Abstract
Background
Principal component analysis (PCA) is a standard method to correct for population stratification in ancestry-specific genome-wide association studies (GWASs) and is used to cluster individuals by ancestry. Using the 1000 genomes project data, we examine how non-linear dimensionality reduction methods such as t-distributed stochastic neighbor embedding (t-SNE) or generative topographic mapping (GTM) can be used to provide improved ancestry maps by accounting for a higher percentage of explained variance in ancestry, and how they can help to estimate the number of principal components necessary to account for population stratification. GTM generates posterior probabilities of class membership which can be used to assess the probability of an individual to belong to a given population - as opposed to t-SNE, GTM can be used for both clustering and classification.
Results
PCA only partially identifies population clusters and does not separate most populations within a given continent, such as Japanese and Han Chinese in East Asia, or Mende and Yoruba in Africa. t-SNE and GTM, taking into account more data variance, can identify more fine-grained population clusters. GTM can be used to build probabilistic classification models, and is as efficient as support vector machine (SVM) for classifying 1000 Genomes Project populations.
Conclusion
The main interest of probabilistic GTM maps is to attain two objectives with only one map: provide a better visualization that separates populations efficiently, and infer genetic ancestry for individuals or populations. This paper is a first application of GTM for ancestry classification models. Our code (https://github.com/hagax8/ancestry_viz) and interactive visualizations (https://lovingscience.com/ancestries) are available online.
Electronic supplementary material
The online version of this article (10.1186/s12859-019-2680-1) contains supplementary material, which is available to authorized users.
Keywords: Generative topographic mapping, Ancestry, Genetics, Population stratification
Background
As of 2018, most genome-wide association studies (GWASs) have used populations of European ancestry. However, larger sample sizes are now available and both societal need and funders are mandating more studies focused on other populations. Visualizing and accurately defining complex population structure is therefore of paramount importance. In this paper, we have three aims: to find a better way to visualize population substructures, to define a new procedure to estimate the optimal number of principal components accounting for population stratification, and to obtain an ancestry classification algorithm which can also estimate probabilities to belong to different ancestry groups. This paper focuses on global (genome-wide) ancestry rather than local ancestry defined within chromosome segments.
Principal component analysis (PCA) is widely used to investigate population structure in genetics [1], and to account for population stratification in GWASs (cf. EIGENSTRAT software [2]). However, the 2 or 3 principal components used to build a PCA plot generally account for a small percentage of variance explained and lead to a simplified visualization of population substructures, focused on major continental ancestry, with only partial sensitivity for the identification of admixed individuals or more complex ancestry. Model-based methods such as STRUCTURE [3] and ADMIXTURE [4] provide maximum likelihood estimations of ancestry based on ancestry proportions and allele frequencies but do not provide the simple 2D maps that can be obtained with PCA, multidimensional scaling (MDS), and other multivariate analysis methods.
A PCA ancestry map is constructed from a genotype matrix G of dimension N×D, where the N instances are individuals and the D features correspond to genetic variants - typically single nucleotide polymorphisms (SNPs) which are pruned to remove SNPs in high linkage disequilibrium with each other so that the identified principal components do not reflect local haplotype structure, but instead reflect genome-wide ancestry. For example, Gnd could be the minor allele count for SNP d in individual n. For visualization purposes, PCA is used to map G to a more interpretable latent or hidden space of 2 or 3 dimensions: G→X, where X has dimension N×2 or N×3. The new variables - typically two for a PCA plot - are the first principal components, which account for the highest percentage of the overall variance. However, the total percentage of variance explained by such a small number of principal components can be low for high-dimensional genotype matrices.
More complex visualization methods such as t-distributed stochastic neighbor embedding (t-SNE) [5] or generative topographic mapping (GTM) [6], which are manifold-based and non-linear dimensionality reduction algorithms, are able to capture more information by embedding a D-dimensional space in a low-dimensional latent space, where D can be any number of features. Instead of two or three principal components, any number of principal components can be used with these methods. To assess the percentage of variance to account for population substructures, we propose to execute two mappings, first carrying out PCA to select principal components and then using t-SNE or GTM: G→X′→X, where X’ is the matrix of F principal components (F>2), and X is the final t-SNE or GTM projection in a 2-dimensional space. The performance of ancestry classification models built with X or the visual assessment of clusters in X could then provide a way to estimate the number of principal components to account for population stratification.
