Abstract
Background
We aim to generate a line of “universal donor” human induced pluripotent stem cells (hiPSCs) that are nonimmunogenic and, therefore, can be used to derive cell products suitable for allogeneic transplantation.
Methods and Results
hiPSCs carrying knockout mutations for 2 key components (β2 microglobulin and class II major histocompatibility class transactivator) of major histocompatibility complexes I and II (ie, human leukocyte antigen [HLA] I/II knockout hiPSCs) were generated using the Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)/CRISPR associated protein 9 (Cas9) gene‐editing system and differentiated into cardiomyocytes. Pluripotency‐gene expression and telomerase activity in wild‐type (WT) and HLAI/II knockout hiPSCs, cardiomyocyte marker expression in WT and HLAI/II knockout hiPSC‐derived cardiomyocytes, and assessments of electrophysiological properties (eg, conduction velocity, action‐potential and calcium transient half‐decay times, and calcium transient increase times) in spheroid‐fusions composed of WT and HLAI/II knockout cardiomyocytes, were similar. However, the rates of T‐cell activation before (≈21%) and after (≈24%) exposure to HLAI/II knockout hiPSC‐derived cardiomyocytes were nearly indistinguishable and dramatically lower than after exposure to WT hiPSC‐derived cardiomyocytes (≈75%), and when WT and HLAI/II knockout hiPSC‐derived cardiomyocyte spheroids were cultured with human peripheral blood mononuclear cells, the WT hiPSC‐derived cardiomyocyte spheroids were smaller and displayed contractile irregularities. Finally, expression of HLA‐E and HLA‐F was inhibited in HLAI/II knockout cardiomyocyte spheroids after coculture with human peripheral blood mononuclear cells, although HLA‐G was not inhibited; these results are consistent with the essential role of class II major histocompatibility class transactivator in transcriptional activation of the HLA‐E and HLA‐F genes, but not the HLA‐G gene. Expression of HLA‐G is known to inhibit natural killer cell recognition and killing of cells that lack other HLAs.
Conclusions
HLAI/II knockout hiPSCs can be differentiated into cardiomyocytes that induce little or no activity in human immune cells and, consequently, are suitable for allogeneic transplantation.
Keywords: 3‐dimensional culture, β2 microglobulin, class II major histocompatibility class transactivator, immunology, stem cell
Subject Categories: Electrophysiology, Cell Therapy, Genetically Altered and Transgenic Models
Clinical Perspective
What Is New?
Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)/CRISPR associated protein 9 (Cas9) mediated knockout of β2 microglobulin and class II major histocompatibility class transactivator, which are crucial for the display of human leukocyte antigen I/II proteins at the cellular surface.
What Are the Clinical Implications?
Human leukocyte antigen I/II knockout human induced pluripotent stem cell‐derived cells may be less susceptible to immune rejection when administered to patients.
Introduction
Induced pluripotent stem cells (iPSCs) are among the most promising therapeutics for regenerative myocardial therapy because they can be differentiated into theoretically unlimited numbers and types of cells. Multiple lineages of iPSC‐derived cardiac cells have been administered via direct intramyocardial injection1, 2 or as a patch of engineered tissue,3, 4 but the effectiveness of these approaches has been limited by a variety of factors,5, 6, 7 including, perhaps most important, the exceptionally small proportion of cells that are engrafted by the native tissues at the site of administration. Because these experiments are generally performed with allogeneic or xenogeneic cells, the low engraftment rate may be at least partially attributable to the host animal's immune response. Immunogenicity is expected to be less of a problem in clinical practice because human iPSCs (hiPSCs) can be generated from each individual patient's own somatic cells.8 However, when iPSCs were generated from a line of inbred (C57BL/6) mice and transplanted back into adult mice of the same inbred strain, most of the tumors formed from the transplanted iPSCs were immune rejected, regardless of whether the iPSCs were generated via the standard retroviral reprogramming method or a more novel episomal approach that did not alter the cellular genome. Subsequent analyses found evidence of necrosis and T‐cell infiltration, which the authors attributed to the abnormal expression of minor antigens.9 Thus, the potential immunogenicity of autologous hiPSC‐derived cardiac cells continue to be one of the barriers to their clinical use.
Immune rejection is mediated by molecules in major histocompatibility classes (MHCs) I and II, which, in humans, are encoded by the human leukocyte antigen system (HLAI and HLAII, respectively). MHC‐I proteins form a complex with β2 microglobulin (B2M) on the cell surface, where they are recognized and destroyed by T cells; thus, several studies10, 11, 12, 13, 14 have been conducted to determine whether the immunogenicity of hiPSCs can be reduced by disrupting the expression of B2M. As expected, the B2M‐deficient hiPSCs were not recognized by T cells, but they remained susceptible to elimination via the “missing self” response, which is mediated by natural killer (NK) cells. Furthermore, human endothelial cells can activate alloantigen reactive memory CD4+ T cells, which may explain why cell‐mediated rejection of vascularized human allografts can occur even after professional antigen‐presenting cells are depleted. However, the endothelial cell–induced activation of reactive memory T cells requires the expression of MHC‐II molecules,15 and when both alleles of the class II MHC transactivator (CIITA) gene were knocked out in human endothelial cells, the cells did not elicit an immune response after transplantation into mice.16
This report presents results of studies that were conducted to determine whether the immunogenicity of hiPSC‐derived cells could be reduced, or even eliminated, by knocking out the expression of both B2M and CIITA. The knockout mutations were induced in hiPSCs via CRISPR/Cas9 gene editing, and both wild‐type (WT) and B2M/CIITA double‐knockout (HLAI/II knockout) hiPSCs were differentiated into cardiomyocytes. Our results confirmed that the HLAI/II knockout mutation did not alter the expression of pluripotency genes in hiPSCs or the electrical properties of hiPSC‐derived cardiomyocyte spheroids; however, measures of T‐cell activation were 3‐fold higher when human peripheral blood mononuclear cells (PBMCs) were incubated with WT hiPSC‐derived cardiomyocytes rather than HLAI/II knockout hiPSC‐derived cardiomyocytes, and spheroids of HLAI/II knockout hiPSC‐derived cardiomyocytes were more resistant than WT hiPSC‐derived cardiomyocyte spheroids to the activity of CD8+ T cells and CD4+ immune cells.
Methods
The data, analytical methods, and study materials are available to other researchers for the purposes of reproducing the results or replicating the procedure, as described in this article or on contacting the corresponding author. All experimental protocols were approved by the Institutional Review Board of the University of Alabama at Birmingham. Adult human blood samples were obtained at the University of Alabama at Birmingham's Alabama Vaccine Research Clinic after informed consent. Detailed methods on hiPSC culture and differentiation and characterization, spheroid culture and spheroid fusions, telomerase activity, Western blot analysis, transmission electron microscopy and statistical analysis are provided in Data S1.
