Abstract
BACKGROUND
Adiponectin (ADIPOQ) is an important factor involved in the regulation of both carbohydrate and lipid metabolism. Polymorphisms in the ADIPOQ gene are known to influence an individual’s predisposition to metabolic syndrome and type 2 diabetes. Moreover, women with gestational diabetes mellitus (GDM) are at an increased risk of developing type 2 diabetes. Several studies have been conducted previously to assess the association between ADIPOQ polymorphisms and GDM; however, the results of the association are inconclusive.
AIM
To quantitatively evaluate the association between ADIPOQ +45T/G, +276G/T, and -11377C/G polymorphisms and the risk of GDM.
METHODS
A systematic search of EMBASE, PubMed, CNKI, Web of Science, and WANFANG DATA was conducted up to October 20, 2018. We calculated merged odds ratios (ORs) with 95% confidence intervals (CIs) using a fixed-effects or random-effects model depending on the between-study heterogeneity to evaluate the association between AIDPOQ +45T/G, +276G/T, and -11377C/G polymorphisms and the risk of GDM. Subgroup analysis was performed by ethnicity. Publication and sensitivity bias analyses were performed to test the robustness of the association. All statistical analyses were conducted using Stata12.0.
RESULTS
Nine studies of +45T/G included 1024 GDM cases and 1059 controls, five studies of +276G/T included 590 GDM cases and 595 controls, and five studies of -11377C/G included 722 GDM cases and 791 controls. Pooled ORs indicated that +45T/G increased GDM risk in Asians (allelic model: OR = 1.47, 95%CI: 1.27-1.70, P = 0.000; dominant model: OR = 1.54, 95%CI: 1.27-1.85, P = 0.000; recessive model: OR=2.00, 95%CI: 1.43-2.85, P = 0.000), not in South Americans (allelic model: OR = 1.21, 95%CI: 0.68-2.41, P = 0.510; dominant model: OR = 1.13, 95%CI: 0.59-2.15, P = 0.710; recessive model: OR = 2.18, 95%CI: 0.43-11.07, P = 0.350). There were no significant associations between +276G/T (allelic model: OR = 0.88, 95%CI: 0.74-1.05, P = 0.158; dominant model: OR = 0.91, 95%CI: 0.65-1.26, P = 0.561; recessive model: OR = 0.82, 95%CI: 0.64-1.05, P = 0.118) or -11377C/G (allelic model: OR = 0.96, 95%CI: 0.72-1.26, P = 0.750; dominant model: OR = 1.00, 95%CI: 0.73-1.37, P = 0.980; recessive model: OR = 0.90, 95%CI: 0.61-1.32, P = 0.570) and the risk of GDM.
CONCLUSION
Our meta-analysis shows the critical role of the ADIPOQ +45T/G polymorphism in GDM, especially in Asians. Studies focused on delineating ethnicity-specific factors with larger sample sizes are needed.
Keywords: Gestational diabetes mellitus, Single nucleotide, Polymorphism, Adiponectin, Gene, Meta-analysis
Core tip: No consensus is available in the literature about the association of adiponectin gene polymorphisms and the risk of gestational diabetes mellitus (GDM). As far as we know, only +45T/G was involved in a previous meta-analysis with a small sample size and obvious heterogeneity. We evaluated the association between ADIPOQ +45T/G, +276G/T, and -11377C/G polymorphisms and GDM with a bigger sample size, less heterogeneity. Moreover, subgroup analysis was performed by ethnicity.
INTRUDUCTION
Gestational diabetes mellitus (GDM) is a condition of impaired glucose tolerance during pregnancy in women without a previous diagnosis of diabetes. It is associated with serious complications for both mother and child in the pre- and postnatal periods[1]. Many kinds of risk factors contribute to GDM, such as ethnicity, genetics, family history, dietary habits, and physical activity[1]. Obesity is a usual risk factor for GDM and can cause insulin resistance. Many biochemical mediators compounded in the adipose tissue and secreted in the circulatory system, such as resistin, adiponectin (ADIPOQ), and leptin, are suggested to correlate with obesity and insulin resistance[2].
ADIPOQ is produced in the adipose tissue and modulates various metabolic processes, including lipid metabolism, glucose and fatty acid oxidation. This hormone decreases insulin resistance, improves lipid metabolism, and exerts anti-inflammatory properties. Decreased plasma ADIPOQ levels were observed in patients with type 2 diabetes (T2D), metabolic syndrome, and obesity[3]. During normal pregnancy, ADIPOQ levels progressively decline, with its plasma concentration reaching even lower in GDM women[1]. Previous studies suggested that ADIPOQ gene single nucleotide polymorphisms (SNPs) could influence the concentration of plasma ADIPOQ and subsequently insulin sensitivity[4-6].
Studies have paid attention to the SNPs +45T/G in exon 2 and +276G/C in intron 2, -11391G/A, and -11377C/G in the promoter region. The two ADIPOQ linkage disequilibrium blocks are where these four variants located within. Block 1 comprises the promoter sequence spanning the region -14811 to -4120, and block 2 encompasses the exons in the region -450 to +4545[2]. The conclusions of these studies have been disputed regarding whether the metabolic phenotypes of GDM are influenced by the variability at this locus and which polymorphisms contribute to this effect. For example, Low et al[7] reported that a significant association was found between SNP 45T/G and GDM, and normal patients with the TT genotype had significantly higher plasma ADIPOQ levels compared to those with the TG or GG genotype. Beltcheva et al[1] reported that -11377C/G is associated with GDM. According to Daher et al[8], GDM is not associated with +45T/G and -11377C/G polymorphisms. Reasons for the conflicting results are small sample sizes in a single study and the hereditary difference of ethnicity.
As the results are discrepant and +45T/G was the only polymorphism which participated in the meta-analysis, our study was meant to evaluate whether and to what extent ADIPOQ gene polymorphisms contribute to GDM.
MATERIALS AND METHODS
Literature search strategy
Electronic databases PubMed, EMBASE, Web of Science, WANFANG DATA, and CNKI were used to search possibly association articles on human genetic studies of ADIPOQ and GDM that had been published up to 20 October 2018. The search terms used were: “Gestational diabetes mellitus” or “GDM” and “adiponectin” or “ADIPOQ” and “single nucleotide polymorphism” or “polymorphism”.
