Short abstract
Over the last several decades, cardiovascular diseases largely increase the morbidity and mortality especially in developed countries, affecting millions of people worldwide. Although extensive work over the last two decades attempted to decipher the molecular network of regulating the pathogenesis and progression of these diseases, evidences from clinical trials with newly revealed targets failed to show more evidently salutary effects, indicating the inefficiency of understanding the complete regulatory landscape. Recent studies have shifted their focus from coding genes to the non-coding ones, which consist of microRNAs (miRNAs), long non-coding RNAs (lncRNAs) and the lately re-discovered a unique group of RNAs—circular RNAs (circRNAs). As the focus now has been shifted to the newly identified group of non-coding RNAs, circRNAs exhibit stability, highly conservation and relative enriched expression abundance in some cases, which are distinct from their cognate linear counterparts—lncRNAs. So far, emerging evidence begins to support the critical role of circRNAs in organogenesis and pathogenesis as exemplified in the central nervous system, and could be just as implicative in the cardiovascular system, suggesting a therapeutic perspective in related diseases.
Impact statement
Circular RNAs are important regulators of multiple biological processes such as organogenesis and oncogenesis. Although the bulk of concerning studies focused on revealing their diversified roles in various types of cancers, reports began to accumulate in cardiovascular field these days. We summarize circular RNAs implicated in cardiovascular diseases, aiming to highlight the advances in the knowledge of such diseases and their potential of being promising target for diagnosis and therapy.
Keywords: Circular RNA, cardiology, vascular, pathology, biomarkers, therapy
Introduction
Cardiovascular diseases (CVD) represent a constellation of disorders in the circulatory system, which include atherosclerosis (AS), cardiac hypertrophy, myocardial infarction (MI), and heart failure (HF). Conventional theory has implicated a group of well-defined risk factors in cardiovascular pathogenesis, including high glucose levels, dyslipidemia, elevated blood pressure, overweight, and pro-inflammatory state.1 For one thing, preventive measures, such as lifestyle modification, are recommended to lower the risk. For another, pharmacological treatments using traditional drugs like renin-angiotensin blocking agents, lipids-modulating drugs, beta-receptor inhibitors, and antithrombotic agents, have failed to hold back the progression of AS. Moreover, surgical interventions, like percutaneous coronary intervention (PCI) and bypass grafting surgery, are highly invasive measures. Above all, painful efforts have long been paid to stall the initiation and progression of acute coronary artery disease (CAD) whereas none of these therapies really addresses in the rising prevalence of CAD. Basic science studies are concentrating in identifying potential therapeutic targets of these diseases, which highlights the crucial need for an in-depth and comprehensive understanding of the underlying molecular mechanisms.
To unravel the pathological mechanisms on a wider scale, systematic and unbiased research techniques are designated at the gene, RNA or protein levels. Early proteomic approaches have been previously employed to discover proteins with pathogenic potential in CAD. A case in point is a study employing an unbiased lipoproteomic-based approach offering a deeper insight into how statin therapy affects lipoproteins and their protein cargo.2,3 However, the extremely low protein concentrations, often in the nano or picomolar range, making it harder to be an ideal disease-related biomarker.4 The genome-wide association studies’ (GWAS) findings are mostly focusing on identifying the disease-related single-nucleotide polymorphisms (SNPs), which may underscore the importance of genetic variants in various kinds of diseases. Chromosome 9p21 is one well-known genetic locus tightly linked to CAD.5,6 As population-based studies, standard statistical analysis of SNP researches are highlighted to find possible pathogenic genetic loci.4 The latest transcriptome sequencing technology has revealed multiple functional roles of RNAs distinct from coding genes and are widely implicated in diseases. Assisted by the Encyclopedia of DNA Elements project which aims to decipher gene’s all functional elements and together with the support from the Functional Annotation of the Mammalian genome program, non-coding transcripts (known as non-coding RNAs, ncRNAs) have been recognized in varied biological roles.7
Two categories of ncRNAs are generally classified according to the nucleotide size: small ncRNAs (<200 nt), and long non-coding RNAs (lncRNAs, >200 nt), represented by microRNAs (miRNAs), tRNA, rRNA, snRNA, snoRNA as well as long lncRNAs. They take an active part during various CVD. For instance, microRNA-21 contributes to myocardial dysfunction and microRNA-155 inhibits neovascularization.8,9 As for lncRNAs, uc022bqs.1 (LIPCAR), confined to mitochondria, promotes cardiac remodeling after MI.10 And Myheart, associated with the heavy chain of myosin, is cardioprotective with an anti-hypertrophic effect.11 Braveheart, a heart-associated lncRNA affects the cardiovascular lineage during its development.12 Moreover, manipulating lincRNA-p21 results in altered proliferation of vascular smooth muscle cell (VSMC) as well as their apoptotic rate, which underlies an important pathological mechanism of AS.13 However, in spite of the intensive research performed on miRNAs and lncRNAs, biological relevance of other types of non-coding molecules began to emerge, as exemplified by the re-discovery of circular RNAs (circRNAs) originally identified about two decades ago, adding more complexity to epigenetic regulation.14
In this review, we explore the circRNAs involved in the pathogenesis of CVD, aiming to highlight their potential in the physiological and pathological function of the cardiovascular system.
Characteristics, biogenesis, and classification of circRNAs
Considered as a “splicing error” or “splicing noise” for a long time, circRNAs have been re-discovered for their biological importance in tissue development and diseases. Unlike the canonically spliced linear RNA, circRNA lacks both 5' and 3' termini and form covalently closed loop when the latter exon’s 3' tail end is linked to the former exon’s 5' head, resulting in its unique circular structure. Accumulating evidence shows these “back spliced” circRNAs are ubiquitously transcribed with diversified spliced isoforms creating variations by the cell type.15 RNA-seq data show that more than 1000 circRNAs have been detected in adult tissues involving the colon, heart, kidney, liver, lung, and stomach. About one-third to one-half is tissue-specific and 33 circRNAs are comprehensively shared. Fetal tissues generate far more circRNAs than those from adults, which is further supported by their kinetic variations at different time points in human and mouse tissues.14,16 Therefore, circRNAs have a dynamic, spatiotemporal specificity in common with their linear counterparts.15,17,18 However, the unique circular structure enables them to have a longer half-life perhaps due to an insusceptibility to ribonuclease R (RNase R), which makes them more stable and optimal for being a biomarker. Moreover, they are generally more conserved across species and more abundant in expression, indicating a functional importance in life.19 Since the first well-studied circRNA, ciRS-7 (CDR1as) came to show its participation in brain development, more studies on functional circRNAs are accumulating.14 Notably, a circRNA profiling found an enrichment of more than 9000 circular species among the hearts of different species (mouse, rat and human) of which as much as 1288 are shared, implicating a functional importance in this system.20
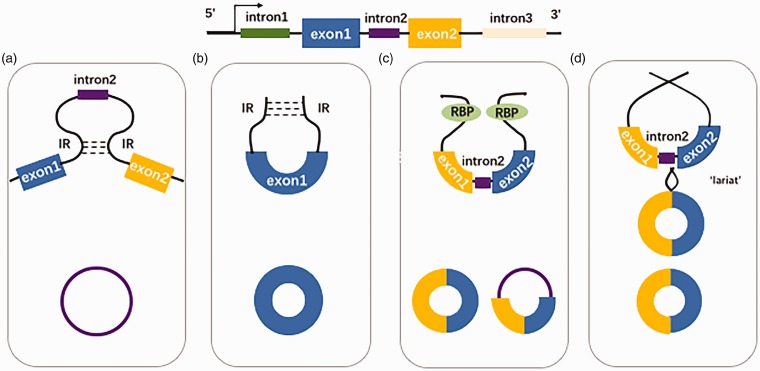
The mechanism of circRNAs biogenesis is currently supported by hypothetical theories: (1) Direct backsplicing: the formation of varied types of circRNAs is assisted by several different mechanisms. Generally, these regulatory elements include reversed complementary sequences which are usually contained within flanking introns (e.g. IRAlus; “intron-pairing”), RNA binding proteins (RBPs; e.g. Quaking) or spliceosome by forming a base pairing induced close proximity within these flanking sequences.21–24 (2) lariat-driven circularization (“exon skipping”): Circularization is promoted by a “lariat” structure usually followed by debranching and a fast exonuclease-mediated degradation. The repeated complementary flanking elements (e.g. ALU) in introns accelerate this process by reverse complementary matches.25,26 Therefore, circRNAs transcribed from distinct genomic loci consist of different elements and are categorized accordingly. Exon-derived circRNA constitutes the most discovered or studied type yet but mostly for its conduction of post-transcriptional regulation in the cytoplasm.27 In contrast, intronic circRNAs (ciRNAs) are a niche group, retained in the nucleus and affect gene transcription.28 Moreover, the intron–exon circularization gives rise to exon-intronic circRNA (EIciRNA), which is suggested to be involved in the promoted transcription of parent mRNA.29 The biogenesis and classification of circRNA are summarized in Figure 1.
