Abstract
Non-small cell lung cancer (NSCLC) is one of the most frequent cancers worldwide. The abnormal expression of long non-coding RNAs (lncRNAs) has been reported to be closely associated with the progression of human cancers, including NSCLC. Here, we demonstrated that differentiation antagonizing noncoding RNA (DANCR) was overexpressed in NSCLC tissues. Upregulation of DANCR expression was significantly associated with larger tumor size, advanced TNM stage and lymph node metastasis, and also predicted poor prognoses of patients with NSCLC. Functional experiments showed that DANCR enhanced NSCLC growth and metastasis both in vitro and in vivo. Further investigation revealed that DANCR could compete with the Sox4 mRNA to bind with miR-138, thus affecting Sox4 expression. In addition, we found that Sox4 bound to the promoter regions of DANCR gene to activate DANCR expression, suggesting a positive feedback loop of DANCR/miR-138/Sox4 in NSCLC. Taken together, these results provide a comprehensive analysis of the roles of DANCR as a competing endogenous RNA (ceRNA) in NSCLC progression.
Keywords: DANCR, metastasis, ceRNA, Sox4
Introduction
Lung cancer is the most frequent cause of cancer-related death worldwide [1]. Non-small cell lung cancer (NSCLC) accounts for approximately 80% of all lung cancer cases [2]. Though the therapeutic advances in diagnosis and clinical treatment for NSCLC have been achieved, the 5-year overall survival rate of NSCLC patients is still less than 15% due to tumor metastasis and recurrence [3]. Therefore, better understanding of tumor progression is of great importance for the advance of early detection, prevention, and exploring novel effective therapies for NSCLC patients.
Long noncoding RNAs (lncRNAs) are a class of RNA transcripts larger than 200 nucleotides in length, which cannot encode proteins. Increasing evidence demonstrates a critical role for lncRNAs in various different cellular processes, including cell proliferation, migration, invasion and stemness [4,5]. MicroRNAs (miRNAs) are an abundant class of endogenous and short (21-24 nucleotides in length) non-coding RNAs, which can induce translational repression or degradation of target mRNAs via binding their 3’-untranslated region (3’-UTR) [6]. Emerging studies demonstrated that lncRNAs could function as competing endogenous RNAs (ceRNA) to regulate gene expression through competitively binding common microRNAs [7]. For example, LINC01133 suppresses gastric cancer progression through acting as a ceRNA of APC and suppressing the Wnt/β-catenin pathway [8]. HCC-associated lncRNA (HCAL) functions as a ceRNA of LAPTM4B to facilitate growth and metastasis of liver cancer [9]. Recently, the lncRNA differentiation antagonizing noncoding RNA (DANCR), locates on chromosome 4, was shown to be upregulated in several human cancers and modulate the growth and metastasis of cancer cells [10,11]. DANCR exerts oncogenic effects by sponging some microRNAs, such as miR-758, miR-335, miR-577 and miR-33a [12-14]. However, the biological functions and influences of DANCR as a ceRNA in NSCLC progression remain largely unclear.
Increasing evidence reported that miR-138 is dramatically downregulated and functions as an important tumor suppressor in several human cancers. In specific, miR-138 inhibits proliferation, migration and invasion through suppression of Sox12 expression [15]. miR-138 also targets EZH2 and then inactivates PI3K-AKT signaling pathway to regulate growth in laryngeal squamous cell carcinoma [16]. Downregulation of miR-138 expression was found in NSCLC tissues and indicated poor prognosis of patients. miR-138 suppresses proliferation, migration, invasion, epithelial-mesenchymal transition (EMT), chemoresistance and gefitinib resistance via targeting GPR124, GIT1, SEMA4C and ZEB2 [17-19]. Hence, the suppressive influence of miR-138 in NSCLC progression deserves further investigation.
Members of the Sox (Sry-related high-mobility group box) family of transcription factors are critical for many developmental processes regulating both the maintenance of stem cells and controlling terminal differentiation of a wide variety of cell types [20]. Sox4 acts as a oncogene in a wide variety of solid tumors, such as colorectal cancer, prostate cancer, hepatocellular carcinoma (HCC), gastric cancer, lung cancer and endometrial cancer [21]. Increased expression of Sox4 modulates malignant phenotypes of cancer cells, including inhibition of apoptosis, increased cell invasion and metastasis, and maintenance of cancer-initiating cells [22]. Recent studies showed that deregulation of miR-138 are responsible for the overexpression of Sox4 in human cancers [23-26]. However, to date, the regulatory mechanism of Sox4 via lncRNAs in NSCLC remains largely unknown. Here, online prediction driving DANCR to arouse our attention for its similar microRNA response elements (MREs) which might be provided for miR-138 as Sox4 did. Moreover, we found that DANCR interacted with miR-138 to regulate Sox4 expression via the ceRNA mechanism. In the present study, we focused on the ceRNA network which is comprising of DANCR, miR-138 and Sox4, and revealed the functional roles of DANCR in NSCLC growth and metastasis.
Materials and methods
Tissue samples
Paired NSCLC and adjacent normal lung tissues were obtained from 64 patients who underwent primary surgical resection in the Department of Respiratory, The First Affiliated Hospital of Zhengzhou University. Diagnosis was based on pathological evidence, and collected tissue samples were immediately snap-frozen in liquid nitrogen and stored at -80°C until used. All patients were not received preoperative treatments for cancer, such as radiotherapy, chemotherapy or immunotherapy. Written informed consent was obtained from all participants. The study was approved by the Ethics Committee of The First Affiliated Hospital of Zhengzhou University.
Cell culture
The normal human bronchial epithelial cells (NHBE) and HEK-293T cells were purchased from the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). The NSCLC cell lines (A549, H1299, H460, SK-MES-1, and Calu-3) were purchased from the American Type Culture Collection (ATCC, Manassas, USA). All cells were cultured in Dulbecco’s modified Eagle medium (DMEM, Hyclone) supplemented with 10% fetal bovine serum (FBS, Invitrogen) and 1% penicillin/streptomycin. All cells were cultured in a humidified incubator at 37°C with 5% CO2.
Transfection
The miR-138 mimics, miR-138 inhibitor (anti-miR-138) and corresponding negative control miR (miR-NC, anti-miR-NC) were purchased from RiboBio (Guangzhou, China). The Sox4 siRNA (siSox4) and corresponding negative control siRNA (siNC) were also purchased from RiboBio (Guangzhou, China). The target sequence of siSox4 was shown as follow: siSox4-1: GCGACAAGATCCCTTTCAT, siSox4-2: CGACAAGATCCCTTTCATT. Transfection was performed by using Lipofectamine 2000 (Invitrogen) following the manufacturer’s protocol.
Overexpression or knockdown of DANCR
For overexpression of DANCR, the full-length human DANCR gene was PCR amplified and subcloned into the lentiviral vector pLV (Addgene). Lentiviral preparations were generated by transient transfection of HEK-293T cells by using pLV-DANCR, psPAX2 and pMD2.G. For knockdown of DANCR, DANCR shRNA was inserted into lentiviral vector pLKO.1 (Addgene). The target sequences for DANCR were shown as follows: shDANCR-1: AGCCAACTATCCCTTCAGT; shDANCR-2: GAGCTAGAGCAGTGACAAT. Lentiviral preparations were generated as above. Lentiviruses were harvested at 48 hr after transfection and then filtered. Cells were infected with above lentiviruses in the presence of 8 μg/ml polybrene (Sigma-Aldrich). After 48 h, stable cells were selected by puromycin (2 μg/mL) for 1 week.
