Abstract
The BET bromodomain protein BRD4 is a chromatin reader that regulates transcription, including in cancer. In prostate cancer, specifically, the anti-tumor activity of BET bromodomain inhibition has been principally linked to suppression of androgen receptor (AR) function. MYC is a well-described BRD4 target gene in multiple cancer types, and prior work demonstrates that MYC plays an important role in promoting prostate cancer cell survival. Importantly, several BET bromodomain clinical trials are ongoing, including in prostate cancer. However, there is limited information about pharmacodynamic markers of response or mediators of de novo resistance. Using a panel of prostate cancer cell lines, we demonstrated that MYC suppression—rather than AR suppression—is a key determinant of BET bromodomain inhibitor sensitivity. Importantly, we determined that BRD4 was dispensable for MYC expression in the most resistant cell lines and that MYC RNAi + BET bromodomain inhibition led to additive anti-tumor activity in the most resistant cell lines. Our findings demonstrate that MYC suppression is an important pharmacodynamic marker of BET bromodomain inhibitor response and suggest that targeting MYC may be a promising therapeutic strategy to overcome de novo BET bromodomain inhibitor resistance in prostate cancer.
Introduction
Although treatment options for patients with castration-resistant prostate cancer (CRPC) are expanding, over 29,000 American men are still predicted to die from prostate cancer in 20181. This demonstrates the urgent need to develop more effective therapies to treat this disease. We and others determined that inhibition of BET bromodomain proteins represents a promising strategy to treat CRPC2–4. BET bromodomain proteins are chromatin readers that recognize and bind to acetylated lysines on histone tails, such as lysine 27 on Histone H3 (H3K27Ac)5. BET bromodomain proteins then recruit other transcriptional machinery that promote expression of important genes in cancer6. BET bromodomain inhibition blocks proliferation and promotes differentiation7, which contributes to cancer cell death. However, key markers of response are unknown, and very little is known about mechanisms of de novo resistance to BET bromodomain inhibition in CRPC.
MYC is a key transcription factor regulated by multiple cellular pathways and transcriptional regulators, including BET bromodomain proteins5,8. Importantly, re-activation of MYC mRNA expression has been implicated as an acquired resistance mechanism to BET bromodomain inhibition in several cancer types, demonstrating MYC’s importance9–12. Herein, using a panel of CRPC cell lines, we demonstrate that suppression of MYC expression by BET bromodomain inhibition strongly correlates with sensitivity to the BET bromodomain inhibitor JQ1. Further, we demonstrate that co-targeting MYC together with BET bromodomain inhibition in cells in which JQ1 fails to suppress MYC expression leads to additive anti-tumor activity. Thus, our results demonstrate the utility of measuring MYC expression as a pharmacodynamic marker of BET bromodomain inhibitor response and demonstrate the need to develop strategies to co-target MYC along with BET bromodomain inhibition in resistant cancer cells.
Results
MYC maintenance contributes to de novo BET bromodomain inhibitor resistance
We first treated a panel of nine CRPC cell lines with dose-escalation of the BET bromodomain inhibitor JQ1 for 72 hours in order to determine the drug’s GR50 value (dose corresponding to 50% growth rate inhibition) in each cell line (Supplementary Table S1). This panel included both AR-dependent and AR-independent CRPC cell lines as well as AR-dependent LNCaP cells engineered to stably overexpress an ectopic MYC cDNA not under the control of BET bromodomain proteins (LNCaP-MYC). AR-independent PC3 and DU145 cells, and LNCaP-MYC cells were the least sensitive to JQ1 (Fig. 1a, y-axis). Further, LNCaP-MYC cells had a GR50 more than 3-fold higher than LNCaP cells overexpressing an empty control vector (LNCaP-EV, Fig. 1a, y-axis). Based on these results, we hypothesized that the degree of suppression of MYC expression by JQ1 may influence sensitivity to JQ1. To test this question, we examined our RNA-seq data from the same cell line panel treated +/−500 nM JQ1 for 24 hours. For each cell line, we plotted MYC expression change with JQ1 (Fig. 1a, x-axis) vs. GR50 (Fig. 1a, y-axis). Importantly, MYC was not repressed by JQ1 in PC3 and DU145 cells, and we observed a significant correlation between MYC mRNA decline by JQ1 and JQ1 sensitivity (Fig. 1a). Higher baseline MYC mRNA expression weakly correlated with JQ1 sensitivity, but was not significant (Fig. S1). To verify lack of MYC suppression by JQ1 in resistant PC3 and DU145 cells as compared to sensitive LNCaP and MR49F cells, we treated cells +/−500 nM JQ1 for 24 hours and then performed western blots to assess MYC protein levels. JQ1 treatment strongly suppressed MYC protein levels in the highly sensitive LNCaP and MR49F lines. However, JQ1 failed to suppress MYC levels in PC3 and DU145 (Fig. 1b). Altogether, our data suggests that reduction of MYC expression is a marker of sensitivity to BET bromodomain inhibition and that persistent MYC expression may confer de novo resistance.
Figure 1.
