Abstract
Background/Aims
Various alterations of microRNA (miRNA) expression have been reported in myelodysplastic syndrome (MDS). We aimed to investigate the unique patterns and prognostic significance of miRNA expression in Korean patients with MDS.
Methods
Bone marrow mononuclear cells were collected from eight healthy controls and 26 patients with MDS, and miRNAs were isolated and assessed via quantitative real-time polymerase chain reaction for selected miRNAs, including miR-21, miR-124a, miR-126, miR-146b-5p, miR-155, miR-182, miR-200c, miR-342-5p, miR-708, and Let-7a.
Results
MiR-124a, miR-155, miR-182, miR-200c, miR-342-5p, and Let-7a were significantly underexpressed in patients with MDS, compared to healthy controls. MiR-21, miR-126, 146b-5p, and miR-155 transcript levels were significantly lower in international prognostic scoring system lower (low and intermediate-1) risk MDS than in higher (intermediate-2 and high) risk MDS. Higher expression levels of miR-126 and miR-155 correlated with significantly shorter overall survival and leukemia-free survival. Higher miR-124a expression also tended to be related to shorter survivals.
Conclusions
Although our study was limited by the relatively small number of patients included, we identified several miRNAs associated with pathogenesis, leukemic transformation, and prognosis in MDS.
Keywords: Myelodysplastic syndromes; MIRN126 microRNA, human; hsa-146b-5p; MIRN155 microRNA, human; MIRN200 microRNA, human
INTRODUCTION
Myelodysplastic syndrome (MDS) is a genetically heterogeneous clonal disorder that is characterized by dysplasia and ineffective hematopoiesis [1]. The disease is more prevalent in older age [2], and it is usually stratified according to clinical variables that predict survival times and risk of transformation to acute myeloid leukemia (AML) [3,4]. Although the pathogenesis of MDS has not been fully understood, various alterations of microRNAs (miRNAs) have been reported in MDS [5]. MiRNAs are short non-coding RNAs that are 18 to 25 nucleotides in length and are encoded as precursor hairpin intermediate RNAs [6]. Mature miRNAs are produced by endonuclease- mediated steps and incorporated into the RNA, where miRNAs can cleave messenger RNA and repress protein translation [6]. MiRNAs also reportedly control transcription by directly regulating DNA meth-ylation [7]. Some miRNAs are associated with myeloid development and function [8], and some miRNAs have tumor-suppressor and oncogenic activities. Recently, several miRNAs, including miR-17-5p, miR-20a, miR-21, and miR-194-5p, were proposed as prognostic markers in MDS [9-11]. We aimed to investigate the disease-specific expression patterns of selected miRNAs in MDS and to evaluate the clinical implications of miRNA expression as it relates to predicting outcomes of patients with MDS.
METHODS
Patients
A total of 26 patients with MDS and eight healthy controls were included in this study. A diagnosis of MDS was made according to standard criteria [12]. This study was approved by the Institutional Review Board of Asan Medical Center, Seoul, Korea (2012-0217), and written informed consent was obtained from each patient. Clinical and laboratory data for patients with MDS were retrospectively obtained. Risk was assessed at the time of MDS diagnosis according to the international prognostic scoring system (IPSS) and the revised IPSS (R-IPSS) [3,4].
Quantitative polymerase chain reaction for miRNA
All bone marrow samples were collected before the patient received any treatment other than transfusion, androgens, or growth factors. Total miRNAs were isolated from bone marrow mononuclear cells using the miRNeasy Mini Kit (Qiagen, Hilden, Germany), according to the manufacturer’s instructions. We performed real-time quantitative polymerase chain reaction (PCR) using a miScript SYBR green PCR kit (Qiagen) for 10 miRNAs: miR-21, miR-124a, miR-126, miR-146b-5p, miR-155, miR-182, miR-200c, miR-342-5p, miR-708, and Let-7a. The reaction proceeded at 95°C for 15 minutes, followed by 40 cycles of 96°C for 15 seconds, 55°C for 30 seconds, 70°C for 30 seconds, on a LightCycler 96 (Roche, Indianapolis, IN, USA). The cycle threshold (Ct) values for all samples were normalized to Ct values of RNU6. Relative expression levels were calculated using the comparative Ct (2-△△Ct) method. All reactions were performed in triplicate.
Statistical analysis
Relative expressions of miRNAs between samples were compared using the nonparametric Mann-Whitney U test or Kruskal-Wallis test. Spearman rank correlation coefficient (rs) was calculated for statistical dependence of expression levels between miRNAs. Overall survival (OS) was calculated from the time of MDS diagnosis to the date of death or last follow-up. Leukemia-free survival (LFS) was calculated from the time of MDS diagnosis to the date of leukemic transformation or death from any cause. Survival probabilities were estimated by the Kaplan-Meier method, and differences in survival distributions were compared using the log-rank test. Statistical analyses were performed using SPSS version 21.0 software (IBM Corp., Armonk, NY, USA). For all analyses, the p values were two-tailed, and p < 0.05 (p < 0.01 for Spearman rank correlation analysis) was considered statistically significant.