Knockout of MHC Class I and II by Ablation of B2M and CIITA Genes
hiPSCs (GRiPS)3 were generated by transfecting male human cardiac fibroblasts with Sendai viruses coding for Octamer‐binding transcription factor 4 (OCT4); SRY (sex determining region Y)‐box 2 (SOX2); Kruppel Like Factor 4 (KLF4); avian myelocytomatosis viral oncogene homolog (C‐MYC)3, and then the genes coding for B2M and class II MHC transactivator (CIITA) were knocked out using the CRISPR/Cas9 genome editing system; the pLentiCRISPR V2 plasmid (Addgene plasmid No. 52961) was a gift from Feng Zhang. Two guide sequences (single guide RNA 1 (sgRNA1), 5′‐GAAAATGTTTCCTGACTCAG‐3′; and sgRNA2, 5′‐CCCCGGACGATATTGAACAA‐3′) flanking the start codon for human B2M and three guide sequences (sgRNA1, 5′‐ TCCTACACAATGCGTTGCC‐3′; sgRNA2, 5′‐TGGCACACTGTGAGCTGCCT‐3′; and sgRNA2, 5′‐GCCCCTAGAAGGTGGCTACC‐3′) flanking the start codon for human CIITA were designed with an online program (http://crispr.mit.edu/), and 2 oligos were synthesized for each guide sequence (Invitrogen Life Technologies). The pLentiCRISPR V2 was digested with BsmBI, and the annealed oligos were cloned into the vector; then, the pLentiCRISPR (with cloned sgRNA) was cotransfected into the 293FT Cell Line (Thermo Fisher Scientific, catalog No. R700‐07) with packaging plasmids pCMV‐VSV‐G, pRSV‐Rev, and pMDLg/pRRE (Addgene plasmids 8454, 12253, and 60488, respectively). The virus‐containing supernatants were collected 48 hours after transfection and centrifuged at 1500×g and 4°C for 10 minutes to pellet the cell debris; then, the supernatants were filtered through a 0.45‐μm low‐protein–binding membrane (Millipore) and used immediately to transduce the hiPSCs. After transduction, the hiPSCs were expanded on Matrigel‐coated dishes for 4 days with puromycin (5 μg/mL) selection, and individual puromycin‐resistant single‐cell–derived colonies were harvested and expanded in culture. B2M and CIITA knockouts were verified via Sanger sequencing and Western blot analysis.
Generation of hiPSC‐Derived Cardiomyocyte Spheroid Fusions
Spheroid fusion was performed, as previously described.17 Briefly, spheroids were grown for 7 days, and then 2 spheroids of diameter ≈800 μm were placed in a 2‐mL, round‐bottom test tube to ensure that the spheroids were in physical contact. Spheroids were cultured in maintenance medium and spontaneous beating was typically observed within 1 day of fusion; the fusions contracted into small, compact, tissuelike structures by day 7.
Optical Mapping of Vm and Ca2+
Optical mapping of spheroids was performed, as described previously.17 Briefly, spheroids were double stained with a low‐affinity Ca2+‐sensitive dye (Cal‐520FF, 10 μmol/L for 1 hour)18 and a transmembrane voltage (Vm)‐sensitive dye (RH‐237, 2.5 μmol/L for 5 minutes), held in place with 2 nylon meshes (280‐μm pore), and perfused with Hank's balanced salt solution (36°C–37°C) supplemented with 10 μmol/L of blebbistatin to eliminate motion artifacts caused by spheroid contractions. Spheroids were stimulated with 2‐ms rectangular pulses delivered from a bipolar electrode placed near the edge of the spheroid, and action potentials (APs) and Ca2+ transients (CaTs) were recorded with a 16×16 photodiode array (Hamamatsu) at a spatial resolution of 110 μm per diode.19 Activation times were measured at 50% of the AP amplitude and used to construct isochronal maps of activation spread. Conduction velocity was calculated at each recording site from local activation times and averaged across the whole map. AP and CaT durations were measured at 50% of signal recovery.
PBMC Isolation
Human whole blood was collected into BD Vacutainer sodium heparin tubes, diluted 1:1 with Dulbecco's PBS plus 2% fetal bovine serum (Stemcell Technologies), and then layered over Lymphoprep (Stemcell Technologies) density gradient medium in SepMate‐50 (Stemcell Technologies) centrifugation tubes. Tubes were centrifuged at 1200g for 10 minutes at room temperature; then, the topmost portion of the upper liquid layer was aspirated, and the remaining liquid (containing the PBMCs) was poured into 50‐mL collection tubes and washed twice (300g, 10 minutes, room temperature) in Dulbecco's PBS+fetal bovine serum. PBMC concentrations were determined with a hemocytometer and adjusted to 1.5×106 cells/mL.
CD8+ and CD4+ Cell Isolation
PBMCs were isolated, as previously described. CD8+ cells were then isolated from PBMCs using anti‐CD8 magnetic beads (EasySep Human CD8 Positive Selection Kit II; Stemcell Technologies), and CD4+ cells were sequentially isolated from the CD8+‐depleted PBMC pour‐off using anti‐CD4 magnetic beads (EasySep Human CD4 Positive Selection Kit II; Stemcell Technologies).
Gross Phenotyping of Immune Cell Challenge Response
A total of 2×105 PBMCs, CD8+ cells, or CD4+ cells were added to 96‐well round‐bottom plate wells containing beating WT cardiomyocyte or knockout cardiomyocyte spheroids and incubated at 37°C in 5% CO2 and humidified air. Spheroids were observed at day 3 and day 5 for gross morphologic changes.
T‐Cell Activation
PBMCs (1.5×106 cells/well) and CD28/CD49d costimulatory molecules (BD Biosciences, 346049) were incubated for 28 hours alone (negative control), with phorbol 12‐myristate 13‐acetate (10 ng/mL, positive control), with WT hiPSC‐derived cardiomyocytes, or with HLAI/II knockout hiPSC‐derived cardiomyocytes at 37°C in 5% CO2 and humidified air; phorbol 12‐myristate 13‐acetate was added for only the last 4 hours of incubation. PBMCs were then collected into 5‐mL cell‐strainer tubes, washed, resuspended in 100 μL of Dulbecco's PBS+fetal bovine serum, and stained with anti‐CD3 and anti‐CD69 antibodies for flow cytometric analysis using a BD LSRFortessa cytometer and FACSDiva software. The lymphocyte population was defined by forward and side scatter characteristics and then gated on the basis of Allophycocyanin (CD3‐APC) and phycoerythrin (CD69‐PE) signal intensities to ultimately produce a histogram showing the proportion of CD3+ cells that also express CD69. Human IgG was added as an Fc‐blocker to all samples. Antibodies, which were used according to the manufacturers’ recommendations, were as follows: CD28/49d (BD Biosciences No. 346049), CD3 (BD Biosciences No. 346049), CD69 (BD Biosciences No. 346049), CD8 (Stemcell Technologies No. 187853) and CD4 (Stemcell Technologies No. 17852).