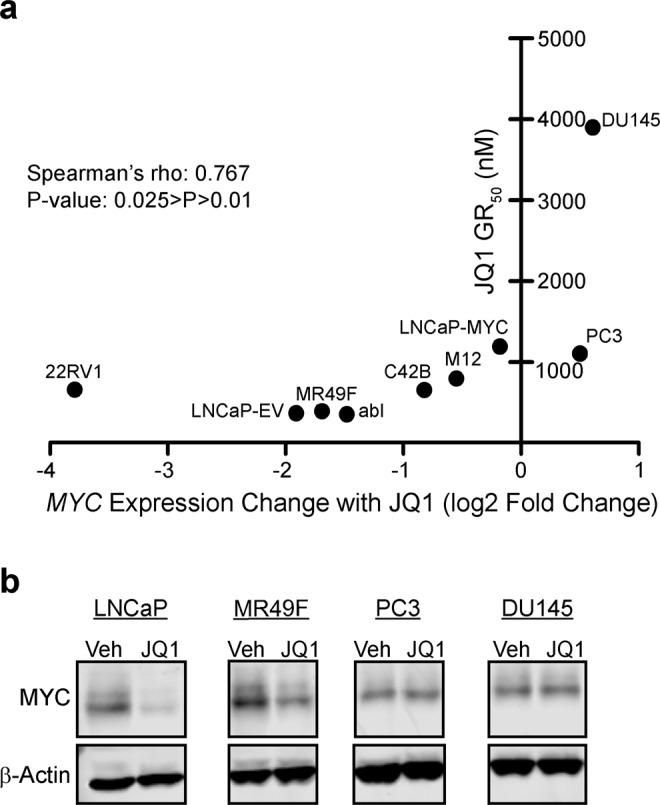
Suppression of MYC expression correlates with JQ1 sensitivity. (a) The indicated cell lines were treated with dose escalation of JQ1 or DMSO vehicle in triplicate. Plotted GR50 values (y-axis) are the mean of at least two independent experiments, except for M12 (see Supplementary Table S1). RNA-seq was performed on the same cell lines (one replicate per condition) treated with either DMSO or 500 nM JQ1. The log2-transformed normalized expression of MYC for each cell line is indicated on the x-axis. A Spearman’s Rank-Order Correlation was performed on the two datasets. (rho = 0.767, P-value is in range of 0.01–0.025 as determined by Spearman’s Rho table). (b) Western blot of MYC protein expression in multiple CRPC cell lines.
Confirmation of the importance of AR-independent effects for the anti-tumor activity of BET bromodomain inhibition
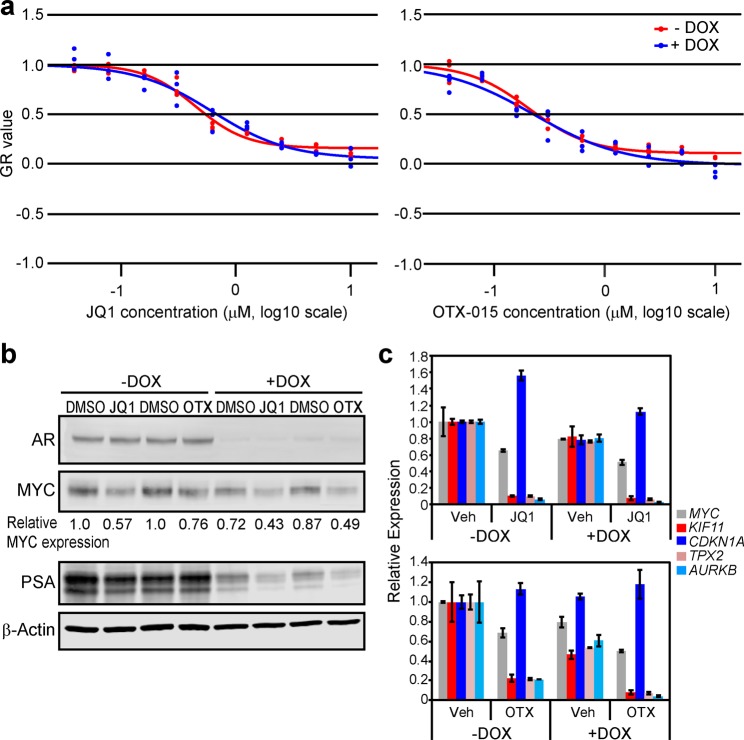
Prior work, including our own, suggested that suppression of AR function is a key mediator of BET bromodomain inhibitor response in CRPC3,4. The cells most resistant to JQ1 in our panel—PC3 and DU145—are AR-independent CRPC cell lines, but our results also suggested that a lack of suppression of MYC expression by JQ1 in these cell lines may be more important. Therefore, to determine the importance of suppression of AR function on BET bromodomain inhibitor response, we used a CRPC cell line (C4-2 cells) engineered to express a doxycycline-inducible AR shRNA13 (C4-2 shAR). We treated the cells +/−doxycycline and subjected them to a dose range of JQ1 or OTX-015, a BET bromodomain inhibitor that has been used clinically (Fig. 2a). We confirmed that doxycycline induced AR knockdown in these cells and blocked AR function as determined by a reduction in PSA (Fig. 2b). By measuring relative growth rate of the cells after 72 h of treatment, we determined that knockdown of AR did not change the sensitivity of the cells to either BET bromodomain inhibitor (Fig. 2a).
Figure 2.
AR suppression does not abrogate the effects of BET bromodomain inhibition. (a) C4-2 shAR cells13 were plated at proper density for logarithmic growth in 96-well plates and treated the next day with dose escalation of JQ1 or OTX-015 [+/−2 µg/mL doxycycline (DOX) to knock down AR expression] in triplicate wells. Relative cell number was measured at time 0 and 72 hours later with the CellTiter-Glo 2.0 Assay Kit (Promega) in order to determine relative growth rate. Individual replicates for each dose are plotted. (b) Western blot of AR, MYC, and PSA protein expression in cells treated +/−DOX as in (a) and with 500 nM JQ1 or 230 nM OTX-015. To quantify MYC protein levels, the intensity of each band as measured by ImageJ was normalized to its corresponding β-Actin control. Relative MYC expression is represented as fold change vs. control (−DOX, DMSO). (c) RT-qPCR of MYC mRNA and MYC target gene expression in samples corresponding to (b). Data is mean of 2 biological replicates. Error bars represent +/−SEM.
We previously demonstrated that AR can regulate MYC expression and function in several CRPC cell lines2. In this current study, using the C4-2 shAR model, we found that BET bromodomain inhibition had a greater effect on reducing MYC protein levels compared to AR knockdown (Fig. 2b). Moreover, BET bromodomain inhibition was equally capable of reducing MYC levels in the AR-expressing or AR-deficient cells (Fig. 2b). We then investigated the effects of BET bromodomain inhibition and/or AR knockdown on MYC function, by measuring expression of MYC target genes identified previously in CRPC2. AR knockdown alone only modestly reduced MYC function, while both bromodomain inhibitors strongly reduced MYC function regardless of AR expression (Fig. 2c). Altogether, our data suggest that suppression of MYC—rather than suppression of AR—may be a key factor that determines BET bromodomain inhibitor sensitivity in CRPC.
