Graphical abstract
Keywords: Babesia, Apicomplexa, Vaccine, Serology, Diagnostics
Highlights
-
•
An open resource of recombinant Babesia microti cell surface and secreted proteins.
-
•
Proteins are expressed in a mammalian expression system and are immunoreactive.
-
•
Concordance of immunoreactive proteins in sera from infected humans and mice.
-
•
Proteins will be useful for diagnostics, vaccines and host-pathogen interactions.
Abstract
Human babesiosis is an emerging tick-borne parasitic disease and blood transfusion-transmitted infection primarily caused by the apicomplexan parasite, Babesia microti. There is no licensed vaccine for B. microti and the development of a reliable serological screening test would contribute to ensuring the safety of the donated blood supply. The recent sequencing of the B. microti genome has revealed many novel genes encoding proteins that can now be tested for their suitability as subunit vaccine candidates and diagnostic serological markers. Extracellular proteins are considered excellent vaccine candidates and serological markers because they are directly exposed to the host humoral immune system, but can be challenging to express as soluble recombinant proteins. We have recently developed an approach based on a mammalian expression system that can produce large panels of functional recombinant cell surface and secreted parasite proteins. Here, we use the B. microti genome sequence to identify 54 genes that are predicted to encode surface-displayed and secreted proteins expressed during the blood stages, and show that 41 (76%) are expressed using our method at detectable levels. We demonstrate that the proteins contain conformational, heat-labile, epitopes and use them to serologically profile the kinetics of the humoral immune responses to two strains of B. microti in a murine infection model. Using sera from validated human infections, we show a concordance in the host antibody responses to B. microti infections in mouse and human hosts. Finally, we show that BmSA1 expressed in mammalian cells can elicit high antibody titres in vaccinated mice using a human-compatible adjuvant but these antibodies did not affect the pathology of infection in vivo. Our library of recombinant B. microti cell surface and secreted antigens constitutes a valuable resource that could contribute to the development of a serological diagnostic test, vaccines, and elucidate the molecular basis of host-parasite interactions.
1. Introduction
Parasites of the genus Babesia are prevalent apicomplexan pathogens transmitted by ticks and infect many mammalian and avian species (Yabsley and Shock, 2013). Tick-borne diseases have a major impact on both human and animal health, with annual economic losses to the global cattle industry estimated at over US$ 17 billion (de Castro, 1997, Homer et al., 2000); consequently, there is a drive to understand the biology of these understudied parasitic diseases. Several Babesia spp. parasites are zoonotic and there are a growing number of reported incidences of human infection, several of which fall outside the established endemic regions of the US northeastern and upper midwestern states (Westblade et al., 2017), including Europe and Asia (Ord and Lobo, 2015). The increase in reported cases has consequently led the Center for Disease Control (USA) to classify Babesia microti as a nationally notifiable disease since 2011 (Herwaldt et al., 2011). Human babesiosis is primarily caused by the parasite, B. microti, and is most commonly tick transmitted; however, it can also be acquired following transfusion of infected blood and occasionally transplacental passage (Levin and Krause, 2016). The infection is characterised by fever and haemolytic anaemia, and can result in death from complications such as heart failure, respiratory distress and pulmonary oedema in severe cases (Vannier et al., 2015). The antibiotics atovaquone and azithromycin are frontline treatments but resistance to these drugs has been reported (Wormser et al., 2010). In the US, babesiosis is the most frequently reported transfusion transmitted microbial infection, and the systematic testing of donated blood has been strongly advocated (Levin and Krause, 2016, Jajosky and Jajosky, 2017). Prevention is therefore limited to questionnaire-based deferral which is largely ineffective as infections are often cryptic and asymptomatic in healthy individuals. Added to this, the parasite can persist in donor blood, and because patients who receive blood transfusions are often immunocompromised (neonates, the elderly, and those in post-operative care) these circumstances increase the likelihood of serious pathological outcomes from the transmission of infected blood such that mortality can be as high as 20% (Herwaldt et al., 2011). There is no licensed vaccine for B. microti, but recent progress in the molecular understanding of how other apicomplexan parasites, particularly the blood stages of the malaria parasite Plasmodium falciparum, recognise and invade host erythrocytes has provided fresh impetus in the development of vaccines that target the blood stages of these parasites (Bustamante et al., 2017, Cowman et al., 2017).
Babesia spp. parasites are comparatively understudied zoonotic pathogens, and B. microti is a distinct and early-arising clade, historically referred to as the small Babesia due to the morphology of the parasite in the parasitized erythrocyte, and is most closely related to Theileria spp. parasites (Cornillot et al., 2012). Babesia microti is transmitted by hard-bodied ixodid ticks with the predominant vector being the deer tick (Ixodes scapularis), and the natural hosts are rodents, mainly the white-footed mouse (Peromyscus leucopus) in the endemic regions of the northeastern US (Vannier et al., 2015). Babesia microti infection occurs while the tick takes a blood meal. Similar to related Babesia parasites, the transmitted sporozoite is thought to recognise and invade erythrocytes directly. Once inside the erythrocyte, B. microti matures into the trophozoite form, completes its asexual reproductive cycle to develop into four merozoites, before lysing the erythrocyte so that the released blood-stage parasites can invade further erythrocytes. Erythrocyte invasion is an essential step in the parasite life-cycle and, as the merozoite stage is directly exposed to circulating host antibodies, targeting invasion by eliciting antibodies to essential parasite invasion ligands is considered a rational approach for vaccine design; however, because very little is known regarding the molecular basis of B. microti erythrocyte invasion, there are a limited number of possible targets. Babesia microti does contain orthologues of the known molecular components that are important for erythrocyte invasion in the more widely-studied malaria parasite P. falciparum, including the AMA-1 and RON-2 proteins (Ord et al., 2016, Wang et al., 2017), implying the conservation of at least some aspects of the invasion mechanism although B. microti lacks clear orthologues of the EBL and RH protein families that are central to the invasion machinery of P. falciparum, suggesting that species-specific innovations have also evolved (Cornillot et al., 2012).
The genome of B. microti has been sequenced recently and is remarkable due to its small size of only ∼6.4 Mbp, encoding just ∼3500 proteins (Cornillot et al., 2012, Silva et al., 2016). It is thought that the B. microti genome evolved by significant gene loss, possibly because its cellular tropism became restricted to erythrocytes in its mammalian hosts (Cornillot et al., 2012). With its compact genome, B. microti perhaps contains the minimal molecular machinery required for erythrocyte invasion which, together with the availability of a tractable rodent infection model, represents an opportunity to expand our understanding of the fundamental mechanisms of erythrocyte invasion and explore the vulnerability of the system to inhibition by vaccine-elicited antibodies. Identifying the direct interactions between the proteins that are secreted and displayed at the surface of P. falciparum with host receptors has been important in furthering our molecular understanding of how other parasites invade erythrocytes, and this has led directly to the identification of new vaccine targets (Bartholdson et al., 2013). Expressing these proteins in a recombinant form provides experimental tractability; however, extracellular proteins are challenging to express recombinantly because their hydrophobic transmembrane-spanning regions make them difficult to solubilise in solvents that retain their native folding, and they often contain structurally critical disulfide bonds that are sometimes not faithfully formed by some heterologous expression systems (Crosnier et al., 2013).
We have developed an approach based on a mammalian expression system that enables production of large panels of secreted recombinant proteins consisting of the entire predicted ectodomains of parasite proteins that are likely to retain their native folding. We have used this approach to compile protein libraries that represent the cell surface receptor repertoire and secretome of the blood stages of P. falciparum (Crosnier et al., 2013, Zenonos et al., 2014) and Plasmodium vivax (Hostetler et al., 2015). We have shown that these proteins can retain their biochemical binding functions and immunoreactivity (Crosnier et al., 2011, Wanaguru et al., 2013, Osier et al., 2014). Here, we compile a library of recombinant proteins representing the predicted cell surface receptor repertoire of the blood stage of B. microti and use them to characterise the host antibody response to infections in both humans and a murine infection model. Finally, we show that high titres of antibodies elicited against the B. microti major coat protein do not affect parasitaemia in mice. These proteins form an important resource for the Babesia research community.
2. Materials and methods
2.1. Babesia microti parasite and mouse strains
All animal experiments were performed in accordance with local ethical and UK Home Office regulations according to the UK Animals (Scientific Procedures) Act 1986. Babesia microti was obtained from BEI Resources, 10,801 University Boulevard, Manassas, VA 20110-2209, USA: Greenwich Yale Lab Strain 1 (LS1), a tick isolate (ATCC: PRA-401) and Nan_Hs_2011, (N11-50), a clinical isolate from blood collected from a babesiosis patient in Nantucket, USA, in 2010 (PRA-399). Babesia microti strains were maintained by serial passage in immunodeficient SCD (NOD.Cg-Prkdc) mice. Vaccine and infection challenge studies were conducted in BALB/c inbred mice.
