Summary:
Expansion segments (ESs) are enigmatic insertions within the eukaryotic ribosome, the longest of which resemble tentacle-like extensions that vary in length and sequence across evolution, with a largely unknown function. By selectively engineering rRNA in yeast, we find that one the largest ES, ES27L, has an unexpected function in translation fidelity. Ribosomes harboring a deletion in the distal portion of ES27L increase amino acid misincorporation, as well as read-through and frameshifting errors. By employing quantitative mass spectrometry, we further find that ES27L acts as an RNA scaffold to facilitate binding of a conserved enzyme, methionine amino peptidase (MetAP). We show that MetAP unexpectedly controls the accuracy of ribosome decoding, which is coupled to an increase in its enzymatic function through its interaction with ES27L. These findings reveal that variable ESs of the ribosome serve important functional roles and act as platforms for the binding of proteins that modulate translation across evolution.
Graphical Abstract
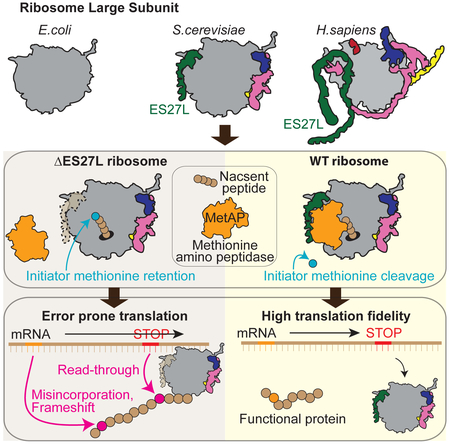
In Brief:
Fujii et al. reveal the function of enigmatic eukaryote specific expansion segments (ES) in the ribosome. They show a critical function for ES27L in translation fidelity by acting as an RNA scaffold to recruit the N-terminal processing enzyme, MetAP, which unexpectedly controls the accuracy of ribosome decoding.
INTRODUCTION
The ribosome is an ancient molecular machine responsible for translating the genetic code, which has a core ribosomal RNA (rRNA) secondary structure that is universally shared across all kingdoms of life. However, there have been large expansions in the size of the ribosome across evolution. For example, the eukaryotic ribosome is over 1MDa larger than the bacterial ribosome, due in part to the insertions of up to 30 eukaryote specific expansion segments (ESs) in the rRNA (Figures. 1A and S1A) (Susan A. Gerbi, 1998). Interestingly, ESs exhibit high variability in terms of length and sequence across different species. The longest of ESs are more than 700nt in H. sapiens and resemble tentacle-like extensions (Anger et al., 2013). Parts of these extensions are also highly mobile and do not appear to be associated with ribosomal proteins (RPs) (Armache et al., 2010), which may preferentially allow for interactions with additional, yet unknown proteins. ESs also have nucleotide level sequence variability within species (Kuo, 1996; Leffers and Andersen, 1993; Parks et al., 2018), consistent with rDNA heterogeneity first noted over 40 years ago (Arnheim and Southern, 1977). For example, there are distinct maternal-type and somatic-type 45S rDNA transcription units in zebrafish and the majority of these differences can be attributed to ESs variability at the level of sequence variation in the form of nucleotide substitutions as well as indels (Locati et al., 2017). Furthermore, rRNA variants may also be expressed between different tissues in the same organism (Parks et al., 2018; Tseng et al., 2008). Interestingly, one of the largest estimated differences in rRNA between tissues are found in sequence variation within ESs (Parks et al., 2018).
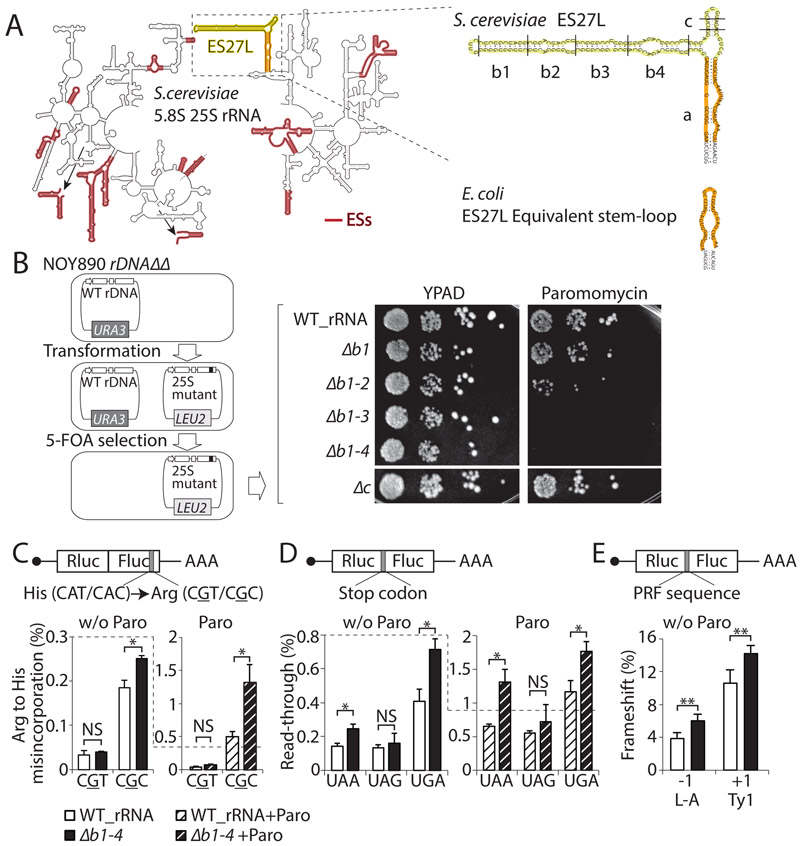
Figure. 1. A eukaryotie-specific rRNA expansion segment, ES27L, has a role in translation fidelity.
(A) Left: The secondary structure of S. cerevisiae 25S and 5.8S rRNAs are shown. ESs are highlighted in red. The arrows are showing the placement of ESs that cannot fit in the 2D rRNA representation. Right: Enlargement of ES27L. ES27L "b" and "c" stem-loops are in yellow. The E. coli ES27L equivalent stem-loop and ES27L "a" helix are in orange. (B) Left: Schematic workflow to make rDNA mutant strains is shown. Right: The spot assay indicates the sensitivity of ES27L mutant strains to Paromomycin, which induces translation error. The picture of the ES27L Δc strain was taken from the same plate shown in Figure S1G. (C) The percentage of amino acid misincorporation was measured by a tandem Rluc-Fluc reporters, in which each has various point mutations at Fluc active site (grey) under normal condition (w/o Paro) or Paromomycin treatment (Paro). (D) The percentage of stop codon read-through detected by reporters with an insertion of a stop codon (grey). (E) The percentage of frameshift induced by the PRF (Programed ribosomal frameshifting) sequence measured by a construct having the −1 PRF sequence or +1 PRF sequence (grey). Data are represented as mean + standard deviation (SD) (t-test, **P < 0.01; *P < 0.05; NS, not significant, n ≥ 3). See also Figure S1.
Whether ESs have a functional role in translational control as well as the significance of their dramatic expansions in evolution remain largely unknown. Large deletions of ESs in their entirety are associated with ribosome biogenesis defects (Jeeninga et al., 1997; Ramesh and Woolford, 2016; Sweeney et al., 1994), which has precluded a more detailed analysis of ES function in translational control. The nature of translational control, such as translation initiation, termination, fidelity and speed, has also changed across evolution (Drummond and Wilke, 2009; Myasnikov et al., 2009; Petry et al., 2008) and it is unknown whether any of these steps in protein synthesis are modulated by the variable ESs of the ribosome. The faithful translation of mRNA into the corresponding protein requires accurate tRNA charging as well as precision in ribosome decoding and translocation. The requirement for translation fidelity also varies between species and cell types, with neurons being the most sensitive (Kapur and Ackerman, 2018). For instance, the fidelity of protein production has increased from prokaryotes to eukaryotes; while amino acid misincorporation varies widely depending on the codon, the higher end of amino acid misincorporation rates are reduced from 35×10−4 per codon in E. coli (Kramer and Farabaugh, 2007) to 7×10−4 in S. cerevisiae (Kramer et al., 2010). Moreover, there is an increase in mean protein length from 310aa in E. coli to 560aa in H. sapiens, extending the probability of translation error occurring in the polypeptide (Balchin et al., 2016). Low translation fidelity and accumulation of translation error can reduce the levels of functional proteins, foster protein aggregation, and may underlie neurological disease (Lee et al., 2006; Meyer-Schuman and Antonellis, 2017). How cells maintain the integrity of their proteomes remains of outstanding relevance for both normal cellular functions as well as in disease states ranging from age-related decline to acute trauma and neurodegenerative disease. Hence, control of translation fidelity is a critical process and whether ESs have any functional role in such a key step of translational control, as well as how the ribosome itself has increased the precision of protein synthesis across evolution, remain outstanding questions.
RESULTS
A eukaryote specific expansion segment in rRNA controls translation fidelity.