BET bromodomain proteins are dispensable for MYC expression in the cell lines most resistant to BET bromodomain inhibition
Because we observed that MYC was not repressed by JQ1 in PC3 and DU145 cells, we next sought to confirm that BET bromodomain proteins were not important for regulating MYC expression in these cells. First,we examined published ChIP-seq data in the VCaP CRPC cell line4, in which MYC expression was shown to decrease with JQ1 treatment4. This analysis demonstrated that BRD4 is enriched at the MYC locus in VCaP cells and that BRD4 binding is reduced by JQ1 (Fig. 3a). The region of BRD4 enrichment is consistent with ChIP-seq data from ENCODE that shows elevated H3K27Ac in a panel of seven cell lines (Fig. 3a). Next, we performed ChIP-seq of BRD4 in PC3 cells treated +/−JQ1. Interestingly, we also observed a significant enrichment of BRD4 at the MYC locus in this cell line, which was also depleted by JQ1 treatment (Fig. 3b). That BRD4 depletion from this locus in PC3 did not reduce MYC expression suggests that other regulatory elements and factors may be more important for MYC expression in PC3 cells.
Figure 3.
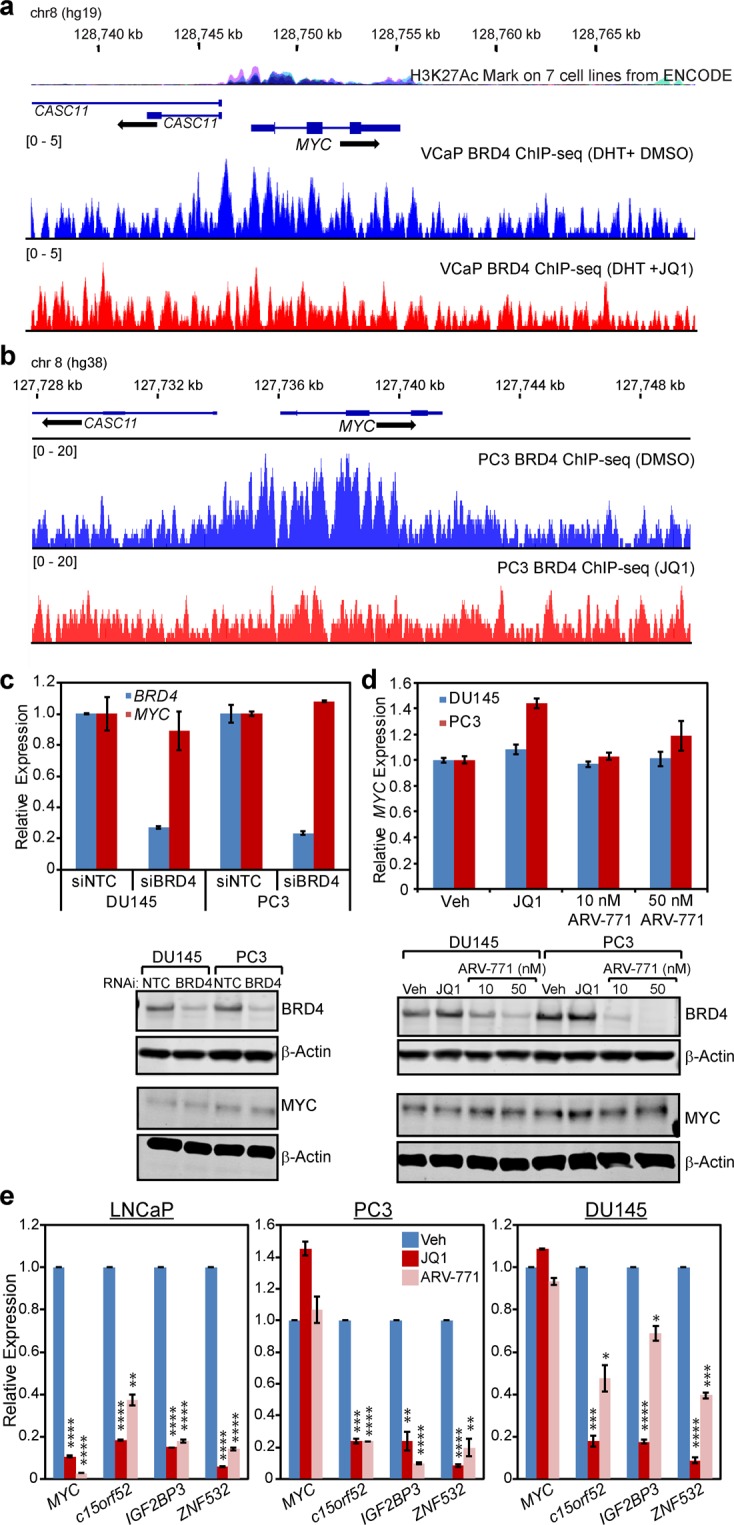
MYC expression is not dependent on BET bromodomain proteins in the cell lines most resistant to BET bromodomain inhibition. (a) ChIP-seq for BRD4 in VCaP cells treated for 12 h with dihydrotestosterone (DHT) plus either vehicle (mock) or 500 nM JQ14. (b) ChIP-seq for BRD4 in PC3 cells treated for 12 h with either vehicle (mock) or 500 nM JQ1. BRD4 binding at the MYC locus is shown. Two independent ChIP-seq experiments were performed for each treatment which yielded comparable results. One representative experiment is shown. (c) RT-qPCR (top) and western blot (bottom) of MYC mRNA and MYC protein expression, respectively, in PC3 and DU145 cells transfected with either control or BRD4 siRNA. Data is mean of 2 biological replicates. Error bars represent +/−SEM. (d) RT-qPCR (top) and western blot (bottom) of MYC mRNA and MYC protein expression, respectively, in PC3 and DU145 cells treated with either JQ1 (500 nM) or the small-molecule pan-BET bromodomain protein degrader ARV-77116. Data is mean of 2 biological replicates. Error bars represent +/−SEM. (e) RT-qPCR of mRNA expression of MYC and several other BET bromodomain target genes in LNCaP, PC3, and DU145 cells treated with vehicle, JQ1 (500 nM) or ARV-771 (50 nM). Data is mean of 2 biological replicates. Error bars represent +/−SEM. *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001 (two-tailed t-test).
