords: venom, toxicity, individual variation, pygmy rattlesnakes, brown anoles, molecular adaptive phenotype
1. Introduction
Assessing the functional significance of phenotypic variation is a key step in inferring its adaptive significance [1]. Snake venom proteins are a complex molecular adaptation where realistic tests of performance (e.g. whole organism lethality studies) can be used to study the functional significance of variation [2]. Our understanding of venom evolution would benefit from functional analyses of the strikingly high level of variation observed both within and between species [3]. In general, this variation is argued to be adaptive, allowing snakes with different venoms to efficiently kill and digest preferred prey [4,5], although non-adaptive explanations have also been proposed (e.g. [6,7]).
At the intraspecific level, the repeated finding of population differentiation in venom composition has been argued to represent a local adaptation to local prey assemblages resulting from a divergent selection [8]. Evidence for this explanation comes from lethality studies that directly test for local adaptation using local versus foreign combinations of pooled venom samples on prey [9]. Less attention has been paid to assessing the functional significance (defined in terms of lethality towards prey) of within-population venom variation despite repeated examples showing that individual variation of major classes of snake venom proteins is of similar magnitude to or greater than that found at the population level [10]. Identifying high levels of functional variation within populations would argue that mechanisms that maintain phenotypic diversity, such as balancing selection (e.g. [11]), need to be incorporated into comprehensive evolutionary explanations for within-species venom variation in snakes.
Here we conduct the first direct assessment of the functional significance of individual venom toxicity for a small pit viper, the pygmy rattlesnake (Sistrurus miliarius), sampled from populations in central Florida. Pygmy rattlesnakes are generalist predators whose diet includes frogs, lizards, mice and centipedes [6]. Both individual- and population-level variations in venom composition have been documented for this snake ([12]; S. Smiley-Walters, unpublished data). Here, we use a novel approach for assessing the lethality of individual venoms towards a common lizard prey, the brown anole (Anolis sagrei). Our results suggest that individual-level venom variation in S. miliarius has a significant effect on prey and is a previously unrecognized source of functional variation.
2. Material and methods
(a). Study organisms
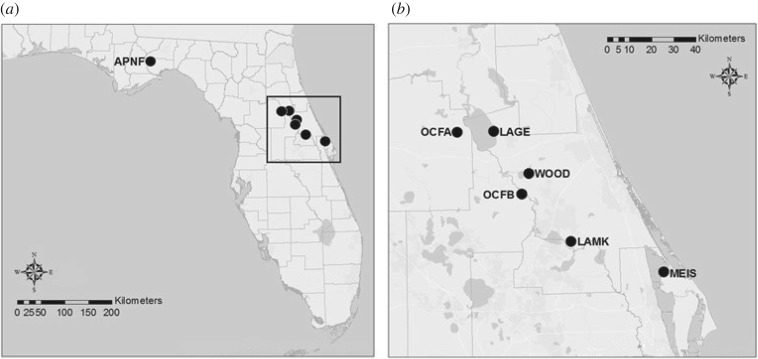
Venom was collected from adult pygmy rattlesnakes (n = 32) from seven different field sites in Florida: Lake Woodruff National Wildlife Refuge (WOOD, n = 12), Lake George Wildlife Management Area (LAGE, n = 10), Lake Monroe Conservation Area (LAMK, n = 5), Ocala National Forest (OCFA and OCFB, n = 3), Merritt Island National Wildlife Refuge (MEIS, n = 1), and Apalachicola National Forest (APNF, n = 1) (figure 1). All sites were within 100 km of one another in central Florida except for the single sample from northern Florida (APNF). At each site, we located adult snakes using visual searching and then extracted venom from them using standard protocols [13]. The protein concentrations were then standardized for subsequent median lethal dose (LD50) testing using a Bio-Rad Protein Assay kit (Bio-Rad Laboratories).
Figure 1.
