Abstract
Background
Infections due to carbapenem-resistant Gram-negative rods (CR-GNR) are increasing in frequency and result in high morbidity and mortality. Appropriate initial antibiotic therapy is necessary to reduce adverse consequences and shorten length of stay.
Methods
To determine risk factors for recovery on culture of CR-GNR, cases were retrospectively analyzed at a major academic hospital system from 2011 to 2016. Ertapenem resistance (ER-GNR) and antipseudomonal (nonertapenem) carbapenem resistance (ACR-GNR) patterns were analyzed separately. A total of 30951 GNR isolates from 12370 patients were analyzed, 563 of which were ER and 1307 of which were ACR.
Results
In multivariate analysis, risk factors for ER-GNR were renal disease, admission from another health care facility, ventilation at any point before culture during the index hospitalization, receipt of any carbapenem in the prior 30 days, and receipt of any anti-methicillin-resistant Staphylococcus aureus (anti-MRSA) agent in the prior 30 days (c-statistic, 0.74). Risk factors for ACR-GNR were male sex, admission from another health care facility, ventilation at any point before culture during the index hospitalization, receipt of any carbapenem in the prior 30 days, and receipt of any anti-MRSA agent in the prior 30 days (c-statistic, 0.76).
Conclusions
A straightforward scoring system derived from these models can be applied by providers to guide empiric antimicrobial therapy; it outperformed use of a standard hospital antibiogram in predicting infections with ER-GNR and ACR-GNR.
Keywords: antimicrobial resistance, antimicrobial stewardship, carbapenems, Gram-negative rods, predictive scoring
Rising worldwide prevalence of human infections with multidrug-resistant organisms (MDROs) is associated with increasing morbidity, mortality, and cost [1]. In the United States, there are approximately 23000 yearly attributable deaths and $50 million in yearly attributable costs from MDRO infections [2]. Appropriate initial antibiotic therapy decreases mortality and hospital length of stay [3, 4], whereas overuse of broad-spectrum antibiotics has been linked with increased prevalence of MDROs [5–9]; the initial choice of antibiotic remains a challenging and high-stakes decision.
Carbapenem resistance among Gram-negative rods (CR-GNR) has been increasing over the past several decades, particularly in Enterobacteriales species [10–13]. Infection with CR-GNR species is associated with higher mortality [10, 12–14], hospital costs [11, 13], and increased risk of inappropriate antibiotic therapy [15] compared with infection with carbapenem-susceptible (CS) isolates. Delayed antimicrobial therapy (DAT) of CR-GNR has been shown to directly impact patient survival, highlighting the need for rapid identification of patients at high risk for CR-GNR.
Prior literature has identified multiple risk factors for the development of CR in various GNR, including receipt of mechanical ventilation [13, 14, 16–18], presence of various indwelling devices [10, 14, 15, 18, 19], more severe illness at the time of culture (intensive care unit [ICU] stay, comorbidities, or septic shock) [12–14, 18, 20], length of hospital stay or recent hospitalization [13, 15, 18, 19], receipt of immunosuppression [15, 20], and recent exposure to various antibiotics [10, 14, 17–20]. Other risk factors for development of MDROs in general included prior residence in a nursing home, hemodialysis, ICU admission [16], increased medical comorbidity [21, 22], prior antibiotic usage, and invasive surgery [22].
Antipseudomonal carbapenems (defined as meropenem, imipenem, and doripenem) likely have a different risk factor profile from ertapenem. Several organisms (most notably Pseudomonas aeruginosa and Acinetobacter baumannii) have intrinsic resistance to ertapenem but not antipseudomonal carbapenems. As such, we performed 2 separate analyses, 1 examining ertapenem resistance (ER) and 1 for antipseudomonal carbapenem resistance (ACR), to examine the similarities and differences in risk factors for recovery of a nonsusceptible isolate. We hypothesized that a large, adequately powered study would provide sufficient observations to identify easily obtainable clinical factors that could serve as prediction tool for identifying patients at high risk for acquiring CR-GNR.
METHODS
We conducted a retrospective study of all patients with positive cultures from any source over a 6-year period to develop a comprehensive model for risk of infection or colonization with CR-GNR, with separate analyses for ER-GNR and ACR-GNR. The study was performed at 2 hospitals in metropolitan Los Angeles, California. Ronald Reagan UCLA Medical Center is a 520-bed tertiary care center with 5 adult intensive care units totaling 109 beds, and Santa Monica–UCLA Medical Center has 266 beds total with 22 mixed intensive care beds in a single unit. Both are part of UCLA Health and serve patients with solid organ and bone marrow transplants, cancer, and various medical and surgical conditions. The Integrated Clinical and Research Data Repository (xDR) serves as a warehouse for all clinical data in the UCLA system since 2006. The data set contained information from all admissions with start dates from January 2011 through November 2016 to either hospital for patients ≥18 years of age with at least 1 positive culture from any source (blood, urine, sputum, wound cultures, or other fluids).
As the end point of this analysis was prediction of development of the first carbapenem-nonsusceptible isolate, once a patient had a culture growing a CR-GNR organism, defined using Clinical and Laboratory Standards Institute (CLSI) breakpoints current to the year of testing, all cultures from that patient occurring at a later time than the original culture were removed from the data set.
