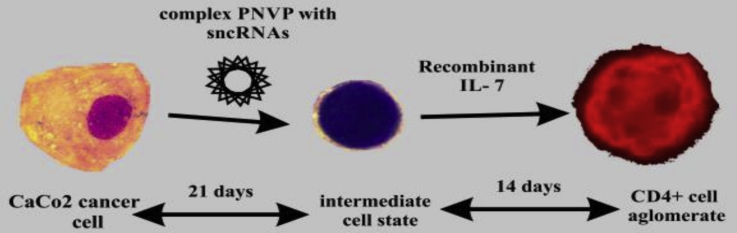
Graphical abstract
Abbreviations: SncRNAs, small non-coding RNAs; miR, micro-RNA; piR, piwi-interacting RNA, P-element induced wimpy testis interacting RNA; IL, interleukin; CD, cluster of differentiation; DTT, dithyothreitol; MKI-67, marker of proliferation ki-67; OCT4, octamer-binding transcription factor 4; mTOR, mechanistic target of rapamycin; VMAF, musculoaponeurotic fibrosarcoma; PIWIL1, piwi-like protein 1; BACH1, BTB domain and CNC homolog 1; HMOX1, heme oxygenase 1; RB1, retinoblastoma 1; DICER1, ribonuclease III; AGO2, argonaute 2; HOXA10, homebox A10; KIR1DL2, CD158b, expressed on natural killer cells and a subset of T cells; TGFBR2, transforming growth factor beta receptor 2; ICOS1B, inducible T-cell co-stimulator; GITR3A, glucocorticoid-induced TNFR-related protein; PNVP, poly-(N-vinylpyrrolidone); TNFRS6B, TNF receptor superfamily 6B; Wnt-1, wingless type MMTV integration site family, member 1; DNMT1, DNA methyltransferase 1; ERK1/2, extracellular signal regulated kinase ½; FGF2, fibroblast growth factor 2; iPS, induced pluripotent stem cells; H3K9me3, tri-methyl lysine 9 of histone H3; TSS, transcriptional start sites; HILI, human piwi; TE, transposon elements
Keywords: miRNA-152, piRNA-30074, Polymer carriers, CaCo2 colorectal adenocarcinoma, Reprogramming, Amphiphilic poly-(N-vinylpyrrolidone)
Highlights
-
•
Morphological and genetic changes of colorectal adenocarcinoma cells after treatment with complex of mix piR-30074 and miR-152 and PNVP carrier were obtained.
-
•
The combination mix of sncRNAs for the transformation and reprogramming proved to be the most preferable.
-
•
MiR-152 proved to be a promoter of apoptosis in CaCo2 cells and regulator of an anti-tumorigenic effect.
-
•
piR-30074 induces transformation of cancer cells into non-cancerous stem cells.
Abstract
Small non-coding RNAs control normal development and differentiation in the embryo. These regulatory molecules play a key role in the development of human diseases and are used often today for researching new treatments for different pathologies. In this study, CaCo2 colorectal adenocarcinoma cells were initially epigenetically reprogrammed and transformed into CD4+ cells with nano-sized complexes of amphiphilic poly-(N-vinylpyrrolidone) (PVP) with miRNA-152 and piRNA-30074. The transformation of cells was confirmed by morphological and genetic changes in the dynamic of reprogramming. CD4+ lymphocytes marker was detected using immunofluorescence. Amphiphilic poly-(N-vinylpyrrolidone)/small non-coding RNAs complexes were investigated for transfection efficiency and duration of transfection of CaCo2 colorectal adenocarcinoma cells using fluorescence.
1. Introduction
Recently a substantial number of articles have been published about different small non-coding RNAs (sncRNAs) due to their wide influence on cell biology and physiology. Despite the great interest in this class of small regulatory molecules, the properties of sncRNAs for modifications of cellular genome have not been studies as much. In this study, genomic reprogramming of CaCo2 adenocarcinoma cells into CD4+ cells were undertaken. Three major types of sncRNAs, small interfering RNAs (siRNAs), micro-RNAs (miRNAs), and piwi-interacting RNAs (piRNAs), associated with proteins in the Argonaute/piwi family. Among the three types of small RNAs, piRNAs are the most numerous and are the least investigated [1]. SncRNAs, which regulate normal stem cells physiology, also support cancer cell reprogramming [[2], [3], [4], [5]].
From the numerous families of sncRNAs we selected separate sequences via bioinformatics tools to be the best candidates for the transformation of CaCo2 cells. Small RNA targets were predicted by three computational algorithms [[6], [7], [8]].
Previous articles had observed transformations of different cancer cells therefore we transformed and investigated cell lines that are more commonly connected with cancers in humans [[9], [10], [11]]. A-549 lung adenocarcinoma cells were reprogrammed into CD4+ cells after incubation of cells with a complex of a DDMC vector with piRNA-30074 and antago-miRNA-155 followed by further treatment of the cells with IL-7 [9]. Girardi Heart cells (combined cervix cancer with write atrium cancer) were transformed into CD4+ cells after treatment with a complex of the DDMC vector with an antagonist of piRNA-30074, miRNA-155 and miRNA-125b [10]. Acute myeloid leukemia cells were transformed into platelet-like cells after using a complex of PNVP with antago-miRNA-155 [11].
This study continues previous investigations about the possibility of transforming cancer cells into other types of cells. In this research nearly 40 sncRNAs and theirs complexes were investigated for their ability to modify CaCo2 colorectal adenocarcinoma cells. The influence that piRNA-30074 and miRNA-152 complexes may have on CaCo2 adenocarcinoma cells, and whether this combination is the best mix of sncRNAs for the transformation/reprogramming of this type of cells into CD4+ cells is described. MiRNA-152 was used for possible induction of apoptosis in CaCo2 cells and for its anti-tumorigenic effect.
PiRNA-30074 was used as a factor for regulating transformation of stem cells. In the first series of experiments, after adding the mixture of piRNA-30074 and miRNA-152 to the CaCo2 adenocarcinoma cells, the transitional form of cells was obtained. In the second series of experiments, IL-7 was added to the obtained cells. After 14 days of incubating the cells with IL-7, CD4+ cells were detected using immunofluorescence techniques.
