Abstract
Hand, foot and mouth disease (HFMD) is responsible for a heavy economic and social burden in the Asia-Pacific region. Previous studies have shown that coxsackievirus A6 (CVA6) and coxsackievirus A10 (CVA10) have become the predominant agents of HFMD in mainland China in recent years, replacing enterovirus 71 (EV71) and coxsackievirus A16 (CVA16), although it is unclear if this is consistent throughout China. In this study, samples from 253 HFMD cases were collected in Shenzhen, China, from May 2013 through April 2014 to identify the etiological agent of HFMD. In total, 64.8% (164/253) of HFMD cases were enterovirus positive, in which 81.1% (133/164) were determined to be CVA6. The phylogenetic tree of the partial viral protein 1 sequence showed that the CVA6 isolates were divided into four clusters (Clusters A to D), and cluster D was further divided into four sub-clusters (Clusters D1 to D4). The 133 CVA6 samples isolated in our study were classified into cluster D4, in which the first identified sequence was isolated in Shenzhen in 2008. This study demonstrated that the CVA6 cluster D4, which is predominantly circulating in HFMD in mainland China, may have originated from a local strain identified in 2008 in Shenzhen.
Introduction
Hand, foot and mouth disease (HFMD) is a common communicable disease in children under 5 years old. Typical manifestations of HFMD include fever, vesicular rash in hand, foot, and mouth1. HFMD outbreaks have been reported in Japan, Australia, Malaysia, Singapore, Vietnam, Hong Kong, Taiwan, and mainland China since the 1970 s2-11. As a result, it has become a public health concern with a heavy economic burden across the world12. In 2008, a large outbreak of HFMD occurred in China that involved 6,049 clinically diagnosed cases including 22 deaths9.
HFMD cases are mostly mild and self-limited. However, it is difficult to distinguish severe and mild cases at an early stage, since very few cases have obvious symptoms. Previous studies reported that severe HFMD cases are mostly caused by enterovirus 71 (EV71), which has been proven to be associated with severe manifestations, such as pulmonary edema, brain stem encephalitis, acute flaccid paralysis (AFP), meningoencephalitis, aseptic meningitis, and cerebellitis11,13–16. Therefore, it is crucial to determine the serotype of enterovirus to identify potentially severe HFMD cases.
EV71 and coxsackievirus A16 (CVA16) had been confirmed to be the most common agents of HFMD in China when HFMD surveillance initiated in 200816–19. However, the most prevalent serotypes of enterovirus have shifted to coxsackievirus A6 (CVA6) and A10 (CVA10) in China starting in 201220,21. Additionally, CVA6 and CVA10 have been responsible for HFMD outbreaks in Finland (2008) and France (2010)22,23. These studies indicate that the epidemic trend of enterovirus may be shifting. Therefore, from May 2013 through April 2014, we conducted a study in a tertiary maternal and child hospital in Shenzhen, China to determine the serotype distribution of enterovirus in HFMD cases and to further study the phylogenetics of CVA6.
Results
HFMD cases
A total of 253 HFMD cases were included in our study, in which 83.8% (212/253) were diagnosed between June and September 2013. There were 161 male and 92 female cases, with a male-to-female ratio of 1.75. The median age of cases was 1 year old, ranging from 4 months to 23 years old. The majority (98.4%, 249/253) were under 6 years old, except one 14-year-old teenager and one 23-year-old nursing mother.
Clinical presentation
The typical clinical presentation included fever, rash and vesicles. In total, 81.0% (205/253) of the cases had a fever and 22.5% (57/253) had a high fever (≥39 °C). Rash was intraoral (48.6%, 123/253), on the limbs (99.6%, 252/253), on the buttocks (67.2%, 170/253), on the trunk (24.1%, 61/253), and on the head and face (7.9%, 20/253). The majority of the cases (96.8%, 245/253) had a rash accompanied by vesicles. Additionally, a total of 13 cases reported skin itchiness (Table 1).
Table 1.