Locations of snake collection sites in Florida. Each population is indicated by a circle. (a) Site APNF (n = 1) and central Florida region indicated by square. (b) Enlarged region of central Florida showing sites OCFA (n = 1), LAGE (n = 10), WOOD (n = 12), OCFB (n = 2), LAMK (n = 5) and MEIS (n = 1). The distance between LAGE and LAMK is approximately 60 km.
For toxicity trials, we collected lizards (Anolis sagrei) from the campus of Stetson University in DeLand, Florida. We used this invasive species as a model prey because it is readily obtained, is preyed upon by pygmy rattlesnakes (S.S.-W. and T.M.F., personal observation) and has been shown to have an LD50 for S. miliarius venom comparable to a congeneric native prey species, the green anole (Anolis carolinensis) [13].
(b). Toxicity trials
Because of the large number of animals needed for traditional LD50 trials with enough sensitivity to detect differences in function of individual venoms (e.g. [13]), we designed an alternative methodology to assess individual venom toxicity. We used a fully crossed design that tested all individual venoms across the same range of fixed doses on individual lizards, which allowed us to use total number of lizards killed as a measure of overall toxicity (see electronic supplementary material, figure S1). We based the range of doses for trials on the LD50 for A. sagrei estimated using a pooled venom sample [13]. To estimate the toxicity of a single snake, diluted venom was injected into 12 randomly assigned lizards using 12 mass-dependent doses ranging from 0.6 to 2.2 mg protein per kg lizard body weight (electronic supplementary material, figure S1). We used an injection volume of 20 µl for each lizard, which was delivered intraperitoneally to the ventral side. We recorded the status of each lizard (alive or dead) at 24 and 48 h after injection. We chose these time points because rattlesnakes typically strike and release prey and then search for prey for over 24 h after the initial envenomation [14]. In total, we injected 384 A. sagrei with a venom solution (32 individual venoms × 12 lizards each) and 37 lizards with a saline control solution.
(c). Data analyses
We tested the significance of individual snake on lizard mortality status using likelihood ratio tests (function anova or function lrtest of the epiDisplay package) that compared full and reduced logistic regression models created with the glm function in R (ver. 3.3.1). To determine if other parameters (dose, weight, snout–vent length (SVL)) influenced lizard mortality, we used mixed logistic regression models (glmer function in package lme4) with the significant effect of individual snake modelled as a random effect. Because the sample size of some populations was small, we repeated this analysis on data from only the two largest populations: WOOD (n = 12) and LAGE (n = 10), where increased sample size permitted a better understanding of within-population variation. Next, we used JMP version 10.0.0 to run a nested logistic model to determine if the individual snake was significant when nested within population of origin. Finally, to estimate the relative amount of variation in lizard mortality explained by individual versus population, we performed an ANOVA in JMP on the mortality data comparing the per cent variation explained by the population of origin to the per cent explained by individual snake nested within population of origin. As several populations were represented by only a few snakes, in this analysis, we included only snakes from the three most sampled populations (WOOD, LAGE and LAMK).
3. Results
Of the 384 lizards injected with venom, 129 (33.6%) were dead after 24 h and 135 (35.2%) were dead after 48 h. None of the saline treatment controls died in the 48 h following injection. We used total number of dead lizards at 48 h as the most inclusive measure of toxicity. We observed a highly significant relationship between the lowest dose at which an individual venom killed a lizard and the total number of lizards that venom killed at 48 h (r2 = 0.78, p < 0.001).
‘Individual snake’ (the model term signifying the effect of venom of individuals) had a significant impact on lizard mortality (χ2 = 155.07, p < 0.001) based on a likelihood ratio test comparing logistic regression models. When only the data from the two largest populations were analysed, individual snake effects remained significant in both the WOOD population (χ2 = 40.41, p < 0.001) and the LAGE population (χ2 = 55.09, p < 0.001). Similarly, ‘individual snake’ nested within population was a significant predictor of mortality in a logistic regression run in JMP (χ2 = 113.17, p < 0.001). Overall, individual venoms displayed a bi-modal distribution with respect to the number of lizards killed (figure 2). This suggests that there are two broad classes of venoms with respect to toxicity to lizards: high and low toxicity.