RNA isolation and quantitative real-time PCR (qRT-PCR)
TRIzol reagent (Invitrogen) was used to extract total RNA from tissues or cells in accordance with the manufacturer’s instructions. One microgram total RNA was reverse transcribed using TransScript first-strand cDNA synthesis SuperMix (TransGen, Beijing, China). qRT-PCR assay was performed by SYBR green qPCR SuperMix (Applied Biosystems Life Technologies, Foster, CA, USA) in ABI prism 7500 sequence detection system (Applied Biosystems Life Technologies). The results were normalized to GAPDH. Fold changes of each gene were calculated by 2-ΔΔCt (cycle threshold). The specific primers used for qRT-PCR were as following: Sox4-forward: CATGGTGTGGTCGCAGAT, Sox4-reverse: GGGATCTTGTCGCTGTCTTT; DANCR-forward: CTGCATTCCTGAACCGTTATCT, DANCR-reverse: GGGTGTAATCCACGTTTCTCAT.
Cell proliferation detection
Cell proliferation was examined by Cell Counting Kit-8 (Dojindo, Beijing, China). 2 × 103 cells in 100 μL medium were seeded into a 96-well plate in quadruplicate. 10 μL of Cell Counting Kit-8 (CCK-8) (Dojindo; Kumamoto, Japan) was added to each well at 24, 48, 72 and 96 h after the cells were seeded. The absorbance was measured at a wavelength of 450 nm.
Transwell assays
For the cell invasion assay, 24-well Transwell™ plates with 8.0-μm-pore Matrigel™-coated membranes were used (Corning, NY). 1 × 105 cells were seeded into the upper chambers with serum-free medium. The lower chambers were filled with medium containing 20% FBS. After 24 h of incubation, cells were fixed by 4% paraformaldehyde and stained with crystal violet. The migration assay was performed similarly without coating the membranes with Matrigel™. The cells that had invaded through the membrane to the lower surface were photographed and counted under a microscope in 10 random fields.
Apoptosis analysis
Cells were resuspended at a concentration of 1 × 106 cells/mL and then stained with 5 μL of propidium iodide (PI) and 5 μL of Annexin V-FITC from the FITC-Annexin V Apoptosis Detection Kit (BD Biosciences). Apoptosis were analyzed with a BD FACS Calibur™ flow cytometer. Data analysis was performed using FlowJo (Tree Star, San Carlos, CA).
Cell cycle analysis
For cell cycle analysis, cells were harvested and fixed with 70% cold ethanol at 4°C overnight. Then, cells were incubated with PI for 10 min, and the DNA content was determined by flow cytometry. Data analysis was performed using FlowJo (Tree Star, San Carlos, CA).
Xenograft model
5 × 106 cells were separately subcutaneously inoculated into the left and right flank in the dorsal of the nude mice for in vivo xenograft assay. Tumor size was measured every 5 days for 30 days. The tumor volume was calculated by the formula (length × width2)/2. For lung metastasis detection, 5 × 105 cells were injected into the lateral tail vein of the nude mice. After 8 weeks, mice were sacrificed, and the lung tissues of each mouse were obtained and followed by H&E staining. The number of pulmonary metastasis in each mouse was counted under a microscope. All experimental procedures were approved by the Animal Ethics Committee of The First Affiliated Hospital of Zhengzhou University.
RNA immunoprecipitation (RIP)
A549 cells were co-transfected with pCMV-MS2, pCMV-DANCR-MS2 or pCMV-DANCR-mut-MS2 and pMS2-GFP (Addgene). After 48 hrs, RIP assay was performed by using a GFP antibody (Abcam) or negative IgG antibody (Millipore) and the Magna RIP™ RNA-Binding Protein Immunoprecipitation Kit (Millipore, Bedford, MA) as the manufacturer’s instructions.
Chromatin immunoprecipitation assay (ChIP)
The binding of Sox4 in DANCR promoter was detected by ChIP assay. ChIP assays were performed by EZ-ChIP-Chromatin Immunoprecipitation (Millipore) as the manufacturer’s instructions. Briefly, cells were cross-linked in 1% formaldehyde and then terminated by the addition of 125 mM (final concentration) glycine. Sox4 antibodies (Abcam) or IgG antibodies (Millipore) were mixed with clear nuclear lysates for immunoprecipitation. Coprecipitated DNA was purified and the level of target genes was quantified using qRT-PCR.
Luciferase reporter assay
The wild-type or mutant DANCR or 3’-UTR of Sox4 mRNA were PCR-amplified and subcloned into pmirGLO vector. pmirGLO, pmirGLO-DANCR or pmirGLO- DANCR-mut was cotransfected with miR-138 mimics into cells by Lipofectamine 2000 (Invitrogen) following the manufacturer’s protocol. pmirGLO or pmirGLO-Sox4 was transfected into different stable cells by Lipofectamine 2000 (Invitrogen) following the manufacturer’s protocol. After 48 h, the luciferase activity was detected with the Dual Luciferase Assay Kit (Promega). Cells were lysed with lysis buffer. After centrifuge, the luciferase activity was determined by a Modulus TD20/20 Luminometer (Turner Biosystems, CA). The relative luciferase activity was normalized to Renilla luciferase activity.
Western blot
Cells were lysed in RIPA Lysis Buffer (Beyotime, Beijing, China) supplemented with PMSF. Equal amounts of protein were separated by SDS-PAGE and then transferred to PVDF membranes (Millipore). The membranes were blocked with 5% non-fat milk and then incubated with primary antibodies for Sox4 (Cell Signal Technology), and GAPDH (Cell Signal Technology) at 4°C overnight. Subsequently, the membranes were exposed to horseradish peroxidase-labeled IgG for 1 h, and the bands were visualized using a Bio-Rad imaging system.
Statistical analysis
Statistical analysis was performed using SPSS 19 software package (IBM SPSS Inc; Chicago, IL, USA). Student’s t test or ANOVA test was used to analyze the results expressed as mean ± SD. The χ2 test was used to analyze the correlation of DANCR expression and clinicopathological characteristics of NSCLC patients. The survival curves were plotted by Kaplan-Meier analysis, and the survival differences were compared using the log-rank test. P<0.05 was regarded to be statistically significant.
Results
Upregulation of DANCR is associated with a poor overall survival time of NSCLC patients
To identify the role of DANCR in NSCLC progression, qRT-PCR analysis was used to investigate DANCR expression in 64 pairs of NSCLC tissues compared with adjacent normal tissues. Our results showed that DANCR expression in tumor tissues was significantly higher than those in the corresponding normal tissues (Figure 1A, P = 0.0018). We also examined the expression levels of DANCR in NSCLC cell lines. As shown in Figure 1B, DANCR expression was also significantly increased in NSCLC cell lines (A549, H1299, H460, SK-MES-1, and Calu-3) compared with that in normal human bronchial epithelial cells (NHBE).
Figure 1.
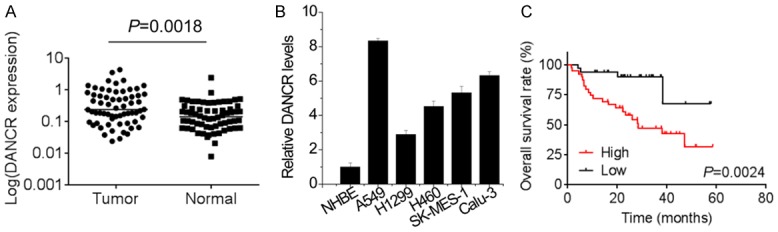
The DANCR is upregualted in NSCLC tissues and predicts poor prognosis of NSCLC patients. A. qRT-PCR analysis was used to investigate DANCR expression in 64 pairs of NSCLC tumor tissues compared with adjacent normal tissues. B. qRT-PCR analysis was used to investigate DANCR expression in NSCLC cell lines (A549, H1299, H460, SK-MES-1, and Calu-3) compared with that in normal human bronchial epithelial cells (NHBE). C. Kaplan-Meier analyses of the correlations between DANCR expression level and overall survival of 64 patients with NSCLC. The median expression level was used as the cutoff. Patients with DANCR expression values below the 50th percentile were classified as high group. Patients with DANCR expression values above the 50th percentile were classified as low group.
