Abstract
The honey bee, Apis mellifera, pollinates a wide variety of essential crops in numerous ecosystems around the world but faces many modern challenges. Among these, the microsporidian pathogen Nosema ceranae is one of the primary detriments to honey bee health. Nosema infects the honey bee gut, which harbors a highly specific, coevolved microbiota heavily involved in bee immune function and nutrition. Here, we extend previous work investigating interactions between the honey bee gut microbiome and N. ceranae by studying experimentally infected bees that were returned to their colonies and sampled 5, 10, and 21 days post-infection. We measured Nosema load with quantitative PCR and characterized microbiota with 16S rRNA gene amplicon sequencing. We found significant colony level variation in infection levels, and subtle differences between the microbiota of colonies with high infection levels versus those with low infection levels. Two exact sequence variants of Gilliamella, a core gut symbiont that has previously been associated with gut dysbiosis, were significantly more abundant in bees from colonies with high Nosema loads versus those with low Nosema loads. These bacteria deserve further study to determine if they facilitate more intense infection by Nosema ceranae.
Introduction
Flower-visiting pollinators, primarily bees (such as Apis mellifera) and other insects1, are estimated to be responsible for approximately 35% of global food production directly consumed by humans2,3. U.S. honey bee pollination is estimated to be worth between $12.3 to $16.4 billion4, with crops like California almonds relying primarily on honey bee pollinators1. From 2006–2014, winter colony losses have been approximately 30% in the USA5, an unsettling trend also observed in other countries6. While the cause for the recent increase in colony losses has not been fully elucidated, it is partially attributed to Nosema ceranae, exposure to agricultural pesticides, environmental variation, and the synergistic effects between these factors7.
Nosema ceranae, originally a parasite of the Asian honey bee Apis cerana, underwent a host shift to infect A. mellifera and has rapidly replaced Nosema apis as the primary honey bee parasite worldwide8–10. Nosema ceranae reduces the number of foragers in a colony, degenerates digestive tissue, and results in an overall reduction of colony strength, particularly in combination with other factors detrimental to honey bee health11–14. Nosema solely infects the bee midgut, inhibiting genes involved in tissue renewal, leading to malnutrition and, in certain studies, increased mortality15–17. The virulence of N. ceranae in A. mellifera, however, varies widely, and the underlying causes of this variation are still open to debate18. Several studies have suggested that colonies differ in their susceptibility to Nosema infection19. However, separate research in the USA20 and Spain21 failed to support any link between N. ceranae infection and host genetics. Therefore, colony-level variation in N. ceranae susceptibility and the contribution of the microbiota to this variability remains an open question.
Surveys of A. mellifera gut microbes from four different continents have revealed that honey bee guts harbor representatives of 5–8 bacterial phylotypes involved in host nutrition, toxicity, immune function, and possibly pathogen resistance22–25. Interestingly, this microbiota is highly conserved despite distinct differences in environment, geography, and bee sub-species. Honey bee-specific microbes are most abundant in the hindgut, but can also be found in appreciable numbers in the midgut26, the sole location of Nosema infection16. The microbiota and Nosema interaction may account for some of the variation in responses to infection and could be manipulatable to improve bee resistance to Nosema.
Understanding how Nosema infection alters the gut microbiota has implications for developing ways to restore a healthy gut community in infected bees and, perhaps, to develop microbial treatments which combat the effects of infection. The microbiota may interact with gut pathogens in multiple ways. The honey bee microbiota stimulates expression of host antimicrobial peptides27, which may prime the honey bee to fight off Nosema infection. Other possible defensive mechanisms include competition for resources between commensal microbes and gut pathogens, secretion of chemicals that inhibit or kill pathogens, and creation of physical barriers (i.e. biofilms) on host gut epithelia28,29.
Short generation times allow the microbiota to respond to selection more rapidly than the bee host, and engineering the microbiota can be a rapid manner to increase host health30. Here, we aim to better understand whether and how the microbiota might interact with Nosema to add to the long-term goal of increasing honey bee health. First, we examined variation in disease load, at a colony level, to workers deliberately infected with N. ceranae. Next, we determined whether specific microbes increase or decrease in abundance with Nosema infection. Finally, we tested for differences in the microbiota of bees from colonies challenged with the same Nosema inocula, but which exhibited opposite intensities of infection. We aimed to determine if specific bacteria correlated with N. ceranae infection are targetable predictive markers of colony health for future studies.
Results
Nosema infection
As expected, bees became significantly more infected over time (F2,136 = 8.254, P < 0.0004, Fig. 1). Moreover, there was a significant colony effect (F8,136 = 11.84, P < 0.0001). In the same ANOVA, treatment was nearly significant (F1,136 = 3.724, P = 0.055, Fig. 1). We found two colony groupings with respect to Nosema infection load (Fig. 1). Highly infected colonies (3 colonies) had bees with significantly more estimated spores than bees in low infection colonies (6 colonies, Tukey HSD test, P < 0.05).
Figure 1.