Results
CRISPR/Cas9‐Mediated HLAI/II‐Knockout
hiPSCs carrying knockout mutations for both HLA class I and class II (HLAI/II knockout hiPSCs) were generated via CRISPR/Cas9 gene editing technology with guide RNA sequences flanking B2M (Figure 1A) and CIITA (Figure 1C). After editing, the B2M gene contained a 4‐bp insertion, a 14‐bp deletion, and a 10‐bp deletion, and the CIITA gene contained a 1‐bp insertion (Figure 1B and 1D and Figures S1 and S2), which introduced new stop codons that prematurely terminated the translation of both genes (Figure 1F and 1H). Successful knockout was confirmed via Western blot analysis (Figure 1E and 1G) and immunofluorescent staining (Figure 2E and 2F): 2 clones lacked the expression of B2M alone, 2 clones lacked CIITA expression alone, and 2 clones lacked both B2M and CIITA expression. Notably, assessments of pluripotency gene expression (Figure 2A) and telomerase activity >50 passages (Figure 2B) in the double‐knockout (HLAI/II knockout) hiPSCs and in WT hiPSCs were similar, indicating that the HLAI/II knockout hiPSCs remained pluripotent.
Figure 1.
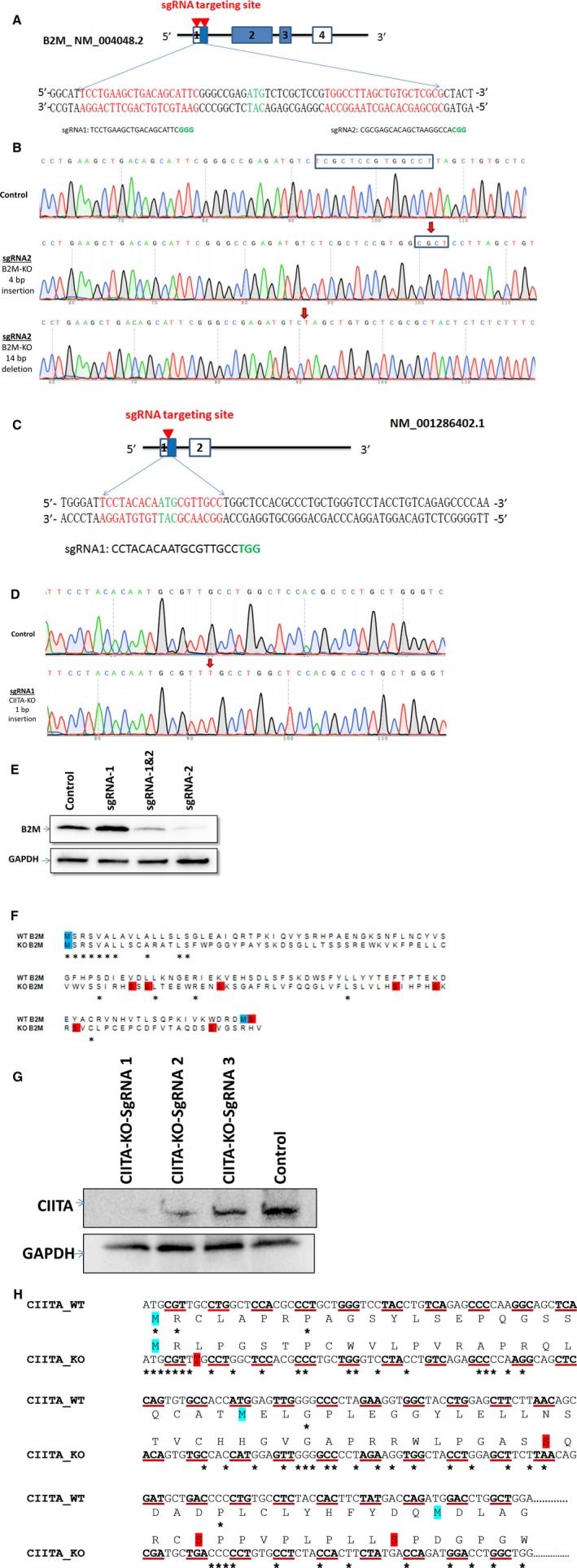
Schematic representation of the β2 microglobulin (B2M) and class II major histocompatibility class transactivator (CIITA) knockout. A, CRISPR‐Cas9 design of B2M gene knockout. The red arrow indicates 2 guide RNA (sgRNA) binding in the first exon of the B2M gene. sgRNA 1 binds in the 5′‐untranslated region and sgRNA 2 binds in the translational start region of the gene. B, sgRNA 2 targeting B2M knockout sequence chromatogram, and red arrows indicate insertions/deletions (indels) of 4‐bp insertion and 14‐bp deletion. C, CRISPR‐Cas9 design of CIITA gene knockout. D, The red arrow indicates sgRNA binding in the first exon of the CIITA gene. sgRNA 1 binds in the translational start region of the gene, and red arrows indicate indels of 1‐bp insertion. E, Western blot data for the following: lane 1, control; lane 2, B2M targeting with sgRNA 1; lane 3, B2M targeting with sgRNA 1 and sgRNA 2; and lane 4, B2M targeting with sgRNA 2. F, Protein sequence alignment of wild‐type (WT) B2M and knockout B2M. G, Western blot data for the following: lane 1, control; lane 2, CIITA targeting sgRNA 3; lane 3, CIITA targeting sgRNA 2; and lane 4, CIITA targeting sgRNA 1. H, Protein and DNA sequence alignment of WT CIITA and knockout CIITA. *Aligned sequence.
Figure 2.
