Abstract
Cotton leaf curl Multan virus (CLCuMuV) belonging to begomoviruses (Family Geminiviridae) can infect cotton and many other agricultural crops. Betasatellite associated with CLCuMuV i.e., cotton leaf curl Multan betasatellite (CLCuMuB) is a small circular single-stranded deoxyribose nucleic acid (ssDNA) molecule that is essential for CLCuMuV to induce disease symptoms. Betasatellite molecule contains a ßC1 gene encoding for a pathogenicity determinant multifunctional protein, which extensively interacts with host plant machinery to cause virus infection. In this study the interaction of ßC1 with selected plant flavonoids has been studied. The study was focused on sequence analysis, three-dimensional structural modeling and docking analysis of ßC1 protein of CLCuMuB. Sequence analysis and physicochemical properties showed that ßC1 is negatively charged protein having more hydrophilic regions and is not very stable. Three-dimensional model of this protein revealed three helical, four beta pleated sheets and four coiled regions. The score of docking experiments using flavonoids as ligand indicated that plant flavonoids robinetinidol-(4alpha,8)-gallocatechin, quercetin 7-O-beta-D-glucoside, swertianolin, 3′,4′,5-trihydroxy-3-methoxyflavon-7-olate, agathisflavone, catiguanin B, 3′,4′,5,6-tetrahydroxy-3,7-dimethoxyflavone, quercetin-7-O-[alpha-L-rhamnopyranosyl(1->6)-beta-D-galactopyranoside], prunin 6″-O-gallate and luteolin 7-O-beta-D-glucosiduronic acid have strong binding with active site of ßC1 protein. The results obtained from this study clearly indicate that flavonoids are involved in defense against the virus infection, as these molecules binds to the active site of ßC1 protein. This information might be interesting to study plant defense mechanism based on the special compounds produced by the plants.
Keyword: Structural biology
1. Introduction
Cotton leaf curl disease is a major threat to cotton crop throughout South Asia and Africa [1]. This infection is caused by a single-stranded deoxyribose nucleic acid (ssDNA) monopartite begomovirus i.e., cotton leaf curl virus and associated satellite molecules. This virus is a member of family Geminiviridae and associated with two satellites i.e., betasatellite and alphasatellite [2]. Cotton leaf curl Multan betasatellite (CLCuMuB) is a member of a recently proposed family Tolecusatellitidae from genus Betasatellite [2]. The genome of betasatellites is comprised of a single ßC1 in the complementary sense orientation, an adenine rich (A-rich) region and a satellite-conserved region (SCR). ßC1 gene encodes a protein having numerous functions, mainly acts as suppressor of post transcriptional gene silencing (PTGS) [3]. It is also involved with development of infection in plant and in increasing the level of viral DNA [4].
ßC1 is involved in intracellular transport of proteins by co-restricting with the endoplasmic reticulum in cell [5]. ßC1 also interfere with autophagy related ubiquitin like protein (ATG8) [6]. It interacts with various proteins in the cell like carboxyl methyl transferase, retinoblastoma-related protein, receptor kinases, salicylic acid, ubiquitin-conjugating enzyme and ATP binding cassette (ABC) transporter proteins [7] proposing a significant part of ßC1 for infection progression and movement.
Flavonoids are poly-phenolic compounds present in plants, animals and bacteria [8]. In plants these compounds are responsible for growth and development of seedling, aroma of flowers [9], color [10], fruit to attract pollinators consequently fruit dispersion and spore germination [11]. Flavonoids also help to protect plant from a biotic and biotic stresses [12]. Up-till now more than 6500 different flavonoids have been discovered [13] from different plant species. Flavonoids play a vital role in the interaction between the plants and their environment [14]. The precursor of all flavonoids is phenylalanine and malonyl-CoA and all flavonoids are produced from a parent structure flavones present in the cell [15]. The enzymes involved in biosynthetic pathway of flavonoids belong to different families like oxoglutarate dependent dioxygenases, cytochromes P450 and glycosyl transferases [14]. Flavonoids perform a critical role as signaling molecules in plant micro-organism interactions [16]. Flavonoids act as detoxification agents, scavenge ROS, H2O2 and toxic metals [17]. These compounds also reduce radiation stress [18]. Flavonoids function as phytoalexins, the chemicals that are produced in plants in response to the infection of microorganism [19, 20].
The function of flavonoids in defense mechanism against bacterial attack has been explored recently [21], but their role in plant defense against viral infection specially ssDNA plant viruses has not been studied yet. In the present study we have performed computational analysis based on docking method to find out the binding of flavonoids with viral pathogenicity determinant ßC1 protein from cotton leaf curl Multan virus.
2. Materials and methods
2.1. Sequence, domain and motif analysis
The protein sequence of ßC1 was downloaded from NCBI using accession number HF567946. The physicochemical properties like amino acid composition, theoretical isoelectric point (pI), molecular mass, atomic composition, extinction coefficient, instability index, estimated half-life, and aliphatic index were obtained by ProtParam from EXPASY server (www.expasy.ch/tools) [22]. For domain and motif analysis tools like NCBI CDD [23], Interproscan [24], Smart [25], Pfam [26] and Motif scan [22] were used.
2.2. Homology modeling
Three-dimensional structure of ßC1 protein was not available in Protein Data Bank therefore homology modeling [27] was used to obtain the three dimensional structure of ßC1 protein. The templates were selected from template identification wizard of Swiss model [28]. The template structure and protein alignment was done utilizing Modeller v9.19 software using align2d facility of Modeller [29]. The output file was obtained in PIR format that was used to construct models of ßC1 proteins by Modeller v9.19. The model built was then subjected to energy minimization with the MOE software. Model quality was evaluated by using ProSA-web Z-score [30], Errat, and verified by 3D Qmean plot [31]. Ramachandran plot was obtained by PROCHECK [32] for model evaluation. Additionally, Root Mean Squared Deviation (RMSD) was obtained by the superimposition of query and template structure, by UCSF Chimera 1.11.2 [33].