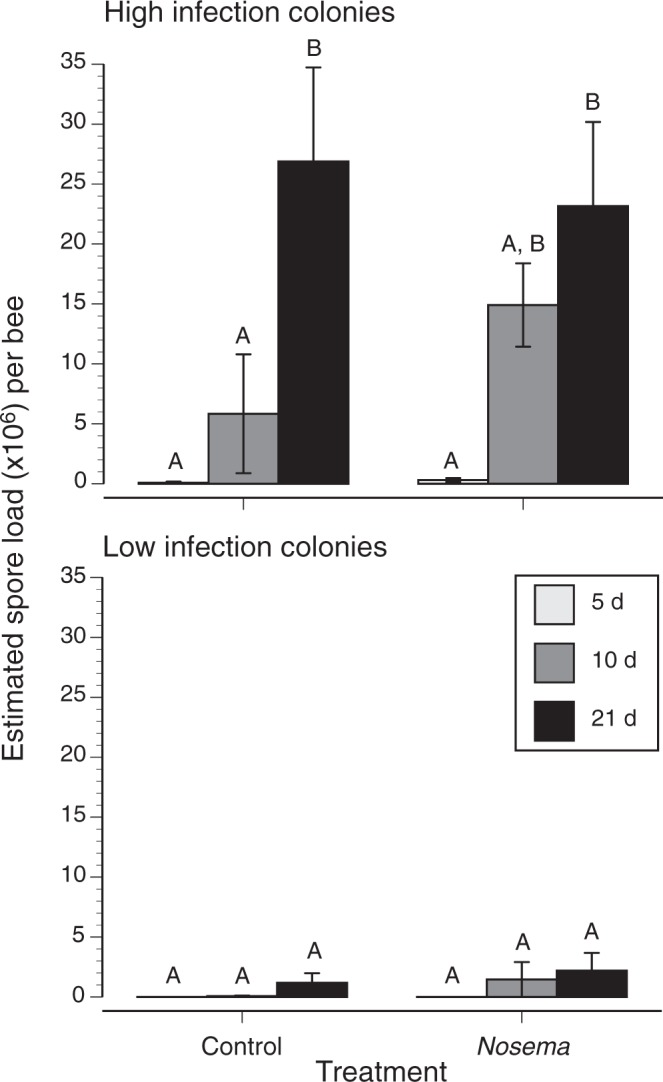
Effects of colony identity, treatment, and bee age (5, 10, or 21 day old adults) on estimated N. ceranae spore loads. Colonies are divided into two groups based upon bee infection levels (high = 3 colonies, N = 66 and low = 6 colonies, N = 82). Different letters indicate significant differences (Tukey HSD tests, P < 0.05).
Initially (5 days post-inoculation), controls and bees fed live Nosema spores had similar, low infection levels (Fig. 1). By day 21 infection levels significantly increased in the high infection colonies regardless of treatment (Tukey HSD, P < 0.02, Fig. 1), but did not significantly differ from original levels in the low infection colonies (Fig. 1). When all samples are considered, however, bees that were fed Nosema spores had significantly higher infection loads as measured by qPCR when compared to controls fed solely sucrose (Fig. 2, t on ranked data = −2.478, df = 146, P = 0.014). Despite infection in some control bees, likely due to interactions with treated bees in the colony, the average infection load in control bees was half that of treated bees.
Figure 2.
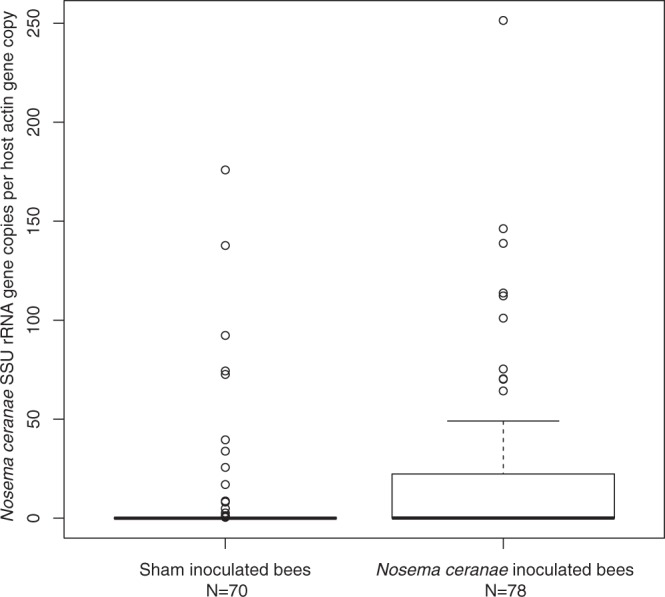
Pooled estimated spore loads (relativized to host Actin counts) for sham inoculated vs. Nosema inoculated bees over all colonies. While most control bees were infected with Nosema, given that they lived and interacted in the same colonies, the treated bees had significantly higher spore counts (more than twice as high as control bees, (t on ranked data = −2.478, df = 146, P = 0.014). Boxplots are shown.
Microbiota
After quality filtering, removal of sequences that were assigned to chloroplast or mitochondria (from either pollen in the bee’s gut or host mitochondria), removal of sequences that occurred in no-template control blank samples (buffer contaminants Shewanella and Halomonas and human-associated Propionibacterium), and removal of samples for which host actin qPCR was below our detection threshold, we retained 1,770,123 sequences across 148 individual bee samples. Based on the number of sequences per sample and the sampling depth at which rarefaction curves leveled off, we rarified each sample to 2,500 sequences, which allowed us to include 144 samples in our downstream analyses. This rarified matrix was used for all alpha and beta diversity analyses, either in its entirety or as two subsets of the entire matrix broken apart by treatment or control bees.