Routine susceptibility testing was performed by the either the CLSI reference broth microdilution method (BMD) or using a Vitek 2 with BMD confirmation using panels prepared in-house. Only data from 2011 onwards were used in this study, as a changeover in clinical data warehousing methods corresponded to significantly more robust clinical information after that time. All antimicrobial susceptibility data were interpreted using current (post-2012) CLSI breakpoints for carbapenems. Organisms demonstrating intrinsic resistance to a given class of antibiotics were removed from predictive models for that antibiotic class (eg, Pseudomonas species were removed from the ER-GNR model but not the ACR-GNR model). An organism was considered ACR-GNR if it showed resistance to any antipseudomonal carbapenem (meropenem, imipenem, or doripenem). Only 27 isolates were tested for doripenem sensitivity; these had a ~95% concordance with both imipenem and meropenem sensitivity. Of the approximately 18000 isolates tested for both meropenem and imipenem sensitivity, there was a ~99% concordance in sensitivity.
Predictor variables were chosen on the basis of prior studies, as well as those with biologic plausibility, which were readily obtained from the medical record. Data collected for each patient included admission hospital, days since admission, location before admission (home vs long-term care facility or other hospital), demographic information, comorbidities (grouped into categories based on Elixhauser score designations) [23], laboratory results from the date of the culture, vital signs on the date the culture was collected (maximum temperature, heart rate, respiratory rate, and minimum blood pressure), vital signs from initial hospital presentation, oxygen/ventilation method, presence of a tracheostomy, presence of a urinary catheter, administration of antibiotics and other selected medications (vasopressors, probiotics, blood products, immunosuppressants, and acid suppressants), culture source, and prior culture positivity for carbapenem-resistant GNR. Administration of antibiotics and the medications listed above was coded as the number of days since last receipt of the medication, Winsorized to a maximum value of 100 (received within 24 hours of the time of culture = 0; 100 days or longer since receipt and never receiving the medication were both coded as 100 days since receipt). Winsorization was chosen as some patients had data from the index hospitalization or prior hospitalizations indicating hundreds of days since the last receipt of a relevant medication. Given the long tail, truncation was the only approach that allowed linearization in the predictors of the outcome.
“Anti-methicillin-resistant Staphylococcus aureus” (anti-MRSA) refers to vancomycin, linezolid, and daptomycin, as these were used at both hospitals in cases of suspected hospital-acquired MRSA. Receipt of antibiotics was by any route, including oral, intravenous, and inhaled. An infection was coded as “hospital acquired” if the culture was submitted to the laboratory >48 hours after the time of first presentation to the hospital. The construct of advanced ventilatory support includes patients receiving either noninvasive or invasive mechanical ventilation.
In cases where laboratory tests were not performed before cultures were performed (typically at the beginning of a patient’s admission), the first set of laboratory results was used for that patient, provided they were performed on specimens collected within 24 hours of culture positivity. For laboratory tests not typically performed daily (eg, liver function tests, measures of coagulation, and protein/prealbumin), the most recent result within the 48-hour period before culture positivity was used.
To facilitate model interpretability, linear variables (time since receipt of medications) were recategorized as binary variables using cutoffs. Various cutoffs were tested against each other in the final model (30 vs 60 vs 90 days since receipt of last antibiotic), and the cutoff that led to the highest c-statistic was chosen for inclusion in the scoring system. In the case of time since medication receipt, the binary cutoffs also outperformed non-Winsorized data for model discrimination.
Statistical Analysis
Two separate analyses were performed, 1 comparing ertapenem-susceptible GNR (ES-GNR) against ER-GNR and 1 comparing antipseudomonal carbapenem-susceptible GNR (ACS-GNR) against ACR-GNR. These 2 analyses were chosen to aid decision-making at the point of initial antibiotic choice, when the consequences for inappropriate antibiotic therapy are the greatest [24–27]. The measured variables in each case were compared between the cases and controls by a 2-sided Mann-Whitney U test, Student t test, or chi-square test, as appropriate. In each case, after bivariate associations were examined, variables with P < .10 or strong biologic plausibility were included in the model selection process; a P value of <.10 was chosen as a threshold to include variables in the model to ensure that marginally significant variables with strong predictive effects were not inappropriately excluded. Only complete cases were included in model selection.
For both models, data from the last 2 years of the data period (2015 and 2016) were withheld as a validation set, whereas data from 2011–2014 were used as a training set for the models.
In each case, the predictor variables were divided into several categories, comprising medical comorbidities, demographics (age, sex, race, location before admission, and social history), indwelling devices, vital signs, laboratory variables, and received medications. For each category, the predictors were tested for collinearity and interactions; in cases of collinearity, the variable providing the most explanatory power was included in the model. At each stage, model selection was performed with LASSO regression (testing a full range of lambdas for parsimony). Model selection was also performed with straightforward logistic regression. Model discrimination was determined by area under the receiver operating characteristic curve (c-statistic), and models were compared by chi-square test if they were nested, or Akaike information criterion if they were not. Variables were generally dropped from a logistic regression model if they had a P value of >.05. In each case, the optimal model reached by LASSO regression matched the optimal model selected by iterative logistic regression. All analyses were performed using the Stata statistical software package, version 14.2 [28].
RESULTS
The complete data set included 30951 GNR isolates from 12370 patients, 563 of which were ER and 1307 of which were ACR. As only complete cases without intrinsic resistance were analyzed for the multivariate model, the final models comprised 14292 cultures, 274 of which were ER-GNR and 618 of which were ACR-GNR. This was the result of 16659 observations with incomplete data. The majority of ER-GNR were Klebsiella species, whereas the most common ES-GNR were Escherichia coli; the most common ACR-GNR were Pseudomonas species, whereas the most common ACS-GNR were again E. coli (Table 1). Respiratory culture source was predictive of both ER-GNR and ACR-GNR, whereas urinary source was predictive of both ES-GNR and ACS-GNR (Table 2).
Table 1.