Selection criteria and data extraction
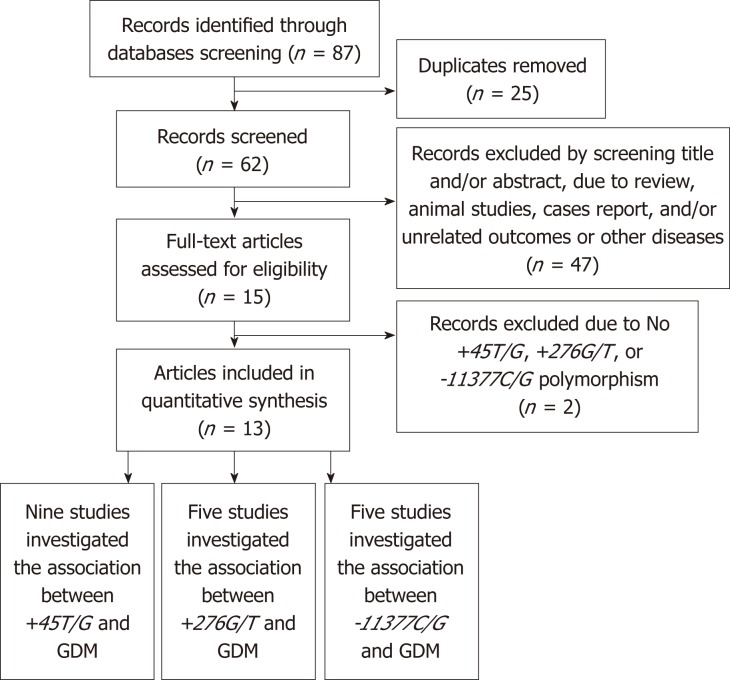
We searched the database and identified 87 articles. The selection criteria of the publications were as follow: (1) well-designed case control studies on genetic association of ADIPOQ and the risk of GDM; (2) clear diagnostic criteria for GDM; (3) independent and sufficient genotype data must be contained in the original papers and the data can calculate odds ratios (ORs) and 95% confidence intervals (CIs); and (4) there should be at least two articles that have studied each polymorphism that we used in our meta-analysis. During the selection, we removed 25 articles for duplicate publication and excluded 49 articles for review, animal studies, case reports with unrelated outcomes or other diseases, and articles with no ADIPOQ +45T/G, +276G/T, or -11377C/G reported.
Finally, 13 articles were adopted in this meta-analysis. Among them, nine studies investigated +45T/G, five studies investigated +276G/T, and five studies investigated -11377C/G. A flow diagram of study selection is presented in Figure 1.
Figure 1.
Flow diagram of study selection.
Data were extracted by two researchers independently. We have extracted the following information from every included study: first author, year of publication, country, ethnicity, matching criteria, genotyping method, numbers of cases and controls, minor allele frequency in controls, and Hardy-Weinberg equilibrium (HWE) status. We obtained the HWE status of controls by calculating from genotype distributions using STATA12.0. The Newcastle-Ottawa quality assessment scale (NOS) was used for quality assessment of primary studies. The study would be regarded as a high-quality study when it had an NOS scores ≥ 6 (Table 1)[9].
Table 1.
Quality assessment of included case control studies using the Newcastle-Ottawa scale
| Author | Yr |
Selection |
Comparability |
Exposure |
Score | ||||||
| Case definition | Case representativeness | Control selection | Control definition | Important confounders | Every confounders | Ascertainment | Consistency | Non-response rate | |||
| Low et al[7] | 2011 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 7 |
| Takhshid et al[2] | 2015 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 7 |
| Daher et al[8] | 2011 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 8 |
| Han et al[4] | 2012 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 7 |
| Gao et al[10] | 2016 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 8 |
| Luan et al[13] | 2015 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 8 |
| Li et al[12] | 2017 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 8 |
| Li et al[11] | 2013 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 7 |
| Zhang et al[14] | 2014 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 8 |
| Beltcheva et al[1] | 2014 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 7 |
| Pawlik et al[3] | 2017 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 7 |
| Chen et al[15] | 2011 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 7 |
| Wang et al[16] | 2016 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 7 |
The quality evaluation content mainly includes three aspects: case selection, baseline comparability, and exposure factors with nine evaluation items. If the evaluation item is met, 1 point is obtained, otherwise 0, and the score ranges from 0 to 9.
Statistical analysis
We used Stata12.0 software for statistical analyses. A fixed-effects or random-effects model was used to merge OR and 95%CI based on allelic models, recessive models, and dominant models to evaluate the association between each genetic variant and the risk of GDM. The Z-test was used for determining the significance of the merged OR. P < 0.05 was considered statistically significant.
We used the Cochran Q test to assess the heterogeneity among the studies and Higgins I2 statistic for quantifying the heterogeneity. We used the random-effects model as the merging method when the variant association presented significant interstudy heterogeneity (Q test, P-value < 0.05, or I2 > 50%), otherwise, we used the fixed-effects model. Subgroup analysis was performed based on the ethnicity of the study population to evaluate ethnic-specific effects. Publication bias was tested by Begg’s funnel plot.
The statistical methods of this study were reviewed by Shi-Min Hu from Department of Epidemiology and Health Statistics, Xiangya School of Public Health, Central South University.
RESULTS
Main characteristics of all the included studies
Thirteen studies were adopted in this meta-analysis; among them, nine studies were about the association of +45T/G and GDM[2,4,7,8,10-14], five studies were about the association of +276G/T and GDM[4,10,12-14], and five studies were about the association of -11377C/G and GDM[1,3,8,15,16]. In total, 1667 GDM cases and 1682 controls were included; ten studies were from Asian descendants[2,4,7,10-15], two studies were from European descendants[1,3], and one study was from a South American descendant[8]. Detailed characteristics of all studies included are shown in Table 2.
Table 2.