Demographics and manifestations of the 253 HFMD cases included in the study.
| CVA6 | EV71 | CA16 | other enterovirus | non-enterovirus | |
|---|---|---|---|---|---|
| Cases (%) | 133 (52.6) | 11 (4.3) | 13 (5.1) | 7 (2.8) | 89 (35.2) |
| Male/female ratio | 1.9 (87/46) | 1.8 (7/4) | 1.6 (8/5) | 1.3 (4/3) | 1.6 (55/34) |
| Age (year), mean ± SD | 1.47 ± 1.16a | 2.91 ± 1.30 | 1.54 ± 1.17 | 2.42 ± 1.63 | 1.59 ± 1.10 |
| Fever ( ≥ 39 °C) (%) | 28.6 (38/133) | 0.0 (0/11) | 0.0 (0/13) | 14.3 (1/7) | 20.2 (18/89) |
| Intraoral rash (%) | 48.9 (65/133) | 36.4 (4/11) | 53.8 (7/13) | 71.4 (5/7) | 47.2 (42/89) |
| Rash on limbs (%) | 99.2 (132b/133) | 100.0 (11/11) | 100.0 (13/13) | 100.0 (7/7) | 100.0 (89/89) |
| Rash on trunk (%) | 27.8 (37/133) | 9.1 (1/11) | 0.0 (0/13) | 0.0 (0/7) | 25.8 (23/89) |
| Rash on buttocks (%) | 71.4 (95/133) | 72.7 (8/11) | 92.3 (12/13) | 57.1 (4/7) | 57.3 (51/89) |
| Rash on head and face (%) | 9.8 (13/133) | 0.0 (0/11) | 0.0 (0/13) | 14.3 (1/7) | 6.7 (6/89) |
| Vesicles (%) | 97.8 (130/133) | 100.0 (11/11) | 100.0 (13/13) | 100.0 (7/7) | 94.4 (84/89) |
| Skin itchiness (%) | 7.5 (10/133) | 0.0 (0/11) | 0.0 (0/13) | 0.0 (0/7) | 3.4 (3/89) |
aTwo older HFMD cases (14-year-old and 23-year-old) were excluded.
bOne case had an intraoral rash and rash on trunk and buttocks.
Enterovirus serotyping
Overall, 64.8% (164/253) of the HFMD cases were enterovirus positive. Among male cases, it was 65.8% (106/161), compared to 63.0% (58/92) in female cases (P = 0.61) (Table 2). All the cases were divided into five groups by age. There were only 4 cases in those ≥6 years old. In the other four age groups, the prevalence of enterovirus varied between 53.8% and 67.5% (P = 0.46) (Table 2).
Table 2.
Enterovirus serotypes by sex and age.
| No. cases | No. enterovirus-positive cases (%) | No. enterovirus serotypes (%) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| CVA6a | EV71b | CVA16 | CVA9 | CVA10 | CVB2 | Undeterminedc | |||
| Sex | |||||||||
| male | 161 | 106 (65.8) | 87 (82.1) | 7 (6.6) | 8 (7.6) | 1 (0.9) | 2 (1.9) | 1 (0.9) | 0 |
| female | 92 | 58 (63.0) | 46 (79.3) | 4 (6.9) | 5 (8.6) | 0 | 0 | 0 | 3 (5.2) |
| Age (year) | |||||||||
| <1 | 53 | 34 (64.2) | 30 (88.2) | 0 | 3 (8.8) | 0 | 1 (3.0) | 0 | 0 |
| 1 | 114 | 77 (67.5) | 67 (87.0) | 2 (2.6) | 6 (7.8) | 0 | 1 (1.3) | 1 (1.3) | 0 |
| 2 | 39 | 21 (53.8) | 16 (76.2) | 2 (9.5) | 2 (9.5) | 1 (4.8) | 0 | 0 | 0 |
| 3–5 | 43 | 29 (67.4) | 17 (58.6) | 7 (24.1) | 2 (6.9) | 0 | 0 | 0 | 3 (10.4) |
| ≥6 | 4 | 3 (75.0) | 3 (100.0) | 0 | 0 | 0 | 0 | 0 | 0 |
aCases under 3 years old vs. cases 3–5 years old, P = 0.001.
bCases under 3 years old vs. cases 3–5 years old, P = 0.001.
cUndetermined serotype by VP1 nested PCR.
Of the 164 enterovirus positive cases, 161 (98.2%) were determined based on an examination of a 336-nt partial viral protein 1 (VP1) sequence, whereas the other 3 cases were undetermined (Table 3). A total of six enterovirus serotypes were determined in our study. CVA6 was the predominant serotype, followed by CVA16 and EV71 (Table 3).
Table 3.