2.2. Selection and synthesis of a library of B. microti cell surface and secreted proteins
Predicted protein coding regions (3609) from B. microti were collated from the B. microti (R1) genome (Cornillot et al., 2012) and existing database sequences identified in expression cloning studies (Lodes et al., 2000, Nishisaka et al., 2001, Homer et al., 2003, Yokoyama et al., 2003, Luo et al., 2011, Cao et al., 2013). These protein sequences were analysed for the presence of predicted transmembrane (TM) domains, glycosylphosphatidylinositol (GPI) anchor and signal peptide (SP) sequences using Phobius (290 proteins were SP-positive and 622 proteins were SP-negative, TM-positive) (Kall et al., 2004), PredGPI (62 proteins) (Pierleoni et al., 2008) and SignalP v4.1 (252 proteins) using the default D-cut-off value (Petersen et al., 2011), respectively. Proteins predicted to be membrane-embedded but for which the ectodomain was unlikely to consist of a contiguous protein sequence such as multi-pass membrane proteins that lacked a signal peptide and those with sequence homologies to proteins localising to membranes of intracellular organelles such as the endoplasmic reticulum or mitochondria were excluded. Systematic protein similarity searches to ensure inclusion of orthologues of known cell surface and secreted proteins from the blood stages of other apicomplexan parasites including Babesia and Plasmodium spp. identified 13 orthologues. Searches for proteins containing domains of interest including thrombospondin and EGF-domain identified 11 proteins that were included in the library and the orthologous protein sequence used to aid manual annotation. RNAseq transcriptome data from mice and hamster blood stage B. microti infections were used to exclude 75 genes not transcribed during the blood stages (Silva et al., 2016). For those selected genes, the loci were manually annotated using transcriptome data aligned to the B. microti genome. In brief, transcriptome data was retrieved from the European Nucleotide Archive (ENA) (accession numbers: PRJNA218917-PRJNA218922) and mapped to the reference genome using TopHat (Trapnell et al., 2009) with a minimum intron length of 20 bp, as B. microti genes were found to include many small introns. Artemis (Rutherford et al., 2000) was used to visualise data for manual annotation of gene architecture. The sequences corresponding to the entire ectodomains were identified by using software tools to predict the location of the signal peptides, transmembrane and GPI-anchor regions as above. Synthetic gene constructs corresponding to these sequences were made by gene synthesis (GeneartAG, Germany), essentially as described (Crosnier et al., 2013). Briefly, signal peptides were removed since a high-scoring exogenous signal peptide from a mouse antibody kappa light chain is encoded on the expression plasmid (Crosnier et al., 2010); sequences were codon-optimised for expression in human cells; and N-linked glycosylation consensus sites (N-X-S/T) were modified by substituting the serine or threonine residue with an alanine residue to prevent the inappropriate addition of large glycans which are likely to be absent from B. microti since, as for Plasmodium spp. parasites, the genes encoding enzymes required for N-glycosylation are absent from the B. microti genome (Lombard, 2016). The coding sequences were cloned into a mammalian expression plasmid containing the rat Cd4 domains 3 and 4 antigenic tag, an enzymatic biotinylatable sequence, and 6-His tags as previously described for Plasmodium parasite protein libraries (Crosnier et al., 2013). All B. microti expression plasmids and full sequences are available from Addgene (www.addgene.org/express). Where proteins were used for immunisation, the coding sequences were subcloned into a vector lacking the Cd4 tag to aid quantification of specific antibody responses.
2.3. Recombinant protein expression, ELISA and purification
Proteins were expressed by transient transfection in suspension-grown HEK293E (Durocher et al., 2002) and HEK293-6E cells (Loignon et al., 2008), essentially as described (Kerr and Wright, 2012). 6-His-tagged proteins were purified from spent tissue culture supernatant using either a 1 mL HisTrap HP column (GE Healthcare, UK) using an ÄKTAxpress or ÄKTApure instrument (GE Healthcare), or using Ni-NTA-Agarose (Jena Bioscience, Germany); 50 μL of Ni-NTA-Agarose bead suspension was added per 50 mL of cell culture supernatant, incubated overnight at 4 °C with rotation before purification according to the manufacturer’s instructions. To determine protein integrity, 2 to 6.5 µL of transfection supernatant, or 2 µg of purified protein was resolved by SDS-PAGE under reducing conditions, transferred to nitrocellulose membrane (Invitrogen, UK), blocked with 2% BSA in Tris-buffered saline/0.1% Tween-20 (TBST), and probed with 0.02 µg/mL of streptavidin-horseradish peroxidase (Jackson Immunoresearch, Europe Ltd., UK) diluted in 2% TBS-BSA. Biotinylated proteins were detected with Supersignal West pico chemiluminescent substrate (Perbio, UK). Expression levels in Table 1 are given as a guide as batch-to-batch variability is observed using this approach, but as a guide, “high” corresponds to 5–50 μg/mL, “medium” (0.5–5 μg/mL) and “low” (0.005–0.5 μg/mL). Protein levels were quantified by ELISA after immobilising dilutions of the biotinylated Cd4-tagged protein in individual wells of streptavidin-coated microtitre plates and using the mouse anti-rat Cd4 monoclonal antibody (OX68) as a detection antibody and an anti-mouse-alkaline phosphatase conjugate as a secondary antibody (Sigma, UK), essentially as described (Crosnier et al., 2013). To quantify elicited host antibody titres, proteins were immobilised directly on streptavidin-coated plates as above, or heat treated for 10 min at 80 °C prior to immobilisation on the streptavidin-coated plate. Sera was diluted 1:1000 in 2% (w/v) BSA-PBS and total host IgG and IgM antibody binding was detected using the appropriate alkaline phosphatase conjugated secondary antibody. Seropositivity is defined as a mean response greater than 3 S.D. above the control protein. Human sera were obtained from individuals who were diagnosed as infected by B. microti by Giemsa staining and/or PCR (18s rRNA) and indirect fluorescence assay using sera from infected hamsters under Institutional Review Board approval from the New York Blood Center, USA, project number: 618-10. Antibody binding was detected using HRP-conjugated anti-human secondary antibody and 3,3′,5,5′-tetramethylbenzidine substrate.
Table 1.
Details of the library of Babesia microti cell surface and secreted proteins. Each protein is grouped according to its predicted architecture (multi-spanning transmembrane protein, single-pass transmembrane protein, glycosylphosphatidylinositol-anchored or secreted). A description of the protein is provided, if known, together with the amino acid numbering of the regions expressed, an indication of the level of expression (Exp.; see Section 2.3; n.d. = not detected), the predicted molecular mass of the protein including protein tags, and the plasmid number in the Addgene plasmid repository.