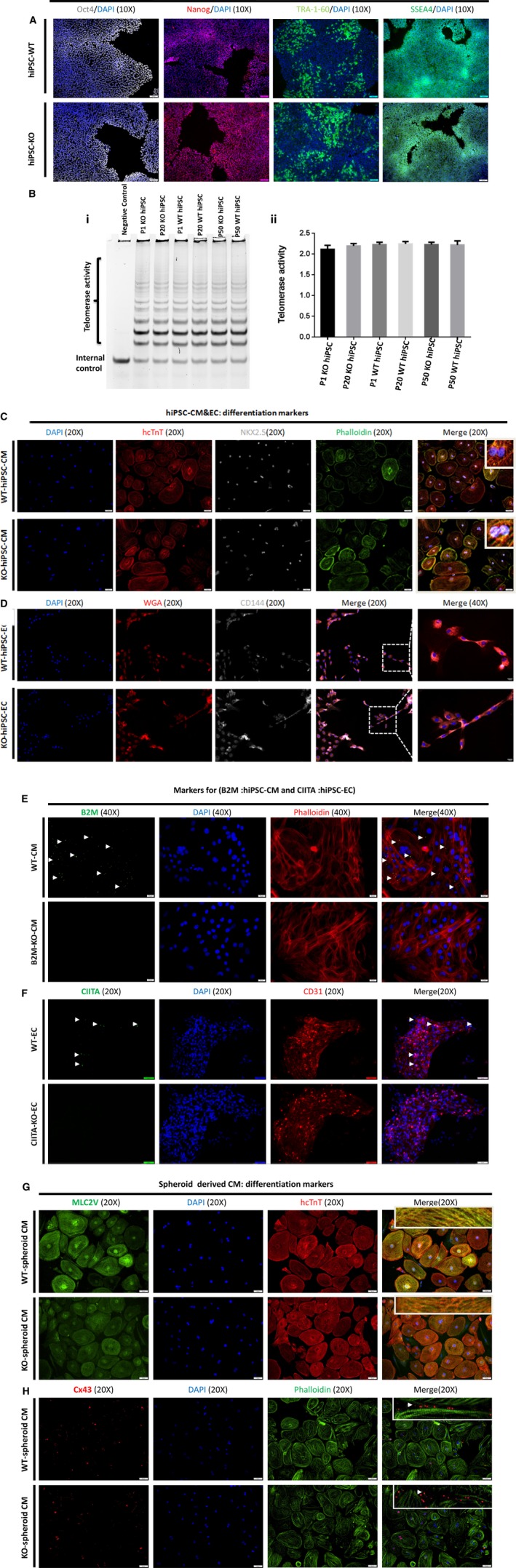
Generation and characterization of pluripotency, telomerase activity, and differentiation of human cardiac lineage‐induced pluripotent stem cells (hiPSCs) in cardiomyocytes, endothelial cells (ECs), and spheroid‐derived cardiomyocytes from wild‐type (WT) and hiPSC‐derived human leukocyte antigen (HLA) I/II knockout cells. A, hiPSC‐derived WT and hiPSC‐HLAI/II knockout cells were cultured on glass coverslips and were immunolabeled for pluripotent markers octamer‐binding transcription factor 4 (Oct4), Nanog homeobox (Nanog), tumour rejection antigen (Tra‐1‐60), and stage‐specific embryonic antigen‐4 (SSEA4) counterstained with 4’,6‐diamidino‐2‐phenylindole (DAPI) staining for nuclei. B, hiPSC‐derived WT and hiPSC‐derived HLAI/II knockout were cultured for 50 passages, and passages 1, 20, and 50 were collected. (i) telomerase activity was evaluated with a nondenaturing polyacrylamide gel and (ii) quantified at the indicated time points via densitometry analysis. C&D, WT and knockout hiPSC‐derived cardiomyocytes stained for human cardiac troponin T (hcTnT; red), phalloidin (green), and NK2 homeobox 5 (NKX2.5) (gray); counterstained nuclei with DAPI and WT and knockout hiPSC‐derived ECs stained for wheat germ agglutinin (WGA; red) CD144 (gray); and counterstained nuclei with DAPI. E&F, WT and knockout hiPSC‐derived cardiomyocytes stained for β2 microglobulin (B2M; green) and phalloidin (red) and counterstained nuclei with DAPI and WT, and knockout hiPSC‐derived ECs stained for class II major histocompatibility class transactivator (CIITA; green) and CD31 (red) and counterstained nuclei with DAPI. H&G, hiPSC‐derived WT and hiPSC‐derived HLAI/II knockout differentiated cardiomyocytes were cultured on glass coverslips and were immunolabeled for cardiomyocyte marker proteins myosin light chain 2 (MLC2v), hcTnT, phalloidin, and connexin 43 (Cx43); nuclei were counterstained with DAPI markers.
HLAI/II Knockout Did Not Alter the Differentiation of hiPSC‐Derived Cardiomyocytes or the Electrophysiological Parameters of hiPSC‐Derived Cardiomyocyte Fusions
WT and HLAI/II knockout hiPSCs were differentiated into cardiomyocytes via an established protocol.17 Spontaneously contracting cells typically appeared 8 to 10 days after differentiation was initiated, and cardiomyocyte identity was confirmed via immunofluorescent assessments of the expression of cardiomyocyte markers, such as ventricular myosin light chain 2, cardiac troponin T, phalloidin, and connexin 43 (Figure 2C, 2D, 2G & 2H). Furthermore, when spheroids composed of WT or HLAI/II hiPSC‐derived cardiomyocytes were fused, the fusions contracted spontaneously and synchronously within 7 days of the fusion event, and electron micrographs confirmed that the HLAI/II knockout spheroids were structurally normal, with well‐defined sarcomeres and Z‐lines (Figure 3C, 3E & 3F). Optical maps of action‐potential propagation and traces of individual APs and CaTs (Figure 3A and 3B) indicated that cardiomyocytes were electrically coupled both within and between the fused spheroids with no detectable conduction delay at the interspheroid boundary and that measurements of conduction velocity, the durations of both APs (AP duration measured at 50% of signal recovery) and CaTs (CaT measured at 50% of signal recovery), and CaT increase times in WT and HLAI/II knockout spheroids were not statistically different (Figure 3D and 3G).
Figure 3.