Figure 1.
Schematic representation illustrating circRNA biogenesis. The generation of circRNAs can be attained via a direct backsplicing path, in which circularization is driven by intronic reverse complementary (IR) sequence (“intron pairing”) (a, b) or RNA-binding protein (RBP) pairing (c) that brings the sequences in proximity of each other to promote circularization. CircRNAs can also be formed through a lariat driven path characterized by a lariat structure whose flanking sequences facilitate the generation of circRNAs (d).
Functional models of circRNAs
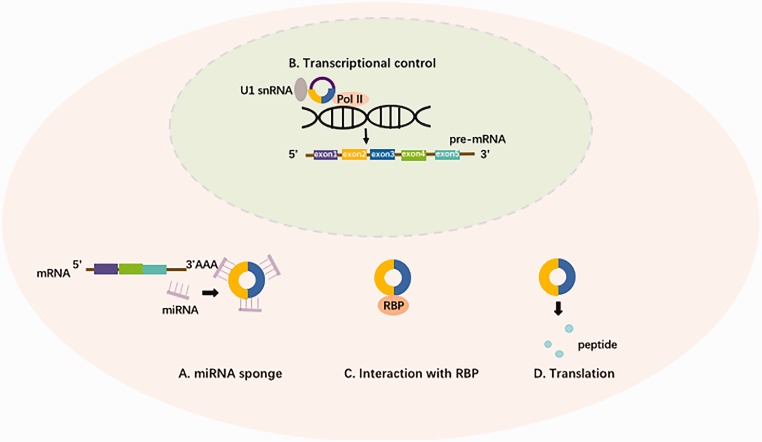
CircRNAs exert their biological functions through different ways depending on their types. Generally, most circRNAs are exon-derived and thus often regulate gene expression post-transcriptionally in the cytoplasm mainly by exerting an “miRNA sponge” effect. As a result, these circular transcripts are marked as a new kind of ceRNAs able to silence miRNAs by complementary base paring. Therefore, they “rescue” the expression of target gene by inhibiting miRNAs-mediated mRNA degradation, e.g. CDR1as, whose binding sites for miR-7 can be added up to 63, setting an example for this circRNA-miRNA interaction in brain development.14,21,30 In contrast, intron-retained circRNAs (including ciRNAs and EIciRNAs) usually convey a cis-regulatory effect on gene transcription within the nucleus. This process involves an interaction with nuclear proteins like RNA polymerase II (Pol II) or U1 small nuclear ribonucleoproteins (snRNPs).31 In addition, circRNAs could also bind to protein for further biological function. For example, the binding between circFoxo3 and transcription factors which include ID1, E2F1, and hypoxia inducible factor 1 alpha (HIF-1a) makes contribution to their nuclear translocation, and thereby, promotes both cell proliferation and survival.32 In recent years, internal ribosome entry site (IRES) sequence, which has been reported to be capable of binding to ribosome or infinite open reading frame (ORF), has been discovered in circRNAs, initiating investigations into the coding potential of circRNAs. For instance, circ-ZNF609, a 753nt ORF in this circRNA could be translated into a protein and thus play roles in myogenesis.33 Meanwhile, circ-FBXW is found to encode a 21-kDa protein, FBXW7-185aa in human brain cells, which takes part in maintaining cancer malignancy.34 A summary of different functional models of circRNAs is shown in Figure 2.
Figure 2.
Mechanism underlying how circRNA functions. (A) MiRNA sponge: harboring binding sites for certain miRNAs, circRNAs can affect miRNA-targeted gene expression by inhibition of target gene mRNA degradation. (B) Transcriptional control: exon–intron circRNAs (EIcircRNAs) regulate gene transcription in the nucleus mainly through binding to polymerase II (Pol II) and U1 snRNA. (C) Interaction with RBPs: circRNAs can interact with proteins through direct binding. For example, circFoxo binds to transcription factors like ID1, E2F1, and HIF-1a and therefore retain them in the cytoplasm. (D) Translation: some circRNAs may translate small peptides through a rolling circle amplification mechanism.
Role of circRNAs in CVD
Clinical value of circRNAs in the diagnosis of CVD
Conventional diagnostic methods for CVD are either non-invasive or invasive. For one thing, electrocardiogram (ECG) and Holter monitoring are convenient non-invasive techniques which have been widely acceptable but are limited by poor sensitivity and specificity. Coronary computed tomography angiography (CTA) is economically prohibitive. For another, invasive examinations which include: coronary arteriography and intravascular ultrasound are not beneficial during the onset of pathological change. As a result, there is a desperate need to seek a non-invasive biomarker which is highly sensitive, specific and effective in the diagnosing coronary heart disease (CHD) during its early stage.
Optimal biomarkers may be non-invasive indicators of CHD if they are consistent with accessibility, high stability, and reliability. For circRNAs, the circular structure promotes stability by eliminating RNase R-mediated degradation and thus extending their half-life in fluids. Additionally, circRNAs are dynamically and differentially expressed in various pathological conditions, which highlight their high sensitivity and specificity in diseases. Furthermore, circRNAs are recently found to be highly enriched in human’s saliva, body fluids, and even exosomes, whose collection and extraction could be easily attained through a relatively non-invasive method. Their wide distribution enhances the accessibility of detecting with diversified test materials.35,36 Multiple studies have evaluated the diagnostic performance of circRNAs in various types of tumor such as ciRS-7 which is suggested in hepatocellular carcinoma and SRY in cholangiocarcinoma.37 Therefore, it is intriguing to study the diagnostic potential as well as assess the clinical value of harnessing circRNAs in detecting CVD.