RESULTS
Patient characteristics
There were 15 males and 11 females included in this study. The median patient age was 57 years (range, 28 to 81) (Table 1). Bone marrow blast percentages were < 5% in 13 patients and ≥ 5% in 13 patients. Fifteen patients had lower risk (low or intermediate-1), and 11 had higher risk (intermediate-2 or high) MDS, according IPSS risk groups.
Table 1.
Patient characteristics at the time of diagnosis
| Characteristic | Value |
|---|---|
| Sex, male/female | 15 (57.7)/11 (42.3) |
| Age, yr | 58.0 (28–81) |
| Hemoglobin, g/dL (n = 26) | 8.0 (4.2–10.4) |
| WBC count, × 103/μL (n = 26) | 3.7 (0.8–35.9) |
| ANC, /μL (n = 26) | 1,332 (96–31,959.9) |
| Circulating blast count, % (n = 26) | 0 (0–12) |
| Platelet count, × 103/μL (n = 26) | 66.0 (7–697) |
| LDH (conversion), mg/dL (n = 26) | 272 (109–846) |
| Bone marrow cellularity, % (n = 25) | 70.0 (25–100) |
| Bone marrow blasts, % (n = 26) | 4.6 (0–18.2) |
| MDS subtype (n = 26) | |
| RCUD | 3 (11.5) |
| RARS | 1 (3.8) |
| RCMD | 7 (26.9) |
| RAEB-1 | 4 (15.4) |
| RAEB-2 | 9 (34.6) |
| MDS-U | 2 (7.7) |
| De novo/secondary MDS | 25 (96.2)/1 (3.8) |
| Cytogenetic risk group (n = 26)a | |
| Good | 15 (57.7) |
| Normal | 13 |
| –Y alone | 1 |
| del (20q) alone | 1 |
| Intermediate | 6 (23.1) |
| +add(1p) | 1 |
| +8,add(17q) | 1 |
| t(3;5) | 1 |
| del(13q) | 1 |
| Others | 2 |
| Poor (complex [≥ 3 abnormalities]) | 5 (19.2) |
| IPSS risk category (n = 26) | |
| Low | 2 (7.7) |
| Intermediate-1 | 13 (50.0) |
| Intermediate-2 | 6 (23.1) |
| High | 5 (19.2) |
| R-IPSS risk category (n = 26) | |
| Very low | 0 |
| Low | 5 (19.2) |
| Intermediate | 9 (34.6) |
| High | 4 (15.4) |
| Very high | 8 (30.8) |
| ECOG performance status, 0/1/2 | 1 (3.8)/22 (84.6)/3 (11.5) |
Values are presented as number (%) or median (range).
WBC, white blood cells; ANC, absolute neutrophil count; LDH, ---; MDS, myelodysplastic syndrome; RCUD, refractory cytopenia with unilineage dysplasia; RARS, refractory anemia with ringed sideroblasts; RCMD, refractory cytopenia with multilineage dysplasia; RAEB, refractory anemia with excess of blasts; MDS-U, myelodysplastic syndrome-unclassifiable; IPSS, international prognostic scoring system; R-IPSS, revised IPSS; ECOG, Eastern Cooperative Oncology Group.
According to IPSS cytogenetic risk groups.
MicroRNA expression in patients with MDS
Relative expression of each miRNA was analyzed using RNU6 as an endogenous control in patients with MDS and normal controls (Fig. 1). Compared to normal controls, patients with MDS showed significantly lower levels of miR-124a (median, 1.471 vs. 0.090; p = 0.010), miR- 155 (median, 0.910 vs. 0.165; p < 0.001), miR-182 (median, 0.924 vs. 0.290; p = 0.012), miR-342-5p (median, 1.546 vs. 0.311; p < 0.001), and Let-7a (median, 1.032 vs. 0.319; p = 0.004).
Figure 1.
