Abstract
Secreted Frizzled-Related Protein 4 (SFRP4), a member of secreted frizzled-related protein family, has been found as a vital modulator in cell proliferation, cell self-renew and apoptosis through Wnt signaling transduction pathway. In the present study, we re-analyzed the expression pattern of SFRPs in Gene Expression Omnibus (GEO) datasets and evaluated the expression of SFRP4 at protein level in both KrasG12D/+; Trp53R172H/+; Pdx1-Cre; (KPC) mice and human pancreatic ductal adenocarcinoma (PDAC) tissue. We found that the expression of SFRP4 increased gradually in PanINs and PDAC lesions in KPC mice and high expression of SFRP4 was much more common in tumor lesions compared to the adjacent non-tumor tissues. Then we performed Kaplan-Meier survival and Cox regression analysis and found that high expression of SFRP4 in the serum and tumor lesions predicted poor prognosis for pancreatic cancer patients. Furthermore, we demonstrated that SFRP4 positively correlated with FOXP3+ Treg cells infiltration while the down-regulation of SFRP4 in tumor cells impaired the production of cytokines and the recruitments of T cells. This study suggested that SFRP4 can be a novel prognostic biomarker and potential therapeutic target for pancreatic cancer.
Keywords: SFRP4, pancreatic ductal adenocarcinoma, prognosis, regulatory T-cell, biomarkers
Introduction
Pancreatic ductal adenocarcinoma (PDAC) is considered to be one of the most lethal disease and predicted to become the second common cause of cancer-related mortality by 2030 [1]. The 5-year survival rate remains at 6% in USA and patients diagnosed with PDAC own mortality that closely equals incidence [2,3]. Surgery seems to be the only cure for PDAC, but only approximately 10-20% of patients are resectable at the time of diagnosis [4]. Since the late diagnosis and treatment resistance, novel biological markers which can accurately predict the patient prognosis are urgently needed.
PDAC is characterized by gene alteration in core signaling pathways, among which the Wnt pathway acts as a key determinant of tumor fate within the pancreas [5,6]. Generally, Wnt family performs specific function via the transcriptional co-activator β-catenin transmitting from cytoplasm into nuclear and regulates embryonic development and adult homeostasis which is called canonical Wnt signaling pathway or β-catenin dependent pathway [7,8]. A family of five secreted frizzled-related glycoproteins (SFRP1-5) have been identified as antagonist of Wnt signaling by directly binding both Wnt proteins and frizzled receptors [9]. Given the oncogenic potential of abnormal activated Wnt signaling, SFRPs have been postulated to act as tumour suppressor genes.
Recently, growing evidence suggests a controversial role of SFRPs. For instance, low expression of SFRP2 was found in gastric cancer, colorectal cancer and oral squamous cell carcinoma suggesting a tumor suppressor role in those tumors [10-12], while high expression of SFRP2 was reported in renal cancer, angiosarcoma, breast cancer and osteosarcoma associated with oncogenic functions [13-16]. Overexpression of SFRP4 associated with decreased tumor growth as well as invasion [17] and predict good prognosis in prostate cancer [18]. Low expression of SFRP4 in mesothelioma promote cell growth and inhibit apoptosis [19], And SFRP4 demonstrate a chemo-sensitization effect in cancer stem cells [20]. However, Sandsmark, Elise and colleges reported that SFRP4 expression is increased in aggressive prostate cancer and predict tumor recurrence [21].
In this study, we re-analyzed the expression pattern of SFRPs in published data and recognized SFRP2 and SFRP4 as potential oncogenes and focus on the role of SFRP4 in PDAC. Then, we confirmed the over-expression of SFRP4 at both mRNA and protein level and extended the association between SFRP4 expression and clinical parameters, including overall survival. From The Cancer Genome Atlas (TCGA) Database, we found that high expression of SFRP4 was strongly correlated with immune response especially the chemotaxis and T cell differentiation. To confirm this result, we analyzed the relationship between SFRP4 expression and infiltration of FOXP3+ Regulatory T-cells (Treg cells) and tested the influence of SFRP4 on the production of cytokines and recruitment of T cells. Based on our data, we proposed that the over-expression of SFRP4 correlated with Treg cell infiltration and poor prognosis in PDAC patients.
Materials and methods
Ethics statement
This study was approved by the Ethics Committee of Ren Ji Hospital, School of Medicine, Shanghai Jiao Tong University. And all patients involved in this study provided written informed consent.
Clinical tissue sample
Human PDAC tissue microarrays contained 205 paired cases of tumor and matched adjacent tissue and all the specimens were obtained from the patients diagnosed with PDAC between January 2002 and June 2014 consecutively from Ren Ji Hospital, School of Medicine, Shanghai Jiao Tong University, China. There were 117 males and 88 females, aging from 38 to 90 years with a median age of 65 years. Clinicopathologic characteristics of patients were available in Table 1. The histology and clinical classification were accordance to the seventh edition of the American Joint Committee on Cancer (AJCC) staging system. None of follow-up cases had received radiotherapy, chemotherapy, hormone therapy or other related anti-tumor therapy before surgery. The serum samples were from another cohort of 31 patients diagnosed with PDAC and 5 healthy donors. Serum samples, freshly frozen PDAC tissues and matched adjacent tissues were also obtained from Ren Ji Hospital, School of Medicine, Shanghai Jiao Tong University, China. After surgery, all of the patients had received a regular follow-up with physical, blood and CT-radiography examinations every 2 months during the first 6 months and then every 3 months. Vital status was obtained prospectively from the medical records and regular contact with patients. The follow-up time was calculated from the date of surgery to death, or June 19, 2016. The median follow-up was 30.3 months, with a range from 0 to 169.8 months. All patients provided written informed consent and all the experiments were approved by the Hospital Research Ethics Committees of Ren Ji Hospital, School of Medicine, Shanghai Jiao Tong University. All methods were carried out in accordance with appropriate guidelines while all the experimental protocols were approved by School of Medicine, Shanghai Jiao Tong University.
Table 1.
Correlations of SFRP4 with clinical characteristics in PDAC patients
| Characteristics | SFRP4 Expression | FOXP3 Infiltration | |||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| Total (n=205) | High (n=116) | Low (n=89) | P value (χ2 test) | High (n=105) | Low (n=100) | P value (χ2 test) | |
| Age (years) | 0.802 | 0.848 | |||||
| <65 | 97 | 54 | 43 | 49 | 48 | ||
| ≥65 | 108 | 62 | 46 | 56 | 52 | ||
| Gender | 0.426 | 0.586 | |||||
| Male | 117 | 69 | 48 | 58 | 59 | ||
| Female | 88 | 47 | 41 | 47 | 41 | ||
| Tumor location | 0.979 | 0.925 | |||||
| Head | 138 | 78 | 60 | 71 | 67 | ||
| Body/tail | 67 | 38 | 29 | 34 | 33 | ||
| Size | 0.594 | 0.629 | |||||
| ≤3 cm | 27 | 14 | 13 | 15 | 12 | ||
| >3 cm | 178 | 102 | 76 | 90 | 88 | ||
| Tumor differentiation | 0.267 | 0.007 | |||||
| Well | 11 | 8 | 3 | 10 | 1 | ||
| Moderate/poor | 194 | 108 | 86 | 95 | 99 | ||
| T classification | 0.017 | 0.026 | |||||
| T1/2 | 30 | 11 | 19 | 21 | 9 | ||
| T3/4 | 175 | 105 | 70 | 84 | 91 | ||
| N classification | 0.336 | 0.361 | |||||
| Absent | 133 | 72 | 61 | 65 | 68 | ||
| Present | 72 | 44 | 28 | 40 | 32 | ||
| AJCC stage | 0.546 | 0.837 | |||||
| Stage I/II | 169 | 94 | 75 | 86 | 83 | ||
| Stage III/IV | 36 | 22 | 14 | 19 | 17 | ||
| Liver metastasis | 0.246 | 0.311 | |||||
| Absent | 191 | 106 | 85 | 96 | 95 | ||
| Present | 14 | 10 | 4 | 9 | 5 | ||
| Vascular invasion | 0.473 | 0.114 | |||||
| Absent | 178 | 99 | 79 | 95 | 83 | ||
| Present | 27 | 17 | 10 | 10 | 17 | ||
| Neural invasion | 0.577 | 0.843 | |||||
| Absent | 106 | 58 | 48 | 55 | 51 | ||
| Present | 99 | 58 | 41 | 50 | 49 | ||
AJCC staging is according to the 7th edition of the American Joint Committee on Cancer (AJCC) staging system. The bold number represents the p-values with significant differences. P value was calculated by χ2 test or Fisher’s exact test.