| Accession | Protein Description | Region | Exp. | MM (kDa) | Addgene |
|---|---|---|---|---|---|
| Transmembrane – multipass | |||||
| BMR1_01g00210 | Cysteine Rich Modular Protein, CRMP | S22-I1938 | low | 239 | 107665 |
| BMR1_03g04695 | Rhoptry neck protein 2 | D31-G1257 | low | 162 | 107666 |
| BMR1_01g03335 | Conserved, similar to T-cell immunomodulatory protein in Theileria spp. | E21-S665 | med | 98 | 107667 |
| BMR1_02g04181 | Conserved, putative nuclear fusion protein | T19-S287 | low | 55 | 107668 |
| BMR1_01g03280 | Putative copper/ion transporter, BMN1-20 | G23-T427 | med | 70 | 107669 |
| Transmembrane – single | |||||
| BMR1_04g07915 | Serine and proline-rich protein | S21-P992 | n.d. | 131 | 107670 |
| BMR1_01g02975 | Conserved, putative Thrombospondin-related protein 3 | V23-P358 | high | 62 | 107671 |
| BMR1_03g01156 | Apical merozoite antigen 1 (AMA-1) | A39-G529 | low | 79 | 107672 |
| BMR1_04g08630 | Unknown | N29-G728 | high | 102 | 107673 |
| BMR1_03g02090 | Erythrocyte membrane-associated plasmodium like protein | K27-M482 | n.d. | 75 | 107674 |
| BMR1_01g00985 | Conserved, unknown | E24-L952 | low | 128 | 107675 |
| BMR1_03g04335 | Conserved, unknown | G26-T1079 | n.d. | 143 | 108000 |
| BMR1_03g00437 | Conserved, unknown | L20-I1451 | med | 187 | 108001 |
| BMR1_04g06950 | Conserved, unknown | T16-I545 | med | 86 | 108002 |
| BMR1_01g01210 | Nicalin | A37-A590 | n.d. | 89 | 108003 |
| GPI-anchored | |||||
| BMR1_02g04260 | GPI9, BMN1 family | G24-S260 | med | 51 | 108004 |
| BMR1_03g00785 | BmSA1, GPI12, BMN1-9 | G37-S308 | high | 55 | 108005 |
| BMR1_04g07810 | BmGPI19, Sexual stage antigen, Pfam s48/45 | S25-S753 | low | 107 | 108006 |
| BMR1_04g09190 | Unknown, EGF-domain containing protein | E24-V190 | med | 43 | 108007 |
| BMR1_02g04275 | GPI10, N1-21a, BMN1 family | G24-P289 | low | 55 | 108008 |
| BMR1_01g00435 | GPI4, 24 tandem repeat protein | H24-L648 | med | 95 | 108009 |
| BMR1_01g01875 | GPI5, Sexual stage antigen, Pfam s48/45 | A18-S862 | n.d. | 120 | 108010 |
| BMR1_01g00425 | GPI2, B. microti-specific | L23-S490 | high | 77 | 108011 |
| BMR1_04g05790 | GPI18, acid phosphatase | H19-S360 | high | 63 | 108012 |
| BMR1_01g00430 | GPI3, B. microti-specific | A19-S516 | high | 80 | 108013 |
| BMR1_03g00790 | GPI13 B. microti-specific | K23-A297 | high | 56 | 108023 |
| BMR1_03g01685 | GPI14, conserved, unknown, | S16-A209 | n.d. | 47 | 108014 |
| BMR1_03g03430 | GPI17, conserved, unknown | V23-S434 | low | 70 | 108015 |
| Secreted | |||||
| BMR1_04g07535 | Maltese cross antigen N1-15 | A27-H2396 | low | 287 | 108016 |
| BMR1_01g00095 | Conserved, unknown | F16-N376 | low | 66 | 108017 |
| BMR1_01g01510 | Unknown | S27-A451 | med | 72 | 108019 |
| BMR1_01g03475 | Unknown | A22-S313 | low | 56 | 108020 |
| BMR1_01g00945 | Unknown | S21-N773 | low | 109 | 108021 |
| BMR1_03g02875 | Unknown | Y17-S446 | n.d. | 77 | 108022 |
| BMR1_03g04855 | BMN2 family | S19-V271 | high | 53 | 108024 |
| BMR1_03g00690 | Unknown | I23-L349 | high | 62 | 108025 |
| BMR1_03g04550 | Conserved, unknown | A20-P139 | high | 38 | 108026 |
| BMR1_04g05295 | Unknown | H24-T575 | med | 88 | 108027 |
| BMR1_02g00320 | Sporozoite microneme protein 2 | R21-N879 | low | 120 | 108028 |
| BMR1_02g00615 | Cell-traversal protein, putative | R21-I173 | high | 42 | 108029 |
| BMR1_01g02875 | MAC/Perforin domain containing protein | I21-E614 | n.d. | 91 | 108030 |
| BMR1_02g00700 | Conserved, unknown | N21-S338 | n.d. | 61 | 108031 |
| BMR1_03g03090 | LCCL domain-containing protein | E14-L1572 | n.d. | 200 | 108032 |
| BMR1_03g00020 | BMN2 family | M1-K474 | low | 82 | 108033 |
| BMR1_04g09435 | Conserved, unknown | K19-F322 | high | 59 | 108034 |
| BMR1_04g08470 | Conserved, unknown | R18-S362 | n.d. | 65 | 108035 |
| BMR1_03g02515 | LCCL domain-containing protein, CCp2 | L21-L1599 | low | 200 | 108036 |
| BmR1_04g07470 | Sporozoite invasion-associated protein 1 | L28-G952 | low | 130 | 108035 |
| BMR1_01g02876 | Unknown | K21-G357 | n.d | 61 | 108038 |
| BMR1_03g02385 | Cathepsin C | D21-T503 | med | 80 | 108040 |
| BMR1_02g04075 | Conserved, unknown | Q24-P572 | n.d. | 90 | 108041 |
| BMR1_01g02031 | Rhoptry neck protein 4, RON4 | K17-T1428 | n.d. | 185 | 108116 |
| BMR1_02g02691 | High molecular weight rhoptry protein 2, RhopH2 | I20-N1330 | n.d. | 176 | 108042 |
| BMR1_03g01540 | Unknown | L21-T419 | med | 68 | 108043 |
2.4. Immunisation, B. microti infection and quantification of parasitaemia by flow cytometry
Nine-week-old female BALB/c mice were immunised i.p. with 100 µg of recombinant protein in sterile PBS adjuvanted with alhydrogel 1:1 by volume (Invivogen, France) according to the manufacturer's instructions, followed by two further boosts of 20 µg. The protein sequence of the B. microti R1 BmSA1 vaccine antigen differs from the LS1 challenge strain at a single amino acid (R133T) (Silva et al., 2016). Peripheral blood samples were taken to determine the antibody titre approximately 16 days following the final immunisation. Mice were rested for 4 weeks prior to infection with 1 × 107 infected erythrocytes from heparinized blood taken from an infected donor SCD mouse (at day 9 of infection during the ascending parasite growth phase), diluted in sterile dPBS (with Ca2+/Mg2+, Sigma) immediately prior to infection challenge. Parasitaemia was quantified by flow cytometry using blood biopsies taken from a peripheral vessel in 50 µL of Alsever’s solution (Sigma) in a 96-well microtitre plate and fixed for greater than 24 h with 100 µL of 0.01% v/v glutaldehyde at 4 °C. For parasite quantification, samples were centrifuged, resuspended in dPBS (with Ca2+/Mg2+, Sigma). Samples (50 µL) were resuspended in 1:5000 SYBR Green I (Invitrogen, UK) in dPBS and incubated for 1 h at 37 °C before washing and resuspending in 200 µL of dPBS. Samples were analysed in a BD Fortessa 2 (BD Biosciences, UK) equipped with a high-throughput sampler. SYBR Green I was detected by a 530/30 filter and autofluorescence of erythrocytes with a 450/50 filter. A BD FACS Diva (BD Biosciences, UK) was used to collect 10,000 events for each sample, gated on the erythrocyte population. The data were analysed using FlowJo software.
3. Results
3.1. A library of recombinant B. microti blood stage cell surface and secreted proteins
To identify proteins expressed on the surface of the blood stage merozoite, we systematically analysed 3609 predicted protein coding regions from B. microti: 3565 from the published B. microti R1 genome (Cornillot et al., 2012, Silva et al., 2016), and an additional 44 that had been previously identified in expression cloning studies (see Section 2.2). Proteins likely to be embedded within the parasite plasma membrane or secreted were identified based on the presence of either a predicted transmembrane sequence or N-terminal signal peptide which resulted in a list of 346 proteins. These proteins were manually examined and those that were predicted to be located in intracellular organelles, encode small peptides (<10 kDa), or spanned the membrane multiple times without a predicted contiguous N-terminal extracellular region, were excluded. Genes not expressed during the blood stages of infection were excluded by mapping transcriptome data from blood stage mouse infections (Silva et al., 2016) to the B. microti R1 strain reference genome sequence. This list was augmented by searching the B. microti genome for homologues of merozoite surface proteins from other apicomplexan parasites that have been shown to have a role in invasion, and for the presence of protein domains that have roles in adhesion such as thrombospondin-like and EGF domains. From this short list, we again used the transcriptome data to manually refine the gene structures resulting in 54 genes that included 15 encoding a protein with one or more transmembrane-spanning regions, 12 containing a C-terminal hydrophobic sequence for the addition of a GPI lipid anchor, and 27 that had no obvious way of being tethered to a membrane and so were predicted to be secreted (Table 1).
The B. microti proteins were produced as soluble recombinant forms using a mammalian expression system that was previously successful in expressing a panel of blood stage proteins from Plasmodium spp. parasites and shown to retain antigenic activity and extracellular binding functions (Crosnier et al., 2013). The entire predicted ectodomains were defined as the full-length sequence for secreted proteins, and for those proteins which are predicted to be tethered to the plasma membrane through either a transmembrane region or GPI-anchor, the sequence was truncated just prior to these sequences (Table 1). Babesia, similar to Plasmodium spp. parasites, appear to lack the full complement of enzymes required for N-linked glycosylation, and so all potential N-linked glycosylation consensus sequences were mutated to prevent the addition of large glycans when expressed in mammalian cells (Lombard, 2016). The coding regions were made by gene synthesis which permitted codon optimization for high level expression in human cells and cloned into a plasmid which contained an efficient secretion signal peptide sequence, a C-terminal enzymatically biotinylatable peptide sequence and hexa-his tags for immobilisation and purification. Proteins were expressed by transient transfection of suspension-grown HEK293 cells (Durocher et al., 2002).