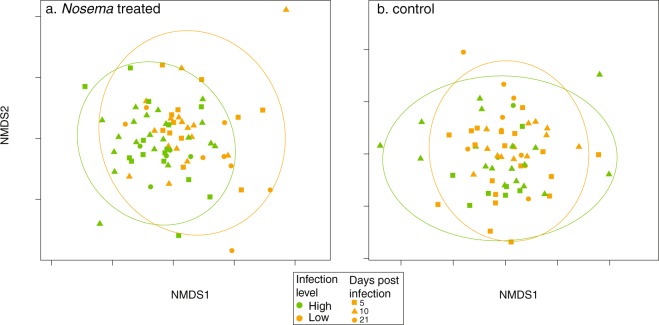
The presence of N. ceranae in control bees may explain why Adonis analysis (permutational MANOVA) of Bray-Curtis distances of gut microbiota composition showed no significant difference in control versus infected bees (Fig. S1, F1,142 = 0.91, P = 0.56). Alpha diversity (observed ESVs) approached significance, but did not differ by treatment (Kruskal-Wallis H134 = 3.54, P = 0.059). To account for infection in control bees, we tested for differences in the gut microbiota of bees from colonies that were classified as having low or high Nosema infection (as described in the Nosema infection section above). Initial graphing of an NMDS ordination of Bray-Curtis distances identified one outlier sample, which we removed for clarity of visualization in Fig. 3. Inclusion or exclusion of this sample did not affect the results. First, we repeated alpha (observed ESVs) and beta diversity analyses (Bray-Curtis distances) using only Nosema treated bees. For Nosema treated samples, high infection level colonies exhibited more ESVs per sample compared to low infection colonies (Fig. S2, Kruskal-Wallis H71 = 12.07, P < 0.0006) and beta diversity differed between colonies with high and low infection levels (Fig. 3, F1,70 = 2.28, P < 0.01). Next, when we considered only control bees alpha diversity did not differ between high and low infection level colonies (Fig. S2, Kruskal-Wallis H64 = 0.12, P = 0.75) and beta-diversity approached significance but no longer differed between colonies with low and high infection levels (Fig. 3, F1,62 = 1.53, P < 0.057).
Figure 3.
NMDS ordination coded by whether the colony of origin of the bee had high or low Nosema infection (color) and time points (shape). Ellipses represent 95% confidence intervals for colony infection level. There are subtle but significant differences in the composition of the gut microbiota in Nosema treated bees (a, F1,70 = 2.28, P < 0.01) but not in control bees (b, F1,62 = 1.53, P < 0.057).
An Ancom test to determine the differential abundance of microbes between colonies with low and high infection levels identified two ESVs that were significantly associated with infection. According to BLAST searches (max scores of 612 and 616 and sequence identities of 100%) and Naive Bayesian Classifier analyses, both ESVs were strains of the core gut symbiont Gilliamella. Both ESVs were positively associated with increasing N. ceranae abundance (Fig. 4). These two ESVs were also differentially abundant when only Nosema treated samples were considered, but not when only control bees were analyzed.
Figure 4.
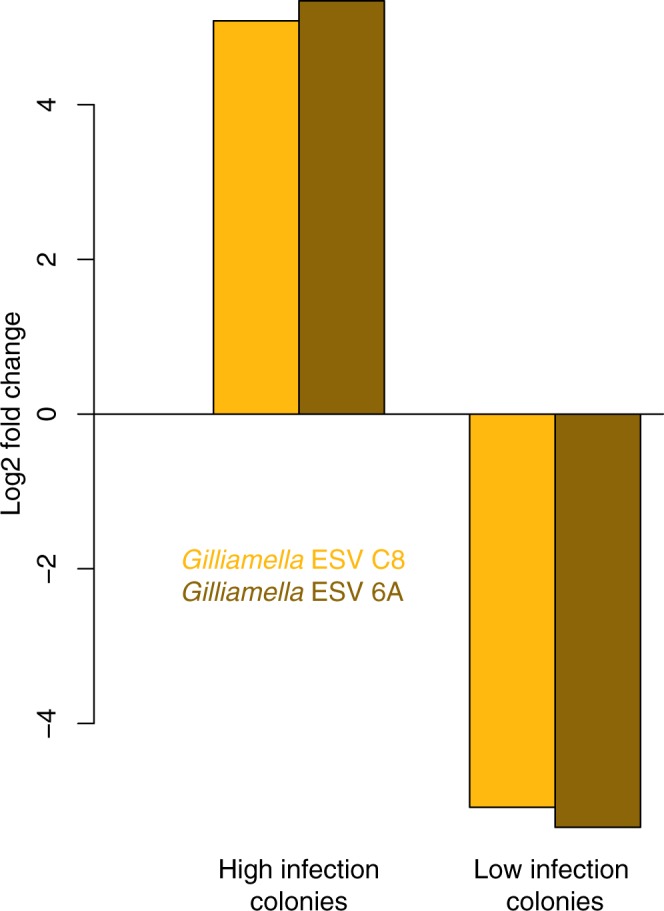
Differential abundance (log2 fold) of two Gilliamella ESVs in colonies with high infection levels versus colonies with low infection (high = 3 colonies, N = 66 and low = 6 colonies, N = 82). Ancom analysis identified these two ESVs as being significantly different across colonies with different infection statuses (False Discovery Rate adjusted P < 0.05).
Discussion
The intensity of Nosema ceranae infection has only subtle effects on the overall composition of the honey bee gut microbiome. Underlying this compositional difference are two Gilliamella ESVs that were significantly associated with Nosema ceranae load. These Gilliamella ESVs are members of the core honey bee microbiota and are thus likely important for host health23,24.