Here, we focused on characterizing one of the largest ESs of the 25S rRNA, the 159 nt ES27L in S. cerevisiae, having three helices called “a”, “b”, and “c” (Armache et al., 2010) (Figure. 1A). An equivalent helix structure in E. coli has a 33 nt single short stem-loop instead (Figure. 1A and S1A). ES27L is highly dynamic and flexible and therefore difficult to resolve by both X-ray crystal structure and cryoelectron microscopy (cryoEM). ES27L has been modeled in cryoEM studies with one preferred orientation pointing towards the peptide exit tunnel (Armache et al., 2010; Greber et al., 2012). The genetic modification of ribosomal DNA (rDNA) is challenging because of the repetitive nature of the rDNA loci found on multiple chromosomes in higher eukaryotes (Romanova et al., 2006). S. cerevisiae has a single rDNA locus, which contains hundreds of tandem-repeated rDNA copies, allowing us to delete the entire rDNA locus and complement it with rDNA expression from an exogenous plasmid (Figure. 1B) (Nemoto et al., 2010; Wai et al., 2000). As large deletions of ESs in their entirety, including ES27L, affect ribosome biogenesis and are lethal in yeast (Jeeninga et al., 1997; Ramesh and Woolford, 2016; Sweeney et al., 1994), we devised strategies to selectively engineer ribosomes to harbor a series of truncating deletions in only the distal portion of the “b” and a complete deletion of only the “c” stem-loops of ES27L (Figure. 1B). Strikingly, these ES27L mutant strains are viable and did not possess any major defects in ribosome biogenesis or in global protein synthesis as assessed by 35S labeled methionine incorporation (Figures. S1B-D). To further investigate a possible function of ES27L in translation, ES27L Δb1-4 and ES27L Δc ribosome mutant strains were treated with a variety of translation inhibitors such as Cycloheximide, Anisomycin, Hygromycin B, or subjected to stress such as heat and cold shock (Figures. S1E and F). Consistent with the lack of observable overall changes in ribosome biogenesis or protein synthesis, the ES27L Δb1-4 mutants did not show any sensitivity to these treatments, but instead a mild to moderate increased tolerance to the translation elongation inhibitors Cycloheximide and Anisomycin. On the contrary, we observed that the ES27L Δb1-4 mutants had a strong and specific sensitivity to the translation error-inducing drug, Paromomycin (Figures. 1B and S1G). Paromomycin binds to the decoding center of the small subunit of the ribosome and induces amino acid misincorporation and stop codon read-through (de Loubresse et al., 2014; Prokhorova et al., 2017). The ES27L Δc mutant did not display any sensitivity or tolerance to any of the inhibitors tested or stress conditions, showing the specificity for the “b” stem-loop mutants towards the ES27L phenotype (Figures. 1B and S1E, F). These phenotypes suggest that ES27L Δb1-4 ribosomes may be error prone.
To further examine the role of the ES27L “b” stem-loop in translation fidelity, we measured translation error using reporter constructs, which express a fusion protein of Renilla luciferase (Rluc) and Firefly luciferase (Fluc). Point mutations were introduced at the active site of Fluc, enabling Fluc activity only when amino acid misincorporation has occurred, and thereby enables the accurate measurement of translation error (Kramer et al., 2010). With these reporters, we observed that the ES27L Δb1-4 strain had higher amino acid misincorporation rates for specific codons (Figure. 1C). The magnitude of changes in translation fidelity observed in the ES27L Δb1-4 strain is comparable to that of a mutation in RPS23/uS12, which is positioned within the decoding center of the ribosome itself (Paolini et al., 2017). Interestingly, the CGC rare codon is particularly sensitive to higher amino acid misincorporation rate in the ES27L Δb1-4 strain, while the CGT codon does not have a significant change. As both codons are decoded by the same ACG tRNAs with wobble base pairing, mistranslation is not due to a secondary effect on tRNA mis-amino acylation.
To test whether the ES27L “b” stem-loop has a more general role in controlling translation fidelity, we also tested other facets of translation fidelity such as stop codon recognition, and frameshifting using reporters where a stop codon or a frameshifting sequence is placed in between Rluc and Fluc, respectively. The ES27L Δb1-4 strain increased both read-through and frameshifting frequency (Figures. 1D and E). Upon Paromomycin treatment, which stabilizes non-cognate tRNA at A-site, we observed an enhanced trend for amino acid misincorporation rates and read-through frequency (Figures. 1C and D). Based on these results, we can conclude that the eukaryotic specific long stem-loop of ES27L has an unexpected function in translation fidelity, and to our knowledge identifies the first known function of a eukaryotic specific ES in translation control.
ES27L recruits conserved N-terminal processing enzymes on the ribosome.
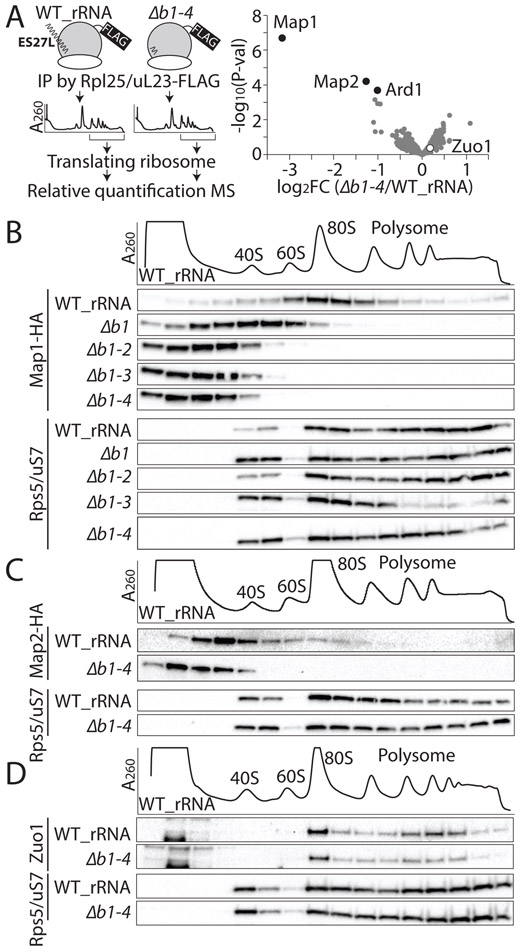
We previously identified hundreds of new ribosome associating proteins (RAPs) that may directly bind to mammalian ribosomes as part of the ‘ribo-interactome’ (Simsek et al., 2017) and we hypothesized that ESs, such as ES27L, act to regulate the binding and recruitment of specific RAPs. To identify specific proteins that could interact with the “b” stem-loop of ES27L, we compared the composition of translating ribosomes containing WT and ES27L Δb1-4 rRNA. To this end, ribosomes were purified from WT and ES27L Δb1-4 strains by affinity-purification with the Rpl25/uL23 C-terminal FLAG tag (Inada et al., 2002) followed by sucrose density gradient (SDG) to purify polysomes (Figure. 2A). We subsequently carried out quantitative mass spectrometry (MS) on WT and ES27L Δb1-4 ribosomes (Figure. S2A). We observed a high correlation between all biological samples (r2 > 0.96 and Figures. S2B and C), with only three proteins showing a significant change in their association between WT and ES27L Δb1-4 translating ribosomes (FDR < 0.05 (FDR, false discovery rate), ∣log2FC∣ > 1 (FC, fold change)). Surprisingly, two of them are the methionine amino peptidase (MetAP) homologs (Map1 and Map2 proteins) (Figure. 2A and Table S1). The third candidate protein identified from quantitative MS of WT and ES27L Δb1-4 translating ribosomes is a subunit of N-terminal acetyltransferase A (NatA) complex, Ard1 protein (Gautschi et al., 2003). In all kingdoms of life, translation starts from initiator methionine (iMet) and iMet is subsequently cleaved off co-translationally from almost a third of all proteins in eukaryotes and prokaryotes by MetAPs (Giglione et al., 2015). These findings suggest that ES27L may be holding the MetAP family of proteins close to the exit tunnel specifically in the eukaryotic ribosome, consistent with the N-terminal processing functions of MetAPs. Indeed, both Map1 and Map2 proteins show dynamic shifts from ribosome fractions to free RNP fractions in the “b” stem-loop mutant strains (Figures. 2B and C), although Zuo1 protein, which is known to be in close proximity to ES27L on the ribosome (Rakwalska and Rospert, 2004; Zhang et al., 2014), does not change. These findings reveal a strong and specific reliance of MetAPs on recruitment to the ribosome via ES27L (Figures. 2A and D). This suggests that ES27L functions to recruit enzymes important for co-translational processing of proteins to the ribosome.
Figure 2. ES27L recruits conserved N-terminal processing enzymes on the ribosome.
(A) Left: Schematic of the identification of ES27L associating factors. IP: Immunoprecipitation, MS: mass spectrometry Right: Comparison of ES27L Δb1-4 ribosome to the WT ribosome, proteins with log2FC < −1 (FC: fold change) with FDR < 0.05 (FDR, false discovery rate) are defined as significantly depleted (black circle). Zuo1 protein is not significant (white circle). The average FC of two biological replicates is shown. (B) The ES27L-dependent ribosome association of hemagglutinin (HA)-tagged Map1 protein is shown by the shift in association in SDG fractions comparing the WT_rRNA strain to ES27L “b” stem loop mutants. (C) The ES27L dependent co-migration of HA-tagged Map2 protein with the ribosome is also shown. (D) ES27L independent ribosome association of Zuo1 protein is shown. Rps5/uS7, is shown as a control. See also Figure S2 and Table S1.
Methionine aminopeptidase (MetAP) is required for translation fidelity.