Transgenic animal model
KPC mice were purchased from The Jackson Laboratory (Bar Harbor, ME). All animal experiments were undertaken in accordance with the National Institutes of Health Guide for the care and Use of Laboratory Animals. All manipulations were performed under approved protocol number 20141204 assigned by the Research Ethics Committee of East China Normal University. The tumor tissue samples were from 6 KPC mice in this study.
Immunohistochemical staining
KPC mice were euthanized at different stage according to previous study and the tumor was dissected and fixed in paraffin [22,23]. The stages of PanINs and PDAC in KPC mice were evaluated in H&E staining slides by experienced pathologists. The tissue microarray sections were rehydrated and treated with 3% hydrogen peroxide, followed by antigen retrieval. After being blocked with 10% normal goat serum for 30 min, the sections were incubated with primary antibodies at 4°C overnight, followed by incubation with a peroxidase-labeled secondary antibody for 30 min at room temperature. Finally, diaminobenzidine tetrahydrochloride (DAB; Maixin Biotech, China) was used for the color-reaction followed by nucleus counterstaining with hematoxylin. The following antibodies were used: rabbit anti-SFRP4 polyclonal antibody (15328-1-AP, ProteinTech), and rabbit anti-FOXP3 polyclonal antibody (22228-1-AP, ProteinTech). Scoring of SFRP4 expression was conducted according to the percentage of positive cell: 0-5% scored 0; 6%-35% scored 1; 36%-70% scored 2; more than 70% scored 3 and staining intensity: no staining scored 0; weakly staining scored 1; moderately staining scored 2 and strongly staining scored 3, respectively. Recent study reported that SFRP4 is present in both α and β cells and is released from islets during the course of type 2 diabetes [24]. Based on the fact that PDAC is associated with dysfunction of islets, some of the islets in the adjacent non-tumor tissue are supposed to express SFRP4 and the staining positive islets are used as inner positive control. The staining score of 3 was evaluated according to the strongest staining of islets tissue and PDAC tissue. The final score was designated as low or high expression group using the percentage of positive cell score multiplied by the staining intensity score: “-” for a score of 0-1, “+” for a score of 2-3, “++” for a score of 4-6 and “+++” for a score of >6; low expression was defined as a total score <4 while high expression with a total score ≥4. The antibody of SFRP4 and FOXP3 were from ProteinTech (Chicago, Illinois, USA). The density of FOXP3+ Tregs was measured in four high power field from each tumor in TMA by experienced pathologist and the average density was calculated. Low infiltration of Tregs was defined as less than 5 FOXP3+ lymphocytes in a high power field. The scoring was done in a blinded manner by two experienced pathologists.
Real-time quantitative PCR
Total RNA from tumor and non-tumor adjacent tissue was extracted using Trizol reagent (Takara, Japan), and reversely transcribed using a PrimeScript RT-PCR Kit (Takara, Japan) according to the manufacturer’s instructions. Quantitative real-time PCR was performed using a 7500 Real-time PCR system (Appiled Biosystem, Inc. USA). Primer sequence are as Table 3.
Table 3.
The primer sequence of target genes
| Gene name | Primer sequence (5’→3’) | |
|---|---|---|
| SFRP4 | Forward | 5’-CCTGGAACATCACGCGGAT-3’ |
| Reverse | 5’-CGGCTTGATAGGGTCGTGC-3’ | |
| CCL4 | Forward | 5’-CTGTGCTGATCCCAGTGAATC-3’ |
| Reverse | 5’-TCAGTTCAGTTCCAGGTCATACA-3’ | |
| CCL5 | Forward | 5’-CCAGCAGTCGTCTTTGTCAC-3’ |
| Reverse | 5’-CTCTGGGTTGGCACACACTT-3’ | |
| CXCL9 | Forward | 5’-CCAGTAGTGAGAAAGGGTCGC-3’ |
| Reverse | 5’-AGGGCTTGGGGCAAATTGTT-3’ | |
| CXCL10 | Forward | 5’-GTGGCATTCAAGGAGTACCTC-3’ |
| Reverse | 5’-TGATGGCCTTCGATTCTGGATT-3’ | |
| GAPDH | Forward | 5’-GCATTGCCCTCAACGACCAC-3’ |
| Reverse | 5’-CCACCACCCTGTTGCTGTAG-3’ | |
Western blot
Total and nuclear protein were extracted using protein extraction buffer (Beyotime, Shanghai, China) and nucleoprotein extraction kit (Sangon Biotech, C500009). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis was used to separate different proteins and then proteins were transferred onto a nitrocellulose (NC) membrane. The NC membrane was blocked with 5% skimmed milk, and then incubated with specific antibodies as follow: anti-SFRP4 (1:1000), anti-β-catenin (1:1000), anti-β-actin (1:2000), anti-lamin A/C (1:1000), followed by species-specific secondary antibodies (1:10000). After incubating with the secondary antibodies for 60 mins, the bands were detected using the Odyssey imaging system (LI-COR, Lincoln, NE, USA).
TCGA and GEO sample acquisition and analysis
The reference series used in this study were GSE15471 and GSE28735 downloaded from the Gene Expression Omnibus (GEO) [25,26]. The project of TCGA-PAAD was downloaded from GDC Data Portal (https://gdc-portal.nci.nih.gov/).
Gene set enrichment analysis (GSEA) was performed to gain an insight into the biological process influenced by SFRP4. The GO gene sets biological process database (c5.bp.v4.0) were used for enrichment analysis of the expression database from TCGA-PAAD.
The correlation between SFRP4 expression and immune cell signature genes was analyzed from the TCGA-PAAD database. The immune cell signature genes were summarized by Bindea, G. etc [27], and the results were demonstrated by Cytoscape [28].
ELISA assay
ELISA assays were performed using SFRP4-specific ELISA kits (SEF878Hu, Cloud-clone, USA) according to the manufacturer’s instructions. The serum samples were diluted 1:20 to meet the detective range of the ELISA kit. We determine the high or low serum level of SFRP4 by the median number in the PDAC patient cohort. The data was analyzed by the CurveExpert 1.4 software.