Other studies have explored interactions between N. ceranae and gut microbes by either manipulating the presence of specific strains or sequencing the microbiome of infected or uninfected bees. Corby-Harris et al.31 and Baffoni32 found that Parasaccharibacter apium and lactobacilli and bifidobacteria supplemental feedings, respectively, lowered the intensity of N. ceranae infection. Li et al.33 used a cage experiment to show that N. ceranae challenge did not affect the microbiota, but that antibiotic treatment made honey bees more susceptible to N. ceranae challenge. Hubert et al.34 used naturally occurring infection (detected by PCR) to determine that N. ceranae infected bees did not harbor significantly different microbiota compared to uninfected bees.
Our data agree with these previous studies that show that N. ceranae infection does not cause large compositional changes to the gut microbiota34. In contrast to these studies, which characterized the microbiota of entire bees, we dissected out and extracted DNA from midguts and hindguts only. While this allowed us to focus on the gut, future research focusing only on the midgut, where N. ceranae localizes35, may be promising. The midgut harbors some bacteria near the pylorus, but the honey bee rectum harbors the bulk of the gut microbiota26. The different localization of N. ceranae and the bulk of the microbiota may explain why no large changes in the entire gut microbiota co-occur with N. ceranae infection.
Our study also differs from these previous studies in that we returned the inoculated and control bees to their colony of origin post-inoculation. Although returning the treated bees to the colony resulted in N. ceranae exposure in our control bees, we were able to control for this exposure by quantifying N. ceranae infection intensity and determining which colonies had high and low levels of infection. Harboring treated bees in the colony allowed us to maintain field-realistic conditions for our study and control for potential colony-level differences. We are therefore able to conclude that N. ceranae infection subtly alters the overall composition of the gut microbiota in a colony setting. This paves the way for a future lab-based study that tests the effects of Nosema feeding on caged bees. This experiment should eliminate infection of control bees and allow us to confirm the microbiome results obtained in our current findings.
We also explicitly tested for a colony effect on Nosema susceptibility. Our results demonstrated that colonies significantly varied in their susceptibility to Nosema infection despite containing similar number of workers and being each exposed to the same number of Nosema-fed workers, each inoculated with the same dose of live spores. Colonies were initially uninfected, based upon standardized random sampling and mid-gut spore counts of 20 foragers per colony36. Gut microbiota alpha and beta diversity differed significantly, but subtly, between colonies with low or high infection levels. Furthermore, two Gilliamella ESVs were differentially more abundant in colonies with high infection levels. The microbiota may therefore play some role in Nosema infection, but it is likely that a substantial amount of variation in Nosema susceptibility at the colony level is due to host genetics. Future work investigating host population genomics, polyandry37, and Nosema susceptibility may therefore be promising.
Gilliamella is a member of the honey bee “core gut microbiome”38 and specific strains can degrade sugars that are found in floral nectar and are toxic to honey bees24. Within the honey bee-associated Gilliamella, however, a vast diversity of strains exists with different genomic repertoires and possible functions22 including a possible role in gut dysbiosis39. Indeed, gut dysbiosis in worker honey bees has been associated with increased Gilliamella counts, although one study found that Frischella perrara and not Gilliamella was significantly associated with Nosema infection40. Our finding that two specific Gilliamella strains are positively associated with Nosema infection intensity is therefore not unprecedented. The correlations between these strains and Nosema merit further study. For example, whether these ESVs reside in the midgut and affect the function of the protective peritrophic matrix or whether they are found in the ileum along with most gut bacteria and therefore interact with Nosema via the host immune system would provide insight into how these ESVs may help increase Nosema loads.
Because the pathogen and the majority of the bacteria studied here reside in different parts of the honey bee gut, it is perhaps not surprising that Nosema infection and gut microbiome composition are not strongly associated. Instead, subtle differences between colonies with high or low levels of infection allowed us to identify two Gilliamella ESVs that positively associate with Nosema infection intensity. We are therefore closer to identifying microbes that contribute to nosemosis. To be able to protect honey bees from this pathogen, we need further work to identify whether this correlation is indeed causative, and whether these bacteria can be selectively removed to promote a healthy and resistant microbiome.
Methods
Generating N. ceranae stock
To generate N. ceranae spore stock, we placed 25 newly emerged worker bees into a cage equipped with a syringe containing 5 ml of 2.0 M sucrose solution mixed with 1 million freshly extracted N. ceranae spores (40,000 per bee). Bees were fed only 2.0 M sucrose ad libitum after this 5 ml was consumed and were given 10–12 days to develop heavy infections36. Spores were harvested within 12 h before each new trial and stored at 4 °C to ensure viability36. Spores were extracted by dissecting out the midgut of infected bees (workers fed approximately 100,000 spores each for 12 days). Three midguts were placed per 100 µl DD H2O in a 1.5 ml Eppendorf tube, ground with Kimble polypropylene pestles, diluted to 1 ml, vacuum filtered through a Buchner funnel lined with Fisherbrand P8 Filter paper, and concentrated by centrifuging for 15 min at 10,000 rpm. The supernatant was discarded, and the precipitates were pooled and re-suspended in 500 µl DD H2O (modified from41). A hemocytometer was observed at 400x total magnification under a compound light microscope to determine spore concentrations36.