According to the median value of DANCR expression in NSCLC tissues, we defined 32 patients with high level of DANCR expression and 32 patients with low level of DANCR expression. We analyzed the association between DANCR expression and the clinicopathological features of patients with NSCLC. As shown in Table 1, increased DANCR level was correlated to larger tumor size, advanced TNM stage and lymph node metastasis of NSCLC patients. However, no significant correlation between DANCR expression and other clinicopathological factors was observed, including gender, age, histological grade, histological classification or smoking status. Moreover, Kaplan-Meier analysis and log-rank test was performed to evaluate survival rate and the results revealed that NSCLC patients in high-level DANCR group had worse overall survival than those in low-level DANCR group (Figure 1C). Overall, these results demonstrated that lncRNA DANCR was upregulated in NSCLC tissues and cells, and indicated the poor prognosis of NSCLC patients.
Table 1.
The association between DANCR expression and clinicopathological features of NSCLC patients
| Characteristics | Expression of DANCR | P value | |
|---|---|---|---|
|
| |||
| Low (N = 32) | High (N = 32) | ||
| Sex | 0.451 | ||
| Male | 16 | 13 | |
| Female | 16 | 19 | |
| Age | 0.802 | ||
| ≤60 | 15 | 16 | |
| >60 | 17 | 16 | |
| Histological grade | 0.200 | ||
| Middle or low | 24 | 28 | |
| High | 8 | 4 | |
| Histological classification | 0.617 | ||
| Squamous cell carcinoma | 17 | 15 | |
| Adenocarcinoma or other | 15 | 17 | |
| TNM stage | 0.002 | ||
| I and II | 7 | 19 | |
| III and IV | 25 | 13 | |
| Lymph node metastasis | 0.012 | ||
| Negative | 9 | 19 | |
| Positive | 23 | 13 | |
| Tumor size | |||
| ≤3 cm | 20 | 5 | <0.0001 |
| >3 cm | 12 | 27 | |
| History of smoking | 0.316 | ||
| Ever | 19 | 15 | |
| Never | 13 | 17 | |
DANCR promotes proliferation and inhibits apoptosis of NSCLC cells in vitro
To understand the functional role of DANCR in NSCLC progression, both overexpression and knockdown experiments were utilized. Lentiviruses for DANCR overexpression and control lentiviruses (Ctrl) were used to infect the H1299 cells, whose DANCR expression was relatively low. Lentiviruses expressing specific shRNA targeting DANCR (shDANCR) and negative control shRNA (shNC) were used in A549 cells, whose DANCR expression was relatively high. The increased or reduced expression of DANCR in these cell lines were confirmed by qRT-PCR (Figure 2A and 2B). The effect of DANCR on cell proliferation was evaluated by CCK-8 assays and the results showed that DANCR inhibition decreased cellular proliferation compared with the control group in A549 cells (Figure 2C), while overexpression of DANCR increased the proliferative rate of H1299 cells compared with the control cells (Figure 2D). Flow cytometry was performed to investigate the role of DANCR on NSCLC cells cycle. We found that DANCR knockdown induced an increase in the percentage of G0/G1-phase cells and a decrease in the percentage of S-phase cells (Figure 2E). Conversely, DANCR overexpression drove progression beyond the G1/S transition in H1299 cells (Figure 2F). Moreover, we detected apoptosis by flow cytometry analysis in NSCLC cells stained for Annexin V and PI. It was found that DANCR-knockdown A549 cells had a significantly higher percentage of Annexin V-positive cells than did in control cells (Figure 2G), whereas DANCR overexpression suppressed cell apoptosis in H1299 cells (Figure 2H).
Figure 2.
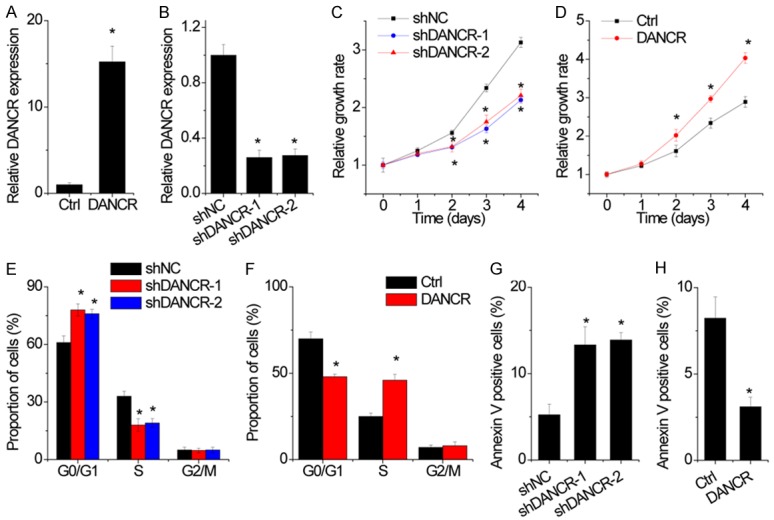
DANCR promotes NSCLC cell proliferation, migration and invasion. A. The DANCR expression was overexpressed in H1299 cells. The relative DANCR expression in H1299 cells with DANCR overexpression was detected by qRT-PCR. B. The DANCR expression was silenced in A549 cells by two different shRNAs. The relative DANCR expression in A549 cells with DANCR knockdown was detected by qRT-PCR. C. DANCR knockdown inhibited cellular proliferation in A549 cells on the basis of CCK8 assays. D. DANCR overexpression promoted cellular proliferation in H1299 cells on the basis of CCK8 assays. E. Cell cycle was analyzed by flow cytometry after knocking down DANCR. The results showed that DANCR depletion led to G1 arrest in A549 cells. F. Cell cycle was analyzed by flow cytometry after overexpression DANCR. The results showed that DANCR overexpression drove progression beyond the G1/S transition in H1299 cells. G. Cell apoptosis was analyzed by flow cytometry after knocking down DANCR. Cells positive for Annexin V staining were counted as apoptotic cells, and the results showed that DANCR depletion induced cell apoptosis in A549 cells. H. Cell apoptosis was analyzed by flow cytometry after overexpressing DANCR. Cells positive for Annexin V staining were counted as apoptotic cells, and the results showed that DANCR overexpression inhibited cell apoptosis in A1299 cells. Date are presented as the means ± SD. *P<0.05.
DANCR enhances cell migration and invasion, and induces EMT in NSCLC cells
A classical transwell assay was further utilized to assess the influence of DANCR on the migration and invasion. The results showed a notable decrease in the number of migrated and invaded cells transfected with DANCR shRNA relative to the negative control shRNA group (Figure 3A). On the contrary, DANCR overexpression significantly increased the migratory and invasive ability of H1299 cell (Figure 3B).
Figure 3.