Figure 2.
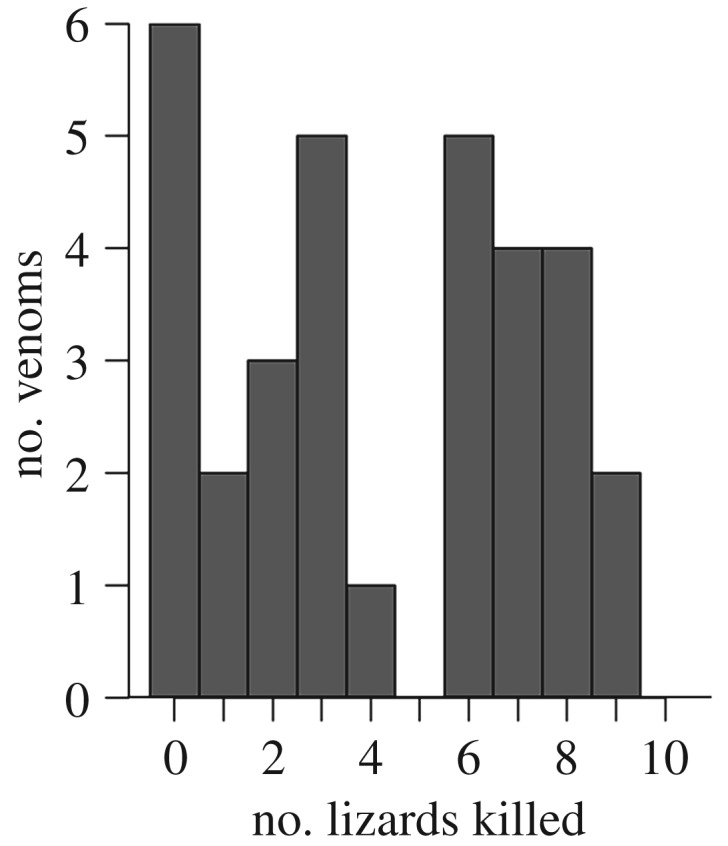
Number of lizards dead at 48 h post-injection (out of 12 tested) for each individual snake venom. Snake venoms show a bi-modal distribution with venoms killing either a low (0–3) or high (6–9) number of lizards.
Other factors also affected mortality. As expected, venom dose was a significant predictor of lizard mortality in our mixed logistic regression (χ2 = 71.38, p < 0.001). Snake SVL (χ2 = 0.34, p = 0.561) and snake weight (χ2 = 0.44, p = 0.508) were not significant predictors of lizard mortality. However, the snake population was a significant predictor (χ2 = 12.67, p = 0.049). Snake venoms from WOOD were the most lethal to lizards, followed by LAMK, LAGE and OCFB (electronic supplementary material, figure S1). Individual snake, nested within population, accounted for 3.6 times more variation in lizard mortality in an ANOVA than snake population of origin (table 1), suggesting much higher levels of functional variation at the level of individual compared with that of population.
Table 1.
Restricted maximum likelihood estimates for the relative amount of variance explained by population of origin of venom and individual nested within population. Dose was modelled as a fixed effect. All variables had a significant association with mortality (p < 0.05; see Results).