2.3. Docking analysis
Docking analysis was performed using the MOE (Molecular Operating Environment) software [30]. The structures of ligands were obtained from chebi (http://www.ebi.ac.uk/chebi/) in .mol format. These structures were modified by the addition of hydrogen atoms and energies were minimized using following parameters Force field: MMFF94X, Chiral constrain: Current geometry, Gradiant: 0.05. Best model obtained from modeler was used for docking analysis. The structure of protein was subjected to 3D protonation and energy minimization using following parameters Force field: MMFF94X + Solvation, Chiral constrain: Current geometry, Gradiant: 0.05. This minimized structure was then used as receptors in docking analysis. The active site of protein was found by site finder module of MOE. Docking was run with default parameters of MOE. Free energies of binding of each ligand structure from a given orientation were obtained by MOE docking score.
2.4. Calculations of ligand interactions
The ligand interactions were calculated by Ligand Interaction module of MOE Program (https://www.chemcomp.com/MOEMolecular_Operating_Environment.htm). It incorporates two-dimensional and three-dimensional representations of ligand and receptor protein interactions and calculated distances among ligand and protein interacting atoms.
3. Results
3.1. Physicochemical properties and sequence analysis
The physicochemical parameters were calculated from Protparam (https://web.expasy.org/protparam/). These results indicated that molecular weight of ßC1 protein is 13.664 KDa. Theoretical isoelectric point (pI) is 4.86, which showed that protein is acidic in nature. Total negatively charged residues were 18 while total positively charged residues were 11. Thus the net charge on protein was negative. The extinction coefficient value was 7450. In-vitro half-life was calculated as 13 hours. While instability index value calculated as 42.26 showing that the protein was quite unstable and short lived in the cell.
3.2. Results of homology modeling
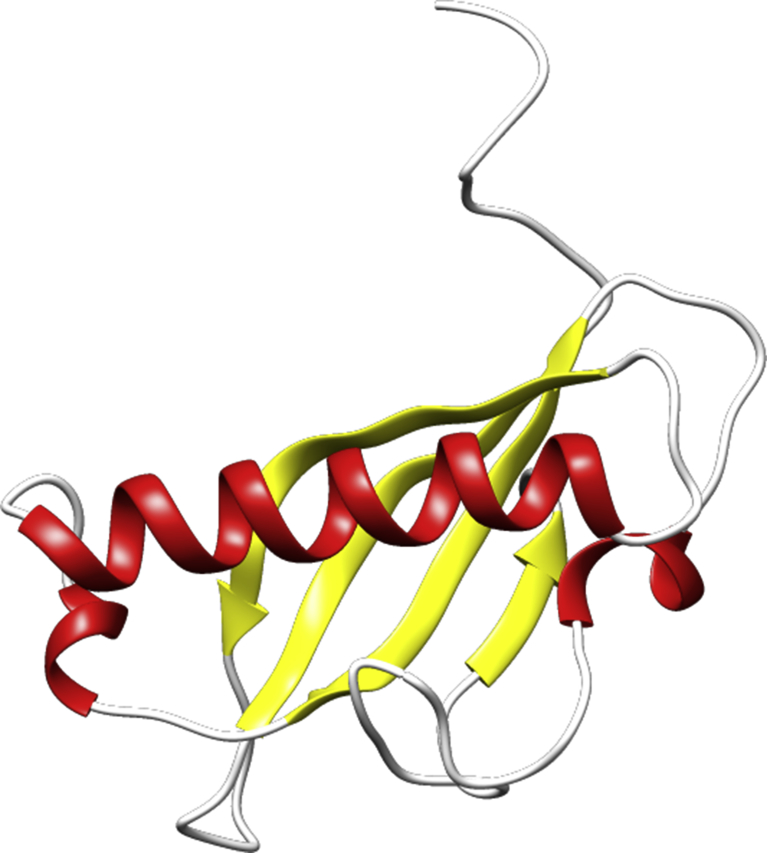
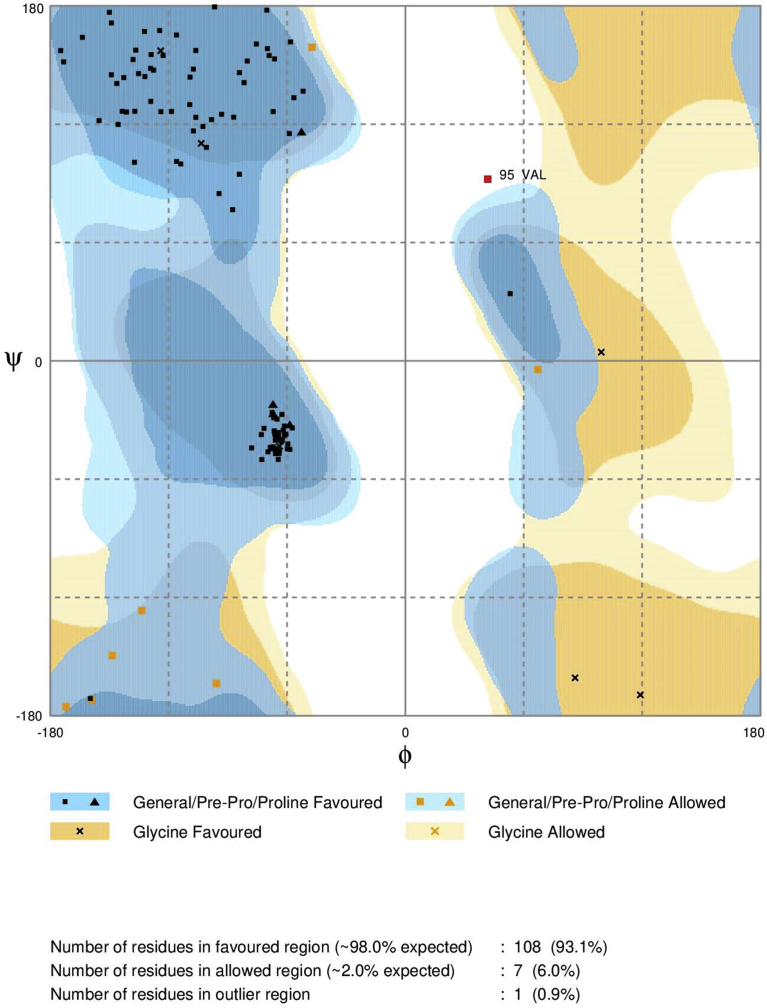
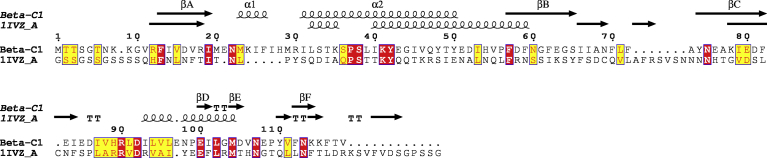
Ten models were generated by means of Modeller v9.19 (https://salilab.org/modeller/). For template search Swiss model was used. The template used to build the model of ßC1 was a protein from Protein Data Bank with ID:1IVZ, which has sequence similarity of 71.4% with ßC1. The minimum similarity required for template selection in homology modeling is 30%. The alignment between template and model along with secondary structure is shown in (Fig. 1). The best model was selected on the basis of model evaluation tools, ProSA-web Z-scores and RAMPAGE Ramachandran plots (Fig. 2). Template model superimposition was done by UCSF Chimera v1.11.2 software showed root mean square deviation value of 0.223 Å, proven a high level of similarity. Ramachandran plot was obtained from RAMPAGE presented that 90% of amino acids were in most favored region (Fig. 2). The best model (Fig. 3) was deposited to Protein Model Database and following PMID: PM0081495 was obtained. According to this model the predicted 3-D protein structure contains three alpha helices (red color), four beta pleated sheets in yellow color and four coiled regions in white color (Fig. 3). The predicted model was used for further docking analysis.
