Abstract
Endometrial cancer (EMCA) is a clinically heterogeneous disease. Previously, we tested the efficacy of Verteporfin (VP) in EMCA cells and observed cytotoxic and anti-proliferative effects. In this study, we analyzed RNA sequencing data to investigate the comprehensive transcriptomic landscape of VP treated Type 1 EMCA cell lines, including HEC-1-A and HEC-1-B. There were 549 genes with differential expression of two-fold or greater and P < 0.05 after false discovery rate correction for the HEC-1-B cell line. Positive regulation of TGFβ1 production, regulation of lipoprotein metabolic process, cell adhesion, endodermal cell differentiation, formation and development, and integrin mediated signaling pathway were among the significantly associated terms. A functional enrichment analysis of differentially expressed genes after VP treatment revealed extracellular matrix organization Gene Ontology as the most significant. CDC23 and BUB1B, two genes crucially involved in mitotic checkpoint progression, were found to be the pair with the best association from STRING among differentially expressed genes in VP treated HEC-1-B cells. Our in vivo results indicate that subcutaneous tumors in mice were regressed after VP treatment by inhibiting cell cycle pathway proteins. The present study revealed multiple key genes of pathological significance in EMCA, thereby improving our understanding of molecular profiles of EMCA cells.
Introduction
Endometrial cancer (EMCA) is a clinically heterogeneous disease. Majority of endometrial carcinomas are generally low grade and low stage with favorable prognoses, however, the high-grade EMCA accounts for a disproportionate number of EMCA deaths1–3. EMCA has been grouped into 2 types. Type 1 is estrogen potentiated, estrogen receptor (ER) and progesterone receptor (PR) positive, and generally carries a favorable prognosis. Type 2 is ER/PR negative tumors, of non-endometrioid histology (mainly serous and clear cell carcinoma), are seen in post-menopausal women, and are associated with atrophic endometrium, and poor outcomes1. High-grade endometrial carcinoma constitutes a biologically, morphologically, genetically, and clinically heterogeneous group of tumors. Recent developments of large-scale genomic studies reveal that this heterogeneity may be a function of the diversity of various molecular alterations during disease progression. The analyses using The Cancer Genome Atlas (TCGA) data have led to an integrated genomic classification of endometrioid endometrial carcinomas (EECs) and serous endometrial carcinomas (SECs) and the identification of the POLE (ultramutated), microsatellite instability (MSI) (hypermutated), copy-number low (endometrioid) and copy-number high (serous-like) subtypes, with distinct combinations of genomic and epigenetic alterations1. Mounting evidence suggests that some molecular alterations are preferentially found in endometrioid endometrial carcinomas (EECs), including mutations in PTEN and CTNNB1, whereas others such as TP53 mutations are more prevalent in serous endometrial carcinomas (SECs)2,3.
The American Cancer Society estimates that 63,230 new cases of cancer of the body of the uterus (uterine body or corpus) will be diagnosed and about 11,350 women will die from cancers of the uterine body4. Although there are many drugs approved for the treatment of ovarian cancer, there is only one FDA-approved drug (Megestrol Acetate) for EMCA, highlighting the need for new therapies to treat advanced, recurrent and metastatic EMCA5,6. Our laboratory identified nuclear expression of the Yes-associated protein (YAP) as a poor prognostic indicator in the overall survival of patients with EMCA7. YAP, the main downstream target of the Hippo pathway, plays an important role in the balance between cell proliferation and apoptosis8–10. Verteporfin (VP)11, an FDA approved drug used in photodynamic therapy (PDT) for adult macular degeneration was recently identified as an inhibitor of YAP and its binding partner TEA Domain Transcription Factor 1 (TEAD) binding12. Since the identification of VP as a YAP/TEAD inhibitor, several in vitro and in vivo studies have revealed the new potential of YAP1 in different cancers, where YAP is overexpressed13–16. We tested the efficacy of VP treatment in Type 1 EMCA cells (HEC-1-A and HEC-1-B) and observed cytotoxic and anti-proliferative effects17. Based on the molecular heterogeneity observed in EMCA patients and the effects of VP on EMCA cells, we hypothesized that VP might alter the biological processes and pathways associated with progression of EMCA cells. The aims of this study were to study the effects of VP on (1) genes or gene expression modules representative of biological processes known to play a role in EMCA, and (2) to define the association of these genes and/or pathways in the progression of EMCA, using RNA sequencing data. Here, we have used RNA sequencing (RNA-seq) to develop transcriptome data set of control and VP treated EMCA cells. We preferred RNA-seq compared to microarray technologies, as RNA-seq has a better dynamic range in estimates of gene expression and better precision18.
Materials and Methods
EMCA cell lines and culture conditions
We used Type 1 EMCA cells for the RNA-seq analysis portion of this study. HEC-1-A (ATCC, HTB-112) and HEC-1-B (ATCC, HTB-113) were obtained from the American Type Culture Collection (ATCC) (Manassas, VA). Both these cell lines were isolated from a patient with stage IA endometrial cancer. HEC-1-A cells were cultured in McCoy’s 5A medium (ATCC, Manassas, VA) supplemented with 10% (v/v) fetal bovine serum (FBS) (Thermo Fisher Scientific, Waltham, MA), HEC-1-B in Eagle’s minimum essential medium (EMEM) (ATCC, Manassas, VA) supplemented with 10%(v/v) FBS. Antibiotics (10 units/ml of penicillin and 10 mg/ml of streptomycin) were added to all culture media. Both cell lines were incubated at 37 °C in a humidified atmosphere containing 5% carbon dioxide.
Verteporfin (VP) treatment
Verteporfin (Sigma, Cat. No. SML0534) was dissolved in DMSO and added to the medium for a final concentration of 10 nM and the cells were treated for 3 h. Equal concentration of DMSO was added to the control cells.
Sample and library preparation
After VP treatment for 3 h at 10 nM, total RNA was isolated from EMCA cells using RNeasy Plus Mini Kit (Qiagen) with DNase treatment. The RNA concentration and purity were measured using the Nanodrop spectrophotometer (Thermo Fisher Scientific), and RNA integrity was evaluated with the Bioanalyzer RNA 6000 Nano Chip. RNA quality was uniformly excellent and met the following criteria; Nanodrop, 260/280 ratio >1.8; Bioanalyzer, RIN > 6.6. The samples were prepared using Illumina TruSeq Stranded Total RNA Sample Prep Kit with Ribo-Zero Gold and then subjected to 125 cycle paired-end sequencing. The average insert size of libraries constructed with the Illumina TruSeq Stranded Total RNA Sample Prep Kit is approximately 150 bp with inserts ranging from 100 to 400 bp. For the RNA-seq analysis, there were two replicates of the control samples (C) and VP-treated samples (VP) for each cell line (HEC-1-A and HEC-1-B) for a total of 8 samples. Sequencing and analysis were done by High-Throughput Genomics and Bioinformatic Analysis, University of Utah Shared Resources (https://healthcare.utah.edu/huntsmancancerinstitute/research/shared-resources/center-managed/bioinformatics/).
Sequence reads alignment and transcript assembly
After processing on the Illumina HiSeq 2500 instrument, FASTQ files containing the nucleotide scores and quality scores for each position were generated; these reads were aligned to the human reference genome (Ensembl release 75) using the STAR read aligner19, specifying that the sequencing experiment was paired-end. The alignment step produced a SAM file. SAMtools was used to generate BAM files from these for the sequencing runs required to generate count matrices20. After reading in the genomic features model (Ensembl’s GTF file) to count reads/fragments and ensuring none of our sequences were circular, a count of all of the exons grouped by gene was calculated.