Diagnosis of CAD using the up-regulated has-circ-0124644 enhances the sensitivity to 0.861, the specificity to 0.626 while the odds ratio (OR) is adjusted to 1.931 [1.511–2.467] and the overall area under the curve (AUC) is up to 0.804 [0.751–0.857]. Notably, when hsa_circ_0098964 is also included, the diagnostic value further increases resulting in a highly sensitive and specified method (0.825 and 0.730) and showing an advantage over ECG (0.290 and 0.670) and TET (0.731 and 0.693).38 In addition to the detection of CAD as a single disease entity, hsa-circRNA11783-2 has been found to be down-regulated in both CAD and type 2 diabetes mellitus (T2DM) patients. The result was based on a cohort study with a large population which found that its diagnostic value was not satisfactory in the detection of CAD alone though subgroup analysis of this circRNA showed good diagnostic value in CAD when combined with T2DM. Considering a persistent growth of CAD patients with metabolic problems, hsa-circRNA11783-2 may be an ideal biomarker for this group of patients.39 Alternatively, circRNA circR-284 is identified in human VSMC as well as in carotid plaques. The increased circR-284 to miR-221 ratio indicates a high potential of being an efficient biomarker whose sensitivity is as much as 0.93 which is consistent with a remarkable specificity of 0.97, and as well as a significant AUC of 0.98 [0.96–1.00] for the prediction of plaque rupture and stroke.40
MI has long been recognized as a serious and even a fatal complication of CAD. Patients suffering from MI are susceptible to end-stage HF. Circulating proteins, like the brain natriuretic peptide (BNP), are widely used for its clinical diagnosis. However, HF-associated BNP favors a delayed determination which is not ideal for tailoring therapeutic strategies during their early stages.41 Based on a blood transcriptome investigation, a newly identified circRNA named MI-associated circular RNA (MIRCA) correlates with the left ventricular dysfunction post-MI. MIRCA is significantly down-regulated in patients with a reduced ejection fraction (EF) of less than 40%, which helps a more stratified risk classification. Compared to a slower reaction of protein in serum, the fast dynamic changes in the transcriptome make it possible for MIRCA to be a better indicator of distinguishing MI patients who have high risks of developing HF. Furthermore, utilization of MIRCA together with NT-pro-BNP adds more credibility to prognostication as dictated by the Akaike Information Criteria.42 And circRNAs with diagnostic potential are summarized in Table 1.
Table 1.
circRNAs of clinical value in diagnosis of CVD.
| circRNA | Expression | Disease | Sensitivity(crude/adjusted) | Specificity(crude/adjusted) | OR(crude/adjusted) | AUC(crude/adjusted) | References |
|---|---|---|---|---|---|---|---|
| hsa_circ_0124644 | Up | CAD | 0.861/0.759 | 0.626/0.704 | 1.856 [1.494–2.306]/1.931[1.511–2.467] | 0.769 [0.710–0.827]/0.804[0.751–0.857] | 38 |
| hsa_circ_0124644+hsa_circ_0098964 | 0.825/0.832 | 0.73/0.696 | – | 0.811 [0.756–0.865]/0.843 [0.796–0.891] | |||
| hsa-circRNA11783-2 | Down | CAD and T2DM | – | – | 0.740 [0.630–0.869]/0.688 [0.571–0.829] in CAD, 0.778 [0.659–0.919]/0.723 [0.591–0.883] in T2DM | – | 39 |
| circR-284/miR-221 | Up | Plaque rupture | 0.93/- | 0.97/- | – | 0.98[0.96–1.00]/- | 40 |
| MIRCA | Down | MI | – | – | 0.53 [0.29–0.97] | – | 42 |
OR:odds ratio; AUC: overall area under the curve; CAD: coronary artery disease; T2DM: type 2 diabetes mellitus; MI: myocardial infarction.
Functional circRNAs in cardiovascular disease
Atherosclerosis and coronary heart disease
It has been long accepted that the endothelium takes a critical part during the pathogenesis of AS. It is well established that endothelia damage initiates the onset of AS and facilitates atheromatous plaque formation. Several circRNAs involved in vascular endothelium dysfunction have been reported recently. One example is hsa_circ_0003575 which is found in human umbilical vein endothelial cell (HUVEC) and is increasingly expressed after ox-LDL treatment. In vitro studies suggest the apoptosis of HUVEC was reduced after hsa_circ_0003575 knockdown. Mechanically, it is speculated that hsa_circ_0003575 “sinks” the expression of miRNAs like miR-199-3p and miR-9-5p, all predicted based on bioinformatics though presently without validation.43 Additionally, hsa_circ_0010729, up-regulated in hypoxic HUVECs, promotes endothelial cell apoptosis after knockdown in vitro. Further analysis found that hsa_circ_0010729 could suppress the expression of miR-186, which has an inhibitory effect on HIF-1α.44 Additionally, circHIPK3 enhances endothelium viability by “sponging” miR-30 and therefore increasing the expressions of its target genes like VEGFC, FZD4, and WNT2.45
Phenotype switching of VSMC is an important molecular mechanism in AS via promoting cellular proliferation.46 Alpha-smooth muscle actin (α-SMA, alpha-actin-2 or ACTA2) constitutes the contractile apparatus which regulates cell motility, structure, and integrity. Loss of α-SMA is correlated with the phenotypical transition of VSMC from a mature state to a proliferating one, accelerating the development of AS. Recently, it was found that circRNAs also have a regulatory role on their host genes’ expression. A case in point is circACTA2 (alpha-actin-2). Transcribed from α-SMA gene, circACTA2 is able to maintain the contractile phenotype of VSMC. Specifically, circACTA2 “sponges” miR-548f-5p, which in turn enhances the mRNA level of its target gene, α-SMA.47 In addition, circWDR77 stimulates the proliferation and migration of VSMC and contributes to a thickened vessel wall due to an increased fibroblast growth factor 2 expression via down-regulating miR-124.48
ANRIL, a well-studied antisense lncRNA, is transcribed from the coronary diseases associated INK4/ARF locus on chromosome 9p21 and can suppress vascular intimal over-proliferation of thereby delaying AS progression by affecting p15INK4b and inhibiting CDK4.49 Recently, some circular transcripts of ANRIL (circANRILs) have been identified to assume varying biological roles in the development of AS. For example, the transcript consisting of primate-specific exons 5, 6, and 7 shows a protective effect on AS by increasing apoptosis and decreasing the proliferation of human VSMCs and macrophages. This leads to increased nucleolar stress and stabilized p53 expression, which accounts for its mechanism.50 Alternatively, the exon 4–6 circularization results in a transcript up-regulation in high-risk patients who are very likely to develop atherosclerotic vascular disease (ASVD) whereas circularized exon 14–5 is less expressed in risk individuals. However, evidence is lacking concerning the underlying mechanism.51 Thus, it is intriguing to study the functional differences of these differently transcribed transcripts for a better understanding of their potency in AS.
Myocardial infarction
MI constitutes a serious complication of CAD due to the acute occlusion of the coronary artery. Prolonged myocardial ischemia triggers the death of cardiomyocytes, resulting in cardiac insufficiency.52 Current therapies aim to protect remaining cardiomyocytes by reperfusion of the injured myocardium. So far increasing circRNAs have been reported to affect cardiomyocyte apoptosis, which makes them promising for serving as a novel therapeutic target.