Obtaining bees
Workers of A. mellifera ligustica (Spinola, 1806) bees were obtained from nine colonies (each with approximately 20,000 bees, based upon visual estimation) that were all re-queened in May 2015. The colonies were housed at UCSD’s Biology Field Station and Elliot Chaparral Reserve from June 2016 to June 2017. Colonies were initially uninfected, based upon standardized random sampling and mid-gut spore counts of 20 foragers per colony36. For each colony, we collected comb frames with at least 200 capped brood cells, removed all adult bees, and transferred the frames into a nuc box that was placed into incubators at 33 °C and 60–80% relative humidity. Frames were kept in incubators for no longer than 5 days and checked daily for newly emerged worker bees.
Bee treatments
Within 24 h of emergence, 100 newly emerged workers per colony were painted on their thoraces with one of two colors of non-toxic enamel paint. Each bee was placed inside its own plastic vial into which a pipette with the treatment was inserted. Newly emerged bees are often reluctant to feed, and thus the vials were placed into racks in which strips of ultraviolet led diodes (SMD 3528, 240 lumens/m, 395–405 nm) illuminated the pipettes, attracting the bees and encouraging feeding. The majority of bees consumed the treatment in less than 1 h, as determined by visual inspection of the pipettes. In separate trials we determined that there was negligible evaporation of the solution over the course of several hours. Half of the bees were treated with 10,000 freshly harvested N. ceranae spores in 7 μL of 1.8 M sucrose solution, the rest with pure 1.8 M sucrose solution. After ingestion, we returned bees to the colony of origin. We gathered five bees from each treatment when they were 5 days old (to establish a mature microbiota26), 10 days old, and 21 days old, dissected out the midgut and hindgut, froze them in an RNase free microcentrifuge tube at −80 °C, and then delivered the guts in liquid nitrogen to UC Riverside for analyses. From these 270 bees, 148 yielded successful 16S rRNA gene, host Actin qPCR, and Nosema ceranae qPCR data (see Table S1 for sample details and raw data).
Molecular characterization of Nosema load and microbiota composition
The midgut and hindgut were transferred to sterile 96 well sample extraction plates from the DNeasy extraction kit (Qiagen, Valencia, CA). To extract DNA from the gut, two sterile 3.2 mm steel beads were added to ~50 µL of 0.1 mm glass beads. To thoroughly lyse the samples, we added 180 µL of Qiagen (Valencia, CA) buffer ATL to each well, and then bead beat the samples for six minutes at 30 hertz with the Tissue Lyser (Qiagen, Valencia, CA). We added 20 µL proteinase K to each sample and incubated them at 56 °C overnight. To complete the extractions, we followed the protocol recommended by the DNeasy Blood and Tissue kit (Qiagen, Valencia CA).
To quantify N. ceranae infection, we followed a modified version of the qPCR protocol of Bourgeois et al.42. For each 15 μl reaction we used 7.5 μl of SsoAdvanced Universal SybR Green Supermix (BioRad, Hercules, CA), 3.9 μl Ultrapure water (Invitrogen, Carlsbad, CA), 3 μl of template DNA, and 0.2 μM of each primer. We used the N. ceranae specific primers NcF (AAGAGTGAGACCTATCAGCTAGTTG) and NcR (CCGTCTCTCAGGCTCCTTCTC). We first used these primers for a standard PCR on a N. ceranae positive sample and sequenced the PCR product to verify the identity as N. ceranae. To create a standard curve, we cloned this PCR product using the TopoTA cloning kit (Invitrogen, Carlsbad, CA) and then created serial dilutions of the recombinant clones ranging from 108–102 copies. To quantify the absolute abundance of N. ceranae, we ran these dilutions on every plate as quantification standards using the CFX96 real time system (BioRad, Hercules, CA). We used the following thermocycler program: 95 °C for 3 minutes followed by 40 cycles of 95 °C for 10 seconds and 58 °C for 30 seconds. Each sample and standard were run in triplicate, including negative controls, and we used the average of the three reactions to determine N. ceranae load. Amplification efficiency was calculated by the CFX manager software, with all efficiencies ranging between 90–110% and most in the mid 90% range (for an example standard curve, see Fig. S3).
To relativize our Nosema qPCR data to a host gene, we performed quantitative PCR with the host housekeeping gene Actin43. We used the same reagent mixtures and thermocycler protocol (except with a 57 °C annealing/extension step) as described above with the primers ActinF (TGCCAACACTGTCCTTTCTG) and ActinR (AGAATTGACCCACCAATCCA). For absolute quantification, we used a gBlock custom oligonucleotide spanning the length of the fragment (156 bp, Integrated DNA Technologies, Skokoe, IL) diluted from 107 to 103 copies to create a standard curve. Amplification efficiency was calculated by the CFX manager software, with all efficiencies ranging between 90–110% and most in the mid 90% range (for an example standard curve, see Fig. S2).
To characterize microbiota composition, we used standard protocols to conduct 16S rRNA gene analyses44. For PCR reactions we used the plastid-excluding 16S rRNA gene 799F45 (CMGGGTATCTAATCCKGTT) and 1115R46 (AGGGTTGCGCTCGTTG) indexed primers that amplifies the V5 and V6 variable regions. Briefly, we used a dual barcoding approach with two primers sets to build the Illumina sequencing construct, as in Kembel et al.46 and McFrederick and Rehan47. To sequence these amplicons, we cleaned up the PCR reactions using the PureLink Pro PCR clean up kit (ThermoFisher Scientific, Waltham, MA), normalized each sample to be equimolar with SequalPrep normalization plates (ThermoFisher Scientific, Waltham, MA) and then pooled and sequenced the reactions using a 2 × 300 bp paired end run with V3 reagents on the Illumina MiSeq platform.