Fig. 1.
Alignment of template protein i.e., 1IVZ and ßC1 protein along with secondary structure units. ß1-ß7 are beta-pleated sheets, α1-α3 are three alpha helices and three coiled regions (TT).
Fig. 2.
Ramachandran plot for the model of ßC1 protein showing 98% residues in most favored, 2% in allowed region while 1% residues in outlier region.
Fig. 3.
Three dimensional model of ßC1 Protein; showing three alpha helices in red color, four beta pleated sheets in yellow color and four coiled regions in white color.
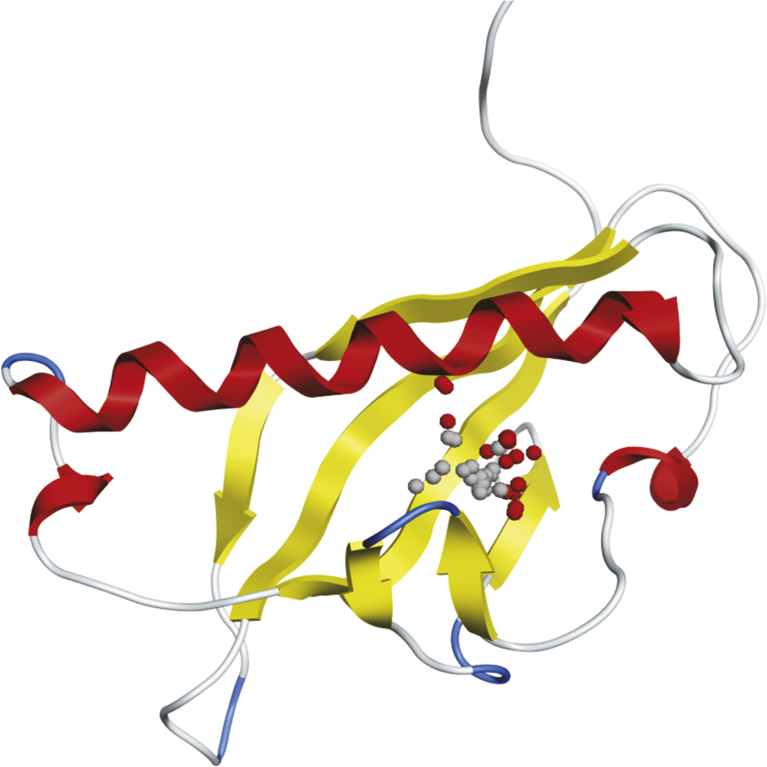
3.3. Docking analysis
For docking analysis active site of ßC1 was obtained by site finder in MOE (Fig. 4). Docking analysis of ßC1 was done by MOE Simulate module. The best model chosen from homology modeling was subjected to docking run. The stringency parameters used for docking were Placement: Triangular Matcher, Rescoring function 1:London dG, Retains: 10. 140 flavonoid structures were used as ligand for docking examination. Docking analysis provided a number of configurations that were scored to determine favorable binding modes. Ten flavonoid structures presenting high scores of docking than previously reported data [34, 35]were shown in Fig. 5. The docking scores and plant sources of each flavonoid are shown in Table 1.
Fig. 4.
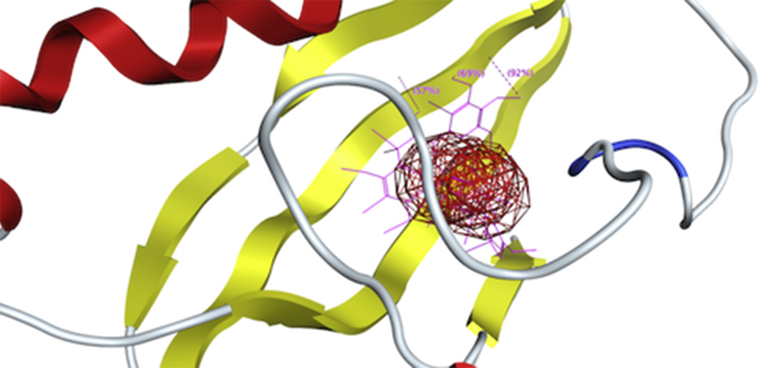
Position of active site in ßC1 three-dimensional protein structure; The region with small red and grey balls showing the active site.
Fig. 5.
Flavonoid structures showing best results of docking analysis; a) robinetinidol-(4alpha,8)-gallocatechin, b) quercetin 7-O-beta-D-glucoside, c) Swertianolin, d) 3′,4′,5-trihydroxy-3-methoxyflavon-7-olate, e) Agathisflavone, f) catiguanin B, g) 3′,4′,5,6-tetrahydroxy-3,7-dimethoxyflavone, h). quercetin-7-O-[alpha-L-rhamnopyranosyl(1->6)-beta-D-galactopyranoside], i) prunin 6″-O-gallate, j) luteolin 7-O-beta-D-glucosiduronic acid.
Table 1.