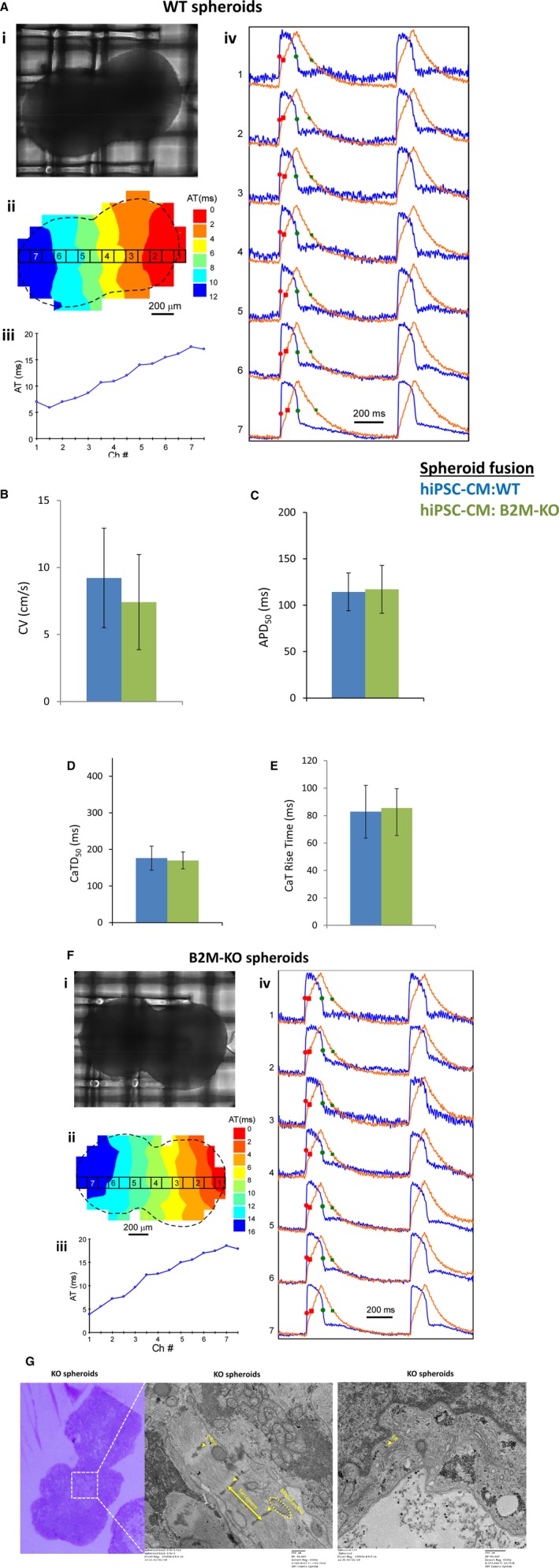
Generation and characterization of spheroid fusions derived from wild‐type (WT) and human induced pluripotent stem cells (hiPSC)–derived human leukocyte antigen (HLA) I/II knockout cells. A, hiPSC‐derived WT cells were differentiated cardiomyocytes that generated spheroid fusion (i) at 7 days of culture to generate (ii) optical maps of action potential propagation, and (iii) traces of individual action potentials (blue) and calcium transients (red); the number of each trace in ii corresponds to the location identified in the map shown in i. B, Conduction velocity. The durations of the action potentials (C) and calcium transients (D) at 50% (ie, APD 50 and CaTD 50, respectively) and CaT increase time recovery (E) were determined for fusions of 2 large spheroids after 7 days in culture (WT, blue) and (HLAI/II knockout, green). F, hiPSC‐derived HLAI/II knockout cells were differentiated cardiomyocytes that generated spheroid fusion (i) at 7 days of culture to generate (ii) optical maps of action potential propagation, and (iii) traces of individual action potentials (blue) and CaTs (red); the number of each trace in ii corresponds to the location identified in the map shown in i. G, The ultrastructure of the spheroid fusion was analyzed by transmission electron microscope to identify sarcomere, Z‐lines, mitochondria, and endoplasmic reticulum (ER). AT indicates activation time; B2M, β2 microglobulin; CV, conduction velocity.
HLAI/II Knockout Reduces the Immunogenicity of hiPSC‐Derived and Spheroid Cardiomyocytes
The immunogenicity of HLAI/II knockout and WT hiPSC‐derived cardiomyocytes were compared by first culturing monolayers of the cells with allogeneic human PBMCs, then afterward collecting the PBMCs for antibody staining (Table S1) against the T‐cell marker CD3 and the T‐cell activation marker CD69 (Figure 4A). Assessments were also performed in PBMCs that had been cultured in the absence of hiPSC‐derived cardiomyocytes (negative control) and in PBMCs that had been cultured without hiPSC‐derived cardiomyocytes but with the T‐cell–activator phorbol 12‐myristate 13‐acetate (positive control). The proportion of T cells that were activated by WT hiPSC‐derived cardiomyocytes (>75%) (Figure 4B) was 3‐fold greater than the proportion that was activated by HLAI/II knockout hiPSC‐derived cardiomyocytes (<25%), and the extent of T‐cell activation by HLAI/II knockout hiPSC‐derived cardiomyocytes was similar to the extent observed in the negative control (≈21%) (Figure 4 and 4C). Furthermore, when WT spheroids were cocultured with human PBMCs for 5 days, WT cardiomyocyte spheroids were significantly reduced in size compared with the knockout cardiomyocyte spheroids, and the contractions of the WT cardiomyocyte spheroids were visibly less forceful than those of the knockout cardiomyocyte spheroids (Videos S1 through S4). Similar results were observed in 3‐day coculture experiments with purified human CD4+ and CD8+ T cells (Figure 4D). Consistent with these data, T‐cell activation markers were significantly elevated in coculture experiments with WT spheroids but not with HLAI/II knockout spheroids. Last, HLA‐G is known to inhibit NK cell recognition and the killing of cells that lack other HLAs. Expression of HLA‐E and HLA‐F was inhibited in HLAI/II knockout cardiomyocyte spheroids after coculture with human PBMCs, but HLA‐G expression was not inhibited. These results are consistent with the essential role of CIITA in transcriptional activation of HLA‐E and HLA‐F genes, but not the HLA‐G gene.20 Although Western blot analysis indicated that interferon‐γ did not induce the NK‐cell inhibitory ligands HLA‐E and HLA‐F in HLAI/II knockout hiPSC‐derived cardiomyocytes, the expression of HLA‐G, which protects placental trophoblasts from lysis by maternal NK cells,20 was unchanged (Figure 4E).
Figure 4.