Statistical analyses
To determine the effects of colony, treatment, and days since infection (all fixed, nominal variables) on ranked spore loads, we used Analysis of Variance (ANOVA) and Tukey Honestly Significant Difference (HSD) tests for post-hoc analyses48. To identify colonies with low or high infection, we used the Tukey HSD results to identify colonies that significantly differed in intensity of infection.
We used QIIME2 version 2017–1149 and the R package vegan50 for all other data analyses. For quality control of the 16S rRNA gene data, we viewed quality scores of the DNA sequence and trimmed reads of low-quality regions. We used DADA251 to infer exact sequence variants (ESVs; bacteria that share the exact DNA sequence over the 16S rRNA gene region that we sequenced). To assign taxonomy to each ESV, we trained the Silva database (v. 12852) to our primer set in QIIME2, then assigned taxonomy using the sklearn classifier53. For alpha and beta diversity analyses, we first aligned representative sequences with MAFFT54 and filtered out poorly aligned sections with QIIME’s alignment mask. We built a phylogeny from the resulting alignment using FastTree55 and conducted alpha and beta diversity analyses with QIIME2’s core diversity metrics. To determine if the microbiota of N. ceranae treated versus control bees differed, we performed Permutational MANOVA (PerMANOVA) and Non-metric Multidimensional Scaling (NMDS) analyses in vegan.
To detect differences in the microbiota, we ran several different Permutational MANOVA analyses in vegan, all with Bray-Curtis distances as the dependent variables. First, we used treatment, the day post-infection, and the interaction between treatment and day post-infection as explanatory variables, and colony of origin as strata. Next, we used colony infection level, day post infection, and the interaction between colony infection level and day post infection as explanatory variables with colony of origin as strata. To understand whether treatment affected the results of the colony infection level test, we split the data set into two: one dataset with control bees only and one with treated bees only. We then ran separate Permutational MANOVA analyses with colony infection level, day post infection, and the interaction between colony infection level and day post infection as explanatory variables with colony of origin as strata. To check for differences in dispersion between groups, we used vegan’s betadisper function.
To understand whether N. ceranae infection was associated with increased or decreased abundance of specific bacteria, we tested for differential abundance of specific ESVs using an Ancom analysis56 in QIIME2 (False Discovery Rate adjusted P < 0.05, N = 144). We additionally performed separate Ancom analysis on feature tables broken out for control bees only (N = 68) or Nosema treated bees only (N = 76).
Supplementary information
Acknowledgements
We would like to thank Joshua Kohn, David Holway and two anonymous reviewers for their advice and helpful comments that have improved this manuscript. We are grateful for assistance from Felipe Hueso, Daniela Zárate, Danielle Nghiem, and Edmund Lau. Funding was made possible by Project ApisM Grant Sub S-000837 (20163523) and Madhava Natural Sweeteners.
Author Contributions
A. Rubanov conducted the colony infection experiments. K. Russell and J. Rothman ran the microbiome and qPCR analyses. A. Rubanov, J. Nieh, and Q. McFrederick analyzed the data and wrote the paper.
Data Availability
Raw 16S rRNA gene sequences are freely available from NCBI’s Sequence Read Archive (SRA accession number SRP144635). All other data are available in Supplemental Table S1.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
James C. Nieh, Email: jnieh@ucsd.edu
Quinn S. McFrederick, Email: quinn.mcfrederick@ucr.edu
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-40347-6.
References
- 1.Morse RA, Calderone NW. The value of honey bees as pollinators of US crops in 2000. Bee Cult. 2000;128:1–15. [Google Scholar]
- 2.Winfree R, Gross BJ, Kremen C. Valuing pollination services to agriculture. Ecol. Econom. 2011;71:80–88. doi: 10.1016/j.ecolecon.2011.08.001. [DOI] [Google Scholar]
- 3.Klein A-M, et al. Importance of pollinators in changing landscapes for world crops. Proc. Roy. Soc. B. 2007;274:303–313. doi: 10.1098/rspb.2006.3721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Losey JE, Vaughan M. The economic value of ecological services provided by insects. BioScience. 2006;56:311–323. doi: 10.1641/0006-3568(2006)56[311:TEVOES]2.0.CO;2. [DOI] [Google Scholar]
- 5.Lee KV, et al. A national survey of managed honey bee 2013–2014 annual colony losses in the USA. Apidologie. 2015;46:292–305. doi: 10.1007/s13592-015-0356-z. [DOI] [Google Scholar]
- 6.McMenamin AJ, Genersch E. Honey bee colony losses and associated viruses. Curr. Opin. Insect Sci. 2015;8:121–129. doi: 10.1016/j.cois.2015.01.015. [DOI] [PubMed] [Google Scholar]
- 7.Goulson D, Nicholls E, Botías C, Rotheray EL. Bee declines driven by combined stress from parasites, pesticides, and lack of flowers. Science. 2015;347:1255957–1255957. doi: 10.1126/science.1255957. [DOI] [PubMed] [Google Scholar]