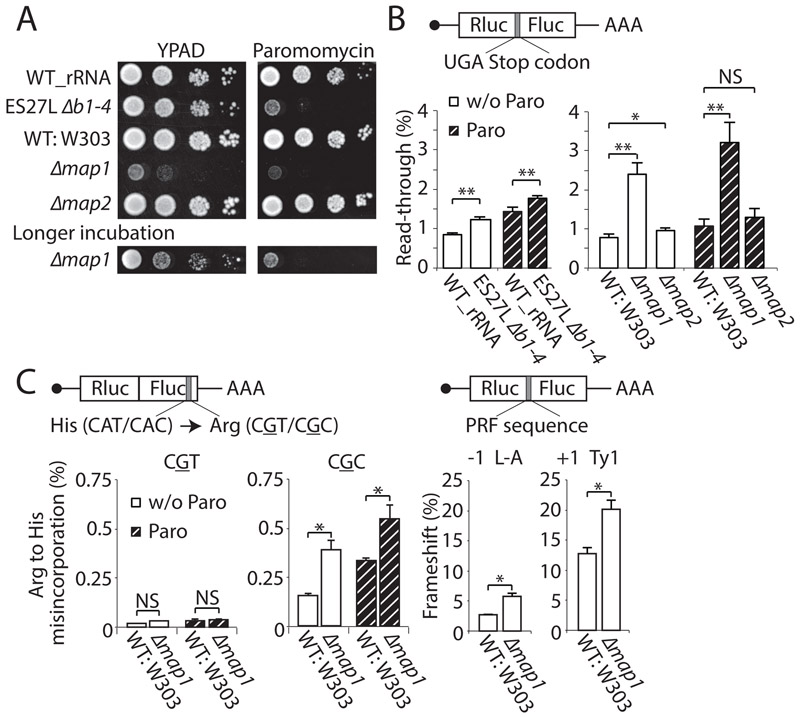
Although MetAPs have never been previously implicated in control of translation fidelity, given the association of MetAP with the ribosome via ES27L (Figures. 2B and C) we next tested whether there may be a previously unappreciated function for MetAPs in increasing the ribosome decoding accuracy. Indeed, we found that although the MAP1 deletion strain grows slower than the ES27L mutants (double MAP1 and MAP2 deletion is lethal (Li and Chang, 1995)), the Paromomycin sensitivity of ES27L mutants (Figure. 1B) is completely phenocopied by the MAP1 deletion but interestingly not MAP2 deletion (Figures. 3A and S3A). The MAP1 deletion strain not only displays sensitivity to translation error-inducing drugs, but also shows increased read-through, more so than MAP2 deletion strain (Figure. 3B), as well as amino acid misincorporation and frameshifting errors similar to the ES27L Δb1-4 strain (Figure. 3C). Interestingly, the Ard1 protein that also has a function in N-terminal processing and is recruited to the ribosome via ES27L (see above), did not show any defects in translation fidelity (Figure. S3B) when deleted. Therefore, MAP1 has a further unexpected function in control of translation fidelity that is phenocopied by ES27L mutants.
Figure. 3. MetAP couples N-terminal processing and translation fidelity.
(A) Spot assay reveals that the MAP1 deletion strain phenocopies the ES27L Δb1-4 strain but the MAP2 deletion strain does not. Since Δmap1 has slower growth, a longer incubation is also shown. (B) The translation fidelity of wild type (WT: W303), Δmap1, and Δmap2 strains were evaluated with reporters containing an insertion of a stop codon between Rluc and Fluc gray under normal conditions (w/o Paro) or Paromomycin treatment (Paro). (C) Translation fidelity in WT: W303 and Δmap1 strains was also evaluated by amino acid misincorporation and both −1 and +1 frameshifting induced by the PRF sequence gray. Data are represented as mean + SD (t-test, **P < 0.01; *P < 0.05; NS, not significant, n ≥ 3). See also Figure S3.
ES27L acts as RNA scaffold to recruit MetAP to increase in vivo activity
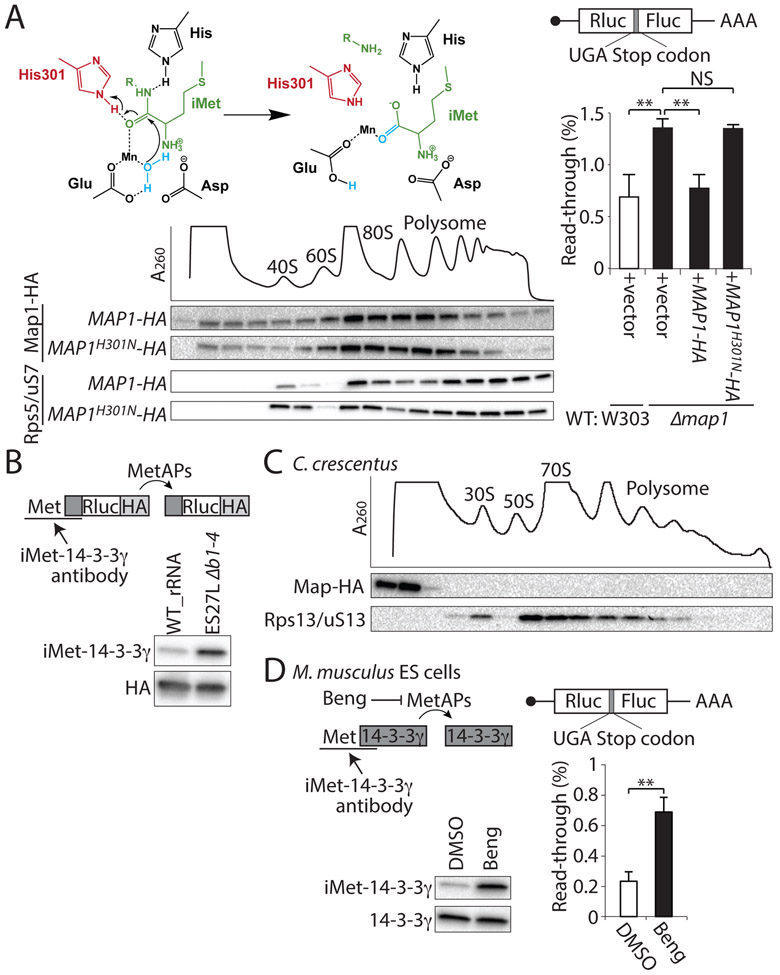
To test whether the enzymatic activity of Map1 protein that is required for iMet processing is also important for control of translation fidelity, we inserted a point mutation into the catalytic amino acid of Map1 protein (Ye et al., 2006) (Figure. 4A), which is perfectly conserved in all organisms from E. coli to H. sapiens in both Map1 (Addlagatta and Matthews, 2006) and Map2 (Liu et al., 1998) proteins (Figure. S4A). The mutation at the catalytic Histidine (H301N) inactivates E. coli Map (Copik et al., 2003; Lowther et al., 1999) and H. sapiens Map2 completely (Griffith et al., 1998). Our results show that this non-active mutant of Map1H301N protein still strongly associates with the ribosome (Figure. 4A). This allowed us to ask whether the Map1H301N protein has a function in controlling ribosome decoding accuracy. Importantly, Map1H301N could not rescue the defect in translation fidelity in the MAP1 deletion strain (Figure. 4A), hence showing an important role for the enzymatic activity of Map1 protein in translation fidelity. We next tested whether binding of MetAPs to the ribosome via ES27L influences their enzymatic properties by measuring MetAP activity in the ES27L Δb1-4 strain. We therefore designed reporter constructs by employing the N-terminal peptide of 14-3-3γ, a model substrate, wherein retained iMet that has not been processed can be detected with an antibody (Figure. 4B). Strikingly, when Map1 and Map2 proteins can no longer directly associate with the ribosome in the ES27L Δb1-4 strain, despite being abundantly present within the cytosol (Figures. 2B and C), iMet processing of this reporter was greatly reduced (Figure. 4B).
Figure. 4. Function of MetAP enzymatic activity for translation fidelity across evolution.
(A) The enzymatic activity of MetAP is required for translation fidelity. The cleavage of iMet (green) is catalyzed through a conserved His 301 (red). H2O is in blue. SDG shows co-migration of the mutant Map1 protein at the catalytic Histidine (MAP1H301N) with the ribosome. The RP (Rps5/uS7) is shown as a control. Right: percentage of the UGA stop codon gray readthrough in WT: W303 (white bar) and Δmap1 (black bar) strains with the overexpression of wild type Map1 protein (+MAP1-HA) or Map1 catalytic Histidine mutant (+MAP1H301N-HA) is shown. (B) MetAP activity in vivo is assessed by iMet retention measured with a Rluc-HA reporter conjugated with the N-terminal peptide of human 14-3-3γ(dark gray box)r(dark gray box), a model substrate, wherein retained iMet that has not been processed can be detected with an iMet-14-3-3γ antibody. The HA antibody detects the entire reporter protein. (C) The SDG fractionation of the bacterial ribosome in C. crescentus, which does not possess ES27L followed by the western blotting for HA-tagged Map protein and ribosomal protein (Rps13/uS13) are shown. (D) The MetAPs activity and the percentage of the UGA stop codon gray read-through upon treatment with the MetAPs inhibitor Bengamide B (Beng) in mouse embryonic stem (ES) cells. Data are represented as mean + SD (t-test, **P < 0.01; *P < 0.05; NS, not significant, n ≥ 3). See also Figure S4.
Evolutionary significance of MetAP association with ES27L in eukaryotes
To gain insight into the evolutionary significance of ES27L in eukaryotes, MetAP association to the ribosome was further examined in prokaryotes, which only have a single MetAP enzyme. While prokaryotic MetAP is known to have a transient interaction with the ribosome for co-translational iMet cleavage (Sandikci et al., 2013), the strong association of MetAP to the ribosome was not observed in the Gram negative dimorphic bacterium C. crescentus which does not possess ES27L (Figure. 4C). We further tested whether this new function of MetAP for translation fidelity in yeast is conserved in mammalian cells. We therefore expanded our analysis to mouse embryonic stem cells (ESCs), which revealed that both MetAP1 and MetAP2 associate with the ribosome (Figures. S4B). MetAP1 also gains a Zinc finger-like domain in eukaryotes via an N-terminal extension (Figure. S4C), which is required for the normal processing function of MetAP1 and may influence ribosome binding (Vetro and Chang, 2002). These findings suggest that the co-evolution of ES27L and the Zinc finger domain of MetAP1 may have enabled MetAP1 association with the ribosome and thus increased translation fidelity.
We next inhibited MetAP activity in ESCs using the inhibitor Bengamide B, which blocks substrate entrance into the catalytic pocket of MetAPs (Lu et al., 2012; Towbin et al., 2003; Xu et al., 2012). In mouse ESCs, treatment with Bengamide B both impedes MetAP enzymatic activity and decreases translation fidelity (Figure. 4D). These results reveal an important function for the evolution of ribosome ESs, in providing both a platform for the binding of additional factors with key functions in translational control, as well as serving as an RNA scaffold for increasing the enzymatic activity of associated proteins.