Enterovirus serotypes by calendar time.
| No. enterovirus-positive cases | No. enterovirus serotypes (%) | |||||||
|---|---|---|---|---|---|---|---|---|
| CVA6 | EV71 | CVA16 | CVA9 | CVA10 | CVB2 | Undetermined | ||
| May, 2013 | 9 | 6 (66.7) | 1 (11.1) | 0 | 1 (11.1) | 0 | 1 (11.1) | 0 |
| Jun, 2013 | 14 | 8 (57.1) | 1 (7.1) | 2 (14.3) | 0 | 1 (7.1) | 0 | 2 (14.3) |
| Jul, 2013 | 39 | 36 (92.3) | 1 (25.6) | 2 (51.3) | 0 | 0 | 0 | 0 |
| Aug, 2013 | 43 | 42 (97.7) | 0 | 0 | 0 | 1 (2.3) | 0 | 0 |
| Sep, 2013 | 37 | 36 (97.3) | 1 (2.7) | 0 | 0 | 0 | 0 | 0 |
| Oct, 2013 | 6 | 4 (66.7) | 0 | 2 (33.3) | 0 | 0 | 0 | 0 |
| Nov, 2013 | 1 | 1 (100.0) | 0 | 0 | 0 | 0 | 0 | 0 |
| Dec, 2013 | 1 | 0 | 0 | 1 (100.0) | 0 | 0 | 0 | 0 |
| Mar, 2014 | 8 | 0 | 5 (62.5) | 3 (37.5) | 0 | 0 | 0 | 0 |
| Apr, 2014 | 6 | 0 | 2 (33.3) | 3 (50.0) | 0 | 0 | 0 | 1 (16.7) |
| Total | 164 | 133 (81.1) | 11 (67.1) | 13 (7.9) | 1 (0.6) | 2 (12.2) | 1 (0.6) | 3 (1.8) |
The prevalence of CVA6 in the male cases (82.1%, 87/106) and female cases (79.3%, 46/58) was similar (P = 0.67) (Table 2). Furthermore, CVA6 was also predominant across all five age groups. However, it was significantly higher among cases under 3 years old (85.6%, 113/132) than among cases 3–5 years old (58.6%, 17/29) (P = 0.001). Additionally, a significantly higher prevalence of EV71 was observed among cases 3–5 years old (24.1%, 7/22), compared to cases under 3 years old (3.0%, 4/132) (P = 0.001).
Phylogenetics of CVA6
A total of 133 CVA6 366-nt partial VP1 sequences isolated in our study and 1212 sequences retrieved from GenBank were combined together for further phylogenetic analysis. The phylogenetic tree showed that CVA6 was divided into four clusters (A to D). Cluster A contained only one strain isolated in the USA in 1949, which could be the origin of HFMD. Several strains isolated between 1992 and 2007 in mainland China formed cluster B. Similarly, cluster C contained only strains isolated in mainland China, India, and the USA during 1996–2009. Cluster D was the largest one that was further divided into four sub-clusters (D1 to D4), suggesting gradual expansion and dissemination of HFMD in recent years, especially in mainland China. All of the 133 CVA6 sequences obtained in our study were classified into cluster D4 (Fig. 1 and Fig. S1).
Figure 1.