Distribution of Organisms for ER-GNR and ACR-GNR Cultures (P < .001 for X 2 Test)
| Genus | ER-GNR | ACR-GNR |
|---|---|---|
| Acinetobacter | n/a | 20.1% |
| Enterobacter | 28.4% | 2.1% |
| Escherichia | 14.8% | 1.7% |
| Klebsiella | 40.5% | 17.3% |
| Pseudomonas | n/a | 54.4% |
| Other | 21.7% | 20.1% |
Acinetobacter and Pseudomonas express intrinsic resistance to ertapenem and were not included in the ER-GNR data set.
Abbreviations: ACR-GNR, antipseudomonal (nonertapenem) carbapenem-resistant Gram-negative rods; ER-GNR, ertapenem-resistant Gram-negative rods.
Table 2.
Distribution of Culture Source for ES-GNR, ER-GNR, ACS-GNR, and ACS-GNR (P < .001 for X2 Test for Both ER and ACR)
| Source | ES-GNR | ER-GNR | ACS-GNR | ACR-GNR |
|---|---|---|---|---|
| Blood | 11.2% | 10.8% | 12.1% | 8.4% |
| Urine | 45.6% | 32.5% | 42.1% | 11.2% |
| Respiratory | 20.4% | 36.8% | 23.7% | 61.6% |
| Externala | 8.3% | 8.4% | 8.1% | 7.9% |
| Other | 14.5% | 11.6% | 14.0% | 10.9% |
Abbreviations: ACR, antipseudomonal (nonertapenem) carbapenem resistance; ACR-GNR, antipseudomonal (nonertapenem) carbapenem-resistant Gram-negative rods; ACS-GNR, antipseudomonal carbapenem-susceptible Gram-negative rods; ER, ertapenem resistance; ER-GNR, ertapenem-resistant Gram-negative rods; ES-GNR, ertapenem-susceptible Gram-negative rods.
aExternal: skin or wound source.
Bivariate Analyses
Selected bivariate associations are reported in Tables 3 and 4. Risk factors were generally similar for ER-GNR and ACR-GNR. As isolates with intrinsic resistance were excluded from analysis, the numbers for the bivariate associations differed between the ER-GNR and ACR-GNR sections. In some cases (such as days since last aminoglycoside receipt), the median, 25th percentile, and 75th percentile were identical for the susceptible and nonsusceptible groups, but the U test still showed a significant difference in the overall distributions.
Table 3.
Selected Bivariate Associations for Demographics and Comorbidities
| ES-GNR | ER-GNR | P | ACS-GNR | ACR-GNR | P | |
|---|---|---|---|---|---|---|
| No. | 16 861 | 563 | 29 664 | 1307 | ||
| Age, y | 64.5 (18.8) | 63.8 (17.2) | .370 | 64 (19.1) | 64.7 (19) | .188 |
| Male sex | 0.451 | 0.536 | <.001 | 0.464 | 0.592 | <.001 |
| Race | .015 | <.001 | ||||
| White | 51.1% | 46.5% | 52.7% | 53.0% | ||
| Asian | 9.1% | 8.8% | 8.7% | 5.9% | ||
| Black | 11.4% | 16.1% | 11.5% | 16.1% | ||
| Latino | 21.9% | 20.7% | 21.0% | 20.9% | ||
| Other | 6.5% | 7.0% | 6.2% | 7.3% | ||
| BMI | 26.3 (6.8) | 26.3 (7.2) | .949 | 26.1 (6.8) | 25.1 (6.9) | <.001 |
| Admitted from health care facility | 14.5% | 22.2% | <.001 | 14.6% | 38.0% | <.001 |
| Hospital (RRMC vs SMH) | 65.7% | 57.4% | .012 | 63.3% | 46.6% | <.001 |
| Log days to culture | 0.13 [–1.84 to 1.99] | 2.14 [–0.33 to 3.11] | <.001 | 0.44 [–1.48 to 2.09] | 1.29 [–0.78 to 2.87] | <.001 |
| Hospital acquired | 41.2% | 61.3% | <.001 | 48.7% | 0.5(0.5) | <.001 |
| In ICU at the time of culture | 18.5% | 29.3% | <.001 | 17.5% | 28.4% | <.001 |
| Any ICU stay during index hosp. | 32.4% | 59.6% | <.001 | 36.1% | 54.6% | <.001 |
| Presence of indwelling urinary catheter | 42.5% | 58.3% | <.001 | 43.7% | 63.2% | <.001 |
| Ventilated during index hosp. | 24.2% | 50.7% | <.001 | 28.0% | 55.3% | <.001 |
| Tracheostomy present on day of culture | 6.9% | 20.4% | <.001 | 9.6% | 28.8% | <.001 |
| Elixhauser score | 14 [5 to 25] | 18 [8 to 29] | <.001 | 16 [6 to 26] | 22 [12 to 31] | <.001 |
| Congestive heart failure | 19.1% | 23.8% | .006 | 19.8% | 26.6% | <.001 |
| Arrhythmia | 40.0% | 49.0% | <.001 | 41.6% | 53.0% | <.001 |
| Valvular disease | 22.4% | 22.7% | .830 | 24.3% | 24.6% | .862 |
| Pulmonary vascular disease | 14.6% | 20.6% | <.001 | 16.1% | 20.1% | <.001 |
| Peripheral vascular disease | 21.4% | 26.6% | .003 | 23.4% | 27.4% | <.001 |
| Paralysis | 7.2% | 6.9% | .778 | 7.8% | 10.1% | .003 |
| Neurologic disease | 25.9% | 39.6% | <.001 | 28.5% | 45.8% | <.001 |
| Chronic pulmonary disease | 22.0% | 27.7% | .001 | 24.7% | 33.4% | <.001 |
| Renal disease | 31.8% | 45.5% | <.001 | 33.3% | 41.4% | <.001 |
| Liver disease | 23.7% | 34.6% | <.001 | 24.3% | 26.5% | .070 |