DISCUSSION
The speed and accuracy of protein synthesis are fundamental for the fitness of living cells and the quality of translation control. These parameters have evolved to fit the individual demands of protein synthesis within organisms. How the ribosome, which is a highly conserved fossil of the RNA world, has evolved to accommodate such species-specific adaptations, including the increase in translation fidelity from prokaryotes to eukaryotes, remains poorly understood. Here we show that an evolutionary change on the ribosome can itself modulate these parameters, specifically how the acquisition of an ES27L in eukaryotic rRNA increases translation fidelity. ES27L enables several enzymes critical for N-terminal processing of proteins, such as MetAPs and NatA, to associate with the eukaryotic ribosome. Since ES27L adopts a conformation close to the exit tunnel, this long expansion can provide a platform for precise positioning of the key enzymes responsible for processing newly synthesized polypeptides. Interestingly, the ribosome export factor Arx1 which is an inactive paralog of MetAP involved in export of the pre-60S subunit from the nucleus to the cytoplasm has been found in cryoEM studies to be situated between ES27L and the ribosomal surface, forming contacts with RPs that are around the polypetide exit tunnel (Greber et al., 2012). Due to the homology of Arx1 and MetAPs, they may bind to the ribosome at the same location. Indeed, we show that binding of MetAPs to the ES27L increases their enzymatic activity in eukaryotes, which can augment the effective concentration of enzyme to substrate.
The unexpected association of MetAP activity with increased translation fidelity raises the question of how an N-terminal processing factor is capable of altering the translation state of a ribosome. One possibility is that the Map1 protein may function in trans by processing ribosome components and translation factors required for accurate protein synthesis. Indeed retention of iMet on several RPs has been observed by proteomics analysis of the Δmap1 Hap1 cell line (Jonckheere et al., 2018). Moreover, the close proximity of Map1 protein via ES27L to the exit tunnel may provide more efficient processing especially for small proteins such as RPs, which only have a short time window for their co-translational processing. iMet retention in such translation components may influence their structure, expression level, or association with the ribosome, thereby leading to increased miscoding of the proteome. In addition, iMet processing and modification are tightly connected with protein stability through the N-end rule, which predicts the half-life of a protein based on its N-terminal amino acid residue (Varshavsky, 2011). Retained iMet is usually N-terminal acetylated and works as a degradation signal for recognition by E3-ubiquitin ligases that target the protein to the ubiquitin-proteasome system (Hwang et al., 2010). Thereby, it is possible that ES27L Δb1-4 ribosomes are error prone by missing specific RPs critical for the translation fidelity or destabilizing translation factors or chaperons required for accurate protein synthesis. Together, these studies highlight that the variable, ESs of the ribosome serve important functional roles, and act as platforms for the binding of proteins that modulate ribosome function. The enigmatic changes of the additional 30 ESs with respect to sequence and length across evolution as well as within species at the level of nucleotide variation may endow greater regulation to this ancient molecular machinery.
STAR Methods
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Maria Barna (mbarna@stanford.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Saccharomyces cerevisiae:
Strains used in this paper are listed at the Experimental Models: Organisms/Strains in the KEY RESOURCES TABLE. Yeast strains were grown in YPAD medium (10 g/L yeast extract, 20 g/L peptone, 40 mg/L adenine sulfate, and 20 g/L glucose) or Synthetic Dextrose (SD) Medium (6.7 g/L yeast nitrogen base and 20 g/L glucose plus appropriate amino acids drop out mix). All yeast strains were cultured at 30 °C, unless specified. Samples were harvested at log phase (OD600 = ∼0.8). Caulobacter crescentus: NA1000 and its derivatives were grown in PYE rich medium (1 g/L yeast extract, 2 g/L Bacto peptone, 1 mM MgSO4, 0.5 mM CaCl2) at 28°C. Mouse embryonic stem cells (ESCs): E14 ESCs (male) (Hooper et al., 1987) were cultured in knockout DMEM (Invitrogen, 10829-018) with 15 % ESC grade FBS (EMD Millipore, ES-009-B), 1× L-glutamine (EMD Millipore, TMS-002-C), 1× ES-grade Penicillin/streptomycin (EMD Millipore, TMS-AB2-C), 1× non-essential amino acid (EMD Millipore, TMS-001-C), 0.1 % 2-mercaptoethanol, 1× Leukemia Inhibitory Factor (LIF, EMD Millipore, ESG1107) in a 37 °C, 5 % CO2 incubator.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti -FLAG M2 affinity Gel | Sigma-Aldrich | Cat# A2220, RRID: AB_10063035 |
| Mouse monoclonal anti-HA 2-2.2.14 | Invitrogen | Cat# 26183, RRID: AB_10978021 |
| Mouse monoclonal anti-Rps5/uS7 | Abcam | Cat# ab58345, RRID: AB_2180899 |
| Rabbit polyclonal anti-Zuo1 | Laboratory Elizabeth Craig (Lee et al., 2016) | N/A |
| Mouse monoclonal anti-Met-14-3-3γ | Novus | Cat# NB100-407, RRID: AB_10003122 |
| Rabbit polyclonal anti-HA SG77 | Invitrogen | Cat# 71-5500, RRID: AB_2533988 |
| Mouse monoclonal anti-Rps13/uS13 | Developmental Studies Hybridoma Bank | Cat# 193E11E5B11, RRID: AB_2618084 |
| Rabbit polyclonal anti-14-3-3γ | Cell Signaling technology | Cat# D15B7, RRID: AB_10827887 |
| Mouse monoclonal anti-MetAP1 | R&D | Cat# MAB3537, RRID: AB_2144061 |
| Rabbit polyclonal anti-MetAP2 | Proteintech | Cat# 17040-1-AP, RRID: AB_2144162 |
| Rabbit polyclonal anti-Rpl10a/uL1 | Abcam | Cat# ab174318 |
| Bacterial and Virus Strains | ||
| Biological Samples | ||
| Chemicals, Peptides, and Recombinant Proteins | ||
| EasyTag L-[35S]-Methionine, 5mCi (185MBq) | PERKINELMER | NEG709A005MC |
| Cycloheximide | Sigma-Aldrich | C7698-5G |
| Hygromycin B | Life Technologies | 687010 |
| Anisomycin | Sigma-Aldrich | A9789-5MG |
| Paromomycin | Sigma-Aldrich | P9297-1G |
| 5-FOA | Fisher Scientific | F10501-5.0 |
| Bengamide B | Santa Cruz | sc-397521 |
| Nourseothricin | G-Biosciences | 50-103-7041 |
| Halt™ Protease and Phosphatase Inhibitor Cocktail (EDTA-free) | Thermo Fisher Scientific | 78443 |
| SUPERase In | Ambion | AM2696 |
| TURBO DNase | Ambion | AM2238 |
| 3xFLAG peptide | Sigma-Aldrich | F4799-4MG |
| iScript Reverse Transcription Supermix kit | Bio-Rad | 1708841 |
| SsoAdvanced Universal SYBR Green Supermix | Bio-Rad | 1725274 |
| Chloramphenicol | Sigma-Aldrich | C0378-5G |
| Lipofectamin 2000 | Invitrogen | 11668 |
| Critical Commercial Assays | ||
| Dual-Luciferase Reporter Assay System | Promega | E1910 |
| ProteoExtract protein precipitation kit | EMD Millipore | 539180-1KIT |
| Deposited Data | ||
| Experimental Models: Cell Lines | ||
| Mouse: E14 embryonic stem cells | Laboratory of Barbara Panning (Hooper et al., 1987) | N/A |
| Experimental Models: Organisms/Strains | ||
| S. cerevisiae: KAY488 (NOY890): MATA ura3-1 leu2-3, 112 his3-11 trp1-1 ade2-1 can1-100 rdnaΔΔ::HIS3 carrying pRDN-hyg::URA3 | Laboratory of Katsura Asano (Nemoto et al., 2010) | N/A |
| S. cerevisiae: WT_rRNA: KAY488: MATA ura3-1 leu2-3,112 his3-11 trp1-1 ade2-1 can1-100 rdnaΔΔ::HIS3 carrying WT_rRNA::LEU2 | This study | N/A |
| S. cerevisiae: ES27L Δb1: KAY488: MATA ura3-1 leu2-3, 112 his3-11 trp1-1 ade2-1 can1-100 rdnaΔΔ::HIS3 carrying ES27L Δb1::LEU2 | This study | N/A |
| S. cerevisiae: ES27L Δb1-2: KAY488: MATA ura3-1 leu2-3, 112 his3-11 trp1-1 ade2-1 can1-100 rdnaΔΔ::HIS3 carrying ES27L Δb1-2::LEU2 | This study | N/A |
| S. cerevisiae: ES27L Δb1-3: KAY488: MATA ura3-1 leu2-3,112 his3-11 trp1-1 ade2-1 can1-100 rdnaΔΔ::HIS3 carrying ES27L Δb1-3::LEU2 | This study | N/A |
| S. cerevisiae: ES27L Δb1-4: KAY488: MATA ura3-1 leu2-3,112 his3-11 trp1-1 ade2-1 can1-100 rdnaΔΔ::HIS3 carrying ES27L Δb1-4::LEU2 | This study | N/A |
| S. cerevisiae: ES27L Δc: KAY488: MATA ura3-1 leu2-3,112 his3-11 trp1-1 ade2-1 can1-100 rdnaΔΔ::HIS3 carrying ES27L Δc::LEU2 | This study | N/A |
| S. cerevisiae: WT_rRNA, MAP2-HA: KAY488: MATA ura3-1 leu2-3, 112 his3-11 trp1-1 ade2-1 can1-100 rdnaΔΔ::HIS3 carrying WT_rRNA::LEU2 MAP2-HA | This study | N/A |