| random effect | variance ratio | variance component | 95% CI (lower, upper) | % total variance explained |
|---|---|---|---|---|
| population | 0.0842 | 0.0123 | −0.0184, 0.0431 | 6.07 |
| individual (population) | 0.3039 | 0.04461 | 0.0142, 0.0749 | 21.90 |
| residual | 0.1468 | 0.1273, 0.1711 | 72.04 | |
| total | 0.2036 | 0.1659, 0.2564 | 100.00 |
4. Discussion
Our main result is that the venoms of different individual rattlesnakes are functionally different in terms of lethality to a lizard prey and that the magnitude of this functional variation is almost four times greater than the amount that can be attributed to population differences in venom composition. Several factors need to be considered when interpreting our results. First, as with all dose-dependent assays of venom lethality, LD50 takes no account of the absolute amount of venom that can potentially be delivered by an individual [13]. Second, for practical reasons related to the number of test animals required, we only assayed a single prey species that previous work has shown is highly susceptible to S. miliarius venom [13]. As such, repeating our study on the other prey species, some of which show greater resistance to the venom of this snake [6,13], would be of great interest, especially to see if there are negative tradeoffs in toxicity towards different prey by individual snakes. Third, there are lethality ‘gaps’ in the toxicity spectrum for individual snakes (see electronic supplementary material, figure S1) that indicate that our measure of individual toxicity (number of lizards killed over the same venom concentration spectrum) is not entirely accurate. The fact that we found a highly significant relationship between the lowest dose at which an individual venom killed a lizard and the total number of lizards that venom killed gives us confidence that this measure captures a significant component of individual variation in venom lethality. We speculate that such gaps are caused by variation among individual lizards from the same population in factors such as physiological condition and/or genetically determined resistance which can impact the susceptibility of individuals to venom [15]. Finally, interpreting functional differences between individual venoms as adaptive requires evaluating the plasticity of individual venoms over time in response to environmental cues alone [3] as well as linking genetically based variation in specific venom proteins to differences in prey toxicity.
Our results have important implications for understanding the evolutionary mechanisms that maintain intraspecific variation in this complex molecular adaptation. They strongly suggest that functional differences in venom between individuals coexisting in the same environment and presumably encountering similar prey over their lifetimes are a significant and previously unappreciated component of adaptive variation in venom within species. Further, this component of functional diversity may equal or outweigh functional differences in venom due to local adaptation that have dominated adaptive explanations for intraspecific variation in venom (e.g. [9,10]). Our results argue that, in addition to diversifying selection leading to local adaptation, evolutionary mechanisms that could maintain phenotypic diversity in venom, such as balancing selection on loci and/or alleles that encode prey-specific toxins [11], need to be considered as potentially important drivers of venom diversity within species.
Supplementary Material
Acknowledgements
We thank K. Braeutigam and J. Butler for help monitoring lizards during toxicity assays. We also thank C. Lind, J. Serrao, M. Pilgrim, E. Royal, D. Rokyta, M. Margres, K. Wray, M. Holding, R. Denton and D. Salazar-Valenzuela for help with fieldwork.
Ethics
The study methodology was approved by Stetson University IACUC (protocol no. 2012TF103). Pigmy rattlesnake venoms were collected under a License to Possess Venomous Reptiles from the Florida Fish and Wildlife Conservation Commission.
Data accessibility
Data are available as electronic supplementary material.
Authors' contributions
All authors designed the research; S.S.-W. and T.M.F. collected the data and performed statistical analyses; all authors contributed to the writing of the original manuscript and revisions, approved the final version of the manuscript and agree to be held accountable for the content therein.
Competing interests
The authors have no competing interests.
Funding
S.S.-W. was supported by the Brown Visiting Teacher-Scholar Fellows Program at Stetson University.
