Abstract
SQUAMOSA-promoter binding protein (SBP)-box genes encode a family of plant-specific transcription factors that play vital roles in plant growth and development. In this study, 15 SBP-box genes were identified and isolated from Citrus clementina (CclSBPs), where 10 of these genes were predicted to be putative targets of Citrus clementina microRNA156 (CclmiR156). The 15 CclSBP genes could be classified into six groups based on phylogenetic analysis, diverse intron–exon structure, and motif prediction, similar to the SQUAMOSA promoter binding protein-like (SPL) gene family of Populus trichocarpa and Arabidopsis thaliana. Furthermore, CclSBPs classified into a group/subgroup have similar gene structures and conserved motifs, implying their functional redundancy. Tissue-specific expression analysis of CclSBPs demonstrated their diversified expression patterns. To further explore the potential role of CclSBPs during floral inductive water deficits, the dynamic changes of the 15 CclSBPs were investigated during floral inductive water deficits, and the results showed that some CclSBPs were associated with floral induction. Among these genes, CclSBP6 was not homologous to the Arabidopsis SBP-box gene family, and CclSBP7 was regulated by being alternatively spliced. Therefore, CclSBP6 and CclSBP7 were genetically transformed in Arabidopsis. Overexpression of the two genes changed the flowering time of Arabidopsis.
Keywords: Citrus clementina, SBP-box genes, gene expression, evolutionary comparison, functional diversity
1. Introduction
SQUAMOSA-promoter binding protein (SBP)-box genes are a family of plant-specific transcript factors that play crucial roles in the regulation of plant growth and development [1]. The common feature of SPP genes is that their protein products contain a highly conserved SBP-box DNA-binding domain (approximately 76 amino acid residues). This domain features a zinc finger motif that contains two zinc finger domains. A putative nuclear localization signal (NLS) is located at the C-terminal of the SBP domain, which partly overlaps with the DNA-binding domain, particularly with the second zinc finger domain [2,3]. The SBP-box genes were first identified in snapdragon (Antirrhinum majus L.) [4], followed by other plants, such as Arabidopsis thaliana [5], silver birch (Betula pendula) [6], green alga (Chlamydomonas reinhardtii) [7], rice (Oryza sativa) [8], maize (Zea mays) [9], poplar (Populus trichocarpa) [10], tomato (Solanum lycopersicum) [11], apple (Malus domestica) [12], and grape (Vitis vinifera) [13]. In these plants, these genes were revealed to control flowering and fruit development, as well as other significant physiological processes.
A total of 16 SBP-box genes were identified in the Arabidopsis genome [5]. Ten members of the 16 AtSPLs including, AtSPL2-6, AtSPL9-11, AtSPL13, and AtSPL15, were targets of miR156 [14,15,16,17]. AtSPL3, AtSPL4 and AtSPL5 contain complementary sequences of miR156 in the 3′-UTR region, and they promote vegetative phase change and flowering [5,15,18]. AtSPL2, AtSPL10, and AtSPL11 play important roles in regulating the morphological traits of leaves and flowers [19]. AtSPL9 and AtSPL15 are functionally redundant, and both of them can control the juvenile-to-adult phase transition and the leaf initiation rate [20]. in Arabidopsis, six SBP-box genes, including AtSPL1, AtSPL7, AtSPL8, AtSPL12, AtSPL14, and AtSPL16, are not targets of miR156. Among them, AtSPL7 is involved in the regulation of copper homeostasis by controlling the expression of COPT6 and its role in root and shoot development [21]. AtSPL8 plays pivotal roles in regulating pollen sac development, male fertility, and Gibberellin (GA) biosynthesis and signaling [22,23,24]. AtSPL14 regulates plant development and sensitivity to fumonisin B1 [25]. To date, the functions of AtSPL1, AtSPL6, AtSPL12, AtSPL13, and AtSPL16 are still unknown, and further characterization is required to determine their function.
Currently, many members of the SBP-box family have been functionally characterized in plants other than Arabidopsis, and they have been shown to have diverse functions in plant growth and development. For instance, BpSPL1 from Betula pendula was shown to be capable of binding a cis-element of MADS5, a close homolog of the Arabidopsis FRUITFULL gene, and this indicated a role for birch SPL genes in the regulation of flower development [6]. In the unicellular alga, an SBP-box protein from Chlamydomonas reinhardtii has been identified to regulate nutritional copper signaling [7]. In rice, OsSPL14 regulated by miR156 controls shoot branching in the vegetative phase [26]. Additionally, an SBP-box gene at the Colorless non-ripening (Cnr) locus from tomatoes is a pivotal factor in fruit ripening [27], while the maize tasselsheath4 (tsh4) SBP-box gene regulates bract development and is important for branch meristem initiation and maintenance [28].
Citrus is economically one of the most important fruit crops in the world. To date, the analysis of SBP-box genes has focused mainly on the model plants, such as Arabidopsis and rice. However, little information is known about the evolution, functions, and expression patterns of the SBP-box genes in citrus compared with the model plants. Recently, miR156-SPL modules have been reported to regulate multiple biological processes in citrus, including juvenile-to-adult phase transition, drought stress induced flowering, fruit load, somatic embryogenesis, and starch accumulation of the callus cells [29,30,31,32]. In order to further elucidate the regulatory networks and functional characteristics of SBP-box genes during the process of growth and development in citrus, we identified 15 SBP-box genes from the citrus genome and further cloned these genes in this study, and then we performed a comprehensive analysis of the chromosomal location, phylogeny, gene structures, conserved motifs, and miR156 target sites. The phylogenetic relationship and evolutionary comparison of SBP-box genes were also performed among various plants. Furthermore, the expression pattern of the 15 CclSBPs was studied in various tissues, as well as in the water deficit floral inductive condition. Moreover, transformation assays with two CclSBP genes suggested an important role in controlling the flowering of citrus. These results lay the foundation for further functional analyses of SBP-box genes in citrus and related species.
2. Materials and Methods
2.1. Plant Materials
The citrus samples were collected from the experiment fields at the National Citrus Breeding Center at Huazhong Agricultural University, Wuhan, China (30° 28′ N, 114° 21′ E, 30). In this study, we used 14-year-old Clementine mandarin trees (Citrus clementina cv. Monreal) propagated by bud grafting to trifoliate orange rootstocks (Citrus trifoliata), where a total of 9 adult healthy trees were selected and randomly divided into three groups with three plants per group. All the samples were collected from three groups of trees and used as biological repeats. For spatial expression analysis of the CclSBPs, the samples were collected and total RNA was isolated from healthy and mature leaves, flowers at full bloom, young fruits at 90 days after flowering, mature fruit at the picking period, and roots of C. clementina, respectively. The tissues were named Rt (roots), Sm (stems), Lf (leaves), Pe (petal), St (stamen), Ca (carpel), Re (receptacle), YF (young fruits), YS (young seeds), MF (mature fruits), and MS (mature seeds). All plant tissues were sampled according to the demands of each experiment, and they were immediately frozen in liquid nitrogen and stored at −80 °C until used.
2.2. Identification and Molecular Cloning of CclSBPs
The nucleotide and deduced amino acid sequences of 16 SPL genes from Arabidopsis [5] were obtained from TAIR (The Arabidopsis Information Resource) database [33]. A genome-wide search of CclSBP genes was carried out using Basic Local Alignment Search Tool (BLAST) analyses, with the 16 AtSPLs genes used as queries against the C. clementina genome [34]. All non-redundant protein sequences of the putative citrus SBP-box genes were checked for the SBP domain using the protein families database (Pfam) [35]. To verify the coding regions of CclSBPs, gene-specific primers were designed for amplification of the 15 CclSBP genes using polymerase chain reaction (PCR) with cDNA templates from leaves of C. clementina (Supplementary Table S1). The PCR was performed in a 20 µl system, in a Veriti 96-Well Thermal Cycler. Amplified products were separated in a 1.0% agarose gel electrophoresis. The target bands were recovered, cloned into pMD18-T vectors (TaKaRa Biotech, Dalian, China), and then transformed into the Escherichia coli strain DH5a. Three positive clones were sequenced for each candidate SBP-box gene.
2.3. Phylogenetic Analysis of CclSBPs
Multiple alignments of the SBP-box protein sequences were performed using the ClustalW program [36]. The sequence logo was obtained using the online Weblogo platform [37]. The phylogenetic trees were generated by MEGA 6.0, using the maximum likelihood (ML) algorithm [38]. Bootstrap analysis with 1000 replicates was used to evaluate the significance of the nodes. Jones–Taylor–Thornton (JTT) model was used to ensure that the divergent domains could contribute to the topology of the ML tree [38].
2.4. Gene Structure, Conserved Domain and miR156 Target Site Analysis of CclSBPs
The exon/intron organization of CclSBPs was determined by comparing the coding sequences to their corresponding genomic sequences using the gene structure display server (GSDS) program [39]. The simple modular architecture research tool (SMART) and multiple em for motif elicitation (MEME) were used to identify the conserved motif structures of the SBP-box protein sequences [40,41]. The sequence target of miR156 (miR156a and miR156b) was determined in a previous work on C. trifoliata [42]. The targets of miR156 were predicted by searching the coding regions, as well as the 3′-UTR of all the CclSBP genes, using the psRNATarget tool with default parameters as in Reference [43].
2.5. Identification of the Specific Repetitive Elements of CclSBPs
To determine whether specific repetitive elements drive the sequence divergence of CclSBPs, the tandem repeat (TR), transposable element (TE), low-complexity repeat (LCR), and simple sequence repeat (SSR) were investigated in the region that was 1.5 kb upstream of the 5′ UTR to the 3’-UTR of the genes. TRs were searched using the Tandem Repeat Finder 4.04 [44], with default parameter values, respectively. TEs and LCRs were detected using RepeatMasker [45]. SSRs were examined using the web-based software SSRIT [46]. Putative cis-elements in the promoter regions of CclSBPs were annotated using the PlantCARE software [47]. The motifs putatively involved in plant growth and development, hormone responses, light response, and stress responses were summarized in this study.