| S. cerevisiae: ES27L Δb1-4, MAP2-HA: KAY488: MATA ura3-1 leu2-3, 112 his3-11 trp1-1 ade2-1 can1-100 rdnaΔΔ::HIS3 carrying ES27L Δb1-4::LEU2 MAP2-HA | This study | N/A |
| S. cerevisiae: W303-A: MATA ura3-1 leu2-3, 112 his3-11 trp1-1 ade2-1 can1-100 | Laboratory of Jarosz lab | N/A |
| S. cerevisiae: Δmap1: W303-A: MATA ura3-1 leu2-3, 112 his3-11 trp1-1 ade2-1 can1-100 map1Δ0 | This study | N/A |
| S. cerevisiae: Δmap2: W303-A: MATA ura3-1 leu2-3, 112 his3-11 trp1-1 ade2-1 can1-100 map2Δ::natMX6 | This study | N/A |
| S. cerevisiae: BY4741: MATA ura3Δ0 leu2Δ0 his3Δ1 met15Δ0 | Laboratory of Jarosz lab | N/A |
| S. cerevisiae: Δmap1: BY4741: MATA ura3Δ0 leu2Δ0 his3Δ1 met15Δ0 map1Δ::kanr | Dharmacon | YSC1053 |
| S. cerevisiae: Δard1: BY4741: MATA ura3Δ0 leu2Δ0 his3Δ1 met15Δ0 ard1Δ::kanr | Dharmacon | YSC1053 |
| C. Crescentus: NA1000 | Laboratory of Lucy Sapiro | N/A |
| C. Crescentus: MAP-HA: NA1000: MAP-HA::kanR | This paper | N/A |
| See Table S2 for the list of Cells used in this study | ||
| Oligonucleotides | ||
| qPCR Primer for 25S rRNA: GACCTCAAATCAGGTAGGAGTACCC | This paper | N/A |
| qPCR Primer for 25S rRNA: CACCGAAGGTACCAGATTTC | This paper | N/A |
| qPCR Primer for pre-25S rRNA: CTAGGCGAACAATGTTCTTAAAG | This paper | N/A |
| qPCR Primer for pre-25S rRNA: CACCGAAGGTACCAGATTTC | This paper | N/A |
| Recombinant DNA | ||
| pNOY373: LEU2, 2μ, Pol1-rDNA-WT_rRNA | Laboratory of Katsura Asano (Nemoto et al., 2010) | N/A |
| pNOY373-ES27L Δb1: LEU2, 2μ, Pol1-rDNA-ES27L Δb1 | This paper | N/A |
| pNOY373-ES27L Δb1-2: LEU2, 2μ, Pol1-rDNA-ES27L Δb1-2 | This paper | N/A |
| pNOY373-ES27L Δb1-3: LEU2, 2μ, Pol1-rDNA-ES27L Δb1-3 | This paper | N/A |
| pNOY373-ES27L Δb1-4: LEU2, 2μ, Pol1-rDNA-ES27L Δb1-4 | This paper | N/A |
| pNOY373-ES27L Δc: LEU2, 2μ, Pol1-rDNA-ES27L Δc | This paper | N/A |
| pML104: URA3, 2μ, Cas9 | (Laughery et al., 2015) | Addgene plasmid # 67638 |
| pML 104-MAP1: URA3, 2μ, Cas9, MAP1 guide RNA | This paper | N/A |
| pML 104-MAP2: URA3, 2μ, Cas9, MAP2 guide RNA | This paper | N/A |
| pRPL25/uL23-FLAG-His: URA3, CEN, RPL25/uL23-FLAG-His | Laboratory of Toshifumi Inada (Inada et al., 2002) | N/A |
| pMAP1-HA: pRS316: URA3, CEN, MAP1-HA | This paper | N/A |
| pMAP1-HA: pRS415: LEU2, CEN, MAP1-HA | This paper | N/A |
| pH301N: pRS415: LEU2, CEN, MAP1H301N-HA | This paper | N/A |
| p14-3-3γ-Rluc: pRS316: URA3, 2μ, 14-3-3γ-Rluc | This paper | N/A |
| p245CAT(WT): URA3, 2μ, Rluc-Fluc | Laboratory of Philip Farabaugh (Kramer et al., 2010) | N/A |
| p245CGT: URA3, 2μ, Rluc-Fluc-245CGT | Laboratory of Philip Farabaugh (Kramer et al., 2010) | N/A |
| p245CGC: URA3, 2μ, Rluc-Fluc-245CGC | Laboratory of Philip Farabaugh (Kramer et al., 2010) | N/A |
| pJD375-yDL-RT (0-frame): URA3, 2μ, Rluc-Fluc | Laboratory of Jonathan Dinman (Muldoon-Jacobs and Dinman, 2006) | N/A |
| pJD376-yDL-L-A PRF: URA3, 2μ, Rluc-LA(−1PRF)-Fluc | Laboratory of Jonathan Dinman (Muldoon-Jacobs and Dinman, 2006) | N/A |
| pJD377-yDL-Ty1 PRF: URA3, 2μ, Rluc-Ty1(+1PRF)-Fluc | Laboratory of Jonathan Dinman (Muldoon-Jacobs and Dinman, 2006) | N/A |
| pJD431-yDL-UAA: URA3, 2μ, Rluc-UAA-Fluc | Laboratory of Jonathan Dinman (Harger and Dinman, 2004) | N/A |
| pJD432-yDL-UAG: URA3, 2μ, Rluc-UAG-Fluc | Laboratory of Jonathan Dinman (Harger and Dinman, 2004) | N/A |
| pJD433-yDL-UGA: URA3, 2μ, Rluc-UGA-Fluc | Laboratory of Jonathan Dinman (Harger and Dinman, 2004) | N/A |
| pCMV-WT: CMV promoter, Rluc-Fluc | This paper | N/A |
| pCMV-433-UGA: CMV promoter, Rluc-UGA-Fluc | This paper | N/A |
| pMCS-2: KanR | Laboratory of Lucy Sapiro (Thanbichler et al., 2007) | N/A |
| pMCS-2-MAP-HA: KanR, MAP-HA | This paper | N/A |
| Software and Algorithms | ||
| PyMOL | (Schrödinger, 2015) | https://www.pymol.org/ |
| edgeR | (Robinson et al., 2009) | https://bioconductor.org/packages/release/bioc/html/edgeR.htm |
| limma | (Smyth et al., 2010) | http://bioinf.wehi.edu.au/limma |
| ClustalOmega | EMBL-EBI webtools | https://www.ebi.ac.uk/Tools/msa/clustalo/ |
| Other | ||
| MISSION siRNA Universal Negative Control 2 | Sigma | SIC002 |
| MetAP1-1: GGAUGAAGGAGCACGGAAAUU | Dharmacon | N/A |
| MetAP1-2: CAGGAAUGCUUUAAAGGAAUU | Dharmacon | N/A |
| MetAP2-1: GGGAUGACGAUGAUAGAAAUU | Dharmacon | N/A |
| MetAP2-2: CCAAAGGACAAGAGUGUGAUU | Dharmacon | N/A |
METHOD DETAILS
Construction
Plasmids for Saccharomyces cerevisiae:
Plasmids used in this paper are listed at the Recombinant DNA in KEY RESOURCES TABLE. The S. cerevisiae MAP1 with 1kb upstream (genomic promoter) and downstream (genomic terminator) sequence was cloned into the pRS415 single copy plasmid with C-terminal HA tag. The MAP1S301N point mutation was inserted by overlap extension PCR. MetAP activity reporter was produced by inserting a 12 N-terminal amino acid sequence of the human 14-3-3γ gene (ATG GTG GAC CGC GAG CAA CTG GTG CAG AAA GCC CGG) before the coding sequence (CDS) of the Renilla luciferase (Rluc). Mouse ESC plasmids: Translation fidelity constructs for ESCs were cloned into an expression vector containing the cytomegalovirus (CMV) promoter. Saccharomyces cerevisiae strains: Cells used in this paper are listed at the Experimental Models: Organisms/Strains in the KEY RESOURCES TABLE. The rDNA mutant strains were produced from KAY488 (NOY890) by plasmid shuffling from pRDN-hyg (RDNAhyg URA3) to pNOY373 (RDNA LEU2) or derivatives containing a mutation in ES27L. To remove the pRDN-hyg plasmid, the strain was negatively selected against URA3 marker using 1 mg/ml of 5-Fluoroorotic Acid (5-FOA) (Fisher Scientific, F10501-5.0), which is processed to a toxic product by the Ura3 enzyme. The pNOY373 derivatives contained the truncated ES27L, which were produced by overlap extension PCR and subsequently were introduced into the Mlu1-Xho1 site of pNOY373. Using the CRISPR system (Laughery et al., 2015), the entire CDS was deleted for the MAP1 deletion strain; or the Human influenza hemagglutinin (HA) tag was inserted at the C-terminus for the MAP2-HA strain. The guide RNA for MAP1 or MAP2 (MAP1: ACAGAGAATTAAATAGATCT, MAP2: TCGAAAGGTGATGACTACTG) was inserted into the pML104 plasmid to co-express with CRISPR. The pML104 derivatives (100ng) and double strand recombination template (1μg) were transformed together (Recombination template for MAP1 deletion: acgatctgctatgatacaagttctaatggcaggccatcgctcaggatggaaatgcatggtcatttgtggcgattctgaggaccagtgtgtttccccttgcggcgtttgcaggcagtttatcaatgacagggatatgcgtgtagagttcaatttgaccgggacagtcaaagattaaatattcatcgttgaaatccccgatctcttcgtcaagccagtctagatttttcaaaagatattcgaagcaa, Recombination template for MAP2 HA tagging: GGGACTTTACCGTTCTGTCGCCGATACCTAGACAGACTTGGCCAAGAGAAATACTTATTTGC GTTGAATAACTTGGTTAGACACGGTTTAGTACAGGATTATCCACCATTGAACGATATCCCCG GATCCTACACTGCACAATTCGAACACACCATCTTGTTGCATGCTCACAAAAAGGAAGTCGTTT CGAAAGGTGATGACTACgcggccgttTACCCATACGATGTTCCTGACTATGCGggcTGAggtaaaatg cgctttcaaatggcctcctcactaggtatatgaatactgttgtagttatatagatagtttaatatggcttgccgcccgtttagaatgatgcattac cttatataagacctatgggggtctcaaacagtggtatgacgacagataatataagcactagttcagacgagtaa), and transformed yeasts were selected on a SD minus uracil plate. To remove the pML104 plasmid, negative selection was performed with 1 mg/ml of 5-FOA. The MAP2 deletion strain was generated by replacing the CDS with the Nourseothricin resistance gene by transforming 1 μg of PCR product of the resistance gene with 40 nt of homology arms to the target site (PCR primers: TTT ACAT CAACAAGTACT GTAGAAGCT CTACCGTATT GAAAAATGcgtacgctgcaggtc and TCATATACCTAGTGAGGAGGCCATTTGAAAGCGCATTTTACCTCAatgaattcgagatag). Selection was performed on the 60mg/L Nourseothricin containing YPD plate. Deletion strains for MAP1 or MAP2 were produced from the wild type W303-A background strain. The deletion strains for MAP1 and ARD1 were picked up from the BY4741 background Yeast Knock Out library (YKO: Dharmacon) and the deletion for target genes were confirmed by genomic PCR. Caulobacter crescentus Strain (Thanbichler et al., 2007): To modify the MAP gene in C. crescentus, the MAP gene was cloned into the recombination vector pMCS-2 with the C-terminal HA tag. The pMCS-2-MAP-HA plasmid was electroporated into the NA1000 strain and genomic MAP was tagged with HA at the C-terminus. Genomic integration was selected by Kanamycin and checked by genomic PCR and western blotting.