Microarray analysis of circRNA profiling has identified circ-Amot 1, a circRNA with a favored expression in neonate myocardium and whose host gene Amotl 1 regulates endothelium migration and capillary formation and therefore affects cardiovascular performance. Yang et al. found that circ-Amotl1 conducts cardioprotective effects by improving primary cardiomyocyte survival and decreasing apoptosis. Up-regulation of circ-Amotl 1 in vivo rescues the enlarged left ventricle, increases ejection fraction, and inhibits apoptosis induced by doxorubicin. Specifically, circ-Amotl activates AKT signaling by promoting phosphorylated AKT via binding to kinase PDK1.53 In contrast, a well-known circRNA, Cdr1as (or CiRS-7) previously studied in brain development, has been re-discovered as a detrimental factor for causing myocardial dysfunction post-MI. Increased expression of Cdr1as in cardiomyocytes facilitates apoptosis due to activated caspase-3 as evidenced by an extended infarct size in vivo. Like most circRNAs, Cdr1as “sponges” miR-7 and therefore lifts the inhibitory effect of miR-7 on genes related to hypoxia and apoptosis such as SP1 and poly (ADP-ribose) polymerase.54
Mitochondria are essential for cardiac sufficiency by providing energy to living cardiomyocytes, and their fission and dysfunction are referred to cardiac diseases including MI and HF.55 For the first time, Li and co-workers found that a novel circRNA named MFACR can affect cardiomyocyte apoptosis through regulating mitochondrial dynamics.56 Inhibition of MFACR in mice relieves ischemia/reperfusion-induced mitochondrial fission and cardiac dysfunction, resulting in decreased cardiomyocyte apoptosis and infarct size. Specifically, miR-652-3p is “sponged” by MIRCA and consequently the target gene MTP18, a nuclear-encoded mitochondrial membrane protein promoting mitochondrial fission and cardiomyocyte apoptosis, is up-regulated.56 Taken together, this study highlights the regulatory axis of MFACR/miR-652-3p/MTP18 as a novel mechanism underlying cardiac injury as well as the therapeutic value for cardiac-related diseases.
Cardiomyopathy
Cardiomyopathy represents a spectrum of disorders in the myocardium due to dysfunction of the contractile apparatus, multiple genes, proteins, and signaling pathways.57,58 Cardiac hypertrophy signifies an increased risk of developing HF with numerous signaling cascades involved. An anti-hypertrophic circRNA has been found and named as heart-related circRNA (HRCR), whose overexpression inhibits cardiac hypertrophy in an isoproterenol-induced hypertrophy mouse model as well as HF. Specifically, HRCR “sponges” miR-223, resulting in an up-regulation of its target gene ARC (apoptosis inhibitor with CARD domain). Therefore, activated ARC holds back the progression of cardiac hypertrophy in the regulation of HRCR.59–62
Recently a large number of circRNAs was found to be transcribed from these cardiomyopathy-related host genes and the dysregulation of circRNA transcription is suggested in the onset of cardiomyopathy. Informatics analysis of these active host genes reveals titin (Ttn), NPPA, and MYH7 being involved in the developmental events, such as regulating cytoskeletal structure, affecting cellular or myocardial movement, packaging myofibril.63–65 Other host genes are also suggested in cardiomyopathy including the ryanodine receptor 2 (Ryr2), Ppp2r3a, and Slc8a1.66–68 Considering their active transcription in disease models, it is beneficial to study their pathogenic potential in cardiomyopathy.69
As a further example, Ttn functions as a molecular spring in the muscle and controls the passive elasticity of the myocardium; its mutation may be causative of dilated cardiomyopathy (DCM).70–-72 Recent studies show a differential expression of Ttn-generated circRNAs in the left ventricles of those patients. Meanwhile, Ttn derived circRNAs are found to be lost in DCM model by the cardiac mutation of RNA binding motif protein 20 (RBM20), which is a recognized splicing factor. And considering the fact RBM20 facilitates circRNA production via excluding specific exons from the pre-mRNA as substrates supplied for circRNA formation, the RBM20 null mice demonstrated a down-regulation of Ttn related circRNAs, enlarged left ventricle and decreased cardiac performance. Thus, it is compelling that interfering with certain circRNAs’ expression may incur DCM, though more understanding of their general function still remains evasive.73 Another actively transcribing gene is Ryr2, which regulates the contraction of cardiomyocyte and whose mutation is involved in arrhythmogenic right ventricular cardiomyopathy. Profiling of circRNA repertoire in adult murine hearts has identified a cardiac enrichment of Ryr2-derived circRNAs throughout the whole lifespan of a mouse in contrast to a much lower expression of the linear counterparts.74 Furthermore, genes coupled with cardiomyopathy, such as ATXN10, CHD7, DNAJC6, and SLC8A1, are under active transcription into circRNAs in β-adrenergic stress-stimulated cardiomyocytes which are derived from the human pluripotent stem cells, which has been verified in human DCM samples. And the rodent homologs of these cirRNAs are also verified in rat and mouse disease models.75 Although experiments are lacking in validating their pathogenic potential and mechanism, their abundance and tight association with diseases-related host genes pose interest for further investigation.
Other cardiovascular diseases
Hypertension is highly indicative for developing CVD, and aortic aneurysm marks a life-threatening vascular disease resulting into aortic dissection or rupture. However, only a few circRNA studies are referred to these diseases and are sufficiently studied. Further investigation remains to be performed for a better understanding of these common diseases.
A circRNA microarray has found 13 down-regulated circRNAs in contrast to 46 up-regulated ones in hypertensive group. Among them, four candidates are significantly expressed and hsa-circ-0005870 and are remarkably down-regulated. The pathway analysis related to gene oncology and Kyoto Encyclopedia of Genes and Genomes predict that hsa-circ-0005870 has a regulatory role in hypertension without further validation.76 Case–control study of essential hypertension has distinguished on has-circ-0037911 with a significant up-regulation in hypertensive participants compared to healthy controls. A positive correlation is also suggested between serum creatinine (Scr) and has-circ-0037911, raising the possibility of a higher expression of has-circ-0037911 in hypertensive nephropathy.77
Hsa-circ-000595, up-regulated in the vascular tissues of aortic aneurysm patients, is positively associated with apoptotic VSMC. The “sponged” miR-19a by hsa-circ-000595 is suggested to be relevant to the pathological manifestations of aortic aneurysm.78 A summary of CVD related circRNAs is found in Table 2.
TABLE 2.
Functional circRNAs in cardiovascular disease.
| Disease | circRNA name | Host cell/tissue | Description | Mechanism | References |
|---|---|---|---|---|---|
| AS and CHD | Hsa_circ_0003575 | HUVEC | Increase apoptosis | miRNA sponge of miR-199-3p, miR-9-5p, miR-377-3p and miR-141-3p (predicted) | 43 |
| Hsa_circ_0010729 | HUVEC | Decrease apoptosis | miRNA sponge of miR-186 | 44 | |
| circHIPK3 | Vascular endothelium | Increase cell viability and tube formation in vitro/increase retina acellular capillaries, vascular permeability and inflammation in vivo | miRNA sponge of miR-30 | 45 | |
| circACTA2 | VSMC | Maintain contractile phenotype | miRNA sponge of miR-548f-5p | 47 | |
| circWDR77 | VSMC | Increase proliferation and migration | miRNA sponge of miR-124 | 48 | |
| circANRIL(exon5-6-7) | VSMC/Macrophage | Increase apoptosis/decrease proliferation | Increased nucleolar stress and stabilize p53 | 50 | |
| circANRIL(exon4-6) | Hela/PBTL | Down-regulated in ASVD | Neighboring gene regulation | 51 | |
| circANRIL(exon14-5) | Hela/PBTL | Up-regulated in ASVD | – | ||
| MI | circ-Amotl 1 | Cardiomyocyte | Improve survival/decrease apoptosis | Binding to PDK1 and phosphorylate AKT | 53 |
| Cdr1as | Cardiomyocyte | Increase apoptosis | miRNA sponge of miR-7 | 54 | |
| MFACR | Cardiomyocyte | Increase apoptosis | miRNA sponge of miR-652-3p | 56 | |
| Cardiomyopathy | circTtn | Heart muscle | Dysregulated in disease model | – | 69 |
| circRyr2 | Heart muscle | Enriched in adult murine heart | – | 70 | |
| circATXN10, circCHD7, circDNAJC6, circSLC8A1 | Cardiomyocyte | Enriched in disease model | – | 71 | |
| Hypertension | Hsa-circ-0005870 | Peripheral blood | Down-regulated in hypertension patients | – | 72 |
| hsa_circ_0037911 | Peripheral blood | Up-regulated in hypertension patients | – | 73 | |
| Hsa-circ-000595 | Aortic aneurysm/VSMC | Increase apoptosis | miRNA sponge of hsa-circ-000595 | 74 |
AS: atherosclerosis; CHD: coronary heart disease; PBTL: primary human peripheral blood T-lymphocytes; ASVD: atherosclerotic vascular disease; VSMC: vascular smooth muscle cell; HUVEC: human umbilical vein endothelial cell; CAD: cardiovascular diseases; DM: diabetic myocardium; MI: myocardial infarction.