2.6. Co-Expression Analysis of CclSBPs During Floral Inductive Water Deficits
The published transcriptome from the lemon bud provided a useful complement to understand the expression patterns of the CclSBPs during floral inductive water deficits [32]. The co-expression network of the CclSBPs was constructed, the correlations between two genes above 0.85 were kept, and then visualized in Cytoscape as in Reference [48]. The Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses of genes in the co-expression network were conducted in R, using the packages “clusterProfiler” and “pathview” as described in reference [49,50]. The predicted protein interaction network of CclSBPs proteins in citrus was built according to knowledge obtained from Arabidopsis.
2.7. Real-Time Polymerase Chain Reaction (RT-PCR)
Total RNA was isolated using the Oligotex mRNA mini kit (Qiagen, Gaithersburg, MD, USA), according to the manufacturer’s instructions. The RNA preparation was treated with DNase I (Promega, Madison, WI, USA), and first strand synthesis of the cDNA was performed using RT Primer Mix and Primescript RT Enzyme Mix I (life technology). Three biological replicates were performed in this study. To normalize the variance among samples, the expression level of the citrus β-actin was used as the internal control. The program was performed as described previously in [51]. The gene-specific primers for real-time polymerase chain reaction (RT-PCR) of CclSBPs were listed in Table S1.
2.8. Arabidopsis Transformation
The full-length cDNA sequences were ligated into pBI121 driven by CaMV35S, and then transferred into the Agrobacterium strain GV3101. Arabidopsis (Col) was transformed using the floral dip method as in Reference [52]. The third generation of transgenic lines and wild type were grown under long-day conditions. Plants were arranged in a randomized complete block design with three blocks. Each genotype within a block was represented by five plants. Thus, 15 plants from each genotype were observed. Flowering time was measured by counting the number of rosette leaves and the number of days from sowing to when the first flower bud was seen. Statistical analysis was performed by one-way analysis of variance (ANOVA), taking p < 0.05 as significant.
3. Results
3.1. Identification, Cloning and Sequence Feature Analysis of CclSBPs
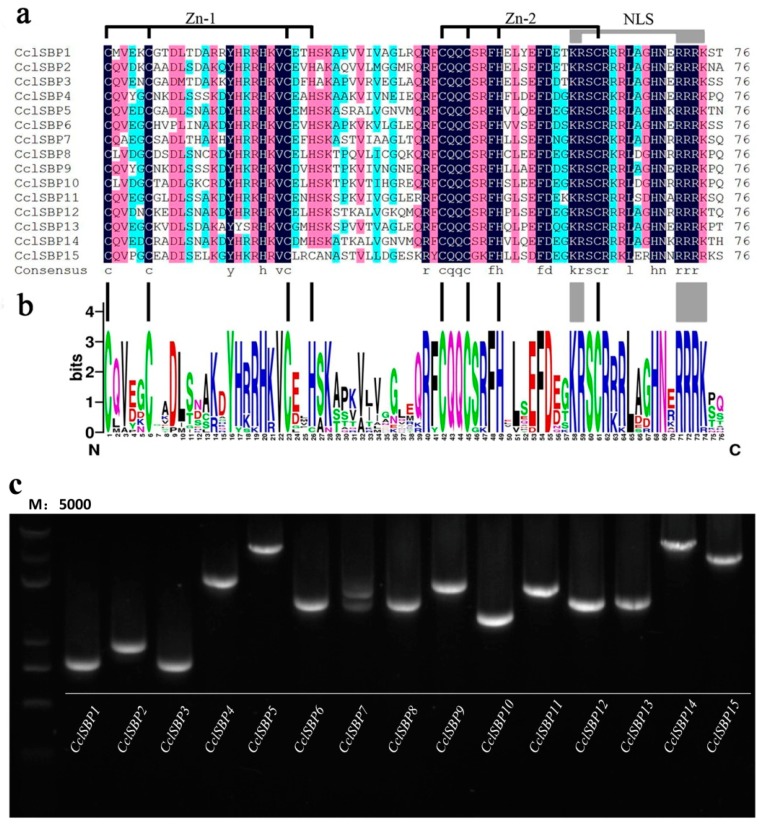
A total of 15 unique genes containing SBP motif were indentified after removing the redundant sequences in this study and they were assigned names from CclSBP1 to CclSBP15 based on their location on seven scaffolds (equivalent to seven chromosomes). The length of the CclSBPs varied from 1247 (CclSBP1) to 7069 bp (CclSBP15), and the length of the coding sequences varied from 393 (CclSBP1) to 3309 bp (CclSBP12). Furthermore, the number of transcripts of each CclSBP gene and the percentage of the expressed sequence tags (EST) that match with the database are shown in Table 1. CclSBPs were unevenly distributed across seven chromosomes: chromosomes 3 and 4 displayed four genes, chromosomes 2 and 9 had three genes, and chromosomes 1, 5, and 6 only showed one gene (Table 1). There was only one conserved domain (the SBP domain) found to be shared by all the CclSBPs. The SBP domains of the CclSBPs were very similar, with a high conservation at certain positions (Figure 1a). All of the CclSBPs shared two zinc finger-like structures (Zn-1, Zn-2) and a highly conserved bipartite nuclear localization signal (NLS). For all the CclSBPs, the second zinc finger within the SBP domain was CysCysHisCys, whilst the first zinc finger was CysCysCysHis, except for the CclSBP15, where the other zinc finger type was CysCysCysCys (Figure 1b). Remarkably, the second zinc finger-like structure of all the CclSBPs was partly overlapped with the nuclear localization signal.
Table 1.
The feature of the SQUAMOSA-promoter binding protein (SBP)-box gene family in citrus.
| Gene Name | Gene Locus | Genomic (bp) | CDs (bp) | No. of Transcript | EST Match | miR156 Target |
|---|---|---|---|---|---|---|
| CclSBP1 | Ciclev10009879m | 1247 | 393 | 1 | +a | 3′-UTR |
| CclSBP2 | Ciclev10016841m | 2716 | 570 | 2 | +a | 3′-UTR |
| CclSBP3 | Ciclev10017104m | 1265 | 432 | 1 | +b | 3′-UTR |
| CclSBP4 | Ciclev10019546m | 3662 | 1674 | 2 | +a | exon |
| CclSBP5 | Ciclev10018697m | 6422 | 2967 | 1 | +c | no |
| CclSBP6 | Ciclev10020532m | 4055 | 1170 | 2 | +a | exon |
| CclSBP7 | Ciclev10021106m | 2387 | 987 | 3 | +a | no |
| CclSBP8 | Ciclev10031834m | 3660 | 1140 | 1 | +a | exon |
| CclSBP9 | Ciclev10031270m | 5184 | 1536 | 2 | +a | exon |
| CclSBP10 | Ciclev10032171m | 3272 | 939 | 1 | +b | exon |
| CclSBP11 | Ciclev10031391m | 5362 | 1443 | 8 | +a | exon |
| CclSBP12 | Ciclev10000100m | 5217 | 3309 | 1 | +a | no |
| CclSBP13 | Ciclev10011938m | 2986 | 1167 | 2 | +a | exon |
| CclSBP14 | Ciclev10004227m | 6489 | 3117 | 6 | +a | no |
| CclSBP15 | Ciclev10004348m | 7069 | 2397 | 2 | +a | no |
Note: The result of the expressed sequence tags (EST) match comes from the HarvEST BLAST(Basic Local Alignment Search Tool) Search on NCBI (National Center for Biotechnology Information, Citrus C52 relaxed), +a represents the percentage of EST match more than 95%, +b represents the percentage match between 80–90%, +c represents the percentage match lower than 80%.
Figure 1.
SBP-domain alignment of the CclSBPs and polymerase chain reaction (PCR) cloning. (a) Multiple alignment of the SBP domains from the citrus SBP-box protein based on ClustalW program, and the two conserved zinc finger structures (Zn-1, Zn-2) and nuclear localization signal (NLS) are demonstrated; (b) sequence logo of the citrus SBP domain. The overall height of each stack represents the degree of conservation at this position, while the height of the letters within each stack indicates the relative frequency of the corresponding amino acids; (c) Detection of 15 the CclSBP family members in Citrus clementina. Lane 1: M, molecular marker, Marker 5000; From Lane 2 to Lane 16 is CclSBP1, CclSBP2, CclSBP3, CclSBP4, CclSBP5, CclSBP6, CclSBP7, CclSBP8, CclSBP9, CclSBP10 CclSBP11, CclSBP12, CclSBP13, CclSBP14, and CclSBP15, respectively.
To further verify the results of the CclSBP gene models obtained from computational prediction, we cloned and sequenced the coding region of all the CclSBP (Figure 1c). For most CclSBP genes, clear single bands were amplified, except CclSBP7. Three positive cloning were sequenced for each gene, and the complete cDNA of each gene was submitted to the GenBank (KT601172–KT601186). Subsequently, the alignment between these cloned cDNA and the coding sequences from the C. clementina genome database were investigated, and the results showed that no differences were found in eight CclSBPs (CclSBP2, CclSBP4, CclSBP5, CclSBP6, CclSBP7, CclSBP8, CclSBP10, and CclSBP12). Only one single nucleotide polymorphism (SNP) site was found in CclSBP1, CclSBP3, CclSBP11, and CclSBP13, respectively; three SNP sites were found in CclSBP15; 36 SNP sites were found in CclSBP14; and one 15 bp deletion was found in CclSBP9.