2. Materials and methods
2.1. Cell culture
The CaCo2 (ATCC HTB-37 ™) is a human colorectal adenocarcinoma cell line with ten common markers, i.e. t(1q;?), 10q-, t(11q17q) and 7 others. The t(1q17q) and M11 were found in a portion of cells. The ins(2), 10q-, and t(15q;?) were generally paired, and t(11q;17q) and t(21q;?) were mostly three-copied. Normal N9 was absent, and N21 was lost in some cells. One to four small acrocentric chromosomes were detected. No Y chromosome with bright distal q-band was detected by Q-observation.
Cells were routinely maintained in accordance with standard protocol [12]. CaCo2 cells cultures routinely were maintained in tissue culture flasks in Dulbecco’s Modified Eagle Medium (DMEM) containing 10% Fetal Bovine Serum, 2 mM l-glutamine, 100 mg/ml penicillin, and 100 ME/ml streptomycin (Pan-Eco, Ltd., Russian Federation) and incubation parameters were set at 37 °C with 5% CO2. After 2 days, cells were prepared for further cultivation with a growth concentration of cells at 0,5 × 106/ml. After accumulation of cells number 1 × 106 / ml, they were prepared for transfection by nanoparticles.
After the medium change, nanoparticles were added in a concentration of 50 μl/ml per milliliter of culture medium (90% DMEM, 10% Fetal Bovine Serum, 2 mM l-glutamine, 100 mg/ml penicillin, and 100 ME/ml streptomycin) and cells were incubated at 37 °C with 5% CO2.
2.2. Cell treatment
Cells were incubated for 30 days after transfection and they were analyzed after 11, 21 and 30 days. One portion of the cells were removed at 11, 21 and 30 days, treated with a lysis buffer from an RNAeasy mini kit (Qiagen, USA), and frozen at -20 °C for further total RNA isolation, reverse transcription reaction, and specific cDNA transcript amplification.
Another portion of the cells was used for staining using the Leishman-Romanowsky method.
2.3. Magnetic separation
On the 31st day of incubation, a third portion of cells was treated with recombinant IL-7 (AbDSerotec, UK, Kidlington), and the cells were further incubated for 14 days, with routine changing of the medium. One part of these cells was used for staining using the Leishman-Romanowsky method, and another part of these cells was treated using the Dynabeads® CD4 Positive Isolation Kit, which utilizes DETACHaBEAD™ DYNAL™ Dynabeads™ (both purchased from Invitrogen, Life Technologies, USA). Separated cells were stained using CD4+/FITC staining reagent (R&D, USA) and were observed using fluorescent microscopy techniques (AxioVertA1, Zeiss, Germany). Negatively selected cells were treated with CD117 purified mouse anti-human antibodies (Caltag Laboratories by Invitrogen, USA), treated with Dynabeads® Pan Mouse IgG (Invitrogen, Life Technologies, USA) and stained using the Leishman-Romanowsky method. The last part of the cells, which were negatively separated, were treated using the Dynal® Monocyte Negative Isolation Kit (Invitrogen, Life Technologies, USA) and stained using the Leishman-Romanowsky method.
2.4. Carriers
An amphiphilic Poly-(N-vinylpyrrolidone), with an Mn = 3500, containing one hydrophobic n-octadecyl end group was used. The side groups of amino acid β-alanin were introduced in 8 mol % of polymer rings by the recently described method [[11], [12], [13], [14], [15], [16], [17], [18], [19], [20], [21], [22]]. The overage diameter of particles was approximately 100 nm.
Oligonucleotides (25% w/w) was added to the polymer. Immobilization of the system was achieved by mix in sterile aqueous solution at room temperature.
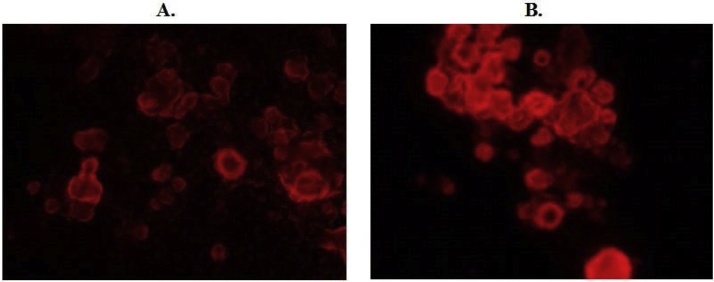
For the nuclear transfection efficiency control, the pmKate2-N vector was used, expressing and encoding far-red fluorescent protein mKate2 (Evrogene, Russia) [[23], [24], [25], [26], [27], [28], [29], [30], [31], [32], [33]]. Photos were made via fluorescent microscopy for quantification of transfection efficiency level (AxioVertA1, Zeiss, Germany). For the control, cells without any treatment were used, and cells which were treated with unloaded nanoparticles (Fig. 1).
Fig. 1.
Microscopic photos of CaCo2 colorectal adenocarcinoma cells treated with complexes of PVP with deep-red Kate2 fluorescent gene vector after 14 (A.) and 28 days (B.) (investigation of transfection efficiency) (Magnification x600).
2.5. Reverse Transcriptase-PCR
Total RNA was extracted from the cell culture using the RNAeasy Mini kit (Qiagen, USA) according to the manufacturer’s protocol. In these experiments a standard two-step reverse transcriptase-PCR procedure was used. All products were from Fermentas (Thermo Fisher). For our investigations we chose gene expression profiles of genes which regulate main functions of cells: apoptosis, proliferation, cell cycle, tumorigenic and mutagenic activity.
Amplification of Caspase-9, MKI-67, OCT4, mTOR, VMAF and PIWIL1, BACH1, HMOX1, RB1, DICER1, AGO2, HOXA10, KIR1DL2, TGFBR2, ICOS1B, GITR3A, and β-actin cDNA (as an internal control) was performed with an automatic thermocycler (TProfessional, Biometra, Germany).
Statistically significant changes in four genes expressions were obtained. The custom primers sequences were: Caspase-9 (NG_029188.1) 5′- TCCAGATTGACGACAAGTGC-3′, 5′- CACTCAGGAAGACGCGTTAC-3′; MKI-67 (ENST00000617118) 5′- TGCAAACAGGTCAGGAAGG-3′, 5′- CTGCCCCCAAGTTCTTGAT-3′; m-TOR (ENST00000376838) 5′- CCACCTATCCCAAGACCTCA-3′, 5′- GTGATCCCCTCTGTGCATCT-3′; PIWIL1 (ENST00000245255) 5′- TGCTATTCACCGGCTTCCTT-3′, 5′- TGCTCACTCCTGAAAGTACGT-3′. Oligonucleotide sequences were for miRNA-152 (MIMAT0026479) 5′-3′: AGG UUC UGU GAU ACA CUC CGA CU, and for piRNA-30074 (DQ569962.1): 5′ – AAAGCTTTAAGTGTGTTGGCGTGCTTC – 3′.