Limitation and perspective
Burgeoning researches on circRNAs have explored various biological roles by serving as promising regulators in various physiological or pathological conditions. The next generation sequencing technology and experimental innovation such as CRISPR/Cas9-mediated gene editing and adeno-associated virus-mediated in vivo delivery techniques hold promise for a deeper understanding of their biology.79 Nevertheless, limits and hurdles are emerging with the surge of circRNA study. First, their coding ability remains unclear. In spite of a vast body of ORFs calculated within circRNAs, only a few has been fully investigated. It is suggested that the actual number of coding circRNAs may be underestimated and the function of the coded peptides is poorly investigated.80 Second, understanding of the way how circRNAs function is far from sufficient. The broadly studied “sponge” effect may not account for the major mechanism considering other possibilities like protein–protein interaction. The variety of mechanisms remains to be investigated more thoroughly. Moreover, technical limits might retard further investigation into their molecular mechanism, which might be overcome by advances or revolution of experimental techniques. For example, conventional in vivo knockout strategy for a coding gene usually targets its ORF. In contrast, deletion of the whole circRNA sequence is much more complicated given the numerous and dispersed miRNA-binding sites within.33,81 It is also rather difficult at present to accurately knockout a circRNA consisting of overlapped exons even in the presence of the latest gene editing technology including CRISPR/Cas9 and CRISPR/Cas13a.82 Furthermore, present cardiovascular studies of circRNAs are overwhelmingly based on the cellular phenotype rather than disease-oriented. Collateral researches are also lacking in certain diseases, such as arrhythmia, diabetes, HF, and congenital heart disease. Other studies into cardiomyopathy and hypertension, for example, are mostly microarray analysis without following experimental supports.
Since the rapid development of in vivo delivery, gene-editing, and modified-RNAs techniques, it is possible that circRNAs can enter clinical trials as therapeutic drugs. Pharmacological utilization of functional circRNAs might serve as promising therapeutic methods in the near future.83,84 Moreover, circRNAs hold advantages over other ncRNAs in the clinical translation into novel diagnostics and therapeutics due to their characteristics as highly conserved, stabilized, enriched and wide distributed properties, which highlights a profound diagnostic and therapeutic potential in the treatment of CVD.
Conclusion
In spite of the increasing knowledge of the roles of circRNA in the genetic regulatory network, more efforts are needed to explore their comprehensive and specific roles in the diversified types of CVD. Since next generation sequencing technology assist to discovering multiple pathogenic circRNAs, and novel gene editing technology has extended our understanding of their molecular biology, individualized treatment of CVD by circRNAs might be realized soon in the near future.
Author contributions
All authors contributed in preparing the manuscript. XG wrote the paper. CYZ and GZW reviewed and edited the manuscript.
Declaration of conflicting interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding
These studies were supported in part by grants from the National Natural Science Foundation of China (31430043, 31730043), National Key R&D Program of China (SQ2018YFC130229), Program of Innovative Research Team by National Natural Science Foundation (81721001).
References
- 1.Matsuzawa Y, Funahashi T, Nakamura T. The concept of metabolic syndrome: contribution of visceral fat accumulation and its molecular mechanism. J Atheroscler Thromb 2011; 18:629–39 [DOI] [PubMed] [Google Scholar]
- 2.Bhandari S, Gupta P, Quinn P, Sandhu J, Hakimi A, Jones D, Ng L. Pleiotropic effects of statins in hypercholesterolaemia: a prospective observational study using a lipoproteomic based approach. Lancet 2015; 385:21. [DOI] [PubMed] [Google Scholar]
- 3.Lindsey ML, Mayr M, Gomes AV, Delles C, Arrell DK, Murphy AM, Lange RA, Costello CE, Jin YF, Laskowitz DT, Sam F, Terzic A, Van Eyk J, Srinivas PR; American Heart Association Council on Functional Genomics and Translational Biology, Council on Cardiovascular Disease in the Young, Council on Clinical Cardiology, Council on Cardiovascular and Stroke Nursing, Council on Hypertension, and Stroke Council. Transformative impact of proteomics on cardiovascular health and disease: a scientific statement from the American Heart Association. Circulation 2015; 132:852–72 [DOI] [PubMed] [Google Scholar]
- 4.Gerszten RE, Wang TJ. The search for new cardiovascular biomarkers. Nature 2008; 451:949–52 [DOI] [PubMed] [Google Scholar]
- 5.Samani NJ, Erdmann J, Hall AS, Hengstenberg C, Mangino M, Mayer B, Dixon RJ, Meitinger T, Braund P, Wichmann HE, Barrett JH, König IR, Stevens SE, Szymczak S, Tregouet DA, Iles MM, Pahlke F, Pollard H, Lieb W, Cambien F, Fischer M, Ouwehand W, Blankenberg S, Balmforth AJ, Baessler A, Ball SG, Strom TM, Braenne I, Gieger C, Deloukas P, Tobin MD, Ziegler A, Thompson JR, Schunkert H; WTCCC and the Cardiogenics Consortium. Genomewide association analysis of coronary artery disease. N Engl J Med 2007; 357:443–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.O'Donnell CJ, Kavousi M, Smith AV, Kardia SL, Feitosa MF, Hwang SJ, Sun YV, Province MA, Aspelund T, Dehghan A, Hoffmann U, Bielak LF, Zhang Q, Eiriksdottir G, van Duijn CM, Fox CS, de Andrade M, Kraja AT, Sigurdsson S, Elias-Smale SE, Murabito JM, Launer LJ, van der Lugt A, Kathiresan S; CARDIoGRAM Consortium, Krestin GP, Herrington DM, Howard TD, Liu Y, Post W, Mitchell BD, O'Connell JR, Shen H, Shuldiner AR, Altshuler D, Elosua R, Salomaa V, Schwartz SM, Siscovick DS, Voight BF, Bis JC, Glazer NL, Psaty BM, Boerwinkle E, Heiss G, Blankenberg S, Zeller T, Wild PS, Schnabel RB, Schillert A, Ziegler A, Münzel TF, White CC, Rotter JI, Nalls M, Oudkerk M, Johnson AD, Newman AB, Uitterlinden AG, Massaro JM, Cunningham J, Harris TB, Hofman A, Peyser PA, Borecki IB, Cupples LA, Gudnason V, Witteman JC. Genome-wide association study for coronary artery calcification with follow-up in myocardial infarction. Circulation 2011; 124:2855–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoeppner MP, Denisenko E, Gardner PP, Schmeier S, Poole AM. An evaluation of function of multicopy non-coding RNAs in mammals using ENCODE/FANTOM data and comparative genomics. Mol Biol Evol 2018; 35:1451–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thum T, Gross C, Fiedler J, Fischer T, Kissler S, Bussen M, Galuppo P, Just S, Rottbauer W, Frantz S, Castoldi M, Soutschek J, Koteliansky V, Rosenwald A, Basson MA, Licht JD, Pena JT, Rouhanifard SH, Muckenthaler MU, Tuschl T, Martin GR, Bauersachs J, Engelhardt S. MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature 2008; 456:980–4 [DOI] [PubMed] [Google Scholar]