Detailed characteristics of all eligible studies for the association between ADIPOQ single nucleotide polymorphism and gestational diabetes mellitus
| SNP | Author | Yr | Country | Ethnicity | Matching criteria | Method |
Sample size |
Genotype1 |
HWE | ||
| Case | Control | Case | Control | ||||||||
| +45T/G | Low et al[7] | 2011 | Malaysia | Asian | NR | Taq PCR | 26 | 53 | 11/13/2 | 35/17/1 | 0.51 |
| Takhshid et al[2] | 2015 | Iran | Asian | Age | PCR-RELF | 65 | 70 | 37/28/0 | 54/16/0 | 0.28 | |
| Daher et al[8] | 2011 | Brazil | SA | Race | PCR-RELF | 79 | 169 | 61/15/3 | 134/32/3 | 0.51 | |
| Han et al[4] | 2012 | China | Asian | NR | PCR-RELF | 152 | 120 | 63/71/18 | 64/50/6 | 0.34 | |
| Gao et al[10] | 2016 | China | Asian | Age, GW | PCR-RELF | 150 | 150 | 59/66/25 | 81/57/12 | 0.66 | |
| Luan et al[13] | 2015 | China | Asian | Age, GW | NR | 60 | 60 | 33/21/6 | 29/26/5 | 0.81 | |
| Li et al[12] | 2017 | China | Asian | Age, GW | PCR-RELF | 130 | 130 | 53/63/14 | 63/60/7 | 0.13 | |
| Li et al[11] | 2013 | China | Asian | NR | Sequencing | 264 | 172 | 134/113/17 | 97/66/9 | 0.6 | |
| Zhang et al[14] | 2014 | China | Asian | Age, BMI, GW | PCR-RELF | 98 | 135 | 38/43/17 | 73/51/11 | 0.62 | |
| +276G/T | Han et al[4] | 2012 | China | Asian | NR | PCR-RELF | 152 | 120 | 12/66/74 | 11/53/56 | 0.34 |
| Gao et al[10] | 2016 | China | Asian | Age, GW | PCR-RELF | 150 | 150 | 15/69/66 | 15/60/75 | 0.66 | |
| Luan et al[13] | 2015 | China | Asian | Age, GW | NR | 60 | 60 | 7/26/27 | 3/25/32 | 0.81 | |
| Li et al[12] | 2017 | China | Asian | Age, GW | PCR-RELF | 130 | 130 | 64/58/8 | 60/56/14 | 0.13 | |
| Zhang et al[14] | 2014 | China | Asian | Age, BMI, GW | PCR-RELF | 98 | 135 | 10/45/43 | 13/54/68 | 0.62 | |
| -11377C/G | Beltcheva et al[1] | 2014 | Bulgaria | European | Age, BMI | TaqMan | 130 | 130 | 80/44/6 | 66/50/14 | 0.34 |
| Pawlik et al[3] | 2017 | Poland | European | NR | TaqMan | 204 | 207 | 92/91/21 | 115/75/17 | 0.34 | |
| Daher et al[8] | 2011 | Brazil | SA | race | PCR-RELF | 79 | 169 | 54/20/5 | 105/50/13 | 0.05 | |
| Chen et al[15] | 2011 | China | Asian | NR | PCR-RELF | 103 | 97 | 55/43/5 | 50/38/9 | 0.65 | |
| Wang et al[16] | 2016 | China | Asian | NR | PCR-RELF | 206 | 189 | 107/84/15 | 106/73/10 | 0.57 | |
Genotype is presented as wild type/heterozygous/homozygous. ADIPOQ: Adiponectin; GDM: Gestational diabetes mellitus; SNP: Single nucleotide polymorphism; SA: South American; GW: Gestational week; PCR-RELF: Polymerase chain reaction-restriction fragment length polymorphism; BMI: Body mass index; HWE: Hardy-Weinberg equilibrium; NR: Not reported.
Association between +45T/G and GDM
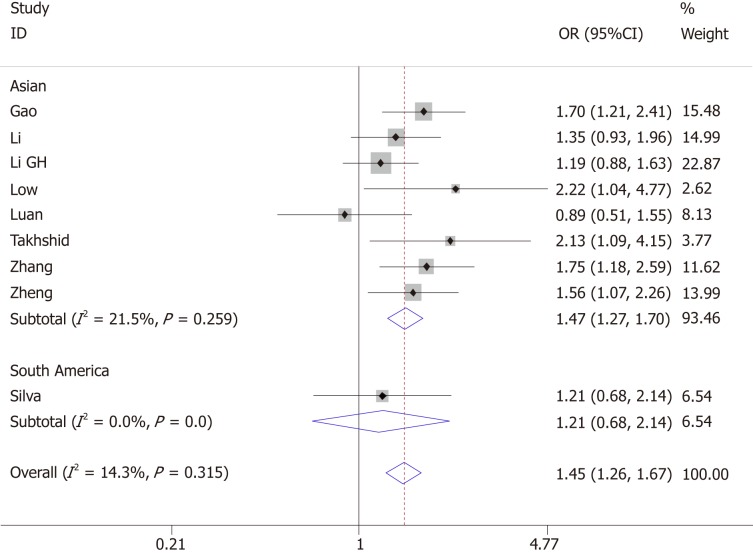
Nine articles evaluated the association of +45T/G and GDM; eight of them were conducted in Asia and 1 in South America, with a total of 1024 GDM cases and 1059 controls. Heterogeneity test revealed P > 0.05 and I² < 50%; the fixed-effects model was used.
The pooled results suggested a significant association between +45T/G and GDM (allelic model: OR = 1.45, 95%CI: 1.26-1.67; dominant model: OR = 1.50, 95%CI: 1.25-1.79; recessive model: OR = 2.00, 95%CI: 1.42-2.84). Ethnicity-based subgroup analysis showed that +45T/G was associated with GDM in Asians (allelic model: OR = 1.47, 95%CI: 1.27-1.70; dominant model: OR = 1.54, 95%CI: 1.27-1.85; recessive model: OR = 2.00, 95%CI: 1.43-2.85). However, there was no association of +45T/G with the risk of GDM in South Americans (allelic model: OR = 1.21, 95%CI: 0.68-2.41; dominant model: OR = 1.13, 95%CI: 0.59-2.15; recessive model: OR=2.18, 95%CI: 0.43-11.07) (Figure 2 and Table 3).
Figure 2.
Forest plot for the association of ADIPOQ +45T/G polymorphism and gestational diabetes mellitus under the allelic model.
Table 3.