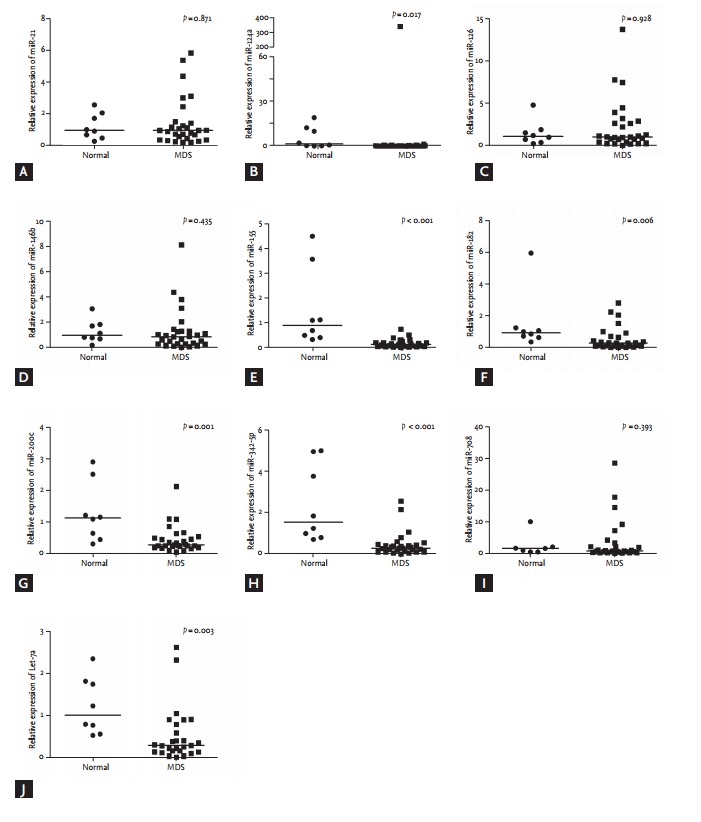
Relative expression of microRNAs using RNU6 as an endogenous control in 26 patients with myelodysplastic syndrome and eight normal controls. Bars indicate median values. (A) miR-21 (B) miR-124a, (C) miR-126, (D) miR-146b, (E) miR-155, (F) miR-182, (G) miR-200c, (H) miR-342-5p, (I) miR- 708, (J) Let-7a. MDS, myelodysplastic syndrome.
When patients were divided into two groups according to IPSS risk stratification (lower [low and intermediate- 1] vs. higher [intermediate-2 and high]), there were significant differences in miR-21 (median, 0.773 vs. 1.212; p = 0.036), miR-126 (median, 0.822 vs. 1.125; p = 0.032), miR-146b (median, 0.560 vs. 1.319; p = 0.006), and miR-155 expression levels (median, 0.098 vs. 0.220; p = 0.002, respectively) between lower and higher risk groups (Fig. 2).
Figure 2.
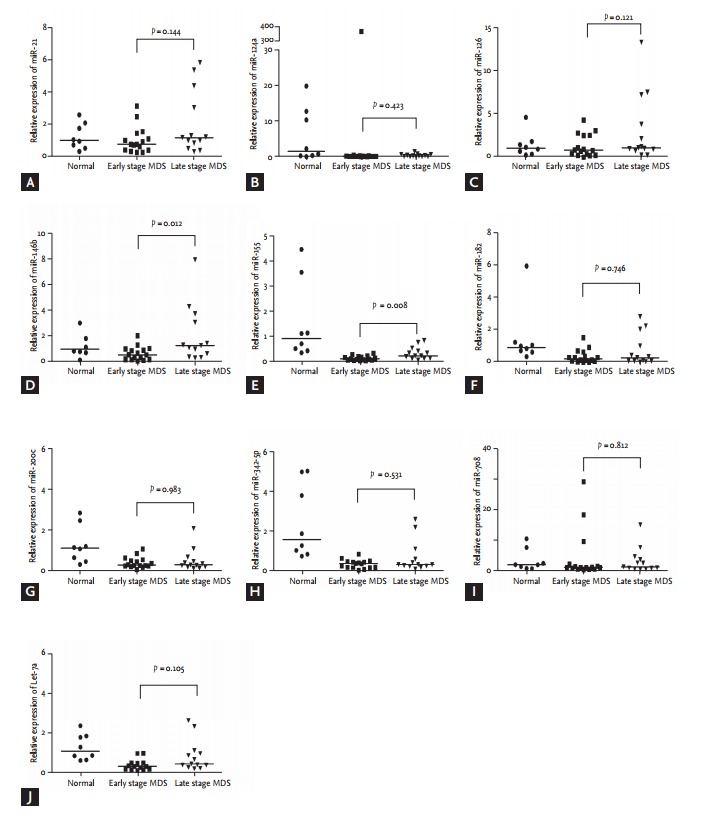
Relative expression of microRNAs using RNU6 as an endogenous control in 26 patients with myelodysplastic syndrome according to international prognostic scoring system (IPSS) risk groups (lower [low and intermediate-1] vs. higher risk [intermediate-2 and high]). The p values were calculated by the non-parametric Mann-Whitney test. Bars indicate median values. (A) miR-21, (B) miR-124a, (C) miR- 126, (D) miR-146b, (E) miR-155, (F) miR-182, (G) miR-200c, (H) miR-342-5p, (I) miR-708, (J) Let-7a. MDS, myelodysplastic syndrome.
MiR-155 expression positively correlated with miR- 342-5p (rs = 0.710, p < 0.001) and Let-7a (rs = 0.650, p < 0.001) expression levels, and there was also a significant positive relationship between miR-200c and miR-342-5p (rs = 0.855, p < 0.001) (Table 2). Table 3 lists common target genes for the positively correlated miRNAs [13].