Analysis of differentially expressed genes
The Bioconductor RNA-seq workflow was followed to detect differential expression21, using the DESeq 2 and other Bioconductor packages in R22. Because counts for transcripts in RNA-seq data can contain many rows with only zeros, rows with little to no information about the amount of gene expression were removed to reduce the size of the data object and to increase computational speed. When analyzing the general effect of VP treatment in both cell lines, standard RNA-seq workflow was followed in removing 292 features with zero reads and 1087 features with 5 of more reads. For the analysis using only HEC-1-B samples, we removed 656 features with zero counts and 1,776 features with fewer than 5 reads. The correlation plots between the replicates of the cell lines (e.g. 2 replicates of HEC-1-A control) show good correlation of normalized transcript counts (measured in log10(FPKM + 1)). DESeq 2 feature counts and other functions were run on the tables of counts to determine differentially expressed genes before and after VP-treatment. Results were considered statistically significant at an adjusted p < 0.05 (DMSO treated vs VP treated). The table of counts for each condition were used as inputs to the DESeq feature counts function to determine differentially expressed genes before and after VP-treatment. Since the expected variance of RNA-seq counts increases with the mean, DESeq 2’s regularized-logarithm transformation (rlog) of the count data was used to steady the variance across the mean21,22. The regularized log transforms (rlog) were taken of the result to generate a matrix of regularized counts for sample visualizations (Fig. 1). To check gene expression after VP treatment, the top 40 most significant genes were sorted by log fold change (logFC) and clustered in a hierarchical fashion using the rlog differences. This was plotted in volcano plots and heatmaps with accompanying dendrograms using pheatmap and ggplot2 packages in R23. An FDR-adjusted P < 0.05 was considered significant24.
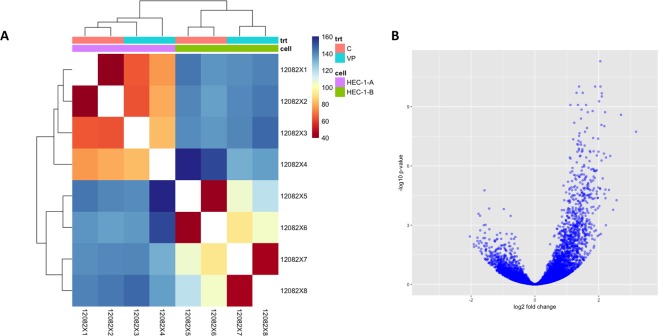
Figure 1.
Gene expression modulated by VP. (A) Expression heat map of sample-to-sample distances on the matrix of variance-stabilized data for overall gene expression. trt = treatment; C = Control; VP = Verteporfin treated. Clustering differentiates between control and VP treated samples. This heatmap was built using DESeq 2 on normalized gene read counts. All of the rlog values of the dispersion estimates were clustered using the R distance function (dist) to calculate the Euclidean distance between samples. Distance plot for HEC-1A and HEC-1B cell lines showing their control and VP-treated versions. (B) Differential gene expression, with fold difference between log2 normalized expression in control (n = 2) and VP treated (n = 2) plotted versus −log10 adjusted P-value. Each gene is colored based on the log10 base mean expression.
To compare VP-treated and untreated samples, a logFC shrinkage method (apeglm) was used to get true low bias logFC estimates for true large differences25. This shrinkage method uses a Bayesian procedure to moderate fold changes from genes with very low and highly variable counts. This serves to reduce noise in differential transcript counts. The logFC after VP treatment was plotted on the y-axis and the average of the counts normalized by size factor plotted on the x-axis to create an MA-plot with no shrinkage and one with logFC shrinkage (Supplementary Fig. S3).
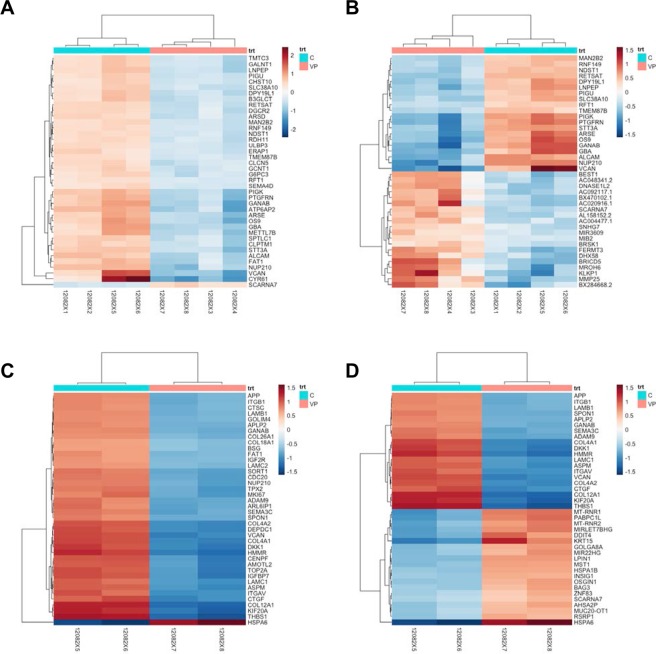
The top 20 up- and down- regulated genes were also sorted by p-value (adjusted for multiple testing using FDR) and plotted those values in heatmaps as well using the pheatmap package wrapped inside the Huntsman Cancer Institute’s hciR package (Fig. 2). False discovery rates (FDR) were calculated by determining the number of control sample differential expression ratios (i.e. ratios of expression values between replicate controls) that exceeded an VP treated sample/control sample ratio and dividing by the rank order of the VP treated sample/control sample ratio. This is the FDR, i.e. fraction of genes that have higher apparent differential expression ratios by chance. 5% of genes with an FDR ≤ 0.05 are expected to be false discoveries. Only genes for which at least one condition had a differential expression ratio with an FDR ≤ 0.05 were included in further analysis. These were converted to log2 values.
Figure 2.
Differential RNA-seq feature counts for both EMCA cell lines. (A) Heatmap showing differential RNA-seq feature counts for both EMCA cell lines (HEC-1-A and HEC-1-B) before and after VP treatment sorted by logFC. Heatmap shows genes with RNA expression most altered after VP treatment. (B) Heatmap showing differential RNA-seq feature counts for both HEC-1A and HEC-1B EMCA cell lines combined, before and after VP treatment sorted by adjusted p-value, top 20 up and top 20 down-regulated genes are shown. (C) Differential RNA-seq feature counts for HEC-1-B cells before and after VP treatment sorted by logFC. Heatmap of genes with RNA expression most altered after VP treatment, sorted by log Fold Change (logFC). There were two replicates of the control samples (C) and VP-treated samples (VP) for 4 total HEC-1B samples. (D) Heatmap showing differential RNA-seq feature counts for HEC-1-B cells before and after VP treatment sorted by adjusted p-value, top 20 up and top 20 down-regulated genes.
Protein-protein interaction network
To investigate the interactions between the protein products of top differentially expressed genes in the HEC-1-B cell line following VP treatment, a STRING network was constructed with the nodes consisting of genes and edges derived from experimentally validated protein-protein interactions. From the list of differentially expressed genes, a hypergeometric score was calculated of the likelihood of co-occurrence of genes (Supplementary Table S3). The Pfam protein domains and KEGG pathways most associated with the differentially expressed genes were also obtained through STRING (Supplementary Table S5). Many of the genes altered in HEC-1-B alone seemed to overlap with genes in the cell cycle pathway; a simple overlap test was performed to produce the list of cell cycle pathway genes differentially expressed between VP-treated and Control.
shRNA treatment
The shRNA targeting the YAP gene was used for downregulating YAP. The control siRNA (sc-37007; Santa Cruz) was used as a negative control. shYAP was transfected into EMCA cells (70% confluency) using Lipofectamine3000 (Invitrogen) according to the manufacturer’s instructions. After 72 hours of transfection, the cells were harvested and used for checking the downregulation of YAP on cell cycle proteins. The knockdown of the YAP was verified by western blotting.
Quantitative real time PCR
All primer sequences were determined using established human or mouse GenBank sequences. Primer sequences were designed using PrimerQuest (IDT) software (Supplementary Table S9). For Real-time polymerase chain reaction (RT-PCR) analysis, total RNA was isolated from control and VP (10 nM, 3 h) treated EMCA cells. Total cellular RNA was extracted with RNeasy kit (Qiagen) with on-column DNase treatment. We used RNA whose A260: A280 ratio is ≥2.0. Total RNA was reverse transcribed into first strand cDNA using iScript cDNA Synthesis Kit (Bio-Rad), as per the manufacturer’s instructions. Quantitative analysis of genes was done by SYBR green based real-time PCR using Applied Biosystems Real-Time PCR Detection System. Each sample was measured in triplicate and normalized to the reference GAPDH/β-actin/PGK1/LDHA/PPIH gene expression. A statistical evaluation of RT-PCR results was performed using one-way analysis of variance (ANOVA) to compare test gene expression between control and VP treated cells.