Main results of the pooled odds ratios in meta-analysis for the association between ADIPOQ polymorphisms and gestational diabetes mellitus
| SNP | N |
Sample size |
Allelic model |
Dominant model |
Recessive model |
||||
| Case | Control | OR (95%CI) | P | OR (95%CI) | P | OR (95%CI) | P | ||
| +45T/G | |||||||||
| Total | 10 | 1024 | 1059 | 1.45 (1.26-1.67) | 0.000a | 1.50 (1.25-1.79) | 0.000a | 2.00 (1.42-2.84) | 0.000a |
| Subgroup | |||||||||
| Ethnicity | |||||||||
| Asian | 8 | 945 | 890 | 1.47 (1.27-1.70) | 0.000a | 1.54 (1.27-1.85) | 0.000a | 2.00 (1.43-2.85) | 0.000a |
| SA | 1 | 79 | 169 | 1.21 (0.68-2.41) | 0.510 | 1.13 (0.59-2.15) | 0.710 | 2.18 (0.43-11.07) | 0.350 |
| +276G/T | |||||||||
| Total | 5 | 590 | 595 | 0.88 (0.74-1.05) | 0.158 | 0.91 (0.65-1.26) | 0.561 | 0.82 (0.64-1.05) | 0.118 |
| -11377C/G | |||||||||
| Total | 5 | 722 | 791 | 0.96 (0.72-1.26) | 0.750 | 1.00 (0.73-1.37) | 0.980 | 0.90 (0.61-1.32) | 0.570 |
| Subgroup | |||||||||
| Ethnicity | |||||||||
| Asian | 2 | 309 | 286 | 1.04 (0.77-1.41) | 0.800 | 1.09 (0.79-1.50) | 0.600 | 0.97 (0.51-1.86) | 0.930 |
| SA | 1 | 79 | 168 | 0.80 (0.50-1.29) | 0.360 | 0.77 (0.44-1.36) | 0.370 | 0.81 (0.28-2.34) | 0.690 |
| European | 2 | 334 | 337 | 0.94 (0.45-1.96) | 0.870 | 1.00 (0.42-2.33) | 0.990 | 0.87 (0.51-1.49) | 0.610 |
P < 0.01. ADIPOQ: Adiponectin; SA: South American; GDM: Gestational diabetes mellitus; SNP: Single nucleotide polymorphism; N: Number of studies; OR: Odds ratio; CI: Confidence interval; NA: Not assessable.
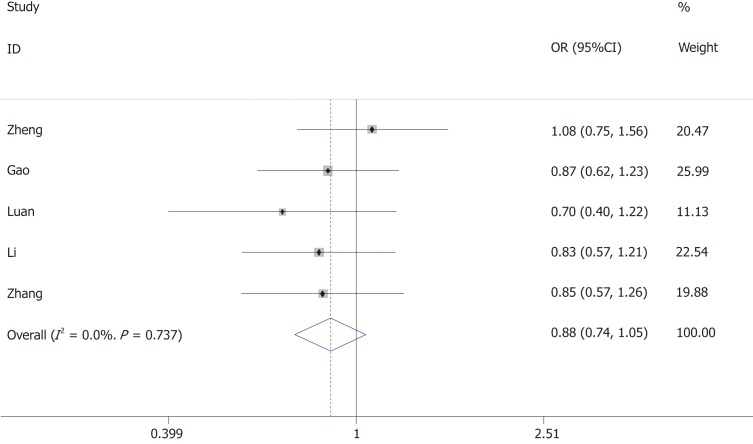
Association between +276G/T and GDM
The association between +276G/T and the risk of GDM was investigated by five studies, including 590 GDM cases and 595 controls. Heterogeneity test revealed P > 0.05 and I² < 50%, and the fixed-effects model was used.
The results showed that +276G/T was not associated with the risk of GDM (allelic model: OR = 0.88, 95%CI: 0.74-1.05; dominant model: OR = 0.91, 95%CI: 0.65-1.26; recessive model: OR = 0.82, 95%CI: 0.64-1.05) (Figure 3 and Table 3).
Figure 3.
Forest plot for the association of ADIPOQ +276G/T polymorphism and gestational diabetes mellitus under the allelic model.
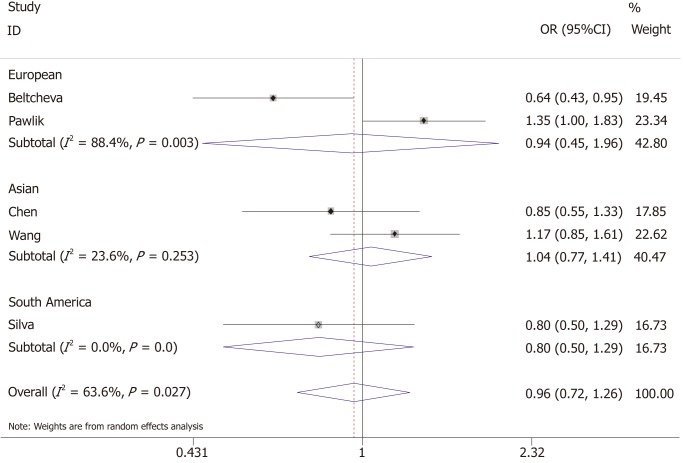
Association between -11377C/G and GDM
The association between -11377C/G and the risk of GDM was investigated by five studies, of which two were conducted in Asia, two conducted in Europe, and one in South America, with a total of 722 GDM cases and 791 controls. Heterogeneity test revealed P < 0.05 and I² > 50%; the random-effects model was used.
The results showed that -11377C/G was not associated with the risk of GDM (allelic model: OR = 0.96, 95%CI: 0.72-1.26; dominant model: OR = 1.00, 95%CI: 0.73-1.37; recessive model: OR = 0.90, 95%CI: 0.61-1.32). Ethnicity-based subgroup analysis also showed that -11377C/G was not associated with GDM in Asian (allelic model: OR = 1.04, 95%CI: 0.77-1.41; dominant model: OR = 1.09, 95%CI: 0.79-1.50; recessive model: OR = 0.97, 95%CI: 0.51-1.86), European (allelic model: OR = 0.94, 95%CI: 0.45-1.96; dominant model: OR = 1.00, 95%CI: 0.42-2.33; recessive model: OR = 0.87, 95%CI: 0.51-1.49) and South American populations (allelic model: OR = 0.80, 95%CI: 0.50-1.29; dominant model: OR = 0.77, 95%CI: 0.44-1.36; recessive model: OR = 0.81, 95%CI: 0.51-1.86) (Figure 4 and Table 3).
Figure 4.
Forest plot for the association of ADIPOQ -11377C/G polymorphism and gestational diabetes mellitus under the allelic model.