Both t-SNE and GTM are used for clustering tasks. However, new instances cannot be projected onto a t-SNE map without training the map once again. GTM, on the other hand, not only allows for the projection of new data points, but comes with a probabilistic framework to build a comprehensive classification model and assign probabilities of class membership. t-SNE is now widely used in genetics, and has already been applied to visual population stratification [7], transcriptome visualization [8], and single-cell analysis [9]. GTM is more popular in cheminformatics, and was used to classify chemical compounds [10] or to compare chemical libraries [11]. GTM could easily be transposed to genetics and used to predict ancestry and relative degree of admixture in an individual or a group.
In this paper, 1000 Genomes Project Phase III [12] data is used to build the genotype matrix G. The 1000 Genomes Project has gathered genotypes from 26 different populations corresponding to 5 superpopulations: Africans (AFR), Admixed Americans (AMR), East Asians (EAS), Europeans (EUR) and South Asians (SAS). We separated these populations into a training set of 20 populations, and an external test set of 6 populations: Americans of African ancestry in Southwest USA (ASW); African Caribbeans in Barbados (ACB); Mexican ancestry from Los Angeles USA (MXL); Gujarati Indian from Houston, Texas (GIH); Sri Lankan Tamil from the UK (STU); and Indian Telugu from the UK (ITU). Ancestry maps are investigated to cluster and visualize superpopulations and populations using PCA, t-SNE, and GTM. t-SNE and GTM maps accounting for 3 to 1000 principal components are compared to a simple PCA plot. We also compare GTM ancestry classification models to two different algorithms: k-nearest neighbors (k-NN) models based on the 2D PCA plot, and linear Support Vector Machine (SVM), a classical machine learning algorithm [13]. We also demonstrate how to assess probabilities of ancestry membership in individuals and populations using GTM.
Results
Classification of 5 superpopulations
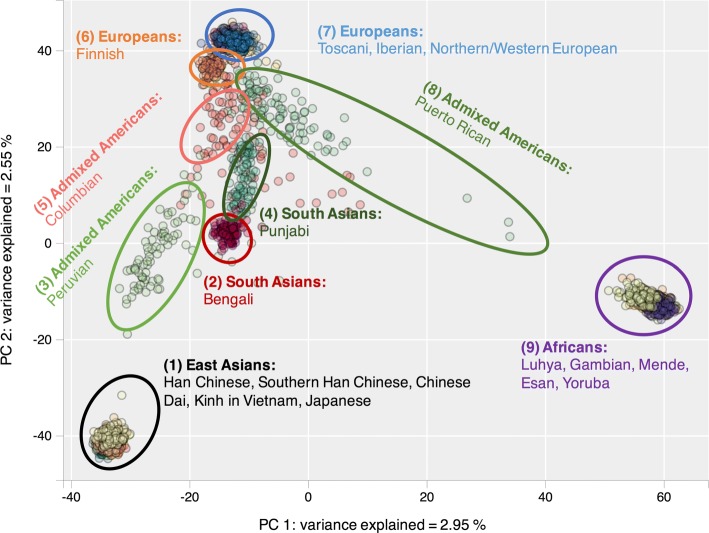
Visualizations and complete model performance statistics can be found in Additional files 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14. PCA clusters and predicts the 5 superpopulations in 1000 Genomes Project efficiently (F1 score = 0.98, cf. Table 1 and Fig. 1): Europeans, Africans, South Asians, East Asians, and Admixed Americans. However, SVM and GTM models with 3 or 10 principal components have higher recall for Admixed Americans and higher precision for South Asians (cf. Additional files 13 and 14). Optimal performances can be achieved by including a third principal component.
Table 1.