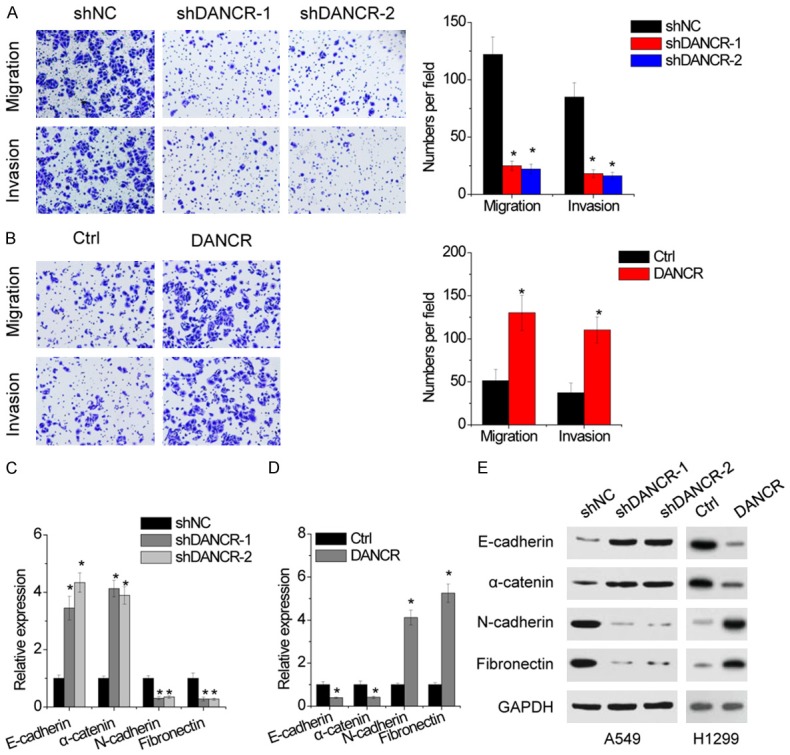
DANCR promotes migration and invasin, and induces EMT. A. DANCR inhibition decreased the migration and invasion of A549 cells as determined by transwell assays. B. DANCR overexpression increased the migration and invasion of H1299 cells as determined by transwell assays. C. qRT-PCR showed a significant increase in the expression of the epithelial markers, E-cadherin and α-catenin, and a corresponding reduction of the mesenchymal markers, N-cadherin, and Fibronectin in DANCR-depleted A549 cells, relative to control. D. qRT-PCR showed a significant decrease in the expression of the epithelial markers, E-cadherin and α-catenin, and a corresponding increase of the mesenchymal markers, N-cadherin, and Fibronectin in DANCR-overexpressed H1299 cells, relative to control. E. The protein expression of mesenchymal and epithelial markers in indicated stable cells was detected by western blot. Date are presented as the means ± SD. *P<0.05.
EMT is associated with tumor metastasis, which converts adherent epithelial cells into migratory cells. Therefore, we determined whether EMT is responsible for DANCR-mediated change in cell motility. Depletion of DANCR in A549 cells induced the mRNA and protein expression of epithelial markers E-cadherin and α-catenin that was accompanied by a concomitant reduction of mesenchymal markers N-cadherin and Fibronectin (Figure 3C and 3E). Conversely, overexpression of DANCR decreased epithelial markers and upregulated mesenchymal markers (Figure 3D and 3E).
DANCR promotes NSCLC cell tumorigenicity and metastasis in vivo
To further prove the role of DANCR in vivo, a xenograft mouse model was used. Control and DANCR knockdown A549 cells were subcutaneously injected into nude mice. As shown in Figure 4A, tumors grown from DANCR stable knockdown A549 cells were smaller than tumors grown from control cell. The average tumor weight in the shDANCR group was also much lower than that in the control group (Figure 4B). Conversely, xenograft tumors grown from DANCR overexpressing H1299 cells had larger mean volumes and weights than tumors grown from control cells (Figure 4C and 4D).
Figure 4.
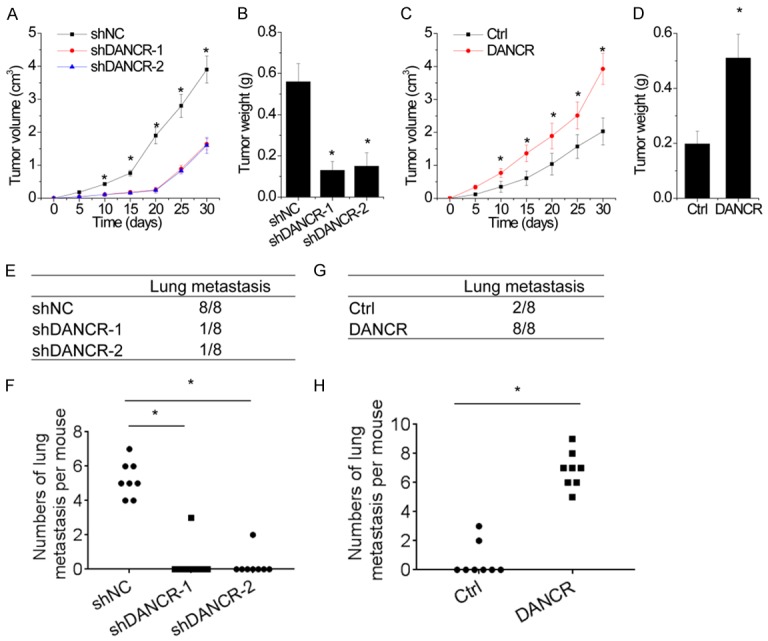
DANCR promotes growth and metastasis of NSCLC cells in vivo. A and B. Effects of DANCR knockdown on tumor growth in vivo. Control and DANCR knockdown A549 cells were injected subcutaneously into nude mice. A. The tumor growth curves. B. Tumor weights. C and D. Effects of DANCR overexpression on tumor growth in vivo. Control and DANCR overexpressing H1299 cells were injected subcutaneously into nude mice. C. The tumor growth curves. D. Tumor weights. E and F. Control and DANCR knockdown A549 cells were injected into the lateral tail vein of the nude mice. After 8 weeks, the lung metastasis was detected. E. Incidence of lung metastasis in each group of nude mice. F. The numbers of hepatic and lung metastases per mouse were calculated. G and H. Control and DANCR overexpressing H1299 cells were injected into the lateral tail vein of the nude mice. After 8 weeks, the lung metastasis was detected. G. Incidence of lung metastasis in each group of nude mice. H. The numbers of hepatic and lung metastases per mouse were calculated. Date are presented as the means ± SD. *P<0.05.
In metastatic models, we injected A549 cells with stably DANCR knockdown into nude mice tail veins. After 8 weeks, both the incidence of lung metastasis and the number of lung metastatic nodules in the A549-shDANCR group was significantly decreased, compared to that in the control group (Figure 4E and 4F). Conversely, in the H1299-DANCR group, all of the mice developed lung metastases; however, only 2 mice in the control group developed lung metastases (100% versus 25%, Figure 4G). The number of lung metastatic nodules in the H1299-DANCR group was also significantly increased, compared to that in the control group (Figure 4H). The results above demonstrate that DANCR positively regulates the tumorigenicity and metastasis of NSCLC cells in vivo.