| Lymphoma | 3.8% | 5.7% | .024 | 4.1% | 5.4% | .021 |
| Metastatic cancer | 9.6% | 9.4% | .862 | 10.5% | 9.0% | .080 |
| Nonmetastatic cancer | 22.8% | 23.6% | .647 | 23.0% | 19.7% | .005 |
| Coagulopathy | 24.6% | 37.3% | <.001 | 26.0% | 35.1% | <.001 |
| Weight loss | 16.4% | 29.5% | <.001 | 18.7% | 33.0% | <.001 |
| Electrolyte disorder | 58.2% | 74.8% | <.001 | 60.3% | 71.8% | <.001 |
| Deficiency anemia | 12.5% | 13.5% | .464 | 13.3% | 13.9% | .514 |
| Drug abuse | 6.6% | 6.2% | .751 | 6.9% | 6.4% | .543 |
| Solid organ transplant | 16.3% | 22.9% | <.001 | 17.0% | 16.9% | .895 |
| Bone marrow transplant | 1.0% | 1.2% | .570 | 1.3% | 1.9% | .042 |
| Renal failure | 13.2% | 22.4% | <.001 | 14.1% | 19.0% | <.001 |
| Cystic fibrosis | 0.2% | 0.2% | .851 | 1.0% | 4.1% | <.001 |
| HIV | 0.7% | 0.4% | .300 | 0.7% | 0.8% | 0.845 |
| Alcohol user | 22.6% | 22.5% | .965 | 22.5% | 17.2% | <.001 |
| Tobacco user | 5.9% | 4.1% | .159 | 5.9% | 6.0% | .989 |
Normally distributed outcomes are reported as mean (SD), and non–normally distributed outcomes are reported as median [interquartile range]. Binary outcomes are reported as percent positive.
Abbreviations: ACR-GNR, antipseudomonal (nonertapenem) carbapenem-resistant Gram-negative rods; ACS-GNR, antipseudomonal carbapenem-susceptible Gram-negative rods; BMI, body mass index; ER-GNR, ertapenem-resistant Gram-negative rods; ES-GNR, ertapenem-susceptible Gram-negative rods; hosp., hospitalization; ICU, intensive care unit; RRMC, Ronald Reagan Medical Center; SMH, Santa Monica UCLA Hospital.
Table 4.
Selected Bivariate Associations for Labs and Medications Received
| ES-GNR | ER-GNR | P | ACS-GNR | ACR-GNR | P | |
|---|---|---|---|---|---|---|
| No. | 16861 | 563 | 29664 | 1307 | ||
| Labs on day of culture | ||||||
| WBC | 12.4 [8.6 to 17.1] | 13.5 [9.4 to 19.3] | <.001 | 12.4 [8.6 to 17.2] | 14.15 [9.7 to 19.45] | <.001 |
| Hemoglobin | 10.1 [8.7 to 11.7] | 9.1 [8.2 to 10.3] | <.001 | 9.9 [8.7 to 11.5] | 9.2 [8.2 to 10.4] | <.001 |
| Hematocrit | 30.8 [26.7 to 35.4] | 28.1 [25 to 31.8] | <.001 | 30.5 [26.6 to 35] | 28.6 [25.3 to 32.3] | <.001 |
| Platelets | 204 [132 to 287] | 207 [103 to 306] | .767 | 205 [132 to 290] | 227 [132 to 329] | <.001 |
| Sodium | 137.3 (5.7) | 137.7 (5.3) | .158 | 137.3 (5.5) | 138.1 (5.6) | <.001 |
| Potassium | 4.1 (0.6) | 4.1 (0.6) | .394 | 4.1 (0.6) | 4.1 (0.6) | .019 |
| Chloride | 102.4 (6.6) | 102.2 (6.4) | .488 | 102.6 (6.5) | 102.5 (7) | .730 |
| Bicarbonate | 24.2 (4.5) | 24.4 (4.9) | .215 | 24.3 (4.7) | 25.3 (5.4) | <.001 |
| Anion gap | 10.7 (4) | 11 (4.3) | .086 | 10.4 (4) | 10.3 (4.3) | .310 |
| Creatinine | 1.4 (1.3) | 1.5 (1.4) | .011 | 1.4 (1.4) | 1.4 (1.3) | .895 |
| BUN | 28 (22.5) | 33.9 (25.1) | <.001 | 28.3 (23.1) | 34.7 (29) | <.001 |
| GFR | 69 [39 to 100] | 63 [33 to 100] | .035 | 71 [39 to 100] | 73 [38 to 100] | .192 |
| Glucose | 137 (60) | 135 (56) | .470 | 135 (58) | 134 (58) | .416 |
| Magnesium | 1.7 (0.3) | 1.7 (0.3) | <.001 | 1.7 (0.3) | 1.7 (0.3) | <.001 |
| Calcium | 8.6 (0.8) | 8.6 (0.9) | .985 | 8.6 (0.8) | 8.6 (0.9) | .673 |
| Phosphorus | 3.3 (1.2) | 3.4 (1.4) | .080 | 3.3 (1.1) | 3.4 (1.3) | .026 |
| Days since: | ||||||
| Last antibiotic | 0 [0 to 12] | 0 [0 to 2] | <.001 | 0 [0 to 8] | 0 [0 to 0] | <.001 |
| Last aminoglycoside | 100 [100 to 100] | 100 [100 to 100] | <.001 | 100 [100 to 100] | 100 [100 to 100] | <.001 |
| Last antipseudomonal carbapenema | 100 [100 to 100] | 100 [29 to 100] | <.001 | 100 [100 to 100] | 100 [6 to 100] | <.001 |
| Last ertapenem | 100 [100 to 100] | 100 [100 to 100] | <.001 | 100 [100 to 100] | 100 [100 to 100] | <.001 |
| Last carbapenem (any) | 100 [100 to 100] | 100 [10 to 100] | <.001 | 100 [100 to 100] | 100 [0 to 100] | <.001 |
| Last fluoroquinolone | 100 [100 to 100] | 100 [44 to 100] | <.001 | 100 [100 to 100] | 100 [42 to 100] | <.001 |
| Last penicillin | 100 [2 to 100] | 27 [1 to 100] | <.001 | 100 [1 to 100] | 67.5 [3.5 to 100] | .005 |
| Last anti-MRSA | 100 [1 to 100] | 7 [0 to 100] | <.001 | 100 [0 to 100] | 5 [0 to 100] | <.001 |
| Last colistin | 100 [100 to 100] | 100 [100 to 100] | <.001 | 100 [100 to 100] | 100 [100 to 100] | <.001 |