Yeast Spot Assay
For the spot assay on the translation inhibitor drug, fresh overnight liquid cultures were adjusted to OD600 = 1 as a starting point, and 10-fold serial dilutions were spotted on a fresh YPAD plate or a YPAD plate containing a translation inhibitor drug. The plates were then incubated at 30 °C for 3 or 4 days. Concentration of Paromomycin is 1mg/ml unless indicated. Hygromycin B is 25 μg/ml, Anisomycin is 5 μg/ml, Cycloheximide is 0.4 μg/ml.
Dual luciferase assay
Saccharomyces cerevisiae:
Stationary yeast cultures transformed with translation fidelity constructs were diluted to OD600 = 0.05 to grow until log phase (OD600 = 0.5). For Paromomycin (Sigma Aldrich, P9297-1G) treatment, the drug (400 μg/ml) was added into the medium at OD600= 0.5. The culture was then incubated for 2 hrs prior to harvest. Mouse ESCs: Translation fidelity constructs were transfected by Lipofectamine 2000 (Invitrogen, 11668) in 12 well plates. For Bengamide B (Santa Cruz, sc-397521A) treatment, the drug (0.1 μM) was added into the medium 3 hrs after transfection. Cells were incubated for 24 hrs, and then samples were harvested by trypsin. Collected yeast pellets and cells were lysed by passive lysis buffer and assayed using the Dual Luciferase kit (Promega, E1980). To measure the mistranslation efficiency, tandem reporters of Renilla luciferase (Rluc) and Firefly luciferase (Fluc) were used and to normalize the input amount, Fluc activity was normalized by Rluc activity. To measure the amino acid misincorporation, each reporter contains various point mutations at Fluc active site and the percentage of the Histidine misincorporation at the Arginine codon was detected (Kramer et al., 2010). To measure the stop codon read-through, a stop codon was inserted between Rluc and Fluc (Muldoon-Jacobs and Dinman, 2006). To measure the frameshift induced by the PRF (Programed ribosomal frameshifting) sequence, the −1 PRF sequence derived from yeast Killer virus (L-A) or +1 PRF sequence derived from Ty1 retrotransposon element was inserted between Rluc and Fluc (Harger and Dinman, 2004). To measure mistranslation efficiency, normalized Fluc activities were further normalized to the “wild type” construct, which does not contain the point mutation at the active site of Fluc and does not have an insertion of the stop codon or frameshift sequence between Rluc and Fluc. Each experiment was performed with a minimum of three biological replicates. Statistical analysis was performed using the Student’s t-test.
Immunoprecipitation of ribosomes
To purify translating ribosomes, C-terminal FLAG-tagged Rpl25/uL23 was expressed from the single copy plasmid in WT_rRNA and ES27L Δb1-4 mutant strains. At log phase (OD600 = ∼0.8), 100 μg/ml Cycloheximide (Sigma Aldrich, C7698-5G) was added into the medium 5 mins before harvest and pellets were flash frozen by liquid nitrogen. To make a lysate, pellets were powderized in liquid nitrogen by mortar and pestle. Powdered yeast lysates were dissolved with lysis buffer (20 mM Hepes, pH 7.5, 100 mM KOAc, 5 mM Mg(OAc)2, 4 mM DTT, 100 μg/ml Cycloheximide, 1× Halt™ Protease and Phosphatase Inhibitor Cocktail (EDTA-free) (Thermo Fisher, 78443), 200 U/ml SUPERase In (Ambion, AM2696), 20 U/ml TURBO DNase (Ambion, AM2239)). Rpl25/uL23-FLAG tagged ribosomes were immuno-precipitated using an ANTI-FLAG M2 affinity Gel (Sigma Aldrich, A2220-5ML) by 2 hr rotation at 4 °C. Beads were washed 3 times and bound ribosomes were eluted by 100 μg/ml 3×FLAG peptide (Sigma-Aldrich, F4799-4MG). To further purify translating ribosomes, immuno-isolated ribosomes were layered onto a 10-40 % sucrose gradient.
Sucrose density gradient
Saccharomyces cerevisiae: To fractionate ribosomes, 5 units of A260 of cell lysate or purified ribosomes were layered onto 10-40 % (w/v) of sucrose gradient (20 mM HEPES, pH 7.5, 100 mM KOAc, 5 mM Mg(OAc)2, 4 mM DTT, 100 μg/ml Cycloheximide) and centrifuged by 40,000 rpm for 2.5 hr at 4 °C using the SW41Ti rotor. Gradients were fractioned using a Brandel gradient fractionator (BR-188) with continuous A260 measurement. Caulobacter crescentus: 100 μg/ml of Chloramphenicol (Sigma Aldrich, C0378-5G) was added 2 min before harvest at log phase (OD600 = ∼0.8). C. crescentus pellets were dissolved with lysis buffer (20 mM HEPES, pH 7.5, 100 mM KOAc, 5 mM Mg(OAc)2, 4 mM DTT, 100 μg/ml Chloramphenicol) and dropped into liquid nitrogen. The C. crescentus lysate was powderized in liquid nitrogen by mortar and pestle. 1x Halt™ Protease and Phosphatase Inhibitor Cocktail (EDTA-free), 200 U/ml SUPERase In, 20 U/ml TURBO DNase was added into the powdered C. crescentus lysates. To fractionate ribosomes, 5 units of A260 of cell lysate were layered onto 10-40 % (w/v) of sucrose gradient (20 mM HEPES, pH 7.5, 100 mM KOAc, 5 mM Mg(OAc)2, 4 mM DTT, 100 μg/ml Chloramphenicol) and centrifuged by 40,000 rpm for 2.5 hr at 4 °C using the SW41Ti rotor. Mouse ESCs: ESCs were treated with the Cycloheximide (Sigma Aldrich, C7698-5G) at 100 mg/ml for 3 min. ESCs were harvested and freeze by liquid nitrogen and thaw by adding lysis buffer (20 mM Tris, pH 7.5, 100 mM NaCl, 15 mM MgCl2, 1 mM DTT, 100 μg/ml Cycloheximide, 1× Halt™ Protease and Phosphatase Inhibitor Cocktail (EDTA-free) (Thermo Fisher, 78443), 200 U/ml SUPERase In (Ambion, AM2696), 20 U/ml TURBO DNase (Ambion, AM2239)). To fractionate ribosomes, cell lysate were layered onto 10-45 % (w/v) of sucrose gradient (20 mM Tris, pH 7.5, 100 mM NaCl, 15 mM MgCl2, 1 mM DTT, 100 μg/ml Cycloheximide) and centrifuged by 40,000 rpm for 2.5 hr at 4 °C using the SW41Ti rotor. For the western blotting from mESCs fractions, first three fractions were five fold diluted.
RT-qPCR
After polysome fractionation, RNA was extracted using Acid-Phenol:Chloroform, pH 4.5 (with IAA, 125:24:1) (Invitrogen, AM9722). RNA was reverse transcribed to cDNA using iScript Reverse Transcription Supermix kit (Bio-Rad, 1708841) and qPCR was performed using the SsoAdvanced Universal SYBR Green Supermix (Bio-Rad, 1725274). RNA levels were quantified using a standard curve method by CFX manager (Bio-Rad), summed across all fractions analyzed and presented as percentage of this total.
Relative quantification mass spectrometry by tandem mass tag labeling (Simsek et al., 2017)
Proteins were purified by the ProteoExtract protein precipitation kit (Calbiochem, 539180-1KIT). Samples were in-gel Trypsin digested and peptides were labeled with tandem mass tag (TMT) reagents. Labeling reactions were combined, cleaned, and dried down. Peptides were resuspended in 5 % Acetonitrile, 5 % formic acid and the sample was shot on an Orbitrap Fusion Mass spectrometer. Peptides were separated using a gradient of 6 to 28 % acetonitrile in 0.125 % formic acid over 180 min. Peptides were detected (MS1) and quantified (MS3) in the Orbitrap. Peptides were sequenced (MS2) in the ion trap. MS2 spectra were searched using the SEQUEST algorithm against a UniProt composite database derived from the yeast proteome containing its reversed complement and known contaminants. Peptide spectral matches were filtered to a 1 % FDR using the target-decoy strategy combined with linear discriminant analysis. Proteins were quantified only from peptides with a summed signal-to-noise (SN) threshold of ≥ 200 and MS2 isolation specificity of 0.5. For the analysis of quantified data, log2 total summed SN intensities were normalized by Trimmed Mean of M-values (TMM) in edgeR Bioconductor package (Robinson et al., 2009) and t-statistics were calculated using voom-limma method (Smyth et al., 2010). Sample similarity was calculated by Multidimensional Scaling (MDS).