miR-138 is a target of DANCR
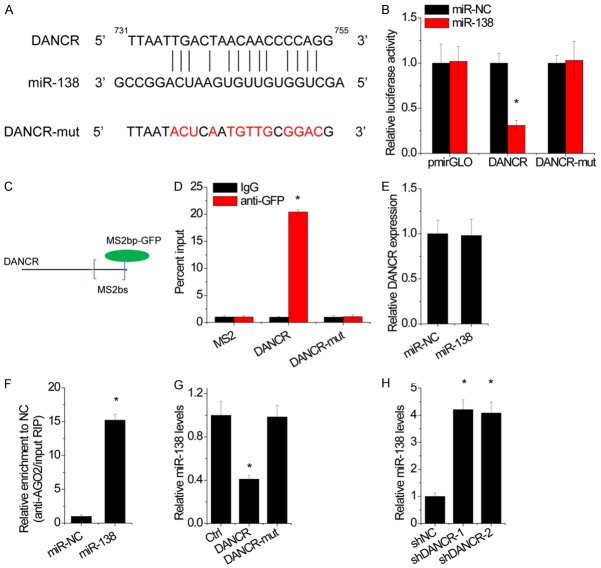
We next explored the underlying mechanism by which DANCR regulates NSCLC progression. Increasing evidence suggested that lncRNA could act as ceRNAs to competitively binding microRNAs. To examine whether DANCR has a similar mechanism in NSCLC, we used starBase online tool to explore the association of DANCR and microRNAs. We found that miR-138, a tumor suppressor microRNA, was predicted to interact with DANCR (Figure 5A). Next, to test the binding sites in DANCR, we subcloned the wild-type DANCR transcript or its mutant sequence (mutant in the binding sites of miR-138) into a luciferase reporter construct, and then transiently cotransfected the reporter constructs with miR-138 or negative control mimics (miR-NC) into A549 cells. Results showed that miR-138 significantly inhibited the luciferase activities in A549 cells transfected with the wild type construct, but did not affect the luciferase activity in cells transfected with the mutant construct (Figure 5B). To further confirm the direct association between DANCR and miR-138, we carried out an anti-MS2 RIP assay. As MS2-binding protein (MS2bp) specifically binds RNA containing MS2-binding sequences (MS2bs), we generated a construct containing DANCR transcripts combined with MS2bs elements and cotransfected into NSCLC cells with a plasmid expressing MS2bp-GFP. Then, the RIP assay was carried out using an anti-GFP antibody (Figure 5C). The result of MS2-RIP showed that the wild-type DANCR could be significantly enriched by miR-138 compared with the empty vector (MS2) and DANCR with mutations in miR-138 targeting sites (DANCR-mut) (Figure 5D). However, we found no significant difference in DANCR levels after overexpression of miR-138, indicating that miR-138 could not induce the degradation of DANCR (Figure 5E). microRNAs bind their target mRNAs and cause posttranscriptional repression in an AGO2-dependent manner. To determine whether DANCR was regulated by miR-138 in this manner, we performed anti-AGO2 RIP in A549 cells transiently transfected with miR-138 or miR-NC mimics. Endogenous DANCR was specifically enriched in A549 cells transfected with miR-138 mimics (Figure 5F), indicating that DANCR and miR-138 were in the same RNA-induced silencing complex (RISC). Moreover, overexpression of wild-type DANCR WT, but not its mutant, decreased the miR-138 expression in H1299 cells (Figure 5G), while knockdown of DANCR upregulated miR-138 level in A549 cells (Figure 5H). Together, these data demonstrate that DANCR physically binds with miR-138 and may function as a ceRNA.
Figure 5.
DANCR interacts with miR-138. A. Schematic representation of the miR-138 site in DANCR. B. Luciferase activity in A549 cells cotransfected with miR-NC or miR-138 and luciferase reporters containing DANCR or mutant DANCR (DANCR-mut). Data are presented as the relative ratio of firefly luciferase activity to renilla luciferase activity. C. A construct containing DANCR transcripts combined with MS2 binding site (MS2bs) elements. D. MS2-RIP followed by miR-138 qRT-PCR to examine miR-138 endogenously associated with DANCR or mutant DANCR. E. The DANCR expression in A549 cells transfected with miR-NC or miR-138. F. AGO2-RIP followed by DANCR qRT-PCR to assay DANCR level after miR-138 overexpression. G. The miR-138 expression in H1299 cells with overexpression of DANCR or mutant DANCR. H. The miR-138 expression in A549 cells with or without DANCR knockdown. Date are presented as the means ± SD. *P<0.05.
The functional roles of miR-138 in NSCLC progression were then determined. A549 and H1299 cells were transfected with miR-138 mimics (Figure S1A). The CCK-8 assays showed that overexpression of miR-138 significantly suppressed proliferation in both A549 and H1299 cells (Figure S1B). Moreover, A549 and H1299 cells with upregulation of miR-138 showed higher migratory and invasive capacities than control cells (Figure S1C).
To explore the underlying mechanism of miR-138-mediated phenotypes, we searched for potential targets of miR-138. The prediction of TargetScan showed that Sox4 may be a direct target of miR-138 (Figure S1D). Then, luciferase reporter vectors containing 3’-UTR of Sox4 mRNA was constructed. It was observed that miR-138 significantly decreased the luciferase activity of Sox4 3’UTR, while miR-138 did not exert an obvious effect on the empty vector or the mutant Sox4 3’UTR (Sox4-mut, mutation in miR-138-targeting sites) (Figure S1E). Additionally, miR-138 suppressed Sox4 expression as proved by qRT-PCR and western blot (Figure S1F and S1G).
DANCR positively regulates Sox4 expression via sponging miR-138
As Sox4 is an important regulators involving in tumor growth and metastasis and a direct target of miR-138, we hypothesized that DANCR could modulate NSCLC progression via functioning as a ceRNA of Sox4. Ectopic expression of DANCR led to an increase in the mRNA and protein level of Sox4 in A1299 cells, whereas overexpression of mutant DANCR did not influence the Sox4 level. Transfection of miR-138 mimics abrogated DANCR-mediated upregulation of Sox4 (Figure 6A and 6B). Inversely, depletion of DANCR repressed Sox4 expression level in A549 cells, while inhibition of miR-138 overcame this decrease (Figure 6C and 6D).
Figure 6.
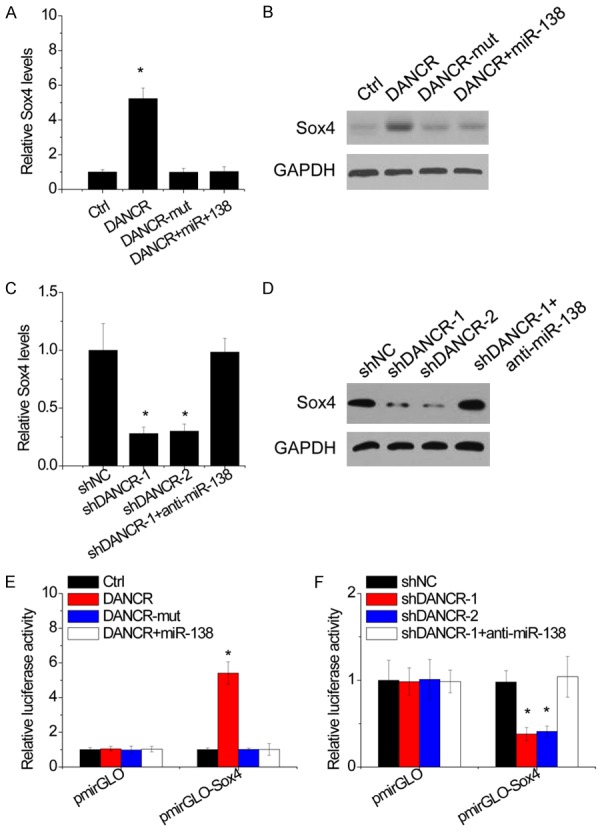
DANCR upregulates Sox4 expression by competitively binding miR-138. A. The H1299 cells were cotranfected with DANCR and miR-138, and the Sox4 mRNA expression was then detected by qRT-PCR. B. The H1299 cells were cotranfected with DANCR and miR-138, and the Sox4 protein expression was then detected by western blot. C. The A549 cells were cotranfected with DANCR shRNA and miR-138 inhibitor, and the Sox4 mRNA expression was then detected by qRT-PCR. D. The A549 cells were cotranfected with DANCR shRNA and miR-138 inhibitor, and the Sox4 protein expression was then detected by western blot. E. Luciferase activity of Sox4-3’UTR in H1299 cells cotransfected with DANCR and miR-138. Data are presented as the relative ratio of firefly luciferase activity to Renilla luciferase activity. F. Luciferase activity of Sox4-3’UTR in A549 cells cotransfected with DANCR shRNA and miR-138 inhibitor. Data are presented as the relative ratio of firefly luciferase activity to Renilla luciferase activity. Date are presented as the means ± SD. *P<0.05.