Cell culture and transfection
Human pancreatic cancer cell lines Patu-8988, Mia Paca-2, SW1990, PANC-1, AsPC-1, CFPAC-1 and immortalized normal human pancreatic ductal epithelial cell line HPDE and HPNE were purchased from ATCC. Cells were maintained in complete medium recommended by the provider containing 10% fetal bovine serum (FBS), 100 U/ml penicillin and 100 μg/ml streptomycin and were incubated at 37°C in a 5% CO2 atmosphere. The small interference RNA (siRNA) targeting human SFRP4 were transfected into the cells using the RNAiMAX transfection reagent (Thermo Fisher, USA), whereas nonspecific siRNA acted as negative controls. The treated cells were cultured for 3 days and replaced the medium with FBS-free culture medium, then culture the cells for another 24 hours. The conditional culture medium was collected and the cells were lysed for RNA extraction.
Chemotaxis assay
Human peripheral blood mononuclear cell (PBMC) were separated by the gradient centrifugation method using lymphocyte separation media (Biosera, France). The 5.0 μm pore size Transwell (Corning, USA) was used to performed the chemotaxis assay. Below the insert, the 1:1 diluted conditional culture medium was used to attract the cells while the PBMC were planted in the Transwell inserts. After 6 hours of culture, the conditional medium was collected for cell counting and staining. Cell counting was performed using precision count beads by flow cytometry. The same amount of beads (10000 particles) were added to each group of cell suspension and the total migrated cell number was calculated by the proportion of beads and cells. And the CD4 positive T cells were stained by CD4 flow cytometry antibody (560650, BD biosciences, USA) for flow cytometry analysis. The flow cytometry was performed by LSR-Fortessa (BD biosciences, USA).
Statistical analysis
Statistical analysis and graphical representations were performed using SPSS 16.0 (SPSS Inc.; Chicago, IL, USA) and Graph Pad Prism 5 (San Diego, CA) software. The Chi-square test was used to analyze the correlations between SFRP4 expression and clinical characteristics in PDAC patients. Survival curves were evaluated by Kaplan-Meier method and the difference between survival curves were tested by log-rank test. Cox regression including univariable and multivariable analyses was used to judge the variable parameters. Only significant different variables in univariable analysis including SFRP4 expression, T classification, N classification, AJCC stage and liver metastasis were entered in the multivariable analysis. A two-sided P-value <0.05 was considered statistically significant.
Results
Up-regulation of SFRP4 at mRNA and protein level in PDAC
To evaluate the expression pattern of SFRPs in human PDAC, we analyzed two independent microarray datasets from Gene Expression Omnibus (GEO) [25,26,29]. The result suggested that the expression of SFRP2, SFRP4 and SFRP5 were consistently altered in two independent PDAC datasets, among which the expression of SFRP2 and SFRP4 were elevated in the PDAC tissues compared to the adjacent non-tumor tissue (Figure 1A). Based on the limited study of SFRP4, we confirmed the expression pattern of SFRP4 in patient cohorts from Renji Hospital. Real-time PCR was applied to determined SFRP4 mRNA expression level in 30 pairs of PDAC tissue and matched adjacent non-tumor tissue. Our results showed a remarkable increase of SFRP4 mRNA expression level in PDAC patients compared to matched adjacent non-tumor tissue (P=0.0072, Figure 1B). Moreover, microarray databases derived from GEO datasets indicated similar SFRP4 mRNA expression pattern in PDAC tissue compared to the adjacent non-tumor tissue (Figure 1C, 1D).
Figure 1.
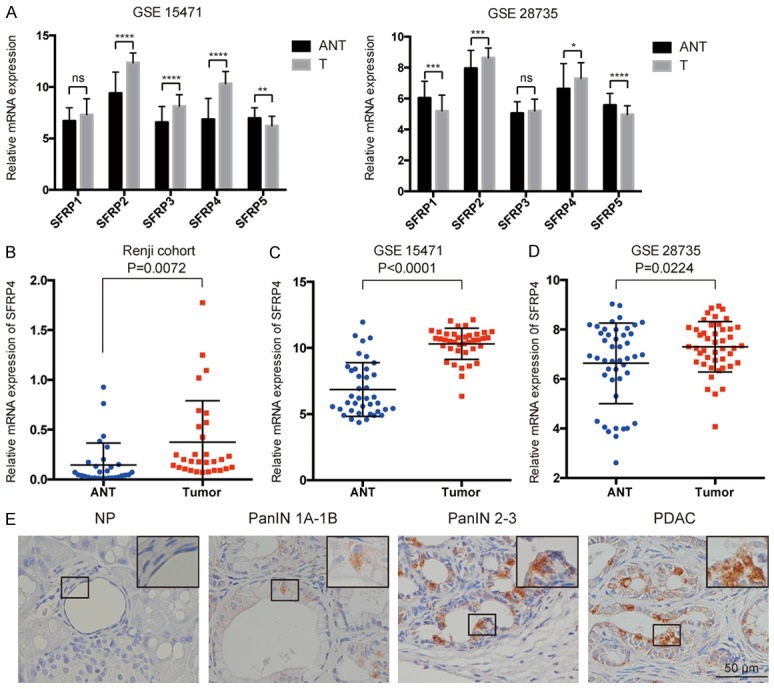
SFRP4 expression in PDAC tissue at mRNA level. A. Re-analysis the expression of SFRPs in GSE 15471 and GSE 28735. Expression of SFRPs in tumor (T) and adjacent non-tumor tissue (ANT) are shown, error bars in the column represent SE. B-D. Expression of SFRP4 was significantly increased in tumor tissues (T) compared to the adjacent non-tumor tissues (ANT) in GSE 15471, GSE 28735 and Renji cohorts. E. Representative IHC staining of SFRP4 in normal pancreas, different stage of PanINs and PDAC lesions.
Then we used KPC mice model to evaluate the expression of SFRP4 in the progression from PanINs to PDAC at protein level. The result suggested that the normal pancreas do not express SFRP4 while the expression of SFRP4 gradually increased along with the disease progression (Figure 1E). We also detected the expression of SFRP4 in 205 cases of paired PDAC specimens by immunohistochemistry (Figure 2). We found that high expression of SFRP4 (++ or +++) was detected in 56.5% (116/205) of PDAC tissue, while only 28.8% (59/205) in the adjacent non-tumor tissue. As shown in Figure 2A, the staining cells in tumor tissue are duct-like tumor cells. While most of the SFRP4 staining positive cells in the adjacent non-tumor tissue were islet cells (Figure 2F).
Figure 2.