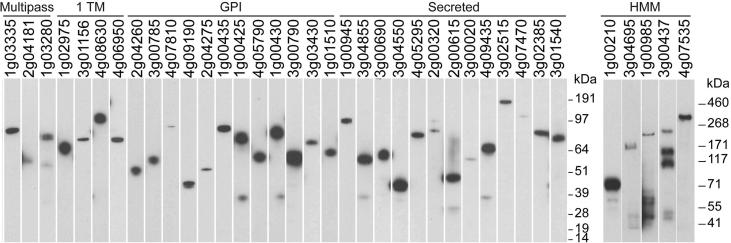
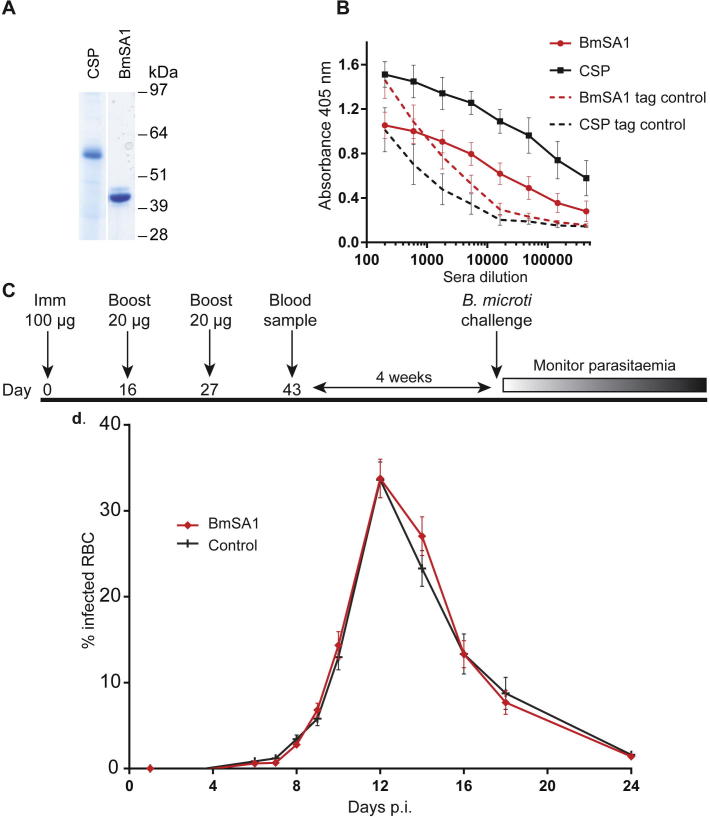
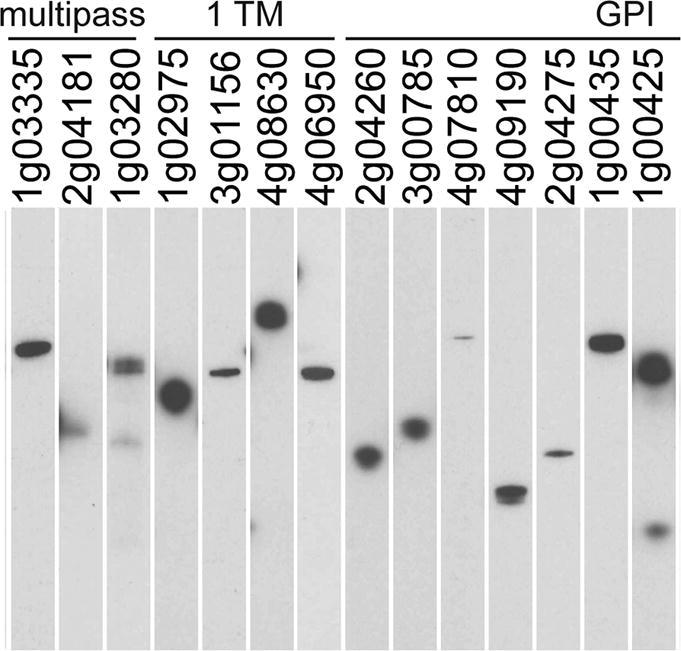
Of the 54 proteins in the library, 41 (76%) were secreted at detectable levels which ranged considerably in yield (Table 1). These parameters are very similar to other apicomplexan parasite protein libraries produced using this approach (Crosnier et al., 2013, Hostetler et al., 2015). Interestingly, the proteins which were not detectably expressed included those that are predicted to be rhoptry-localised (RON4 and RhopH2) and the orthologues of these proteins from P. falciparum proteins also failed to express in this system (Crosnier et al., 2013), suggesting these proteins require parasite-specific chaperones for functional expression. Proteins were analysed by western blotting to verify that the proteins were produced at their expected molecular mass and to determine their integrity (Fig. 1). Essentially all proteins were detected as a single band at their expected mass, with the exception of the major surface antigens BMR1_03g00020 and BMR1_01g00210, which were poorly expressed and migrated quicker than expected, suggesting these proteins are proteolytically processed; similarly, some of the higher molecular mass proteins showed evidence of proteolytic processing which is sometimes observed with this expression system (Galaway et al., 2017). The major coat protein, BmSA1 (BMR1_03g00785), migrated slightly slower than its predicted mass, which may be due to the presence of highly repetitive acidic amino acids in this protein. Together, these data demonstrate that panels of B. microti cell surface and secreted proteins can be expressed as secreted recombinant proteins in a mammalian expression system.
Fig. 1.
A library of soluble recombinant cell surface and secreted Babesia microti proteins. Recombinant B. microti proteins from cell culture supernatant were normalised and resolved by SDS–PAGE under reducing conditions, blotted and biotinylated proteins detected using streptavidin-horseradish peroxidase. High molecular mass proteins were purified prior to SDS-PAGE. All proteins contain a C-terminal rat Cd4 tag (∼25 kDa) and were enzymatically monobiotinylated during expression.
3.2. The B. microti blood stage library proteins are immunoreactive and contain conformational epitopes
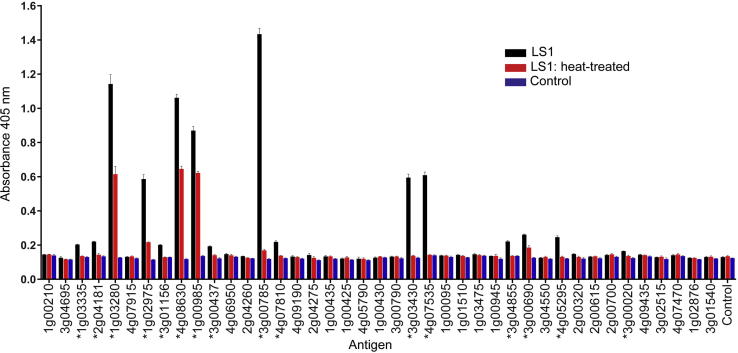
There are very few known biochemical activities for B. microti proteins and so to systematically determine whether the recombinant proteins within our library were folded and contained conformational epitopes, we asked whether the proteins were immunoreactive to sera from a chronically infected host, with the rationale that antibody responses that are able to control the pathology of the infection will generally recognise the parasite proteins in their folded, native conformation. Forty-one proteins from the library were expressed as enzymatically biotinylated proteins, their expression levels normalised and then captured in individual wells of streptavidin-coated microtitre plates before probing them with immune sera from experimentally-infected BALB/c mice. To determine if the host antibodies were recognising conformational epitopes, we denatured the proteins by heat-treatment before immobilising the proteins via their biotin tag. In total, 16 proteins were immunoreactive including BMR1_01g02975 (Thrombospondin-related protein 3), BMR1_04g08630, BMR1_01g00985, BMR1_03g00785 (BmSA1), BMR1_03g03430 (GPI17), BMR1_04g07535 (maltese cross antigen), which have been shown previously to be highly immunoreactive (Yokoyama et al., 2003, Luo et al., 2011, Cornillot et al., 2016, Silva et al., 2016) (Fig. 2). Two immunoreactive transmembrane proteins, BMR1_01g02975 (Thrombospondin-related protein 3) and BMR1_04g08630, which contain a vWA domain and putative Duffy-binding protein like domain (pfam12361), both reveal conformation-dependent and independent epitopes. BMR1_01g03280, a multipass protein, where a partial open reading frame (ORF) was identified previously as BMN1-20 by serological expression cloning using patient sera (Lodes et al., 2000), is highly immunoreactive and likely contains conformational epitopes; interestingly, this immunoreactivity was not detected using recombinant proteins expressed in Escherichia coli. Weak immunoreactivity is seen for AMA-1 (BMR1_03g01156) and RON2 (BMR1_03g04695), which are orthologues of important invasion proteins in other apicomplexans (Silva et al., 2016).
Fig. 2.
The Babesia microti blood stage library proteins are immunoreactive and contain conformational epitopes. The immunoreactivity of the recombinant B. microti proteins was determined by ELISA using sera from BALB/c infected with B. microti LS1 strain. Soluble biotinylated proteins were captured on streptavidin-coated microtitre plates either with or without heat treatment (80 °C for 10 min) before adding an alkaline-phosphatase-conjugated secondary antibody to quantify immunoreactivity. Seropositivity is defined as mean response greater than 3 S.D. above the control protein with absorbance >0.15. Seropositive antigens are marked with an asterisk and bars represent mean ± S.D.; n = 3.