Selected flavonoid molecules downloaded from chebi (http://www.ebi.ac.uk/chebi/) showing maximum docking score.
| Flavonoid no. | Chebi ID | Name | Plant sources | Docking score (Kcal/mol) |
|---|---|---|---|---|
| 1 | 68332 | Robinetinidol-(4alpha,8)-gallocatechin | Acacia | −18.613 |
| 2 | 28529 | Quercetin 7-O-beta-D-glucoside | Galinsoga parviflora Galinsoga quadriradiata |
−18.433 |
| 3 | 65478 | Swertianolin | Swertia chirayita Swertia macrosperma Gentiana campestris |
−17.701 |
| 4 | 57928 | 3′,4′,5-trihydroxy-3-methoxyflavon-7-olate | Chrysosplenium americanum | −17.653 |
| 5 | 2512 | Agathisflavone | Canarium manii | −17.476 |
| 6 | 65602 | Catiguanin B | Trichilia catigua | −17.279 |
| 7 | 27767 | 3′,4′,5,6-tetrahydroxy-3,7-dimethoxyflavone | Lobelia chinensis Trigonostemon reidioides |
−17.167 |
| 8 | 70147 | Quercetin-7-O-[alpha-L-rhamnopyranosyl(1->6)-beta-D-galactopyranoside] | Grevillea “Poorinda Queen” | −17.116 |
| 9 | 73787 | Prunin 6″-O-gallate | Matricaria recutita | −17.128 |
| 10 | 18128 | Luteolin 7-O-beta-D-glucosiduronic acid | Thymus vulgaris Tanacetum parthenium |
−16.844 |
3.4. Calculation of ligand interaction
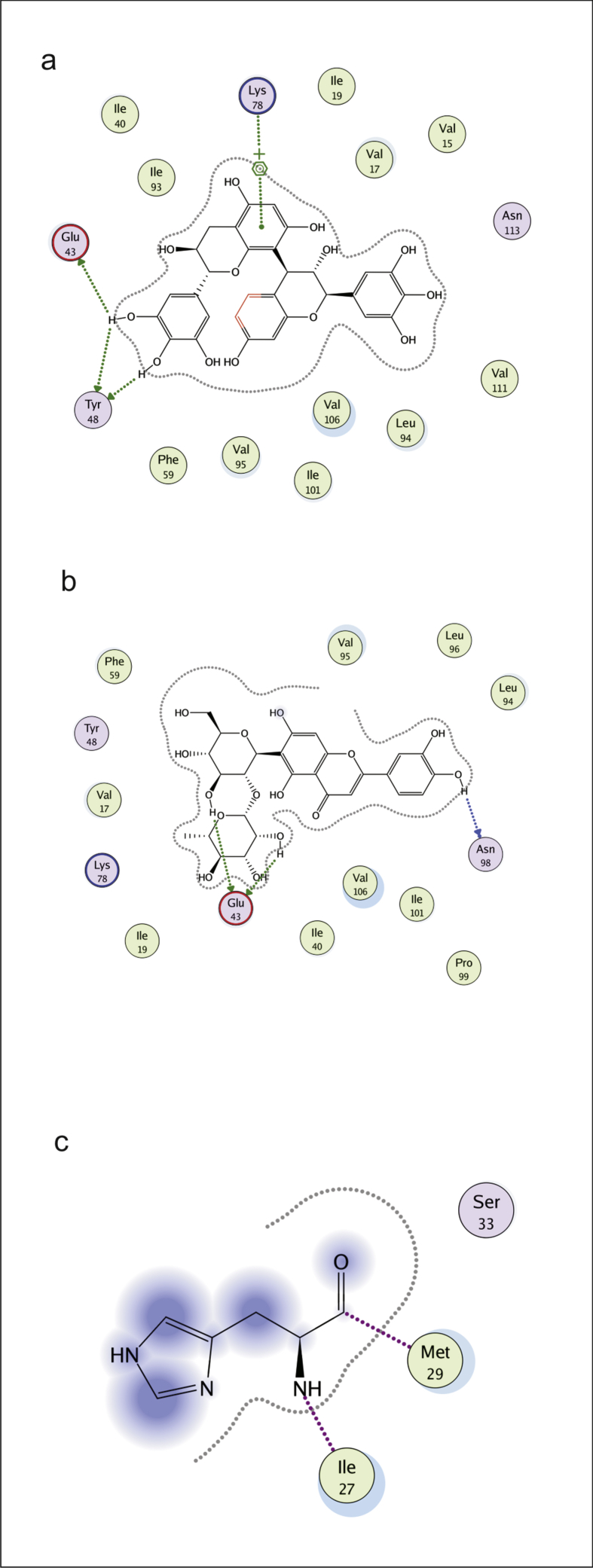
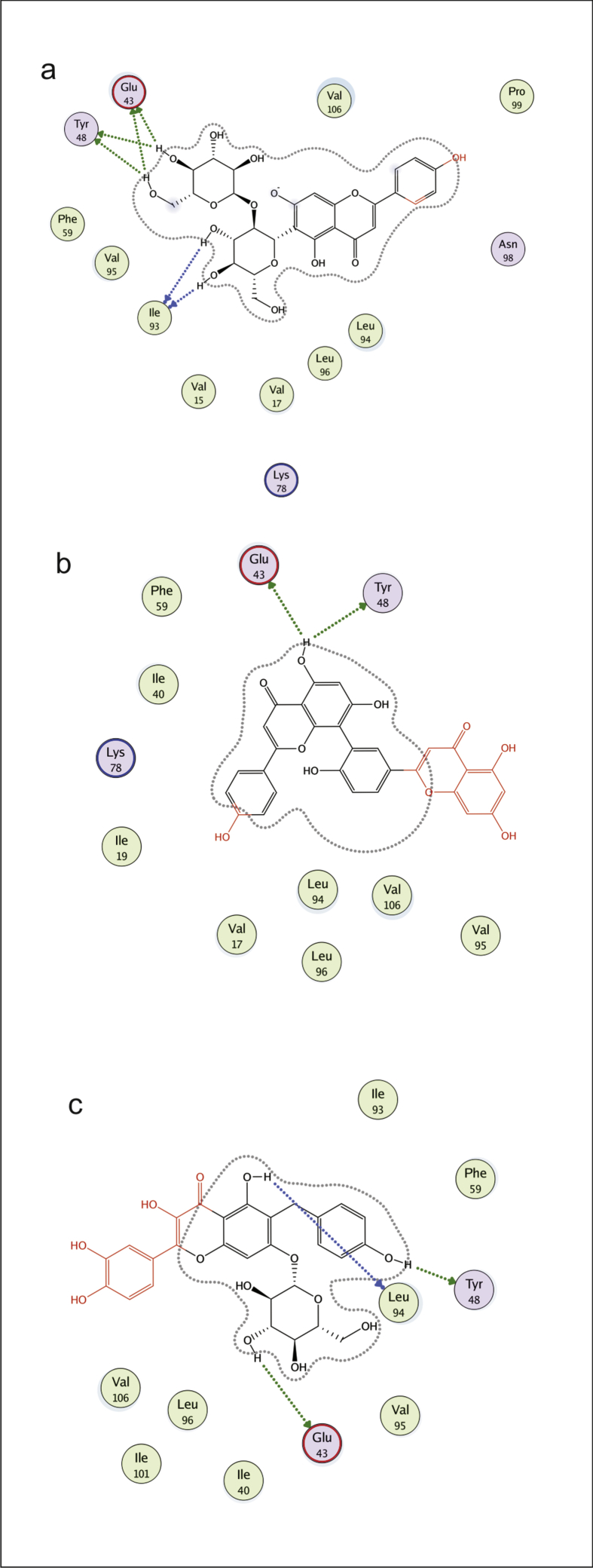
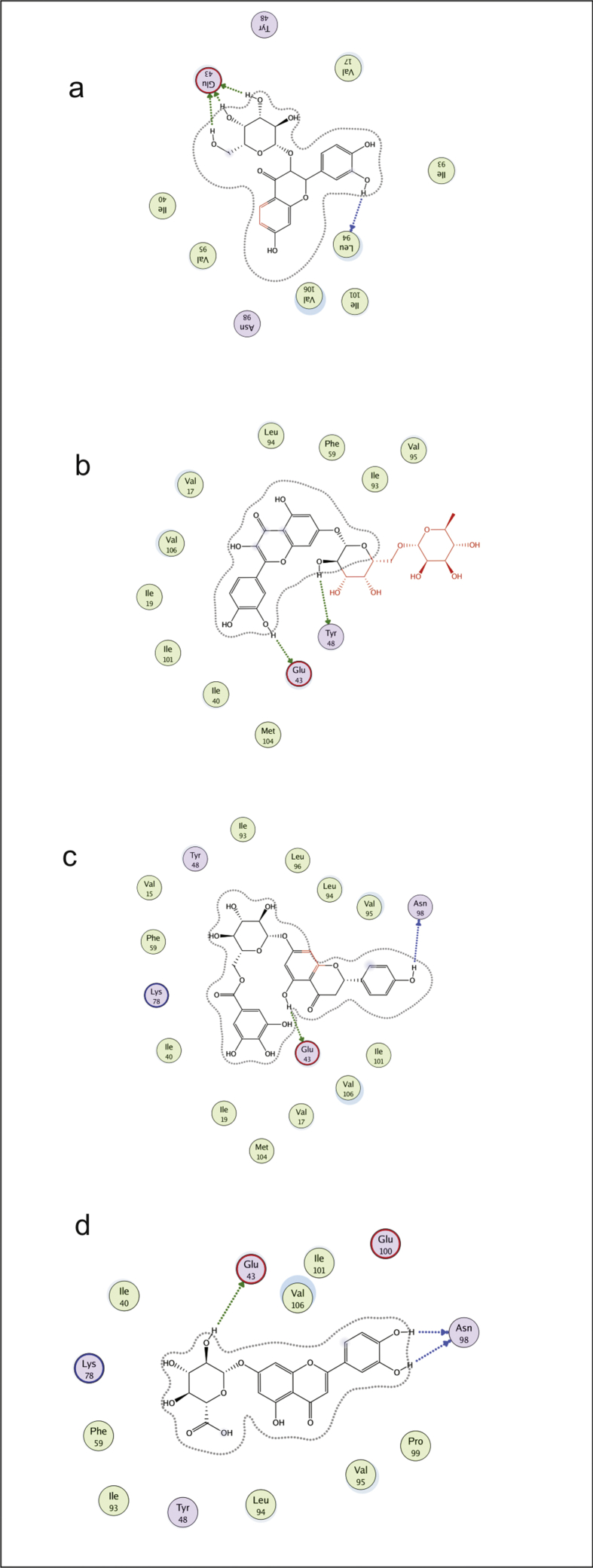
Interactions of ßC1 protein with flavonoids were obtained by MOE program. These flavonoid- ßC1 interaction studies provided us information about each atom of ligand and protein involved in this interaction. Depicted view of 3D interaction is shown in Fig. 6. Flavonoid 1 formed an interaction with three amino acids