Western blot analysis
Control and VP-treated mice tissues or control and VP-treated cells were lysed in RIPA buffer (Boston Bioproducts, Cat. No. BP-115DG) supplemented with protease and phosphatase inhibitors and subjected to SDS-PAGE. Samples were separated electrophoretically on 10% to 12% gels, electroblotted onto nitrocellulose membrane (Bio-Rad), blots were blocked at room temperature for 1 h in 5% (w/v) milk in phosphate-buffered saline and incubated overnight at 4 °C with primary antibodies. Protein bands were visualized with SuperSignal™ West Pico Chemiluminescent Substrate (Thermo Fisher Scientific) and detected using LAS-3000 (Fujifilm, Tokyo, Japan). All the antibodies used in this study and their details were provided in Supplementary Table S10.
Mice experiments
To evaluate the efficacy of VP for inhibition of tumor growth in a mouse model, we injected HEC-1-B GFP cells into NCr nude mice by subcutaneous (SC) administration. Mice experiments were conducted by Anticancer Inc. (http://www.anticancer.com). All animal studies were conducted with an Anticancer Institutional Animal Care and Use Committee (IACUC)-protocol specifically approved for this study and in accordance with the principles and procedures outlined in the National Institute of Health Guide for the Care and Use of Animals. All animal procedures were carried out under specific-pathogen-free (SPF) conditions. Log-phase HEC-1-B GFP cells (5 × 106) were suspended in 100 µl PBS and injected SC in left flank of mouse. After the tumors reached a size >100 mm3, VP was administered IP at a dose 50 mg/kg bodyweight to VP-treated group mice (n = 10). VP was given 3 times a week for 3 weeks. Control group mice (n = 10) were administered DMSO in a similar manner. Tumor size was measured by caliper and by GFP imaging twice a week. Body weight was measured twice per week and Body condition scores (BCS) were taken daily. Mice were sacrificed after 3 weeks of treatment. Primary tumors were excised and weighed at necropsy. Blood samples were collected, immediately processed to prepare plasma and flash frozen. Based on tumor volume, VP-treated mice were divided into Responders (tumor volume <500 mm3) and Non-responders (tumor volume >500 mm3). There were 6 mice in the Responders group (animal numbers 1, 2, 4, 5, 6 and 8) and 4 mice in the Non-responders group (animal numbers 3, 7, 9 and 10) (Supplementary Fig. S4).
LC-MS/MS analysis of Verteporfin in mice blood samples
Verteporfin present in plasma samples of mice at necropsy was analyzed at the Proteomics facility of Cornell University (http://www.biotech.cornell.edu/brc/proteomics-and-mass-spectrometry). The LC-MS method was developed for detecting Verteporfin with high selectivity and sensitivity. We used targeted IDA method for identification of the analyte. There were two forms of Verteporfin (depending on the position of CH3 and H) found in standard as well as in the samples having the retention time 9.86 and 10.2 min respectively. Verteporfin was analyzed in control, Responder and Non-responder samples (n = 3 each).
Results
Transcriptome analysis of control and Verteporfin treated EMCA cells
We treated Type 1 EMCA cells (HEC-1-A and HEC-1-B) with VP at 10 nM for 3 h. This short time frame (half-life of VP is approximately 5–6 hours) and the lower concentration of VP was chosen to allow us to examine immediate responses to VP and to minimize confounding effects associated with severe damage and cell death. Under these conditions, VP causes several changes in gene expression within 3 h of treatment. We used RNA-seq to measure the transcriptomes altered by VP treatment. We generated 220–270 million reads per lane, filtered reads to have high quality scores and mapped 75–80% of those reads to the human genome. In order to get an overview over similarities and dissimilarities between samples, a hierarchical clustering to the heatmap function based on the sample distances was used (Figs 1 and S1). The strict counts of genes after VP-treatment tended to be less than in their non-treated counterparts in both HEC-1-A and HEC-1-B (Supplementary Fig. S2A). The differential expression of genes between the two groups was compared using a negative binomial test (Fig. 2). When considering the effect of VP treatment in both cell lines, 841 genes were upregulated and 251 genes down regulated after VP treatment (Fig. 1B). Treatment with VP was found to modulate many different genes and the top 20 upregulated (Table 1) and top 20 downregulated genes (Table 2) in EMCA cells after VP treatment were tabulated. The GO annotations of the differentially expressed genes for the combined cell line data were provided (Table 3). Positive regulation of TGF-β1 production, regulation of lipoprotein metabolic process, cell adhesion, endodermal cell differentiation, formation and development, and integrin mediated signaling pathway were among the significantly associated terms.
Table 1.
Top 20 upregulated genes in HEC-1-A and HEC-1-B cells after VP treatment, ranked by logFC.
| Gene | Description | log2 Fold Change | lfcSE | padj |
|---|---|---|---|---|
| CYR61 | Cysteine rich angiogenic inducer 61 | 3.170006 | 0.481229 | 1.858732e-08 |
| VCAN | Versican | 2.696764 | 0.386285 | 2.578137e-09 |
| COL12A1 | Collagen type XII alpha 1 chain | 2.558625 | 0.52389 | 5.453066e-05 |
| THBS1 | Thrombospondin 1 | 2.451070 | 0.544776 | 1.680463e-04 |
| ITGAV | Integrin subunit alpha V | 2.361123 | 0.38602 | 3.119408e-07 |
| TFPI2 | Tissue factor pathway inhibitor 2 | 2.320415 | 0.43918 | 1.646115e-05 |
| APP | Amyloid beta precursor protein | 2.227067 | 0.366527 | 3.757190e-07 |
| ANKRD1 | Ankyrin repeat domain 1 | 2.224969 | 0.530091 | 1.007740e-03 |
| GPC4 | Glypican 4 | 2.211857 | 0.437625 | 4.312393e-05 |
| SEMA3C | Semaphorin 3C | 2.209851 | 0.417216 | 1.579824e-05 |
| GANAB | Glucosidase II alpha subunit | 2.200511 | 0.312619 | 1.903285e-09 |
| EGR1 | Early growth response 1 | 2.176855 | 0.41974 | 2.630788e-05 |
| ATP6AP2 | ATPase H+ transporting accessory protein 2 | 2.176744 | 0.321361 | 9.655713e-09 |
| ITGB4 | Integrin subunit beta 4 | 2.164238 | 0.35064 | 2.381589e-07 |
| ADAM9 | ADAM metallopeptidase domain 9 | 2.124249 | 0.380008 | 4.332059e-06 |
| GBA | Glucosyl ceramidase beta | 2.097261 | 0.283565 | 3.036162e-10 |
| NUP210 | Nucleoporin 210 | 2.096090 | 0.281061 | 2.108597e-10 |
| DSG2 | Desmoglein 2 | 2.088867 | 0.347257 | 5.337427e-07 |
| OS9 | OS9, endoplasmic reticulum lectin | 2.085436 | 0.306665 | 8.375584e-09 |
| COL4A2 | Collagen type IV alpha 2 chain | 2.066695 | 0.53173 | 2.780290e-03 |
Standard error of the logFC and p-value adjusted for false discovery rate are also included.
Table 2.
Top 20 downregulated genes in HEC-1-A and HEC-1-B cells after VP treatment.
| Gene | Description | log2 Fold Change | lfcSE | padj |
|---|---|---|---|---|
| KLKP1 | Kallikrein pseudogene 1 | −2.149389 | −2.149771 | 0.000370 |
| MROH2A | Maestro heat like repeat family member 2A | −2.038033 | −2.110868 | 0.003762 |
| PDE1B | Phosphodiesterase 1B | −1.913882 | −2.089721 | 0.009297 |
| SLC5A10 | Solute carrier family 5 member 10 | −1.907402 | −2.038577 | 0.012501 |
| HSPA6 | Heat shock protein family A (Hsp70) member 6 | −1.839579 | −2.031518 | 0.011117 |
| PPP1R27 | Protein phosphatase 1 regulatory subunit 27 | −1.799172 | −1.928942 | 0.006376 |
| DNLZ | DNL-type zinc finger | −1.796807 | −1.914579 | 0.004334 |
| MMP25 | Matrix metallopeptidase 25 | −1.766102 | −1.909672 | 0.000267 |
| CEND1 | Cell cycle exit and neuronal differentiation 1 | −1.759645 | −1.908139 | 0.013744 |
| EFCAB12 | EF-hand calcium binding domain 12 | −1.754972 | −1.898853 | 0.008427 |
| BRICD5 | BRICHOS domain containing 5 | −1.718193 | −1.851035 | 0.000335 |
| IL9RP3 | Interleukin 9 receptor pseudogene 3 | −1.717188 | −1.840025 | 0.005862 |
| AOC3 | Amine oxidase, copper containing 3 | −1.713105 | −1.806662 | 0.006405 |
| FTCD | Formimidoyltransferase cyclodeaminase | −1.711106 | −1.799549 | 0.001850 |
| MSLNL | Mesothelin-like | −1.704571 | −1.797131 | 0.020615 |
| RN7SL472P | RNA, 7SL, cytoplasmic 472, pseudogene | −1.703349 | −1.795745 | 0.028688 |
| MIR4521 | MicroRNA 4521 | −1.699961 | −1.793331 | 0.005985 |
| RBFOX3 | RNA binding protein, fox-1 homolog 3 | −1.678967 | −1.77783 | 0.016016 |
| SRPK3 | SRSF protein kinase 3 | −1.667756 | −1.777185 | 0.007663 |
| C9orf131 | Chromosome 9 open reading frame 131 | −1.667409 | −1.766292 | 0.024349 |
Table 3.