The polymerase chain reaction was made in accordance with standard protocols. All components of reaction were purchased from Invitrogen (USA). PCR products were loaded on 1.5% agarose gel and electrophoresed then colored with Ethidium Bromide, exposed to a gel doc system (Syngene, India) and quantified with Quantity One Software (Bio-Rad, USA). Sequences for inner control β-actin (ENST00000331789) gene primers were: 5′- TCCCTGGAGAAGAGCTACGA-3′, 5′-AGCACTGTGTTGGCGTACAG-3′.
Leishman-Romanowsky staining of the CaCo2 cells was made in accordance with the Blood safety and clinical technology guidelines on standard operating procedures for hematology.
3. Statistics
Data are presented as the mean ± SEM. Two-tailed Student’s t-test was used for analyses comparing the groups. The observed differences between study groups were considered statistically significant if p-values were ≤ 0.05. For the control groups, cells without any treatment, and cells that were treated with unloaded nanoparticles were used. All manipulations with the CaCo2 cell culture were repeated three times. All gene expression data were normalized to gene expression levels of beta-actin. For the external control, culture of cells without any treatment in the same moment of time as experimental cells was used. All samples were prepared in triplets.
4. Results
CaCo2 colorectal adenocarcinoma cells were transformed into CD4+ cells after treatment of the cells with complexes of PNVP with the apoptotic regulator miRNA-152 and piRNA-30074 for 30 days, and subsequent treatment with recombinant IL-7, a differentiation factor for lymphocytes [24,25]. A small number of CD117+ cells was also obtained using manual magnetic separation.
In the pilot studies, transfection activity of poly-(N-vinylpyrrolidone) (PVP) carriers with colorectal CaCo2 cancer cells had been investigated. The highest and most stable transfection activity using PVP particles was from 14 days to 28 days after treatment of the cells (Fig. 1).
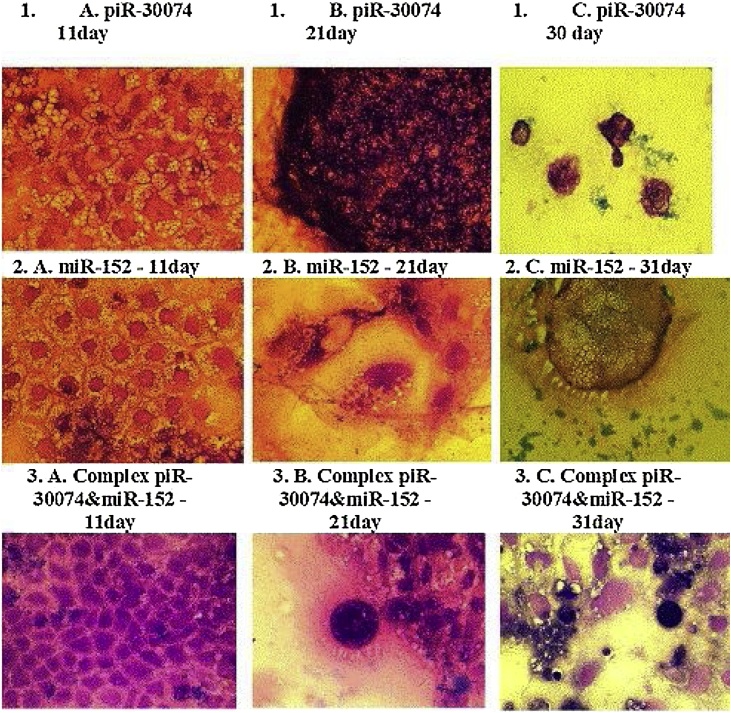
In the first series of experiments, the influence of complex PVP with piRNA-30074, complex PVP with miRNA-152 and complexes of PVP with piRNA-30074 and miRNA-152 on the morphology and genetics of human CaCo2 colorectal adenocarcinoma cells was investigated. The morphology of the adenocarcinoma cells changed during the dynamic transformation. Cellular and nuclear forms, sizes and number were different when compared with control cells (Fig. 2). Cells had apoptotic changes such as: pyknosis of nuclei, apoptotic vesicles in the cytoplasm and in the extracellular medium, and increased cellular size (Fig. 2).
Fig. 2.
Changes of CaCo2 colorectal adenocarcinoma cancer cells morphology on the 11th, 21 st and 31 st days after transfection.
Microscopic photos of CaCo2 colorectal adenocarcinoma cells after using the complex of PVP with piR-30074 (1), complex of PVP with miR-152 (2) and complex of PVP with piR-30074 and miR-152 (3) on the 11th (A), 21 st (B) and 31 st (C) days of incubation.
In photos obtained staining with the Leishmann-Romanowsky method (Magnification x600).
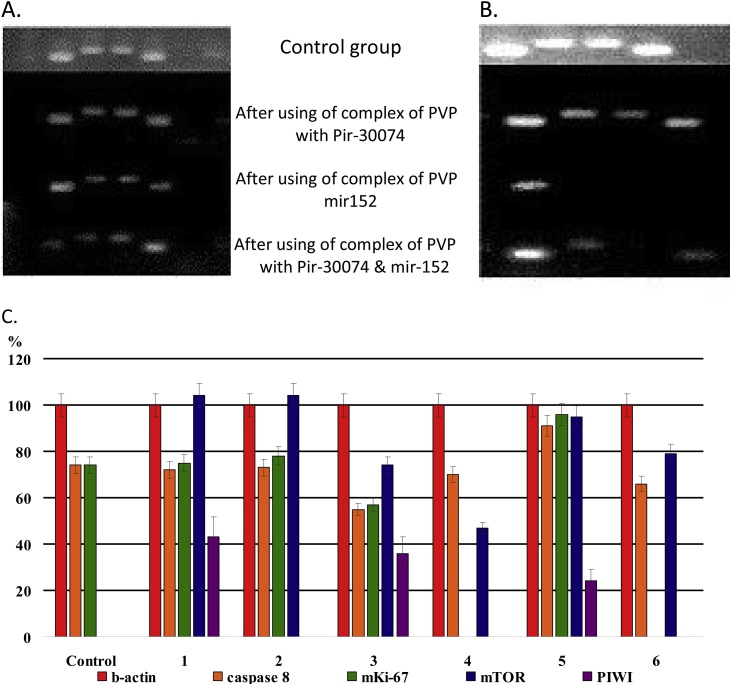
The expression levels of Caspase 9 gene was increased on the 11th day after treatment with complexes of PVP with piRNA-30074 and miRNA-152, compared to control cells. Decreased levels of the proliferation marker MKI-67 gene in cells treated with complexes of PNVP with miRNA-152 were obtained after 11 and 31 days, as well as in cells treated with complexes of PVP with piRNA-30074 and miRNA-152 after 31 days, compared with the control group. The level of mTOR gene expression was increased in all treated cells, compared to the cell control group. Levels of PIWIL1 gene expression were also increased in all treated groups of cells 11 days after the addition of sncRNAs (Fig. 3).