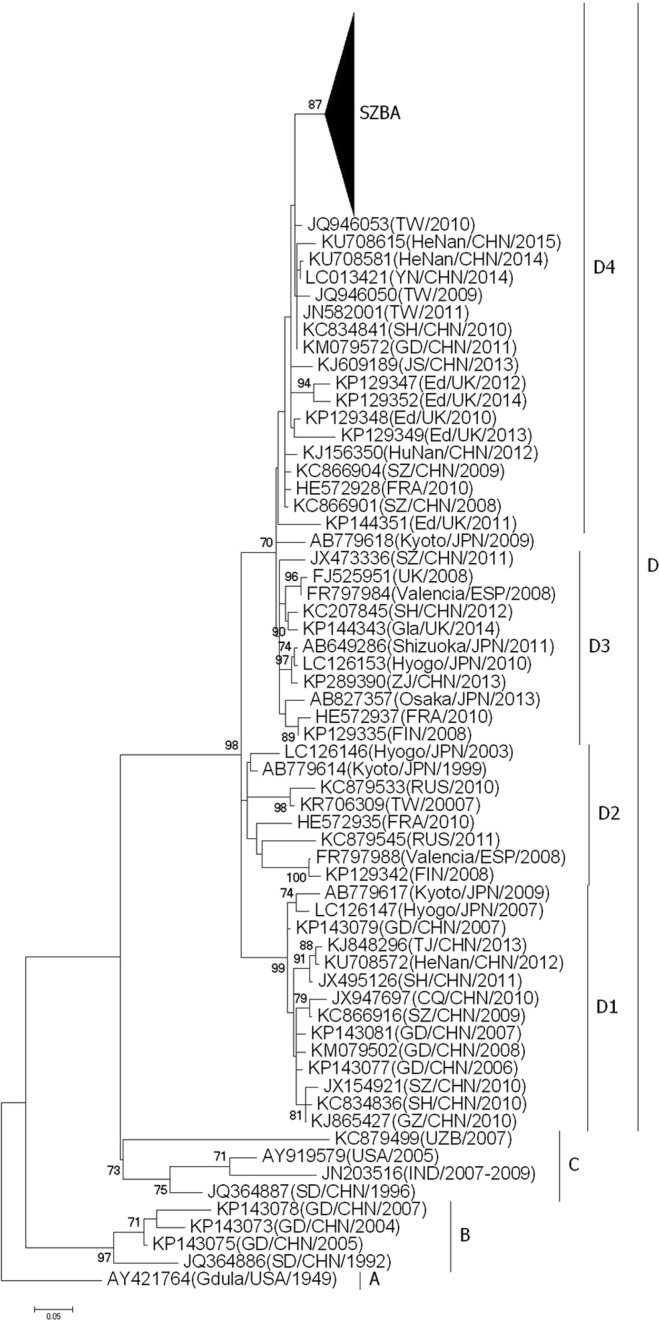
Phylogenetic tree reconstructed based on a 336-nt CVA6 partial VP1 sequence. The tree was reconstructed using the maximum likelihood method (bootstrap of 1000 replications) in MEGA 6.0. SZBA: a total of 133 CVA6 partial sequences isolated in our study.
Discussion
HFMD has a typical seasonality that is associated with meteorological factors, such as temperature, humidity, sunshine, and air pressure16,24,25. The peak of HFMD incidence usually occurs in summer. However, prevalent seasons of HFMD in Shenzhen are both summer and fall. Additionally, it is basically active throughout the year in Shenzhen, which is consistent with seasonal patterns reported in Hong Kong26, Beijing27 and Thailand28.
The clinical presentation of HFMD cases caused by CVA6 was fever and rash on limbs, buttocks and intraoral areas, and 97.8% cases in this study had vesicles. Additionally, CVA6 cases were more likely to have fever (≥39 °C) and rash on trunk, head and face, compared to cases with EV71 and CVA16, as has been previously reported20,22,28,29.
In our study, 64.8% of the HFMD cases were enterovirus positive, which were determined by using a semi-nested PCR serotyping method30. The findings showed that CVA6 was the predominant serotype, accounting for approximately 81% of enterovirus positive cases, which was much higher than the prevalence of EV71 (7.9%) and CVA16 (6.7%). CVA6 was first reported to cause HFMD outbreaks in Finland in fall 200831, with following reports in Singapore in 20088, France22 and Taiwan32 in 2010, USA33, Spain34, Japan35, and Cuba36 in 2011, and Thailand28 in 2012. In mainland China, CVA6 was a minor contributor to HFMD for a long time; however, a dramatic increase of this serotype was found in HFMD cases since fall 201220,37. Our study has also illustrated that enterovirus serotypes in Shenzhen had shifted by 2013; CVA6 had taken the place of both EV71 and CVA16, and had become the predominant serotype in HFMD cases, which confirmed previous studies21,37,38. He et al. reported the variation of enterovirus serotypes from 2008 through 2012 in Shenzhen and suggested that CVA6 and CVA10 had gradually become the most common enterovirus serotype in Shenzhen starting in 201221, and Lu et al. reported that CVA6 was a new epidemic trend in Guangdong province, where Shenzhen is located, as of late 201237.
The majority (98.4%) of cases in our study were less than 6 years old. The prevalence of enterovirus was not significantly different across age groups. Additionally, we compared the distribution of enterovirus serotype across age groups and noticed that CVA6 was the most common serotype regardless of age groups. However, the proportion of CVA6 among cases under 3 years old was significantly higher than that among cases 3–5 years old; instead, EV71 was significantly higher. It indicated that EV71 preferentially infects preschool-aged children, although CVA6 remains most common20. We also compared the difference of enterovirus infection and distribution of serotypes by gender; the findings showed no statistically significant difference between male and female cases. Furthermore, CVA6 was the most common serotype in both male and female cases, suggesting that gender is not related to enterovirus infection or serotype20.