Publication bias
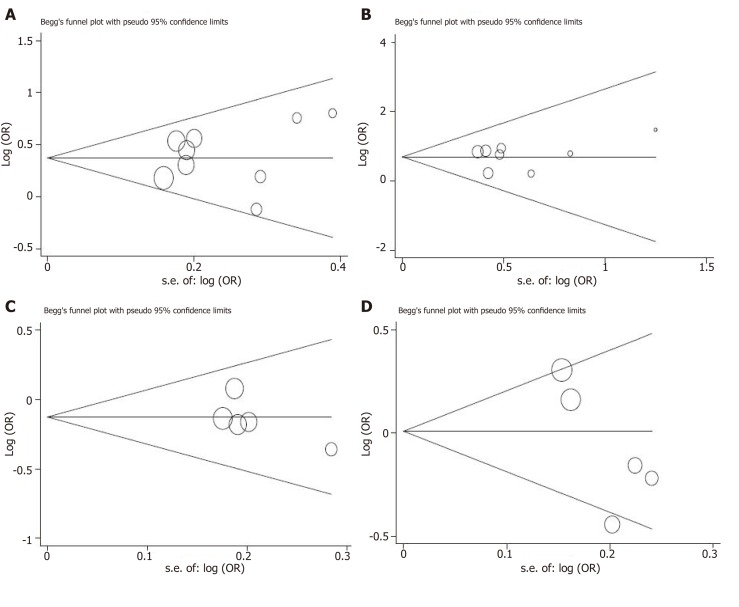
We used the Egger regression asymmetry test and Begg’s funnel plot to assess the public bias of the studies. The evidence of publication bias cannot be found in the meta-analysis of +45T/G (allelic model: continuity corrected P-value = 1.000, Egger regression asymmetry test t = -0.62, P = 0.554; recessive model: continuity corrected P-value = 0.466, Egger regression asymmetry test t = -0.15, P = 0.883), +276G/T (allelic model: continuity corrected P-value = 0.26, Egger regression asymmetry test t = -1.24, P = 0.282) and -11377C/G (allelic model: continuity corrected P-value = 0.221, Egger regression asymmetry test t = -2.48, P = 0.089)(Figure 5).
Figure 5.
Begg’s funnel plots for testing publication bias. A: +45T/G under allelic model; B: +45T/G under recessive model; C: +276G/T under allelic model; D: -11377C/G under allelic model. OR: Odds ratio.
Sensitivity analysis
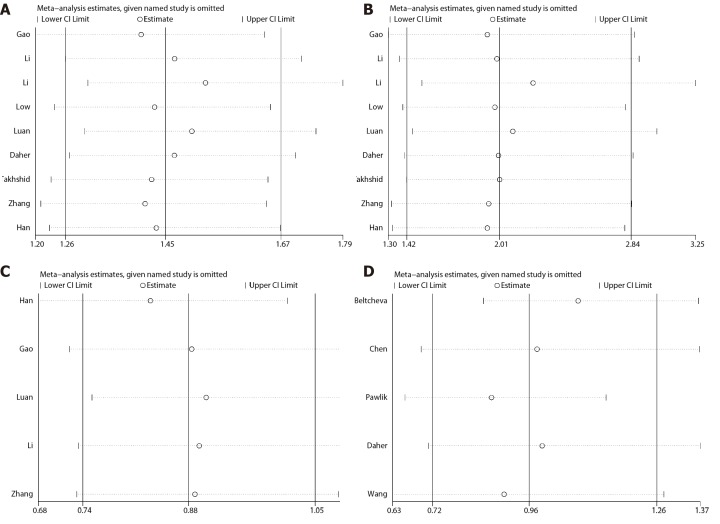
To assess the stability of the results, we performed the sensitivity analysis by sequentially excluding individual studies for each meta-analysis. For the association between +45T/G, +276G/T, or -11377C/G and GDM, there was no significant change of corresponding merged ORs when one study was sequentially excluded from every meta-analysis. Therefore, the results of our meta-analysis are stable and reliable (Figure 6).
Figure 6.
Sensitivity analysis between ADPIOQ polymorphisms and gestational diabetes mellitus in all studies. A: +45T/G under allelic model; B: +45T/G under recessive model; C: +276G/T under allelic model; D: -11377C/G under allelic model. CI: Confidence interval.
DISCUSSION
ADIPOQ has been considered an important factor in regulating glucose and lipid metabolism. It is secreted by adipose tissue, which has a negative correlation with insulin resistance, T2D, and metabolic syndrome. ADIPOQ can increase insulin sensitivity, anti-inflammation, and anti-atherosclerosis, promote glucose uptake in muscle tissue, and inhibit intrahepatic synthetic glucose[17].
In the chromosomal region where the ADIPOQ gene is located, there are susceptible sites of T2D and metabolic syndrome, and its SNPs can affect the level of ADIPOQ in blood, leading to obesity, insulin resistance, and the occurrence of T2D[15].
Plasma ADIPOQ levels gradually decreased with gestational week during pregnancy, consistent with the gradual decrease of insulin sensitivity[3], and plasma adiponectin levels decreased more significantly in GDM women. This phenomenon is closely related to the decreased transcriptional activity of ADIPOQ during pregnancy. Previous studies revealed the association of ADIPOQ SNPs, such as +45T/G[1,2,4,7,8,10-14], +276G/T[1,4,10,12,13,18], -11377C/G[1,3,8,15,16], -3971A/G[13,19], and -11426A/G[15,19], and the risk of GDM. A total of 66.7% (6 of 9) of the studies adopted in this meta-analysis reported that the +45T/G polymorphism increased the risk of GDM, and 40% (2 of 5) reported that the -11377C/G polymorphism was associated with GDM. A higher prevalence of the G allele was observed among women with GDM. All studies regarding +276G/T reported that this polymorphism had no association with the risk of GDM.
Thirteen studies were included in our study; nine studies were about +45T/G, with 1024 cases and 1059 controls, five studies were about +276G/T, with 590 cases and 595 controls, and five studies were about -11377C/G, with 722 cases and 791 controls. We not only had a larger sample size than previous studies but also performed subgroup analysis based on the ethnicity of the study population to evaluate ethnic-specific effects. +45T/G was proved by our meta-analysis to be a risk factor for GDM (allelic model: OR = 1.45, 95%CI: 1.26-1.67, P = 0.000), and 66.7% (6 of 9) of studies reported a positive result[2,4,7,10,12,14]. Subgroup analysis showed that +45T/G was associated with GDM in Asians (allelic model: OR = 1.47, 95%CI: 1.27-1.70, P = 0.000) but not in South Americans. In addition, no association of +276G/T or -11377C/G and the risk of GDM was observed.
Obvious heterogeneity was detected among the -11377C/G studies (allelic model: I2 = 64.0%, P = 0.03, dominant model: I2 = 55.0%, P = 0.06, recessive model: I2 = 32.0%, P = 0.21). We used subgroup analysis based on ethnicity, and the heterogeneity could not be reduced, indicating that a small sample size and other reasons may have influenced the heterogeneity. The association of -11377C/G with the risk of GDM remains to be verified by further studies. No heterogeneity was found in the studies of +45T/G (allelic model: I2 = 14.3%, P = 0.32, dominant model: I2 = 18.1%, P = 0.28, recessive model: I2 = 0.0%, P = 0.90) and +276G/T (allelic model: I2 = 0.0%, P = 0.74, dominant model: I2 = 0.0%, P = 0.78, recessive model: I2 = 0.0%, P = 0.83), so the conclusion that +45T/G has, but +276G/T has no, association with the risk of GDM is relatively reliable. Begg’s funnel plot was used to test publication bias. The test showed that there was no publication bias among the studies. Sensitivity analysis indicated that the results are stable and reliable.