| Last aztreonam | 100 [100 to 100] | 100 [100 to 100] | .005 | 100 [100 to 100] | 100 [100 to 100] | <.001 |
| Last beta-lactam | 1 [0 to 100] | 0 [0 to 10] | <.001 | 0 [0 to 100] | 0 [0 to 11] | <.001 |
| Last acid suppressant | 0 [0 to 100] | 0 [0 to 0] | <.001 | 0 [0 to 100] | 0 [0 to 1] | <.001 |
| Last probiotic | 100 [100 to 100] | 100 [100 to 100] | <.001 | 100 [100 to 100] | 100 [100 to 100] | <.001 |
| Last steroid | 100 [100 to 100] | 100 [2 to 100] | <.001 | 100 [58 to 100] | 100 [68.5 to 100] | .925 |
| Last chemotherapy | 100 [100 to 100] | 100 [100 to 100] | .197 | 100 [100 to 100] | 100 [100 to 100] | .079 |
| Last immunosuppressant | 100 [10 to 100] | 100 [0 to 100] | <.001 | 100 [1 to 100] | 100 [26.5 to 100] | .218 |
| Last blood product | 100 [100 to 100] | 100 [9 to 100] | <.001 | 100 [100 to 100] | 100 [35 to 100] | <.001 |
Normally distributed outcomes are reported as mean (SD), and non–normally distributed outcomes are reported as median [interquartile range]. Binary outcomes are reported as percent positive.
Abbreviations: ACR-GNR, antipseudomonal (nonertapenem) carbapenem-resistant Gram-negative rods; ACS-GNR, antipseudomonal carbapenem-susceptible Gram-negative rods; BUN, blood urea nitrogen; ER-GNR, ertapenem-resistant Gram-negative rods; ES-GNR, ertapenem-susceptible Gram-negative rods; GFR, race-adjusted glomerular filtration rate; MRSA, methicillin-resistant Staphylococcus aureus; WBC, white blood cell count.
aAntipseudomonal carbapenem: meropenem, imipenem, or doripenem.
Multivariate Analyses
Many of the variables that were significant on bivariate analysis were strongly colinear and were tested against each other in groups to determine which predictors were most representative from the various groupings of medical comorbidities, demographics, laboratory values, indwelling devices, and recently administered medications. Next, representative predictors were added together, and the most parsimonious models were chosen. To facilitate model interpretability, the variables representing days since receipt of medications were dichotomized to receipt within the prior 30 days vs not; this did not significantly affect model fit. In both cases, using LASSO regression to assess model fit produced identical final model predictive factors. The coefficients reported are from LASSO regression.
For the model predicting ER-GNR, the predictors in the final model were the presence of renal disease, admission from another health care facility, mechanical ventilation at any point before culture during the index hospitalization, receipt of any anti-MRSA agent in the prior 30 days, and receipt of any carbapenem in the prior 30 days; this model had a c-statistic of 0.74 (Table 5). When applied to the validation set, the model had a c-statistic of 0.73.
Table 5.
Model Specifications for ER-GNR and ACR-GNR
| ER-GNR | Coefficient | Standard Error | P |
|---|---|---|---|
| Renal disease | 0.55 | 0.17 | <.001 |
| Ventilated during index hospitalization | 0.71 | 0.18 | <.001 |
| In facility before admission | 0.47 | 0.19 | .015 |
| Carbapenems within 30 d | 0.75 | 0.19 | <.001 |
| Anti-MRSA agents within 30 d | 0.92 | 0.20 | <.001 |
| ACR-GNR | |||
| Male sex | 0.35 | 0.11 | .001 |
| Ventilated during index hospitalization | 0.72 | 0.11 | <.001 |
| In facility before admission | 1.33 | 0.11 | <.001 |
| Carbapenems within 30 d | 0.93 | 0.12 | <.001 |
| Anti-MRSA agents within 30 d | 0.26 | 0.13 | .035 |
Abbreviations: ACR-GNR, antipseudomonal (nonertapenem) carbapenem-resistant Gram-negative rods; ER-GNR, ertapenem-resistant Gram-negative rods; MRSA, methicillin-resistant Staphylococcus aureus.
For the model predicting ACR-GNR, the predictors in the final model were largely overlapping: male sex, admission from another health care facility, ventilation at any point before culture during the index hospitalization, receipt of any anti-MRSA agent in the prior 30 days, and receipt of any carbapenem in the prior 30 days; this model had a c-statistic of 0.76 (Table 5). When applied to the validation set, the model had a c-statistic of 0.77.