Western Blotting
Protein samples were loaded onto an SDS-PAGE gel and transferred to a PVDF membrane using Trans-Blot Turbo (Bio-Rad, 170-4273) following the manufacture’s protocol. Western blotting signals were developed using Clarity Western ECL Substrate (Bio-Rad, 170-5061) and imaged with the ChemiDoc MP (Bio-Rad, 17001402). Primary antibody dilutions were: 1:1000 for Met-14-3-3γ (Novus, NB100-407), 1:1000 for 14-3-3γ (Cell Signaling, D15B7), 1:250 for HA (Thermo Fisher, SG77), 1:1000 for Rps5/uS7 (Abcam, ab58345), 1:1000 for Zuo1 (Dr. Elizabeth Craig) (Lee et al., 2016), 0.4 μg/ml for bacteria Rps13/uS13 (Developmental Studies Hybridoma Bank, 193E11E5B11), 1:500 for MetAP1 (R&D, MAB3537), 1:1000 for MetAP2 (Proteintech, 17040-1-AP), 1/1000 for Rpl10a/uL1 (Abcam, ab174318).
Structural Model of Ribosome
The structural models of E. coli ribosome (PDB: 3E1B) and S. cerevisiae ribosome (PDB: 4V6I) were downloaded from the Protein Data Bank (PDB) website and edited using PyMOL (Schrödinger, 2015).
[35S]methionine labeling
Cells in log phase were adjusted to OD600 0.5 for each strain and [35S]methionine (Perkinelmer, NEG709A005MC) was added at 10 μCi/ml. Samples were taken at 0, 2, 5, and 10 min. By adding 20 mM NaN3 and 200 μg/ml of Cycloheximide, translation was frozen and samples were put on ice. Cells were washed 3 times with water and lysed by the glass beads method. Radioactivity in the lysate was measured with a scintillation counter. Relative [35S]methionine incorporation (10 min) was calculated against the WT_rRNA strain.
Knockdown in mouse ESCs
Gene knockdown was achieved by transfecting mESCs with small double-stranded interfering RNAs (siRNA) with Lipofectamine 2000 (Invitrogen, 11668). siRNAs used for targeting were custom designed (Dharmacon) as follows: for MetAP1-1: GGAUGAAGGAGCACGGAAAUU, MetAP1-2: CAGGAAUGCUUUAAAGGAAUU, for MetAP2-1: GGGAUGACGAUGAUAGAAAUU, MetAP2-2: CCAAAGGACAAGAGUGUGAUU. MISSION siRNA Universal Negative Control 2 (Sigma, SIC002) was used as a control. Knockdown was performed for 48 hrs with twice transfection in every 24 hrs.
Map1 and Map2 protein sequence alignments
Multiple sequence alignments (MSAs) of Map1 and Map2 protein sequences were done using ClustalOmega from EMBL-EBI webtools with default settings. Neighbor-joining phylogenetic tree data without distance corrections were also obtained from the same MSA output. Eukaryotic Map1 and bacterial MAP sequences used are: P53582 (H. sapiens), Q8BP48 (M. musculus), Q7ZWV9 (X. laevis), Q4QRK0 (D. rerio),Q9VC48 (D. melanogaster), Q965Y1 (C. elegans), Q01662 (S. cerevisiae), Q9A4Z4 (C. crescentus), P19994 (B. subtilis), P9WK21 (M. tuberculosis), and P0AE18 (E. coli). Eukaryotic Map2 and archaic MAP sequences used are: P50579 (H. sapiens), O08663 (M. musculus), P38174 (S. cerevisiae), Q5JGD1 (T. kodakarensis), and P95963 (S. solfataricus).
QUANTIFICATION AND STATISTICAL ANALYSIS
For the analysis of quantified data, log2 total summed SN intensities were normalized by Trimmed Mean of M-values (TMM) in edgeR Bioconductor package (Robinson et al., 2009) and t-statistics were calculated using voom-limma method (Smyth et al., 2010). Sample similarity was calculated by Multidimensional Scaling (MDS).
In all figures, data is presented as mean, SD and **P < 0.01; *P < 0.05; NS, not significant. Blinding and randomization were not used in any of the experiments. Number of independent biological replicates used for experiments are listed in the figure legends. Tests and specific p-values used are indicated in the figure legends.
Supplementary Material
Table1 S1: Relative quantitative mass spec data of WT_rRNA and ES27L Δb1-4 ribosomes. Related to Figure 2.
Highlights:
ES27L functions in translation fidelity
ES27Lrecruits nascent peptide processing enzymes, including MetAP
Ribosome association increases MetAP activity
MetAP activity is required for higher translation fidelity in eukaryotes
Acknowledgements
We thank the Barna lab members and D. Ruggero for constructive suggestions and thoughtful critiques of the work. We thank E. Craig (University of Wisconsin-Madison) for sharing the Zuo1 antibody and F. Ward from Cate lab (UC Berkeley) for the Rps13/uS13 antibody. We thank K. Asano (Kansas state university), M. Kitabatake from Ohno lab (Kyoto university), A. Chakravarty from Jarosz lab (Stanford University) for providing yeast strains. We thank T. Inada (Tohoku University), P. Farabough (University of Maryland), J. Dinman (University of Maryland) for providing plasmid constructs. This work was supported by the New York Stem Cell Foundation NYSCF-R-I36 (M.B.), National Institutes of Health grant 1R01HD086634 (M.B.), Alfred P. Sloan Research Fellowship (M.B.), and a Pew Scholars Award (M.B.). K.F was supported by the Uehara Memorial Foundation and Human Frontier Science Program Fellowship. T.S was supported by National Science Scholarship (PhD) from the Agency for Science, Technology and Research -[of Singapore] M.B. is a New York Stem Cell Foundation Robertson Investigator.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
The authors declare no competing interests.
References
- Addlagatta A, and Matthews BW (2006). Structure of the angiogenesis inhibitor ovalicin bound to its noncognate target, human Type 1 methionine aminopeptidase. Protein Sci. 15, 1842–1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anger AM, Armache J-P, Berninghausen O, Habeck M, Subklewe M, Wilson DN, and Beckmann R (2013). Structures of the human and Drosophila 80S ribosome. Nature 497, 80–85. [DOI] [PubMed] [Google Scholar]
- Armache J-P, Jarasch A, Anger AM, Villa E, Becker T, Bhushan S, Jossinet F, Habeck M, Dindar G, Franckenberg S, et al. (2010). Cryo-EM structure and rRNA model of a translating eukaryotic 80S ribosome at 5.5-A resolution. Proc. Natl. Acad. Sci. U. S. A. 107, 19748–19753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnheim N, and Southern EM (1977). Heterogeneity of the ribosomal genes in mice and men. Cell. [DOI] [PubMed] [Google Scholar]
- Balchin D, Hayer-Hartl M, and Hartl FU (2016). In vivo aspects of protein folding and quality control. Science (80-. ). 353, aac4354–aac4354. [DOI] [PubMed] [Google Scholar]
- Copik AJ, Swierczek SI, Lowther WT, D’Souza VM, Matthews BW, and Holz RC (2003). Kinetic and spectroscopic characterization of the H178A methionyl aminopeptidase from Escherichia coli. Biochemistry 42, 6283–6292. [DOI] [PubMed] [Google Scholar]
- Drummond DA, and Wilke CO (2009). The evolutionary consequences of erroneous protein synthesis. Nat. Rev. Genet. 10, 715–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautschi M, Just S, Mun A, Ross S, Rücknagel P, Dubaquié Y, Ehrenhofer-murray A, Rospert S, Ru P, and Dubaquie Y (2003). The Yeast N α -Acetyltransferase NatA Is Quantitatively Anchored to the Ribosome and Interacts with Nascent Polypeptides. Mol. Cell. Biol. 23, 7403–7414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giglione C, Fieulaine S, and Meinnel T (2015). N-terminal protein modifications: Bringing back into play the ribosome. Biochimie 114, 134–146. [DOI] [PubMed] [Google Scholar]
- Greber BJ, Boehringer D, Montellese C, and Ban N (2012). Cryo-EM structures of Arx1 and maturation factors Rei1 and Jjj1 bound to the 60S ribosomal subunit. Nat. Struct. Mol. Biol. 19, 1228–1233. [DOI] [PubMed] [Google Scholar]
- Griffith EC, Su Z, Niwayama S, Ramsay CA, Chang Y-H, and Liu JO (1998). Molecular recognition of angiogenesis inhibitors fumagillin and ovalicin by methionine aminopeptidase 2. Proc. Natl. Acad. Sci. 95, 15183–15188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harger JW, and Dinman JD (2004). Evidence against a direct role for the Upf proteins in frameshifting or nonsense codon readthrough. RNA 10, 1721–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper M, Hardy K, Handyside A, Hunter S, and Monk M (1987). HPRT-deficient (Lesch-Nyhan) mouse embryos derived from germline colonization by cultured cells. Nature. [DOI] [PubMed] [Google Scholar]
- Hwang C-S, Shemorry A, and Varshavsky A (2010). N-Terminal Acetylation of Cellular Proteins Creates Specific Degradation Signals. Science (80-. ). 327, 973–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inada T, Winstall E, Tarun SZ, Yates JR, Schieltz D, and Sachs AB (2002). One-step affinity purification of the yeast ribosome and its associated proteins and mRNAs. RNA 8, 948–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeeninga RE, Van Delft Y, de Graaff-Vincent M, Dirks-Mulder A, Venema J, and Raué HA (1997). Variable regions V13 and V3 of Saccharomyces cerevisiae contain structural features essential for normal biogenesis and stability of 5.8S and 25S rRNA. RNA 3, 476–488. [PMC free article] [PubMed] [Google Scholar]