In this study, we explored the origin of CVA6 with phylogenetic analysis. All 133 sequences isolated in our study belonged to cluster D4, which has been reported to be circulating in United Kingdom and France. The cluster contained isolates in multiple provinces in mainland China, such as Shanghai, Yunnan, Henan, Jiangsu, Shandong, and Guangdong. Our findings demonstrated that CVA6 within cluster D4 has become the prevalent serotype in mainland China and has spread into Europe. Furthermore, the phylogenic tree has shown that cluster D4 was first isolated in Shenzhen in 2008 (GenBank Accession No. KC866901) and continually circulated in mainland China through 2015 (Fig. 1 and Fig. S1). The cluster was close to strains isolated in Finland in 2008; however, it remained separate from strains isolated in other countries, suggesting that CVA6 within cluster D4 might originate from a local strain, which has been documented in previous studies39,40.
In conclusion, our study demonstrated that CVA6 has become predominant in HFMD cases. Cluster D4 of CVA6 originated in Shenzhen and has been circulating in mainland China since 2008.
Methods
Participants
From May 2013 through April 2014, a total of 253 HFMD cases were included from the Department of Paediatrics and the Department of Dermatology in Baoan Maternal and Child Health Hospital, Jinan University, Shenzhen, China. Manifestations of the cases included eruption rash and/or vesicles on the hand, foot, buttock or trunk. Diagnosis criteria of HFMD are defined according to the National Health and Family Planning Commission of the People’s Republic of China (http://www.nhfpc.gov.cn/jkj/s3577/200805/e73df45b7b1549188b1d4e1efd604da9.shtml). This study was approved by the Ethics Committee of Baoan Maternal and Child Health Hospital, Jinan University, Shenzhen, China. All the examinations were carried out per official guidelines and regulations. Informed consent was obtained from the patients’ parents or legal guardians.
Enterovirus examination
Vesicular fluid, throat swab, stool or anal swabs of cases were collected for enterovirus examination. A total of 253 specimens were collected, one from each HFMD case, by sterile swabs and then put into sterile containers. Specimens were immediately sent to a laboratory and stored in −20 °C prior to examination. All specimens were examined for enterovirus by using a pan-enterovirus 5′ UTR one-step reverse transcript real-time quantitative PCR (RT-qPCR) Kit (Da’an gene Comp. LTD, Guangzhou) according to the manufacturer’s instructions. Briefly, specimens were dissolved in 1 mL of normal saline and then centrifuged at 3500 g for 5 min. A total of 350 μL of supernatant was drawn for RNA extraction using a commercial kit containing TRIzol (Da’an gene Co. Ltd, Guangzhou). One-step RT-qPCR was then performed in the MJ Research Opticon 2TM system (BIORAD, USA).
Enterovirus serotyping
The specimens that were enterovirus positive were further serotyped using a viral protein 1 (VP1) semi-reverse transcription nested PCR, which can identify almost all enterovirus serotypes30. The cDNA was synthesized using PrimeScriptTM II 1st strand cDNA synthesis kit (Takara Biotechnology (Dalian) Co.,Ltd). A 25 µL volume of PCR mix was prepared as follows: 2.5 µL of 10 × PCR buffer (Qiagen), 1.0 µL of MgCl2 (25 mM), 1.0 µL of dNTP (2.5 mM), 1.0 µL of each forward and reverse primers (10 mM; Primers 222 and 224 for the first round, primers AN88 and AN89 for the second round), 0.1 µL of HotStar Taq polymerase (5 U/µL, Qiagen), 3 μL of template DNA (cDNA for the first round and products of the first round PCR for the second round), and molecular biology-grade water. The thermal profile in both sets of PCR was 95 °C for 15 min, 35 cycles of 95 °C for 1 min, 50 °C for 1 min, and 72 °C for 1 min, then 72 °C for 10 min. PCR products were sequenced in both directions by using the BigDye Terminator (version 3.1) cycle sequencing kit and the ABI PRISM 3730xl DNA analyzer (Thermofisher Sientific, USA). Enterovirus serotype was determined with the BLAST search in GenBank.