JQ1 interferes with binding of the bromodomain region of BET bromodomain protein family members such as BRD4 to chromatin7. However, BET bromodomain proteins also have bromodomain domain-independent functions that contribute to gene expression14,15. Therefore, to determine whether BET bromodomain proteins regulate MYC independently of their bromodomain domain-dependent function, we used RNAi to deplete BRD4 (Fig. 3c) or treated cells with the pan-BET bromodomain protein degrader ARV-77116 (Fig. 3d). Importantly, neither method of BRD4 protein depletion reduced levels of MYC mRNA or MYC protein in either cell line. We verified that ARV-771 was effective in reducing MYC protein levels in LNCaP cells at the same doses (Fig. S2). To confirm on-target effects of JQ1 and ARV-771 in PC3 and DU145 cells versus more sensitive cells (LNCaP), we measured expression of genes from our RNA-seq data that were similarly downregulated by JQ1 in all three cell lines. These genes (c15orf52, IGF2BP3, and ZNF532) were chosen because BRD4 was bound to their promoter regions in our PC3 ChIP-seq, and this binding was depleted by JQ1 (Fig. S3). Expression of c15orf52, IGF2BP3, and ZNF532 were significantly reduced by JQ1 and ARV-771 in LNCaP, PC3, and DU145. In contrast, MYC was only reduced in LNCaP (Fig. 3e). Altogether our data demonstrate that BET bromodomain proteins are not required for transcription of MYC in JQ1-resistant PC3 and DU145 cell lines.
Combination of MYC suppression and JQ1 treatment results in additive anti-tumor activity in the cell lines most resistant to BET bromodomain inhibition
Our data demonstrate that persistent MYC expression confers de novo resistance to BET bromodomain inhibition. Therefore, we next sought to determine if combining MYC suppression with JQ1 treatment would lead to additive anti-tumor activity in cells in which JQ1 failed to suppress MYC. To determine that, we used RNAi to deplete MYC in PC3 and DU145 cells and then treated the cells +/−500 nM JQ1 for 96 hours (Fig. 4). JQ1 treatment or MYC knockdown alone had modest effects on growth rate compared to cells transfected with non-targeted control siRNA and vehicle (Fig. 4). However, combined MYC RNAi + JQ1 treatment led to a significant additive effect on suppressing growth of both cell lines. Importantly, this effect was recapitulated with multiple MYC-targeting siRNAs (Fig. 4). These data demonstrate that maintenance of MYC is not only a marker of resistance to BET bromodomain inhibition but also that co-targeting MYC together with BET bromodomain inhibition may be a useful strategy to overcome de novo resistance.
Figure 4.
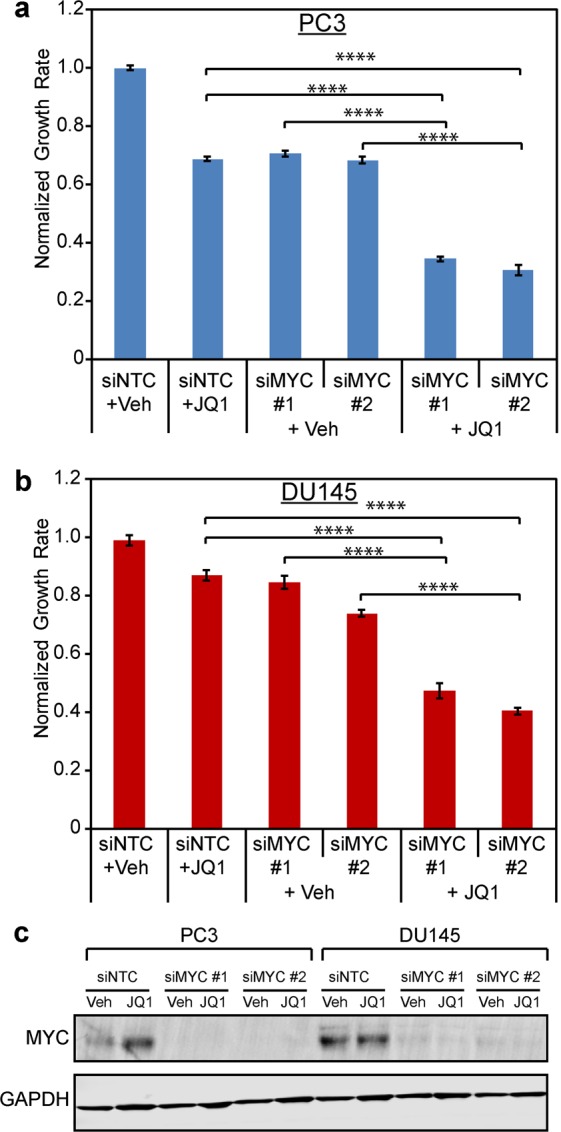
Co-suppression of MYC with BET bromodomain inhibition has additive anti-tumor activity in the cell lines most resistant to BET bromodomain inhibition. (a) PC3 and (b) DU145 cells were transfected with control (siNTC) or MYC siRNAs (siMYC) and treated with 500 nM JQ1 or vehicle. Relative cell number was measured at time 0 and 96 hours later with the CellTiter-Glo 2.0 Assay Kit (Promega). These assays were performed in 10 biological replicates per condition. ****p ≤ 0.0001 (one-way ANOVA with Bonferroni correction). (c) Western blot analysis confirming MYC protein suppression in panels a and b.