- 9.Pankratz F, Bemtgen X, Zeiser R, Leonhardt F, Kreuzaler S, Hilgendorf I, Smolka C, Helbing T, Hoefer I, Esser JS, Kustermann M, Moser M, Bode C, Grundmann S. MicroRNA-155 exerts cell-specific antiangiogenic but proarteriogenic effects during adaptive neovascularization. Circulation 2015; 131:1575–89 [DOI] [PubMed] [Google Scholar]
- 10.Kumarswamy R, Bauters C, Volkmann I, Maury F, Fetisch J, Holzmann A, Lemesle G, de Groote P, Pinet F, Thum T. Circulating long noncoding RNA, LIPCAR, predicts survival in patients with heart failure. Circ Res 2014; 114:1569–75 [DOI] [PubMed] [Google Scholar]
- 11.Han P, Li W, Lin CH, Yang J, Shang C, Nuernberg ST, Jin KK, Xu W, Lin CY, Lin CJ, Xiong Y, Chien H, Zhou B, Ashley E, Bernstein D, Chen PS, Chen HV, Quertermous T, Chang CP. A long non-coding RNA protects the heart from pathological hypertrophy. Nature 2014; 514:102–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klattenhoff CA, Scheuermann JC, Surface LE, Bradley RK, Fields PA, Steinhauser ML, Ding H, Butty VL, Torrey L, Haas S, Abo R, Tabebordbar M, Lee RT, Burge CB, Boyer LA. Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell 2013; 152:570–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu G, Cai J, Han Y, Chen J, Huang ZP, Chen C, Cai Y, Huang H, Yang Y, Liu Y, Xu Z, He D, Zhang X, Hu X, Pinello L, Zhong D, He F, Yuan GC, Wang DZ, Zeng C. LincRNA-p21 regulates neointima formation, vascular smooth muscle cell proliferation, apoptosis, and atherosclerosis by enhancing p53 activity. Circulation 2014; 130:1452–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, Loewer A, Ziebold U, Landthaler M, Kocks C, Le Noble F, Rajewsky N. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013; 495:333–8 [DOI] [PubMed] [Google Scholar]
- 15.Moran JV, Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO. Cell-type specific features of circular RNA expression. PLoS Genet 2013; 9:e1003777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu T, Jing W, Ping H, Zhao Z, Song X. Circular RNA expression profiles and features in human tissues: a study using RNA-seq data. BMC Genomics 2017; 18:680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Capel B, Swain A, Nicolis S, Hacker A, Walter M, Koopman P, Goodfellow P, Lovell-Badge R. Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell 1993; 73:1019–30 [DOI] [PubMed] [Google Scholar]
- 18.Cheng J, Metge F, Dieterich C. Specific identification and quantification of circular RNAs from sequencing data. Bioinformatics 2016; 32:1094–6 [DOI] [PubMed] [Google Scholar]
- 19.Enuka Y, Lauriola M, Feldman ME, Sas-Chen A, Ulitsky I, Yarden Y. Circular RNAs are long-lived and display only minimal early alterations in response to a growth factor. Nucleic Acids Res 2016; 44:1370–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Werfel S, Nothjunge S, Schwarzmayr T, Strom T-M, Meitinger T, Engelhardt S. Characterization of circular RNAs in human, mouse and rat hearts. J Mol Cell Cardiol 2016; 98:103–7 [DOI] [PubMed] [Google Scholar]
- 21.Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B, Luo Y, Lyu D, Li Y, Shi G, Liang L, Gu J, He X, Huang S. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun 2016; 7:11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liang D, Wilusz JE. Short intronic repeat sequences facilitate circular RNA production. Genes Dev 2014; 28:2233–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ivanov A, Memczak S, Wyler E, Torti F, Porath HT, Orejuela MR, Piechotta M, Levanon EY, Landthaler M, Dieterich C, Rajewsky N. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep 2015; 10:170–7 [DOI] [PubMed] [Google Scholar]
- 24.Conn SJ, Pillman KA, Toubia J, Conn VM, Salmanidis M, Phillips CA, Roslan S, Schreiber AW, Gregory PA, Goodall GJ. The RNA binding protein quaking regulates formation of circRNAs. Cell 2015; 160:1125–34 [DOI] [PubMed] [Google Scholar]
- 25.Wilusz J. Circular RNA and splicing: skip happens. J Mol Biol 2015; 427:2411–3 [DOI] [PubMed] [Google Scholar]
- 26.Vicens Q, Westhof E. Biogenesis of circular RNAs. Cell 2014; 159:13–4 [DOI] [PubMed] [Google Scholar]
- 27.Qu S, Yang X, Li X, Wang J, Gao Y, Shang R, Sun W, Dou K, Li H. Circular RNA: a new star of noncoding RNAs. Cancer Lett 2015; 365:141–8 [DOI] [PubMed] [Google Scholar]
- 28.Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, Zhu S, Yang L, Chen LL. Circular intronic long noncoding RNAs. Mol Cell 2013; 51:792–806 [DOI] [PubMed] [Google Scholar]
- 29.Yu B, Shan G. Functions of long noncoding RNAs in the nucleus. Nucleus 2016; 7:155–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li F, Zhang L, Li W, Deng J, Zheng J, An M, Lu J., Zhou Y. . Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/β-catenin pathway. Oncotarget 2015; 6:6001–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, Zhong G, Yu B, Hu W, Dai L, Zhu P, Chang Z, Wu Q, Zhao Y, Jia Y, Xu P, Liu H, Shan G. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol 2015; 22:256–64 [DOI] [PubMed] [Google Scholar]
- 32.Du WW, Yang W, Chen Y, Wu ZK, Foster FS, Yang Z, Li X, Yang BB. Foxo3 circular RNA promotes cardiac senescence by modulating multiple factors associated with stress and senescence responses. Eur Heart J 2017; 38:1402–12 [DOI] [PubMed] [Google Scholar]