To further confirm whether this observed effect depends on the modulation of the Sox4 3’UTR, luciferase reporter assay was performed after cotransfection of luciferase plasmid containing Sox4 3’UTR with DANCR. Overexpression of wild-type DANCR, but not the mutant, increased the luciferase activity of Sox4 3’UTR. Transfection of miR-138 mimics attenuated this increase (Figure 6E). In contrast, knockdown of DANCR reduced the luciferase activity of Sox4 3’UTR, which was abolished by miR-150 inhibitor (Figure 6F). These results suggest that DANCR upregulates Sox4 expression by competitively binding miR-138.
DANCR promotes NSCLC progression partly through miR-138-Sox4 axis
We then investigated whether DANCR regulates NSCLC progression through miR-138-Sox4 axis. Overexpression of wild-type DANCR, but not its mutant, promoted cell proliferation, migration and invasion in H1299 cells. Ectopic expression of miR-138 or Sox4 knockdown partly abolished this effect (Figure 7A and 7B). Inversely, the depletion of DANCR reduced the proliferative, migratory and invasive capacities of A549 cells, which were partially abrogated by inhibition of miR-138 or Sox4 overexpression (Figure 7C and 7D).
Figure 7.
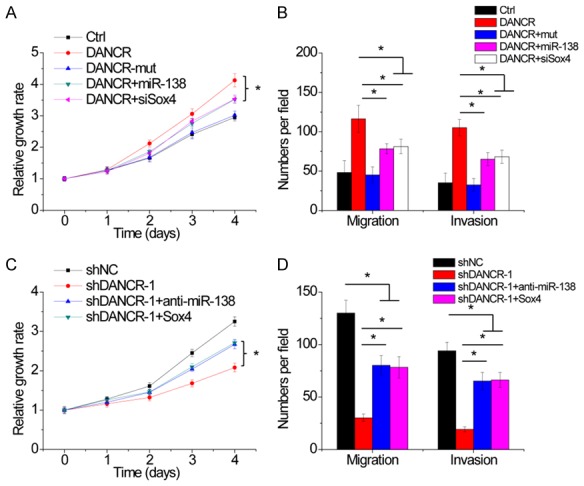
DANCR promotes NSCLC progression partly through miR-138-Sox4 axis. (A and B). Upregulatin of miR-138 or inhibition of Sox4 partly abolished the increase of proliferation (A), migration and invasion (B) mediated by DANCR overexpression in H1299 cells. (C and D). Inhibition of miR-138 or Sox4 overexpression partly rescued the decrease of proliferation (C), migration and invasion (D) mediated by DANCR knockdown in A549 cells. Date are presented as the means ± SD. *P<0.05.
Sox4 activates DANCR transcription
Recently, it has been reported that lncRNA could form a positive feedback loop with transcription factors [27]. To assay whether transcription factor Sox4 could regulate DANCR expression in turn, we silenced Sox4 expression in A549 and H1299 cells. We found that knockdown of Sox4 induced a suppression of DANCR expression (Figure 8A). Using the JASPAR online tool (http://jaspar.genereg.net/), we found that Sox4 is predicted to be bound to the DANCR promoter region. We designed two primers that covered the Sox4 binding sites and performed chromatin immunoprecipitation (ChIP) assays to validate whether Sox4 could bind to these sites. The results showed that Sox4 could bind to both sites (Figure 8B). To clarify which element was critical for the Sox4-mediated upregulation of DANCR expression, the two predicted Sox4-binding sites were individually deleted. We found that upregulation of Sox4 promoted DANCR transcriptional activity, and E1 or E2 absence downregulated DANCR promoter activity in part (Figure 8C), indicating that both E1 and E2 are involved in Sox4-induced DANCR transcription. These findings suggest that there exists a positive feedback regulation between Sox4 and lncRNA DANCR, which may continuously enhance their oncogenic functions.
Figure 8.
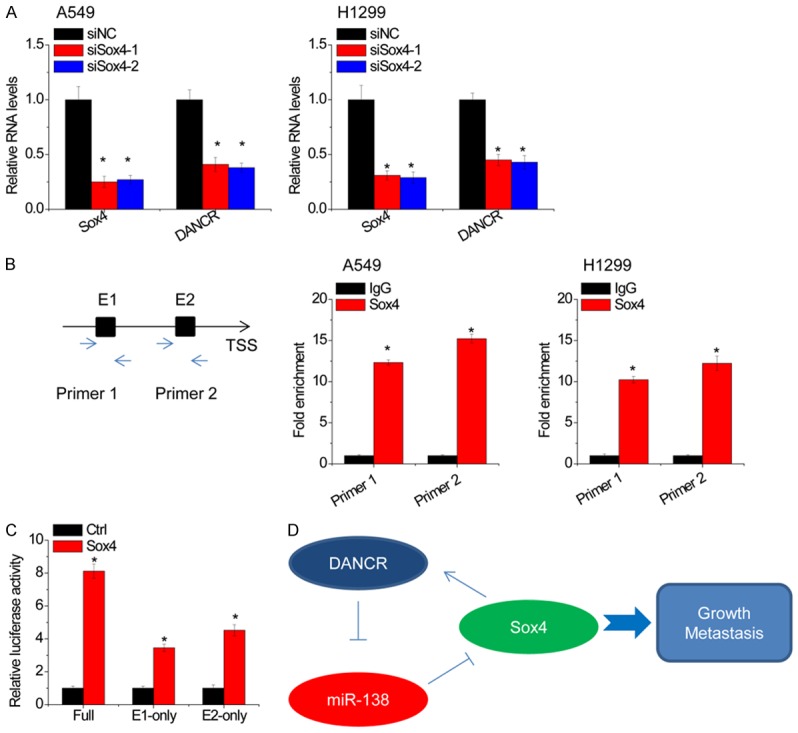
Sox4 activates DANCR transcription. A. siRNAs against Sox4 were transfected into A549 and H1299 cells, and the relative expression of Sox4 and DANCR was detected by qRT-PCR. B. The ChIP was used to determine the binding of Sox4 in DANCR promoter. IgG was taken as a negative control. C. The luciferase activity of full-length, E1-only or E2-only promoter of DANCR in 293T cell transfected with empty vector or Sox4. D. Summary of the positive feedback regulation of DANCR/miR-138/Sox4 in NSCLC progression. Date are presented as the means ± SD. *P<0.05.
Discussion
Dysregulation of lncRNAs has been closely linked to the tumorigenesis and progression of human cancers, including NSCLC [28,29]. In this study, we demonstrated that DANCR expression was frequently increased in NSCLC tissues, relative to adjacent normal tissues. DANCR overexpression was correlated with larger tumor size, advanced TNM stage and lymph node metastasis of NSCLC patients. Additionally, NSCLC patients with high-level DANCR expression had worse prognoses than did patients with low-level DANCR expression. These clinical data suggest that DANCR contributes to the malignant progression of NSCLC and may be used as a promising prognostic biomarker.