Phylogenetic analysis
CVA6 sequences were aligned by using BioEdit 7.0 software. A phylogenetic tree was reconstructed by maximum likelihood method with a GTR + G + I model (bootstrap test of 1000 replications) in MEGA 6.0. A total of 1345 CVA6 sequences, including 133 sequences isolated in our laboratory and 1212 sequences retrieved from GenBank, were combined to reconstruct a phylogenetic tree. Subsequently, a smaller phylogenetic tree was reconstructed with the 133 sequences in our laboratory and 61 representative sequences (out of the 1212) from GenBank. The 133 CVA6 partial VP1 sequences were deposited into GenBank under the following accession numbers: MG195737 to MG195869.
Statistical analysis
Chi-square tests or Fisher’s exact tests were employed to compare differences between groups using SPSS17.0 software (IBM, USA). A P value of less than 0.05 was considered to be statistically significant.
Supplementary information
Figure S1. Phylogenetic analysis of CVA6 from HFDM cases in Shenzhen based on partial VP1 sequences (nt 2573 to 2908 according to Gdula/USA/1949 strain AY421764).
Acknowledgements
This study was supported by Medical Scientific Research Foundation of Guangdong Province (A2018455), Shenzhen Basic Research Grants (JCYJ20150402111430635), and the Sanming Project of Medicine in Shenzhen (SZSM201406007, SZSM201606088). The authors would like to thank the staff of Department of Dermatology and Department of Pediatrics in Baoan Maternal and Child Health Hospital, Jinan University, Shenzhen, China. The authors also acknowledge Dr. Abram L. Wagner’s help in editing this manuscript.
Author Contributions
K.X., L.D., S.C. and Y.L. conceived the study and prepared the manuscript; K.X., Y.P., M.W. and G.M. performed the PCR and sequencing experiments. K.X., Z.Y. and S.C. performed the data analysis. All authors have read and approved the final manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Kelin Xiao and Lian Duan contributed equally.
Contributor Information
Shuiwen Chen, Email: csw319@163.com.
Yihan Lu, Email: luyihan@fudan.edu.cn.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-40402-2.
References
- 1.Aswathyraj S, Arunkumar G, Alidjinou EK, Hober D. Hand, foot and mouth disease (HFMD): emerging epidemiology and the need for a vaccine strategy. Med Microbiol Immunol. 2016;205:397–407. doi: 10.1007/s00430-016-0465-y. [DOI] [PubMed] [Google Scholar]
- 2.McMinn P, Stratov I, Nagarajan L, Davis S. Neurological manifestations of enterovirus 71 infection in children during an outbreak of hand, foot, and mouth disease in Western Australia. Clin Infect Dis. 2001;32:236–242. doi: 10.1086/318454. [DOI] [PubMed] [Google Scholar]
- 3.Gilbert GL, et al. Outbreak of enterovirus 71 infection in Victoria, Australia, with a high incidence of neurologic involvement. Pediatr Infect Dis J. 1988;7:484–488. doi: 10.1097/00006454-198807000-00007. [DOI] [PubMed] [Google Scholar]
- 4.Samuda GM, Chang WK, Yeung CY, Tang PS. Monoplegia caused by Enterovirus 71: an outbreak in Hong Kong. Pediatr Infect Dis J. 1987;6:206–208. doi: 10.1097/00006454-198702000-00013. [DOI] [PubMed] [Google Scholar]
- 5.Lee MS, et al. Incidence rates of enterovirus 71 infections in young children during a nationwide epidemic in Taiwan, 2008-09. PLoS Negl Trop Dis. 2012;6:e1476. doi: 10.1371/journal.pntd.0001476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Craig ME, Vale T, Robertson P, Rawlinson WD, Gould B. Enterovirus 71 infection in Australian expatriate children following an outbreak in Malaysia. J Paediatr Child Health. 1999;35:107–108. doi: 10.1046/j.1440-1754.1999.t01-1-00348.x. [DOI] [PubMed] [Google Scholar]
- 7.Chua KB, Kasri AR. Hand foot and mouth disease due to enterovirus 71 in Malaysia. Virol Sin. 2011;26:221–228. doi: 10.1007/s12250-011-3195-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu Y, et al. The largest outbreak of hand; foot and mouth disease in Singapore in 2008: the role of enterovirus 71 and coxsackievirus A strains. Int J Infect Dis. 2010;14:e1076–1081. doi: 10.1016/j.ijid.2010.07.006. [DOI] [PubMed] [Google Scholar]