3.3. Antibody titres to B. microti blood proteins increase rapidly following infection, and are maintained during chronic infection
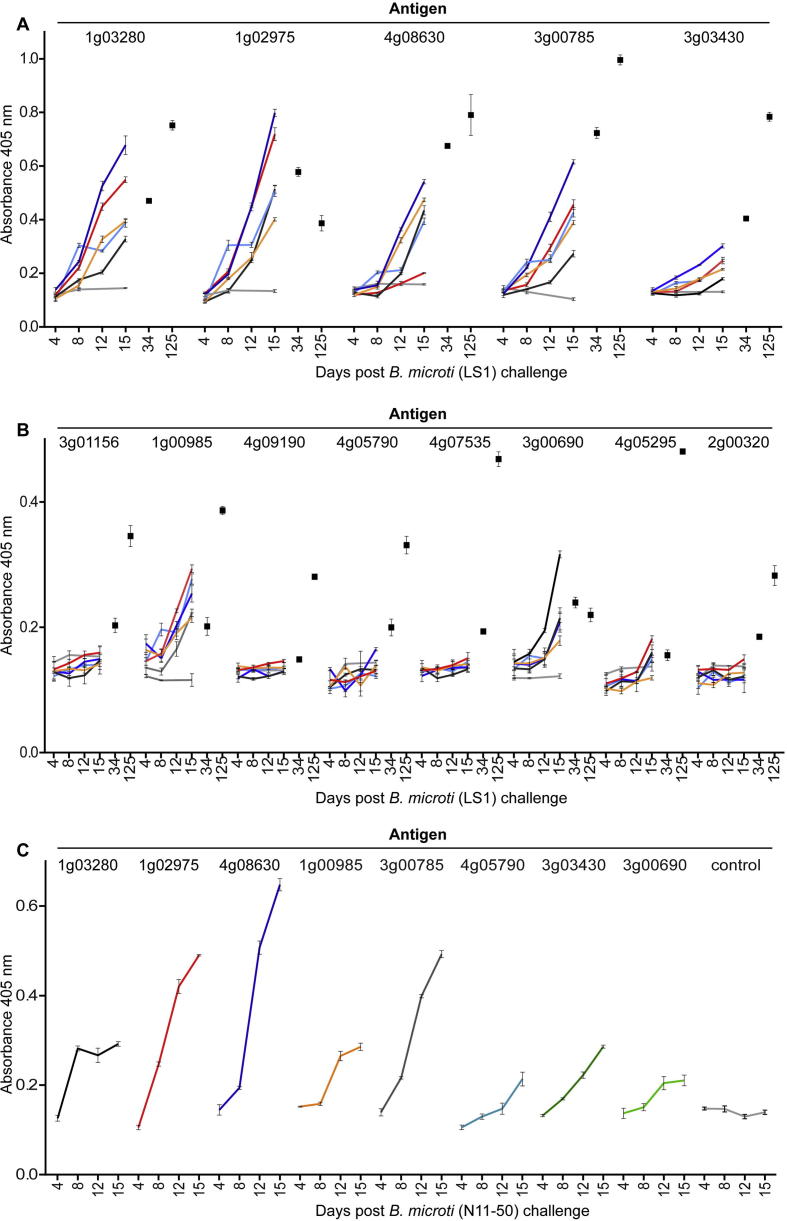
We next sought to establish the specificity and kinetics of the acquired antibody response across the panel of recombinant proteins during experimental B. microti infection. Characterising antigens for detection of early and late or chronic infection in mice allows us to infer utility in blood donation screening, where individual donor infection profiles are likely to exhibit more variation than clinical samples where individuals are symptomatic. In experimental infections of immunocompetent BALB/c mice, parasitaemia increased rapidly before peaking at approximately 2 weeks; after which, parasitaemia dropped rapidly to become cryptic by around 3 weeks p.i. Immunocompetent humans are also able to effectively control parasitaemia, although the kinetics of infection are more varied (Westblade et al., 2017). To determine the kinetics and inter-individual variability of the acquired antibody response, we quantified the immunoreactivity to the antigens during the ascending phase of parasitaemia in individual mice. Five mice were infected with B. microti Greenwich Lab strain 1 (LS1) and blood biopsies taken at days 4, 8, 12 and 15 when animals typically reach the peak of parasitaemia. We observed a rapid and immunoreactive antibody response to several antigens that correlated with the expected timing of the control of the infection. Heterogeneity in the profile and magnitude of response was observed in this highly controlled experimental infection using in-bred mice (Fig. 3A, B). Some individuals were able to raise an antibody response to all immunoreactive antigens, whereas others exhibited immunoreactivity for only a subset. Reactivity to the immunogenic antigens remained high during the cryptic phase of infection at days 34 and 125 after initial infection (Fig. 3A, B).
Fig. 3.
Profiling the immunoreactivity and kinetics of the host humoral antibody response to two different strains of Babesia microti using a panel of recombinant B. microti proteins. (A and B) The kinetics and magnitude of antibody responses to each protein within the library were determined by ELISA from sera taken from individual mice experimentally infected with the LS1 strain of B. microti. Monobiotinylated recombinant proteins were captured on streptavidin-coated plates and antibody responses quantified by ELISA using blood biopsies taken from five individual mice at days 4, 8, 12, and 15 p.i. Responses in individual mice are represented by a different colour and compared with non-infected control sera (grey line). Two post-parasite clearance data points from days 34 and 125 using pooled sera are shown (black squares). Data points represent means ± S.D.; n = 3. The antigens are segregated in those that are strongly (A) and weakly (B) immunoreactive. (C) The antibody responses in mice infected with a different clinical isolate of B. microti (N11-50) are broadly similar. Data shown are from sera samples pooled from five mice. The kinetics of antibody responses in individual mice infected with the N11-50 strain of B. microti correlate well with the responses of mice infected with the LS1 strain. Data points represent means ± S.D.; n = 3.
A further consideration in immunoassay development is strain specificity, with antigens that are immunoreactive to antibodies elicited by different strains ideally required for useful diagnosis. The protein library was based on the genome sequence from the B. microti reference strain R1 which was isolated from a human patient thought to have been infected in Nantucket (Stahl et al., 2018); however, the R1 strain genome has been recently shown to represent a different lineage from other clinically relevant strains found in the New England (USA) area (Silva et al., 2016). As LS1 was originally isolated from a rodent, we felt it important to verify immunoreactive immunogens in a clinical isolate. Five mice were experimentally infected with B. microti N11-50, a strain isolated from a human babesiosis patient in 2010, and again blood biopsies taken on days 4, 8, 12 and 15 p.i. and the antibody responses to proteins within the panel quantified. We observed a broadly similar profile of immunoreactivity to our antigen panel. This demonstrates that, at least between these two strains of B. microti, the same proteins elicit high-titre antibody responses and can be detected using the protein sequence taken from the genome reference strain.
3.4. The profile of antibody responses to B. microti infection is correlated between humans and mice
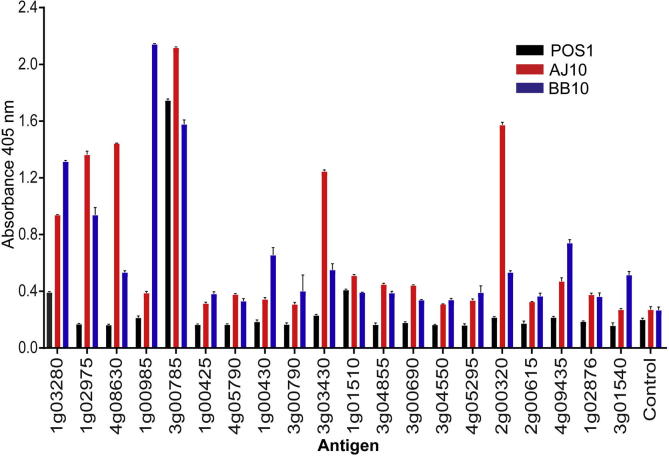
The antigens within our panel of recombinant proteins represent possible serological antigenic biomarkers for human B. microti infections. To profile the antibody responses in humans, we tested sera from three human B. microti infections from New York (USA). These samples all were validated as positive by PCR/IFA as well as Giemsa staining for sample POS1 (∼1% parasitaemia). The immunoreactivity of the sera to a protein panel containing 20 of the most highly expressed antigens was quantified by ELISA (Fig. 4). As expected, the samples showed individual heterogeneity, but 14 antigens were positive in at least two of the samples, with BmSA1 (BMR1_03g00785), BMN1-20 (BMR1_01g03280), GPI17 (BMR1_01g01510), positive in all samples. These data show that while there is some inter-individual variability in human antibody responses to B. microti infections, the profile of antibody responses largely correlates between mice and humans.
Fig. 4.