Fig. 3.
A. Photos of results of electrophoresis in agarose gel of Caspase-9, MKI-67, mTOR, OCT4, and PIWIL1 gene expressions (from left to right) in CaCo2 colorectal adenocarcinoma cells after adding the complex of PVP with piR-30074, complex of PVP with miR-152 and complex of PVP with piR-30074 and miR-152 on the 11th day. B. Photos of results of electrophoresis in agarose gel of genes expressions of beta-actin, Caspase 8, mKi-67, mTOR, and PIWI (from left to right) in CaCo2 cells after using the complexes of PVP with piR-30074, PVP with miR-152 and complex of PVP with piR-30074 and miR-152 on the 31st day. C. Graphs of genes expressions levels (in percent) calculated for Caspase 9, MKI-67, mTOR, OCT4, PIWIL1to compare with the internal beta-actin control (100%).
1 – Complex of PVP and piR-30074 -11 day,
2 – Complex of PVP and piR-30074 31 day,
3 – Complex of PVP with miR-152 - 11 day,
4 – Complex of PVP with miR-152 -31 day,
5 – Complex of PVP with piR-30074 and miR-152 – 11 day,
6 - Complex of PVP with piR-30074 and miR-152 – 31 day.
In the literature, data regarding colorectal cancer have, to some extent, been contradictory. Several studies have reported no prognostic value of MKI-67 expression. One study reported an association between a low tumor cell proliferation rate at the invasive margin and poor prognosis of TNM stage II colorectal cancer, whereas others have reported an adverse prognostic value of a high MKI-67 after curative resection for colorectal cancer [26,27]. In accordance with our data, a study of prognostic markers in colon cancer stage II and III treated with surgery with or without adjuvant 5-FU and leucovorin (calcium folinate) therapy, showed an improved outcome among patients with a high percentage of MKI-67-positive tumor cells [28,29].
One study has delineated a role for mTOR in pancreatic cancer cell lines, confirming that the mTOR pathway is important for stem-like cell functions [30].
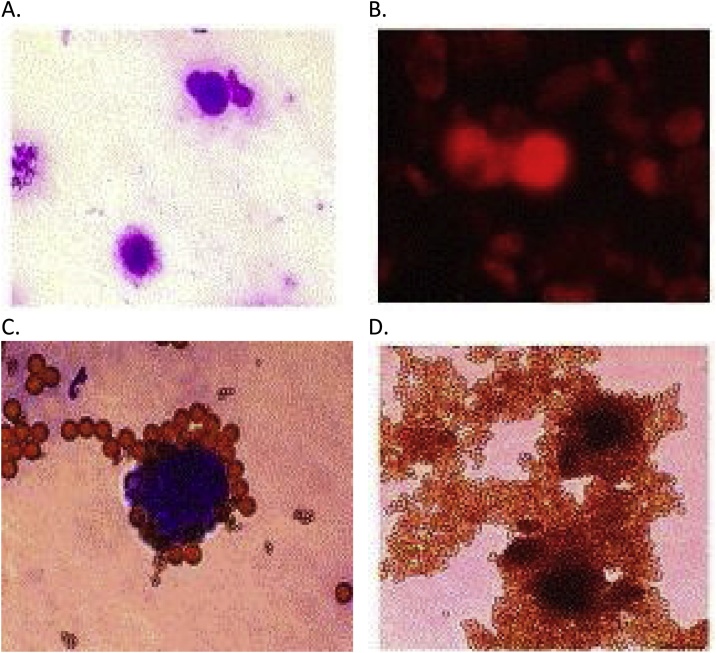
In the next series of experiments CD4+ cells after incubation with IL-7 were obtained, which are previously changed cells after pretreatment with complexes of PNVP with piRNA-30074 and miRNA-152. An absence of adhesive properties and changes in the sizes and morphology of cells and nuclei were obtained following treatment with complexes of PVP with piRNA-30074 and miRNA-152. Cells exhibited sharp forms, and the cytoplasm enriched a large nucleus. In the group of cells treated with complexes of PVP with only piRNA-30074 or only miRNA-152, had observed changes in cellular morphology, but these changes were not as clear as the changes observed in cells that were treated with complexes of PVP with piRNA-30074 and miRNA-152 plus IL-7. A total of 10–15% of CD4+ cells were obtained following cell separation (Fig.4).
Fig. 4.
Microscopic photos of transformed CaCo2 colorectal adenocarcinoma cells on the 31 st day after transfection with complex of PVP with piR-30074 and miR-152, and on the 14th day after adding of recombinant IL-7 (stained using the Leishmann-Romanowsky method) (Magnification x600): A. Cells are stained using the Leishmann-Romanowsky method. B. Cells separated using CD4+ positive magnetic separation and labeled using CD4+/FITC reagent. C. Cells selected using CD117+ positive magnetic separation (stained using the Leishmann-Romanowsky method). D. CD4+ cells before washing with DETACHaBEAD™ DYNAL™ Dynabeads™.
Transformation of CaCo2 adenocarcinoma cells into CD4+ cells was obtained and confirmed. Cells were separated with magnetic nanoparticles in order to isolate CD117 cells (Fig. 4). CD4+ and CD-117+ cells were obtained, which indicate the pro-T stage and the subsequent pre-T stage, respectively, of αβ T cell development [31].