- 33.Legnini I, Di TG, Rossi F, Morlando M, Briganti F, Sthandier O, Fatica A, Santini T, Andronache A, Wade M, Laneve P, Rajewsky N, Bozzoni I. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol Cell 2017; 66:22–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abe N, Matsumoto K, Nishihara M, Nakano Y, Shibata A, Maruyama H, Shuto S, Matsuda A, Yoshida M, Ito Y, Abe H. Rolling circle translation of circular RNA in living human cells. Sci Rep 2015; 5:16435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Memczak S, Papavasileiou P, Peters O, Rajewsky N. Identification and characterization of circular RNAs as a new class of putative biomarkers in human blood. PloS One 2015; 10:e0141214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bahn JH, Zhang Q, Li F, Chan TM, Lin X, Kim Y, Wong DT, Xiao X. The landscape of microRNA, Piwi-interacting RNA, and circular RNA in human saliva. Clin Chem 2015; 61:221–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao ZJ, Shen J. Circular RNA participates in the carcinogenesis and the malignant behavior of cancer. RNA Biol 2017; 14:514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao Z, Li X, Gao C, Jian D, Hao P, Rao L, Li M. Peripheral blood circular RNA hsa_circ_0124644 can be used as a diagnostic biomarker of coronary artery disease. Sci Rep 2017; 7:39918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li X, Zhao Z, Jian D, Li W, Tang H, Li M. Hsa-circRNA11783-2 in peripheral blood is correlated with coronary artery disease and type 2 diabetes mellitus. Diab Vasc Dis Res 2017; 14:510–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bazan HA, Hatfield SA, Brug A, Brooks AJ, Lightell DJ, Jr, Woods TC. Carotid plaque rupture is accompanied by an increase in the ratio of serum circr-284 to mir-221 levels. Circ Cardiovasc Genet 2017; 10:e001720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Talwar S, Squire IB, Downie PF, McCullough AM, Campton MC, Davies JE, Barnett DB, Ng LL. Profile of plasma N-terminal proBNP following acute myocardial infarction. Correlation with left ventricular systolic dysfunction. Eur Heart J 2000; 21:1514–21 [DOI] [PubMed] [Google Scholar]
- 42.Salgadosomoza A, Zhang L, Vausort M, Devaux Y. The circular RNA MICRA for risk stratification after myocardial infarction. Int J Cardiol Heart Vasc 2017; 17:33–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li CY, Ma L, Yu B. Circular RNA hsa_circ_0003575 regulates oxLDL induced vascular endothelial cells proliferation and angiogenesis. Biomed Pharmacother 2017; 95:1514–9 [DOI] [PubMed] [Google Scholar]
- 44.Dang RY, Liu FL, Li Y. Circular RNA hsa_circ_0010729 regulates vascular endothelial cell proliferation and apoptosis by targeting the miR-186/HIF-1alpha axis. Biochem Biophys Res Commun 2017; 490:104–10 [DOI] [PubMed] [Google Scholar]
- 45.Shan K, Liu C, Liu BH, Chen X, Dong R, Liu X, Zhang YY, Liu B, Zhang SJ, Wang JJ, Zhang SH, Wu JH, Zhao C, Yan B. Circular noncoding RNA HIPK3 mediates retinal vascular dysfunction in diabetes mellitus. Circulation 2017; 136:1629. [DOI] [PubMed] [Google Scholar]
- 46.Bochatonpiallat ML, Gabbiani G. Modulation of smooth muscle cell proliferation and migration: role of smooth muscle cell heterogeneity. Handb Exp Pharmacol 2005; 170:645–63 [DOI] [PubMed] [Google Scholar]
- 47.Sun Y, Yang Z, Zheng B, Zhang XH, Zhang ML, Zhao XS, Zhao HY, Suzuki T, Wen JK. A novel regulatory mechanism of smooth muscle alpha-actin expression by NRG-1/circACTA2/miR-548f-5p axis. Circ Res 2017; 121:628–35 [DOI] [PubMed] [Google Scholar]
- 48.Chen J, Cui L, Yuan J, Zhang Y, Sang H. Circular RNA WDR77 target FGF-2 to regulate vascular smooth muscle cells proliferation and migration by sponging miR-124. Biochem Biophys Res Commun 2017; 494:126–32 [DOI] [PubMed] [Google Scholar]
- 49.Jarinova O, Stewart AF, Roberts R, Wells G, Lau P, Naing T, Buerki C, McLean BW, Cook RC, Parker JS, McPherson R. Functional analysis of the chromosome 9p21.3 coronary artery disease risk locus. Arterioscler Thromb Vasc Biol 2009; 29:1671–7 [DOI] [PubMed] [Google Scholar]
- 50.Holdt LM, Stahringer A, Sass K, Pichler G, Kulak NA, Wilfert W, Kohlmaier A, Herbst A, Northoff BH, Nicolaou A, Gäbel G, Beutner F, Scholz M, Thiery J, Musunuru K, Krohn K, Mann M, Teupser D. Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat Commun 2016; 7:12429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Burd CE, Jeck WR, Liu Y, Sanoff HK, Wang Z, Sharpless NE. Expression of linear and novel circular forms of an INK4/ARF-associated non-coding RNA correlates with atherosclerosis risk. Plos Genet 2010; 6:e1001233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Takemura G, Kanoh M, Minatoguchi S, Fujiwara H. Cardiomyocyte apoptosis in the failing heart – a critical review from definition and classification of cell death. Int J Cardiol 2013; 167:2373–86 [DOI] [PubMed] [Google Scholar]
- 53.Zeng Y, Du WW, Wu Y, Yang Z, Awan FM, Li X, Yang W, Zhang C, Yang Q, Yee A, Chen Y, Yang F, Sun H, Huang R, Yee AJ, Li RK, Wu Z, Backx PH, Yang BB. A circular RNA binds to and activates AKT phosphorylation and nuclear localization reducing apoptosis and enhancing cardiac repair. Theranostics 2017; 7:3842–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fan G-C, Geng H-H, Li R, Xiao J, Pan M, Cai XX, Ji XP. The circular RNA Cdr1as promotes myocardial infarction by mediating the regulation of miR-7a on its target genes expression. PloS One 2016; 11:e0151753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sabbah HN, Gupta RC, Singh-Gupta V, Zhang K, Lanfear DE. Abnormalities of mitochondrial dynamics in the failing heart: normalization following long-term therapy with elamipretide. Cardiovasc Drugs Ther 2018; 32:319–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang K, Gan TY, Li N, Liu CY, Zhou LY, Gao JN, Chen C, Yan KW, Ponnusamy M, Zhang YH, Li PF. Circular RNA mediates cardiomyocyte death via miRNA-dependent upregulation of MTP18 expression. Cell Death Differ 2017; 24:1111–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fatkin D, Graham RM. Molecular mechanisms of inherited cardiomyopathies. Physiol Rev 2002; 82:945–80 [DOI] [PubMed] [Google Scholar]
- 58.Seidman JG, Seidman C. The genetic basis for cardiomyopathy. Cell 2001; 104:557–67 [DOI] [PubMed] [Google Scholar]
- 59.Wang K, Long B, Liu F, Wang JX, Liu CY, Zhao B, Zhou LY, Sun T, Wang M, Yu T, Gong Y, Liu J, Dong YH, Li N, Li PF. A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur Heart J 2016; 37:2602–11 [DOI] [PubMed] [Google Scholar]
- 60.Fan X, Weng X, Zhao Y, Chen W, Gan T, Xu D. Circular RNAs in cardiovascular disease: an overview. Biomed Res Int 2017; 2017:5135781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Devaux Y, Creemers EE, Boon RA, Werfel S, Thum T, Engelhardt S, Dimmeler S, Squire I. Cardiolinc network circular RNAs in heart failure. Eur J Heart Fail 2017; 19:701–709 [DOI] [PubMed] [Google Scholar]