The SNP +45T/G is a synonymous mutation (GGTGGG, Gly15Gly) at exon 2. The results of Yang et al[20] indicated that +45T/G polymorphism may influence the expression of ADIPOQ by influencing RNA splicing and stability. Some studies reported that the G allele of +45T/G polymorphism in the ADIPOQ gene is associated with obesity, insulin resistance, and T2D in several populations. Very few studies have investigated the association of ADIPOQ +45T/G polymorphism with GDM and the results of these studies were controversial.
As far as we know, only +45T/G was involved in a previous meta-analysis that reported no association of ADIPOQ +45T/G polymorphism with the risk of GDM (allelic model: OR = 1.17, 95%CI: 0.79-1.76; dominant model: OR = 0.86, 95%CI: 0.50-1.48; recessive model: OR = 1.21, 95%CI: 0.62-2.33)[21]. The reason for this controversy is most likely the following: (1) the small sample size (case number = 875, control number = 884); (2) obvious heterogeneity (all P-values for heterogeneity less than 0.01); and (3) false HWE status of 25% studies (2 of 8) involved in that meta-analysis will cause insufficient power which may lead to the false-negative results.
The role of +276G/T in the pathogenesis of metabolic syndrome and diabetes mellitus has also been reported to be contradictory. Commonly, the T allele has an association with a higher adiponectin level and protection against T2D[22,23], but some studies showed that T carriers have a higher risk of obesity and diabetes[24,25], or +276G/T polymorphism is not associated with T2D or GDM[26].
All the studies on +276G/T polymorphism included in our study came from the Chinese population. This result suggests that +276G/T in the Chinese population may associate with the risk of GDM. The studies about the association between +276G/T and GDM from other countries could not be found. Geographical, environmental, and genetic factors of different ethnic groups lead to different susceptibility to diabetes; therefore, we need more studies about the association between +276G/T and GDM of other ethnic groups to reach reliable conclusions.
Zhang et al[27] found that ADIPOQ gene promoter region has four transcription stimulatory protein (SP1) binding sites, while the G allele of - 11377 C/G in the promoter region can change the DNA sequence of one of the SP1 binding sites, leading to the loss of binding force to SP1. This may reduce the ADIPOQ gene transcription activity, inhibit the expression of genes, and lead to lower plasma ADIPOQ, which could associate with glucolipid metabolic abnormalities and insulin resistance. Consistent with the results of Vasseur et al[28], Petrone et al[29] reported that -11377G haplotype is associated with low plasma ADIPOQ levels and T2D. However, due to ethnic and geographical differences, the results of studies on the association between ADIPOQ gene -11377C/G polymorphism and diabetes mellitus are not completely consistent[30].
According to the literatures on -11377C/G polymorphism included in our studies, Asians accounted for 40%, Europeans for 40%, and South Americans for 20%. Due to the small sample size and large heterogeneity of each ethnic subgroup, our results, which are inconsistent with the previous studies, are unreliable.
The limitations of this study should be considered. First, the number of cases and controls involved in the meta-analysis for exploring the association of ADIPOQ and GDM in different ethnicities may have little power, and studies with larger sample sizes and multiple ethnicities are needed. Second, GDM has complicated cases, with genetic susceptibility, environmental triggers, and acquired dispositions, such as age, gestational weeks, condition of nutrition, and physique. In this meta-analysis, we failed to conduct a multivariate analysis of confounders. Therefore, further comprehensive studies with strict matching criteria for cases and controls are needed. Third, few studies have reported the association between polymorphisms and serum ADIPOQ levels, so genotype-phenotype analysis was prevented[21].
In conclusion, our meta-analysis reveals the association of the ADIPOQ +45T/G polymorphism and the risk of GDM; this polymorphism increases GDM risk in Asian populations. Another two polymorphisms, +276G/T and -11377C/G, seem to have no association with the risk of GDM. Prospective studies of high quality with larger sample sizes are required to reveal the association of ADIPOQ polymorphisms with GDM, the existence of ethnicity-specific factors, and the role that ADIPOQ polymorphisms play in pathology.
ARTICLE HIGHLIGHTS
Research background
Many biochemical mediators that are synthesized in the adipose tissue and secreted in the circulation, such as leptin, adiponectin (ADIPOQ), and resistin, are thought to be involved in obesity and insulin resistance. ADIPOQ is produced in the adipose tissue and regulates a variety of metabolic processes such as lipid metabolism, glucose and fatty acid oxidation. This hormone can reduce insulin resistance, improve lipid metabolism, and exert anti-inflammatory effects. Plasma ADIPOQ levels are decreased in patients with type 2 diabetes, metabolic syndrome, and obesity. Previous studies have shown that ADIPOQ single nucleotide polymorphisms can affect plasma ADIPOQ concentrations, which in turn affect insulin sensitivity.
Research motivation
Previous studies have evaluated the relationship between ADIPOQ polymorphisms and gestational diabetes mellitus (GDM), but the results of the association between ADIPOQ polymorphisms and GDM is uncertain.
Research objectives
We evaluated the association between ADIPOQ +45T/G, +276G/T, and -11377C/G polymorphisms and GDM with a bigger sample size and less heterogeneity. Moreover, subgroup analysis was performed by ethnicity.
Research methods
Potentially related articles on human fat metabolism and GDM gene research published before October 20, 2018 were retrieved through the electronic databases EMBASE, Web of Science, PubMed, WANFANG DATA, and China National Knowledge Infrastructure. A fixed-effects or random-effects model was used to calculate pooled odds ratios (ORs) with 95% confidence intervals (CIs), based on the between-study heterogeneity to evaluate the association between AIDPOQ +45T/G, +276G/T, and -11377C/G polymorphisms and the risk of GDM.