References
- 1.Storz JF, Wheat CW. 2010. Integrating evolutionary and functional approaches to infer adaptation at specific loci. Evolution 64, 2489–2509. ( 10.1111/j.1558-5646.2010.01044.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chippaux JP, Williams V, White J. 1991. Snake venom variability: methods of study, results and interpretation. Toxicon 29, 1279–1303. ( 10.1016/0041-0101(91)90116-9) [DOI] [PubMed] [Google Scholar]
- 3.Casewell NR, Wüster W, Vonk FJ, Harrison RA, Fry BG. 2013. Complex cocktails: the evolutionary novelty of venoms. Trends Ecol. Evol. 28, 219–229. ( 10.1016/j.tree.2012.10.020) [DOI] [PubMed] [Google Scholar]
- 4.Mackessy SP. 2008. Venom composition in rattlesnakes: trends and biological significance. In The biology of rattlesnakes (eds Hayes WK, Beaman KR, Cardwell MD, Bush SP), pp. 495–510. Loma Linda, CA: Loma Linda University Press. [Google Scholar]
- 5.Gibbs HL, Mackessy SP. 2009. Functional basis of molecular adaptation: prey-specific toxic effects of venom from Sistrurus rattlesnakes. Toxicon 53, 672–679 ( 10.1016/j.toxicon.2009.01.034) [DOI] [PubMed] [Google Scholar]
- 6.Sasa M. 1999. Diet and snake venom evolution: can local selection alone explain intraspecifc venom variation? Toxicon 37, 249–252. ( 10.1016/S0041-0101(98)00121-4) [DOI] [PubMed] [Google Scholar]
- 7.Williams V, White J, Schwaner TD, Sparrow A. 1988. Variation in venom properties from isolated population of tiger snakes (Notechis ater niger, N. scutatus) in South Australia. Toxicon 26, 1067–1075. ( 10.1016/0041-0101(88)90205-X) [DOI] [PubMed] [Google Scholar]
- 8.Daltry JC, Wüster W, Thorpe RS. 1996. Diet and snake venom evolution. Nature 379, 537–540. ( 10.1038/379537a0) [DOI] [PubMed] [Google Scholar]
- 9.Smiley-Walters SA, Farrell TM, Gibbs HL. 2017. Evaluating local adaptation of a complex phenotype: reciprocal tests of pigmy rattlesnake venoms on treefrog prey. Oecologia 184, 739–748. ( 10.1007/s00442-017-3882-8) [DOI] [PubMed] [Google Scholar]
- 10.Sunagar K, et al. 2014. Intraspecific venom variation in the medically significant southern Pacific rattlesnake (Crotalus oreganus helleri): biodiscovery, clinical and evolutionary implications. J. Proteomics 99, 68–83. ( 10.1016/j.jprot.2014.01.013) [DOI] [PubMed] [Google Scholar]
- 11.Dowell NL, Giorgianni MW, Griffin S, Kassner S, Selegue VA, Sanchez JE, Carroll SB. 2018. Extremely divergent haplotypes in two toxin gene complexes encode alternative venom types within rattlesnake species. Curr. Biol. 28, 1016–1026. ( 10.1016/j.cub.2018.02.031) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gibbs HL, Sanz L, Calvete JJ. 2009. Snake population venomics: proteomics-based analyses of individual variation reveals gene regulation effects on venom protein expression in Sistrurus rattlesnakes. J. Mol. Evol. 68, 113–125. ( 10.1007/s00239-008-9186-1) [DOI] [PubMed] [Google Scholar]
- 13.Smiley-Walters SA, Farrell TM, Gibbs HL. 2018. The importance of species: pygmy rattlesnake venom toxicity differs between native prey and related non-native species. Toxicon 144, 42–47. ( 10.1016/j.toxicon.2018.01.022) [DOI] [PubMed] [Google Scholar]
- 14.Smith TL, Kardong KV, Lavín-Murcio PA. 2000. Persistence of trailing behavior: cues involved in poststrike behavior by the rattlesnake, Crotalus viridis oreganus. Behaviour 137, 691–703. ( 10.1163/156853900502295) [DOI] [Google Scholar]
- 15.Holding ML, Drabeck DH, Jansa SA, Gibbs HL. 2016. Venom resistance as a model for understanding the molecular basis of complex coevolutionary adaptations. Integr. Comp. Biol. 56, 1032–1043. ( 10.1093/icb/icw082) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are available as electronic supplementary material.