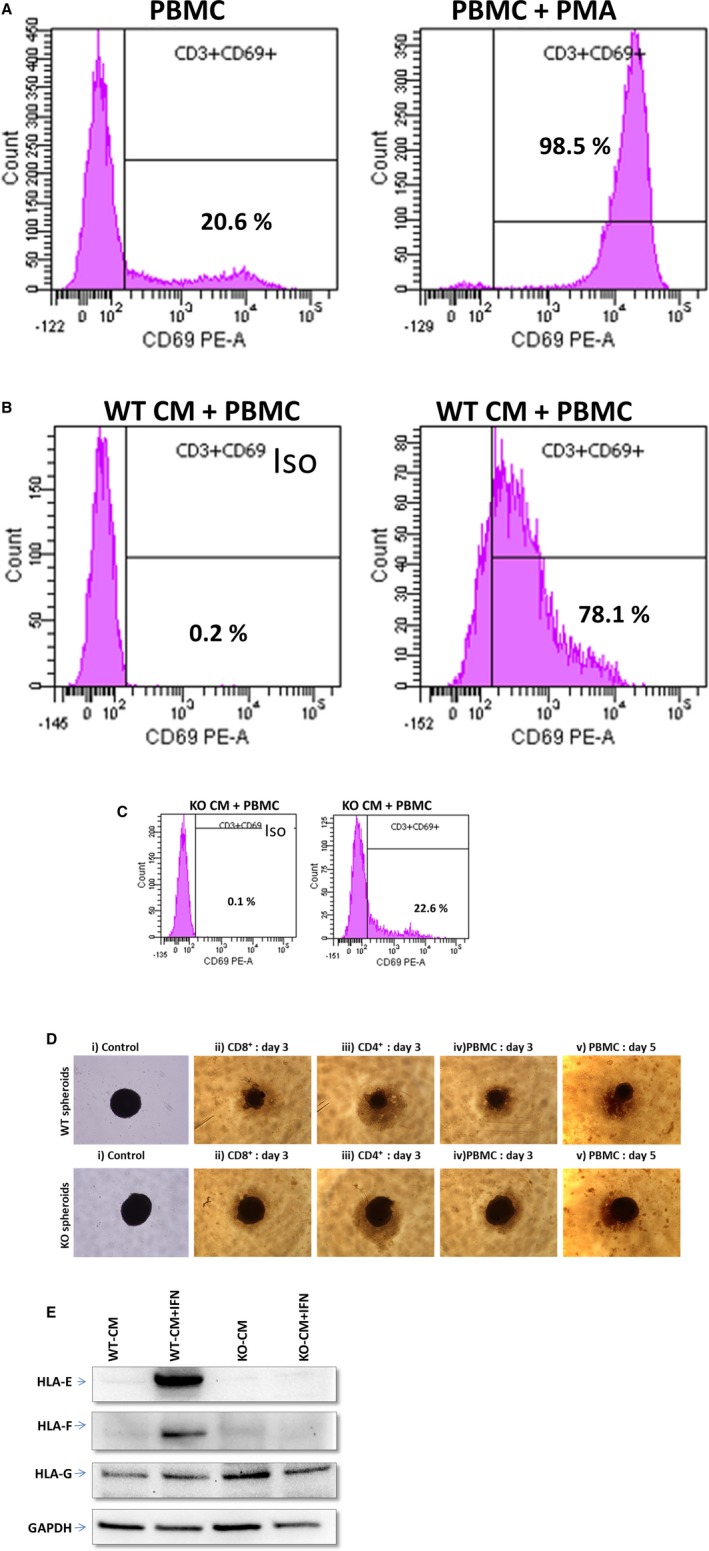
Characterization of immunogenicity in spheroids from wild‐type (WT) and human induced pluripotent stem cell–derived human leukocyte antigen (HLA) I/II knockout cells. A total of 1.5×106 peripheral blood mononuclear cells (PBMCs) plus CD28/CD49d costimulatory molecules were added to 6‐well plates that had wells containing WT cardiomyocytes and wells containing knockout cardiomyocytes. The T‐cell stimulator phorbol 12‐myristate 13‐acetate (PMA) was added to one of the PMBC control wells (PBMC + PMA). After incubation, PBMCs were collected for CD3 (T‐cell marker) and CD69 (T‐cell stimulation marker) antibody staining. A, PBMC controls. B, WT cardiomyocytes + PBMCs. C, Knockout cardiomyocytes + PBMCs. (Percentage values represent the CD3+ cells that are also CD69+.) D, Control, CD8+ cells, CD4+ cells, and PBMCs were added to 96‐well plate wells containing beating WT cardiomyocytes or knockout cardiomyocyte spheroids. Observations from day 3 and day 5 show that WT cardiomyocyte spheroids challenged with immune cells were smaller compared with the knockout cardiomyocyte spheroids; in addition, the contractions of the WT cardiomyocyte spheroids were visibly less forceful than those of the knockout cardiomyocyte spheroids. i) spheroids without immune cells denoted “control”, ii) spheroids co‐cultured with CD8+ cells for 3 days denoted “CD8+: day3”, iii) spheroids co‐cultured with CD4+ cells for 3 days denoted “CD4+: day3” iv) spheroids co‐cultured with PBMC cells for 3 days denoted “PBMC: day3” and v) spheroids co‐cultured with PBMC cells for 3 days denoted “PBMC: day5”. E, Western blot data for the following: lane 1, WT cardiomyocytes; lane 2, WT cardiomyocytes treated with interferon‐γ; lane 3, knockout cardiomyocytes; and lane 4, knockout cardiomyocytes treated with interferon‐γ for HLA class I histocompatibility antigen, α chain (HLA‐E, HLA alpha chain E; HLA‐F, HLA alpha chain F; HLA‐G, HLA alpha chain G). Iso indicates CD69 isotype control; PE‐A, phycoerythrin channel of flow cytometer.
Discussion
hiPSCs are not expected to provoke an immune response during clinical use because they can be generated from the somatic cells of each individual patient.8 Nevertheless, studies suggest that the tumors formed from transplanted murine iPSCs can be rejected by the recipient mice, even if the cells and the mice are genetically identical.9 Furthermore, the time required to produce autologous hiPSC‐derived cells precludes their use for treatment of acute conditions. Thus, techniques for generating nonimmunogenic hiPSCs could be used to increase the engrafted cells and to produce lines of allogeneic “universal donor” hiPSCs. Herein, we show that the CRISPR/Cas9 gene‐editing system can successfully knock out 2 key components (B2M and CIITA) of the HLAI/II system in hiPSCs without altering the cells’ pluripotency or telomerase activity, that HLAI/II knockout hiPSC‐derived cardiomyocytes fail to activate human T cells, and those spheroids of HLAI/II knockout hiPSC‐derived cardiomyocytes retain the electrical properties of WT hiPSC‐derived cardiomyocyte spheroids but are more resistant to the cytotoxic effects of PBMCs.