Gene Ontology for top 20 upregulated genes in HEC-1A and HEC-1B after VP treatment (GO term results with FDR-adjusted p < 0.05), sorted by fold enrichment. The p-value was adjusted for False Discovery Rate (FDR).
| GO biological process complete | H. sapiens - REFLIST (21042) | upregulated (20) | upregulated (expected) | upregulated (Fold enrichment) | upregulated (over/under) | upregulated (raw P-value) | upregulated (FDR) |
|---|---|---|---|---|---|---|---|
| Positive regulation of transforming growth factor beta1 production | 6 | 2 | 0.01 | >100 | + | 2.39E-05 | 9.35E-03 |
| Positive regulation of transforming growth factor beta production | 17 | 2 | 0.02 | >100 | + | 1.45E-04 | 3.07E-02 |
| Regulation of multicellular organismal process | 2827 | 12 | 2.69 | 4.47 | + | 1.55E-06 | 2.01E-03 |
| Positive regulation of multicellular organismal process | 1568 | 9 | 1.49 | 6.04 | + | 5.69E-06 | 3.87E-03 |
| Regulation of transforming growth factor beta1 production | 10 | 2 | 0.01 | >100 | + | 5.62E-05 | 1.63E-02 |
| Positive regulation of osteoblast proliferation | 11 | 2 | 0.01 | >100 | + | 6.64E-05 | 1.85E-02 |
| Regulation of osteoblast proliferation | 23 | 2 | 0.02 | 91.49 | + | 2.54E-04 | 4.56E-02 |
| Regulation of lipoprotein metabolic process | 13 | 2 | 0.01 | >100 | + | 8.93E-05 | 2.37E-02 |
| Regulation of protein metabolic process | 2809 | 10 | 2.67 | 3.75 | + | 9.29E-05 | 2.38E-02 |
| Cell adhesion mediated by integrin | 24 | 3 | 0.02 | >100 | + | 2.11E-06 | 2.36E-03 |
| Cell adhesion | 889 | 9 | 0.84 | 10.65 | + | 4.92E-08 | 2.56E-04 |
| Biological adhesion | 895 | 9 | 0.85 | 10.58 | + | 5.21E-08 | 2.04E-04 |
| Positive regulation of macrophage activation | 23 | 2 | 0.02 | 91.49 | + | 2.54E-04 | 4.51E-02 |
| Endodermal cell differentiation | 41 | 3 | 0.04 | 76.98 | + | 9.46E-06 | 5.10E-03 |
| Cell differentiation | 3636 | 12 | 3.46 | 3.47 | + | 2.27E-05 | 9.09E-03 |
| Cellular developmental process | 3728 | 12 | 3.54 | 3.39 | + | 2.94E-05 | 1.05E-02 |
| Developmental process | 5654 | 16 | 5.37 | 2.98 | + | 1.13E-06 | 1.61E-03 |
| Endoderm formation | 51 | 3 | 0.05 | 61.89 | + | 1.76E-05 | 7.25E-03 |
| Endoderm development | 76 | 3 | 0.07 | 41.53 | + | 5.53E-05 | 1.63E-02 |
| Tissue development | 1704 | 11 | 1.62 | 6.79 | + | 8.51E-08 | 2.22E-04 |
| Anatomical structure development | 5299 | 16 | 5.04 | 3.18 | + | 4.38E-07 | 8.55E-04 |
| Formation of primary germ layer | 116 | 4 | 0.11 | 36.28 | + | 4.52E-06 | 3.72E-03 |
| Anatomical structure formation involved in morphogenesis | 870 | 8 | 0.83 | 9.67 | + | 7.12E-07 | 1.11E-03 |
| Anatomical structure morphogenesis | 2104 | 12 | 2.00 | 6.00 | + | 5.98E-08 | 1.87E-04 |
| Gastrulation | 161 | 4 | 0.15 | 26.14 | + | 1.60E-05 | 7.13E-03 |
| Embryonic morphogenesis | 563 | 5 | 0.54 | 9.34 | + | 1.55E-04 | 3.11E-02 |
| Embryo development | 919 | 7 | 0.87 | 8.01 | + | 1.45E-05 | 6.89E-03 |
| Multicellular organism development | 4918 | 15 | 4.67 | 3.21 | + | 1.56E-06 | 1.87E-03 |
| Multicellular organismal process | 6807 | 16 | 6.47 | 2.47 | + | 1.66E-05 | 7.19E-03 |
| Integrin-mediated signaling pathway | 92 | 3 | 0.09 | 34.31 | + | 9.59E-05 | 2.34E-02 |
| Cellular response to stimulus | 6741 | 15 | 6.41 | 2.34 | + | 1.02E-04 | 2.41E-02 |
| Extracellular matrix organization | 323 | 8 | 0.31 | 26.06 | + | 3.66E-10 | 5.72E-06 |
| Extracellular structure organization | 366 | 8 | 0.35 | 23.00 | + | 9.59E-10 | 7.50E-06 |
| Cell-matrix adhesion | 123 | 3 | 0.12 | 25.66 | + | 2.21E-04 | 4.12E-02 |
| Positive regulation of cellular amide metabolic process | 133 | 3 | 0.13 | 23.73 | + | 2.77E-04 | 4.76E-02 |
| Response to tumor necrosis factor | 218 | 4 | 0.21 | 19.30 | + | 5.10E-05 | 1.53E-02 |
| Response to chemical | 4323 | 12 | 4.11 | 2.92 | + | 1.35E-04 | 3.02E-02 |
| Striated muscle tissue development | 289 | 4 | 0.27 | 14.56 | + | 1.49E-04 | 3.07E-02 |
| Muscle tissue development | 302 | 4 | 0.29 | 13.94 | + | 1.76E-04 | 3.44E-02 |
When comparing DESeq-derived differentially-expressed genes by cell line, HEC-1-B showed the most marked change in RNA expression after VP treatment (Supplementary Fig. S2B, S2C). While many genes with low mean normalized count showed a high logFC after VP treatment, more genes with high mean normalized count showed general downregulation in HEC-1-B (Fig. 3 and Supplementary Tables S1, S2). For the analysis using only HEC-1-B samples, 8582 genes had differential expression with P < 0.05 after false discovery rate (FDR) correction. Of these, 3235 were downregulated and 5271 were upregulated in VP treated cells as compared to control cells. In HEC-1-B, differentially expressed genes with lowest p-values were VCAN, COL4A1, LAMC1, HSPA6, COL4A2, COL12A1, ITGAV, SPON1, KIF20A, ASPM. Notable was the fact that versican (VCAN), a gene that codes for a large extracellular matrix proteoglycan, had the lowest adjusted p-value (2.45e-216) and large absolute log2FC (log2FC = −3.463, lfcSE = 0.109) amongst all differentially expressed genes in HEC-1-B after VP-treatment. The tumor microenvironment has been found to contribute to VCAN mRNA expression (mostly in cleaved forms) in multiple tumor cell lines26,27.
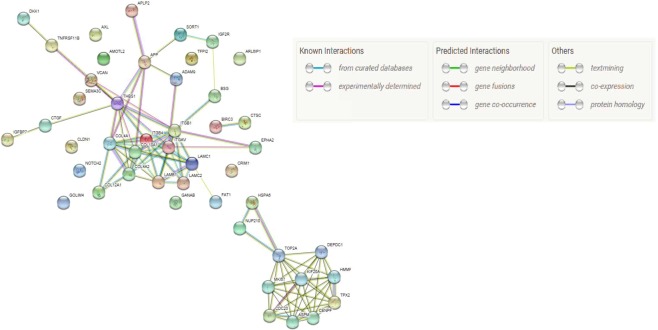
Figure 3.