Table 2.
Correlation of expression levels between microRNAs
| miR-124 | miR-155 | miR-182 | miR-200c | miR-342-5p | Let-7a | |
|---|---|---|---|---|---|---|
| miR-124 | ||||||
| rs | - | 0.512 | 0.478 | 0.005 | 0.028 | 0.682a |
| p value | 0.007 | 0.014 | 0.980 | 0.893 | < 0.001 | |
| miR-155 | ||||||
| rs | 0.512 | - | 0.348 | 0.514 | 0.710a | 0.650a |
| p value | 0.007 | 0.081 | 0.007 | < 0.001 | 0.001 | |
| miR-182 | ||||||
| rs | 0.478 | 0.348 | - | –0.040 | –0.003 | 0.717a |
| p value | 0.014 | 0.081 | 0.847 | 0.988 | < 0.001 | |
| miR-200c | ||||||
| rs | 0.005 | 0.514 | –0.040 | - | 0.855a | 0.221 |
| p value | 0.980 | 0.007 | 0.847 | < 0.001 | 0.299 | |
| miR-342-5p | ||||||
| rs | 0.028 | 0.650a | –0.003 | 0.855a | - | 0.277 |
| p value | 0.893 | 0.001 | 0.988 | < 0.001 | 0.191 | |
| Let-7a | ||||||
| rs | 0.682a | 0.650a | 0.717a | 0.221 | 0.277 | - |
| p value | < 0.001 | 0.001 | < 0.001 | 0.299 | 0.191 |
rs, Spearman rank correlation coefficient.
This indicates the significant positive correlation between two miRNA expressions.
Table 3.
Common target genes for positively correlated microRNAs
| Target gene | Positively correlated microRNAs | ||
|---|---|---|---|
| Eukaryotic translation initiation factor 2C, 4 | miR-155-5p | miR-342-5p | hsa-let-7a-5p |
| Insulin-like growth factor 2 (somatomedin A) | miR-155-5p | miR-342-5p | hsa-let-7a-5p |
| V-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog | miR-155-5p | hsa-let-7a-5p | |
| E2F transcription factor 2 | miR-155-5p | hsa-let-7a-5p | |
| Phosphatase domain containing, paladin 1 | miR-155-5p | hsa-let-7a-5p | |
| Growth differentiation factor 6 | miR-155-5p | hsa-let-7a-5p | |
| Cytochrome b-561 domain containing 1 | miR-155-5p | hsa-let-7a-5p | |
| WW domain binding protein 1-like | miR-155-5p | miR-342-5p | |
| Zinc finger protein 652 | miR-155-5p | miR-342-5p | |
| GATS, stromal antigen 3 opposite strand | miR-155-5p | miR-342-5p | |
| OTU domain, ubiquitin aldehyde binding 2 | miR-155-5p | miR-342-5p | |
| Caspase 3, apoptosis-related cysteine peptidase | miR-182-3p | hsa-let-7a-5p | |
| Ras association (RalGDS/AF-6) domain family member 2 | miR-200c-3p | miR-342-5p | |
| Vac14 homolog (Saccharomyces cerevisiae) | miR-200c-3p | miR-342-5p | |
| Immunoglobulin superfamily, member 10 | miR-200c-3p | miR-342-5p | |
| Sodium channel, voltage gated, type VIII, alpha subunit | miR-200c-3p | miR-342-5p | |
Adapted from National Chiao Tung University [13].
Prognostic significance of miRNA expression in MDS patients
With a median follow up duration of 14.8 months (range, 0.4 to 83.8) for surviving patients, 2-year survival probabilities were 44.0% for OS and 44.6% for LFS. Prognostic factor analysis showed that old age (≥ 65 years) and high bone marrow blast percentage (≥ 5%) were significantly associated with shorter OS and LFS (Table 4). IPSS and R-IPSS were also used to stratify patients into different OS and LFS groups. The 2-year OS probability was significantly more inferior for patients with higher expression levels of miR-126 (24.2% vs. 68.2%, p = 0.039) and miR-155 (0% vs. 60.8%, p = 0.003) than for those with lower expression levels of each miRNA (Fig. 3). The 2-year LFS probability was also significantly more inferior for patients with higher expression levels of miR-126 (25.0% vs. 68.2%, p = 0.038) and miR-155 (0% vs. 61.1%, p = 0.002) than those with lower expression levels (Fig. 3). Higher expression levels of miR-124a also tended to be associated with lower OS (16.7% vs. 54.1% at 2 years, p = 0.084) and lower LFS (16.7% vs. 54.6% at 2 years, p = 0.054), although the differences were not statistically significant.
Table 4.