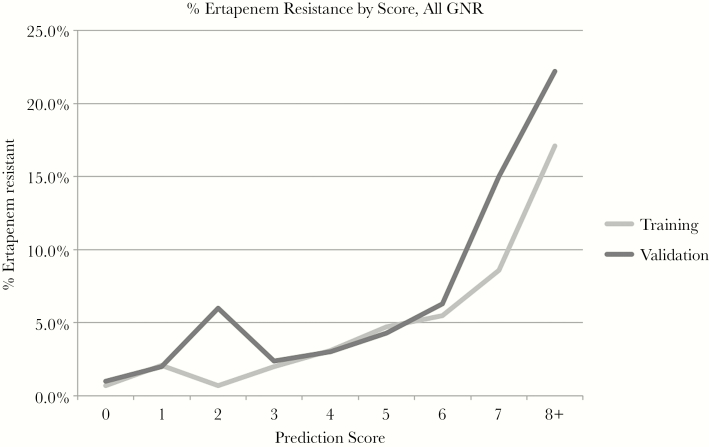
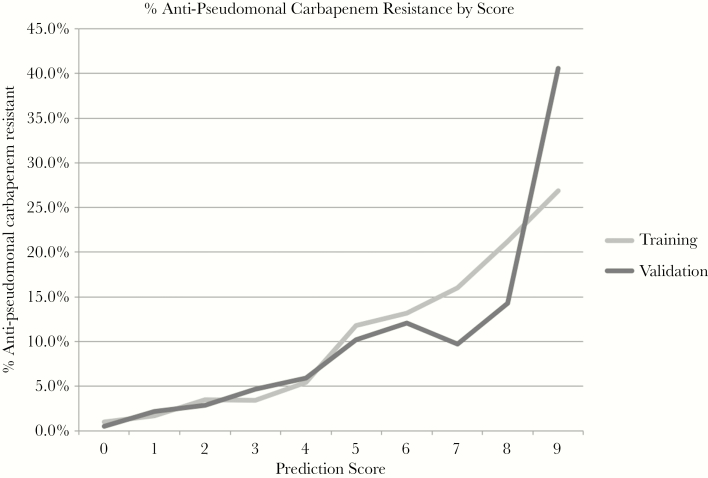
Treating each multivariate model as a score with points assigned based on the coefficients in the model (Table 6), we created a potentially user-friendly tool to predict the probability of ER and ACR. Figures 1 and 2 show the predicted (training set) and observed (validation set) rates of resistance at each score total for ER and ACR, respectively. In the ER-GNR model, only 33 isolates had the highest score of 9, 1 of which was ER-GNR. Due to the small sample size at this score and the unusual rates observed in these small samples, the scores of 8 and 9 were combined into a single category for ease of interpretability.
Table 6.
Points Assigned for Each Factor in the ER-GNR and ACR-GNR Models
| ER-GNR | ACR-GNR | |
|---|---|---|
| Renal disease | 1 | n/a |
| Male sex | n/a | 1 |
| Ventilated during index hospitalization | 2 | 2 |
| In facility before admission | 1 | 3 |
| Carbapenems within 30 d | 2 | 2 |
| Anti-MRSA agents within 30 d | 3 | 1 |
Abbreviations: ACR-GNR, antipseudomonal (nonertapenem) carbapenem-resistant Gram-negative rods; ER-GNR, ertapenem-resistant Gram-negative rods; MRSA, methicillin-resistant Staphylococcus aureus.
Figure 1.
Positive predictive value for ertapenem resistance at each score value for Gram-negative rods for the training and validation sets.
Figure 2.
Positive predictive value for antipseudomonal carbapenem resistance at each score value for Gram-negative rods for the training and validation sets. Abbreviation: GNR, Gram-negative rods.
Model Subset Validation
In the incomplete cases, data were missing largely as a result of 2 factors: periodic outages in transmission from the medical record to the data warehouse (affecting all data types), and negligence in users inputting data (more common with social history elements such as location before admission and some medical comorbidities). As both of these mechanisms are theoretically independent of the clinical state of the patient, there was potential that the data were missing completely at random. The data set from the logistic models (cases that were complete for the selected variables) was compared against the complete data set, and no substantial differences were found in variables that were significant in intermediate steps in model selection (calculations not shown).
DISCUSSION
Infection with CR-GNR is associated with substantially increased cost and risk for mortality, and appropriate antibiotic treatment is paramount in mitigating these risks [10–15]. Our scores can be calculated by providers at the time of decision-making; they may more accurately reflect a patient’s risk for carbapenem-resistant organisms than a hospital-wide or unit-specific antibiogram, which provides a flat percent observed susceptibility for a given organism in the prior year and is not useful for management of rare events. All information used in the models was extracted directly from the medical record without any direct examination of individual patient records, allowing this score to potentially be calculated automatically.
Prior studies of risk factors for CR among GNR have largely focused on the family Enterobacteriales, which includes many commonly treated GNR but excludes several clinically significant genera, including Pseudomonas and Acinetobacter. Additionally, many of these studies have been limited in scope, analyzing a small number of patients (typically in the low hundreds) [10, 12, 14, 15, 17–20, 29] or focusing on a single organism [10, 17–19, 29]. The largest prior study of ~40000 patients with Enterobacteriales infection (including 1227 with CRE) used hospital administrative data, and although it had an extensive list of covariates, it did not include information about non-Enterobacteriales GNR or prior antibiotic exposure and did not include a multivariate analysis or result in a clinical decision rule [13].