Research results
Nine +45T/G studies included 1024 GDM cases and 1059 controls, five +276G/T studies included 590 GDM cases and 595 controls, and five -11377C/G studies included 722 GDM cases and 791 controls. Pooled ORs showed that +45T/G increased Asian GDM risk (allele model OR = 1.47, 95%CI: 1.27-1.70, P = 0.000; dominant model OR = 1.54, 95%CI: 1.27-1.85, P = 0.000; recessive mode: OR = 2.00, 95%CI: 1.43-2.85, P = 0.000), but not in South Americans (equal pattern: OR = 1.21, 95%CI: 0.68-2.41, P = 0.510; dominant model OR = 1.13, 95%CI: 0.59-2.15, P = 0.710; recessive mode OR = 2.18, 95%CI: 0.43-11.07, P = 0.350). There was no significant correlation between +276G/T (allele model OR = 0.88, 95%CI: 0.74-1.05, P = 0.158; dominant model OR = 0.91, 95%CI: 0.65-1.26, P = 0.561; recessive mode: OR = 0.82, 95%CI: 0.64-1.05, P = 0.118) or -11377C/G (equal pattern: OR = 0.96, 95%CI: 0.72-1.26, P = 0.750; dominant model OR = 1.00, 95%CI: 0.73-1.37, P = 0.980; recessive model: OR = 0.90, 95%CI: 0.61-1.32, P = 0.570) and GDM risk.
Research conclusions
Our meta-analysis reveals the association of the ADIPOQ +45T/G polymorphism and the risk of GDM; this polymorphism increases GDM risk in Asian populations.
Research perspectives
In order to reveal the association of ADIPOQ polymorphisms with GDM, the existence of ethnicity-specific factors, and the role that ADIPOQ polymorphisms play in pathology, studies focused on delineating ethnicity-specific factors with larger sample sizes are needed.
Footnotes
Conflict-of-interest statement: The authors deny any conflict of interest.
PRISMA Checklist: The authors have read the PRISMA 2009 Checklist, and the manuscript was prepared and revised according to the PRISMA Checklist.
Manuscript source: Unsolicited manuscript
Peer-review started: November 21, 2018
First decision: December 15, 2018
Article in press: January 3, 2019
Specialty type: Medicine, Research and Experimental
Country of origin: China
Peer-review report classification
Grade A (Excellent): A
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: Pastromas S, Cengiz M S- Editor: Dou Y L- Editor: Wang TQ E- Editor: Tan WW
Contributor Information
Lin-Ting Huang, Department of Epidemiology and Health Statistics, Xiangya School of Public Health, Central South University, Changsha 410083, Hunan Province, China.
Shi-Lan Wu, Department of Epidemiology and Health Statistics, Xiangya School of Public Health, Central South University, Changsha 410083, Hunan Province, China.
Xin Liao, Department of Epidemiology and Health Statistics, Xiangya School of Public Health, Central South University, Changsha 410083, Hunan Province, China.
Shu-Juan Ma, Department of Epidemiology and Health Statistics, Xiangya School of Public Health, Central South University, Changsha 410083, Hunan Province, China.
Hong-Zhuan Tan, Department of Epidemiology and Health Statistics, Xiangya School of Public Health, Central South University, Changsha 410083, Hunan Province, China. tanhz99@qq.com.
References
- 1.Beltcheva O, Boyadzhieva M, Angelova O, Mitev V, Kaneva R, Atanasova I. The rs266729 single-nucleotide polymorphism in the adiponectin gene shows association with gestational diabetes. Arch Gynecol Obstet. 2014;289:743–748. doi: 10.1007/s00404-013-3029-z. [DOI] [PubMed] [Google Scholar]
- 2.Takhshid MA, Haem Z, Aboualizadeh F. The association of circulating adiponectin and + 45 T/G polymorphism of adiponectin gene with gestational diabetes mellitus in Iranian population. J Diabetes Metab Disord. 2015;14:30. doi: 10.1186/s40200-015-0156-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pawlik A, Teler J, Maciejewska A, Sawczuk M, Safranow K, Dziedziejko V. Adiponectin and leptin gene polymorphisms in women with gestational diabetes mellitus. J Assist Reprod Genet. 2017;34:511–516. doi: 10.1007/s10815-016-0866-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Han Y, Zheng YL, Liu MH, Fan YP. Association of adiponectin gene single nucleotide polymorphism with gestational diabetes mellitus and pregnancy outcomes. Shiyong Fuchanke Zazhi. 2012;22:743–746. [Google Scholar]
- 5.Hara K, Boutin P, Mori Y, Tobe K, Dina C, Yasuda K, Yamauchi T, Otabe S, Okada T, Eto K, Kadowaki H, Hagura R, Akanuma Y, Yazaki Y, Nagai R, Taniyama M, Matsubara K, Yoda M, Nakano Y, Tomita M, Kimura S, Ito C, Froguel P, Kadowaki T. Genetic variation in the gene encoding adiponectin is associated with an increased risk of type 2 diabetes in the Japanese population. Diabetes. 2002;51:536–540. doi: 10.2337/diabetes.51.2.536. [DOI] [PubMed] [Google Scholar]
- 6.Menzaghi C, Ercolino T, Di Paola R, Berg AH, Warram JH, Scherer PE, Trischitta V, Doria A. A haplotype at the adiponectin locus is associated with obesity and other features of the insulin resistance syndrome. Diabetes. 2002;51:2306–2312. doi: 10.2337/diabetes.51.7.2306. [DOI] [PubMed] [Google Scholar]
- 7.Low CF, Mohd Tohit ER, Chong PP, Idris F. Adiponectin SNP45TG is associated with gestational diabetes mellitus. Arch Gynecol Obstet. 2011;283:1255–1260. doi: 10.1007/s00404-010-1548-4. [DOI] [PubMed] [Google Scholar]
- 8.Daher S, Torloni MR, Gueuvoghlanian-Silva BY, Moron AF, Mattar R. Inflammatory mediator gene polymorphisms and gestational diabetes: A review of the literature. J Reprod Immunol. 2011;90:111–116. doi: 10.1016/j.jri.2011.04.008. [DOI] [PubMed] [Google Scholar]
- 9.Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol. 2010;25:603–605. doi: 10.1007/s10654-010-9491-z. [DOI] [PubMed] [Google Scholar]
- 10.Gao Y, Wang W, Wang SS. Association of Adiponectin Gene Single Nucleotide Polymorphism with Gestational Diabetes Mellitus. Jilin Yixue. 2016;6:1301–1302. [Google Scholar]