STRING network of protein-protein interactions between top 50 down-regulated and up-regulated genes in HEC1-B by adjusted p-value.
Gene co-expression
The VP-treated HEC-1-A cell line is more similar to VP-treated HEC-1-B in terms of transcriptomic gene expression; in fact, they are closer to each other than the control HEC-1-A and the control HEC-1-B (Figs 1 and 2). Functional enrichment analysis of differentially expressed genes in Type 1 cells after VP revealed that the extracellular matrix (ECM) organization Gene Ontology was the most significant (FDR p-value = 2.88e-15) (Table 3).
Protein product co-expression network
The resulting protein products of differentially expressed transcripts after VP-treatment were investigated for possible interaction with each other using STRING, which uses prior biological data (Figs 3 and S3, Supplementary Table S3)28. As the HEC-1-B cell line exhibited the most differential expression after VP-treatment, it was posited that the most differentially expressed genes would have strong interactions with each other. CDC23 and BUB1B, two genes crucially involved in mitotic checkpoint progression, were found to be the pair that had the best association from STRING among genes that were significantly differentially expressed in HEC-1-B after VP treatment (Supplementary Table S3). We obtained the Pfam protein domains and KEGG (Kyoto Encyclopedia of Genes and Genomes) pathways mostly associated with the differentially expressed genes using STRING (Fig. 3, Supplementary Table S5). Significantly enriched KEGG29 pathway protein products (Top 25) from genes differentially expressed in HEC-1-B after VP treatment were tabulated in Supplementary Table S4. Out of 124 genes in the KEGG cell cycle pathway (hsa-4110), 92 of them exhibited altered expression (adjusted p-value < 0.05) in HEC-1-B after treatment in VP; the top 25 are shown in Table 4. Enrichment in cell cycle genes was not present to the same degree in HEC-1-A. Further, we validated the RNASEQ data using qRTPCR in HEC-1-B cells treated with VP and observed that HSPA6 and C2orf88 genes were upregulated whereas CYR61, THBS1and ANKRD1 were downregulated after VP treatment. Based on the protein product co-expression data, we checked the expression of BUB1B, CDC23, MAD2L1, BUB3 and CDC27 genes and observed that these were downregulated after VP treatment in HEC-1-B cells. (Supplementary Table S6.) Similarly, we checked the expression of cell cycle genes CCRK, CDK2, CCND1, CCNE1 and E2F1 in both VP-treated EMCA cells (Supplementary Table S7) and VP-treated mice tumors (Supplementary Table S8) and observed that VP effectively inhibited the cell cycle genes. The downregulation of cell cycle proteins was established in VP-treated EMCA cells (Supplementary Fig. S5A). Further, we tested the role of YAP in regulating cell cycle proteins. YAP-knockdown in HEC-1-B cells by shYAP treatment resulted in downregulation of cell cycle proteins CCRK, CDK2 and Cyclin D1 (Supplementary Fig. S5B). These results show that VP is effective in inhibiting cell cycle both at transcription and translation levels. Interestingly, we also observed that VP downregulates pluripotency marker Oct4 either in vitro and in vivo (Supplementary Fig. S5C and S5D).
Table 4.
Genes altered with an FDR adjusted p-value < 0.05 that belong to KEGG cell cycle pathways in HEC-1-B.
| ID | Gene | Biotype | Chromosome | Description | Base Mean | log2Fold Change | lfcSE | padj |
|---|---|---|---|---|---|---|---|---|
| ENSG00000164611 | PTTG1 | protein coding | 5 | Pituitary tumor-transforming 1 | 1266.799095 | −2.891629 | 0.153204 | 8.33E-77 |
| ENSG00000157456 | CCNB2 | protein coding | 15 | Cyclin B2 | 712.149481 | −3.302557 | 0.174789 | 1.08E-76 |
| ENSG00000169679 | BUB1 | protein coding | 2 | BUB1 mitotic checkpoint serine/threonine kinase | 749.048122 | −2.559155 | 0.166587 | 5.09E-51 |
| ENSG00000145386 | CCNA2 | protein coding | 4 | Cyclin A2 | 519.014978 | −2.649113 | 0.177117 | 2.50E-48 |
| ENSG00000170312 | CDK1 | protein coding | 10 | Cyclin dependent kinase 1 | 1000.755561 | −2.648046 | 0.1822 | 9.28E-46 |
| ENSG00000166851 | PLK1 | protein coding | 16 | Polo like kinase 1 | 597.264747 | −2.390371 | 0.16569 | 4.52E-45 |
| ENSG00000156970 | BUB1B | protein coding | 15 | BUB1 mitotic checkpoint serine/threonine kinase B | 766.341487 | −2.396273 | 0.177853 | 2.06E-39 |
| ENSG00000100297 | MCM5 | protein coding | 22 | Minichromosome maintenance complex component 5 | 1912.808686 | −1.775605 | 0.145411 | 1.55E-32 |
| ENSG00000076003 | MCM6 | protein coding | 2 | Minichromosome maintenance complex component 6 | 2258.121144 | −1.561124 | 0.128417 | 2.93E-32 |
| ENSG00000166483 | WEE1 | protein coding | 11 | WEE1 G2 checkpoint kinase | 1064.819728 | −1.837664 | 0.154052 | 4.46E-31 |
| ENSG00000104738 | MCM4 | protein coding | 8 | Minichromosome maintenance complex component 4 | 3564.827682 | −1.513408 | 0.128823 | 3.52E-30 |
| ENSG00000164754 | RAD21 | protein coding | 8 | RAD21 cohesion complex component | 5391.58818 | −1.250919 | 0.106903 | 6.02E-30 |
| ENSG00000080839 | RBL1 | protein coding | 20 | RB transcriptional corepressor like 1 | 678.135332 | −1.958297 | 0.167687 | 8.47E-30 |
| ENSG00000164109 | MAD2L1 | protein coding | 4 | Mitotic arrest deficient 2 like 1 | 409.014019 | −2.418097 | 0.209223 | 3.83E-29 |
| ENSG00000094804 | CDC6 | protein coding | 17 | Cell division cycle 6 | 1542.735216 | −1.598088 | 0.143578 | 3.62E-27 |
| ENSG00000149554 | CHEK1 | protein coding | 11 | Checkpoint kinase 1 | 745.52042 | −1.611426 | 0.151025 | 5.26E-25 |
| ENSG00000158402 | CDC25C | protein coding | 5 | Cell division cycle 25C | 161.535519 | −3.008824 | 0.2819 | 1.45E-24 |
| ENSG00000073111 | MCM2 | protein coding | 3 | Minichromosome maintenance complex component 2 | 4500.5434 | −1.518689 | 0.146626 | 1.27E-23 |
| ENSG00000105810 | CDK6 | protein coding | 7 | Cyclin dependent kinase 6 | 568.901075 | −1.704795 | 0.165148 | 1.90E-23 |
| ENSG00000198176 | TFDP1 | protein coding | 13 | Transcription factor Dp-1 | 2356.504771 | −1.25606 | 0.129064 | 6.35E-21 |
Effect of Verteporfin in vivo in nude mice
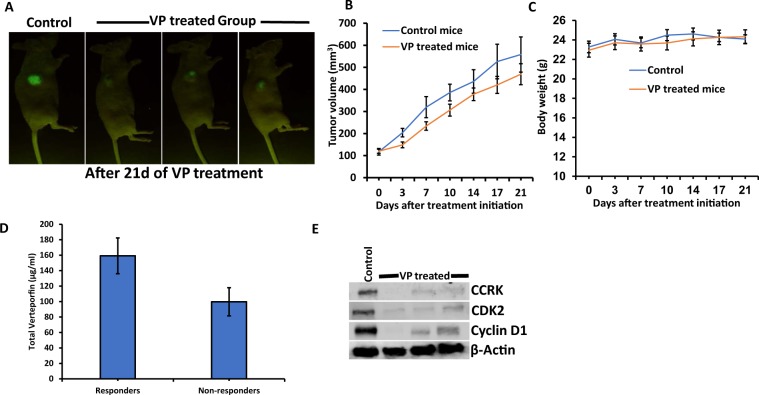
Since we observed significant cytotoxicity and induction of apoptosis by VP in EMCA cells, we employed a preclinical xenograft mouse model of EMCA to investigate the in vivo efficacy of VP on HEC-1-B cells in NCr nude mice. HEC-1-B GFP cells were injected into mice by subcutaneous (SC) administration. IP administration in this model achieved tumor regression (Fig. 4A,B) and importantly did not hasten time to euthanasia or decrease in weight between control and treated animals (Fig. 4C). These results show that VP is not toxic to mice. Based on the tumor volume data, we classified VP-treated mice into Responders and Non-responders (See Materials and Methods). Mean tumor volume in control mice was 558.34 mm3. Responders recorded mean tumor volume of 369.13 mm3 (33.89% decrease compared to control) whereas Non-responders had tumor volume of 619.15 mm3 (10.89% increase compared to control) (Supplementary Fig. S4A, S4C). Similarly, mean tumor weight of control mice was 0.302 g. In case of Responders, mean tumor weight decreased by 45.36% (0.165 g) compared to an increase of 18.54% (0.358 g) in Non-responders (Supplementary Fig. S4B). In order to check the concentration of VP in mouse blood, we analyzed their plasma samples using the LC-MS/MS method. Verteporfin was not detected in control samples and it was detected in Responder and Non-responder samples with good signal intensity. Responders had high amounts of VP (159.25 µg/ml) compared to non-responders (99.75 µg/ml) (Fig. 4D). These results indicate the bio-availability of VP and its effect on tumor regression. For further analyses, we used tumor samples of responders. Western analysis of control and VP treated tissues shows that VP is probably inducing tumor regression by acting upon cell cycle proteins (Fig. 4E). In accordance with our in vitro results, we observed a moderate effect of VP on EMCA tumor growth.