3.2. Gene Structure, Conserved Domain and miR156 Target Site Analysis of CclSBPs
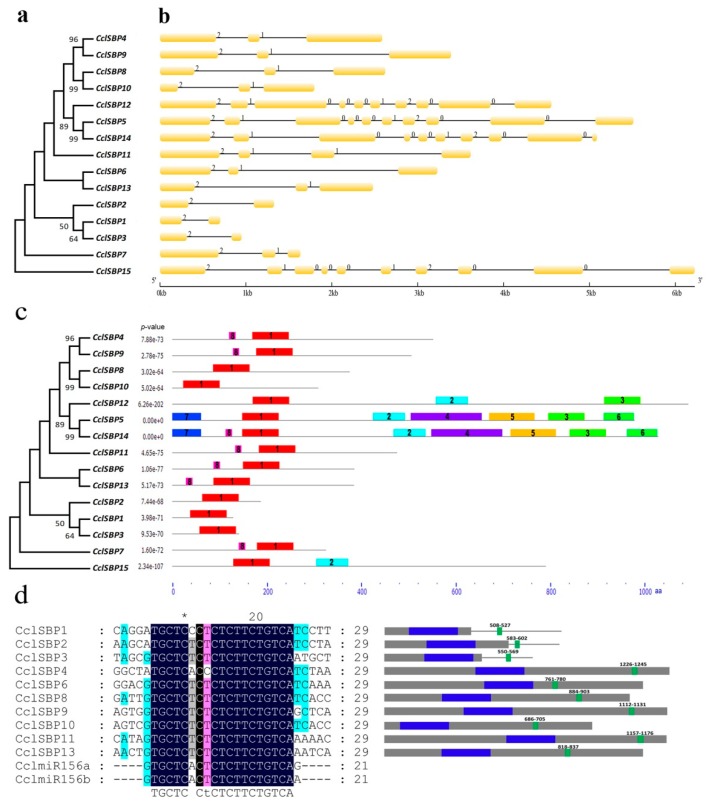
To provide further insights into the evolutionary relationship of CclSBPs, a phylogenetic tree was constructed based on the citrus full-length SBP-box protein sequences. The 15 SBP-box genes were clustered into different groups based on the evolutionary relationship (Figure 2a). The exon/intron organization in the coding sequences of each CclSBP gene was also performed, and the number of introns in the coding region of 15 CclSBPs varied from 1 to 9 (Figure 2b). Most closely related members shared similar exon/intron structures in terms of intron number and exon length. In addition to the SBP domain, other conserved motifs could also be important for the functioning of SBP-box proteins. Hence, we further searched the conserved motifs using the Multiple Em for Motif Elicitation (MEME) online server and we applied an e-value cut-off of 10−10 to the recognition. As a result, eight conserved motifs were discovered among the 15 CclSBPs proteins (Figure 2c). The number of motifs in each CclSBPs varied from 1 to 8 (Figure 2c). Motif 1 was actually the SBP domain and it existed in all the CclSBPs. However, the putative functions of the other motifs are currently unknown and they need to be further investigated. As expected, most of the closely related members had common motif compositions, such as CclSBP4 and CclSBP9, CclSBP8 and CclSBP10, CclSBP5 and CclSBP14, CclSBP1 and CclSBP3. This probably implied functional similarities in plant growth and development amongst these CclSBPs which shared similar gene structures and conserved motifs.
Figure 2.
Phylogenetic tree, gene structures and conserved motifs of the CclSBPs. (a) Phylogenetic tree was constructed based on 15 citrus SBP-box protein sequences; (b) exon-intron structures of the CclSBPs. Introns are represented by lines. Exons are indicated by yellow boxes; (c) distribution of the conserved motifs in the CclSBPs. Motifs represented with boxes are predicted using Multiple Em for Motif Elicitation (MEME). The number in the boxes (1–8) represents motif 1–motif 8, respectively. Box size indicates the length of the motifs; (d) Multiple alignment of the CclmiR156 complementary sequence with 10 CclSBPs and their binding site position.
To understand the miR156-mediated posttranscriptional regulation of CclSBPs, a total of 10 CclSBPs were predicted to be targets of miR156 (Figure 2d). The target sites of miR156 were located in the coding regions for five CclSBPs (CclSBP4, CclSBP6, CclSBP9, CclSBP11, and CclSBP13) and two CclSBPs (CclSBP8 and CclSBP10) belonging to two different subgroups. The target sites for the other three CclSBPs (CclSBP1, CclSBP2, and CclSBP3) belonging to a same subgroup, were located in the 3’-UTR regin. It suggested that miR156-mediated posttranscriptional regulation of SBP-box genes was conserved in these plant species.
3.3. Comparative and Phylogenetic Analysis of the SBP-Box Genes in Various Plants
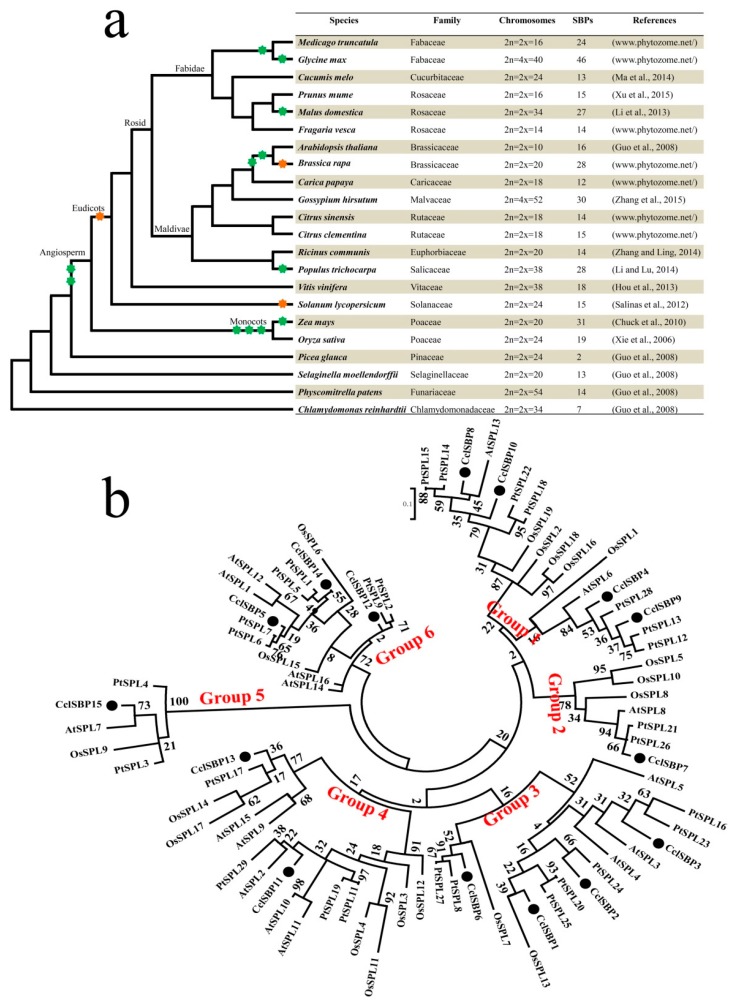
Previous studies and a growing number of fully sequenced plant genomes make it possible to perform a comparative genomic analysis of the SBP-box gene family across a wide range of plant species. In this study, we analyzed some of the major types of model organisms whose genomes have already been sequenced, including green algae, chlorophytes, moss, lycophyte, eudicots and monocots (Figure 3a). The number of SBP-box genes has been identified in 15 plant species, and the SBP genes of seven plant species from the Phytozome database (Medicago truncatula, Glycine max, Fragaria vesca, Brassica rapa, Carica papaya, Citrus sinensis and C. clementina) were checked in our study. We constructed a phylogenetic tree and displayed the duplication events of these 22 species (Figure 3a). In brief, the number of SBP-box genes in algal, lowland species, monocots, and dicots offered further insight into the evolutionary processes of this gene family.
Figure 3.
Comparative and phylogenetic analysis of the SBP genes in various plants. (a) The species tree and number of SBP-box paralogs in each species. The species tree and the duplication events are based on information in phytozome and plant genome duplication database (PGDD). The green star represents whole genome duplication and the yellow star represents whole genome triplication, respectively. Branch lengths do not represent time or the relative amount of character change; (b) phylogenetic analysis of citrus and other plant SBP-box genes. Phylogenetic tree was constructed using SBP domain protein sequences from citrus, Populus (PtSPLs), Arabidopsis (AtSPLs), and rice (OsSPLs), using MEGA 6.0 software [38].
On the other hand, to further investigate the evolutionary relationship between citrus, Arabidopsis, poplar and rice, a total of 79 SBP-box genes were used for phylogenetic analysis based on their conserved SBP domains (Figure 3b). These SBP genes were also clustered into six groups, each of which contained at least one AtSPL and one CclSBP. It was shown that the number of citrus, Arabidopsis, Populus and rice SBP genes varied across the six groups (the CclSBPs in group 1 to group 6 had 4, 1, 4, 2, 1, 3 members, respectively; the PtSPLs in group 1 to group 6 had 7, 2, 7, 4, 2, 6 members, respectively; the OsSPLs in group 1 to group 6 had 5, 3, 2, 6, 1, 2 members, respectively; and the AtSPLs in group 1 to group 6 had 2, 1, 3, 5, 1, 4 members, respectively). Interestingly, the SBP domain of SBP-box genes in group 5 were divergent from the other groups (Figure 3b). The N-terminal zinc finger of group 5 SBP-box genes has four cysteine residues in the SBP domain, while SBP-box genes in other groups mainly contain three cysteines and one histidine. Based on the phylogenetic tree, the miR156-targeted SBP-box genes, including sequences from rice, Arabidopsis and poplar, were distributed into only three of the subgroups (Groups 1, 3 and 4). Generally, most of the CclSBPs showed a closer phylogenetic relationship with PtSPLs than AtSPLs, and they showed the most distant phylogenetic relationship with OsSPLs. These results indicated that orthologous genes between woody plants showed higher similarities than those genes between wood plants and herbaceous plants, and orthologous genes between dicots had a higher similarity than between monocots and dicots. Additionally, it is worth noting that the clusters group together Arabidopsis, rice, poplar, and citrus genes, which are closer to their orthologous counterparts from the other species than to the other family members from their own species. These results indicated that the SBP-box family of genes was present in the ancestor plants that gave rise to the monocot and dicot lineages, making it possible to estimate the minimum number of SBP-box genes in this ancestor.