- 8.Paxton RJ, Klee J, Korpela S, Fries I. Nosema ceranae has infected Apis mellifera in Europe since at least 1998 and may be more virulent than Nosema apis. Apidologie. 2007;38:558–565. doi: 10.1051/apido:2007037. [DOI] [Google Scholar]
- 9.Chaimanee V, et al. Susceptibility of four different honey bee species to Nosema ceranae. Vet. Parasit. 2012;193:260–265. doi: 10.1016/j.vetpar.2012.12.004. [DOI] [PubMed] [Google Scholar]
- 10.Fries I. Nosema ceranae in European honey bees (Apis mellifera) J. Invert. Path. 2010;103(Suppl 1):S73–9. doi: 10.1016/j.jip.2009.06.017. [DOI] [PubMed] [Google Scholar]
- 11.Aufauvre J, et al. Parasite-insecticide interactions: a case study of Nosema ceranae and fipronil synergy on honeybee. Sci. Rep. 2012;2:1–7. doi: 10.1038/srep00326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pettis JS, Van Engelsdorp D, Johnson J, Dively G. Pesticide exposure in honey bees results in increased levels of the gut pathogen. Nosema. Naturwiss. 2012;99:153–158. doi: 10.1007/s00114-011-0881-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Higes M, Meana A, Bartolomé C, Botías C, Martín-Hernández R. Nosema ceranae (Microsporidia), a controversial 21st century honey bee pathogen. Environ. Micro. Rep. 2013;5:17–29. doi: 10.1111/1758-2229.12024. [DOI] [PubMed] [Google Scholar]
- 14.Kralj J, Fuchs S. Nosema sp. influences flight behavior of infected honey bee (Apis mellifera) foragers. Apidologie. 2009;41:21–28. doi: 10.1051/apido/2009046. [DOI] [Google Scholar]
- 15.Martín-Hernández R, et al. Comparison of the energetic stress associated with experimental Nosema ceranae and Nosema apis infection of honeybees (Apis mellifera) Parasit. Res. 2011;109:605–612. doi: 10.1007/s00436-011-2292-9. [DOI] [PubMed] [Google Scholar]
- 16.Huang W-F, Solter LF. Comparative development and tissue tropism of Nosema apis and Nosema ceranae. J. Invert. Path. 2013;113:35–41. doi: 10.1016/j.jip.2013.01.001. [DOI] [PubMed] [Google Scholar]
- 17.Chaimanee V, Chantawannakul P, Chen YP, Evans JD, Pettis JS. Differential expression of immune genes of adult honey bee (Apis mellifera) after inoculated by Nosema ceranae. J. Ins. Phys. 2012;58:1090–1095. doi: 10.1016/j.jinsphys.2012.04.016. [DOI] [PubMed] [Google Scholar]
- 18.Forsgren E, Fries I. Comparative virulence of Nosema ceranae and Nosema apis in individual European honey bees. Vet. Parasit. 2010;170:212–217. doi: 10.1016/j.vetpar.2010.02.010. [DOI] [PubMed] [Google Scholar]
- 19.Martín-Hernández R, et al. Nosema ceranae in Apis mellifera: a 12 years postdetection perspective. Environ. Micro. 2018;20:1302–1329. doi: 10.1111/1462-2920.14103. [DOI] [PubMed] [Google Scholar]
- 20.Villa JD, Bourgeois AL, Danka RG. Negative evidence for effects of genetic origin of bees on Nosema ceranae, positive evidence for effects of Nosema ceranae on bees. Apidologie. 2013;44:511–518. doi: 10.1007/s13592-013-0201-1. [DOI] [Google Scholar]
- 21.Jara L, et al. Linking evolutionary lineage with parasite and pathogen prevalence in the Iberian honey bee. J. Invert. Path. 2012;110:8–13. doi: 10.1016/j.jip.2012.01.007. [DOI] [PubMed] [Google Scholar]
- 22.Engel P, Martinson VG, Moran NA. Functional diversity within the simple gut microbiota of the honey bee. PNAS. 2012;109:11002–11007. doi: 10.1073/pnas.1202970109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zheng H, Powell JE, Steele MI, Dietrich C, Moran NA. Honeybee gut microbiota promotes host weight gain via bacterial metabolism and hormonal signaling. PNAS. 2017;114:4775–4780. doi: 10.1073/pnas.1701819114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zheng H, et al. Metabolism of toxic sugars by strains of the bee gut symbiont Gilliamella apicola. mBio. 2016;7:661–9. doi: 10.1128/mBio.01326-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Engel, P. et al. The bee microbiome: impact on bee health and model for evolution and ecology of host-microbe interactions. mBio7, e02164–15–9 (2016). [DOI] [PMC free article] [PubMed]
- 26.Martinson VG, Moy J, Moran NA. Establishment of characteristic gut bacteria during development of the honeybee worker. Appl. Environ. Micro. 2012;78:2830–2840. doi: 10.1128/AEM.07810-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kwong WK, Mancenido AL, Moran NA. Immune system stimulation by the native gut microbiota of honey bees. Royal Society Open Science. 2017;4:170003–9. doi: 10.1098/rsos.170003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pickard JM, Zeng MY, Caruso R, Núñez G. Gut microbiota: Role in pathogen colonization, immune responses, and inflammatory disease. Immuno. Rev. 2017;279:70–89. doi: 10.1111/imr.12567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hillman K, Goodrich-Blair H. Are you my symbiont? Microbial polymorphic toxins and antimicrobial compounds as honest signals of beneficial symbiotic defensive traits. Curr. Opin. Microbiol. 2016;31:184–190. doi: 10.1016/j.mib.2016.04.010. [DOI] [PubMed] [Google Scholar]
- 30.Mueller UG, Sachs JL. Engineering microbiomes to improve plant and animal health. Trends Microbiol. 2015;23:606–617. doi: 10.1016/j.tim.2015.07.009. [DOI] [PubMed] [Google Scholar]