- 11.Li GH, Kong LJ, Zhang L, Zhang WY. [Association of adiponectin gene polymorphisms +45T/G with gestational diabetes mellitus and neonate birth weight] Zhonghua Yixue Zazhi. 2013;93:3770–3772. [PubMed] [Google Scholar]
- 12.Li JY, Ma S, Zhao J, Duan LJ, Sun HY. The correlation between single nucleotide polymorphism of adiponectin gene and gestational diabetes and its effect on pregnancy outcome. Zhongguo Fuyoubaojian Zazhi. 2017;22:5674–5677. [Google Scholar]
- 13.Luan YY, Guo XH, Yang JH. Study on the correlation between the damage of adiponectin gene polymorphism with gestational impaired glucose. Zhongguo Shiyanzhenduanxue Zazhi. 2015;19:1093–1096. [Google Scholar]
- 14.Zhang C, Liang XX. The relationship between adiponectin gene polymorphism of Guangxi Zhuang ethnic group and gestational diabetes. Zhongguo Yishi Zazhi. 2014;9:1221–1223. [Google Scholar]
- 15.Chen ZY, Du J. Relationship between diponectin gene polymorphism and gestational diabetes mellitus. Xiandai Fuchanke Jinzhan. 2011;20:718–721. [Google Scholar]
- 16.Wang XX, Zhang L, Zhou GF, Pu XM. Study of the correlation between adiponectin gene polymorphism and gestational diabetes mellitus. Zhongguo Fuyoubaojian Zazhi. 2016;31:2546–2549. [Google Scholar]
- 17.Peng JJ, Shi FX, Wang HY, Wang ZP, Yuan P. Correlation between adiponectin and gestaional diabetes mellitus. Zhongguo Fuyoubaojian Zazhi. 2012;27:1314–1316. [Google Scholar]
- 18.Zhang J, Chi H, Xiao H, Tian X, Wang Y, Yun X, Xu Y. Interleukin 6 (IL-6) and Tumor Necrosis Factor α (TNF-α) Single Nucleotide Polymorphisms (SNPs), Inflammation and Metabolism in Gestational Diabetes Mellitus in Inner Mongolia. Med Sci Monit. 2017;23:4149–4157. doi: 10.12659/MSM.903565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen XX, Wei BR, Zhang LF, Yang YM. Association of -3771A/G Polymorphism in Promoter Region of Adiponectin Gene with Gestational Diabetes Mellitus. Nanchangdaxue Xuebao Yixueban. 2013;53:18–21. [Google Scholar]
- 20.Yang WS, Tsou PL, Lee WJ, Tseng DL, Chen CL, Peng CC, Lee KC, Chen MJ, Huang CJ, Tai TY, Chuang LM. Allele-specific differential expression of a common adiponectin gene polymorphism related to obesity. J Mol Med (Berl) 2003;81:428–434. doi: 10.1007/s00109-002-0409-4. [DOI] [PubMed] [Google Scholar]
- 21.Xu F, Zhang H, Qi H. No association of adiponectin +45 T/G polymorphism with the risk of gestational diabetes mellitus: Evidence from a meta-analysis. J Renin Angiotensin Aldosterone Syst. 2016;17:1470320316653283. doi: 10.1177/1470320316653283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liang Z, Dong M, Cheng Q, Chen D. Gestational diabetes mellitus screening based on the gene chip technique. Diabetes Res Clin Pract. 2010;89:167–173. doi: 10.1016/j.diabres.2010.04.001. [DOI] [PubMed] [Google Scholar]
- 23.Pollin TI, Tanner K, O'connell JR, Ott SH, Damcott CM, Shuldiner AR, McLenithan JC, Mitchell BD. Linkage of plasma adiponectin levels to 3q27 explained by association with variation in the APM1 gene. Diabetes. 2005;54:268–274. doi: 10.2337/diabetes.54.1.268. [DOI] [PubMed] [Google Scholar]
- 24.Beebe-Dimmer JL, Zuhlke KA, Ray AM, Lange EM, Cooney KA. Genetic variation in adiponectin (ADIPOQ) and the type 1 receptor (ADIPOR1), obesity and prostate cancer in African Americans. Prostate Cancer Prostatic Dis. 2010;13:362–368. doi: 10.1038/pcan.2010.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bouatia-Naji N, Meyre D, Lobbens S, Séron K, Fumeron F, Balkau B, Heude B, Jouret B, Scherer PE, Dina C, Weill J, Froguel P. ACDC/adiponectin polymorphisms are associated with severe childhood and adult obesity. Diabetes. 2006;55:545–550. doi: 10.2337/diabetes.55.02.06.db05-0971. [DOI] [PubMed] [Google Scholar]
- 26.Urbanek M, Hayes MG, Lee H, Freathy RM, Lowe LP, Ackerman C, Jafari N, Dyer AR, Cox NJ, Dunger DB, Hattersley AT, Metzger BE, Lowe WL., Jr The role of inflammatory pathway genetic variation on maternal metabolic phenotypes during pregnancy. PLoS One. 2012;7:e32958. doi: 10.1371/journal.pone.0032958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang D, Ma J, Brismar K, Efendic S, Gu HF. A single nucleotide polymorphism alters the sequence of SP1 binding site in the adiponectin promoter region and is associated with diabetic nephropathy among type 1 diabetic patients in the Genetics of Kidneys in Diabetes Study. J Diabetes Complications. 2009;23:265–272. doi: 10.1016/j.jdiacomp.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 28.Vasseur F, Helbecque N, Dina C, Lobbens S, Delannoy V, Gaget S, Boutin P, Vaxillaire M, Leprêtre F, Dupont S, Hara K, Clément K, Bihain B, Kadowaki T, Froguel P. Single-nucleotide polymorphism haplotypes in the both proximal promoter and exon 3 of the APM1 gene modulate adipocyte-secreted adiponectin hormone levels and contribute to the genetic risk for type 2 diabetes in French Caucasians. Hum Mol Genet. 2002;11:2607–2614. doi: 10.1093/hmg/11.21.2607. [DOI] [PubMed] [Google Scholar]
- 29.Petrone A, Zavarella S, Caiazzo A, Leto G, Spoletini M, Potenziani S, Osborn J, Vania A, Buzzetti R. The promoter region of the adiponectin gene is a determinant in modulating insulin sensitivity in childhood obesity. Obesity (Silver Spring) 2006;14:1498–1504. doi: 10.1038/oby.2006.172. [DOI] [PubMed] [Google Scholar]
- 30.Enns JE, Taylor CG, Zahradka P. Variations in Adipokine Genes AdipoQ, Lep, and LepR are Associated with Risk for Obesity-Related Metabolic Disease: The Modulatory Role of Gene-Nutrient Interactions. J Obes. 2011;2011:168659. doi: 10.1155/2011/168659. [DOI] [PMC free article] [PubMed] [Google Scholar]