When biallelic CIITA‐knockout mutations were induced in untransformed endothelial cells,16 the CIITA‐knockout cells failed to activate alloreactive CD4+ memory T cells, and their immunogenicity was restored via the overexpression of CIITA, which confirmed that the observed phenotype was attributable to the loss of CIITA, rather than random off‐target mutations. CIITA has also been linked to the induction of HLA‐E and HLA‐F, but does not appear to activate the expression of HLA‐G.21 Notably, although the outermost layer of the human placenta is generally devoid of most HLAI/II molecules, and, consequently, escapes recognition from maternal T cells, HLA‐G is expressed by trophoblast cells that are in direct contact with maternal tissues, where it appears to inactivate NK cells by functioning as a “self” antigen.20 The presence of HLA‐E on the cellular surface also protects against lysis by NK cells because it binds to the NK‐cell inhibitory complex CD94/Natural killer group 2, member A (NGK2A).22, 23, 24 Also, human embryonic stem cells in which the B2M gene had been knocked out and replaced with HLA‐E were not recognized as allogeneic by CD8+ T cells, failed to bind anti‐HLA antibodies, and resisted the cytotoxic activity of NK cells.25
In conclusion, the results presented in this report demonstrate that the CRISPR/Cas9‐mediated knockout of B2M and CIITA, which are crucial for the display of HLAI/II proteins at the cellular surface, does not alter the pluripotency of hiPSCs or the electrophysiological properties of spheroids composed of HLAI/II knockout hiPSC‐derived cardiomyocytes. However, the HLA knockout hiPSC‐derived cardiomyocytes did not activate T cells, as measured by expression of CD69, which is known to be upregulated in acutely activated T cells responding to antigen, and were more resistant than WT hiPSC‐derived cardiomyocytes to the cytotoxic effects of PBMCs, including NK cells. Thus, cells derived from HLAI/II knockout hiPSCs may be less susceptible to immune rejection when administered to patients and, therefore, provide a source of universal donor cells to be used for the treatment of acute conditions.
Sources of Funding
This work was supported in part by National Institutes of Health National Heart, Lung, and Blood Institute RO1 grants HL 95077, HL114120, HL131017, HL 138023, and UO1 HL134764 (Zhang). We thank the State of Alabama and the University of Alabama at Birmingham (UAB) School of Medicine for supporting this work through the UAB Stem Cell Institute (Townes).
Disclosures
None.
Supporting information
Data S1. Supplemental Methods.
Table S1. Antibodies
Figure S1. DNA sequence alignment of (A) wild type B2M 4bp (B2M_WT_4bp) and Knockout B2M 4bp insertion (B2M_KO_4bp); (B) wild type B2M 14bp (B2M_WT_14bp) and Knockout B2M 4bp deletion (B2M_KO_14bp). C, sgRNA 1 targeting B2M‐KO sequence chromatogram and red arrows indicate 10 bp deletion. D, ATGpr used to predict which ATG is the initiation codon for) wild type B2M 10bp (B2M_WT_10bp) and Knockout B2M 10bp deletion (B2M_KO_10bp). *Aligned nucleotide bases.
Figure S2. DNA sequence alignment of (A) wild type CIITA 1bp (CIITA _WT _1bp) and Knockout CIITA 1bp insertion (B2M_KO_1bp); Protein sequence alignment of (B) wild type 3 different isoforms of CIITA of 1bp (CIITA_WT_1bp). (C) CRISPR/Cas9 design of CIITA gene knockout sgRNA 2&3. (D) DNA sequence alignment of wild type 3 different isoforms of CIITA of 1bp (CIITA_WT_1bp). *Aligned nucleotide and protein sequence.
Video S1. WT hiPSC differentiated cardiomyocytes derived spheroid synchronous contractile activity of individual spheroids in culture. Best viewed with Windows Media Player.
Video S2. KO hiPSC differentiated cardiomyocytes derived spheroid synchronous contractile activity of individual spheroids in culture. Best viewed with Windows Media Player.
Video S3. WT spheroid co‐culture with PBMC for 5 days will result in the loss of spontaneous and synchronous contractile activity of individual spheroids in culture. Best viewed with Windows Media Player.
Video S4. KO spheroid co‐culture with PBMC for 5 days will result in no change of spontaneous and synchronous contractile activity of individual spheroids in culture. Best viewed with Windows Media Player.
Acknowledgments
The authors thank Dr Yanwen Liu and Dr Davide Botta for excellent technical assistance and Dr Paul Goepfert for critical discussions and biological reagents.
(J Am Heart Assoc. 2018;7:e010239 DOI: 10.1161/JAHA.118.010239.)
References
- 1. Alcon A, Cagavi Bozkulak E, Qyang Y. Regenerating functional heart tissue for myocardial repair. Cell Mol Life Sci. 2012;69:2635–2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Xiong Q, Ye L, Zhang P, Lepley M, Swingen C, Zhang L, Kaufman DS, Zhang J. Bioenergetic and functional consequences of cellular therapy: activation of endogenous cardiovascular progenitor cells. Circ Res. 2012;111:455–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ye L, Chang YH, Xiong Q, Zhang P, Zhang L, Somasundaram P, Lepley M, Swingen C, Su L, Wendel JS, Guo J, Jang A, Rosenbush D, Greder L, Dutton JR, Zhang J, Kamp TJ, Kaufman DS, Ge Y, Zhang J. Cardiac repair in a porcine model of acute myocardial infarction with human induced pluripotent stem cell‐derived cardiovascular cells. Cell Stem Cell. 2014;15:750–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gao L, Gregorich ZR, Zhu W, Mattapally S, Oduk Y, Lou X, Kannappan R, Borovjagin AV, Walcott GP, Pollard AE, Fast VG, Hu X, Lloyd SG, Ge Y, Zhang J. Large cardiac muscle patches engineered from human induced‐pluripotent stem cell‐derived cardiac cells improve recovery from myocardial infarction in swine. Circulation. 2018;137:1712–1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jawad H, Ali NN, Lyon AR, Chen QZ, Harding SE, Boccaccini AR. Myocardial tissue engineering: a review. J Tissue Eng Regen Med. 2007;1:327–342. [DOI] [PubMed] [Google Scholar]
- 6. Wang F, Guan J. Cellular cardiomyoplasty and cardiac tissue engineering for myocardial therapy. Adv Drug Deliv Rev. 2010;62:784–797. [DOI] [PubMed] [Google Scholar]