Immunoreactivity to Babesia microti proteins in humans correlate with those in mice. Recombinant monobiotinylated B. microti proteins were immobilised on streptavidin-coated microtitre plates and the immunoreactivity of sera from verified human infections was quantified by ELISA. Bar charts show mean ± S.D.; n = 3.
3.5. The B. microti major coat protein adjuvanted in alhydrogel does not affect the pathology of murine infections
The profiling of sera from both B. microti-infected humans and mice across our panel of antigens identified the major coat protein BmSA1, a protein that is localised to the merozoite membrane (Man et al., 2017), as a highly immunoreactive antigen. Because both human and murine hosts, when fully immunocompetent, are able to control B. microti infections, these antibody responses may be associated with controlling parasite multiplication, making BmSA1 a strong subunit vaccine candidate. To test this, we expressed BmSA1 as a secreted recombinant protein in HEK293 cells and purified it from spent tissue culture supernatant (Fig. 5A). To elicit high antibody titres, recombinant proteins are usually administered with adjuvants, but because strong adjuvants can act as general stimulators of the immune response they may cause “non-specific” protective effects (Awate et al., 2013); to mitigate any possible adjuvant effects we used alhydrogel, a human-compatible adjuvant. Using this approach, we were able to elicit relatively high antibody titres to both BmSA1 and a control antigen, with endpoint dilution titres greater than 1:500,000 (Fig. 5B). Again, to avoid any confounding adjuvant effects, we also “rested” the vaccinated animals for 4 weeks after the final immunisation before challenging with B. microti LS1 strain parasites (Fig. 5C). The BmSA1-vaccinated group showed no difference from the control immunised group, demonstrating BmSA1 may not be an effective B. microti vaccine when adjuvanted in human-compatible adjuvants (Fig. 5D).
Fig. 5.
The Babesia microti major coat protein adjuvanted in alhydrogel does not affect the pathology of murine infections. (A) The B. microti BmSA1 and control Plasmodium falciparum circumsporozoite protein were purified and resolved by SDS-PAGE under reducing conditions and visualised using Coomassie staining. The proteins resolved as single bands, demonstrating they were >95% pure. (B) Quantification of the elicited antibody titres in mice immunised with either BmSA1 or control CSP recombinant proteins. Sera from individual mice were serially diluted and their immunoreactivity quantified by ELISA against the corresponding biotinylated protein immobilised in individual wells of a streptavidin-coated microtitre plate. Antibody responses to the hexa-His and enzymatically biotinylatable peptide sequences were determined by using an unrelated control protein containing the same protein tags (tag controls). Data points represent means ± S.D.; n = 13 (BmSA1); n = 12 circumsporozoite protein. (C) Schematic showing the immunisation schedule: 9-week-old female BALB/c mice were immunised three times with either BmSA1 or control circumsporozoite protein as indicated and rested for 4 weeks before challenging with B. microti LS1 strain. (D) Parasitaemia is quantified as the percentage of infected RBCs in BmSA1 (n = 13) and control CSP-vaccinated (n = 12) mice followed the same kinetics, suggesting no protective effects of antibodies to BmSA1.
4. Discussion
Here, we have used the genome sequence of B. microti R1 strain to compile a panel of recombinant proteins that represents the repertoire of cell surface and secreted proteins for the blood stage antigens of B. microti. We have focussed on extracellular proteins because they are directly accessible to the host immune system and thus are considered attractive vaccine targets and serological diagnostic markers. The protein library was produced using a mammalian expression system which promotes the formation of structurally critical posttranslational modifications required for the native folding of many extracellular proteins. This approach ensures the proteins contain a greater number of epitopes, maximising target identification for diagnostic assay development and eliciting a broad spectrum of antibodies for vaccine studies. While it is possible that secreted ectodomains expressed in a heterologous expression system will not adopt the same structure as found on the parasite membrane, this approach has been successfully applied in the study of the cell surface and secreted blood stage antigens of another apicomplexan parasite, P. falciparum, showing utility in seroepidemiology (Osier et al., 2014), systematic malaria vaccine testing (Bustamante et al., 2017), and identification of host receptors (Crosnier et al., 2011, Bartholdson et al., 2012, Crosnier et al., 2016). Here, we report a similar success rate with ∼75% of candidates selected, expressed at detectable levels. It is possible that the number of expressed proteins could be increased by domain boundary optimisation or reannotation, as inaccuracies in the gene models often leads to no or poor expression. Unusually for apicomplexans, B. microti genes contain many small introns which increases the chance of errors in automated gene structure prediction (Silva et al., 2016) which can be improved by manual annotation.
Babesia microti is now the most commonly reported transfusion-transmitted microbial infection in the US with a seroprevalence of up to 2.5% in the general population from endemic regions (Levin and Krause, 2016). The potential problem is not restricted to the US, with pockets of human babesiosis caused by B. microti reported in Germany and Switzerland (Hunfeld et al., 2002). An inherent challenge for blood donation services in tackling transfusion-transmitted babesiosis is that healthy infected donors are typically asymptomatic so deferment by questionnaire, the existing method of screening, is largely ineffective. Recent studies have shown that the systematic testing of donated blood from endemic regions is an effective way of preventing transfusion-transmitted infections (Moritz et al., 2016). Diagnostic and screening methods are based on low throughput hamster infection, microscopy or immunofluorescence assay and PCR, which requires controlled environments and specialised training (Levin and Krause, 2016). An ELISA-based assay has considerable advantages, such as being amenable to both low and high throughput screening, and uses equipment available in many routine diagnostic laboratories. Recent progress towards developing a serological diagnostic assay has used a mixture of four chemically-synthesised peptides derived from the BMN1 family of B. microti antigens, which has been tested in a human blood donor setting (Levin et al., 2014), and screening parasite surface antigens (Silva et al., 2016), including GPI-anchored proteins (Cornillot et al., 2016) expressed in E. coli. Our findings confirm that the major surface antigen BmSA1 is a highly immunogenic protein and additionally show that all immunoreactivity was lost upon heat treatment of the protein, demonstrating that the vast majority of elicited antibodies recognise conformation-specific epitopes. This suggests that a folded protein produced in mammalian cells may contain more informative epitopes and therefore be a sensitive diagnostic antigen. In addition to BmSA1, we also identified several immunoreactive antigens which included two with recognisable protein domains: BMR1_01g02975 (Thrombospondin-related protein 3), and BMR1_04g08630, which contains a vWA and putative Duffy-binding protein like domain. These proteins have not been previously characterised as potential diagnostic antigens and thus inclusion of these proteins in a serological diagnostic test may improve the sensitivity and specificity of testing. Consistent with the lack of genetic diversity observed between the genome sequences of different B. microti strains, we observed few polymorphisms in the genes encoding the protein in our library, including those that were highly immunoreactive (Silva et al., 2016). We observed that the antibody responses to these antigens varied in individual infections, both in mice and humans, and further work will be required to determine if these correlate with infection parameters such as parasitaemia, immunocompetence of the host, and stage of the disease.
Here, we have started to characterise antigens for detection of early and late or chronic infection. In donor blood testing, individual donor infection profiles are likely to exhibit more variation than clinical samples where individuals are, by definition, symptomatic (Levin and Krause, 2016). It is also important to evaluate if a serological approach can distinguish between active and resolved infections, as there is a cost implication of deferring non-infective donors who have been exposed previously to B. microti. In experimental infections of immunocompetent BALB/c mice, parasitaemia increases rapidly before peaking at approximately 2 weeks, after which parasitaemia drops rapidly to become cryptic by around 3 weeks p.i. To determine the kinetics and inter-individual variability of the acquired antibody response, we quantified the immunoreactivity to the antigens during the ascending phase of parasitaemia in individual mice and observed heterogeneity in the serological responses within individuals. This suggests that the reliance on a single diagnostic antigen may lead to false negative diagnoses, and so future work will therefore ascertain whether an optimised smaller subset of this B. microti protein panel will be informative in a serological diagnostics setting.
There has been an increase in the incidence of human babesiosis in the last decade and development of resistance to frontline antibiotics, high rates of drug failure and poor drug tolerance has led to renewed interest in the development of a safe and effective vaccine (Westblade et al., 2017). The broad host range of B. microti is a major advantage towards the development of a vaccine as the rodent model can be used for testing the candidacy of different antigens. A major challenge in vaccine development has been the translation of vaccine efficacy from experimental laboratory animals to humans because some candidates that exhibit strongly protective effects in mice have only limited or no efficacy when tested in humans. One possible reason for this is the common use of experimental adjuvants such as complete/incomplete Freund’s that are strong immune stimulators that may confound any vaccine-specific effects. Indeed, here, we report no in vivo protective effect of the major surface coat protein, BmSA1, despite eliciting a high titre antibody response using alhydrogel, which is a weak but human-compatible adjuvant. Others, however, have recently reported that the same protein can elicit protective effects when administered in Freund’s adjuvant, but additionally observed strong effects on parasitaemia with an adjuvant-only control (Man et al., 2017); further work will therefore be required to resolve these effects. It is also likely that an effective blood stage vaccine will contain multiple components, each targeting a different stage of the erythrocyte invasion process, a strategy that seems effective for P. falciparum (Bustamante et al., 2017) and already suggested for B. microti (Wang et al., 2017). Similarly, where potential vaccine antigens exhibit geographically restricted sequence variation, it could be the case that certain antigens may be less effective in some regions, similar to what has been described for P. falciparum (Takala and Plowe, 2009). The economic case to invest in future research to develop a vaccine, however, must be continually balanced against other treatment options such as the development of new drugs, tick and deer control, as well as the relatively small potential market size. Our library of B. microti proteins identified from the genome sequence now provides a wider selection of antigens that can be systematically tested for vaccine candidacy.