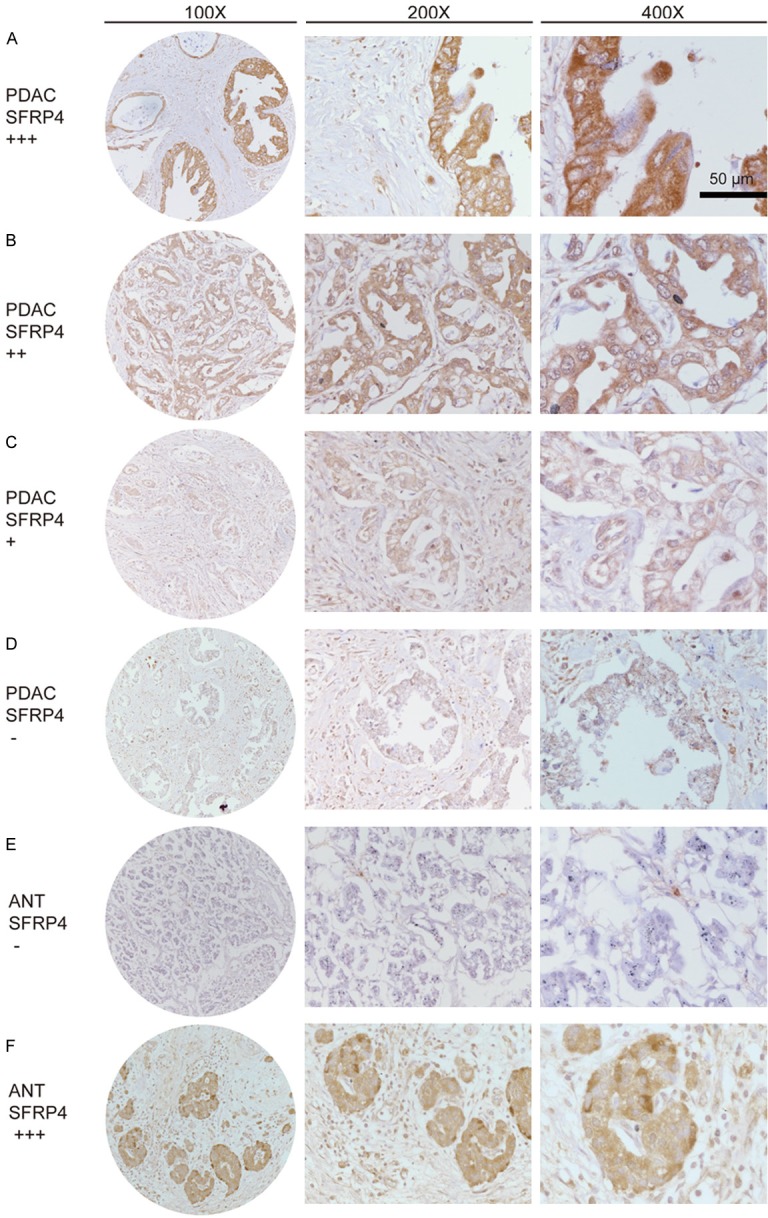
SFRP4 expression in PDAC tissue microarray by IHC. A. PDAC, scored as (+++); B. PDAC, scored as (++); C. PDAC, scored as (+); D. PDAC, scored as (-); E. Adjacent non-tumor tissue, scored as (-); F. Adjacent non-tumor tissue, the islet cells scored as (+++).
Correlations of SFRP4 expression with clinical characteristics
The Chi-square test was used to analyze the relationship between SFRP4 expression and clinical parameters for the purpose of evaluating the clinical significance of SFRP4 expression in PDAC. The result demonstrated that SFRP4 expression in PDAC was correlated with T classification, and it showed that patients in T3/4 group have a higher expression of SFRP4 than patients in T1/2 group (P=0.017, Table 1).
Prognostic significance of SFRP4 in PDAC patients
Based on our previous follow-up work, we further investigated the relationship between SFRP4 expression and clinical prognosis in PDAC patients by Kaplan-Meier survival analysis and log-rank test. As shown in Figure 3A, high level of SFRP4 expression was correlated with poor overall survival (P=0.007). Univariable and multivariable analysis were used to evaluate whether SFRP4 has the potential to be an independent risk factor for poor prognosis in our cohort of PDAC. Univariable analysis showed that SFRP4 expression, T classification, N classification, AJCC stage and liver metastasis were closely associated with overall survival. While multivariable Cox regression analysis confirmed that SFRP4 expression and N classification are independent risk factors of prognosis in PDAC (Table 2).
Figure 3.
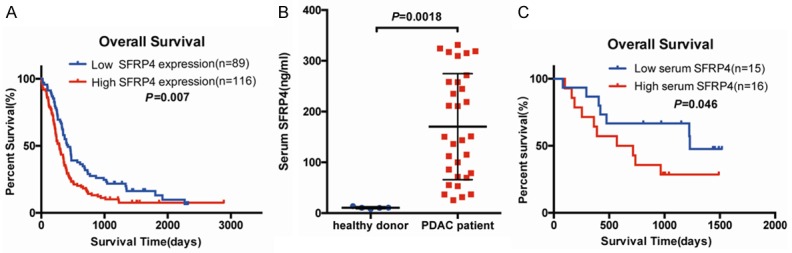
SFRP4 expression is associated with overall survival in PDAC patients. A. Kaplan-Meier survival curves demonstrated that high expression of SFRP4 was significantly correlated with poor survival time of PDAC (P=0.007). p-values were statistically calculated by log-rank test. B. The serum level of SFRP4 in healthy donor and PDAC patients. C. High concentration of SFRP4 at serum level was correlated with poor prognosis for PDAC patients.
Table 2.
Univariate and multivariate analyses of prognosis factors for survival in PDAC patients
| Prognostic parameter | Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|
|
| ||||||
| HR | 95% CI | P value | HR | 95% CI | P value | |
| SFRP4 (low vs. high) | 1.559 | 1.124-2.163 | 0.008 | 1.422 | 1.018-1.986 | 0.039 |
| Age (<65 vs. ≥65) | 1.348 | 0.977-1.859 | 0.069 | |||
| Gender (male vs. female) | 0.881 | 0.637-1.219 | 0.445 | |||
| Tumor location (head vs. body/tail) | 1.046 | 0.746-1.466 | 0.795 | |||
| Size (≤2 cm vs. >2 cm) | 1.551 | 0.957-2.514 | 0.075 | |||
| Tumor differentiation (well vs. moderate/poor) | 1.362 | 0.636-2.917 | 0.426 | |||
| T classification (T3/T4 vs. T1/T2) | 2.098 | 1.244-3.538 | 0.005 | 1.668 | 0.969-2.869 | 0.065 |
| AJCC stage (III/IV vs. I/II) | 1.645 | 1.075-2.517 | 0.022 | 1.250 | 0.712-2.194 | 0.437 |
| N classification (present vs. absent) | 1.761 | 1.264-2.454 | 0.001 | 1.596 | 1.136-2.244 | 0.007 |
| Liver metastasis (present vs. absent) | 2.393 | 1.316-4.353 | 0.004 | 1.566 | 0.710-3.454 | 0.266 |
| Vascular invasion (present vs. absent) | 1.381 | 0.861-2.214 | 0.181 | |||
| Neural invasion (present vs. absent) | 0.924 | 0.672-1.271 | 0.629 | |||
HR: Hazard ratio; CI: confidence interval. The bold number represents the p-values with significant difference.
Serum concentration of SFRP4 is increased in PDAC patients and suggests an unfavorable prognosis
Since SFRP4 is detectable in peripheral blood [30,31], we want to assess whether serum level of secreted SFRP4 is also increased in PDAC patients and the relationship between the concentration of serum SFRP4 and PDAC patients’ prognosis. To address this issue we performed the ELISA assay for serum samples from 5 healthy donors and 31 PDAC patients. As shown in Figure 3B, the serum level of secreted SFRP4 in PDAC patients (170.33 ± 104.43 ng/ml) is higher than healthy donor (10.67 ± 2.02 ng/ml) with statistical difference (P=0.0018). We also used Kaplan-Meier survival analysis to assess the correlation between serum level of SFRP4 and patients’ prognosis (Figure 3C), the result demonstrated that high serum level of SFRP4 is correlated with poor prognosis of PDAC patients consistent with our finding in tissue microarray (P=0.046).