of viral protein. Arene cation link with amino acid Lys 78 and side chain donor link Glu 43 and Tyr 48 with in the active site (Fig. 7a). Flavonoid 2 formed the interaction with two amino acids of viral protein (Glu 43 and Asn 98) with in active site (Fig. 7b). Flavonoid 3 formed a side chain donor link with Ile 27 and Met 29 (Fig. 7c). Flavonoid 4 formed hydrogen bonding with Glu 43, Tyr 48 and Ile 93 (Fig. 8a). Flavonoid 5 formed interaction with Glu 43 and Tyr 48 (Fig. 8b). Flavonoid 6 formed the interaction with Tyr 48 Glu 43 and Leu 94 (Fig. 8c). Flavonoid 7 interacted with Glu 43 and Leu 94 (Fig. 9a). Flavonoid 8 formed interaction with Tyr 48 and Glu 43 (Fig. 9b). Flavonoid 9 has shown an interaction with amino acid Glu 43 and Asn 98 (Fig. 9c). Flavonoid 10 formed interaction with Glu 43 and Asn 98 (Fig. 9d).
Fig. 6.
3D view of docking of ßC1 with reacting region of robinetinidol-(4alpha,8)-gallocatechin. The values i.e., 92%, 69% and 57% show the binding strength of hydrogens present in flavonoid and side chains of active site of ßC1 protein.
Fig. 7.
2D interaction diagram, showing the interacting flavonoid atoms with specific residues in active site of ßC1 protein. Dotted lines are showing specific interaction. a) robinetinidol-(4alpha,8)-gallocatechin, b) quercetin 7-O-beta-D-glucoside, c) Swertianolin.
Fig. 8.
2D interaction diagram, showing the interacting flavonoid atoms with specific residues in active site of ßC1 protein. Dotted lines are showing specific interaction. a) 3′,4′,5-trihydroxy-3-methoxyflavon-7-olate, b) Agathisflavone, c) catiguanin B.