Figure 4.
(A) NCr nude mice showing tumor regression after VP treatments in a subcutaneous model after 21 days. n = 10/group. (B) Tumor volume growth curve and (C) Body weight curves in control and VP treated mice. (D) Quantitation of VP in mice plasma samples by LC-MS/MS analysis. (E) Western blot showing VP-induced inhibition of cell cycle proteins in subcutaneous tumors of mice. Error bars indicate Mean ± SEM.
Discussion
Verteporfin is a porphyrinic photosensitizer clinically used for the photodynamic treatment of age-related macular degeneration. YAP is a transcriptional co-activator and a potent oncogene30–32. Inhibition of the Hippo pathway leads to YAP activation, nuclear localization and cell proliferation in most cell types. The interaction of YAP with the TEAD family of transcriptional activators leads to DNA binding and transcription. YAP is amplified in many human cancers including breast, esophageal, hepatocellular, malignant mesothelioma, medulloblastoma and ovarian30,33–36. Based on the recent reports, VP is found to be a multi-target drug interacting with several proteins implicated in major cellular processes37. However, VP has been shown to inhibit autophagy in the absence of light activation both in vitro and in vivo38–40. Given the highly reactive nature of VP with light, Konstantinou et al.41 investigated the proposed mechanisms of non-light activated VP effects and suggested that VP-induced high molecular weight protein complexes (HMWC) require the presence of light. They observed that both singlet oxygen and radical generation mediate the formation of cross-linked oligomers and HMWC by VP in the presence of light. However, non-light activated, and light-independent VP effects have been demonstrated by several authors42–44. Verteporfin was identified as a potent inhibitor of cell growth in retinoblastoma cells, disrupting YAP-TEAD signaling and pluripotential marker Oct442. Verteporfin can also reverse the paclitaxel resistance induced by YAP over-expression in HCT-8/T cells without photoactivation through inhibiting YAP expression43. Cytotoxic effects of VP were mediated by light-independent production of reactive oxygen species and its anti-leukemic effects were also exerted in vivo without photoirradiation44. Similarly, Donohue et al. proposed that exposure to bright light could be harmful for VP-treated animals39. Based on these reports, in order to study the non-light activated effects of VP, we treated EMCA cells with VP for 3 h in the dark at 37 °C, followed by lysis with overhead fluorescence ambient laboratory lighting. Similar to previous findings, we observed maximum effect of VP in EMCA cells in inhibiting cell cycle and inducing apoptosis17. Based on the present results of RNA-seq analysis, VP induced modification of several genes belonging to different pathways, altering the transcriptomic etiology of EMCA cells.
Currently, VP has been widely used to understand the role of YAP1 in various cancers and is used for the treatment of various cancers17,45. Very few studies focused on the RNASeq analysis of EMCA pathophysiology. Mi et al. identified that the RACGAP1-STAT3-survivin signaling pathway is required for the invasive phenotype of uterine carcinosarcoma by applying RNA-Seq analysis to prospectively collected uterine carcinosarcoma tumor samples from patients46. Chen et al. studied the lncRNA transcriptome of endometrial cancers and adjacent normal endometrium from the same patients and compared with transcriptomes of other gynecologic malignancies including ovarian and cervical cancers47. The RNA-seq data of uterine corpus endometrial carcinoma (UCEC) samples identified potential genes associated with the development of UCEC. This group downloaded UCEC RNA-seq data from The Cancer Genome Atlas database48 and identified multiple key genes in UCEC and clinically relevant small molecule agents, thereby improving the understanding of UCEC and expanding perspectives on targeted therapy for this type of cancer. So far, there have been no reports on transcriptome of EMCA cells under in vitro conditions. In our study, we used RNA-seq and bioinformatic methods to better understand the EMCA cellular response to VP at the transcriptomic level, and possibly gain insight into the molecular mechanisms that might account for the pathophysiology of EMCA. Moreover, the underlying transcriptional response to VP suggests that EMCA cells carry out their function through different gene regulatory mechanisms. The results of the present study indicated that multiple differentially expressed genes are associated with EMCA pathophysiology. Differential expression analysis revealed changes in the expression patterns of many protein-coding genes previously reported to be involved in other cancers (Tables 3–4). Genes modulated by VP treatment include the genes related to positive regulation TGFβ1, lipoprotein metabolic processes, cell adhesion mediated by integrins, endodermal cell differentiation and extracellular matrix organization.
Even though we performed RNA-Seq analysis for both HEC-1-A and HEC-1-B cell lines, combining data related to both cell lines led to a result with not many significant pathways or gene-gene interactions. As in the PCA plot (Supplementary Fig. S1), the transcriptomic distance between HEC-1-A and HEC-1-B is too great to come up with many combined pathways. Hence, we focused our further analysis on HEC-1-B cell line data. Previously, we have shown that VP inhibits cell cycle progression of Type 1 EMCA cells17, hence we analyzed KEGG cell cycle pathways in HEC-1-B. Out of 124 genes in the KEGG cell cycle pathway, 92 of them exhibited altered expression after VP treatment. These results also corroborate with our in vivo studies, as we observed that tumor regression in mice is induced by inhibition of cell cycle proteins. Amongst all genes that were differentially expressed in HEC-1-B after VP treatment, naïve of biological annotation, mRNA transcripts of VCAN were most likely to be downregulated with VP treatment (Supplementary Fig. S2). The role of Versican in cell adhesion, migration, and proliferation has been extensively studied49,50, and in cancer, stromal expression of VCAN was strongly correlated with disease recurrence26. The downregulating effect of VP treatment on VCAN expression for the HEC-1-B cell line is promising, but the precise mechanism of this effect remains to be elucidated, especially considering the fact that a contrary upregulating effect on VCAN expression was found in HEC-1-A, or when both HEC-1-A and HEC-1-B were analyzed together (Table 1). Cyclin-dependent kinase 20 (CDK20) or CCRK, is a member of CDK family with strong linkage to human cancers. Recent studies reported the consistent overexpression of CCRK in cancers arising from brain, colon, liver, lung and ovary. The signaling molecules perturbed by CCRK are divergent and cancer-specific, including the cell cycle regulators CDK2, cyclin D1, cyclin E and RB in glioblastoma, ovarian carcinoma and colorectal cancer and lung cancer51. Overexpression of CCRK increases cyclin D1 expression, which suggests the role of CCRK in the control or cell proliferation via regulation of cyclin D1 expression52. Recently Zapiecki et al. showed that significantly higher expression of cyclin D1 and cyclin E was detected in patients dying from endometrial cancer53. Cyclin D1 plays an important role in LSD1-regulated estrogen-induced endometrial cancer cell proliferation54. This group also verified the positive correlation between LSD1 and cyclin D1 in endometrial cancer tissues by immunohistochemistry. In conformity with previous findings, our studies also indicate that inhibition of cyclin D1, CCRK and CDK2 are involved in inhibition of growth and proliferation in EMCA cells and tumor regression in mice. Normal adult stem cells and cancer stem cells maintain expression of Oct3/4, consistent with the stem cell hypothesis of carcinogenesis. Hence, a strategy to target “cancer stem cells” is to suppress the Oct4 gene expression55. Recently Ding et al. studied the characteristics of CD133+ cells isolated from endometrial cancer56. They proposed that CD133+ cells increased expression of embryonic stem cells markers including Oct4, Nanog, Sox2 than CD133− cells. Verteporfin was shown to downregulate Oct4 expression in retinoblastoma cells41. Our findings corroborate with previous results under both in vitro and in vivo conditions and also suggest a mechanism of action of VP.