- 62.Gomes CPC, Salgado-Somoza A, Creemers EE, Dieterich C, Lustrek M, Devaux Y; Cardiolinc™ network. Circular RNAs in the cardiovascular system. Noncoding RNA Res 2018; 3:1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Herman DS, Lam L, Taylor MR, Wang L, Teekakirikul P, Christodoulou D, Conner L, DePalma SR, McDonough B, Sparks E, Teodorescu DL, Cirino AL, Banner NR, Pennell DJ, Graw S, Merlo M, Di Lenarda A, Sinagra G, Bos JM, Ackerman MJ, Mitchell RN, Murry CE, Lakdawala NK, Ho CY, Barton PJ, Cook SA, Mestroni L, Seidman JG, Seidman CE. Truncations of titin causing dilated cardiomyopathy. N Engl J Med 2012; 366:619–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Taha MF, Javeri A. The expression of NPPA splice variants during mouse cardiac development. DNA Cell Biol 2015; 34:19–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lamont PJ, Wallefeld W, Hilton-Jones D, Udd B, Argov Z, Barboi AC, Bonneman C, Boycott KM, Bushby K, Connolly AM, Davies N, Beggs AH, Cox GF, Dastgir J, DeChene ET, Gooding R, Jungbluth H, Muelas N, Palmio J, Penttilä S, Schmedding E, Suominen T, Straub V, Staples C, Van den Bergh PY, Vilchez JJ, Wagner KR, Wheeler PG, Wraige E, Laing NG. Novel mutations widen the phenotypic spectrum of slow skeletal/β – cardiac myosin (MYH7) distal myopathy. Hum Mutat 2014; 35:868–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yang Y, Guo T, Oda T, Chakraborty A, Chen L, Uchinoumi H, Knowlton AA, Fruen BR, Cornea RL, Meissner G, Bers DM. Cardiac myocyte Z-line calmodulin is mainly RyR2-bound, and reduction is arrhythmogenic and occurs in heart failure. Circ Res 2015; 114:295–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Degrande ST, Little SC, Nixon DJ, Wright P, Snyder J, Dun W, Murphy N, Kilic A, Higgins R, Binkley PF, Boyden PA, Carnes CA, Anderson ME, Hund TJ, Mohler PJ. Molecular mechanisms underlying cardiac protein phosphatase 2A regulation in heart. J Biol Chem 2013; 288:1032–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Luo X, Zhang H, Xiao J, Wang Z. Regulation of human cardiac ion channel genes by microRNAs: theoretical perspective and pathophysiological implications. Cell Physiol Biochem 2010; 25:571–86 [DOI] [PubMed] [Google Scholar]
- 69.Tan WL, Lim BT, Anene-Nzelu CG, Ackers-Johnson M, Dashi A, See K, Tiang Z, Lee DP, Chua WW, Luu TD, Li PY, Richards AM, Foo RS. A landscape of circular RNA expression in the human heart. Cardiovasc Res 2017; 113:298–309 [DOI] [PubMed] [Google Scholar]
- 70.Okuda S, Sufu-Shimizu Y, Kato T, Fukuda M, Nishimura S, Oda T, Kobayashi S, Yamamoto T, Morimoto S, Yano M. CaMKII-mediated phosphorylation of RyR2 plays a crucial role in aberrant Ca2+ release as an arrhythmogenic substrate in cardiac troponin T-related familial hypertrophic cardiomyopathy. Biochem Biophys Res Commun 2018; 496:1250–6 [DOI] [PubMed] [Google Scholar]
- 71.Ware JS, Cook SA. Role of titin in cardiomyopathy: from DNA variants to patient stratification. Nat Rev Cardiol 2018; 15:241–52 [DOI] [PubMed] [Google Scholar]
- 72.Tayal U, Newsome S, Buchan R, Whiffin N, Halliday B, Lota A, Roberts A, Baksi AJ, Voges I, Midwinter W, Wilk A, Govind R, Walsh R, Daubeney P, Jarman JWE, Baruah R, Frenneaux M, Barton PJ, Pennell D, Ware JS, Prasad SK, Cook SA. Phenotype and clinical outcomes of titin cardiomyopathy. J Am Coll Cardiol 2017; 70:2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Khan MA, Reckman YJ, Aufiero S, van den Hoogenhof MM, van der Made I, Beqqali A, Koolbergen DR, Rasmussen TB, van der Velden J, Creemers EE, Pinto YM. RBM20 regulates circular RNA production from the titin gene. Circ Res 2016; 119:996–1003 [DOI] [PubMed] [Google Scholar]
- 74.Jakobi T, Czaja-Hasse LF, Reinhardt R, Dieterich C. Profiling and validation of the circular RNA repertoire in adult murine hearts. Genomics Proteomics Bioinformatics 2016; 14:216–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Siede D, Rapti K, Gorska AA, Katus HA, Altmüller J, Boeckel JN, Meder B, Maack C, Völkers M, Müller OJ, Backs J, Dieterich C. Identification of circular RNAs with host gene-independent expression in human model systems for cardiac differentiation and disease. J Mol Cell Cardiol 2017; 109:48–56 [DOI] [PubMed] [Google Scholar]
- 76.Na W, Ling J, Cai J. Profiling and bioinformatics analyses reveal differential circular RNA expression in hypertensive patients. Clin Exp Hypertens 2017; 39:454–9 [DOI] [PubMed] [Google Scholar]
- 77.Bao X, Zheng S, Mao S, Gu T, Liu S, Sun J, Zhang L. A potential risk factor of essential hypertension in case–control study: circular RNA hsa_circ_0037911. Biochem Biophys Res Commun 2018; 498:789–94 [DOI] [PubMed] [Google Scholar]
- 78.Zheng C, Niu H, Li M, Zhang H, Yang Z, Tian L, Wu Z, Li D, Chen X. Cyclic RNA hsa-circ-000595 regulates apoptosis of aortic smooth muscle cells. Mol Med Rep 2015; 12:6656–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang HX, Li M, Lee CM, Chakraborty S, Kim HW, Bao G, Leong KW. CRISPR/Cas9-based genome editing for disease modeling and therapy: challenges and opportunities for nonviral delivery. Chem Rev 2017; 117:9874. [DOI] [PubMed] [Google Scholar]
- 80.Yang Y, Gao X, Zhang M, Yan S, Sun C, Xiao F, Huang N, Yang X, Zhao K, Zhou H, Huang S, Xie B, Zhang N. Novel role of FBXW7 circular RNA in repressing glioma tumorigenesis. J Natl Cancer Inst 2018; 110:304–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Piwecka M, Glažar P, Hernandezmiranda LR, Memczak S, Wolf SA, Rybak-Wolf A, Filipchyk A, Klironomos F, Cerda Jara CA, Fenske P, Trimbuch T, Zywitza V, Plass M, Schreyer L, Ayoub S, Kocks C, Kühn R, Rosenmund C, Birchmeier C, Rajewsky N. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science 2017; 357:6357. [DOI] [PubMed] [Google Scholar]
- 82.Abudayyeh OO, Gootenberg JS, Essletzbichler P, Han S, Joung J, Belanto JJ, Verdine V, Cox DBT, Kellner MJ, Regev A, Lander ES, Voytas DF, Ting AY, Zhang F. RNA targeting with CRISPR-Cas13. Nature 2017; 550:280–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Janssen HL, Reesink HW, Lawitz EJ, Zeuzem S, Rodriguez-Torres M, Patel K, van der Meer AJ, Patick AK, Chen A, Zhou Y, Persson R, King BD, Kauppinen S, Levin AA, Hodges MR. Treatment of HCV infection by targeting microRNA. N Engl J Med 2013; 368:1685–94 [DOI] [PubMed] [Google Scholar]
- 84.Li Z, Rana TM. Therapeutic targeting of microRNAs: current status and future challenges. Nat Rev Drug Discov 2014; 13:622–38 [DOI] [PubMed] [Google Scholar]