Fig. 9.
2D interaction diagram, showing the interacting flavonoid atoms with specific residues in active site of ßC1 protein. Dotted lines are showing specific interaction. a) 3′,4′,5,6-tetrahydroxy-3,7-dimethoxyflavone, b) quercetin-7-O-[alpha-L-rhamnopyranosyl(1->6)-beta-D-galactopyranoside, c) prunin 6″-O-gallate, d) luteolin 7-O-beta-D-glucosiduronic acid.
4. Discussion
ßC1 is a multifunction protein encoded by a betasatellite molecule associated with majority of Old World begomoviruses causing diseases in many crop plants [5]. Plant defense against viruses, include many mechanisms such as RNA interference (RNAi), ubiquitous antiviral defense, DNA methylation, hypersensitive response, systemic acquired resistance response and production of plant secondary metabolites [2]. The flavonoids compounds are known to produce under stress condition in plants [8]. Flavonoids are being utilized as a part of traditional Eastern medicine, having anti-inflammatory, antioxidant, anti-tumor and anti-proliferative activities [36, 37, 38]. The antiviral effect of naturally occurring flavonoids against human viruses like herpes simplex virus type 1 (HSV-1), polio-virus type 1, para-influenza virus type 3 (Pf-3), and respiratory syncytial virus (RSV) has been well established [39]. In our study we have analyzed the interaction of plant flavonoids with pathogenicity determinant viral protein ßC1 by computational based docking method. For docking analysis we need three-dimensional structure of protein. The structure of ßC1 protein was not determined experimentally so we developed model of this protein by homology modeling method as this method is recognized as most appropriate for protein modeling. The computer generated model of ßC1 was then evaluated by different online servers for its quality and stereochemistry. Protein structure in .pdb format is used for docking analysis. In docking analysis the binding energy of a compound and best pose of molecule for the binding with protein is predicted. In the present study, our focus was to find out the role of natural flavonoids against the viral protein ßC1. For this purpose, more than 100 flavonoid structures that are naturally produced by the plant downloaded from chebi. Then these flavonoids were virtually screened against the ßC1 protein through MOE software. Finally the ten flavonoids i.e., robinetinidol-(4alpha,8)-gallocatechin, quercetin 7-O-beta-D-glucoside, swertianolin, 3′,4′,5-trihydroxy-3-methoxyflavon-7-olate, agathisflavone, catiguanin B, 3′,4′,5,6-tetrahydroxy-3,7-dimethoxyflavone, quercetin-7-O-[alpha-L-rhamnopyranosyl(1->6)-beta-D-galactopyranoside], prunin 6″-O-gallate and luteolin 7-O-beta-D-glucosiduronic acid, were obtained as potential inhibitor of ßC1 on the basis of best docking scores against the active site of ßC1. Previously these flavonoids were known to have alpha amylase and lipase inhibitory activity [40], antioxidant activity inhibits reactions brought about by dioxygen or peroxides [41], acetylcholinesterase and monoamine oxidase inhibitory activities [42], enzymatic activity [43], heptoprotective activity [44], antioxidant activity [45]. Flavonoids 3′,4′,5,6-tetrahydroxy-3,7-dimethoxyflavone and prunin 6″-O-gallate have some unknown function. Flavonoids quercetin-7-O-[alpha-L-rhamnopyranosyl(1->6)-beta-D-galactopyranoside] and luteolin 7-O-beta-D-glucosiduronic acid have antimalarial activity [46], and radical scavengers activity respectively [47]. ßC1 protein not only enhances the infection of Cotton leaf curl virus but also involve in the transmission of virus from cell to cell. It interacts with various proteins of host cell machinery so that viral replication in host cells is enhanced resulting in the severity of symptoms. This protein is also involved the suppression of RNA interference machinery [48].
Our study suggests the potential role of flavonoids as inhibitors of ßC1. If more work is done on the flavonoid inhibition studies and finally the flavonoids can be hyper-produce to develop virus resistant crops. This study provided a new horizon for the scientists working on the plant defense mechanism against viruses and engineering resistance against these viruses.
5. Conclusion
Our study provided an insight of structure and interactions of ßC1 protein with plant flavonoids. The role of flavonoids under stress conditions i.e., both biotic and abiotic, is very well documented. This is first time that we are reporting a possible role of plant flavonoids in defense against the begomovirus infection. This information might be interesting to develop resistance against begomoviruses and add new information about plant defense mechanism based on the special compounds produced by the plants.
Declarations
Author contribution statement
Muhammad Waseem Sarwar, Adeel Riaz, Nazia Nahid: Performed the experiments.
Ahmed Al Qahtani, Ayesha Younus: Analyzed and interpreted the data; Wrote the paper.
Nisar Ahmed: Conceived and designed the experiments.
Muhammad Mubin, M. Shah Nawaz-Ul-Rehman: Conceived and designed the experiments; Analyzed and interpreted the data; Wrote the paper.
Funding statement
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Competing interest statement
The authors declare no conflict of interest.
Additional information
Data associated with this study has been deposited at Protein Model Database under the accession number PMID: PM0081495.
Acknowledgements
We are thankful to the members of virology lab for their contributions and help.