The administration of VP in subcutaneous mouse model identified both Responders and Non-responders to this treatment. Our results show that VP is not toxic to mice without significant effect on their body weight. Visudyne®, the commercial liposomal formulation of VP was developed for photodynamic therapy. Based on the calculations of low verteporfin-to-lipid ratio of Visudyne, Donohue et al. administered 20 mg/kg VP and observed that VP tumor accumulation following i.v. administration was very poor in pancreatic ductal adenocarcinoma (PDAC) mouse model39. To increase VP tumor accumulation, they developed an alternate micellar formulation with DSPE-mPEG2000 dissolved in PBS. They observed that VP inhibited autophagy in vivo but did not reduce tumor volume or increase survival as a single agent in PDAC mouse model. They also observed that VP in combination with gemcitabine moderately reduced tumor growth and enhanced survival compared to gemcitabine alone. Similar to their findings Zhao et al. demonstrated that VP in combination with a pan-RAF inhibitor LY3009120 significantly enhanced tumor regression in KRAS-mutant pancreatic cancer57. Our results support their findings as we observed that VP failed to show antitumor activity in non-responders, even though the drug accumulated at the tumor site based on our LC-MS/MS analysis. These results warrant further standardization of solubility of VP, dosing schedule of VP, route of administration, as well as use of VP in combination with other chemotherapeutic agents for effective treatment in EMCA. On-going experiments in our laboratory include development of orthotopic model of EMCA, administration of VP as mPEG-PLGA nanoparticles and IV administration of VP with pharmacokinetic studies. The present study revealed multiple key genes of pathological significance in EMCA as well as mechanism of activity of VP with the ultimate goal of improving our understanding of molecular profile of EMCA which expands our perspective on targeted therapy for this cancer. Taken together our results suggest that VP may be repurposed for the treatment of advanced endometrial cancer.
Supplementary information
Acknowledgements
We gratefully acknowledge the funding support from Geisinger Medical Center (SRC-075) (RG) and Rice Women’s Cancer Research Fund. We are also grateful to Dr. John D Nash and Gyn. Oncology Fund from Geisinger Clinic for their support. This project is funded, in part, under a grant with the Pennsylvania Department of Health (#SAP 4100070267). The Department specifically disclaims responsibility for any analyses, interpretations or conclusions. The project described was supported by Award Number P30CA042014 from the National Cancer Institute. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health. The authors are grateful to University of Utah shared resources (https://healthcare.utah.edu/huntsmancancerinstitute/research/shared-resources/center-managed/high-throughput-genomics.php). We also thank Dr. Brian Dalley and Chris Stubben of the High Throughput Genomics Shares Resources of University of Utah for their help in RNASEQ analysis. We are thankful to Dr. Robert Hoffman, Dr. Yong Zhang and Li Tang of Anticancer for their technical help in conducting animal studies. Our sincere thanks are due to Dr. Sheng Zhang and Dr. Ruchika Bhawal of Proteomics Facility and Metabolomics Facility of Cornell University for their help in LC-MS/MS analysis of blood samples.
Author Contributions
Study conception and design: V.R.D., R.P.G. Acquisition of data: V.R.D., L.G.B. Analysis and interpretation of data: V.R.D., L.G.B. Drafting of manuscript: V.R.D., L.G.B. Critical revision: V.R.D., L.G.B., D.K., R.P.G. Final approval of the version: V.R.D., L.G.B., D.K., R.P.G.
Data Availability
Additional information for reproducing the results described in the article is available upon reasonable request and subject to a data use agreement.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Lisa Gahyun Bang and Venkata Ramesh Dasari contributed equally.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-40495-9.
References
- 1.Levine DA, Network CGAR. Integrated genomic characterization of endometrial carcinoma. Nature. 2013;497:67–73. doi: 10.1038/nature12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Matias‐Guiu X, Prat J. Molecular pathology of endometrial carcinoma. Histopathology. 2013;62:111–123. doi: 10.1111/his.12053. [DOI] [PubMed] [Google Scholar]
- 3.Murali R, Soslow RA, Weigelt B. Classification of endometrial carcinoma: more than two types. The Lancet Oncology. 2014;15:e268–e278. doi: 10.1016/S1470-2045(13)70591-6. [DOI] [PubMed] [Google Scholar]
- 4.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA: a cancer journal for clinicians. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 5.National Cancer Institute. Drugs Approved for Endometrial Cancer. https://www.cancer.gov/about-cancer/treatment/drugs/endometrial (2018).
- 6.Makker V, et al. New therapies for advanced, recurrent, and metastatic endometrial cancers. Gynecologic Oncology Research and Practice. 2017;4:19. doi: 10.1186/s40661-017-0056-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tsujiura, M. et al Yes-associated protein (YAP) modulates oncogenic features and radiation sensitivity in endometrial cancer. PLoS One. 2014 Jun 27;9(6):e100974, 10.1371/journal.pone.0100974. eCollection (2014). [DOI] [PMC free article] [PubMed]
- 8.Oka T, Mazack V, Sudol M. Mst2 and Lats kinases regulate apoptotic function of Yes kinase-associated protein (YAP) Journal of Biological Chemistry. 2008;283:27534–27546. doi: 10.1074/jbc.M804380200. [DOI] [PubMed] [Google Scholar]
- 9.Harvey KF, Zhang X, Thomas DM. The Hippo pathway and human cancer. Nature Rev Cancer. 2013;13:246–257. doi: 10.1038/nrc3458. [DOI] [PubMed] [Google Scholar]
- 10.Wang H, Du Y-C, Zhou X-j, Liu H, Tang SC. The dual functions of YAP-1 to promote and inhibit cell growth in human malignancy. Cancer and Metastasis Reviews. 2014;33:173–181. doi: 10.1007/s10555-013-9463-3. [DOI] [PubMed] [Google Scholar]
- 11.Michels S, Schmidt-Erfurth U. Photodynamic therapy with verteporfin: a new treatment in ophthalmology. Seminars in ophthalmology. 2001;16:201–206. doi: 10.1076/soph.16.4.201.10298. [DOI] [PubMed] [Google Scholar]
- 12.Liu-Chittenden Y, et al. Genetic and pharmacological disruption of the TEAD–YAP complex suppresses the oncogenic activity of YAP. Genes & development. 2012;26:1300–1305. doi: 10.1101/gad.192856.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang L, et al. Overexpression of YAP and TAZ is an independent predictor of prognosis in colorectal cancer and related to the proliferation and metastasis of colon cancer cells. PloS one. 2013;8:e65539. doi: 10.1371/journal.pone.0065539. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14.Edwards DN, et al. The receptor tyrosine kinase EphA2 promotes glutamine metabolism in tumors by activating the transcriptional coactivators YAP and TAZ. Sci. Signal. 2017;10:eaan4667. doi: 10.1126/scisignal.aan4667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin C, et al. Overexpression of Yes‐associated protein and its association with clinicopathological features of hepatocellular carcinoma: A meta‐analysis. Liver International. 2017;37:1675–1681. doi: 10.1111/liv.13428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiao S, et al. Targeting IRF3 as a YAP agonist therapy against gastric cancer. Journal of Experimental Medicine. 2018;215:699–718. doi: 10.1084/jem.20171116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dasari VR, et al. Verteporfin exhibits YAP-independent anti-proliferative and cytotoxic effects in endometrial cancer cells. Oncotarget. 2017;8:28628–28640. doi: 10.18632/oncotarget.15614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.‘t Hoen PA, et al. Deep sequencing-based expression analysis shows major advances in robustness, resolution and inter-lab portability over five microarray platforms. Nucleic acids research. 2008;36:e141–e141. doi: 10.1093/nar/gkn705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Love MI, Anders S, Kim V, Huber W. RNA-Seq workflow: gene-level exploratory analysis and differential expression. F1000Research. 2015;4:1070. doi: 10.12688/f1000research.7035.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq 2. Genome biology. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.R: A language and environment for statistical computing. (R Foundation for Statistical Computing, Vienna, Austria, 2010).