Prognostic implications of clinical variables at the time of diagnosis and microRNAs
| Variable | OS at 2 yr, % | p value | LFS at 2 yr, % | p value |
|---|---|---|---|---|
| Age, yr | 0.013 | 0.016 | ||
| < 65 | 60.7 | 61.8 | ||
| ≥ 65 | 15.6 | 16.0 | ||
| Sex | 0.354 | 0.405 | ||
| Male | 34.4 | 35.4 | ||
| Female | 56.3 | 57.1 | ||
| Bone marrow blast, % | 0.010 | 0.008 | ||
| < 5 | 68.8 | 68.8 | ||
| ≥ 5 | 21.6 | 22.0 | ||
| IPSS | 0.289 | 0.222 | ||
| Low | 100 | 100 | ||
| Intermediate-1 | 52.9 | 52.9 | ||
| Intermediate-2 | 22.2 | 25.0 | ||
| High | 30.0 | 30.0 | ||
| IPSS | 0.051 | |||
| Lower risk | 57.1 | |||
| Higher risk | 28.3 | |||
| R-IPSS | 0.066 | 0.054 | ||
| Very low/low | 100 | 100 | ||
| Intermediate | 50.8 | 51.9 | ||
| High | 37.5 | 37.5 | ||
| Very high | 18.8 | 18.8 | ||
| miRNA expression | ||||
| miR-124a (75% quartile) | 0.084 | 0.054 | ||
| Lower | 54.1 | 54.6 | ||
| Higher | 16.7 | 16.7 | ||
| miR-126 (median) | 0.039 | 0.038 | ||
| Lower | 68.2 | 68.2 | ||
| Higher | 24.2 | 25.0 | ||
| miR-155 (75% quartile) | 0.003 | 0.002 | ||
| Lower | 60.8 | 61.1 | ||
| Higher | 0 | 0 | ||
| miR-708 (75% quartile) | 0.225 | 0.144 | ||
| Lower | 51.4 | 51.9 | ||
| Higher | 20.0 | 20.0 |
OS, overall survival; LFS, leukemia-free survival; IPSS, international prognostic scoring system; R-IPSS, revised IPSS.
Figure 3.
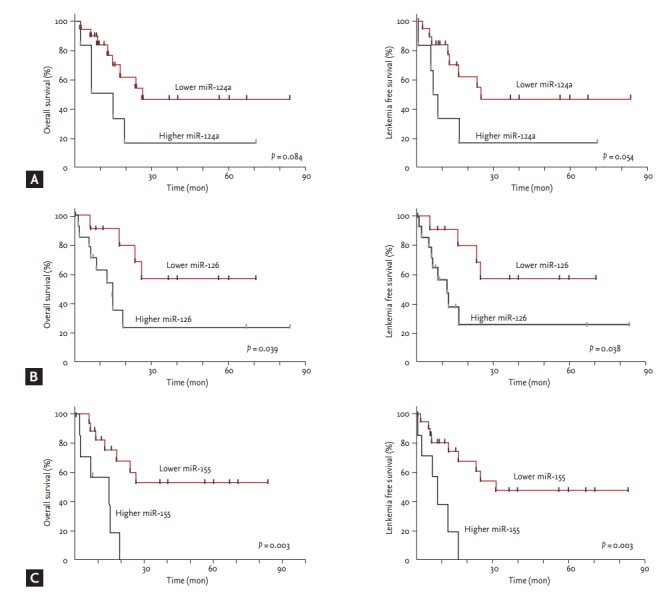
Association of expression levels of microRNA and survivals. (A) miR-124a, (B) miR-126, (C) miR-155.
DISCUSSION
In the present study, we found that MDS patients had significantly lower expression levels of several miRNAs, including miR-124a, miR-155, miR-182, miR-200c, miR-342-5p, and Let-7a, than normal healthy volunteers. Although dysregulation of miR-200c has never been reported in MDS, it has been noted in solid tumors, including colorectal cancer, renal cell carcinoma, and ovarian cancer [14-16]. MiR-200c regulates proliferation, migration, and invasion in breast and ovarian cancers [17], and the expression of miR-200c in endometrial, breast, and ovarian cancer cells inversely correlates with ZEB1 expression [18]. Cyclin-dependent kinase (CDK) 2 is a potential target of miR-200c, which directly suppresses CDK2 expression in renal clear cell carcinoma cell lines and xenografts [16]. Cell cycle control genes, such as CDK2, CDK6, and cyclin A1, are reportedly aberrantly overexpressed in patients with MDS [19,20]. Thus, the underexpression of miR-200c may lead to cell growth and cell cycle progression in MDS by increasing levels of CDK2. Regarding miR-342-5p levels in patients with MDS, there have been controversial results. MiR- 342 is reported to be overexpressed in MDS with del(5q) [21], while it was underexpressed in another study [22]. Down-regulation of miR-124 in patients with MDS has been demonstrated in a previous study [23], and there was an inverse correlation between miR-124 expression and the degree of promoter methylation [24]. The un-derexpression of Let-7a in patients with MDS has also been noted in previous studies [11,23]. Let-7a has been negatively correlated with RAS, and down-regulation of Let-7 family miRNAs might lead to overexpression of RAS [11].