- 9.Zhang Y, et al. An emerging recombinant human enterovirus 71 responsible for the 2008 outbreak of hand foot and mouth disease in Fuyang city of China. Virol J. 2010;7:94. doi: 10.1186/1743-422X-7-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Geoghegan JL, et al. Phylodynamics of Enterovirus A71-Associated Hand, Foot, and Mouth Disease in Viet Nam. J Virol. 2015;89:8871–8879. doi: 10.1128/JVI.00706-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ishimaru Y, Nakano S, Yamaoka K, Takami S. Outbreaks of hand, foot, and mouth disease by enterovirus 71. High incidence of complication disorders of central nervous system. Arch Dis Child. 1980;55:583–588. doi: 10.1136/adc.55.8.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Christian KA, et al. What we are watching–five top global infectious disease threats, 2012: a perspective from CDC’s Global Disease Detection Operations Center. Emerg Health Threats J. 2013;6:20632. doi: 10.3402/ehtj.v6i0.20632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shimizu H, et al. Enterovirus 71 from fatal and nonfatal cases of hand, foot and mouth disease epidemics in Malaysia, Japan and Taiwan in 1997-1998. Jpn J Infect Dis. 1999;52:12–15. [PubMed] [Google Scholar]
- 14.Ooi MH, et al. Human enterovirus 71 disease in Sarawak, Malaysia: a prospective clinical, virological, and molecular epidemiological study. Clin Infect Dis. 2007;44:646–656. doi: 10.1086/511073. [DOI] [PubMed] [Google Scholar]
- 15.Ho M. Enterovirus 71: the virus, its infections and outbreaks. J Microbiol Immunol Infect. 2000;33:205–216. [PubMed] [Google Scholar]
- 16.Xing W, et al. Hand, foot, and mouth disease in China, 2008-12: an epidemiological study. Lancet Infect Dis. 2014;14:308–318. doi: 10.1016/S1473-3099(13)70342-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen L, et al. Detection of human enterovirus 71 and coxsackievirus A16 in children with hand, foot and mouth disease in China. Mol Med Rep. 2012;5:1001–1004. doi: 10.3892/mmr.2012.742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yan XF, et al. Epidemic characteristics of hand, foot, and mouth disease in Shanghai from 2009 to 2010: Enterovirus 71 subgenotype C4 as the primary causative agent and a high incidence of mixed infections with coxsackievirus A16. Scand J Infect Dis. 2012;44:297–305. doi: 10.3109/00365548.2011.634433. [DOI] [PubMed] [Google Scholar]
- 19.Fan X, et al. Detection of human enterovirus 71 and Coxsackievirus A16 in an outbreak of hand, foot, and mouth disease in Henan Province, China in 2009. Virus Genes. 2013;46:1–9. doi: 10.1007/s11262-012-0814-x. [DOI] [PubMed] [Google Scholar]
- 20.Di B, et al. Circulation of Coxsackievirus A6 in hand-foot-mouth disease in Guangzhou, 2010–2012. Virol J. 2014;11:157. doi: 10.1186/1743-422X-11-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He YQ, et al. Emergence, circulation, and spatiotemporal phylogenetic analysis of coxsackievirus a6- and coxsackievirus a10-associated hand, foot, and mouth disease infections from 2008 to 2012 in Shenzhen, China. J Clin Microbiol. 2013;51:3560–3566. doi: 10.1128/JCM.01231-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mirand A, et al. Outbreak of hand, foot and mouth disease/herpangina associated with coxsackievirus A6 and A10 infections in 2010, France: a large citywide, prospective observational study. Clin Microbiol Infect. 2012;18:E110–118. doi: 10.1111/j.1469-0691.2012.03789.x. [DOI] [PubMed] [Google Scholar]
- 23.Blomqvist S, et al. Co-circulation of coxsackieviruses A6 and A10 in hand, foot and mouth disease outbreak in Finland. J Clin Virol. 2010;48:49–54. doi: 10.1016/j.jcv.2010.02.002. [DOI] [PubMed] [Google Scholar]