- 24.Camp RL, Dolled-Filhart M, Rimm DL. X-tile: a new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clinical cancer research. 2004;10:7252–7259. doi: 10.1158/1078-0432.CCR-04-0713. [DOI] [PubMed] [Google Scholar]
- 25.Zhu, A., Ibrahim, J. G. & Love, M. I. Heavy-tailed prior distributions for sequence count data: removing the noise and preserving large differences. bioRxiv, 10.1101/303255 (2018). [DOI] [PMC free article] [PubMed]
- 26.Chida S, et al. Stromal VCAN expression as a potential prognostic biomarker for disease recurrence in stage II–III colon cancer. Carcinogenesis. 2016;37:878–887. doi: 10.1093/carcin/bgw069. [DOI] [PubMed] [Google Scholar]
- 27.Asano K, et al. Stromal Versican Regulates Tumor Growth by PromotingAngiogenesis. Scientific reports. 2017;7:17225. doi: 10.1038/s41598-017-17613-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jensen LJ, et al. STRING 8–a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009;37:D412–416. doi: 10.1093/nar/gkn760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45:D353–361. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Overholtzer M, et al. Transforming properties of YAP, a candidate oncogene on the chromosome 11q22 amplicon. Proceedings of the National Academy of Sciences. 2006;103:12405–12410. doi: 10.1073/pnas.0605579103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dong J, et al. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell. 2007;130:1120–1133. doi: 10.1016/j.cell.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao B, et al. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes & development. 2007;21:2747–2761. doi: 10.1101/gad.1602907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fernandez-L A, et al. YAP1 is amplified and up-regulated in hedgehog-associated medulloblastomas and mediates Sonic hedgehog-driven neural precursor proliferation. Genes & development. 2009;23:2729–2741. doi: 10.1101/gad.1824509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Muramatsu T, et al. YAP is a candidate oncogene for esophageal squamous cell carcinoma. Carcinogenesis. 2010;32:389–398. doi: 10.1093/carcin/bgq254. [DOI] [PubMed] [Google Scholar]
- 35.Yokoyama T, et al. YAP1 is involved in mesothelioma development and negatively regulated by Merlin through phosphorylation. Carcinogenesis. 2008;29:2139–2146. doi: 10.1093/carcin/bgn200. [DOI] [PubMed] [Google Scholar]
- 36.Zender L, et al. Identification and validation of oncogenes in liver cancer using an integrative oncogenomic approach. Cell. 2006;125:1253–1267. doi: 10.1016/j.cell.2006.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gibault F, et al. Non-photoinduced biological properties of verteporfin. Current medicinal chemistry. 2016;23:1171–1184. doi: 10.2174/0929867323666160316125048. [DOI] [PubMed] [Google Scholar]
- 38.Donohue E, et al. Inhibition of autophagosome formation by the benzoporphyrin derivative verteporfin. Journal of Biological Chemistry. 2011;286:7290–7300. doi: 10.1074/jbc.M110.139915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Donohue E, et al. The autophagy inhibitor verteporfin moderately enhances the antitumor activity of gemcitabine in a pancreatic ductal adenocarcinoma model. Journal of Cancer. 2013;4:585. doi: 10.7150/jca.7030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Donohue E, Balgi AD, Komatsu M, Roberge M. Induction of covalently crosslinked p62 oligomers with reduced binding to polyubiquitinated proteins by the autophagy inhibitor verteporfin. PloS one. 2014;9:e114964. doi: 10.1371/journal.pone.0114964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Konstantinou EK, et al. Verteporfin-induced formation of protein cross-linked oligomers and high molecular weight complexes is mediated by light and leads to cell toxicity. Scientific reports. 2017;7:46581. doi: 10.1038/srep46581. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 42.Brodowska K, et al. The clinically used photosensitizer Verteporfin (VP) inhibits YAP-TEAD and human retinoblastoma cell growth in vitro without light activation. Experimental eye research. 2014;124:67–73. doi: 10.1016/j.exer.2014.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pan W, et al. Verteporfin can reverse the paclitaxel resistance induced by YAP over-expression in HCT-8/T cells without photoactivation through inhibiting YAP expression. Cellular Physiology and Biochemistry. 2016;39:481–490. doi: 10.1159/000445640. [DOI] [PubMed] [Google Scholar]
- 44.Morishita T, et al. The photosensitizer verteporfin has light-independent anti-leukemic activity for Ph-positive acute lymphoblastic leukemia and synergistically works with dasatinib. Oncotarget. 2016;7:56241. doi: 10.18632/oncotarget.11025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huggett MT, et al. Phase I/II study of verteporfin photodynamic therapy in locally advanced pancreatic cancer. British journal of cancer. 2014;110:1698. doi: 10.1038/bjc.2014.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mi S, et al. RNA-seq identification of RACGAP1 as a metastatic driver in uterine carcinosarcoma. Clinical Cancer Research. 2016;22:4676–4686. doi: 10.1158/1078-0432.CCR-15-2116. [DOI] [PubMed] [Google Scholar]
- 47.Chen BJ, et al. Transcriptome landscape of long intergenic non-coding RNAs in endometrial cancer. Gynecologic oncology. 2017;147:654–662. doi: 10.1016/j.ygyno.2017.10.006. [DOI] [PubMed] [Google Scholar]
- 48.Shen L, Liu M, Liu W, Cui J, Li C. Bioinformatics analysis of RNA sequencing data reveals multiple key genes in uterine corpus endometrial carcinoma. Oncology Letters. 2018;15:205–212. doi: 10.3892/ol.2017.7346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wight TN, Kinsella MG, Evanko SP, Potter-Perigo S, Merrilees MJ. Versican and the Regulation of Cell Phenotype in Disease. Biochimica et biophysica acta. 2014;1840:2441–2451. doi: 10.1016/j.bbagen.2013.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim S, et al. Carcinoma Produced Factors Activate Myeloid Cells via TLR2 to Stimulate Metastasis. Nature. 2009;457:102–106. doi: 10.1038/nature07623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mok MT, et al. CCRK is a novel signalling hub exploitable in cancer immunotherapy. Pharmacology & therapeutics. 2018;186:138–151. doi: 10.1016/j.pharmthera.2018.01.008. [DOI] [PubMed] [Google Scholar]
- 52.Wu GQ, et al. Cell cycle‐related kinase supports ovarian carcinoma cell proliferation via regulation of cyclin D1 and is a predictor of outcome in patients with ovarian carcinoma. International journal of cancer. 2009;125:2631–2642. doi: 10.1002/ijc.24630. [DOI] [PubMed] [Google Scholar]
- 53.Zapiecki K, Manahan KJ, Miller GA, Geisler JP. Cyclin E is overexpressed by clear cell carcinomas of the endometrium and is a prognostic indicator of survival. European journal of gynaecological oncology. 2015;36:114–116. [PubMed] [Google Scholar]
- 54.Chen C, et al. LSD1 sustains estrogen-driven endometrial carcinoma cell proliferation through the PI3K/AKT pathway via di-demethylating H3K9 of cyclin D1. International journal of oncology. 2017;50:942–952. doi: 10.3892/ijo.2017.3849. [DOI] [PubMed] [Google Scholar]
- 55.Trosko JE. From adult stem cells to cancer stem cells. Annals of the New York Academy of Sciences. 2006;1089:36–58. doi: 10.1196/annals.1386.018. [DOI] [PubMed] [Google Scholar]
- 56.Ding D-C, Liu H-W, Chang Y-H, Chu T-Y. Expression of CD133 in endometrial cancer cells and its implications. Journal of Cancer. 2017;8:2142–2153. doi: 10.7150/jca.18869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhao X, et al. A combinatorial strategy using YAP and pan-RAF inhibitors for treating KRAS-mutant pancreatic cancer. Cancer letters. 2017;402:61–70. doi: 10.1016/j.canlet.2017.05.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Additional information for reproducing the results described in the article is available upon reasonable request and subject to a data use agreement.