Mounting evidences discovered the role of lncRNAs in various cancers. DANCR was firstly found to increase stemness features of hepatocellular carcinoma by association of DANCR with CTNNB1 mRNA and blocking the repressing effect of miR-214, miR-320a, and miR-199a on CTNNB1 mRNA [30]. Moreover, DANCR has been proved to exert oncogenic effect in other cancers, such as colorectal cancer, prostate cancer, and osteosarcoma [13,14,31,32]. In consistent with our observations, Wang et al. [12] found that DANCR acts as a oncogene in NSCLC cells. Using loss-of-function and gain-of-function assays, we demonstrated that DANCR plays a critical role in NSCLC cell proliferation, migration and invasion. Knockdown of DANCR significantly decreased NSCLC cell growth, induced apoptosis and cell cycle arrest, and inhibited migration and invasion, whereas overexpression of DANCR exerted the opposite effects. These findings suggest that DANCR functions as an oncogene in NSCLC, and its overexpression contributes to the initiation and progression of NSCLC.
Increasing studies showed that lncRNAs could act as ceRNAs to posttranscriptionally regulate mRNA expression. The association between lncRNAs and miRNAs suppresses the inhibitory effect on target mRNAs of microRNAs. For example, lncRNA ZFAS1 upregulated ZEB1, MMP14 and MMP16 expression to facilitate growth and metastasis by sponging miR-138 in liver cancer [33]. Li et al. [34] found that lncRNA MALAT1 acted as a ceRNA by targeting miR-124 and increased STAT3 expression in NSCLC. However, whether DANCR affected NSCLC progression through ceRNA mechanism has not yet been reported. In the present study, the results of luciferase reporter and RIP assays proved that miR-138 acted as an inhibitory target of DANCR. Moreover, our data showed that DANCR functioned as a ceRNA of Sox4 through competitively binding miR-138. However, overexpression of miR-138 or knockdown of Sox4 did not completely abolish the proliferation, migration and invasion increased by DANCR overexpression, indicating that other mechanisms were involved in this procedure. A previous study demonstrated that DANCR could sponge miR-738 to promote NSCLC cell proliferation, migration and invasion [12], suggesting that DANCR may regulate NSCLC progression through inhibition of a number of different miRNAs.
Recent studies have demonstrated that lncRNAs expression can be regulated in a manner which is similar to protein coding genes. For example, transcription factor E2F1 activates lncRNA ANRIL expression in gastric cancer [35], and SP1 triggers lncRNA LINC00673 transcription [36]. However, the regulators responsible for lncRNA DANCR upregulation in human cancers remain unclear. Here, the results of luciferase and ChIP assay showed that transcription factor Sox4 could bind to the promoter region of DANCR gene and then activate its transcription. Collectively, DANCR upregulates Sox4 expression through acting as a ceRNA, and Sox4 trigger DANCR transcription in turn, revealing a positive feedback regulatory loop between lncRNA DANCR and Sox4 (Figure 8D). Similar regulatory manner between lncRNA and its target was also observed in other cancers. For example, in gastric cancer, lncRNA PVT1 directly interacts with the signal transducer activator phospho-STAT3 to increase its protein stability. Meanwhile, activated STAT3 can occupy the PVT1 promoter to facilitate its transcription [27]. LINC01296 harbored miR-598 to upregulate Twist1, and Twist1 can transactivate LINC01296 expression in turn, indicating a positive feedback loop of LINC01296/miR-598/Twist1 [37].
In conclusion, we elucidated that DANCR can stimulate growth and metastasis via regulating miR-138-Sox4 axis in NSCLC. Thus, these findings shed new light on the oncogenic roles of DANCR, and DANCR may be a novel therapeutic target for NSCLC treatment.
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Herbst RS, Heymach JV, Lippman SM. Lung cancer. N Engl J Med. 2008;359:1367–1380. doi: 10.1056/NEJMra0802714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wood SL, Pernemalm M, Crosbie PA, Whetton AD. Molecular histology of lung cancer: from targets to treatments. Cancer Treat Rev. 2015;41:361–375. doi: 10.1016/j.ctrv.2015.02.008. [DOI] [PubMed] [Google Scholar]
- 4.He A, He S, Li X, Zhou L. ZFAS1: a novel vital oncogenic lncRNA in multiple human cancers. Cell Prolif. 2019;52:e12513. doi: 10.1111/cpr.12513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang L, Peng D, Sood AK, Dang CV, Zhong X. Shedding light on the dark cancer genomes: long noncoding RNAs as novel biomarkers and potential therapeutic targets for cancer. Mol Cancer Ther. 2018;17:1816–1823. doi: 10.1158/1535-7163.MCT-18-0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ding L, Lan Z, Xiong X, Ao H, Feng Y, Gu H, Yu M, Cui Q. The dual role of microRNAs in colorectal cancer progression. Int J Mol Sci. 2018;19 doi: 10.3390/ijms19092791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yuan JH, Yang F, Wang F, Ma JZ, Guo YJ, Tao QF, Liu F, Pan W, Wang TT, Zhou CC, Wang SB, Wang YZ, Yang Y, Yang N, Zhou WP, Yang GS, Sun SH. A long noncoding RNA activated by TGF-beta promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell. 2014;25:666–681. doi: 10.1016/j.ccr.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 8.Yang XZ, Cheng TT, He QJ, Lei ZY, Chi J, Tang Z, Liao QX, Zhang H, Zeng LS, Cui SZ. LINC01133 as ceRNA inhibits gastric cancer progression by sponging miR-106a-3p to regulate APC expression and the Wnt/beta-catenin pathway. Mol Cancer. 2018;17:126. doi: 10.1186/s12943-018-0874-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xie CR, Wang F, Zhang S, Wang FQ, Zheng S, Li Z, Lv J, Qi HQ, Fang QL, Wang XM, Yin ZY. Long noncoding RNA HCAL facilitates the growth and metastasis of hepatocellular carcinoma by acting as a ceRNA of LAPTM4B. Mol Ther Nucleic Acids. 2017;9:440–451. doi: 10.1016/j.omtn.2017.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shi H, Shi J, Zhang Y, Guan C, Zhu J, Wang F, Xu M, Ju Q, Fang S, Jiang M. Long non-coding RNA DANCR promotes cell proliferation, migration, invasion and resistance to apoptosis in esophageal cancer. J Thorac Dis. 2018;10:2573–2582. doi: 10.21037/jtd.2018.04.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu D, Yu J, Gao G, Lu G, Zhang Y, Ma P. LncRNA DANCR functions as a competing endogenous RNA to regulate RAB1A expression by sponging miR-634 in glioma. Biosci Rep. 2018;38 doi: 10.1042/BSR20171664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang S, Jiang M. The long non-coding RNA-DANCR exerts oncogenic functions in non-small cell lung cancer via miR-758-3p. Biomed Pharmacother. 2018;103:94–100. doi: 10.1016/j.biopha.2018.03.053. [DOI] [PubMed] [Google Scholar]