Our bivariate analysis is consistent with prior studies, confirming associations between CR and receipt of mechanical ventilation and presence of various indwelling devices [10, 13–19], more severe illness at the time of culture [12–14, 18, 20], length of hospital stay or recent hospitalization [13, 15, 18, 19], and recent exposure to various antibiotics [10, 14, 17–20]. Several factors that featured prominently in other analyses were found not to contribute to the optimal prediction model. Length of stay, most medical comorbidities, and receipt of immunosuppressive medications, although individually important, do not directly contribute on multivariate analysis. This is most likely because they are related to a construct of chronic medical illness that is mediated through other concepts (such as frequent contact with the medical system, exposure to MDROs, and susceptibility to infection) that are better proxied by other variables.
Although it is improbable that exposure to all antibiotics mechanistically leads to development of CR, some of these exposures likely proxy recent infection with MDR GNR, whereas anti-MRSA receipt proxies recent concern for sepsis, as nearly all patients with suspicion for sepsis receive at least 1 dose of vancomycin at our institutions. Our multivariate analysis suggests that variables associated with acute illness are less of a determining factor than chronic illness and recent antibiotic exposure in determining risk for CR-GNR.
Some of the risk factors observed in this study, particularly neurologic disease, weight loss, and recent receipt of carbapenems, were also shown to be predictive of resistance to colistin [30] and aminoglycosides (Richter, unpublished data). Additionally, these risk factors did not cluster temporally, reducing the probability that an outbreak isolated to a particular population or unit was responsible for the high prevalence of these risk factors in the affected population.
Our study has limitations. It examines patients from only 2 hospitals within a single hospital system. Approximately 50% of ER- and ACR-GNR cases could not be included in the final analysis due to a lack of complete data across the relevant domains. Sensitivity analyses did not show a significant effect from missing data, although it is possible that the point estimates would have varied slightly if full data were available. Additionally, we only had access to data from inpatient hospitalizations within our hospital system, potentially excluding relevant information from outpatient encounters or treatment at other facilities. These limitations reflect the real-world data that are available at the time of decision-making or the eventual integration of a similar score into an electronic health record. However, it is the largest investigation to date in terms of number of subjects and spans a period of 6 years, allowing us to examine far more potential explanatory variables than prior investigations of risk factors for development of ER-GNR and ACR-GNR. By performing a cohort study of patients with positive cultures, we eliminated potential selection bias in choosing controls and strengthened the validity of observed associations [31].
Finally, there is the inherent weakness of logistic regression models in that they are unable to account for incomplete cases, and such cases are dropped from the analysis. This introduces the possibility of bias in the model if the missing data are not at random. As the primary mechanisms for data loss (intermittent outages of data transfer and negligence regarding data entry by providers) are theoretically independent of the clinical state of the patient, there is hope that data are missing completely at random. The data set from the final models (cases that were complete for the selected variables) was compared against the complete data set, and no substantial differences were found in variables that were significant in intermediate steps in model selection; this supports the possibility that the data were missing at random and did not bias the model.
Both the ER-GNR and ACR-GNR models can effectively rule out risk for carbapenem resistance (<5% chance) at scores <6 for the ER-GNR model and scores <4 for the ACR-GNR model, allowing reasonably confident treatment with carbapenems as a first option without waiting for definitive carbapenem susceptibility testing, which can take up to several days.
Our study demonstrates the potential to harness currently available information from an existing electronic medical record to inform clinical decision-makers. Our simplified scoring system improved on the traditional antibiogram approach of offering a single hospital-wide percentage rate of susceptibility by creating an individualized score than can be used and interpreted by individual clinicians without computer assistance. In the current era of data-intensive medical care, we should harness all available information to better manage our patients. Further research should focus on validating this score in other populations and other hospital systems and analyzing the cost–benefit thresholds for initiating specific antibiotic regimens in cases of uncertainty.
Acknowledgments
Financial support. This work was supported by the National Institutes of Health (U54GM114833-01) and the National Center for Advancing Translational Sciences (UL1TR001881).
Potential conflicts of interest. All authors: no reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. CDC. Antibiotic resistance threats in the United States, 2013 2013. https://www.cdc.gov/drugresistance/pdf/ar-threats-2013-508.pdf Accessed 10 November 2016.
- 2. O’Neill J. AMR review paper - tackling a crisis for the health and wealth of nations 2014. https://amr-review.org/sites/default/files/AMR Review Paper - Tackling a crisis for the health and wealth of nations_1.pdf Accessed 10 November 2016.