We identified that the expression of miR-21, miR- 146b-5p, miR-126 and miR-155 were significantly higher in higher risk (intermediate-2/high) than in lower risk (low and intermediate-1) according to the IPSS risk groups (lower vs. higher risk) (Fig. 2), and the results suggest that the miRNAs might be associated with MDS progression and leukemic transformation. miR-21 mediates hematopoietic suppression in MDS by activating transforming growth factor β signaling [25].
The apoptotic features of early stage MDS are known to be lost during disease progression to late stage MDS [26]. MiR-146b-5p targets platelet-derived growth factor receptor α, which negatively regulates erythropoiesis and megakaryocytopoiesis [27], and TRAF6 in dendrit-ic cell apoptosis [28]. An in vitro study using K562 cells demonstrated that miR-146b-5p within BCR-ABL1-positive microvesicles promoted leukemic transformation of hematopoietic cells [29]. MiR-146b expression tended to increase according to increment of bone marrow fibrosis grade in patients with myeloproliferative neoplasms [30]. These results suggest that miR-146b-5p inhibits the apoptotic pathway and may have an important role in MDS progression.
We identified that higher expression levels of miR- 126 and miR-155 could not only distinguish higher from lower risk MDS but also significantly correlated with shorter OS and transformation risk to AML in MDS patients. As consistent to our result, Sokol et al. [23] also showed that overexpression of miR-126 in higher risk MDS comparing to lower risk MDS using miRNA array. MiR-126 is known to be associated with angiogenesis and megakaryocytopoiesis [31,32], as it regulates HOXA9 by binding to the homeobox [33]. The cluster of HOX genes has been implicated in early hematopoiesis as well as leukemogenesis [33]. MiR-126 is upregulated by epigenetic therapy with azacitidine or histone deacetylase inhibitor 4-phenylbutyric acid [32].
Our data revealed that miR-155 expressions both in lower and higher risk MDS were lower in normal controls. But, the depth of miR-155 underexpression in MDS did not correlate with the depth of disease. Among MDS patients, the relatively higher expressions of miR- 155 had an adverse prognostic significance. Higher miRNA-155 levels in higher risk MDS compared to lower- risk MDS have also been demonstrated in a previous study [23]. Moreover, miR-155 expression in AML has been also reported to be overexpressed [34]. MiR-155 is a translational repressor of several myeloid transcription factors including PU.1, C/ERBβ, and CSF1R [35,36], and mice transplanted with miR-155-transfected stem cells developed a myeloproliferative disorder with abnormal granulocyte morphology analogous to MDS [35]. The dysregulation of miR-155 within the primitive bone marrow compartment may promote aberrant hematopoietic stem cell self-renewal and progression to AML [5]. MiR-155 targets SHIP-1 gene which was expressed in MDS progenitor cells. Loss of SHIP-1 protein expression may result in myeloid leukemia growth [26]. These findings suggest a role of miR-155 in MDS progression. Thus, we suspect that underexpression of miR-155 may be associated with the development of early stage MDS. And the reincrease of underexpressed miR-155 levels may be a possible mechanism of MDS progression and the leukemic transformation. There may be some genetic alterations other than miR-155 having more important role in the pathogenesis of advanced MDS.
As shown in Fig. 1, miR-124a underexpression may be a role in pathogenesis of MDS. In addition, miR-124a tended to be associated with OS and LFS, although there were no differences in miR-124a levels between higher and lower risk MDS patients. MiR-124a expression inversely correlates with promoter methylation [24]. Thus, relatively higher miR-124a expressions in MDS patients might lead to poor response to hypomethylating agents and this might have an adverse impact on progression to AML and survivals. Further investigations are needed to define the mechanism of miR-124a and miR-155 in MDS pathogenesis and prognosis. Although our study was limited by the relatively small number of patients included, we identified several miRNAs associated with pathogenesis, leukemic transformation, and prognosis in MDS.
KEY MESSAGE
1. MiR-124a, miR-155, miR-182, miR-200c, miR- 342-5p, and Let-7a expressions might be associated with myelodysplastic syndrome (MDS) pathogenesis.
2. MiR-21, miR-126, 146b-5p, and miR-155 might be associated with MDS progression.
3. The higher expressions of miR-126 and miR-155 may predict significantly worse prognosis.
Acknowledgments
The biospecimen and data used in this study were provided by Asan Bio-Resource Center, Korea Biobank Network (2010-0093). This study was supported by a grant from the Korean Health Technology R&D Project, Ministry of Health and Welfare, Republic of Korea (HI12C0129).