5. Discussion
The transformation of human colorectal adenocarcinoma CaCo2 cells into CD4+ cells 30 days after reprogramming with complexes of amphiphilic PVP with miRNA-152 and piRNA-30074 and 14 days following IL-7 addition was observed. SncRNAs play an important role in many intracellular processes such as regulation of gene transcription, protein translation, epigenetic modification, genomic stability, and chromatin organization [[32], [33], [34], [35], [36]].
Eventually, almost all cells could be reprogrammed to pluripotency, although with different latency periods. Induction of the apoptosis pathway and overexpression of pluripotency factors accelerate the kinetics of reprogramming by increasing the cell division rate, which may facilitate the acquisition of DNA and/or histone modifications [[37], [38], [39]].
At the 11th day after transfection with complexes of PVP with piRNA-30074 and miRNA-152, an increase was found in the proliferative factor MKI-67, which may indicate changes in the proliferative potential of transfected cells and the influence of added sncRNAs on cell genetics [[40], [41], [42]]. Certainly, primary activation and genetic transformation of cells begins in the first two weeks after addition of complexes of PVP with piRNA-30074 and miRNA-152.
After this period, the expression of specific proteins is followed up because of the appearance of new types of cells, which is where CD4+ cells develop. What happened with primary cancer cells? First, one type of the cells is “dying” due to that they can’t transform and adapt to new environmental conditions. Another population then of cells can be ready for further transformation. Potential candidates for this observation could be cancer cells after division and cancer stem cells. These underlying mechanisms need further investigation.
Three possible mechanisms could be proposed that result in transformation of cells into CD4+ progenitor cells. First, miRNA-152 in combination with piRNA can induce apoptosis, blockade of mobile transposable elements, or caspase nucleus elimination. Piwi proteins, whose gene expression was observed 11 days after transfection, may affect the post-transcriptional regulation of oncogenes and tumor suppressor genes [43,44]. MiRNA-152 is a member of the miRNA-148/152 family. These family members potentially act as oncogenes and tumor suppressors [45,46].
MiRNA-152 mimics can induce caspases activity, suppress cell growth and inhibit cell motility in hepatocellular cancer by down-regulation of TNFRS6B, Wnt-1, DNMT1, ERK1/2, AKT, FGF2 [47,48].
Low miRNA-148/152 expression is associated with a significantly shorter life expectancy, a decrease in therapeutic response, and a poor prognosis in patients with colorectal cancer [49]. Increased levels of Caspase 8 were obtained after transfection of colorectal adenocarcinoma cells with complexes of piRNA-30074 and miRNA-152. In this case, miRNA-152 could play a role similar to master-regulator miRNA in changing gene expression of cell lifetime.
A second possible mechanism to explain the observed transformation into CD4+ cells is regulation of chromatin acetylation and histone methylation in cells by piRNAs, in order to support chromatin stability in the genomes of cells in post-transformation stage [50]. Recent insights have been gained by treating reprogrammed cells with agents that affect the chromatin state. It is likely that a key step in the generation of induced pluripotent stem cells (iPS) cells is the reopening of the somatic cell chromatin.
However, piRNAs subsequently recruit piwi proteins and H3K9me3 to the promoter region to form large transcriptional silencing complexes, which cover transcriptional start sites (TSS), hindering the ability of RNA polymerase II to recognize the TSS, leading to silencing of target gene expression [[51], [52], [53]].
PIWI proteins accumulate γ-H2Av foci and can form a complex with RecQ1, suggesting a positive contribution of PIWI proteins to the repair of DNA damage [[54], [55], [56], [57], [58], [59]]. In an ovarian cancer cell line, HILI repaired cisplatin–induced DNA damage to help cancer cells survive platinum-based chemotherapy [60]. Most piRNAs are complementary to transcripts and are able to inhibit expression of retrotransposon mRNAs, as well as repress the mobile genomic transposon elements (TE) that protect the integrity of the genome [61,62]. Components of TEs, both transcripts and proteins, are elevated in cancers compared to normal tissues. In gastric cancer and multiple myeloma cell lines, piRNA-823 levels affect tumor aggressiveness, although strangely, in opposite directions: in gastric cancer cells, piRNA-823 has an overall tumor suppressor activity in multiple myeloma, it promotes cancer development, suggesting perhaps functional interactions with cell-specific factors [63].
MiRNA inhibitor molecules and siRNA-based therapeutics are currently being developed as potential therapeutic avenues [[64], [65], [66], [67], [68], [69]]. However, piRNAs have not been explored as a therapeutic target [70].
A third, possibility is regulation of reprogramming and partial cell differentiation in an IL-7-dependent way orchestrated by a combination of piRNA, miRNA and IL-7, the powerful lymphocyte progenitor differentiation factor.
6. Conclusions
PiRNAs and miRNAs edit intra-nuclear and extra-nuclear genetic program of CaCo2 colorectal adenocarcinoma cells caused in theirs modification and transformation into stem-cell-like state, and prepare this cell to further changes after using the recombinant IL-7. Treatment of transitory form of cells with IL-7, support maturation of stem-cell-like form of cells into CD4+ lymphocyte-like morphologic and genotypic cell-type. Applying a sncRNAs mix with a nanosize polymer carrier may be a useful tool in the future as combined therapy of colorectal cancer.