10 times repeated 5-fold cross-validated F1 score in five 1000 Genomes Project superpopulations using SVM, PCA or GTM
| Ancestry | 1000G code | PCA 8-NN | SVM 10 PCs | GTM 3 PCs | GTM 10 PCs |
|---|---|---|---|---|---|
| Africans | AFR | 1.00±0.00 | 1.00±0.00 | 1.00±0.00 | 1.00±0.00 |
| Admixed Americans | AMR | 0.93±0.00 | 1.00±0.00 | 1.00±0.00 | 1.00±0.00 |
| East Asians | EAS | 1.00±0.00 | 1.00±0.00 | 1.00±0.00 | 1.00±0.00 |
| Europeans | EUR | 0.99±0.00 | 1.00±0.00 | 1.00±0.00 | 1.00±0.00 |
| South Asians | SAS | 0.93±0.01 | 1.00±0.00 | 1.00±0.00 | 1.00±0.00 |
| Overall F1 score | 0.98±0.00 | 1.00±0.00 | 1.00±0.00 | 1.00±0.00 |
SVM10 = support vector machine classification model using 10 principal components, PCA = k-nearest neighbours model based on 2D PCA map (k = 7), GTM{3,10,100} = bayesian classification model based on generative topographic mapping using 3, 10 or 100 principal components. Each value is an average with 95% confidence interval
Fig. 1.
PCA clustering Principal Component Analysis (PCA) plot of 20 populations from 1000 Genomes Project, built using 2 first principal components. The following populations were not used to build the map: ASW = Americans of African Ancestry in SW USA; ACB = African Caribbeans in Barbados; MXL = Mexican Ancestry from Los Angeles USA; GIH = Gujarati Indian from Houston, Texas; STU = Sri Lankan Tamil from the UK; ITU = Indian Telugu from the UK
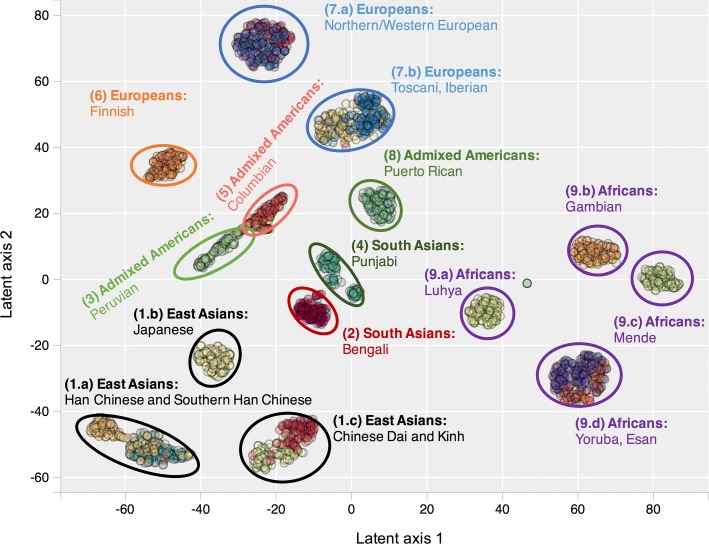
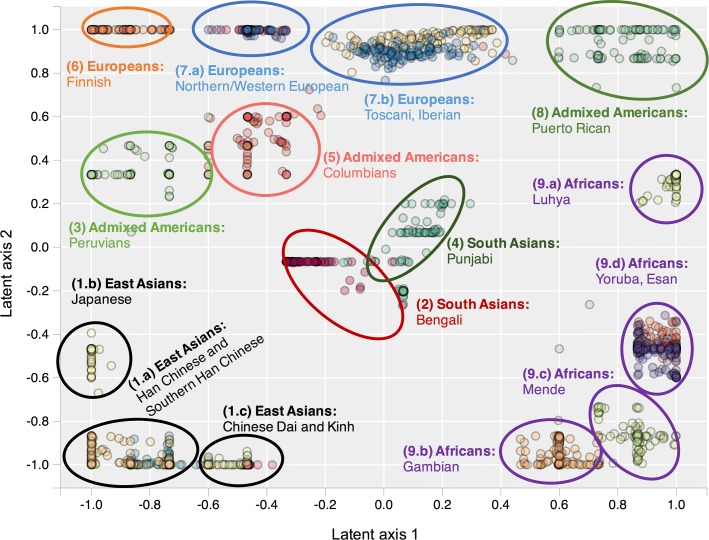
From Figs. 2 and 3, it can be seen that t-SNE and GTM recognize the same clusters. However, GTM suffers from a packing effect, which results in data points being packed together on a map. t-SNE remedies this situation with Student’s t-distributions in the latent space, which allow small distances between data points in the original space to be translated into larger distances in the 2D latent space.