- 31.Corby-Harris, V. et al. Parasaccharibacter apium, gen. nov., sp. nov., improves honey bee (Hymenoptera: Apidae) resistance to Nosema. Ecotox. 1–7, 10.1093/jee/tow012 (2016). [DOI] [PubMed]
- 32.Baffoni L, et al. Effect of dietary supplementation of Bifidobacterium and Lactobacillus strains in Apis mellifera L. against Nosema ceranae. Benef. Microbes. 2015;7:1–8. doi: 10.3920/BM2015.0085. [DOI] [PubMed] [Google Scholar]
- 33.Li JH, et al. New evidence showing that the destruction of gut bacteria by antibiotic treatment could increase the honey bee’s vulnerability to Nosema infection. PLOS ONE. 2017;12:e0187505. doi: 10.1371/journal.pone.0187505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hubert J, et al. Changes in the bacteriome of honey bees associated with the parasite Varroa destructor, and pathogens Nosema and Lotmaria passim. Microbial Ecology. 2016;73:1–14. doi: 10.1007/s00248-016-0869-7. [DOI] [PubMed] [Google Scholar]
- 35.Bourgeois AL, Beaman LD, Holloway B, Rinderer TE. External and internal detection of Nosema ceranae on honey bees using real-time PCR. J. Invert. Path. 2012;109:1–3. doi: 10.1016/j.jip.2012.01.002. [DOI] [PubMed] [Google Scholar]
- 36.Fries I, et al. Standard methods for Nosema research. J. Apic. Res. 2015;52:1–28. doi: 10.3896/IBRA.1.52.1.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baer B, Schmid-Hempel P. Experimental variation in polyandry affects parasite loads and fitness in a bumble-bee. Nature. 1999;397:151–154. doi: 10.1038/16451. [DOI] [Google Scholar]
- 38.Moran NA, Hansen AK, Powell JE, Sabree ZL. Distinctive gut microbiota of honey bees assessed using deep sampling from individual worker bees. PLOS ONE. 2012;7:e36393–10. doi: 10.1371/journal.pone.0036393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Anderson KE, Ricigliano VA. Honey bee gut dysbiosis: a novel context of disease ecology. Curr. Opin. Insect Sci. 2017;22:125–132. doi: 10.1016/j.cois.2017.05.020. [DOI] [PubMed] [Google Scholar]
- 40.Maes, P. W., Rodrigues, A P, Oliver, P., Mott, B. M. & Anderson, K. E. Diet related gut bacterial dysbiosis correlates with impaired development, increased mortality and Nosema disease in the honey bee (Apis mellifera). Mol. Ecol. 1–31, 10.1111/mec.13862 (2016). [DOI] [PubMed]
- 41.Webster TC, Pomper KW, Hunt G, Thacker EM, Jones SC. Nosema apis infection in worker and queen Apis mellifera. Apidologie. 2004;35:49–54. doi: 10.1051/apido:2003063. [DOI] [Google Scholar]
- 42.Bourgeois AL, Rinderer TE, Beaman LD, Danka RG. Genetic detection and quantification of Nosema apis and N. ceranae in the honey bee. J. Invert. Path. 2010;103:53–58. doi: 10.1016/j.jip.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 43.Ricigliano, V. A. et al. Honey bees overwintering in a southern climate: longitudinal effects of nutrition and queen age on colony-level molecular physiology and performance. Sci. Rep. 1–11, 10.1038/s41598-018-28732-z (2018). [DOI] [PMC free article] [PubMed]
- 44.Engel P, et al. Standard methods for research on Apis mellifera gut symbionts. J. Apic. Res. 2015;52:1–24. doi: 10.3896/IBRA.1.52.4.07. [DOI] [Google Scholar]
- 45.Hanshew AS, Mason CJ, Raffa KF, Currie CR. Minimization of chloroplast contamination in 16S rRNA gene pyrosequencing of insect herbivore bacterial communities. Journal of Microbiological Methods. 2013;95:149–155. doi: 10.1016/j.mimet.2013.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kembel SW, et al. Relationships between phyllosphere bacterial communities and plant functional traits in a neotropical forest. PNAS. 2014;111:13715–13720. doi: 10.1073/pnas.1216057111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McFrederick QS, Rehan SM. Characterization of pollen and bacterial community composition in brood provisions of a small carpenter bee. Mol. Ecol. 2016;25:2302–2311. doi: 10.1111/mec.13608. [DOI] [PubMed] [Google Scholar]
- 48.R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing (2018).
- 49.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Oksanen J, et al. Vegan: community ecology package. R package version 2.4-3. R package version. 2017;2:4–3. [Google Scholar]
- 51.Callahan BJ, et al. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods. 2016;13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Quast C, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Research. 2012;41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bokulich, N. A. et al. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. 1–17, 10.1186/s40168-018-0470-z, (2018). [DOI] [PMC free article] [PubMed]
- 54.Katoh K, Misawa K, Kuma K-I, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Research. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Price MN, Dehal PS, Arkin AP. FastTree 2–approximately maximum-likelihood trees for large alignments. PLOS ONE. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mandal S, et al. Analysis of composition of microbiomes: a novel method for studying microbial composition. Microb. Ecol. Health Dis. 2015;26:27663. doi: 10.3402/mehd.v26.27663. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw 16S rRNA gene sequences are freely available from NCBI’s Sequence Read Archive (SRA accession number SRP144635). All other data are available in Supplemental Table S1.