Discussion
BET bromodomain inhibition is a promising therapeutic strategy for cancer, and there are several recently completed or ongoing BET bromodomain inhibitor clinical trials, including in prostate cancer (NCT02259114, NCT02711956, NCT02705469, NCT02391480). However, there is a dearth of information on biomarkers of response. Numerous pre-clinical studies in a wide array of cancer types have identified MYC as a key target of BET bromodomain inhibition5,8,17,18, and our results corroborate those findings. Importantly, while JQ1 treatment suppressed MYC mRNA in most CRPC cell lines, JQ1 failed to suppress MYC in the most resistant cell lines. These data suggest that serial measurements of MYC expression in tumor cells may serve as a useful biomarker of response to BET bromodomain inhibition in the clinic.
Suppression of AR function has been previously described as a key mechanism contributing to the anti-tumor activity of BET bromodomain inhibitors in CRPC3,4. Importantly, the two cell lines in our panel that were least sensitive to JQ1 were AR-independent CRPC cell lines (PC3 and DU145). However, it is noteworthy that AR-independent M12 cells had a GR50 similar to the more sensitive AR-expressing lines. Additionally, knockdown of AR in C4-2 shAR cells did not alter their sensitivity to BET bromodomain inhibition. We previously demonstrated that AR has a role in regulating MYC expression2. However, AR knockdown in C4-2 cells did not impact MYC levels as strongly as BET bromodomain inhibition, and BET bromodomain inhibition reduced MYC expression even when AR was absent from these cells. Thus, it is clear that BET bromodomain proteins regulate MYC via AR-independent mechanisms in these CRPC cells. Altogether, our data suggests that targets other than the AR—such as MYC—may be important determinants of response to BET bromodomain inhibition in CRPC. This is especially important since AR-independent forms of prostate cancer may be increasing in frequency19, and a recent report demonstrated that MYC was an essential gene for driving progression of normal basal prostatic epithelial cells to AR-independent, neuroendocrine tumors that have features similar to clinical neuroendocrine prostate cancer20.
We determined that treatment of PC3 cells with JQ1 does displace BRD4 from the chromatin and suppresses expression of several BRD4 target genes, demonstrating target engagement in these cell lines. However, BRD4 eviction was not sufficient to reduce MYC expression. Furthermore, we confirmed that BET bromodomain protein depletion using RNAi or with the pan-BET bromodomain protein degrader ARV-771 was insufficient to reduce expression of MYC in PC3 and DU145 cells. Interestingly, JQ1 treatment increased MYC levels in PC3 cells, which has been observed in this cell line previously4. MYC upregulation as a result of JQ1 treatment has also been observed in cell line models of other cancer types, including lung cancer21. It is possible that BET bromodomain inhibition activates pathways that promote MYC expression or stability in these cell lines. Altogether, our data strongly suggest that BET bromodomain proteins are not required for activation of MYC mRNA expression in BET bromodomain inhibitor resistant CRPC cells such as PC3 and DU145. However, that RNAi-mediated knockdown of MYC combined with JQ1 led to additive anti-tumor activity in PC3 and DU145 cells, demonstrates MYC’s importance to growth of these JQ1-resistant cells.
Altogether, our findings support the utility of MYC as an important pharmacodynamic marker of BET bromodomain inhibitor treatment response in CRPC. Further, our work demonstrates that targeting other signaling pathways that regulate MYC22–24, or targeting MYC, itself, may be a promising approach to overcome de novo BET bromodomain inhibitor resistance and to treat MYC-driven cancers. Historically, MYC has been regarded as “undruggable.” However investigation of MYC-targeting strategies remains an extremely active area of current research25–29. Our results demonstrate the potential application of those agents to overcome de novo resistance to BET bromodomain inhibition in CRPC.
Methods
Cell Culture
LNCaP (clone FGC), 22RV1, PC3, and DU145 cells were purchased from American Type Culture Collection (ATCC, Manassas, VA). C4-2B cells were purchased from ViroMed Laboratories. Abl30 cells were a kind gift from Dr. Zoran Culig (Department of Urology, Innsbruck Medical University). M12 cells31 were a kind gift from Dr. Stephen Plymate (University of Washington Medicine). MR49F32,33 cells were a kind gift from Dr. Martin Gleave (Vancouver Prostate Centre and Dept. of Urologic Sciences, University of British Columbia). To generate stable MYC overexpressing cells, we transfected LNCaP-FGC cells with pcDNA3-2XFLAG-cMYC plasmid34 (a kind gift from Dr. Mu-Shui Dai, OHSU Molecular & Medical Genetics) or empty pcDNA3 vector with Lipofectamine 2000 (Life Technologies) and selected with 300 µg/mL G418 (Sigma-Aldrich). C4-2 shAR cells13 were a kind gift from Dr. Paul Rennie (Vancouver Prostate Centre and Dept. of Urologic Sciences, University of British Columbia). LNCaP (FGC parental, EV, and MYC), 22RV1, C42B, PC3, DU145, MR49F, and C4-2 shAR cells were grown in RPMI1640 + 10% FBS (Premium Select grade, Atlanta Biologicals). Enzalutamide-resistant MR49F cells were cultivated in media containing 10 µM Enzalutamide (MedchemExpress #HY-70002). C4-2 shAR cells were supplemented with 1 µg/mL puromycin (Sigma-Aldrich #P8833) and 1.25 µg/mL blasticidin (Sigma-Aldrich #15205) to maintain integration of the stable shRNA construct. These cells were cultured without antibiotic selection for all experiments. Abl cells were grown in RPMI with 10% charcoal/dextran-treated FBS (Atlanta Biologicals). M12 cells were grown in RPMI 1640 + 5% FBS, 0.01 µM dexamethasone (MP Biomedicals #ICN19456125), 10 ng/ml EGF (Corning #CB40052), and 10 mL/L ITS (Corning #25-800-CR). LNCaP-FGC, 22RV1, C42B, PC3, and DU145 cells were authenticated by Short Tandem Repeat (STR) profiling (DDC Medical). Other cell lines were not authenticated due to a lack of published ATCC STR profile. Mycoplasma testing was regularly performed on cells using a DNA-based PCR test35 and were found to be negative in all cases.