- 7. Zimmermann WH, Melnychenko I, Wasmeier G, Didie M, Naito H, Nixdorff U, Hess A, Budinsky L, Brune K, Michaelis B, Dhein S, Schwoerer A, Ehmke H, Eschenhagen T. Engineered heart tissue grafts improve systolic and diastolic function in infarcted rat hearts. Nat Med. 2006;12:452–458. [DOI] [PubMed] [Google Scholar]
- 8. Tapia N, Scholer HR. Molecular obstacles to clinical translation of iPSCs. Cell Stem Cell. 2016;19:298–309. [DOI] [PubMed] [Google Scholar]
- 9. Zhao T, Zhang ZN, Rong Z, Xu Y. Immunogenicity of induced pluripotent stem cells. Nature. 2011;474:212–215. [DOI] [PubMed] [Google Scholar]
- 10. de Almeida PE, Ransohoff JD, Nahid A, Wu JC. Immunogenicity of pluripotent stem cells and their derivatives. Circ Res. 2013;112:549–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Riolobos L, Hirata RK, Turtle CJ, Wang PR, Gornalusse GG, Zavajlevski M, Riddell SR, Russell DW. HLA engineering of human pluripotent stem cells. Mol Ther. 2013;21:1232–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lu P, Chen J, He L, Ren J, Chen H, Rao L, Zhuang Q, Li H, Li L, Bao L, He J, Zhang W, Zhu F, Cui C, Xiao L. Generating hypoimmunogenic human embryonic stem cells by the disruption of beta 2‐microglobulin. Stem Cell Rev. 2013;9:806–813. [DOI] [PubMed] [Google Scholar]
- 13. Wang D, Quan Y, Yan Q, Morales JE, Wetsel RA. Targeted disruption of the beta2‐microglobulin gene minimizes the immunogenicity of human embryonic stem cells. Stem Cells Transl Med. 2015;4:1234–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Feng Q, Shabrani N, Thon JN, Huo H, Thiel A, Machlus KR, Kim K, Brooks J, Li F, Luo C, Kimbrel EA, Wang J, Kim KS, Italiano J, Cho J, Lu SJ, Lanza R. Scalable generation of universal platelets from human induced pluripotent stem cells. Stem Cell Reports. 2014;3:817–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pober JS, Merola J, Liu R, Manes TD. Antigen presentation by vascular cells. Front Immunol. 2017;8:1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Abrahimi P, Chang WG, Kluger MS, Qyang Y, Tellides G, Saltzman WM, Pober JS. Efficient gene disruption in cultured primary human endothelial cells by CRISPR/Cas9. Circ Res. 2015;117:121–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mattapally S, Zhu W, Fast VG, Gao L, Worley C, Kannappan R, Borovjagin AV, Zhang J. Spheroids of cardiomyocytes derived from human‐induced pluripotent stem cells improve recovery from myocardial injury in mice. Am J Physiol Heart Circ Physiol. 2018;315:H327–H339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kong W, Fast VG. The role of dye affinity in optical measurements of cai(2+) transients in cardiac muscle. Am J Physiol Heart Circ Physiol. 2014;307:H73–H79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sowell B, Fast VG. Ionic mechanism of shock‐induced arrhythmias: role of intracellular calcium. Heart Rhythm. 2012;9:96–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pazmany L, Mandelboim O, Vales‐Gomez M, Davis DM, Reyburn HT, Strominger JL. Protection from natural killer cell‐mediated lysis by HLA‐G expression on target cells. Science. 1996;274:792–795. [DOI] [PubMed] [Google Scholar]
- 21. Gobin SJ, van den Elsen PJ. Transcriptional regulation of the MHC class ib genes HLA‐E, HLA‐F, and HLA‐G. Hum Immunol. 2000;61:1102–1107. [DOI] [PubMed] [Google Scholar]
- 22. Braud VM, Allan DS, O'Callaghan CA, Soderstrom K, D'Andrea A, Ogg GS, Lazetic S, Young NT, Bell JI, Phillips JH, Lanier LL, McMichael AJ. HLA‐E binds to natural killer cell receptors CD94/NKG2A, B and C. Nature. 1998;391:795–799. [DOI] [PubMed] [Google Scholar]
- 23. Lee N, Llano M, Carretero M, Ishitani A, Navarro F, Lopez‐Botet M, Geraghty DE. HLA‐E is a major ligand for the natural killer inhibitory receptor CD94/NKG2A. Proc Natl Acad Sci USA. 1998;95:5199–5204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Horowitz A, Strauss‐Albee DM, Leipold M, Kubo J, Nemat‐Gorgani N, Dogan OC, Dekker CL, Mackey S, Maecker H, Swan GE, Davis MM, Norman PJ, Guethlein LA, Desai M, Parham P, Blish CA. Genetic and environmental determinants of human NK cell diversity revealed by mass cytometry. Sci Transl Med. 2013;5:208ra145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gornalusse GG, Hirata RK, Funk SE, Riolobos L, Lopes VS, Manske G, Prunkard D, Colunga AG, Hanafi LA, Clegg DO, Turtle C, Russell DW. HLA‐E‐expressing pluripotent stem cells escape allogeneic responses and lysis by NK cells. Nat Biotechnol. 2017;35:765–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1. Supplemental Methods.
Table S1. Antibodies
Figure S1. DNA sequence alignment of (A) wild type B2M 4bp (B2M_WT_4bp) and Knockout B2M 4bp insertion (B2M_KO_4bp); (B) wild type B2M 14bp (B2M_WT_14bp) and Knockout B2M 4bp deletion (B2M_KO_14bp). C, sgRNA 1 targeting B2M‐KO sequence chromatogram and red arrows indicate 10 bp deletion. D, ATGpr used to predict which ATG is the initiation codon for) wild type B2M 10bp (B2M_WT_10bp) and Knockout B2M 10bp deletion (B2M_KO_10bp). *Aligned nucleotide bases.
Figure S2. DNA sequence alignment of (A) wild type CIITA 1bp (CIITA _WT _1bp) and Knockout CIITA 1bp insertion (B2M_KO_1bp); Protein sequence alignment of (B) wild type 3 different isoforms of CIITA of 1bp (CIITA_WT_1bp). (C) CRISPR/Cas9 design of CIITA gene knockout sgRNA 2&3. (D) DNA sequence alignment of wild type 3 different isoforms of CIITA of 1bp (CIITA_WT_1bp). *Aligned nucleotide and protein sequence.
Video S1. WT hiPSC differentiated cardiomyocytes derived spheroid synchronous contractile activity of individual spheroids in culture. Best viewed with Windows Media Player.
Video S2. KO hiPSC differentiated cardiomyocytes derived spheroid synchronous contractile activity of individual spheroids in culture. Best viewed with Windows Media Player.
Video S3. WT spheroid co‐culture with PBMC for 5 days will result in the loss of spontaneous and synchronous contractile activity of individual spheroids in culture. Best viewed with Windows Media Player.
Video S4. KO spheroid co‐culture with PBMC for 5 days will result in no change of spontaneous and synchronous contractile activity of individual spheroids in culture. Best viewed with Windows Media Player.