SFRP4 correlates with Treg cells infiltration in PDAC
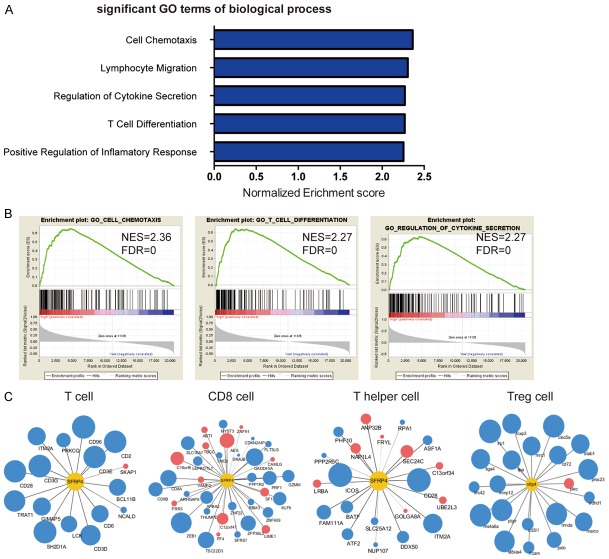
To determine the biological process influenced by SFRP4, we performed GSEA in TCGA-PAAD project. As shown in Figure 4A and 4B, the significant GO pathways enriched in high SFRP4 expression group were related to immune response especially the lymphocytes migration and differentiation. This finding demonstrated that secreted protein SFRP4 may play an important role in the immune microenvironment. Since the GSEA highlighted the relationship between T cell differentiation and SFRP4, we analyzed the correlation between the signature genes of different T cell subsets and SFRP4 expression [27]. The blue or red color represent the positive or negative correlation respectively while the size of the dot represents the correlation coefficient. We found that SFRP4 was positive correlated with the Treg cells signature genes and relationship between SFRP4 and signature genes of other cells remains unclear (Figure 4C).
Figure 4.
Correlation between SFRP4 expression and Treg cell infiltration in PDAC. A, B. GSEA shows the presentative GO term of biological process that positive correlates with SFRP4. C. The correlation between the expression of signature genes of T cell, CD8 cell, T helper cell, Treg cell and the expression of SFRP4. The size of the dots represents the correlation coefficient and the blue dots represent the positive correlation while the red dots represent the negative correlation. The data was analyzed from TCGA-PAAD database and demonstrated by Cytoscape.
IHC of FOXP3 protein in PDAC tissue microarray was performed to access the density of FOXP3+ Treg cells infiltration in tumor microenvironments (Figure 5A). Then we analyzed the correlation of Treg cells infiltration and clinical characteristics in PDAC patients. The result suggested that Treg cells are more likely to present in well differentiation and low T classification group (Table 1). While the Figure 5B confirmed that the patients with high expression of SFRP4 have a higher ratio of Treg cells infiltration. We also performed IHC for SFRP4 and FOXP3 in tumor sample from KPC mice, the result suggested that the density of FOXP3+ Treg cells increased in the area with high expression of SFRP4 (Figure 5C).
Figure 5.
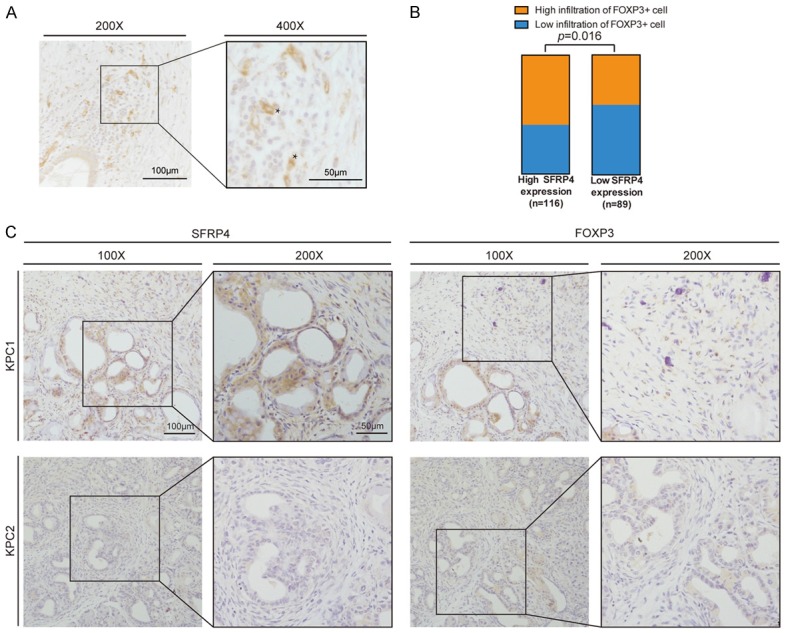
Down-regulation of SFRP4 impairs the recruitment of T cells. A. The presentative image of FOXP3 staining by IHC. The asterisks marked the FOXP3+ Treg cells. B. The patients with high SFRP4 expression demonstrated a greater proportion of high infiltration of FOXP3+ Treg cells. C. The presentative image of SFRP4 and FOXP3 staining by IHC in the tumor tissue from KPC mice.
Down-regulated SFRP4 impairs the production of cytokines and the recruitment of CD4 positive cells
Treg cells generated in the thymus or differentiated from Naïve CD4+ T cells in the periphery. It is demonstrated that tumor cells can secrete many factors to induce Tregs, either by direct action on T cells or producing factors that attract Tregs locally [32]. Our previous bioinformatic analysis suggested that SFRP4 influenced the cytokine secretion (Figure 4A, 4B), so we want to confirm that whether the SFRP4 influenced the production of cytokines and the recruitment of T cells, especially CD4 positive T cells. To this aim, we selected SW1990 as the SFRP4 high expression cell line and used the small interference RNA to down-regulate the expression of SFRP4 in SW1990 (Figure 6A, 6B). Then we tested the expression of classical cytokines responsible for the recruitment of T cells by qRT-PCR. We found the expression of CCL4, CCL5, CXCL9 and CXCL10 were all decreased while the down-regulation of CXCL10 is more significant (Figure 6C). At the same time, the conditional culture medium (CM) from those treated tumor cells was collected to performed the chemotaxis assay for the peripheral blood mononuclear cell. We found that the down-regulation of SFRP4 significantly impaired the recruitment of total migrated cells (Figure 6D). Furthermore, we used flow cytometry to analyze the proportion of and CD4 positive T cells (Figure 6E). Then we calculated the precision number of total migrated cells and migrated CD4 positive cells, the results suggested that SFRP4 down-regulation decreased the total migrated cells and CD4 positive cells significantly (Figure 6F). Based on those results, we supposed that SFRP4 promoted the secretion of T cell specific cytokines and increased the recruitment of CD4 positive T cells which may promote the process of Treg differentiation.
Figure 6.