Footnotes
No potential conflict of interest relevant to this article was reported.
REFERENCES
- 1.List AF, Vardiman J, Issa JP, DeWitte TM. Myelodysplastic syndromes. Hematology Am Soc Hematol Educ Program. 2004:297–317. doi: 10.1182/asheducation-2004.1.297. [DOI] [PubMed] [Google Scholar]
- 2.Park EH, Lee H, Won YJ, et al. Nationwide statistical analysis of myeloid malignancies in Korea: incidence and survival rate from 1999 to 2012. Blood Res. 2015;50:204–217. doi: 10.5045/br.2015.50.4.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Greenberg P, Cox C, LeBeau MM, et al. International scoring system for evaluating prognosis in myelodysplastic syndromes. Blood. 1997;89:2079–2088. [PubMed] [Google Scholar]
- 4.Greenberg PL, Tuechler H, Schanz J, et al. Revised international prognostic scoring system for myelodysplastic syndromes. Blood. 2012;120:2454–2465. doi: 10.1182/blood-2012-03-420489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rhyasen GW, Starczynowski DT. Deregulation of microRNAs in myelodysplastic syndrome. Leukemia. 2012;26:13–22. doi: 10.1038/leu.2011.221. [DOI] [PubMed] [Google Scholar]
- 6.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 7.Khraiwesh B, Arif MA, Seumel GI, et al. Transcriptional control of gene expression by microRNAs. Cell. 2010;140:111–122. doi: 10.1016/j.cell.2009.12.023. [DOI] [PubMed] [Google Scholar]
- 8.O’Connell RM, Zhao JL, Rao DS. MicroRNA function in myeloid biology. Blood. 2011;118:2960–2969. doi: 10.1182/blood-2011-03-291971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choi JS, Nam MH, Yoon SY, Kang SH. MicroRNA-194-5p could serve as a diagnostic and prognostic biomarker in myelodysplastic syndromes. Leuk Res. 2015;39:763–768. doi: 10.1016/j.leukres.2015.04.013. [DOI] [PubMed] [Google Scholar]
- 10.Kim Y, Cheong JW, Kim YK, et al. Serum microRNA-21 as a potential biomarker for response to hypomethylating agents in myelodysplastic syndromes. PLoS One. 2014;9:e86933. doi: 10.1371/journal.pone.0086933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vasilatou D, Papageorgiou SG, Kontsioti F, et al. Expression analysis of mir-17-5p, mir-20a and let-7a microRNAs and their target proteins in CD34+ bone marrow cells of patients with myelodysplastic syndromes. Leuk Res. 2013;37:251–258. doi: 10.1016/j.leukres.2012.11.011. [DOI] [PubMed] [Google Scholar]
- 12.Vardiman JW, Thiele J, Arber DA, et al. The 2008 revision of the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia: rationale and important changes. Blood. 2009;114:937–951. doi: 10.1182/blood-2009-03-209262. [DOI] [PubMed] [Google Scholar]
- 13.National Chiao Tung University . Hsinchu (TW): National Chiao Tung University; c2017. miRTarBase [Internet] [cited 2016 Apr 20]. Available from: http://mirtarbase.mbc.nctu.edu.tw. [Google Scholar]
- 14.Shelygin YA, Shubin VP, Frolov SA, et al. The analysis of microRNAs miR-200C and miR-145 expression in colorectal cancer of different molecular subtypes. Dokl Biochem Biophys. 2015;463:243–246. doi: 10.1134/S1607672915040122. [DOI] [PubMed] [Google Scholar]
- 15.Ibrahim FF, Jamal R, Syafruddin SE, et al. MicroRNA-200c and microRNA-31 regulate proliferation, colony formation, migration and invasion in serous ovarian cancer. J Ovarian Res. 2015;8:56. doi: 10.1186/s13048-015-0186-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang X, Chen X, Han W, et al. miR-200c targets CDK2 and suppresses tumorigenesis in renal cell carcinoma. Mol Cancer Res. 2015;13:1567–1577. doi: 10.1158/1541-7786.MCR-15-0128. [DOI] [PubMed] [Google Scholar]
- 17.Wang J, Zhao H, Tang D, Wu J, Yao G, Zhang Q. Overexpressions of microRNA-9 and microRNA-200c in human breast cancers are associated with lymph node metastasis. Cancer Biother Radiopharm. 2013;28:283–288. doi: 10.1089/cbr.2012.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cochrane DR, Spoelstra NS, Howe EN, Nordeen SK, Richer JK. MicroRNA-200c mitigates invasiveness and restores sensitivity to microtubule-targeting chemotherapeutic agents. Mol Cancer Ther. 2009;8:1055–1066. doi: 10.1158/1535-7163.MCT-08-1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shi J, Shao ZH, Liu H, et al. Expression of cell cycle control genes in myelodysplastic syndromes. Zhonghua Xue Ye Xue Za Zhi. 2005;26:10–14. [PubMed] [Google Scholar]