3.4. Specific Repetitive Elements in the CclSBPs
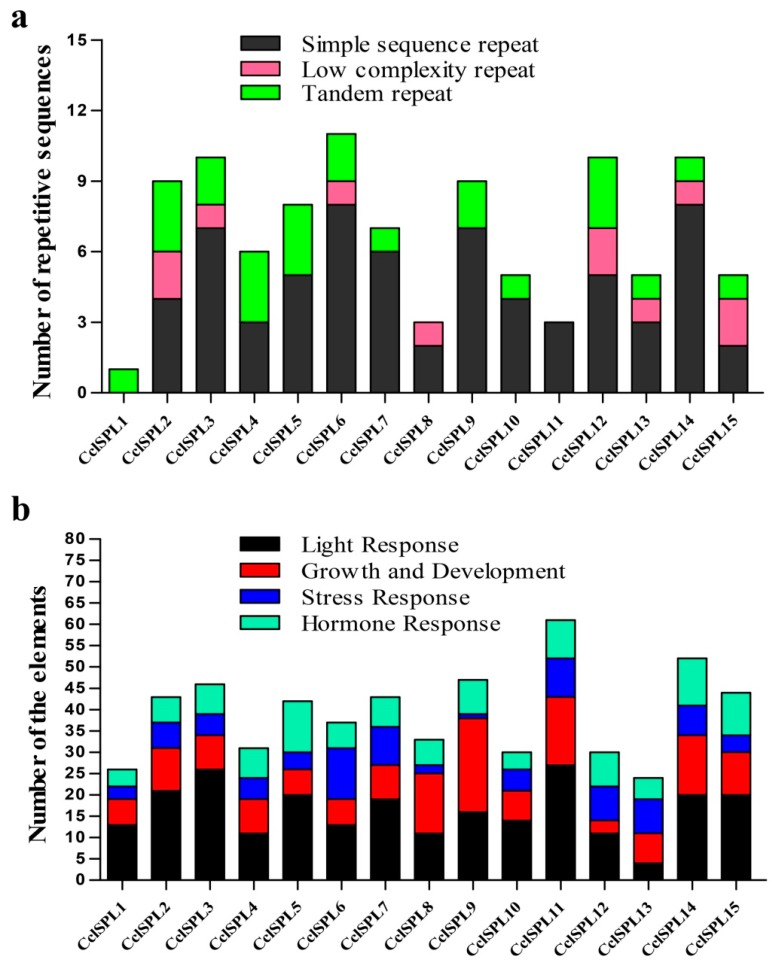
To characterize the sequence divergence of CclSBPs, the spatial distribution of repetitive sequences with respect to the genomic position of the CclSBP was also examined. We investigated the distribution of four types of repetitive sequences that are frequently found in the promoter and coding regions of the CclSBP (TR, TE, LCR, and SSR). The results indicated that all the CclSBP had repetitive sequence insertions and the repetitive sequences were frequent in the promoter and genome DNA regions (Figure 4a). Different types of repetitive sequences frequency were different in the 15 CclSBP genes, and SSR had the largest number and was present in all the CclSBPs except CclSBP1. TR were also found to be present in the genome regions of all the CclSBPs, except CclSBP8 and CclSBP11. LCR was only found in eight CclSBP genes. It is worth noting that TE was not found in any of the CclSBP sequences (Figure 4a).
Figure 4.
The number statistics repetitive sequences of the full-length sequence and Cis-elements in the promoter regions of CclSBPs. (a) The number of repetitive sequences was counted for every 1.5 kb of promoter and DNA. (b) Elements were identified in the 1.5 kb sequences upstream of the start codon of the CclSBP genes using the PlantCARE database. The elements associated with specific functions are indicated by different colors for each gene.
Cis-elements play important roles in the regulation of gene transcription during plant growth, development, and stress responses. To understand the transcriptional regulation mechanisms, the cis-elements in the promoter regions of CclSBP genes were identified through the PlantCARE database (Figure 4b). Except for the common cis-acting elements, such as CAAT-box and TATA-box, many cis-elements were identified in the promoter regions of the 15 CclSBP genes (Table S2). According to their putative functions, these elements were categorized into four classes. The results showed that light-responsive elements had the largest number and were present in all the promoter regions of CclSBP. The hormone responsive elements, plant growth and development, and stress responsive elements were also found to be present in the promoter regions of all the CclSBP genes. In addition, other rarely distributed cis-elements in CclSBP were also found to be functionally involved in transcription regulation, circadian control, protein binding, and stress responsiveness. Therefore, the transcription of the CclSBP genes could be regulated by various environmental and developmental changes, which implied that CclSBP genes were involved in important physiological processes and developmental events. Furthermore, limited similar cis-elements distribution was observed amongst these CclSBP genes, even for those CclSBP in the same phylogenetic group.
3.5. Analysis of the Expression Patterns of the CclSBPs Gene Family
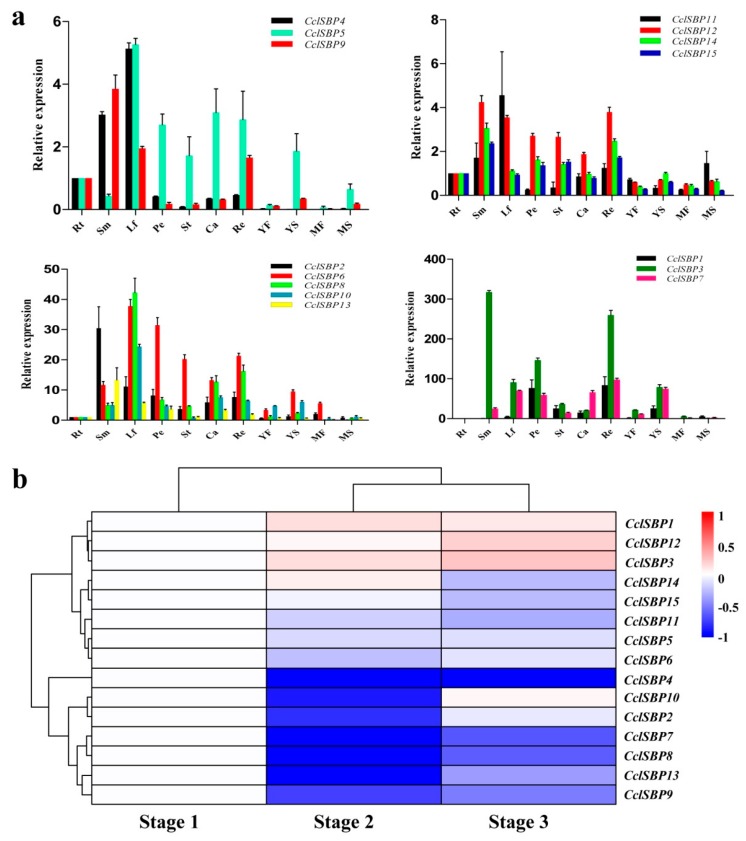
To preliminarily elucidate the roles of CclSBPs in citrus growth and development, we examined the relative expression levels of 15 CclSBPs in 11 different tissues (Figure 5a). All the CclSBPs were detected in at least one of the tissues examined, but a differential expression was observed. The expression data showed a high variability in the transcript abundance of CclSBPs in various tissues and organs, strongly indicating the diversified functions of CclSBPs in citrus growth and development. It is worth noting that many genes have shown similar expression patterns, such as CclSBP4 and CclSBP9, CclSBP8 and CclSBP10, CclSBP12 and CclSBP14, and CclSBP2 and CclSBP3 belonging to the same subgroup (Figure 2a), indicating their redundant functions. Nevertheless, the expression patterns of a few similar gene pairs, including CclSBP1 and CclSBP3, CclSBP6 and CclSBP13, and CclSBP5 and CclSBP14 belonging to the same subgroup (Figure 2a) are distinct. This suggests that these genes may play different roles in citrus growth and development, although they have high sequence similarity.
Figure 5.
Analysis of the expression patterns of the CclSBPs gene family. (a) Relative quantities of CclSBPs in various tissues. Fold changes of the transcript levels in the root (Rt), stem (Sm), leaf (Lf), petal (Pe), stamen (St), carpel (Ca), receptacle (Re), young fruits (YF), young seeds (YS), mature fruits (MF), mature seeds (MS) of citrus are shown. Error bars represent the standard deviations of mean value from at least three biological replicates. (b) Cluster analysis of the expression profiles of CclSBPs at three stages (stage 1: one week before water deficit; stage 2: one week after the beginning of water deficit; and stage 3: one week after release from the water deficit), as shown in a previous study [32]. Each column represents a sample, and each row represents a single citrus transcript sequence. The bar represented the scale of relative expression levels of differentially expressed genes (DEGs), and the colors indicate relative signal intensities.
To explore the potential role of CclSBPs during floral inductive water deficits, RNA sequencing was performed in a previous study on lemon buds at three stages (Stage 1: one week before water deficit; Stage 2: one week after the beginning of water deficit; and Stage 3: one week after release from water deficit) [32]. We classified the 15 CclSBPs into four clusters based on the similarity of the expression patterns (Figure 5b). Cluster 1 genes (including CclSBP3 and CclSBP12) were induced immediately at stage 1 and mostly maintained high expression levels at Stage 3(Figure 5b). These genes were significantly induced at the beginning of the water deficit, indicating that the gene cluster might play a key role in the necessary growth and development of citrus. Cluster 2 genes (including CclSBP11, and CclSBP15) were suppressed immediately at stage 1 and mostly maintained low expression levels at Stage 3 (Figure 5b). Cluster 3 genes (including CclSBP2, CclSBP6, CclSBP7, CclSBP8, CclSBP9, CclSBP10, CclSBP12, and CclSBP13) were transiently suppressed at stage 2 and were then induced at stage 3 (Figure 5b). This cluster showed up-regulated expression at later stages of treatment, indicating the expression of genes involved in the flowering and recovery of vegetative growth. Cluster 4 genes (including CclSBP1, CclSBP5, and CclSBP14) were transiently induced at stage 2 and were then suppressed at stage 3 (Figure 5b). The suppression of the gene cluster may have implied possible involvement in the drought stress response, floral induction, and flower bud differentiation of lemon. A total of four genes (CclSBP4, CclSBP7, CclSBP8, and CclSBP13) were considered to be differentially expressed based on a probability ≥0.8 and an absolute value of log2Ratio ≥ 1 as a threshold. These findings suggested that these genes may play important roles during the floral inductive water deficit process.