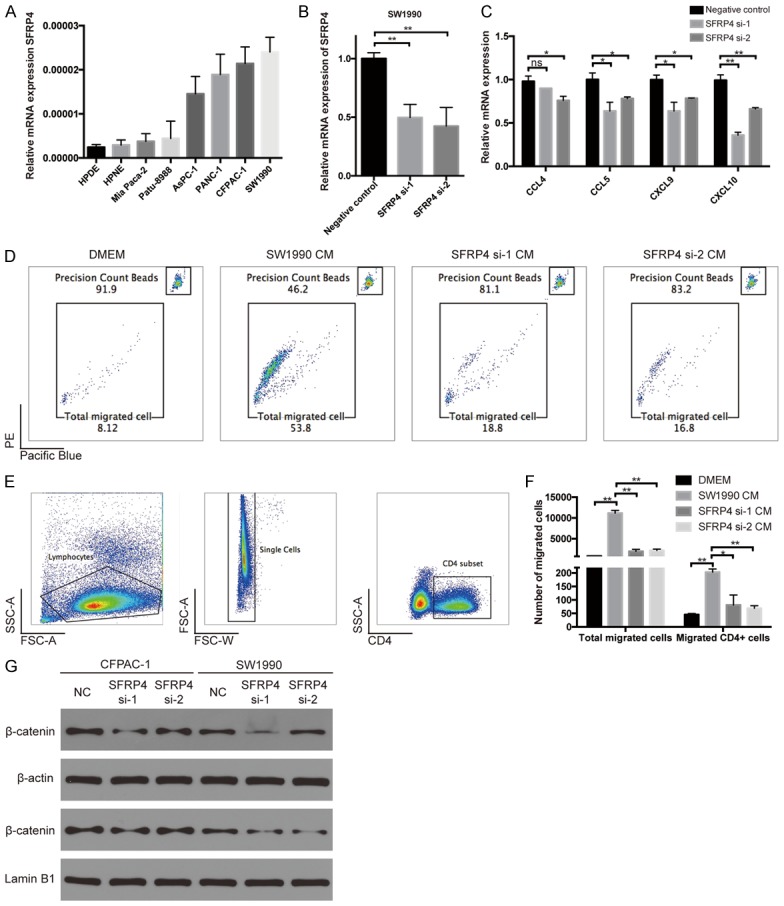
Down-regulation of SFRP4 decreased chemokine secretion. A. The mRNA expression of SFRP4 in cell lines. B. The down-regulation of SFRP4 by small interference RNA. C. The expression of cytokines at mRNA level influenced by SFRP4 siRNA. D. The proportion of counting beads and total migrated cell by flow cytometry. E. The gating strategy for the CD4 positive T cells. F. The precision number of total migrated cells and migrated CD4 positive cells. G. The level of β-catenin in the cell plasma and in the nuclear determined by western blot.
Furthermore, we also performed western blot to determine the relationship between SFRP4 and activation of canonical Wnt signaling. We found that downregulation of SFRP4 in PDAC cell lines decreased the level of β-catenin in the cell plasma and in the nuclear (Figure 6G).
Discussion
Despite the great endeavors on early diagnosis of pancreatic cancer, efforts failed to improve the survival of this fatal malignancy [33-35]. SFRPs tended to play a role of tumor suppressor in a plenty of previous literature research [36,37], however the wispy difference in expression and biological function among their family members remains to be determined [38,39]. In the present work, we re-analyzed the expression pattern of SFRPs in published database, while the result suggested that SFRP2 and SFRP4 may act as oncogene in PDAC. SFRP4, an unique and multifunctional Wnt signaling component, emerged as a potential biological marker in PDAC. Here, SFRP4 expression and its clinical significance were reported in a more comprehensive manner.
First, we assessed SFRP4 expression at mRNA and protein level. Our results from real-time PCR and data from two independent GEO datasets consistently confirmed that SFRP4 expression was elevated in pancreatic cancer compared to adjacent non-tumor tissues at mRNA level. At protein level, similar results were obtained by IHC in KPC mice sample and tissue microarray of Renji cohort. KPC mouse, which could mimic the disease progression, was the most widely used PDAC transgenic animal model. We noticed that the expression of SFRP4 correlated with the pathological stage of the lesions while the data from PDAC TMA also confirmed the relationship between SFRP4 expression and T classification. Together, we found that SFRP4 expression was increased significantly in PDAC tissue, suggesting a potential oncogene role of SFRP4. Then, by using the Kaplan-Meier analysis we found that high expression of SFRP4 in both PDAC tissue and serum indicated a remarkably shorter survival time in PDAC patients. Furthermore, univariable and multivariable cox regression confirmed that high expression of SFRP4 could act as an independent risk factor for poor prognosis in PDAC patients.
Given the different expression pattern of SFRP4 reported among previous study, there may be several possible explanations for these conflicts. First, SFRP4 demonstrates the least structural homology to frizzled receptors compared to other members of SFRP family [40]. Besides the well-known cysteine-rich domain (CRD), the netrin-like domain (NLD) located in C-terminal of SFRP4 may lead the activation of Wnt/β-catenin independent signaling pathway [41]. Second, Suzuki H et al. found over-expression SFRP4 had little influence with the cytoplasmic and nuclear β-catenin [42]. Simultaneously, He B et al. reported that SFRP4 may induce cell fate not only through canonical Wnt pathway but also through β-catenin independent non-canonical pathway [43]. Lastly, the hypermethylation of SFRPs’ promoter region has been identified as an important epigenetic gene silencing mechanism for the low expression pattern of SFRPs in a variety of cancers [44,45]. However, SFRP4 tended to display a lower frequency of promoter hypermethylation compared to other SFRPs [46,47].
It’s widely accepted that tumors can be eliminated by immune system. This process is termed immune surveillance. However, cancer cells escape from the immune surveillance process by multiple mechanisms, including regulatory immune cells and immune check-point pathways [48]. Recent study demonstrated that the blockade of immune check-point pathway has achieved noteworthy benefit in multiple cancers, while the PDAC remains to be resistance to the immunotherapy [49]. Treg cells contribute to the immunosuppressive microenvironment in a variety of tumors including PDAC [50], while the mechanism of Treg cell differentiation and migration remained to be investigated. In this study, we demonstrated that high expression of SFRP4 was strongly correlated with Treg cell infiltration in PDAC and proved that SFRP4 promoted the production of cytokines and recruitment of CD4 positive T cells. Our work provides a potential target for cancer therapy.
Our study demonstrated that SFRP4 expression was increased in PDAC and high expression of SFRP4 correlated with Treg cell infiltration and poor survival in PDAC patients. Since SFRP4 is detectable in peripheral blood [30,31], our work highlights that SFRP4 expression can function as an important prognosis marker for PDAC.
Acknowledgements
This work was supported by grants from National Natural Science Foundation of China Project 81401931, Project 81502489, Project 81602414, Project 81802317, Science and Technology development found incubator fund innovation project of Pudong New Area of Shanghai Project PKR2014-E09 and Cultivating Funds of South Campus, Renji Hospital, School of Medicine, Shanghai Jiaotong University Project 2017PYQA06.
Disclosure of conflict of interest
None.