- 24.Huang Y, et al. Effect of meteorological variables on the incidence of hand, foot, and mouth disease in children: a time-series analysis in Guangzhou, China. BMC Infect Dis. 2013;13:134. doi: 10.1186/1471-2334-13-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ma E, Lam T, Wong C, Chuang SK. Is hand, foot and mouth disease associated with meteorological parameters? Epidemiol Infect. 2010;138:1779–1788. doi: 10.1017/S0950268810002256. [DOI] [PubMed] [Google Scholar]
- 26.Ma E, Chan KC, Cheng P, Wong C, Chuang SK. The enterovirus 71 epidemic in 2008–public health implications for Hong Kong. Int J Infect Dis. 2010;14:e775–780. doi: 10.1016/j.ijid.2010.02.2265. [DOI] [PubMed] [Google Scholar]
- 27.Hongyan G, et al. Hand, foot and mouth disease caused by coxsackievirus A6, Beijing, 2013. Pediatr Infect Dis J. 2014;33:1302–1303. doi: 10.1097/INF.0000000000000467. [DOI] [PubMed] [Google Scholar]
- 28.Puenpa J, et al. Hand, foot, and mouth disease caused by coxsackievirus A6, Thailand, 2012. Emerg Infect Dis. 2013;19:641–643. doi: 10.3201/eid1904.121666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li JL, et al. Epidemic characteristics of hand, foot, and mouth disease in southern China, 2013: coxsackievirus A6 has emerged as the predominant causative agent. J Infect. 2014;69:299–303. doi: 10.1016/j.jinf.2014.04.001. [DOI] [PubMed] [Google Scholar]
- 30.Nix WA, Oberste MS, Pallansch MA. Sensitive, seminested PCR amplification of VP1 sequences for direct identification of all enterovirus serotypes from original clinical specimens. J Clin Microbiol. 2006;44:2698–2704. doi: 10.1128/JCM.00542-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Osterback R, et al. Coxsackievirus A6 and hand, foot, and mouth disease, Finland. Emerg Infect Dis. 2009;15:1485–1488. doi: 10.3201/eid1509.090438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wei SH, et al. An outbreak of coxsackievirus A6 hand, foot, and mouth disease associated with onychomadesis in Taiwan, 2010. BMC Infect Dis. 2011;11:346. doi: 10.1186/1471-2334-11-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.CDC. Notes from the field: severe hand, foot, and mouth disease associated with coxsackievirus A6 - Alabama, Connecticut, California, and Nevada, November 2011-February 2012. MMWR. Morbidity and mortality weekly report61, 213–214 (2012). [PubMed]
- 34.Montes M, et al. Hand, Foot, and Mouth Disease Outbreak and Coxsackievirus A6, Northern Spain, 2011. Emerg Infect Dis. 2013;19:676–678. doi: 10.3201/eid1904.121589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fujimoto T, et al. Hand, foot, and mouth disease caused by coxsackievirus A6, Japan, 2011. Emerg Infect Dis. 2012;18:337–339. doi: 10.3201/eid1802.111147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fonseca MC, et al. Coxsackievirus A6 and enterovirus 71 causing hand, foot and mouth disease in Cuba, 2011-2013. Arch Virol. 2014;159:2451–2455. doi: 10.1007/s00705-014-2071-x. [DOI] [PubMed] [Google Scholar]
- 37.Lu J, et al. Hand, foot and mouth disease in Guangdong, China, in 2013: new trends in the continuing epidemic. Clinical Microbiology and Infection. 2014;20:O442–O445. doi: 10.1111/1469-0691.12468. [DOI] [PubMed] [Google Scholar]
- 38.Zhang C, et al. Phylogenetic analysis of the major causative agents of hand, foot and mouth disease in Suzhou City, Jiangsu province, China, in 2012-2013. Emerg Microbes Infect. 2015;4:e12. doi: 10.1038/emi.2015.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zeng H, et al. The Epidemiological Study of Coxsackievirus A6 revealing Hand, Foot and Mouth Disease Epidemic patterns in Guangdong, China. Sci Rep. 2015;5:10550. doi: 10.1038/srep10550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Feng X, et al. A novel recombinant lineage’s contribution to the outbreak of coxsackievirus A6-associated hand, foot and mouth disease in Shanghai, China, 2012-2013. Sci Rep. 2015;5:11700. doi: 10.1038/srep11700. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Phylogenetic analysis of CVA6 from HFDM cases in Shenzhen based on partial VP1 sequences (nt 2573 to 2908 according to Gdula/USA/1949 strain AY421764).