In summary, we have compiled a library of recombinant proteins produced in a mammalian expression system that represent the cell surface repertoire and secreted proteins of the blood stage of B. microti. We envisage that, together with the availability of a murine B. microti infection model, these proteins could be systematically tested using a “reverse vaccinology” approach to identify potential vaccine candidates as well as informative serological diagnostic markers. Beyond these clinical applications, we also believe that these proteins will be a valuable resource to investigate the basic biology of the parasite and especially the molecular basis of erythrocyte recognition and invasion by Babesia spp. parasites, of which very little is currently known.
Acknowledgements
This work was support by the Wellcome Trust, UK (grant number 206194) and a Daphne Jackson Fellowship, Daphne Jackson Trust, UK to C.M.E. We thank Ulrika Boehme for manual curation of genes and the Sanger Institute, UK, flow cytometry facility.
References
- Awate S., Babiuk L.A., Mutwiri G. Mechanisms of action of adjuvants. Front. Immunol. 2013;4:114. doi: 10.3389/fimmu.2013.00114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartholdson S.J., Bustamante L.Y., Crosnier C., Johnson S., Lea S., Rayner J.C., Wright G.J. Semaphorin-7A is an erythrocyte receptor for P. falciparum merozoite-specific TRAP homolog, MTRAP. PLoS Path. 2012;8 doi: 10.1371/journal.ppat.1003031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartholdson S.J., Crosnier C., Bustamante L.Y., Rayner J.C., Wright G.J. Identifying novel Plasmodium falciparum erythrocyte invasion receptors using systematic extracellular protein interaction screens. Cell. Microbiol. 2013;15:1304–1312. doi: 10.1111/cmi.12151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustamante L.Y., Powell G.T., Lin Y.C., Macklin M.D., Cross N., Kemp A., Cawkill P., Sanderson T., Crosnier C., Muller-Sienerth N., Doumbo O.K., Traore B., Crompton P.D., Cicuta P., Tran T.M., Wright G.J., Rayner J.C. Synergistic malaria vaccine combinations identified by systematic antigen screening. Proc. Natl. Acad. Sci. USA. 2017;114:12045–12050. doi: 10.1073/pnas.1702944114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao S., Luo Y., Aboge G.O., Terkawi M.A., Masatani T., Suzuki H., Igarashi I., Nishikawa Y., Xuan X. Identification and characterization of an interspersed repeat antigen of Babesia microti (BmIRA) Exp. Parasitol. 2013;133:346–352. doi: 10.1016/j.exppara.2012.12.015. [DOI] [PubMed] [Google Scholar]
- Cornillot E., Dassouli A., Pachikara N., Lawres L., Renard I., Francois C., Randazzo S., Bres V., Garg A., Brancato J., Pazzi J.E., Pablo J., Hung C., Teng A., Shandling A.D., Huynh V.T., Krause P.J., Lepore T., Delbecq S., Hermanson G., Liang X., Williams S., Molina D.M., Ben Mamoun C. A targeted immunomic approach identifies diagnostic antigens in the human pathogen Babesia microti. Transfusion. 2016;56:2085–2099. doi: 10.1111/trf.13640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornillot E., Hadj-Kaddour K., Dassouli A., Noel B., Ranwez V., Vacherie B., Augagneur Y., Bres V., Duclos A., Randazzo S., Carcy B., Debierre-Grockiego F., Delbecq S., Moubri-Menage K., Shams-Eldin H., Usmani-Brown S., Bringaud F., Wincker P., Vivares C.P., Schwarz R.T., Schetters T.P., Krause P.J., Gorenflot A., Berry V., Barbe V., Ben Mamoun C. Sequencing of the smallest Apicomplexan genome from the human pathogen Babesia microti. Nucleic Acids Res. 2012;40:9102–9114. doi: 10.1093/nar/gks700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowman A.F., Tonkin C.J., Tham W.H., Duraisingh M.T. The molecular basis of erythrocyte invasion by malaria parasites. Cell Host Microbe. 2017;22:232–245. doi: 10.1016/j.chom.2017.07.003. [DOI] [PubMed] [Google Scholar]
- Crosnier C., Bustamante L.Y., Bartholdson S.J., Bei A.K., Theron M., Uchikawa M., Mboup S., Ndir O., Kwiatkowski D.P., Duraisingh M.T., Rayner J.C., Wright G.J. Basigin is a receptor essential for erythrocyte invasion by Plasmodium falciparum. Nature. 2011;480:534–537. doi: 10.1038/nature10606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosnier C., Iqbal Z., Knuepfer E., Maciuca S., Perrin A.J., Kamuyu G., Goulding D., Bustamante L.Y., Miles A., Moore S.C., Dougan G., Holder A.A., Kwiatkowski D.P., Rayner J.C., Pleass R.J., Wright G.J. Binding of Plasmodium falciparum merozoite surface proteins DBLMSP and DBLMSP2 to human immunoglobulin M is conserved among broadly diverged sequence variants. J. Biol. Chem. 2016;291:14285–14299. doi: 10.1074/jbc.M116.722074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosnier C., Staudt N., Wright G.J. A rapid and scalable method for selecting recombinant mouse monoclonal antibodies. BMC Biol. 2010;8:76. doi: 10.1186/1741-7007-8-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosnier C., Wanaguru M., McDade B., Osier F.H., Marsh K., Rayner J.C., Wright G.J. A library of functional recombinant cell-surface and secreted P. falciparum merozoite proteins. Mol. Cell Proteom. 2013;12:3976–3986. doi: 10.1074/mcp.O113.028357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Castro J.J. Sustainable tick and tickborne disease control in livestock improvement in developing countries. Vet. Parasitol. 1997;71:77–97. doi: 10.1016/s0304-4017(97)00033-2. [DOI] [PubMed] [Google Scholar]
- Durocher Y., Perret S., Kamen A. High-level and high-throughput recombinant protein production by transient transfection of suspension-growing human 293-EBNA1 cells. Nucleic Acids Res. 2002;30:E9. doi: 10.1093/nar/30.2.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galaway F., Drought L.G., Fala M., Cross N., Kemp A.C., Rayner J.C., Wright G.J. P113 is a merozoite surface protein that binds the N terminus of Plasmodium falciparum RH5. Nature Commun. 2017;8:14333. doi: 10.1038/ncomms14333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herwaldt B.L., Linden J.V., Bosserman E., Young C., Olkowska D., Wilson M. Transfusion-associated babesiosis in the United States: a description of cases. Annal Intern. Med. 2011;155:509–519. doi: 10.7326/0003-4819-155-8-201110180-00362. [DOI] [PubMed] [Google Scholar]
- Homer M.J., Aguilar-Delfin I., Telford S.R., 3rd, Krause P.J., Persing D.H. Babesiosis. Clin. Microbiol. Rev. 2000;13:451–469. doi: 10.1128/cmr.13.3.451-469.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homer M.J., Lodes M.J., Reynolds L.D., Zhang Y., Douglass J.F., McNeill P.D., Houghton R.L., Persing D.H. Identification and characterization of putative secreted antigens from Babesia microti. J. Clin. Microbiol. 2003;41:723–729. doi: 10.1128/JCM.41.2.723-729.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hostetler J.B., Sharma S., Bartholdson S.J., Wright G.J., Fairhurst R.M., Rayner J.C. A library of Plasmodium vivax recombinant merozoite proteins reveals new vaccine candidates and protein-protein interactions. PLoS Negl. Trop. Dis. 2015;9 doi: 10.1371/journal.pntd.0004264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunfeld K.P., Lambert A., Kampen H., Albert S., Epe C., Brade V., Tenter A.M. Seroprevalence of Babesia infections in humans exposed to ticks in midwestern Germany. J. Clin. Microbiol. 2002;40:2431–2436. doi: 10.1128/JCM.40.7.2431-2436.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jajosky R.P., Jajosky A.N. Is babesiosis the most common transfusion transmitted infection in the United States of America? The answer is not simple! Transfus Apher. Sci. 2017;56:609–610. doi: 10.1016/j.transci.2017.08.002. [DOI] [PubMed] [Google Scholar]
- Kall L., Krogh A., Sonnhammer E.L. A combined transmembrane topology and signal peptide prediction method. J. Mol. Biol. 2004;338:1027–1036. doi: 10.1016/j.jmb.2004.03.016. [DOI] [PubMed] [Google Scholar]