- 20.Jia JS, Xu SR. Expression of cyclin A1 mRNA in patients with myelodysplastic syndrome and its clinical significance. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2009;17:377–381. [PubMed] [Google Scholar]
- 21.Hussein K, Theophile K, Busche G, et al. Aberrant microRNA expression pattern in myelodysplastic bone marrow cells. Leuk Res. 2010;34:1169–1174. doi: 10.1016/j.leukres.2010.04.012. [DOI] [PubMed] [Google Scholar]
- 22.Votavova H, Grmanova M, Dostalova Merkerova M, et al. Differential expression of microRNAs in CD34+ cells of 5q- syndrome. J Hematol Oncol. 2011;4:1. doi: 10.1186/1756-8722-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sokol L, Caceres G, Volinia S, et al. Identification of a risk dependent microRNA expression signature in myelodysplastic syndromes. Br J Haematol. 2011;153:24–32. doi: 10.1111/j.1365-2141.2011.08581.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dickstein J, Senyuk V, Premanand K, et al. Methylation and silencing of miRNA-124 by EVI1 and self-renewal exhaustion of hematopoietic stem cells in murine myelodysplastic syndrome. Proc Natl Acad Sci U S A. 2010;107:9783–9788. doi: 10.1073/pnas.1004297107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bhagat TD, Zhou L, Sokol L, et al. miR-21 mediates hematopoietic suppression in MDS by activating TGF-β signaling. Blood. 2013;121:2875–2881. doi: 10.1182/blood-2011-12-397067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee DW, Futami M, Carroll M, et al. Loss of SHIP-1 protein expression in high-risk myelodysplastic syndromes is associated with miR-210 and miR-155. Oncogene. 2012;31:4085–4094. doi: 10.1038/onc.2011.579. [DOI] [PubMed] [Google Scholar]
- 27.Zhai PF, Wang F, Su R, et al. The regulatory roles of microRNA-146b-5p and its target platelet-derived growth factor receptor α (PDGFRA) in erythropoiesis and megakaryocytopoiesis. J Biol Chem. 2014;289:22600–22613. doi: 10.1074/jbc.M114.547380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park H, Huang X, Lu C, Cairo MS, Zhou X. MicroRNA- 146a and microRNA-146b regulate human dendritic cell apoptosis and cytokine production by targeting TRAF6 and IRAK1 proteins. J Biol Chem. 2015;290:2831–2841. doi: 10.1074/jbc.M114.591420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang HM, Li Q, Zhu X, et al. miR-146b-5p within BCR-ABL1-positive microvesicles promotes leukemic transformation of hematopoietic cells. Cancer Res. 2016;76:2901–2911. doi: 10.1158/0008-5472.CAN-15-2120. [DOI] [PubMed] [Google Scholar]
- 30.Ha JS, Jung HR. Up-regulation of MicroRNA 146b is associated with myelofibrosis in myeloproliferative neoplasms. Ann Clin Lab Sci. 2015;45:308–314. [PubMed] [Google Scholar]
- 31.Garzon R, Pichiorri F, Palumbo T, et al. MicroRNA fingerprints during human megakaryocytopoiesis. Proc Natl Acad Sci U S A. 2006;103:5078–5083. doi: 10.1073/pnas.0600587103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saito Y, Friedman JM, Chihara Y, Egger G, Chuang JC, Liang G. Epigenetic therapy upregulates the tumor suppressor microRNA-126 and its host gene EGFL7 in human cancer cells. Biochem Biophys Res Commun. 2009;379:726–731. doi: 10.1016/j.bbrc.2008.12.098. [DOI] [PubMed] [Google Scholar]
- 33.Shen WF, Hu YL, Uttarwar L, Passegue E, Largman C. MicroRNA- 126 regulates HOXA9 by binding to the homeobox. Mol Cell Biol. 2008;28:4609–4619. doi: 10.1128/MCB.01652-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garzon R, Volinia S, Liu CG, et al. MicroRNA signatures associated with cytogenetics and prognosis in acute myeloid leukemia. Blood. 2008;111:3183–3189. doi: 10.1182/blood-2007-07-098749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O’Connell RM, Rao DS, Chaudhuri AA, et al. Sustained expression of microRNA-155 in hematopoietic stem cells causes a myeloproliferative disorder. J Exp Med. 2008;205:585–594. doi: 10.1084/jem.20072108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Georgantas RW, 3rd, Hildreth R, Morisot S, et al. CD34+ hematopoietic stem-progenitor cell microRNA expression and function: a circuit diagram of differentiation control. Proc Natl Acad Sci U S A. 2007;104:2750–2755. doi: 10.1073/pnas.0610983104. [DOI] [PMC free article] [PubMed] [Google Scholar]