Dose-response curve experiments
Cells were plated at proper density for logarithmic growth in 96-well plates and treated the next day with serial dilutions of JQ1 (BPS Bioscience #27402) or OTX-015 (Selleck S7360), in addition to DMSO vehicle, in triplicate wells. Cell viability was measured at time 0 and 72 hours post-treatment using the CellTiter-Glo 2.0 (CTG) assay (Promega) per manufacturer’s instructions. Nonlinear regression curves were fitted and GR50 values were calculated using the GRMetrics package in R36.
RNA-seq library preparation and data processing
All cell lines were treated with 500 nM JQ1, and cells were harvested after 24 hours. Total RNA was extracted with Trizol/CHCl3 (Life Technologies). Library preparation for RNA-seq was performed as previously described3. Libraries were sequenced on an Illumina HiSeq as single-end 50 bp reads. RNA-seq data analysis was performed using the Tuxedo Suite37–39. Each sample was mapped independently to the human genome build GRCh37/hg19 using TopHat v2.0.937,38. Transcript assembly and quantification was done with HTSeq v0.6.140 to generate a gene-level read count value.
Drug treatments
For all in vitro experiments using JQ1 (excluding dose-response curves), DMSO stock of 1 mM JQ1 was diluted 1:2000 to 500 nM in cell culture media. For in vitro experiments using OTX-015 (excluding dose-response curves), DMSO stock of 230 µM OTX-015 was diluted 1:1000 to 230 nM in cell culture media.100 µM and 20 µM DMSO stocks of ARV-771 (Arvinas, LLC)16 were diluted 1:2000 in cell culture media to 50 nM and 10 nM, respectively. Vehicle-only controls were used for all experiments, and vehicle concentrations were normalized across treatments for each experiment.
RNAi experiments
Transient knockdowns were performed using the following siRNAs: non-targeted control (custom synthesis, 5′-CGUACGCGGAAUACUUCGAdTdT-3′), BRD4 (Dharmacon L-004937-00), MYC #1 (Ambion #4392421), MYC #2 (Dharmacon #J003282-26). Cells were transfected with DharmaFECT 3 (GE Dharmacon) transfection reagent. siRNA was transfected at a concentration of 15 nM for experiments depicted in Fig. 4, and 50 nM for experiments depicted in Fig. 3c. Cells used for RNA and protein harvest were seeded/transfected in either 12 or 6-well plates. Cells used in viability assays were seeded/transfected in 96-well plates. Cell viability was measured at time 0 and endpoint using the CTG assay described above, these values were used to calculate relative growth rate as described previously36.
RNA preparation and RT-qPCR
We performed RNA preparation and RT-qPCR as described previously3. Briefly, RNA was extracted as described above and reverse-transcribed into cDNA. Quantitative reverse transcription PCR (RT-qPCR) was performed using TaqMan probes/reagents and a QuantStudio 3 thermocycler (Thermo Fisher). TaqMan probes were used to detect human BRD4 (Hs04188087_m1), MYC (Hs00153408_m1), c15orf52 (Hs05010185_g1), IGF2BP3 (Hs00559907_g1), KIF11 (Hs00189698_m1), CDKN1A (Hs00355782_m1), TPX2 (Hs00201616_m1), AURKB (Hs00177782_m1), and ZNF532 (Hs01557225_g1). Human UBC TaqMan probe (Hs00824723_m1) was used as an endogenous control. Ct data was analyzed with QuantStudio Design & Analysis Software 1.3.1 and DataAssist Software v3.0 (Thermo Fisher).
Immunoblotting
Immunoblotting experiments were performed by running protein lysates on NuPAGE Protein gels (Thermo Fisher) and transferring onto PVDF membranes. 4–12% Bis-Tris gels were used for data in Figs 1b and 2b, 3–8% Tris-Acetate gels were used for Figs 3c,d, and S2. Antibodies used were: MYC (Abcam #ab32072), β-actin (Sigma-Aldrich #A5441), AR (Millipore 06-680), PSA (Abcam #ab53774), BRD4 (Bethyl #A301-985A) and GAPDH (Santa Cruz #sc-32233). Blots were imaged using an Odyssey imaging system (LI-COR) according to the manufacturer’s instructions.
Chromatin immunoprecipitation (ChIP)
ENCODE data for H3K27Ac depicted in Fig. 3a was obtained as a publicly-available track on the UCSC Genome Browser. BRD4 ChIP-seq data depicted in Fig. 3a is from a publically-available dataset originally described previously4. ChIP-seq data depicted in Figs 3b and S3 were generated by our group using the following methods: ChIP experiments were performed with formaldehyde cross-linked cells using anti-BRD4 (Bethyl #A301-985A) or normal rabbit IgG (Millipore #12-370) antibodies. Input and ChIP samples were processed using Diagenode iDeal ChIP-seq for Transcription Factors Kit. Library preparation for ChIP-seq samples (BRD4 ChIP and input, two biological replicates per treatment) was performed using the Illumina TruSeq ChIP Sample Prep Kit, using 10 ng of input material. The samples were then sequenced using an Illumina NextSeq. 500 sequencing system. Sequences were trimmed to 100 bp, and sequencing quality was confirmed using the FastQC utility41. FASTQ files were aligned to the full Ensembl hg38 human genome release 8542 using the bwa utility with the bwa-mem algorithm43, and subsequently converted to sorted.bam files using the samtools view and samtools sort utilities44. The sequencing quality was further confirmed at this point using samtools stats44. For later analysis, output.bam files were translated to sorted.bed files using bedtools45. Peaks were called using the epic peak caller46. Epic is a reimplementation of the SICER algorithm47 which was designed to capture broad peaks associated with histone marks. For peaks called in Fig. S3, the JQ1-treated BRD4 ChIP served as the negative control in the analysis. This method allowed us to perform differential analysis by determining regions in which BRD4 is enriched in the mock-treated cells and depleted by JQ1.