- Jonckheere V, Fijałkowska D, and Van Damme P (2018). Omics assisted N-terminal proteoform and protein expression profiling upon methionine aminopeptidase 1 (MetAP 1) deletion. Mol. Cell. Proteomics 1–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapur M, and Ackerman SL (2018). mRNA Translation Gone Awry: Translation Fidelity and Neurological Disease. Trends Genet. 34, 218–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer EB, and Farabaugh PJ (2007). The frequency of translational misreading errors in E. coli is largely determined by tRNA competition. Rna 13, 87–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer EB, Vallabhaneni H, Mayer LM, and Farabaugh PJ (2010). A comprehensive analysis of translational missense errors in the yeast Saccharomyces cerevisiae. RNA 16, 1797–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo B (1996). Human ribosomal RNA variants from a single individual and their expression in different tissues. Nucleic Acids Res. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laughery MF, Hunter T, Brown A, Hoopes J, Ostbye T, Shumaker T, and Wyrick JJ (2015). New vectors for simple and streamlined CRISPR-Cas9 genome editing in Saccharomyces cerevisiae. Yeast 32, 711–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JW, Beebe K, Nangle LA, Jang J, Longo-Guess CM, Cook SA, Davisson MT, Sundberg JP, Schimmel P, and Ackerman SL (2006). Editing-defective tRNA synthetase causes protein misfolding and neurodegeneration. Nature 443, 50–55. [DOI] [PubMed] [Google Scholar]
- Lee K, Sharma R, Shrestha OK, Bingman CA, and Craig EA (2016). Dual interaction of the Hsp70 J-protein cochaperone Zuotin with the 40S and 60S ribosomal subunits. Nat. Struct. Mol. Biol. 23, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leffers H, and Andersen, a H. (1993). The sequence of 28S ribosomal RNA varies within and between human cell lines. Nucleic Acids Res. 21, 1449–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, and Chang YH (1995). Amino-terminal protein processing in Saccharomyces cerevisiae is an essential function that requires two distinct methionine aminopeptidases. Proc. Natl. Acad. Sci. 92, 12357–12361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Widom J, Kemp CW, Crews CM, and Clardy J (1998). Structure of human methionine aminopeptidase-2 complexed with fumagillin. Science (80-. ). 282, 1324–1327. [DOI] [PubMed] [Google Scholar]
- Locati MD, Pagano JFB, Girard G, Ensink WA, Van Olst M, Van Leeuwen S, Nehrdich U, Spaink HP, Rauwerda H, Jonker MJ, et al. (2017). Expression of distinct maternal and somatic 5.8S, 18S, and 28S rRNA types during zebrafish development. RNA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Loubresse NG, Prokhorova I, Holtkamp W, Rodnina MV, Yusupova G, and Yusupov M (2014). Structural basis for the inhibition of the eukaryotic ribosome. Nature 513, 517–522. [DOI] [PubMed] [Google Scholar]
- Lowther WT, Orville AM, Madden DT, Lim S, Rich DH, and Matthews BW (1999). Escherichia coli methionine aminopeptidase: Implications of crystallographic analyses of the native, mutant, and inhibited enzymes for the mechanism of catalysis. Biochemistry 38, 7678–7688. [DOI] [PubMed] [Google Scholar]
- Lu JP, Yuan XH, and Ye QZ (2012). Structural analysis of inhibition of Mycobacterium tuberculosis methionine aminopeptidase by bengamide derivatives. Eur. J. Med. Chem. 47, 479–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer-Schuman R, and Antonellis A (2017). Emerging mechanisms of aminoacyl-tRNA synthetase mutations in recessive and dominant human disease. Hum. Mol. Genet. 26, R114–R127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muldoon-Jacobs K. L., and Dinman JD. (2006). Specific effects of ribosome-tethered molecular chaperones on programmed-1 ribosomal frameshifting. Eukaryot. Cell 5, 762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myasnikov AG, Simonetti A, Marzi S, and Klaholz BP (2009). Structure-function insights into prokaryotic and eukaryotic translation initiation. Curr. Opin. Struct. Biol. 19, 300–309. [DOI] [PubMed] [Google Scholar]
- Nemoto N, Singh CR, Udagawa T, Wang S, Thorson E, Winter Z, Ohira T, Ii M, Valášek L, Brown SJ, et al. (2010). Yeast 18 S rRNA Is directly involved in the ribosomal response to stringent AUG selection during translation initiation. J. Biol. Chem. 285, 32200–32212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paolini NA, Attwood M, Sondalle SB, Vieira CM dos S, van Adrichem AM, di Summa FM, O’Donohue MF, Gleizes PE, Rachuri S, Briggs JW, et al. (2017). A Ribosomopathy Reveals Decoding Defective Ribosomes Driving Human Dysmorphism. Am. J. Hum. Genet. 100, 506–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks MM, Kurylo CM, Dass RA, Bojmar L, Lyden D, Vincent CT, and Blanchard SC (2018). Variant ribosomal RNA alleles are conserved and exhibit tissue-specific expression. Sci. Adv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petry S, Weixlbaumer A, and Ramakrishnan V (2008). The termination of translation. Curr. Opin. Struct. Biol. 18, 70–77. [DOI] [PubMed] [Google Scholar]
- Prokhorova I, Altman RB, Djumagulov M, Shrestha JP, Urzhumtsev A, Ferguson A, Chang CWT, Yusupov M, Blanchard SC, and Yusupova G (2017). Aminoglycoside interactions and impacts on the eukaryotic ribosome. Proc. Natl. Acad. Sci. 201715501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakwalska M, and Rospert S (2004). The ribosome-bound chaperones RAC and Ssb1/2p are required for accurate translation in Saccharomyces cerevisiae. Mol. Cell. Biol. 24, 9186–9197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramesh M, and Woolford JL (2016). Eukaryote-specific rRNA expansion segments function in ribosome biogenesis. Rna 22, 1153–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, and Smyth GK (2009). edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanova L, Korobova F, Noniashvilli E, Dyban A, and Zatsepina O (2006). High Resolution Mapping of Ribosomal DNA in Early Mouse Embryos by Fluorescence Collection of Embryos. Biol. Reprod. 815, 807–815. [DOI] [PubMed] [Google Scholar]
- Sandikci A, Gloge F, Martinez M, Mayer MP, Wade R, Bukau B, and Kramer G (2013). Dynamic enzyme docking to the ribosome coordinates N-terminal processing with polypeptide folding. Nat. Struct. Mol. Biol. 20, 843–850. [DOI] [PubMed] [Google Scholar]
- Schrödinger (2015). The PyMOL Molecular Graphics System. Schrödinger LLC Wwwpymolorg. [Google Scholar]
- Simsek D, Tiu GC, Flynn RA, Xu AF, Chang HY, Barna M, Simsek D, Tiu GC, Flynn RA, Byeon GW, et al. (2017). The Mammalian Ribo-interactome Reveals Ribosome Functional Diversity and Heterogeneity Article The Mammalian Ribo-interactome Reveals Ribosome Functional Diversity and Heterogeneity. Cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK, Ritchie M, and Thorne N (2010). Linear Models for Microarray Data User ’ s Guide. [Google Scholar]
- Gerbi Susan A. (1998). Expansion segments: regions of variable size that interrupt the universal core secondary structure of ribosomal RNA. In Ribosomal RNA: Structure, Evolution, Processing, and Function in Protein Biosynthesis, pp. 71–87. [Google Scholar]
- Sweeney R, Chen L, and Yao MC (1994). An rRNA variable region has an evolutionarily conserved essential role despite sequence divergence. Mol. Cell. Biol. 14, 4203–4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanbichler M, Iniesta AA, and Shapiro L (2007). A comprehensive set of plasmids for vanillate - And xylose-inducible gene expression in Caulobacter crescentus. Nucleic Acids Res. 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin H, Bair KW, DeCaprio JA, Eck MJ, Kim S, Kinder FR, Morollo A, Mueller DR, Schindler P, Song HK, et al. (2003). Proteomics-based target identification: Bengamides as a new class of methionine aminopeptidase inhibitors. J. Biol. Chem. 278, 52964–52971. [DOI] [PubMed] [Google Scholar]
- Tseng H, Chou W, Wang J, Zhang X, Zhang S, and Schultz RM (2008). Mouse ribosomal RNA genes contain multiple differentially regulated variants. PLoS One. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshavsky A (2011). The N-end rule pathway and regulation by proteolysis. Protein Sci. 20, 1298–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetro JA, and Chang YH (2002). Yeast methionine aminopeptidase type 1 is ribosome-associated and requires its N-terminal zinc finger domain for normal function in vivo. J. Cell. Biochem. 85, 678–688. [DOI] [PubMed] [Google Scholar]
- Wai HH, Vu L, Oakes M, and Nomura M (2000). Complete deletion of yeast chromosomal rDNA repeats and integration of a new rDNA repeat: use of rDNA deletion strains for functional analysis of rDNA promoter elements in vivo. Nucleic Acids Res. 28, 3524–3534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Lu JP, and Ye QZ (2012). Structural analysis of bengamide derivatives as inhibitors of methionine aminopeptidases. J. Med. Chem. 55, 8021–8027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Q-Z, Xie S-X, Ma Z-Q, Huang M, and Hanzlik RP (2006). Structural basis of catalysis by monometalated methionine aminopeptidase. Proc. Natl. Acad. Sci. U. S. A. 103, 9470–9475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Ma C, Yuan Y, Zhu J, Li N, Chen C, Wu S, Yu L, Lei J, and Gao N (2014). Structural basis for interaction of a cotranslational chaperone with the eukaryotic ribosome. Nat. Struct. Mol. Biol. 6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table1 S1: Relative quantitative mass spec data of WT_rRNA and ES27L Δb1-4 ribosomes. Related to Figure 2.