- 3. Kollef MH, Sherman G, Ward S, Fraser VJ. Inadequate antimicrobial treatment of infections: a risk factor for hospital mortality among critically ill patients. Chest 1999; 115:462–74. [DOI] [PubMed] [Google Scholar]
- 4. Kumar A, Roberts D, Wood KE, et al. Duration of hypotension before initiation of effective antimicrobial therapy is the critical determinant of survival in human septic shock. Crit Care Med 2006; 34:1589–96. [DOI] [PubMed] [Google Scholar]
- 5. MacKenzie FM, Bruce J, Struelens MJ, et al. ; ARPAC Steering Group Antimicrobial drug use and infection control practices associated with the prevalence of methicillin-resistant Staphylococcus aureus in European hospitals. Clin Microbiol Infect 2007; 13:269–76. [DOI] [PubMed] [Google Scholar]
- 6. Neuhauser MM, Weinstein RA, Rydman R, et al. Antibiotic resistance among gram-negative bacilli in US intensive care units: implications for fluoroquinolone use. JAMA 2003; 289:885–8. [DOI] [PubMed] [Google Scholar]
- 7. Polk RE, Johnson CK, McClish D, et al. Predicting hospital rates of fluoroquinolone-resistant Pseudomonas aeruginosa from fluoroquinolone use in US hospitals and their surrounding communities. Clin Infect Dis 2004; 39:497–503. [DOI] [PubMed] [Google Scholar]
- 8. Torres-Gonzalez P, Cervera-Hernandez ME, Niembro-Ortega MD, et al. Factors associated to prevalence and incidence of carbapenem-resistant Enterobacteriaceae fecal carriage: a cohort study in a Mexican tertiary care hospital. PLoS One 2015; 10:e0139883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fortin E, Platt RW, Fontela PS, et al. Predicting antimicrobial resistance prevalence and incidence from indicators of antimicrobial use: what is the most accurate indicator for surveillance in intensive care units? PLoS One 2015; 10:e0145088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Akgul F, Bozkurt I, Sunbul M, et al. Risk factors and mortality in the carbapenem-resistant Klebsiella pneumoniae infection: case control study. Pathog Glob Health 2016; 110:321–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bartsch SM, McKinnell JA, Mueller LE, et al. Potential economic burden of carbapenem-resistant Enterobacteriaceae (CRE) in the United States. Clin Microbiol Infect 2017; 23:48.e9–48.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. de Maio Carrilho CM, de Oliveira LM, Gaudereto J, et al. A prospective study of treatment of carbapenem-resistant Enterobacteriaceae infections and risk factors associated with outcome. BMC Infect Dis 2016; 16:629–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zilberberg MD, Nathanson BH, Sulham K, et al. Carbapenem resistance, inappropriate empiric treatment and outcomes among patients hospitalized with Enterobacteriaceae urinary tract infection, pneumonia and sepsis. BMC Infect Dis 2017; 17:279–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mariappan S, Sekar U, Kamalanathan A. Carbapenemase-producing Enterobacteriaceae: risk factors for infection and impact of resistance on outcomes. Int J Appl Basic Med Res 2017; 7:32–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Leibman V, Martin ET, Tal-Jasper R, et al. Simple bedside score to optimize the time and the decision to initiate appropriate therapy for carbapenem-resistant Enterobacteriaceae. Ann Clin Microbiol Antimicrob 2015; 14:31–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Shorr AF, Zilberberg MD, Micek ST, Kollef MH. Prediction of infection due to antibiotic-resistant bacteria by select risk factors for health care-associated pneumonia. Arch Intern Med 2008; 168:2205–10. [DOI] [PubMed] [Google Scholar]
- 17. Lodise TP Jr, Miller C, Patel N, et al. Identification of patients with Pseudomonas aeruginosa respiratory tract infections at greatest risk of infection with carbapenem-resistant isolates. Infect Control Hosp Epidemiol 2007; 28:959–65. [DOI] [PubMed] [Google Scholar]
- 18. Yang D, Xie Z, Xin X, et al. A model for predicting nosocomial carbapenem-resistant Klebsiella pneumoniae infection. Biomed Rep 2016; 5:501–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Meng X, Liu S, Duan J, et al. Risk factors and medical costs for healthcare-associated carbapenem-resistant Escherichia coli infection among hospitalized patients in a Chinese teaching hospital. BMC Infect Dis 2017; 17:82–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Miller BM, Johnson SW. Demographic and infection characteristics of patients with carbapenem-resistant Enterobacteriaceae in a community hospital: development of a bedside clinical score for risk assessment. Am J Infect Control 2016; 44:134–7. [DOI] [PubMed] [Google Scholar]
- 21. Viale P, Giannella M, Lewis R, et al. Predictors of mortality in multidrug-resistant Klebsiella pneumoniae bloodstream infections. Expert Rev Anti Infect Ther 2013; 11:1053–63. [DOI] [PubMed] [Google Scholar]
- 22. Tacconelli E, Cataldo MA, De Pascale G, et al. Prediction models to identify hospitalized patients at risk of being colonized or infected with multidrug-resistant Acinetobacter baumannii calcoaceticus complex. J Antimicrob Chemother 2008; 62:1130–7. [DOI] [PubMed] [Google Scholar]
- 23. van Walraven C, Austin PC, Jennings A, et al. A modification of the Elixhauser comorbidity measures into a point system for hospital death using administrative data. Med Care 2009; 47:626–33. [DOI] [PubMed] [Google Scholar]
- 24. Lee SS, Kim Y, Chung DR. Impact of discordant empirical therapy on outcome of community-acquired bacteremic acute pyelonephritis. J Infect 2011; 62:159–64. [DOI] [PubMed] [Google Scholar]
- 25. Raman G, Avendano E, Berger S, Menon V. Appropriate initial antibiotic therapy in hospitalized patients with gram-negative infections: systematic review and meta-analysis. BMC Infect Dis 2015; 15:395–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shorr AF, Micek ST, Welch EC, et al. Inappropriate antibiotic therapy in Gram-negative sepsis increases hospital length of stay. Crit Care Med 2011; 39:46–51. [DOI] [PubMed] [Google Scholar]
- 27. Thom KA, Schweizer ML, Osih RB, et al. Impact of empiric antimicrobial therapy on outcomes in patients with Escherichia coli and Klebsiella pneumoniae bacteremia: a cohort study. BMC Infect Dis 2008; 8:116–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. StataCorp. Stata Statistical Software: Release 14. College Station, TX: StataCorp LP; 2015. [Google Scholar]
- 29. Falcone M, Russo A, Iacovelli A, et al. Predictors of outcome in ICU patients with septic shock caused by Klebsiella pneumoniae carbapenemase-producing K. pneumoniae. Clin Microbiol Infect 2016; 22:444–50. [DOI] [PubMed] [Google Scholar]
- 30. Richter SE, Miller L, Uslan DZ, et al. Risk factors for colistin resistance among gram-negative rods and Klebsiella pneumoniae isolates. J Clin Microbiol. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mann CJ. Observational research methods. Research design II: cohort, cross sectional, and case-control studies. Emerg Med J 2003; 20:54–60. [DOI] [PMC free article] [PubMed] [Google Scholar]