References
- 1.Rajagopalan P.A., Naik A., Katturi P., Kurulekar M., Kankanallu R.S., Anandalakshmi R. Dominance of resistance-breaking cotton leaf curl Burewala virus (CLCuBuV) in northwestern India. Arch. Virol. 2012;157:855–868. doi: 10.1007/s00705-012-1225-y. [DOI] [PubMed] [Google Scholar]
- 2.Zubair M., Zaidi S.S., Shakir S., Amin I., Mansoor S. An insight into cotton leaf curl multan betasatellite, the most important component of cotton leaf curl disease complex. Viruses. 2017:9. doi: 10.3390/v9100280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pandi G., Purushothaman P., Bhaskaran S., Jose J., Anandam K.Y., Usha R. Differential roles of C4 and beta C1 in mediating suppression of post-transcriptional gene silencing: evidence for transactivation by the C2 of Bhendi yellow vein mosaic virus, a monopartite begomovirus. Virus Res. 2007 doi: 10.1016/j.virusres.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 4.Qazi J., Amin I., Mansoor S., Iqbal M.J., Briddon R.W. Contribution of the satellite encoded gene betaC1 to cotton leaf curl disease symptoms. Virus Res. 2007;128:135–139. doi: 10.1016/j.virusres.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 5.Saeed M., Zafar Y., Randles J.W., Rezaian M.A. A monopartite begomovirus-associated DNA beta satellite substitutes for the DNA B of a bipartite begomovirus to permit systemic infection. J. Gen. Virol. 2007;88:2881–2889. doi: 10.1099/vir.0.83049-0. [DOI] [PubMed] [Google Scholar]
- 6.Haxim Y., Ismayil A., Jia Q., Wang Y., Zheng X., Chen T., Qian L., Liu N., Wang Y., Han S., Cheng J., Qi Y., Hong Y., Liu Y. Autophagy functions as an antiviral mechanism against geminiviruses in plants. Elife. 2017:6. doi: 10.7554/eLife.23897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tiwari N., Sharma P.K., Malathi V.G. Functional characterization of betaC1 gene of Cotton leaf curl Multan betasatellite. Virus Genes. 2013;46:111–119. doi: 10.1007/s11262-012-0828-4. [DOI] [PubMed] [Google Scholar]
- 8.Samanta A., Das G., Das S. 2011. Roles of Flavonoids in Plants. [Google Scholar]
- 9.Panche A.N., Diwan A.D., Chandra S.R. Flavonoids: an overview. J. Nutr. Sci. 2016;5:e47. doi: 10.1017/jns.2016.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Forkmann G. Flavonoids as flower pigments: the formation of the natural spectrum and its extension by genetic engineering. Plant Breed. 1991;106:1–26. [Google Scholar]
- 11.Ramírez D., Naranjo B., Duchicela J. 2017. Stimulation of germination of spores and root colonization of Diversispora trimulares by flavonoids in Nicotiana glauca root exudates. [Google Scholar]
- 12.Pourcel L., Routaboul J.M., Cheynier V., Lepiniec L., Debeaujon I. Flavonoid oxidation in plants: from biochemical properties to physiological functions. Trends Plant Sci. 2007;12:29–36. doi: 10.1016/j.tplants.2006.11.006. [DOI] [PubMed] [Google Scholar]
- 13.Corradini E., Foglia P., Giansanti P., Gubbiotti R., Samperi R., Lagana A. Flavonoids: chemical properties and analytical methodologies of identification and quantitation in foods and plants. Nat. Prod. Res. 2011 doi: 10.1080/14786419.2010.482054. [DOI] [PubMed] [Google Scholar]
- 14.Tanaka Y., Sasaki N., Ohmiya A. Biosynthesis of plant pigments: anthocyanins, betalains and carotenoids. Plant J. 2008;54:733–749. doi: 10.1111/j.1365-313X.2008.03447.x. [DOI] [PubMed] [Google Scholar]
- 15.Kumar S., Pandey A.K. Chemistry and biological activities of flavonoids: an overview. Sci. World J. 2013;2013:16. doi: 10.1155/2013/162750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kikuchi K., Matsushita N., Suzuki K., Hogetsu T. Flavonoids induce germination of basidiospores of the ectomycorrhizal fungus Suillus bovinus. Mycorrhiza. 2007;17:563–570. doi: 10.1007/s00572-007-0131-8. [DOI] [PubMed] [Google Scholar]
- 17.Yamasaki H., Sakihama Y., Ikehara N. Flavonoid-peroxidase reaction as a detoxification mechanism of plant cells against H2O2. Plant Physiol. 1997;115:1405–1412. doi: 10.1104/pp.115.4.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jansen M.A., van den Noort R.E., Tan M.Y., Prinsen E., Lagrimini L.M., Thorneley R.N. Phenol-oxidizing peroxidases contribute to the protection of plants from ultraviolet radiation stress. Plant Physiol. 2001;126:1012–1023. doi: 10.1104/pp.126.3.1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gottstein D., Gross D. Phytoalexins of woody plants. Trees. 1992;6:55–68. [Google Scholar]
- 20.Parniske M., Ahlborn B., Werner D. Isoflavonoid-inducible resistance to the phytoalexin glyceollin in soybean rhizobia. J. Bacteriol. 1991;173:3432–3439. doi: 10.1128/jb.173.11.3432-3439.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Falcone Ferreyra M.L., Rius S.P., Casati P. Flavonoids: biosynthesis, biological functions, and biotechnological applications. Front. Plant Sci. 2012;3:222. doi: 10.3389/fpls.2012.00222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gasteiger E., Gattiker A., Hoogland C., Ivanyi I., Appel R.D., Bairoch A. ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003;31:3784–3788. doi: 10.1093/nar/gkg563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marchler-Bauer A., Lu S., Anderson J.B., Chitsaz F., Derbyshire M.K., DeWeese-Scott C., Fong J.H., Geer L.Y., Geer R.C., Gonzales N.R., Gwadz M., Hurwitz D.I., Jackson J.D., Ke Z., Lanczycki C.J., Lu F., Marchler G.H., Mullokandov M., Omelchenko M.V., Robertson C.L., Song J.S., Thanki N., Yamashita R.A., Zhang D., Zhang N., Zheng C., Bryant S.H. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2011;39:D225–D229. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Finn R.D., Attwood T.K., Babbitt P.C., Bateman A., Bork P., Bridge A.J., Chang H.-Y., Dosztányi Z., El-Gebali S., Fraser M., Gough J., Haft D., Holliday G.L., Huang H., Huang X., Letunic I., Lopez R., Lu S., Marchler-Bauer A., Mi H., Mistry J., Natale D.A., Necci M., Nuka G., Orengo C.A., Park Y., Pesseat S., Piovesan D., Potter S.C., Rawlings N.D., Redaschi N., Richardson L., Rivoire C., Sangrador-Vegas A., Sigrist C., Sillitoe I., Smithers B., Squizzato S., Sutton G., Thanki N., Thomas P.D., Tosatto Silvio C.E., Wu C.H., Xenarios I., Yeh L.