3.6. Co-Expression Analysis of the CclSBPs Under Floral Inductive Water Deficits Conditions
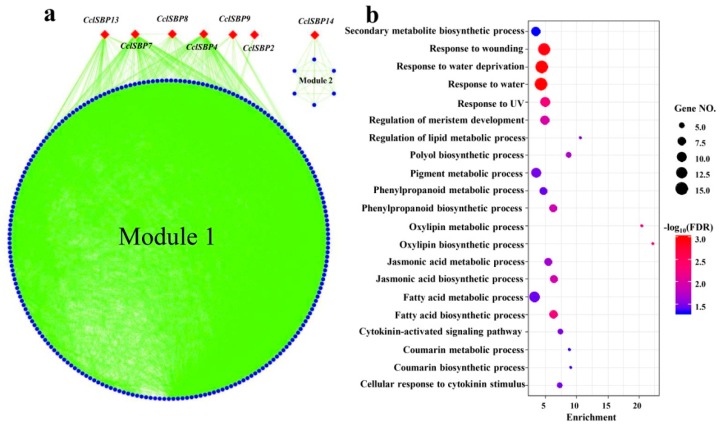
To uncover the possible roles that CclSBPs played in citrus, a co-expression analysis was performed under the floral inductive water deficit conditions. There were 15 CclSBPs together with the 1638 differentially expressed genes (DEGs) forming a co-expression network via 20,450 interactions (edges). The co-expression network naturally clustered into two modules with certain CclSBPs (Figure 6a). For instance, module 1 represented the largest module in the co-expression network containing six CclSBPs (including CclSBP2, CclSBP4, CclSBP7, CclSBP8, CclSBP9, and CclSBP13). Genes from module 2 were uniquely co-expressed with CclSBP14. Gene ontology (GO) term analysis of the CclSBPs centered co-expression network uncovered the possible roles of CclSBPs in the processes of growth and stress response (Figure 6b). For instance, the genes from the co-expression network were enriched in terms of the “response to water/water deprivation”, “response to wounding”, “regulation of meristem development”, “photosynthesis”, as well as in terms of hormone synthesis and metabolism (Figure 6b). As mentioned above, the co-expression network was naturally separated into two dependent modules; and it was interesting to check the specific roles enacted by the different modules. The GO term analysis further confirmed that the genes from the different modules had distinct functions involved in the different aspects of plant growth and development.
Figure 6.
Co-expression network analysis of the CclSBPs using the data from a previous study [32]. (a) CclSBPs centered the gene co-expression network under floral inductive water deficit conditions. (b) The biological processes of the GO term that were significantly enriched in the CsSBP centered network.
3.7. Phenotypes of CclSBP6 Over-Expression in Arabidopsis
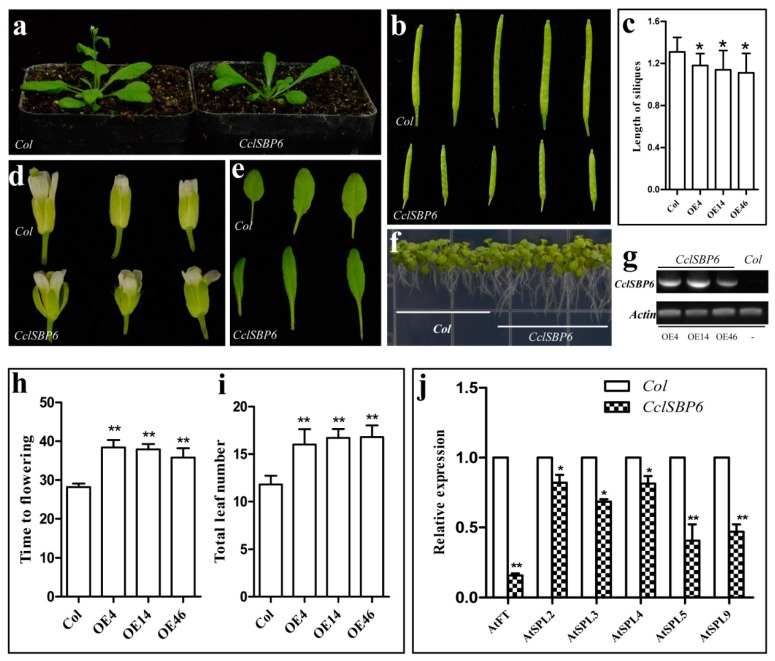
CclSBP6 did not exhibit homology with SPL gene family members of Arabidopsis (Figure 3b), and we speculated that this gene may perform special functions during the growth and development of citrus. Therefore, the function of CclSBP6 was investigated by introduction into Arabidopsis (Figure 7). Twenty-three transgenic plants were obtained in the T1 generation; where all of the lines flowered later than their wild-type counterparts. For further analysis of the CclSBP6 function, three independent transgenic lines in the third generation were selected for phenotypic observation (Figure 7g). Three CclSBP6 transgenic lines flowered significantly later than the wild-type plants in terms of both days to flowering and the number of leaves. The average time to flowering of the transgenic plants ranged from 35.8 to 38.4 days, while that of the wild-type plants was 28.2 days (Figure 7h). The average number of leaves at flowering ranged from 16.0 to 16.8 in the transgenic plants, and was 11.8 in the wild-type plants (Figure 7g). In addition, the transgenic plants of CclSBP6 showed multiple morphological changes, such as smaller flowers, slender leaves, shorter siliques, and extended root systems under long-days (Figure 7c–f).
Figure 7.
The phenotypes of transgenic Arabidopsis with CclSBP6 under long-day conditions. (a) Phenotypes of transgenic Arabidopsis with CclSBP6; (b) the silique length of the control and CclSBP6 transgenic Arabidopsis; (c) the statistics of silique length of the control and CclSBP6 transgenic Arabidopsis; (d) the flower morphologies of the control and CclSBP6 transgenic Arabidopsis; (e) the leaf morphologies of the control and CclSBP6 transgenic Arabidopsis; (f) phenotypes of root length of the control and CclSBP6 transgenic Arabidopsis; (g) PCR identification of transgenic Arabidopsis, wild type Arabidopsis as a negative control; OE4, OE14, and OE46 are the CclSBP6 transgenic lines; (h) times to flowering of the T3 plants of three independent transgenic lines from CclSBP6; (i) number of leaves to flowering of the T3 plants of three independent transgenic lines from CclSBP6; (j) transcript levels of endogenous flowering related genes in the control and transgenic Arabidopsis.
To further elucidate the physiological functions of CclSBP6 during the flowering of transgenic Arabidopsis, we analyzed the abundance of Arabidopsis early flowering related genes at 1-cm inflorescence stages of the wild-type. The levels of Arabidopsis endogenous FLOWERING LOCUS T (FT) and SPL2/3/4/5/9 transcripts were clearly reduced in the transgenic lines (Figure 7j). These data suggested that CclSBP6 functions may act as a floral repressor and might be involved in the citrus flowering. Previously, the transgenic Arabidopsis of citrus miR156a was developed [53]. The results showed that over-expression of citrus miR156a resulted in an extended juvenile phase in transgenic plants compared to the control plants. The phenotypes of CclSBP6 and citrus miR156a transgenic plants were the same by genetic transformation of the Arabidopsis, where we speculated that CclSBP6 may not be the target gene of miR156a in citrus.
3.8. Alternative Splicing and Functional Analysis of CclSBP7 in Transgenic Arabidopsis
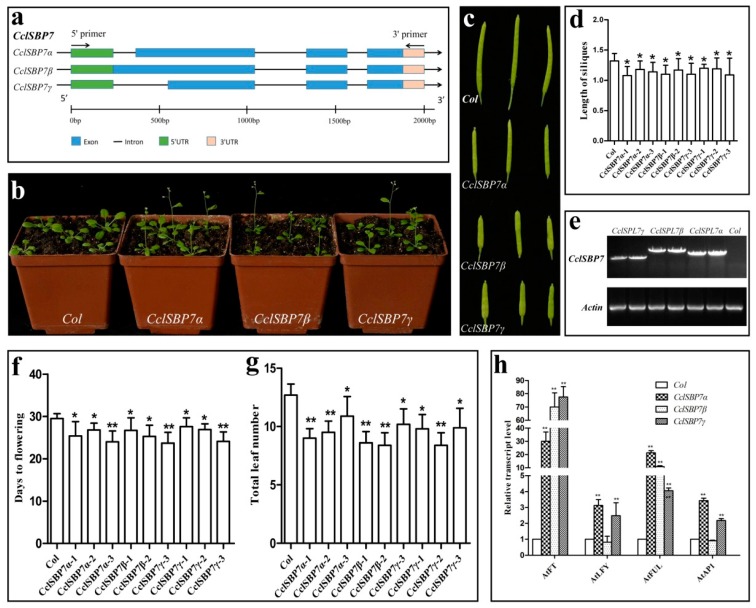
Further expression analysis of the CclSBP7 was analyzed using RT-PCR, with the primers designed according to the open reading frame (ORF) of the CclSBP7 from the citrus database, where they were named CclSBP7α. The templates from the adult plants’ mRNA were prepared. However, three bands were discovered in the RT-PCR analyses observation (Figure 1c). These amplification products were recovered and sequenced. Other than CclSBP7α, two different transcripts were also isolated. During comparisons of these cDNA sequences with each other, three transcripts of CclSBP7 showed high identities with each other. They encoded the same 5′- and 3′-UTRs, whereas a region of these different transcripts was strongly divergent from the transcription initiation site (TSS) to 739 in the ORF through some nucleotide acids deletion and insertion. To further investigate whether these different transcripts were due to alternative splicing or they were transcripts from different genes, we isolated the full-length DNA of CclSBP7. Sequence analysis revealed that these CclSBP7 transcripts came from the same genomic DNA, which was about 2 kb in size and had three introns and three exons with reference to the nucleotide sequence of CclSBP7α. CclSBP7β contained 1098 nucleotides of an ORF because of the first intron retention. CclSBP7γ contained 651 nucleotides of an ORF because of the partial deletion of the first exon (Figure 8a). The three unique transcripts of CclSBP7 were conceptually translated and they showed three unique peptide sequences, and the three sequences were CclSBP7α (329aa), CclSBP7β (365aa), and CclSBP7γ (216aa).
Figure 8.