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Siomi M.C., Sato K., Pezic D., Aravin A.A. PIWI-interacting small RNAs: the vanguard of genome defence. Nat. Rev. 2011;12:246–258. doi: 10.1038/nrm3089. [DOI] [PubMed] [Google Scholar]
- 2.Klimenko O.V., Onishi Y. The disappeared cancer cell by sncRNAs: application of DDMC vector/sncRNAs complex for transformation of cancer cells into non-cancerous cells. J. Nanomed. Biother. Discov. 2018;8:1–2. [Google Scholar]
- 3.Klimenko O.V. Small non-coding RNAs as regulators of structural evolution and carcinogenesis. Noncoding RNA Res. 2017;2:88–92. doi: 10.1016/j.ncrna.2017.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pulito C., Donzelli S., Muti P., Puzzo L., Strano S., Blandino G. microRNAs and cancer metabolism reprogramming: the paradigm of metformin. Ann. Transl. Med. 2014;2:58. doi: 10.3978/j.issn.2305-5839.2014.06.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hatziapostolou M., Polytarchou C., Iliopoulos D. miRNAs link metabolic reprogramming to oncogenesis, Trends Endocrinol. Metabolism. 2013;24:361–373. doi: 10.1016/j.tem.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 6.Lewis B.P., Shih I.H., Jones-Rhoades M.W., Bartel D.P., Burge C.B. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 7.Krek A., Grun D., Poy M.N., Wolf R., Rosenberg L., Epstein E.J. Combinatorial microRNA target predictions. Nat. Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 8.Griffiths-Jones S., Grocock R.J., van A., Bateman D.S., Enright A.J. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klimenko O.V. Joint action of the nano-sized system of small non-coding RNAs with DDMC vector and recombinant IL-7 reprograms A-549 lung adenocarcinoma cells into CD4+ cells. Immunother. (Los Angel) 2017;3:1–8. [Google Scholar]
- 10.Klimenko O.V. Complex of small non-coding RNAs piR-30074 and Antago-miR-155 and miR-125b with DDMC carrier transforms girardi heart cells into CD4+ cells. J. Cancer Tumor. Int. 2016;4:1–8. [Google Scholar]
- 11.Klimenko O.V., Shtilman M.I. Transfection of Kasumi-1 cells with a new type of polymer carriers loaded with miR-155 and antago-miR-155. Cancer Gene Ther. 2013;20:237–241. doi: 10.1038/cgt.2013.11. [DOI] [PubMed] [Google Scholar]
- 12.Torchilin V.P., Levchenko T.S., Whiteman K.R., Yaroslavov A.A., Tsatsakis A.M., Rizos A.K., Michailova E.V., Shtilman M.I. Amphiphilic poly-N-vinylpyrrolidones: synthesis, properties and liposome surface modification. Biomaterials. 2001;22:3035–3044. doi: 10.1016/s0142-9612(01)00050-3. [DOI] [PubMed] [Google Scholar]
- 13.Rizos A.K., Tsikalis I., Tsatsakis A.M., Shtilman M.I. Characterization of amphiphilic poly-N-vinylpyrrolidone derivatives by dynamic light scattering. J. Non-Crystall. Solids. 2006;352:5055–5059. [Google Scholar]
- 14.Kuskov A.N., Shtilman M.I., Goryachaya A.V., ashmuhamedov R.I.T., Yaroslavov A.A., Torchilin V.P., Tsatsakis A.M., Rizos A.K. Self-assembling nanoscaled drug delivery systems composed of amphiphilic poly-N-vinylpyrrolidones. J. Non-Crystall. Solids. 2007;353:3969–3975. [Google Scholar]
- 15.Kuskov A.N., Villemson A.L., Larionova N.I., Tsatsakis A.M., Shtilman M.I. Amphiphilic poly-N-vinylpyrrolidone nanocarriers with incorporated model proteins. J. Phys.: Condens. Mater. 2007;19:459–468. [Google Scholar]
- 16.Kuskov A.N., Voskresenskaya A.A., Goryachaya A.V., Artyukhov A.A., Shtilman M.I., Tsatsakis A.M. Preparation and characterization of amphiphilic poly-N-vinylpyrrolidone nanoparticles containing indomethacin. J. Mater. Sci.: Mater. Med. 2010;21:1521–1530. doi: 10.1007/s10856-010-4029-1. [DOI] [PubMed] [Google Scholar]
- 17.Luss A.L., Andersen C.L., Benito I.G., Marzo R.C., Medina Z.H., Rosenlund M.B., Romme S.B., Kulikov P.P., Pennisi C.P., Shtilman M.I., Gurevich L. Drug delivery platform based on amphiphilic Poly-N-Vinyl-2-Pyrrolidone: the role of size distribution in cellular uptake. Biophys. J. 2018;114:278–279. [Google Scholar]
- 18.Kuskov A.N., Voskresenskaya A.A., Goryachaya A.V., Shtilman M.I., Spandidos D.A., Rizos A.K., Tsatsakis A.M. Amphiphilic poly-N-vinylpyrrolidone nanoparticles as carriers for non-steroidal anti-inflammatory drugs: characterization and in vitro controlled release of indomethacin. Int. J. Mol. Med. 2010;26:85–94. doi: 10.3892/ijmm_00000438. [DOI] [PubMed] [Google Scholar]
- 19.Kuskov A.N., Kulikov P.P., Shtilman M.I., Rakitskii V.N., Tsatsakis A.M. Amphiphilic poly-N-vynilpyrrolidone nanoparticles: cytotoxicity and acute toxicity study. Food Chem. Toxicol. 2016;96:273–279. doi: 10.1016/j.fct.2016.08.017. [DOI] [PubMed] [Google Scholar]
- 20.Kuskov A.N., Kulikov P.P., Goryachaya A.V., Tzatzarakis M., Docea A.O., Velonia K., Shtilman M.I., Tsatsakis A.M. Amphiphilic poly-N-vinylpyrrolidonee nanoparticles as carriers for nonsteroidal, anti-inflammatory drugs: in vitro cytotoxicity and in vivo acute toxicity study, Nanomedicine: nanotechnology. Biol. Med. 2017;13:1021–1030. doi: 10.1016/j.nano.2016.11.006. [DOI] [PubMed] [Google Scholar]
- 21.Basak E., Neagu M., Nikitovich D., Henrich-Noack P., Docea A., Shtilman M., Golokhvast K., Tsatsakis A. Mechanistic understanding of nanoparticles’ interactions with extracellular matrix: the cell and immune system Ayse. Part. Fibre Toxicol. 2017;14:22–28. doi: 10.1186/s12989-017-0199-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuskov A.N., Kulikov P.P., Goryachaya A.V., Tsatzarakis M.N., Tsatsakis A.M., Velonia K., Shtilman M.I. Self-assembled amphiphilic poly-N-vinylpyrrolidone nanoparticles as carriers for hydrophobic drugs: stability aspects. J. Appl. Polym. Sci. 2018;135:45673. [Google Scholar]
- 23.Shcherbo E.M., Merzlyak T.V., Chepurnykh A.F., Fradkov G.V., Ermakova E.A. Solovieva. Bright far-red fluorescent protein for whole body imaging. Nat. Methods. 2007;4:741–746. doi: 10.1038/nmeth1083. [DOI] [PubMed] [Google Scholar]