- Kerr J.S., Wright G.J. Avidity-based extracellular interaction screening (AVEXIS) for the scalable detection of low-affinity extracellular receptor-ligand interactions. J. Visual Exp. 2012;e3881 doi: 10.3791/3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin A.E., Krause P.J. Transfusion-transmitted babesiosis: is it time to screen the blood supply? Curr. Opin. Hematol. 2016;23:573–580. doi: 10.1097/MOH.0000000000000287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin A.E., Williamson P.C., Erwin J.L., Cyrus S., Bloch E.M., Shaz B.H., Kessler D., Telford S.R., 3rd, Krause P.J., Wormser G.P., Ni X., Wang H., Krueger N.X., Caglioti S., Busch M.P. Determination of Babesia microti seroprevalence in blood donor populations using an investigational enzyme immunoassay. Transfusion. 2014;54:2237–2244. doi: 10.1111/trf.12763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodes M.J., Houghton R.L., Bruinsma E.S., Mohamath R., Reynolds L.D., Benson D.R., Krause P.J., Reed S.G., Persing D.H. Serological expression cloning of novel immunoreactive antigens of Babesia microti. Infect. Immun. 2000;68:2783–2790. doi: 10.1128/iai.68.5.2783-2790.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loignon M., Perret S., Kelly J., Boulais D., Cass B., Bisson L., Afkhamizarreh F., Durocher Y. Stable high volumetric production of glycosylated human recombinant IFNalpha2b in HEK293 cells. BMC Biotech. 2008;8:65. doi: 10.1186/1472-6750-8-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard J. The multiple evolutionary origins of the eukaryotic N-glycosylation pathway. Biol. Direct. 2016;11:36. doi: 10.1186/s13062-016-0137-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y., Jia H., Terkawi M.A., Goo Y.K., Kawano S., Ooka H., Li Y., Yu L., Cao S., Yamagishi J., Fujisaki K., Nishikawa Y., Saito-Ito A., Igarashi I., Xuan X. Identification and characterization of a novel secreted antigen 1 of Babesia microti and evaluation of its potential use in enzyme-linked immunosorbent assay and immunochromatographic test. Parasitol. Int. 2011;60:119–125. doi: 10.1016/j.parint.2010.11.001. [DOI] [PubMed] [Google Scholar]
- Man S., Fu Y., Guan Y., Feng M., Qiao K., Li X., Gao H., Cheng X. Evaluation of a major surface antigen of Babesia microti merozoites as a vaccine candidate against Babesia infection. Front. Microbiol. 2017;8:2545. doi: 10.3389/fmicb.2017.02545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moritz E.D., Winton C.S., Tonnetti L., Townsend R.L., Berardi V.P., Hewins M.E., Weeks K.E., Dodd R.Y., Stramer S.L. Screening for Babesia microti in the U.S Blood Supply. New Engl. J. Med. 2016;375:2236–2245. doi: 10.1056/NEJMoa1600897. [DOI] [PubMed] [Google Scholar]
- Nishisaka M., Yokoyama N., Xuan X., Inoue N., Nagasawa H., Fujisaki K., Mikami T., Igarashi I. Characterisation of the gene encoding a protective antigen from Babesia microti identified it as eta subunit of chaperonin containing T-complex protein 1. Int. J. Parasitol. 2001;31:1673–1679. doi: 10.1016/s0020-7519(01)00278-8. [DOI] [PubMed] [Google Scholar]
- Ord R.L., Lobo C.A. Human babesiosis: pathogens, prevalence, diagnosis and treatment. Curr. Clin. Microbiol. Rep. 2015;2:173–181. doi: 10.1007/s40588-015-0025-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ord R.L., Rodriguez M., Cursino-Santos J.R., Hong H., Singh M., Gray J., Lobo C.A. Identification and characterization of the Rhoptry Neck Protein 2 in Babesia divergens and B. microti. Infect. Immun. 2016;84:1574–1584. doi: 10.1128/IAI.00107-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osier F.H., Mackinnon M.J., Crosnier C., Fegan G., Kamuyu G., Wanaguru M., Ogada E., McDade B., Rayner J.C., Wright G.J., Marsh K. New antigens for a multicomponent blood-stage malaria vaccine. Sci. Transl. Med. 2014;6 doi: 10.1126/scitranslmed.3008705. 247ra102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen T.N., Brunak S., von Heijne G., Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nature Meth. 2011;8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- Pierleoni A., Martelli P.L., Casadio R. PredGPI: a GPI-anchor predictor. BMC Bioinform. 2008;9:392. doi: 10.1186/1471-2105-9-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford K., Parkhill J., Crook J., Horsnell T., Rice P., Rajandream M.A., Barrell B. Artemis: sequence visualization and annotation. Bioinformatics. 2000;16:944–945. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- Silva J.C., Cornillot E., McCracken C., Usmani-Brown S., Dwivedi A., Ifeonu O.O., Crabtree J., Gotia H.T., Virji A.Z., Reynes C., Colinge J., Kumar V., Lawres L., Pazzi J.E., Pablo J.V., Hung C., Brancato J., Kumari P., Orvis J., Tretina K., Chibucos M., Ott S., Sadzewicz L., Sengamalay N., Shetty A.C., Su Q., Tallon L., Fraser C.M., Frutos R., Molina D.M., Krause P.J., Ben Mamoun C. Genome-wide diversity and gene expression profiling of Babesia microti isolates identify polymorphic genes that mediate host-pathogen interactions. Sci. Rep. 2016;6:35284. doi: 10.1038/srep35284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl P., Poinsignon Y., Pouedras P., Ciubotaru V., Berry L., Emu B., Krause P.J., Ben Mamoun C., Cornillot E. Case report of the patient source of the Babesia microti R1 reference strain and implications for travelers. J. Travel Med. 2018;25 doi: 10.1093/jtm/tax073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takala S.L., Plowe C.V. Genetic diversity and malaria vaccine design, testing and efficacy: preventing and overcoming 'vaccine resistant malaria'. Parasite Immunol. 2009;31:560–573. doi: 10.1111/j.1365-3024.2009.01138.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Pachter L., Salzberg S.L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vannier E.G., Diuk-Wasser M.A., Ben Mamoun C., Krause P.J. Babesiosis. Infect. Dis. Clin. North America. 2015;29:357–370. doi: 10.1016/j.idc.2015.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanaguru M., Crosnier C., Johnson S., Rayner J.C., Wright G.J. Biochemical analysis of the Plasmodium falciparum erythrocyte-binding antigen-175 (EBA175)-glycophorin-A interaction: implications for vaccine design. J. Biol. Chem. 2013;288:32106–32117. doi: 10.1074/jbc.M113.484840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G., Efstratiou A., Adjou Moumouni P.F., Liu M., Jirapattharasate C., Guo H., Gao Y., Cao S., Mo Z., Suzuki H., Igarashi I., Xuan X. Expression of truncated Babesia microti apical membrane protein 1 and rhoptry neck protein 2 and evaluation of their protective efficacy. Exp. Parasitol. 2017;172:5–11. doi: 10.1016/j.exppara.2016.11.001. [DOI] [PubMed] [Google Scholar]
- Westblade L.F., Simon M.S., Mathison B.A., Kirkman L.A. Babesia microti: from mice to ticks to an increasing number of highly susceptible humans. J. Clin. Microbiol. 2017;55:2903–2912. doi: 10.1128/JCM.00504-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wormser G.P., Prasad A., Neuhaus E., Joshi S., Nowakowski J., Nelson J., Mittleman A., Aguero-Rosenfeld M., Topal J., Krause P.J. Emergence of resistance to azithromycin-atovaquone in immunocompromised patients with Babesia microti infection. Clin. Infect. Dis. 2010;50:381–386. doi: 10.1086/649859. [DOI] [PubMed] [Google Scholar]
- Yabsley M.J., Shock B.C. Natural history of Zoonotic Babesia: role of wildlife reservoirs. Int. J. Parasitol. Parasites Wildlife. 2013;2:18–31. doi: 10.1016/j.ijppaw.2012.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama N., Bork S., Nishisaka M., Hirata H., Matsuo T., Inoue N., Xuan X., Suzuki H., Sugimoto C., Igarashi I. Roles of the Maltese cross form in the development of parasitemia and protection against Babesia microti infection in mice. Infect. Immun. 2003;71:411–417. doi: 10.1128/IAI.71.1.411-417.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenonos Z.A., Rayner J.C., Wright G.J. Towards a comprehensive Plasmodium falciparum merozoite cell surface and secreted recombinant protein library. Malaria J. 2014;13:93. doi: 10.1186/1475-2875-13-93. [DOI] [PMC free article] [PubMed] [Google Scholar]