References
- 1.Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014;74:2913–2921. doi: 10.1158/0008-5472.CAN-14-0155. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 3.Kamisawa T, Wood LD, Itoi T, Takaori K. Pancreatic cancer. Lancet. 2016;388:73–85. doi: 10.1016/S0140-6736(16)00141-0. [DOI] [PubMed] [Google Scholar]
- 4.Muniraj T, Jamidar PA, Aslanian HR. Pancreatic cancer: a comprehensive review and update. Dis Mon. 2013;59:368–402. doi: 10.1016/j.disamonth.2013.08.001. [DOI] [PubMed] [Google Scholar]
- 5.Jones S, Zhang X, Parsons DW, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Kamiyama H, Jimeno A, Hong SM, Fu B, Lin MT, Calhoun ES, Kamiyama M, Walter K, Nikolskaya T, Nikolsky Y, Hartigan J, Smith DR, Hidalgo M, Leach SD, Klein AP, Jaffee EM, Goggins M, Maitra A, Iacobuzio-Donahue C, Eshleman JR, Kern SE, Hruban RH, Karchin R, Papadopoulos N, Parmigiani G, Vogelstein B, Velculescu VE, Kinzler KW. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heiser PW, Cano DA, Landsman L, Kim GE, Kench JG, Klimstra DS, Taketo MM, Biankin AV, Hebrok M. Stabilization of beta-catenin induces pancreas tumor formation. Gastroenterology. 2008;135:1288–1300. doi: 10.1053/j.gastro.2008.06.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clevers H, Nusse R. Wnt/beta-catenin signaling and disease. Cell. 2012;149:1192–1205. doi: 10.1016/j.cell.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 8.Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127:469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 9.MacDonald BT, Tamai K, He X. Wnt/beta-catenin signaling: components, mechanisms, and diseases. Dev Cell. 2009;17:9–26. doi: 10.1016/j.devcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kinoshita T, Nomoto S, Kodera Y, Koike M, Fujiwara M, Nakao A. Decreased expression and aberrant hypermethylation of the SFRP genes in human gastric cancer. Hepatogastroenterology. 2011;58:1051–1056. [PubMed] [Google Scholar]
- 11.Takeda M, Nagasaka T, Dong-Sheng S, Nishie H, Oka T, Yamada E, Mori Y, Shigeyasu K, Morikawa T, Mizobuchi S, Fujiwara T. Expansion of CpG methylation in the SFRP2 promoter region during colorectal tumorigenesis. Acta Med Okayama. 2011;65:169–177. doi: 10.18926/AMO/46628. [DOI] [PubMed] [Google Scholar]
- 12.Paluszczak J, Hemmerling D, Kostrzewska-Poczekaj M, Jarmuż-Szymczak M, Grenman R, Wierzbicka M, Baer-Dubowska W. Frequent hypermethylation of WNT pathway genes in laryngeal squamous cell carcinomas. J Oral Pathol Med. 2014;43:652–657. doi: 10.1111/jop.12178. [DOI] [PubMed] [Google Scholar]
- 13.Yamamura S, Kawakami K, Hirata H, Ueno K, Saini S, Majid S, Dahiya R. Oncogenic functions of secreted Frizzled-related protein 2 in human renal cancer. Mol Cancer Ther. 2010;9:1680–1687. doi: 10.1158/1535-7163.MCT-10-0012. [DOI] [PubMed] [Google Scholar]
- 14.Fontenot E, Rossi E, Mumper R, Snyder S, Siamakpour-Reihani S, Ma P, Hilliard E, Bone B, Ketelsen D, Santos C, Patterson C, Klauber-DeMore N. A novel monoclonal antibody to secreted frizzled-related protein 2 inhibits tumor growth. Mol Cancer Ther. 2013;12:685–695. doi: 10.1158/1535-7163.MCT-12-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Siamakpour-Reihani S, Caster J, Bandhu Nepal D, Courtwright A, Hilliard E, Usary J, Ketelsen D, Darr D, Shen XJ, Patterson C, Klauber-Demore N. The role of calcineurin/NFAT in SFRP2 induced angiogenesis--a rationale for breast cancer treatment with the calcineurin inhibitor tacrolimus. PLoS One. 2011;6:e20412. doi: 10.1371/journal.pone.0020412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Techavichit P, Gao Y, Kurenbekova L, Shuck R, Donehower LA, Yustein JT. Secreted Frizzled-Related Protein 2 (sFRP2) promotes osteosarcoma invasion and metastatic potential. BMC Cancer. 2016;16:869. doi: 10.1186/s12885-016-2909-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Horvath LG, Lelliott JE, Kench JG, Lee CS, Williams ED, Saunders DN, Grygiel JJ, Sutherland RL, Henshall SM. Secreted frizzled-related protein 4 inhibits proliferation and metastatic potential in prostate cancer. Prostate. 2007;67:1081–90. doi: 10.1002/pros.20607. [DOI] [PubMed] [Google Scholar]
- 18.Horvath LG, Henshall SM, Kench JG, Saunders DN, Lee CS, Golovsky D, Brenner PC, O’Neill GF, Kooner R, Stricker PD, Grygiel JJ, Sutherland RL. Membranous expression of secreted frizzled-related protein 4 predicts for good prognosis in localized prostate cancer and inhibits PC3 cellular proliferation in vitro. Clin Cancer Res. 2004;10:615–25. doi: 10.1158/1078-0432.ccr-0707-03. [DOI] [PubMed] [Google Scholar]
- 19.He B, Lee AY, Dadfarmay S, You L, Xu Z, Reguart N, Mazieres J, Mikami I, McCormick F, Jablons DM. Secreted frizzled-related protein 4 is silenced by hypermethylation and induces apoptosis in β-catenin-deficient human mesothelioma cells. Cancer Res. 2005;65:743–8. [PubMed] [Google Scholar]
- 20.Deshmukh A, Kumar S, Arfuso F, Newsholme P, Dharmarajan A. Secreted frizzled-related protein 4 (sFRP4) chemo-sensitizes cancer stem cells derived from human breast, prostate, and ovary tumor cell lines. Sci Rep. 2017;7:2256. doi: 10.1038/s41598-017-02256-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sandsmark E, Andersen MK, Bofin AM, Bertilsson H, Drabløs F, Bathen TF, Rye MB, Tessem MB. SFRP4 gene expression is increased in aggressive prostate cancer. Sci Rep. 2017;7:14276. doi: 10.1038/s41598-017-14622-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hingorani SR, Petricoin EF, Maitra A, Rajapakse V, King C, Jacobetz MA, Ross S, Conrads TP, Veenstra TD, Hitt BA, Kawaguchi Y, Johann D, Liotta LA, Crawford HC, Putt ME, Jacks T, Wright CV, Hruban RH, Lowy AM, Tuveson DA. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell. 2003;4:437–450. doi: 10.1016/s1535-6108(03)00309-x. [DOI] [PubMed] [Google Scholar]
- 23.Hingorani SR, Wang L, Multani AS, Combs C, Deramaudt TB, Hruban RH, Rustgi AK, Chang S, Tuveson DA. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell. 2005;7:469–483. doi: 10.1016/j.ccr.2005.04.023. [DOI] [PubMed] [Google Scholar]
- 24.Mahdi T, Hänzelmann S, Salehi A, Muhammed SJ, Reinbothe TM, Tang Y, Axelsson AS, Zhou Y, Jing X, Almgren P, Krus U, Taneera J, Blom AM, Lyssenko V, Esguerra JL, Hansson O, Eliasson L, Derry J, Zhang E, Wollheim CB, Groop L, Renström E, Rosengren AH. Secreted frizzled-related protein 4 reduces insulin secretion and is overexpressed in type 2 diabetes. Cell Metab. 2012;16:625–633. doi: 10.1016/j.cmet.2012.10.009. [DOI] [PubMed] [Google Scholar]
- 25.Badea L, Herlea V, Dima SO, Dumitrascu T, Popescu I. Combined gene expression analysis of whole-tissue and microdissected pancreatic ductal adenocarcinoma identifies genes specifically overexpressed in tumor epithelia. Hepatogastroenterology. 2008;55:2016–2027. [PubMed] [Google Scholar]