- 24.Guo S.-L., Peng Z., Yang X., Fan K.-J., Ye H., Li Z.-H. miR-148a promoted cell proliferation by targeting p27 in gastric cancer cells. Int. J. Biol. Sci. 2011;7:567–574. doi: 10.7150/ijbs.7.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang H., Li Y., Huang Q., Ren X., Hu H., Sheng H. MiR-148a promotes apoptosis by targeting Bcl-2 in colorectal cancer. Cell Death Differ. 2011;18:1702–1710. doi: 10.1038/cdd.2011.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Palmqvist R., Sellberg P., Oberg A., Tavelin B., Rutegard J.N., Stenling R. Low tumour cell proliferation at the invasive margin is associated with a poor prognosis in Dukes’ stage B colorectal cancers. Br. J. Cancer. 1999;79:577–581. doi: 10.1038/sj.bjc.6690091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kimura T., Tanaka S., Haruma K., Sumii K., Kajiyama G., Shimamoto F. Clinical significance of MUC1 and E-cadherin expression, cellular proliferation, and angiogenesis at the deepest invasive portion of colorectal cancer. Int. J. Oncol. 2000;16:55–64. doi: 10.3892/ijo.16.1.55. [DOI] [PubMed] [Google Scholar]
- 28.Allegra C.J., Paik S., Colangelo L.H., Parr A.L., Kirsch I., Kim G. Prognostic value of thymidylate synthase, Ki-67, and p53 in patients with Dukes’ B and C colon cancer: a National Cancer institute-National Surgical Adjuvant Breast and Bowel Project collaborative study. J. Clin. Oncol. 2003;21:241–250. doi: 10.1200/JCO.2003.05.044. [DOI] [PubMed] [Google Scholar]
- 29.Fluge Q., Gravdal K., Carlsen E., Vonen B., Kjellevold K., Refsum S. Expression of EZH2 and Ki-67 in colorectal cancer and associations with treatment response and prognosis. Br. J. Cancer. 2009;101:1282–1289. doi: 10.1038/sj.bjc.6605333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matsubara S., Ding Q., Miyazaki Y., Kuwahata T., Tsukasa K., Takao S. mTOR plays critical roles in pancreatic cancer stem cells through specific and stemness-related functions. Sci. Rep. 2013;3:3230. doi: 10.1038/srep03230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Abbas A.K., Lichtman A.H., Pillai S. Cellular and Molecular Immunology. 6-E. Elsevier Inc.; 2007. Lymphocyte development and the rearrangement and expression of antigen receptor genes; p. 177. [Google Scholar]
- 32.Bagga S., Bracht J., Hunter S., Massier K., Holtz J., Eachus R. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122:553–563. doi: 10.1016/j.cell.2005.07.031. [DOI] [PubMed] [Google Scholar]
- 33.Lytle J.R., Yario T.A., Steitz J.A. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5’UTR as in the 3’UTR. PNAS. 2007;104:9667–9672. doi: 10.1073/pnas.0703820104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Folco H.D., Pidoux A.L., Urano T., Allshire R.C. Heterochromatin and RNAi are required to establish CENP-A chromatin at centromeres. Science. 2008;319:94–97. doi: 10.1126/science.1150944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tay Y., Zhang J., Thomson A.M., Lim B., Rigoutsos I. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature. 2008;455:1124–1128. doi: 10.1038/nature07299. [DOI] [PubMed] [Google Scholar]
- 36.Varambally S., Cao Q., Mani R.S., Shankar S., Wang X., Ateeq B. Genomic loss of microRNA-101 leads to overexpression of histone methyltransferase EZH2 in cancer. Science. 2008;322:1695–1699. doi: 10.1126/science.1165395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hanna J., Saha K., Pando B., van Zon J., Lengner C.J., Creyghton M.P., van Oudenaarden A., Jaenisch R. Direct cell reprogramming is a stochastic process amenable to acceleration. Nature. 2009;462:595–601. doi: 10.1038/nature08592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cox D.N., Chao A., Baker J., Chang L., Qiao D., Lin H. A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes Dev. 2008;12:3715–3727. doi: 10.1101/gad.12.23.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cox D.N., Chao A., Lin H. Piwi encodes a nucleoplasmic factor whose activity modulates the number and division rate of germline stem cells. Development. 2000;127:503–514. doi: 10.1242/dev.127.3.503. [DOI] [PubMed] [Google Scholar]
- 40.Yang C., Su H., Liao X., Han C., Yu T., Zhu G., Wang X., Winkler C.A., O’Brien S.J., Peng T. Marker of proliferation Ki-67 expression is associated with transforming growth factor beta 1 and can predict the prognosis of patients with hepatic B virus-related hepatocellular carcinoma. Cancer Manag. Res. 2018;10:679–696. doi: 10.2147/CMAR.S162595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao L., Chen H., Hu B., Zhang H., Lin Q. Prognostic significance of Ki67 expression and the derived neutrophil-lymphocyte ratio in nasopharyngeal carcinoma. Cancer Manag. Res. 2018;10:1919–1926. doi: 10.2147/CMAR.S167626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miller L., Min M., Yang C., Tian C., Gookin S., Carter D., Spencer S.L. Ki67 is graded rather than a binary marker of proliferation versus quiescence. Cell Rep. 2018;24:1105–1112. doi: 10.1016/j.celrep.2018.06.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grivna S.T., Pyhtila B., Lin H. MIWI associates with translational machinery and PIWI-interacting RNAs (piRNAs) in regulating spermatogenesis. PNAS. 2006;103:13415–13420. doi: 10.1073/pnas.0605506103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grivna S.T., Beyret E., Wang Z., Lin H. A novel class of small RNAs in mouse spermatogenic cells. Genes Dev. 2006;20:1709–1714. doi: 10.1101/gad.1434406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen Y., Song Y.-X., Wang Z.-N. The microRNA-148/152 family: multi-faceted players. Mol. Cancer. 2013;12:43–51. doi: 10.1186/1476-4598-12-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guan Z., Song B., Liu F., Sun D., Wang K., Qu H. TGF-b induces HLA-G expression through inhibiting miR-152 in gastric cancer cells. J. Biomed. Sci. 2015;22:107–113. doi: 10.1186/s12929-015-0177-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dang Y.W., Zeng J., He R.Q., Rong M.H., Luo D.Z., Chen G. Effects of miR-152 on cell growth inhibition, motility suppression and apoptosis induction in hepatocellular carcinoma cells. Asian Pac. J. Cancer Prev. 2014;15:4969–4976. doi: 10.7314/apjcp.2014.15.12.4969. [DOI] [PubMed] [Google Scholar]