- 13.Wang Y, Zeng X, Wang N, Zhao W, Zhang X, Teng S, Zhang Y, Lu Z. Long noncoding RNA DANCR, working as a competitive endogenous RNA, promotes ROCK1-mediated proliferation and metastasis via decoying of miR-335-5p and miR-1972 in osteosarcoma. Mol Cancer. 2018;17:89. doi: 10.1186/s12943-018-0837-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang N, Wang X, Xie X, Liao Y, Liu N, Liu J, Miao N, Shen J, Peng T. lncRNA DANCR promotes tumor progression and cancer stemness features in osteosarcoma by upregulating AXL via miR-33a-5p inhibition. Cancer Lett. 2017;405:46–55. doi: 10.1016/j.canlet.2017.06.009. [DOI] [PubMed] [Google Scholar]
- 15.Qu M, Zhu Y, Jin M. MicroRNA-138 inhibits SOX12 expression and the proliferation, invasion and migration of ovarian cancer cells. Exp Ther Med. 2018;16:1629–1638. doi: 10.3892/etm.2018.6375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Si F, Sun J, Wang C. MicroRNA-138 suppresses cell proliferation in laryngeal squamous cell carcinoma via inhibiting EZH2 and PI3K/AKT signaling. Exp Ther Med. 2017;14:1967–1974. doi: 10.3892/etm.2017.4733. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 17.Gao Y, Fan X, Li W, Ping W, Deng Y, Fu X. miR-138-5p reverses gefitinib resistance in non-small cell lung cancer cells via negatively regulating G protein-coupled receptor 124. Biochem Biophys Res Commun. 2014;446:179–186. doi: 10.1016/j.bbrc.2014.02.073. [DOI] [PubMed] [Google Scholar]
- 18.Li J, Wang Q, Wen R, Liang J, Zhong X, Yang W, Su D, Tang J. MiR-138 inhibits cell proliferation and reverses epithelial-mesenchymal transition in non-small cell lung cancer cells by targeting GIT1 and SEMA4C. J Cell Mol Med. 2015;19:2793–2805. doi: 10.1111/jcmm.12666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jin Z, Guan L, Song Y, Xiang GM, Chen SX, Gao B. MicroRNA-138 regulates chemoresistance in human non-small cell lung cancer via epithelial mesenchymal transition. Eur Rev Med Pharmacol Sci. 2016;20:1080–1086. [PubMed] [Google Scholar]
- 20.Harley V, Lefebvre V. Twenty Sox, twenty years. Int J Biochem Cell Biol. 2010;42:376–377. doi: 10.1016/j.biocel.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 21.Parvani JG, Schiemann WP. Sox4, EMT programs, and the metastatic progression of breast cancers: mastering the masters of EMT. Breast Cancer Res. 2013;15:R72. doi: 10.1186/bcr3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vervoort SJ, van Boxtel R, Coffer PJ. The role of SRY-related HMG box transcription factor 4 (SOX4) in tumorigenesis and metastasis: friend or foe? Oncogene. 2013;32:3397–3409. doi: 10.1038/onc.2012.506. [DOI] [PubMed] [Google Scholar]
- 23.Li D, He C, Wang J, Wang Y, Bu J, Kong X, Sun D. MicroRNA-138 inhibits cell growth, invasion, and EMT of non-small cell lung cancer via SOX4/p53 feedback loop. Oncol Res. 2018;26:385–400. doi: 10.3727/096504017X14973124850905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yeh YM, Chuang CM, Chao KC, Wang LH. MicroRNA-138 suppresses ovarian cancer cell invasion and metastasis by targeting SOX4 and HIF-1alpha. Int J Cancer. 2013;133:867–878. doi: 10.1002/ijc.28086. [DOI] [PubMed] [Google Scholar]
- 25.Pang L, Li B, Zheng B, Niu L, Ge L. miR-138 inhibits gastric cancer growth by suppressing SOX4. Oncol Rep. 2017;38:1295–1302. doi: 10.3892/or.2017.5745. [DOI] [PubMed] [Google Scholar]
- 26.Liu F, Wu L, Wang A, Xu Y, Luo X, Liu X, Hua Y, Zhang D, Wu S, Lin T, He D, Wei G, Chen S. MicroRNA-138 attenuates epithelial-to-mesenchymal transition by targeting SOX4 in clear cell renal cell carcinoma. Am J Transl Res. 2017;9:3611–3622. [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao J, Du P, Cui P, Qin Y, Hu C, Wu J, Zhou Z, Zhang W, Qin L, Huang G. LncRNA PVT1 promotes angiogenesis via activating the STAT3/VEGFA axis in gastric cancer. Oncogene. 2018;37:4094–4109. doi: 10.1038/s41388-018-0250-z. [DOI] [PubMed] [Google Scholar]
- 28.Chen C, Zhang Z, Li J, Sun Y. SNHG8 is identified as a key regulator in non-small-cell lung cancer progression sponging to miR-542-3p by targeting CCND1/CDK6. Onco Targets Ther. 2018;11:6081–6090. doi: 10.2147/OTT.S170482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang H, Wang J, Liang CF, Zhou T. Expression of long non-coding RNA PRAL as a potential biomarker for diagnosis in non-small-cell lung cancer patients is associated with the inhibition of cell proliferation and metastasis. Clin Lab. 2018;64:1341–1348. doi: 10.7754/Clin.Lab.2018.171237. [DOI] [PubMed] [Google Scholar]
- 30.Yuan SX, Wang J, Yang F, Tao QF, Zhang J, Wang LL, Yang Y, Liu H, Wang ZG, Xu QG, Fan J, Liu L, Sun SH, Zhou WP. Long noncoding RNA DANCR increases stemness features of hepatocellular carcinoma by derepression of CTNNB1. Hepatology. 2016;63:499–511. doi: 10.1002/hep.27893. [DOI] [PubMed] [Google Scholar]
- 31.Liu Y, Zhang M, Liang L, Li J, Chen YX. Over-expression of lncRNA DANCR is associated with advanced tumor progression and poor prognosis in patients with colorectal cancer. Int J Clin Exp Pathol. 2015;8:11480–11484. [PMC free article] [PubMed] [Google Scholar]
- 32.Jia J, Li F, Tang XS, Xu S, Gao Y, Shi Q, Guo W, Wang X, He D, Guo P. Long noncoding RNA DANCR promotes invasion of prostate cancer through epigenetically silencing expression of TIMP2/3. Oncotarget. 2016;7:37868–37881. doi: 10.18632/oncotarget.9350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li T, Xie J, Shen C, Cheng D, Shi Y, Wu Z, Deng X, Chen H, Shen B, Peng C, Li H, Zhan Q, Zhu Z. Amplification of Long Noncoding RNA ZFAS1 Promotes Metastasis in Hepatocellular Carcinoma. Cancer Res. 2015;75:3181–3191. doi: 10.1158/0008-5472.CAN-14-3721. [DOI] [PubMed] [Google Scholar]
- 34.Li S, Mei Z, Hu HB, Zhang X. The lncRNA MALAT1 contributes to non-small cell lung cancer development via modulating miR-124/STAT3 axis. J Cell Physiol. 2018;233:6679–6688. doi: 10.1002/jcp.26325. [DOI] [PubMed] [Google Scholar]
- 35.Zhang EB, Kong R, Yin DD, You LH, Sun M, Han L, Xu TP, Xia R, Yang JS, De W, Chen J. Long noncoding RNA ANRIL indicates a poor prognosis of gastric cancer and promotes tumor growth by epigenetically silencing of miR-99a/miR-449a. Oncotarget. 2014;5:2276–2292. doi: 10.18632/oncotarget.1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang M, Hou J, Wang Y, Xie M, Wei C, Nie F, Wang Z, Sun M. Long noncoding RNA LINC00673 is activated by SP1 and exerts oncogenic properties by interacting with LSD1 and EZH2 in gastric cancer. Mol Ther. 2017;25:1014–1026. doi: 10.1016/j.ymthe.2017.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 37.Xu L, Wei B, Hui H, Sun Y, Liu Y, Yu X, Dai J. Positive feedback loop of lncRNA LINC01296/miR-598/Twist1 promotes non-small cell lung cancer tumorigenesis. J Cell Physiol. 2019;234:4563–4571. doi: 10.1002/jcp.27235. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.