The gene structure of CclSBP7 and phenotypes of transgenic Arabidopsis with CclSBP7 under long-day conditions. (a) Schematic representation of the alternative processing of CclSBP7; rectangular boxes indicate exons and lines indicate introns; (b) phenotypes of transgenic Arabidopsis with CclSBP7; (c) the silique length of the control and CclSBP7 transgenic Arabidopsis; (d) the statistical analysis of silique length transgenic Arabidopsis and the control; (e) PCR identification of transgenic Arabidopsis, wild-type Arabidopsis as a negative control; (f) times to flowering of the T3 plants of nine independent transgenic lines from three alternatively spliced transcripts of CclSBP7; (g) number of leaves to flowering of the T3 plants of nine independent transgenic lines from three alternatively spliced transcripts of CclSBP7; (h) transcript levels of the endogenous flowering gene in transgenic Arabidopsis and the control.
To further analyze the function of the CclSBP7, three transgenic lines were randomly selected for each alternatively spliced transcript (Figure 8e). We selected 15 T3 plants for each transgenic line. Compared with control plants, the CclSBP7α, CclSBP7β, and CclSBP7γ transgenic lines flowered significantly earlier than the control plants in terms of both the number of leaves and days to flowering (Figure 8b). In the CclSBP7α, CclSBP7β and CclSBP7γ, the average time to flowering ranged from 23.7 to 27.6 days in six transgenic lines, whereas that of the control plants was 29.5 days (Figure 8f). The average number of leaves at flowering ranged from 8.4 to 10.9, and it was 12.7 in the control plants (Figure 8g). In addition, it is worth noting that the transgenic siliques were approximately 80% as long as the siliques of the control plants (Figure 8c–d). To evaluate the possible relation between the expression of CclSBP7 and the early flowering phenotype of the transgenic Arabidopsis, the expression of some endogenous flowering-related genes from Arabidopsis was also assessed at 1-cm inflorescence stages of transgenic Arabidopsis. The levels of FT, FRUITFULL (FUL), APETALA1 (AP1) and LEAFY (LFY) transcripts were clearly elevated in the transgenic plants compared to the wild-type (Figure 8h). These findings further supported our conclusion that the early flowering phenotype of the transgenic Arabidopsis was attributable to the expression of CclSBP7. Meanwhile, these data suggested that CclSBP7 functions may act as a floral activator and might be involved in citrus flowering.
4. Discussion
The SBP-box genes are plant-specific transcription factors encoding proteins that contain a highly conserved SBP domain. This can specifically bind to the promoters of the floral meristem identity gene and it plays significant regulatory roles in plant growth and development, including sporogenesis, leaf development, vegetative and reproductive phase transitions, response to copper and fungal toxins and hormone signaling [4,5,15,21,22,23,25,54]. In a previous study, Shalom et al. identified the members of the SBP gene family in citrus, and they studied their seasonal expression patterns in buds and leaves, and in response to de-fruiting [30]. In this study, a comprehensive overview of the SBP-box gene family was undertaken, including the gene structures, phylogeny, chromosome locations, conserved motifs, and cis-elements in the promoter sequences as compared with the previous study. Meanwhile, we also performed a functional analysis of some of the members of the SBP gene family in Arabidopsis. The roles of SPLs and miR156 as regulators of flowering have been extensively studied in Arabidopsis [15,17,24]. However, considerably less research has been done with woody plants. In the current work, similar to other plants, about two-thirds of the CclSBP contained sequences that were complementary to miR156. Furthermore, three (CclSBP1, CclSBP2, and CclSBP3) of 10 citrus SBP-box genes contained sequences that were complementary to miR156 in the 3’-UTR region, except for the seven other members contained in the miR156-binding site located in the exon region, which comprised a relatively short protein size, consistent with previous reports on Arabidopsis [15,55]. The results provided a basis for elucidating the functions of the SBP-box genes in citrus.
It is believed that the SBP-box gene family has undergone gene duplications in many plants, such as Arabidopsis [55], rice [8], maize [9], Populus [10], grape [13], and apple [12]. The number of SBP-box genes identified in citrus was less than in Arabidopsis, which is inconsistent with the three-fold larger genome size of the Citrus clementina (367 Mb) versus that of Arabidopsis (125 Mb). This finding was similar to the results of a previous study that analyzed the CCCH gene family [51]. Evidence suggests that citrus is rather ancient and has an infrequent reproductive cycle in some taxa due to apomixis, male or female sterility, long juvenility and vegetative propagation [56]. These properties may greatly contribute to the restriction of genome expansion and evolution. Likewise, previous studies have demonstrated that there were no whole genome duplication events (WGDs) in citrus except an ancient triplication, called the γ event, which was shared by all the core eudicots [56]. There are, however, additional recent WGDs that have occurred in Arabidopsis, rice, maize, Populus and apple. Therefore, recent WGDs are likely to be the reason for a larger SBP-box gene number in these plants. It is noteworthy that CclSBP6 and CclSBP7 tended to be putative tandem duplicated genes based on the comprehensive analysis of gene locations and sequence properties. Several SBP-box gene pairs in citrus shared high-sequence similarity and were likely to be putative segmental duplicated genes (CclSBP4/9, CclSBP8/10 and CclSBP5/14). The phylogenetic tree demonstrated that most citrus SBP-box genes were clustered more tightly with Populus and Arabidopsis rather than rice SBP-box genes. This is consistent with the fact that citrus, Populus and Arabidopsis are dicots and diverged more recently from a common ancestor than plants from the lineage leading to monocots. These results indicate that although plant SBP-box genes may be derived from a common ancestor and appeared after the divergence of plants and animals, many of them have undergone distinct patterns of differentiation and played different roles after the separation of each lineage.
To further reveal the possible roles of CclSBP genes, we constructed a phylogenetic tree based on 15 CclSBPs, 16 AtSPLs, and several SBP-box genes, where the functions have been characterized in other plant species (Figure S1). The phylogenetic tree was further divided into eight groups, and the members where functions have been characterized in each group were annotated using different colored circles. Within group I, aside from the miR156-binding site unknown of AmSBP1, all the other members were contained in the miR156-binding site. AmSBP1 and AtSPL3 have also been reported to bind cis-elements in the promoters of the floral organ identity genes SQUAMOSA and APETALA1(AP1), respectively [4], and AtSPL3/4/5, as well as AmSBP1, have all been implicated in the vegetative phase change and floral induction [4,18]. Another SBP-box gene within this group, CNR, has been reported to be pivotal for normal fruit ripening in tomato [27]. Surprisingly, a recent study has shown that the CclSBP1 was able to promote flowering independently of the photoperiod in Arabidopsis, while miR156 repressed its flowering-promoting activity [30]. Meanwhile, CclSBP1, CclSBP2, and CclSBP3 have a close relationship with these functional investigated genes, and they showed high expression levels in flowering. These results suggest strongly that CclSBP1, CclSBP2, and CclSBP3 have putative significant roles in the citrus flower and/or fruit development.
By contrast with Group I, the members of Group II were relatively large and lacked negative regulation by miR156. Only PpSBP2 and AtSPL14 were functionally characterized within this group. Although PpSBP2 plays important roles in the regulation of copper homeostasis in Barbula unguiculata [57], it showed a relatively more distant relationship with other members from citrus and Arabidopsis compared to AtSPL14. AtSPL14 has been reported to play significant roles in plant architecture [25]. As the ortholog gene of AtSPL14, CclSBP12 probably has a similar function among them. Within group III, all the SBP-box members contained the miR156-binding site. CclSBP6, CclSBP13, AtSPL9, AtSPL15, and OsSPL14 were clustered in one subgroup close together. In Arabidopsis, AtSPL9 and AtSPL15 play redundant roles in vegetative phase change and reproductive transition [20]. In rice, OsSPL14 can repress vegetative branching and promote inflorescence branching [26]. Recently, new SBP-box genes (NbSPL6) have been identified from Nicotiana benthamiana, which is essential for the N-mediated resistance to the Tobacco mosaic virus. Similarly, AtSPL6 functions in resistance to the bacterial pathogen Pseudomonas syringae, expressing the AvrRps4 effector [58]. Therefore, CclSBP4 and CclSBP9 may have similar function because of their closely phylogenetic relationship in the other subgroup.
Although they lack regulation by miR156, the members of group IV were characterized by functional diversity. Two SBP-box genes, PpSBP1 and PpSBP4, were divided into one subgroup and were involved in regulating phase change and the circadian clock in moss [59]. In the other subgroup, CclSBP7, AtSPL8, OsSPL8, and ZmLG1 were clustered closely together. The AtSPL8 was the first SBP-box gene to be functionally characterized in Arabidopsis [22]. Although mutations of AtSPL8 did not play roles in phase change, they have a profound effect on the seed set, petal trichome production, root growth, and male fertility [22,23,24]. In our study, the sequence analysis of several clones from CclSBP7 obtained using RT-PCR resulted in the discovery of three alternative splicing transcripts. Transgenic Arabidopsis over-expressing these transcripts flowered earlier than the control. A previous study indicated that CclSBP7 played a role in the floral inductive water deficit process. These results further suggested that CclSBP7 acts as a floral inducer in citrus. One possible explanation for this observation is that the regulatory mechanism of CclSBP7 differs between Arabidopsis and woody plants. Within group V, CclSBP8 and CclSBP10 were highly expressed in the leaf and homologous to AtSPL13, teosinte glume architecture 1 (tga1), and OsSPL6. AtSPL13 has been shown to affect the initiation of the first true leaves [54], maize tga1 is involved in the ear glume development [60], and OsSPL16 controls the grain size, shape, and quality in rice [61]. Therefore, we hypothesized that CclSBP8 and CclSBP10 may provide functions in controlling the characteristics of leaves and/or fruit.