- 26.Zhang G, He P, Tan H, Budhu A, Gaedcke J, Ghadimi BM, Ried T, Yfantis HG, Lee DH, Maitra A, Hanna N, Alexander HR, Hussain SP. Integration of metabolomics and transcriptomics revealed a fatty acid network exerting growth inhibitory effects in human pancreatic cancer. Clin Cancer Res. 2013;19:4983–4993. doi: 10.1158/1078-0432.CCR-13-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bindea G, Mlecnik B, Tosolini M, Kirilovsky A, Waldner M, Obenauf AC, Angell H, Fredriksen T, Lafontaine L, Berger A, Bruneval P, Fridman WH, Becker C, Pagès F, Speicher MR, Trajanoski Z, Galon J. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity. 2013;39:782–795. doi: 10.1016/j.immuni.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 28.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang G, Schetter A, He P, Funamizu N, Gaedcke J, Ghadimi BM, Ried T, Hassan R, Yfantis HG, Lee DH, Lacy C, Maitra A, Hanna N, Alexander HR, Hussain SP. DPEP1 inhibits tumor cell invasiveness, enhances chemosensitivity and predicts clinical outcome in pancreatic ductal adenocarcinoma. PLoS One. 2012;7:e31507. doi: 10.1371/journal.pone.0031507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garufi G, Seyhan AA, Pasarica M. Elevated secreted frizzled-related protein 4 in obesity: a potential role in adipose tissue dysfunction. Obesity (Silver Spring) 2015;23:24–27. doi: 10.1002/oby.20915. [DOI] [PubMed] [Google Scholar]
- 31.Liu F, Qu H, Li Y, Tang Q, Yang Z, Wang H, Deng H. Relationship between serum secreted frizzled-related protein 4 levels and the first-phase of glucose-stimulated insulin secretion in individuals with different glucose tolerance. Endocr J. 2015;62:733–740. doi: 10.1507/endocrj.EJ15-0212. [DOI] [PubMed] [Google Scholar]
- 32.Zou W. Regulatory T cells, tumour immunity and immunotherapy. Nat Rev Immunol. 2006;6:295–307. doi: 10.1038/nri1806. [DOI] [PubMed] [Google Scholar]
- 33.Turtoi A, Musmeci D, Wang Y, Dumont B, Somja J, Bevilacqua G, De Pauw E, Delvenne P, Castronovo V. Identification of novel accessible proteins bearing diagnostic and therapeutic potential in human pancreatic ductal adenocarcinoma. J Proteome Res. 2011;10:4302–4313. doi: 10.1021/pr200527z. [DOI] [PubMed] [Google Scholar]
- 34.Sheng W, Dong M, Zhou J, Li X, Liu Q, Dong Q, Li F. The clinicopathological significance and relationship of Gli1, MDM2 and p53 expression in resectable pancreatic cancer. Histopathology. 2014;64:523–535. doi: 10.1111/his.12273. [DOI] [PubMed] [Google Scholar]
- 35.Tian M, Cui YZ, Song GH, Zong MJ, Zhou XY, Chen Y, Han JX. Proteomic analysis identifies MMP-9, DJ-1 and A1BG as overexpressed proteins in pancreatic juice from pancreatic ductal adenocarcinoma patients. BMC Cancer. 2008;8:241. doi: 10.1186/1471-2407-8-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chung MT, Lai HC, Sytwu HK, Yan MD, Shih YL, Chang CC, Yu MH, Liu HS, Chu DW, Lin YW. SFRP1 and SFRP2 suppress the transformation and invasion abilities of cervical cancer cells through Wnt signal pathway. Gynecol Oncol. 2009;112:646–653. doi: 10.1016/j.ygyno.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 37.Linhart HG, Lin H, Yamada Y, Moran E, Steine EJ, Gokhale S, Lo G, Cantu E, Ehrich M, He T, Meissner A, Jaenisch R. Dnmt3b promotes tumorigenesis in vivo by gene-specific de novo methylation and transcriptional silencing. Genes Dev. 2007;21:3110–3122. doi: 10.1101/gad.1594007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bu XM. Hypermethylation and aberrant expression of secreted fizzled-related protein genes in pancreatic cancer. World J Gastroenterol. 2008;14:3421–4. doi: 10.3748/wjg.14.3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chung MT, Sytwu HK, Yan MD, Shih YL, Chang CC, Yu MH, Chu TY, Lai HC, Lin YW. Promoter methylation of SFRPs gene family in cervical cancer. Gynecol Oncol. 2009;112:301–306. doi: 10.1016/j.ygyno.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 40.Jones SE, Jomary C. Secreted Frizzled-related proteins: searching for relationships and patterns. Bioessays. 2002;24:811–820. doi: 10.1002/bies.10136. [DOI] [PubMed] [Google Scholar]
- 41.Perumal V, Krishnan K, Gratton E, Dharmarajan AM, Fox SA. Number and brightness analysis of sFRP4 domains in live cells demonstrates vesicle association signal of the NLD domain and dynamic intracellular responses to Wnt3a. Int J Biochem Cell Biol. 2015;64:91–96. doi: 10.1016/j.biocel.2015.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Suzuki H, Watkins DN, Jair KW, Schuebel KE, Markowitz SD, Chen WD, Pretlow TP, Yang B, Akiyama Y, Van Engeland M, Toyota M, Tokino T, Hinoda Y, Imai K, Herman JG, Baylin SB. Epigenetic inactivation of SFRP genes allows constitutive WNT signaling in colorectal cancer. Nat Genet. 2004;36:417–422. doi: 10.1038/ng1330. [DOI] [PubMed] [Google Scholar]
- 43.He B, Lee AY, Dadfarmay S, You L, Xu Z, Reguart N, Mazieres J, Mikami I, McCormick F, Jablons DM. Secreted frizzled-related protein 4 is silenced by hypermethylation and induces apoptosis in beta-catenin-deficient human mesothelioma cells. Cancer Res. 2005;65:743–748. [PubMed] [Google Scholar]
- 44.Goeppert B, Konermann C, Schmidt CR, Bogatyrova O, Geiselhart L, Ernst C, Gu L, Becker N, Zucknick M, Mehrabi A, Hafezi M, Klauschen F, Stenzinger A, Warth A, Breuhahn K, Renner M, Weichert W, Schirmacher P, Plass C, Weichenhan D. Global alterations of DNA methylation in cholangiocarcinoma target the Wnt signaling pathway. Hepatology. 2014;59:544–554. doi: 10.1002/hep.26721. [DOI] [PubMed] [Google Scholar]
- 45.Jost E, Gezer D, Wilop S, Suzuki H, Herman JG, Osieka R, Galm O. Epigenetic dysregulation of secreted Frizzled-related proteins in multiple myeloma. Cancer Lett. 2009;281:24–31. doi: 10.1016/j.canlet.2009.02.002. [DOI] [PubMed] [Google Scholar]
- 46.Zeng G, Germinaro M, Micsenyi A, Monga NK, Bell A, Sood A, Malhotra V, Sood N, Midda V, Monga DK, Kokkinakis DM, Monga SP. Aberrant Wnt/beta-catenin signaling in pancreatic adenocarcinoma. Neoplasia. 2006;8:279–289. doi: 10.1593/neo.05607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Feng Han Q, Zhao W, Bentel J, Shearwood AM, Zeps N, Joseph D, Iacopetta B, Dharmarajan A. Expression of sFRP-4 and beta-catenin in human colorectal carcinoma. Cancer Lett. 2006;231:129–137. doi: 10.1016/j.canlet.2005.01.026. [DOI] [PubMed] [Google Scholar]
- 48.Topalian SL, Taube JM, Anders RA, Pardoll DM. Mechanism-driven biomarkers to guide immune checkpoint blockade in cancer therapy. Nat Rev Cancer. 2016;16:275–287. doi: 10.1038/nrc.2016.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Royal RE, Levy C, Turner K, Mathur A, Hughes M, Kammula US, Sherry RM, Topalian SL, Yang JC, Lowy I, Rosenberg SA. Phase 2 trial of single agent Ipilimumab (anti-CTLA-4) for locally advanced or metastatic pancreatic adenocarcinoma. J Immunother. 2010;33:828–833. doi: 10.1097/CJI.0b013e3181eec14c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tang Y, Xu X, Guo S, Zhang C, Tang Y, Tian Y, Ni B, Lu B, Wang H. An increased abundance of tumor-infiltrating regulatory T cells is correlated with the progression and prognosis of pancreatic ductal adenocarcinoma. PLoS One. 2014;9:e91551. doi: 10.1371/journal.pone.0091551. [DOI] [PMC free article] [PubMed] [Google Scholar]