Statistical Methods
P-values for experiments depicted in Figs 1a and S1 were determined using the Spearman Rank-Order Correlation method and Spearman’s Rho table. P-values depicted in Fig. 3e were determined using a 2-tailed t-test. P-values depicted in Fig. 4a,b were determined using a one-way ANOVA with Bonferroni correction. Values for n are indicated in the corresponding figure legends for these data.
Data Availability
RNA-seq and ChIP-seq data depicted in Figs 1a and 3b and Supplementary Figs S1 and S3 were generated by our group and deposited to the Gene Expression Omnibus (GEO) with accession number GSE98069. ChIP-seq data depicted in Fig. 3a is from a publically-available dataset originally described previously4. BigWig (.bw) tracks were obtained via GEO, accession GSE55062.
Supplementary information
Supplementary Material (Figures S1-S3, full blot images)
Supplementary Table S1 - dose-response curve metrics
Acknowledgements
The authors wish to thank other members of the Alumkal and Heiser labs for critical feedback and review. We also thank Sunil Joshi for technical assistance with western blotting, and the OHSU Massively Parallel Sequencing Shared Resource for performing short-read sequencing assays. We also thank the following funding sources: (1) National Institutes of Health (NIH)/National Cancer Institute (NCI) R01 award CA178610, (2) Pacific Northwest Prostate Cancer SPORE/NCI (P50 CA097186), (3) Cancer Center Support Grant (CCSG) (P30 CA069533), (4) Oregon Clinical and Translational Research Institute (OCTRI) (UL1TR000128) from the National Center for Advancing Translational Sciences (NCATS), a component of the NIH; (5) Department of Defense Synergistic Idea Award (W81XWH-13-1-0420), (6) Department of Defense Impact Awards (W81XWH-16-1-0597, W81XWH-16-1-1601), (7) Hope Foundation Award, (8) Stand Up to Cancer - Prostate Cancer Foundation - Prostate Dream Team Translational Cancer Research Grant (SU2C-AACR-DT0409), this research grant is made possible by the generous support of the Movember Foundation (Stand Up To Cancer is a program of the Entertainment Industry Foundation administered by the American Association for Cancer Research (7465sc)); (9) Prospect Creek Foundation, and (10) Wayne D. Kuni and Joan E. Kuni Foundation.
Author Contributions
D.J.C., L.G., J.S., L.M.H. and J.J.A. designed the research; D.J.C., L.G., J.S., J.E.K., D.S. and J.U. performed the experiments; D.J.C., J.S., J.E.K., D.S.D., A.B., J.B., C.J.K., K.E.C., J.U., L.M.H. and J.J.A. analyzed the data; D.J.C. and J.J.A. wrote the paper.
Competing Interests
Dr. Alumkal has performed consulting or held an advisory role with Astellas Pharma, Bayer, and Janssen Biotech, Inc. OHSU has received institutional research funding from Aragon Pharmaceuticals Inc., Astellas Pharma, Norvartis, Zenith Epigenetics Ltd, and Gilead Sciences Inc. These interests had no role in the design or conduct of this study.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Laura M. Heiser, Email: heiserl@ohsu.edu
Joshi J. Alumkal, Email: alumkalj@ohsu.edu
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-40518-5.
References
- 1.Cancer Facts and Figures 2018. (American Cancer Society, 2018).
- 2.Gao L, et al. Androgen receptor promotes ligand-independent prostate cancer progression through c-Myc upregulation. PLoS One. 2013;8:e63563. doi: 10.1371/journal.pone.0063563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coleman, D. J. et al. Cellular androgen content influences enzalutamide agonism of F877L mutant androgen receptor. Oncotarget, 10.18632/oncotarget.9816 (2016). [DOI] [PMC free article] [PubMed]
- 4.Asangani, I. A. et al. Therapeutic targeting of BET bromodomain proteins in castration-resistant prostate cancer. Nature, 10.1038/nature13229 (2014). [DOI] [PMC free article] [PubMed]
- 5.Delmore JE, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904–917. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rahman S, et al. The Brd4 extraterminal domain confers transcription activation independent of pTEFb by recruiting multiple proteins, including NSD3. Mol Cell Biol. 2011;31:2641–2652. doi: 10.1128/MCB.01341-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Filippakopoulos P, et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mertz JA, et al. Targeting MYC dependence in cancer by inhibiting BET bromodomains. Proceedings of the National Academy of Sciences. 2011;108:16669–16674. doi: 10.1073/pnas.1108190108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shu, S. et al. Response and resistance to BET bromodomain inhibitors in triple-negative breast cancer. Nature (2016). [DOI] [PMC free article] [PubMed]
- 10.Fong CY, et al. BET inhibitor resistance emerges from leukaemia stem cells. Nature. 2015;525:538–542. doi: 10.1038/nature14888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rathert P, et al. Transcriptional plasticity promotes primary and acquired resistance to BET inhibition. Nature. 2015;525:543–547. doi: 10.1038/nature14898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kumar K, et al. GLI2-dependent c-MYC upregulation mediates resistance of pancreatic cancer cells to the BET bromodomain inhibitor JQ1. Scientific reports. 2015;5:9489. doi: 10.1038/srep09489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Snoek R, et al. In vivo knockdown of the androgen receptor results in growth inhibition and regression of well-established, castration-resistant prostate tumors. Clinical cancer research: an official journal of the American Association for Cancer Research. 2009;15:39–47. doi: 10.1158/1078-0432.CCR-08-1726. [DOI] [PubMed] [Google Scholar]
- 14.Wu S-Y, Lee A-Y, Lai H-T, Zhang H, Chiang C-M. Phospho switch triggers Brd4 chromatin binding and activator recruitment for gene-specific targeting. Molecular cell. 2013;49:843–857. doi: 10.1016/j.molcel.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Winter GE, et al. BET bromodomain proteins function as master transcription elongation factors independent of CDK9 recruitment. Molecular cell. 2017;67:5–18. e19. doi: 10.1016/j.molcel.2017.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raina K, et al. PROTAC-induced BET protein degradation as a therapy for castration-resistant prostate cancer. Proceedings of the National Academy of Sciences. 2016;113:7124–7129. doi: 10.1073/pnas.1521738113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bandopadhayay, P. et al. BET-bromodomain inhibition of MYC-amplified Medulloblastoma. Clinical Cancer Research, clincanres. 2281.2013 (2013). [DOI] [PMC free article] [PubMed]
- 18.Shao Q, et al. BET protein inhibitor JQ1 attenuates Myc-amplified MCC tumor growth in vivo. Cancer research. 2014;74:7090–7102. doi: 10.1158/0008-5472.CAN-14-0305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aggarwal, R. R. et al. (American Society of Clinical Oncology, 2018).