-S., Young S.-Y., Mitchell A.L. InterPro in 2017—beyond protein family and domain annotations. Nucleic Acids Res. 2017;45:D190–D199. doi: 10.1093/nar/gkw1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Letunic I., Bork P. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 2018;46:D493–D496. doi: 10.1093/nar/gkx922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Finn R.D., Coggill P., Eberhardt R.Y., Eddy S.R., Mistry J., Mitchell A.L., Potter S.C., Punta M., Qureshi M., Sangrador-Vegas A., Salazar G.A., Tate J., Bateman A. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 2016;44:D279–D285. doi: 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sarwar M.W., Saleem I.B., Ali A., Abbas F. Insilico characterization and homology modeling of Arabitol Dehydrogenase (ArDH) from Candida albican. Bioinformation. 2013;9:952–957. doi: 10.6026/97320630009952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Benkert P., Biasini M., Schwede T. Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics. 2011;27:343–350. doi: 10.1093/bioinformatics/btq662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marti-Renom M.A., Stuart A.C., Fiser A., Sanchez R., Melo F., Sali A. Comparative protein structure modeling of genes and genomes. Annu. Rev. Biophys. Biomol. Struct. 2000;29:291–325. doi: 10.1146/annurev.biophys.29.1.291. [DOI] [PubMed] [Google Scholar]
- 30.Wiederstein M., Sippl M.J. ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007;35:W407–W410. doi: 10.1093/nar/gkm290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Benkert P., Kunzli M., Schwede T. QMEAN server for protein model quality estimation. Nucleic Acids Res. 2009;37:W510–W514. doi: 10.1093/nar/gkp322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Laskowski R.A., Rullmannn J.A., MacArthur M.W., Kaptein R., Thornton J.M. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 33.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera – a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 34.Hira Iftikhar S.R. Molecular docking studies of flavonoids for their inhibition pattern against β-catenin and pharmacophore model generation from experimentally known flavonoids to fabricate more potent inhibitors for Wnt signaling pathway. Phcog. Mag. 2014;10:S264. doi: 10.4103/0973-1296.133269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shen W., Lu Y.-H. Molecular docking of citrus flavonoids with some targets related to diabetes. Bangladesh J. Pharmacol. 2013;8:156–170. [Google Scholar]
- 36.Williams R.J., Spencer J.P., Rice-Evans C. Flavonoids: antioxidants or signalling molecules? Free Radic. Biol. Med. 2004;36:838–849. doi: 10.1016/j.freeradbiomed.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 37.Singh R.P., Gu M., Agarwal R. Silibinin inhibits colorectal cancer growth by inhibiting tumor cell proliferation and angiogenesis. Cancer Res. 2008;68:2043–2050. doi: 10.1158/0008-5472.CAN-07-6247. [DOI] [PubMed] [Google Scholar]
- 38.Sung B., Pandey M.K., Aggarwal B.B. Fisetin, an inhibitor of cyclin-dependent kinase 6, down-regulates nuclear factor-kappaB-regulated cell proliferation, antiapoptotic and metastatic gene products through the suppression of TAK-1 and receptor-interacting protein-regulated IkappaBalpha kinase activation. Mol. Pharmacol. 2007;71:1703–1714. doi: 10.1124/mol.107.034512. [DOI] [PubMed] [Google Scholar]
- 39.Kaul T.N., Middleton E., Jr., Ogra P.L. Antiviral effect of flavonoids on human viruses. J. Med. Virol. 1985;15:71–79. doi: 10.1002/jmv.1890150110. [DOI] [PubMed] [Google Scholar]
- 40.Kusano R., Ogawa S., Matsuo Y., Tanaka T., Yazaki Y., Kouno I. α-Amylase and lipase inhibitory activity and structural characterization of acacia bark proanthocyanidins. J. Nat. Prod. 2011;74:119–128. doi: 10.1021/np100372t. [DOI] [PubMed] [Google Scholar]
- 41.Bazylko A., Stolarczyk M., Derwińska M., Kiss A.K. Determination of antioxidant activity of extracts and fractions obtained from Galinsoga parviflora and Galinsoga quadriradiata, and a qualitative study of the most active fractions using TLC and HPLC methods. Nat. Prod. Res. 2012;26:1584–1593. doi: 10.1080/14786419.2011.582469. [DOI] [PubMed] [Google Scholar]
- 42.Urbain A., Marston A., Grilo L.S., Bravo J., Purev O., Purevsuren B., Batsuren D., Reist M., Carrupt P.A., Hostettmann K. Xanthones from Gentianella amarella sp. acuta with acetylcholinesterase and monoamine oxidase inhibitory activities. J. Nat. Prod. 2008;71:895–897. doi: 10.1021/np070690l. [DOI] [PubMed] [Google Scholar]
- 43.De Luca V., Ibrahim R.K. Enzymatic synthesis of polymethylated flavonols in Chrysosplenium americanum. I. Partial purification and some properties of S-adenosyl-L-methionine:flavonol 3-, 6-, 7-, and 4′-O-methyltransferases. Arch. Biochem. Biophys. 1985;238:596–605. doi: 10.1016/0003-9861(85)90205-x. [DOI] [PubMed] [Google Scholar]
- 44.Anand K.K., Gupta V.N., Rangari V., Singh B., Chandan B.K. Structure and hepatoprotective activity of a biflavonoid from Canarium manii. Planta Med. 1992;58:493–495. doi: 10.1055/s-2006-961533. [DOI] [PubMed] [Google Scholar]
- 45.Tang W., Hioki H., Harada K., Kubo M., Fukuyama Y. Antioxidant phenylpropanoid-substituted epicatechins from Trichilia catigua. J. Nat. Prod. 2007;70:2010–2013. doi: 10.1021/np0703895. [DOI] [PubMed] [Google Scholar]
- 46.Ovenden S.P.B., Cobbe M., Kissell R., Birrell G.W., Chavchich M., Edstein M.D. Phenolic glycosides with antimalarial activity from Grevillea “Poorinda queen”. J. Nat. Prod. 2011;74:74–78. doi: 10.1021/np100737q. [DOI] [PubMed] [Google Scholar]
- 47.Dapkevicius A., van Beek T.A., Lelyveld G.P., van Veldhuizen A., de Groot A., Linssen J.P.H., Venskutonis R. Isolation and structure elucidation of radical scavengers from Thymus vulgaris leaves. J. Nat. Prod. 2002;65:892–896. doi: 10.1021/np010636j. [DOI] [PubMed] [Google Scholar]
- 48.Zubair M., Zaidi S., Shakir S., Amin I., Mansoor S. An insight into Cotton leaf curl Multan betasatellite, the most important component of cotton leaf curl disease complex. Viruses. 2017;9:280. doi: 10.3390/v9100280. [DOI] [PMC free article] [PubMed] [Google Scholar]