Surprisingly, only CclSBP11 was classified together with AtSPL2, AtSPL10, and AtSPL11 into Group VI. AtSPL10/11/2 are involved in the development of lateral organs, shape of the cauline leaves, and the number of trichomes on cauline leaves and flowers [19]. The results from spatial expression showed that CclSBP11 was expressed at relatively higher levels in the leaves. Therefore, the CclSBP11 probably had similar functions with the three Arabidopsis SBP-box genes in citrus. Within group VII, the AtSPL7 and its ortholog gene CclSBP15 were characterized by their large size and lack of miR156-binding site. Evidence reveals that AtSPL7 can bind to the Cu-response element (CuRE) and be involved in copper homeostasis [62]. Furthermore, the COPPER RESPONSE REGULATOR 1 (CRR1) in Chlamydomonas reinhardtii is the only one classified in group VIII, and it is homologous to AtSPL7. Similar to AtSPL7, CRR1 recognizes and binds to the GTAC core sequence of CuRE in Chlamydomonas reinhardtii, and it has a similar function as the copper-responsive gene [7]. In addition, CclSBP15 also exhibited responsiveness to abiotic stress, being up-regulated by drought. The results strongly suggest that CclSBP15 probably has a similar function involved in copper homeostasis. The phylogenetic tree helped to predict the putative functions of the CclSBP genes based on the functions of SBP-box proteins in other species clustered in the same group. However, these findings only provide helpful information for understanding the function and regulation mechanism of CclSBPs in citrus. Further efforts will be exerted to find more direct evidence, including ectopic expression of different transcripts by transformation in citrus.
5. Conclusions
Function analysis has shown that SBP-box genes play crucial roles in the regulation of plant growth and development, especially in terms of flowering time, meristem identity and architecture, and fruit development. Citrus, as a perennial fruit tree, has distinct growth habits and quantitative traits in contrast to annual plant species. Comparative analysis of the SBP-box genes among various plants suggests that CclSBPs probably plays some unique roles and has undergone distinct evolutionary processes. In the present work, a total of 15 CclSBPs were identified from the whole genome sequence of Citrus, and the complete cDNA of all the members were isolated. Subsequently, a clear nomenclature was provided for them based on their chromosomal locations. Comprehensive analysis including gene structures, phylogenetic relationships, conserved motifs, expression patterns and miR156-mediated transcriptional regulation was conducted. We also performed a functional analysis of some CclSBPs in Arabidopsis. The results showed that it was able to change the flowering time of transgenic plants. The results provide useful information on CclSBPs, which should facilitate further research on elucidating the functions of SBP-box genes in citrus.
Supplementary Materials
The supplementary materials are available online at https://www.mdpi.com/2218-273X/9/2/66/s1. Figure S1. Functional diversity analysis of SBP-box genes in other plants, and phylogenetic relationship with citrus SBP-box genes. Phylogeny of SBP-box genes based on an alignment of the SBP-box domain, only genes for which functional data were available selected from other plant species. Green circles, presence; white circles, absence; black circles, unknown. Table S1 Gene-specific primers used in this study. Table S2. Cis-regulatory elements predicted in promoter region of CclSBPs.
Author Contributions
Methodology, R.-F.Z. and S.-R.L.; data curation, R.-F.Z., J.-J.Z. and Z.-M.G.; writing-original draft preparation, S.-R.L.; writing-review and editing, J.-Z.Z.; supervision, J.-Z.Z. and C.-G.H.; project administration, J.-Z.Z. and C.-G.H.; funding acquisition, J.-Z.Z.
Funding
This research was supported financially by the National Major Research and Development Plan (2018YFD100004), the National Natural Science Foundation of China (grant nos. 31672110 and 31772252) and the Fundamental Research Funds for the Central Universities (2662018JC044).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Chen X., Zhang Z., Liu D., Kai Z., Li A., Long M. SQUAMOSA Promoter-Binding Protein-Like transcription factors: Star players for plant growth and development. J. Integr. Plant Biol. 2010;52:946–951. doi: 10.1111/j.1744-7909.2010.00987.x. [DOI] [PubMed] [Google Scholar]
- 2.Yamasaki K., Kigawa T., Inoue M., Tateno M., Yamasaki T., Yabuki T., Aoki M., Seki E., Matsuda T., Nunokawa E. A novel zinc-binding motif revealed by solution structures of DNA-binding domains of Arabidopsis SBP-family transcription factors. J. Mol. Biol. 2004;337:49–63. doi: 10.1016/j.jmb.2004.01.015. [DOI] [PubMed] [Google Scholar]
- 3.Birkenbihl R.P., Jach G., Saedler H., Huijser P. Functional dissection of the plant-specific SBP-domain: Overlap of the DNA-binding and nuclear localization domains. J. Mol. Biol. 2005;352:585–596. doi: 10.1016/j.jmb.2005.07.013. [DOI] [PubMed] [Google Scholar]
- 4.Klein J., Saedler H., Huijser P. A new family of DNA binding proteins includes putative transcriptional regulators of the Antirrhinum majus floral meristem identity gene SQUAMOSA. Mol. Gen. Genet. 1996;250:7–16. doi: 10.1007/BF02191820. [DOI] [PubMed] [Google Scholar]
- 5.Cardon G., Höhmann S., Klein J., Nettesheim K., Saedler H., Huijser P. Molecular characterisation of the Arabidopsis SBP-box genes. Gene. 1999;237:91–104. doi: 10.1016/S0378-1119(99)00308-X. [DOI] [PubMed] [Google Scholar]
- 6.Lännenpää M., Jänönen I., HölttäUori M., Gardemeister M., Porali I., Sopanen T. A new SBP-box gene BpSPL1 in silver birch (Betula pendula) Physiol. Plant. 2004;120:491–500. doi: 10.1111/j.0031-9317.2004.00254.x. [DOI] [PubMed] [Google Scholar]
- 7.Kropat J., Tottey S., Birkenbihl R.P., Depege N., Huijser P., Merchant S. A regulator of nutritional copper signaling in Chlamydomonas is an SBP domain protein that recognizes the GTAC core of copper response element. Proc. Natl. Acad. Sci. USA. 2005;102:18730–18735. doi: 10.1073/pnas.0507693102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xie K., Wu C., Xiong L. Genomic organization, differential expression, and interaction of SQUAMOSA promoter-binding-like transcription factors and microRNA156 in rice. Plant Physiol. 2006;142:280. doi: 10.1104/pp.106.084475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hultquist J.F., Dorweiler J.E. Feminized tassels of maize mop1 and ts1 mutants exhibit altered levels of miR156 and specific SBP-box genes. Planta. 2008;229:99–113. doi: 10.1007/s00425-008-0813-2. [DOI] [PubMed] [Google Scholar]
- 10.Li C., Lu S. Molecular characterization of the SPL gene family in Populus trichocarpa. BMC Plant Biol. 2014;14:131. doi: 10.1186/1471-2229-14-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Salinas M., Xing S., Höhmann S., Berndtgen R., Huijser P. Genomic organization, phylogenetic comparison and differential expression of the SBP-box family of transcription factors in tomato. Planta. 2012;235:1171–1184. doi: 10.1007/s00425-011-1565-y. [DOI] [PubMed] [Google Scholar]
- 12.Li J., Hou H., Li X., Jiang X., Yin X., Gao H., Zheng Y., Bassett C.L., Wang X. Genome-wide identification and analysis of the SBP-box family genes in apple (Malus × domestica Borkh.) Plant Physiol. Biochem. 2013;70:100–114. doi: 10.1016/j.plaphy.2013.05.021. [DOI] [PubMed] [Google Scholar]
- 13.Hou H., Li J., Gao M., Singer S.D., Wang H., Mao L., Fei Z., Wang X. Genomic organization, phylogenetic comparison and differential expression of the SBP-Box family genes in grape. PLoS ONE. 2013;8:e59358. doi: 10.1371/journal.pone.0059358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schwab R., Palatnik J.F., Riester M., Schommer C., Schmid M., Weigel D. Specific effects of microRNAs on the plant transcriptome. Dev. Cell. 2005;8:517. doi: 10.1016/j.devcel.2005.01.018. [DOI] [PubMed] [Google Scholar]
- 15.Wu G., Poethig R.S. Temporal regulation of shoot development in Arabidopsis thaliana by miR56 and its target SPL3. Development. 2006;133:3539–3547. doi: 10.1242/dev.02521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang J.-W., Czech B., Weigel D. miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell. 2009;138:738–749. doi: 10.1016/j.cell.2009.06.014. [DOI] [PubMed] [Google Scholar]
- 17.Yu N., Cai W.J., Wang S., Shan C.M., Wang L.J., Chen X.Y. Temporal control of trichome distribution by microRNA156-targeted SPL genes in Arabidopsis thaliana. Plant Cell. 2010;22:2322–2335. doi: 10.1105/tpc.109.072579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jung J.-H., Seo P.J., Kang S.K., Park C.-M. miR172 signals are incorporated into the miR156 signaling pathway at the SPL3/4/5 genes in Arabidopsis developmental transitions. Plant Mol. Biol. 2011;76:35–45. doi: 10.1007/s11103-011-9759-z. [DOI] [PubMed] [Google Scholar]
- 19.Shikata M., Koyama T., Mitsuda N., Ohmetakagi M. Arabidopsis SBP-Box genes SPL10, SPL11 and SPL2 control morphological change in association with shoot maturation in the reproductive phase. Plant Cell Physiol. 2009;50:2133. doi: 10.1093/pcp/pcp148. [DOI] [PubMed] [Google Scholar]
- 20.Schwarz S., Grande A.V., Bujdoso N., Saedler H., Huijser P. The microRNA regulated SBP-box genes SPL9 and SPL15 control shoot maturation in Arabidopsis. Plant Mol. Biol. 2008;67:183–195. doi: 10.1007/s11103-008-9310-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yamasaki H., Hayashi M., Fukazawa M., Kobayashi Y., Shikanai T. SQUAMOSA Promoter Binding Protein-Like7 is a central regulator for copper homeostasis in Arabidopsis. Plant Cell. 2009;21:347–361. doi: 10.1105/tpc.108.060137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Unte U.S., Sorensen A.M., Pesaresi P., Gandikota M., Leister D., Saedler H., Huijser P. SPL8, an SBP-box gene that affects pollen sac development in Arabidopsis. Plant Cell. 2003;15:1009–1019. doi: 10.1105/tpc.010678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Y., Schwarz S., Saedler H., Huijser P. SPL8, a local regulator in a subset of gibberellin-mediated developmental processes in Arabidopsis. Plant Mol. Biol. 2007;63:429. doi: 10.1007/s11103-006-9099-6. [DOI] [PubMed] [Google Scholar]