- 20.Park JW, et al. Reprogramming normal human epithelial tissues to a common, lethal neuroendocrine cancer lineage. Science. 2018;362:91–95. doi: 10.1126/science.aat5749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klingbeil O, Lesche R, Gelato KA, Haendler B, Lejeune P. Inhibition of BET bromodomain-dependent XIAP and FLIP expression sensitizes KRAS-mutated NSCLC to pro-apoptotic agents. Cell death & disease. 2016;7:e2365. doi: 10.1038/cddis.2016.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yashiro-Ohtani Y, et al. Long-range enhancer activity determines Myc sensitivity to Notch inhibitors in T cell leukemia. Proceedings of the National Academy of Sciences. 2014;111:E4946–E4953. doi: 10.1073/pnas.1407079111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Palomero T, et al. NOTCH1 directly regulates c-MYC and activates a feed-forward-loop transcriptional network promoting leukemic cell growth. Proceedings of the National Academy of Sciences. 2006;103:18261–18266. doi: 10.1073/pnas.0606108103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaidi A, Williams AC, Paraskeva C. Interaction between β-catenin and HIF-1 promotes cellular adaptation to hypoxia. Nature cell biology. 2007;9:210. doi: 10.1038/ncb1534. [DOI] [PubMed] [Google Scholar]
- 25.Koehler AN. A complex task? Direct modulation of transcription factors with small molecules. Current opinion in chemical biology. 2010;14:331–340. doi: 10.1016/j.cbpa.2010.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McKeown MR, Bradner JE. Therapeutic strategies to inhibit MYC. Cold Spring Harbor perspectives in medicine. 2014;4:a014266. doi: 10.1101/cshperspect.a014266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen H, Liu H, Qing G. Targeting oncogenic Myc as a strategy for cancer treatment. Signal transduction and targeted therapy. 2018;3:5. doi: 10.1038/s41392-018-0008-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fletcher S, Prochownik EV. Small-molecule inhibitors of the Myc oncoprotein. Biochimica Et Biophysica Acta (BBA)-Gene Regulatory Mechanisms. 2015;1849:525–543. doi: 10.1016/j.bbagrm.2014.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Whitfield JR, Beaulieu M-E, Soucek L. Strategies to inhibit Myc and their clinical applicability. Frontiers in cell and developmental biology. 2017;5:10. doi: 10.3389/fcell.2017.00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Culig Z, et al. Switch from antagonist to agonist of the androgen receptor bicalutamide is associated with prostate tumour progression in a new model system. British journal of cancer. 1999;81:242–251. doi: 10.1038/sj.bjc.6690684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun S, et al. Castration resistance in human prostate cancer is conferred by a frequently occurring androgen receptor splice variant. The Journal of clinical investigation. 2010;120:2715–2730. doi: 10.1172/JCI41824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bishop JL, et al. PD-L1 is highly expressed in Enzalutamide resistant prostate cancer. Oncotarget. 2015;6:234–242. doi: 10.18632/oncotarget.2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuruma H, et al. A novel antiandrogen, Compound 30, suppresses castration-resistant and MDV3100-resistant prostate cancer growth in vitro and in vivo. Molecular cancer therapeutics. 2013;12:567–576. doi: 10.1158/1535-7163.MCT-12-0798. [DOI] [PubMed] [Google Scholar]
- 34.Dai MS, Arnold H, Sun XX, Sears R, Lu H. Inhibition of c-Myc activity by ribosomal protein L11. The EMBO journal. 2007;26:3332–3345. doi: 10.1038/sj.emboj.7601776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Uphoff CC, Drexler HG. Detecting mycoplasma contamination in cell cultures by polymerase chain reaction. Methods Mol Biol. 2011;731:93–103. doi: 10.1007/978-1-61779-080-5_8. [DOI] [PubMed] [Google Scholar]
- 36.Hafner M, Niepel M, Chung M, Sorger PK. Growth rate inhibition metrics correct for confounders in measuring sensitivity to cancer drugs. Nature methods. 2016;13:521–527. doi: 10.1038/nmeth.3853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Trapnell C, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nature protocols. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nature biotechnology. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Anders S, Pyl PT, Huber W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Andrews, S. In FastQC: a quality control tool for high throughput sequence data (2010).
- 42.Aken BL, et al. The Ensembl gene annotation system. Database. 2016;2016:baw093. doi: 10.1093/database/baw093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Samtools, http://www.htslib.org/ (2017).
- 45.bedtools: a powerful toolset for genome arithmetic, http://bedtools.readthedocs.io/en/latest/ (2017).
- 46.epic: diffuse domain ChIP-Seq caller based on SICER, https://github.com/biocore-ntnu/epic (2017).
- 47.Xu, S., Grullon, S., Ge, K. & Peng, W. Spatial clustering for identification of ChIP-enriched regions (SICER) to map regions of histone methylation patterns in embryonic stem cells. Stem Cell Transcriptional Networks: Methods and Protocols, 97–111 (2014). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material (Figures S1-S3, full blot images)
Supplementary Table S1 - dose-response curve metrics
Data Availability Statement
RNA-seq and ChIP-seq data depicted in Figs 1a and 3b and Supplementary Figs S1 and S3 were generated by our group and deposited to the Gene Expression Omnibus (GEO) with accession number GSE98069. ChIP-seq data depicted in Fig. 3a is from a publically-available dataset originally described previously4. BigWig (.bw) tracks were obtained via GEO, accession GSE55062.