- 48.Cheng Z., Ma R., Tan W., Zhang L. MiR-152 suppresses the proliferation and invasion of NSCLC cells by inhibiting FGF2. Exp. Mol. Med. 2014;46:e112–e120. doi: 10.1038/emm.2014.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Takahashi M., Cuatrecasas M., Balaguer F., Hur K., Toiyama Yu., Castells A. The clinical significance of miR-148a as predictive biomarker in patients with advanced colorectal cancer. PLoS One. 2012;7 doi: 10.1371/journal.pone.0046684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Peschansky V.J., Wahlestedt C. Non-coding RNAs as direct and indirect modulators of epigenetic regulation. Epigenetics. 2014;9:3–12. doi: 10.4161/epi.27473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Meller V.H., Joshi S.S., Deshpande N. Modulation of chromatin by noncoding RNA. Annu. Rev. Genet. 2015;49:673–695. doi: 10.1146/annurev-genet-112414-055205. [DOI] [PubMed] [Google Scholar]
- 52.Gaspar-Maia A., Alajem A., Meshorer E., Ramalho-Santos M. Open chromatin in pluripotency and reprogramming. Nat. Rev. Mol. Cell Biol. 2011;12:36–47. doi: 10.1038/nrm3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Singhal N., Graumann J., Wu G., Arauzo-Bravo M.J., Han D.W., Greber B. Chromatin-remodeling components of the BAF complex facilitate reprogramming. Cell. 2010;141:943–955. doi: 10.1016/j.cell.2010.04.037. [DOI] [PubMed] [Google Scholar]
- 54.Kuramochi-Miyagawa S., Kimura T., Ijiri T.W., Isobe T., Asada N., Fujita Y. Mili, a mammalian member of piwi family gene, is essential for spermatogenesis. Development. 2004;131:839–849. doi: 10.1242/dev.00973. [DOI] [PubMed] [Google Scholar]
- 55.Carmell M.A., Girard A., van de Kant H.J., Bourc’his D., Bestor T.H., deRooij D.G. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev. Cell. 2007;12:503–514. doi: 10.1016/j.devcel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 56.Klattenhoff C., Bratu D.P., McGinnis-Schultz N., Koppetsch B.S., Cook H.A., Theurkauf W.E. Drosopilarasi RNA pathway mutations disrupt embryonic axis specification through activation of an ATR/Chk2 DNA damage response. Dev. Cell. 2007;12:45–55. doi: 10.1016/j.devcel.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 57.Lau N.C., Seto A.G., Kim J., Kuramochi-Miyagawa S., Nakano T., Barte D.P. Characterisation of the piRNA complex from rat testes. Science. 2006;313:363–367. doi: 10.1126/science.1130164. [DOI] [PubMed] [Google Scholar]
- 58.Whangbo J.S., Hunter C.P. Environmental RNA interference. Trends Genet. 2008;24:297–305. doi: 10.1016/j.tig.2008.03.007. [DOI] [PubMed] [Google Scholar]
- 59.Robert V.J., Davis M.W., Jorgensen E.M., Bessereau J.-L. Gene conversion and End-Joining-Repair double-strand breaks in the Caenorhabditis elegans germline. Genetics. 2008;180:673–679. doi: 10.1534/genetics.108.089698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang Q.E., Han C., Milum K., Wani A.A. Stem cell protein Piwil2 modulates chromatin modifications upon cisplatin treatment. Mutat. Res. 2011;708:59–68. doi: 10.1016/j.mrfmmm.2011.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ishizu H., Siomi H., Siomi M.C. Biology of PIWI-interacting RNAs: new insights into biogenesis and function inside and outside of germlines. Genes Dev. 2012;26:2361–2373. doi: 10.1101/gad.203786.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang Y., Sun T., Wang K., Wang J.-X., Li P.F. PiRNAs link epigenetic modifications to reprogramming. Histol. Histopathol. 2014;29:1–9. doi: 10.14670/HH-29.1489. [DOI] [PubMed] [Google Scholar]
- 63.Moyano M., Giovanni S. piRNA involvement in genome stability and human cancer. J. Hematol. Oncol. 2015;8:38. doi: 10.1186/s13045-015-0133-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Heneghan H.M., Miller N., Lowery A.J., Sweeney K.J., Newell J., Kerin M.J. Circulating microRNAs as novel minimally invasive biomarkers for breast cancer. Ann. Surg. 2010;251:499–505. doi: 10.1097/SLA.0b013e3181cc939f. [DOI] [PubMed] [Google Scholar]
- 65.Brennecke J., Aravin A.A., Stark A., Dus M., Kellis M., Sachidanandam R. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- 66.Gunawardane S., Saito K., Nishida K.M., Miyoshi K., Kawamura Y., Nagami T. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science. 2007;315:1587–1590. doi: 10.1126/science.1140494. [DOI] [PubMed] [Google Scholar]
- 67.Ahmadzada T., Reid G., McKenzie D.R. Fundamentals of siRNA and miRNA therapeutics and a review of targeted nanoparticle delivery systems in breast cancer. Biophys. Rev. 2018;10:69–86. doi: 10.1007/s12551-017-0392-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chakraborty C., Sharma A.R.G., Sarkar B.K., Lee S.-S. The novel strategies for next-generation cancer treatment: miRNA combined with chemotherapeutic agents for the treatment of cancer. Oncotarget. 2018;9:10164–10174. doi: 10.18632/oncotarget.24309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chakraborty C., Sharma A.R., Sharma G., Doss C.G.P., Lee S.-S. Therapeutic miRNA and siRNA: moving from bench to clinic as next generation medicine. Mol. Ther. Nucleic Acids. 2017;8:132–143. doi: 10.1016/j.omtn.2017.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chalbatani G.M., Dana H., Memari F., Gharagozolou E., Ashjaei S., Kheirandish P., Marmari V., Mahmoudzadeh H., Maleki A.R., Sadeghian E., Nia E.Z., Miri S.R., Nia N.Z., Rezaeian O., Eskandary A., Razavi N., hirkhoda M., Rouzbahani F.N. Biological function and molecular mechanism of piRNA in cancer. Pract. Lab. Med. 2018;7(December) doi: 10.1016/j.plabm.2018.e00113. [DOI] [PMC free article] [PubMed] [Google Scholar]