- 24.Xing S., Salinas M., Höhmann S., Berndtgen R., Huijser P. miR156-targeted and nontargeted SBP-Box transcription factors act in concert to secure male fertility in Arabidopsis. Plant Cell. 2010;22:3935–3950. doi: 10.1105/tpc.110.079343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stone J.M., Liang X., Nekl E.R., Stiers J.J. Arabidopsis AtSPL14, a plant-specific SBP-domain transcription factor, participates in plant development and sensitivity to fumonisin B1. Plant J. 2005;41:744–754. doi: 10.1111/j.1365-313X.2005.02334.x. [DOI] [PubMed] [Google Scholar]
- 26.Jiao Y., Wang Y., Xue D., Wang J., Yan M., Liu G., Dong G., Zeng D., Lu Z., Zhu X., et al. Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat. Genet. 2010;42:541–544. doi: 10.1038/ng.591. [DOI] [PubMed] [Google Scholar]
- 27.Manning K., Tör M., Poole M., Hong Y., Thompson A.J., King G.J., Giovannoni J.J., Seymour G.B. A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nat. Genet. 2006;38:948–952. doi: 10.1038/ng1841. [DOI] [PubMed] [Google Scholar]
- 28.Chuck G., Whipple C., Jackson D., Hake S. The maize SBP-box transcription factor encoded by tasselsheath4 regulates bract development and the establishment of meristem boundaries. Development. 2010;137:1243–1250. doi: 10.1242/dev.048348. [DOI] [PubMed] [Google Scholar]
- 29.Liu M.Y., Wu X.M., Long J.M., Guo W.W. Genomic characterization of miR156 and SQUAMOSA promoter binding protein-like genes in sweet orange (Citrus sinensis) Plant Cell Tissue Organ Cult. 2017;130:103–116. doi: 10.1007/s11240-017-1207-6. [DOI] [Google Scholar]
- 30.Shalom L., Shlizerman L., Zur N., Doronfaigenboim A., Blumwald E., Sadka A. Molecular characterization of SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) gene family from Citrus and the effect of fruit load on their expression. Front. Plant Sci. 2015;6:389. doi: 10.3389/fpls.2015.00389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Long J.-M., Liu C.-Y., Feng M.-Q., Liu Y., Wu X.-M., Guo W.-W. miR156-SPL modules regulate induction of somatic embryogenesis in citrus callus. J. Exp. Bot. 2018;69:2979–2993. doi: 10.1093/jxb/ery132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li J.-X., Hou X.-J., Zhu J., Zhou J.-J., Huang H.-B., Yue J.-Q., Gao J.-Y., Du Y.-X., Hu C.-X., Hu C.-G., et al. Identification of genes associated with lemon floral transition and flower development during floral inductive water deficits: A hypothetical model. Front. Plant Sci. 2017;8:1013. doi: 10.3389/fpls.2017.01013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huala E., Dickerman A.W., Garcia-Hernandez M., Weems D., Reiser L., LaFond F., Hanley D., Kiphart D., Zhuang M., Huang W., et al. The Arabidopsis Information Resource (TAIR): A comprehensive database and web-based information retrieval, analysis, and visualization system for a model plant. Nucleic Acids Res. 2001;29:102–105. doi: 10.1093/nar/29.1.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu G.A., Prochnik S., Jenkins J., Salse J., Hellsten U., Murat F., Perrier X., Ruiz M., Scalabrin S., Terol J., et al. Sequencing of diverse mandarin, pummelo and orange genomes reveals complex history of admixture during citrus domestication. Nat. Biotechnol. 2014;32:656. doi: 10.1038/nbt.2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Finn R.D., Bateman A., Clements J., Coggill P., Eberhardt R.Y., Eddy S.R., Heger A., Hetherington K., Holm L., Mistry J., et al. Pfam: The protein families database. Nucleic Acids Res. 2014;42:D222–D230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thompson J.D., Gibson T.J., Higgins D.G. Multiple Sequence Alignment Using ClustalW and ClustalX. Curr. Protoc. Bioinform. 2003;1:2.3.1–2.3.22. doi: 10.1002/0471250953.bi0203s00. [DOI] [PubMed] [Google Scholar]
- 37.Crooks G.E., Hon G., Chandonia J.M., Brenner S.E. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tamura K., Filipski A., Peterson D., Stecher G., Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guo A.-Y., Zhu Q.-H., Chen X., Luo J.-C. GSDS: A gene structure display server. Yi Chuan. 2007;29:1023–1026. doi: 10.1360/yc-007-1023. [DOI] [PubMed] [Google Scholar]
- 40.Letunic I., Doerks T., Bork P. SMART: Recent updates, new developments and status in 2015. Nucleic Acids Res. 2015;43:257–260. doi: 10.1093/nar/gku949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bailey T.L., Williams N., Misleh C., Li W.W. MEME: Discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006;34:W369–W373. doi: 10.1093/nar/gkl198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun L.-M., Ai X.-Y., Li W.-Y., Guo W.-W., Deng X.-X., Hu C.-G., Zhang J.-Z. Identification and comparative profiling of miRNAs in an early flowering mutant of trifoliate orange and its wild type by genome-wide deep sequencing. PLOS ONE. 2012;7:e43760. doi: 10.1371/journal.pone.0043760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dai X., Zhao P.X. psRNATarget: A plant small RNA target analysis server. Nucleic Acids Res. 2011;39:W155–W159. doi: 10.1093/nar/gkr319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Benson G. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 1999;27:573. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bedell J.A., Korf I., Gish W. MaskerAid: A performance enhancement to RepeatMasker. Bioinformatics. 2000;16:1040–1041. doi: 10.1093/bioinformatics/16.11.1040. [DOI] [PubMed] [Google Scholar]
- 46.Temnykh S., DeClerck G., Lukashova A., Lipovich L., Cartinhour S., McCouch S. Computational and experimental analysis of microsatellites in rice (Oryza sativa L.): frequency, length variation, transposon associations, and genetic marker potential. Genome Res. 2001;11:1441–1452. doi: 10.1101/gr.184001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lescot M., Déhais P., Thijs G., Marchal K., Moreau Y., Van de Peer Y., Rouzé P., Rombauts S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002;30:325–327. doi: 10.1093/nar/30.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu G., Wang L.G., Han Y., He Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. Omics-A J. Integr. Biol. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Luo W., Brouwer C. Pathview: An R/Bioconductor package for pathway-based data integration and visualization. Bioinformatics. 2013;29:1830–1831. doi: 10.1093/bioinformatics/btt285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu S., Khan M.R.G., Li Y., Zhang J., Hu C. Comprehensive analysis of CCCH-type zinc finger gene family in citrus (Clementine mandarin) by genome-wide characterization. Mol. Genet. Genom. 2014;289:855–872. doi: 10.1007/s00438-014-0858-9. [DOI] [PubMed] [Google Scholar]
- 52.Clough S.J., Bent A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 53.Wang C.-Y., Liu S.-R., Zhang X.-Y., Ma Y.-J., Hu C.-G., Zhang J.-Z. Genome-wide screening and characterization of long non-coding RNAs involved in flowering development of trifoliate orange (Poncirus trifoliata L. Raf.) Sci. Rep. 2017;7:43226. doi: 10.1038/srep43226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Martin R.C., Asahina M., Liu P.-P., Kristof J.R., Coppersmith J.L., Pluskota W.E., Bassel G.W., Goloviznina N.A., Nguyen T.T., Martinez-Andujar C., et al. The regulation of post-germinative transition from the cotyledon- to vegetative-leaf stages by microRNA-targeted SQUAMOSA PROMOTER-BINDING PROTEIN LIKE13 in Arabidopsis. Seed Sci. Res. 2010;20:89–96. doi: 10.1017/S0960258510000073. [DOI] [Google Scholar]
- 55.Guo A.-Y., Zhu Q.-H., Gu X., Ge S., Yang J., Luo J. Genome-wide identification and evolutionary analysis of the plant specific SBP-box transcription factor family. Gene. 2008;418:1–8. doi: 10.1016/j.gene.2008.03.016. [DOI] [PubMed] [Google Scholar]
- 56.Xu Q., Chen L.-L., Ruan X., Chen D., Zhu A., Chen C., Bertrand D., Jiao W.-B., Hao B.-H., Lyon M.P., et al. The draft genome of sweet orange (Citrus sinensis) Nat. Genet. 2013;45:59–U92. doi: 10.1038/ng.2472. [DOI] [PubMed] [Google Scholar]
- 57.Nagae M., Nakata M., Takahashi Y. Identification of negative cis-acting elements in response to copper in the chloroplastic iron superoxide dismutase gene of the moss Barbula unguiculata. Plant Physiol. 2008;146:1687–1696. doi: 10.1104/pp.107.114868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Padmanabhan M.S., Ma S., Burch-Smith T.M., Czymmek K., Huijser P., Dinesh-Kumar S.P. Novel positive regulatory role for the SPL6 transcription factor in the N TIR-NB-LRR receptor-mediated plant innate immunity. PLoS Pathog. 2013;9:e1003235. doi: 10.1371/journal.ppat.1003235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Riese M., Zobell O., Saedler H., Huijser P. SBP-domain transcription factors as possible effectors of cryptochrome-mediated blue light signalling in the moss Physcomitrella patens. Planta. 2008;227:505–515. doi: 10.1007/s00425-007-0661-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang H., Nussbaum-Wagler T., Li B., Zhao Q., Vigouroux Y., Faller M., Bomblies K., Lukens L., Doebley J.F. The origin of the naked grains of maize. Nature. 2005;436:714–719. doi: 10.1038/nature03863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang S., Wu K., Yuan Q., Liu X., Liu Z., Lin X., Zeng R., Zhu H., Dong G., Qian Q. Control of grain size, shape and quality by OsSPL16 in rice. Nat. Genet. 2012;44:950–954. doi: 10.1038/ng.2327. [DOI] [PubMed] [Google Scholar]
- 62.Zhang H., Zhao X., Li J., Cai H., Deng X.W., Li L. MicroRNA408 Is Critical for the HY5-SPL7 Gene Network That Mediates the Coordinated Response to Light and Copper. Plant Cell. 2014;26:4933–4953. doi: 10.1105/tpc.114.127340. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.