Fig. 2.
GTM clustering with 10 principal components Generative Topographic Mapping (GTM) plot of 20 populations from 1000 Genomes Project, built using 10 first principal components. The following populations were not used to build the map: ASW = Americans of African Ancestry in SW USA; ACB = African Caribbeans in Barbados; MXL = Mexican Ancestry from Los Angeles USA; GIH = Gujarati Indian from Houston, Texas; STU = Sri Lankan Tamil from the UK; ITU = Indian Telugu from the UK
Fig. 3.
t-SNE clustering with 10 principal components t-distributed stochastic neighbor embedding (t-SNE) plot of 20 populations from 1000 Genomes Project, built using 10 first principal components. The following populations were not used to build the map: ASW = Americans of African Ancestry in SW USA; ACB = African Caribbeans in Barbados; MXL = Mexican Ancestry from Los Angeles USA; GIH = Gujarati Indian from Houston, Texas; STU = Sri Lankan Tamil from the UK; ITU = Indian Telugu from the UK
Classification performances for 19 ancestry classes
In Table 2, we report performance measures (10 times repeated 5-fold cross-validated F1 score) for SVM, GTM with 3 or 10 principal components, and PCA classification models based on 19 ancestry classes (CEU and GBR populations were merged) from 1000 Genomes Project. Although the PCA plot performs rather well for the 5 classes problem, it cannot properly classify the 19 finer population classes - except for Finnish (FIN), Puerto Ricans (PUR), Peruvians (PEL), Punjabi (PJL) and Bengali (BEB). On the other hand, GTM and SVM models built from only 10 principal components can efficiently classify individuals from most of the 1000 Genomes Project populations (F1 score = 0.80). Some populations are never properly separated, even in sophisticated models taking into account more principal components; this indicates that these populations have a high genetic overlap. This is the case between the Chinese Dai (CDX) and the Kinh in Vietnam (KHV), between the Yoruba (YRI) and Esan (ESN) populations in Nigeria, and between Toscani (TSI) and Iberian populations (IBS) in Europe.
Table 2.
10 times repeated 5-fold cross-validated F1 score for 19 population classes from 1000 Genomes Project using SVM, PCA or GTM
| Ancestry | 1000G code | Population | PCA 8-NN | SVM 10 PCs | GTM 3 PCs | GTM 10 PCs |
|---|---|---|---|---|---|---|
| EAS | CHB | Han Chinese | 0.20±0.01 | 0.78±0.01 | 0.45±0.04 | 0.75±0.01 |
| EAS | JPT | Japanese | 0.37±0.02 | 1.00±0.00 | 0.80±0.01 | 1.00±0.00 |
| EAS | CHS | Southern Han Chinese | 0.34±0.02 | 0.80±0.01 | 0.54±0.02 | 0.80±0.01 |
| EAS | CDX | Chinese Dai | 0.24±0.02 | 0.10±0.02 | 0.51±0.03 | 0.44±0.08 |
| EAS | KHV | Kinh in Vietnam | 0.44±0.01 | 0.68±0.00 | 0.63±0.01 | 0.71±0.01 |
| EUR | CEU+GBR | Northern/Western Eur. | 0.75±0.01 | 0.99±0.00 | 0.79±0.01 | 0.99±0.00 |
| EUR | TSI | Toscani | 0.46±0.01 | 0.74±0.02 | 0.58±0.01 | 0.54±0.06 |
| EUR | FIN | Finnish | 0.95±0.01 | 0.99±0.00 | 0.91±0.01 | 0.99±0.01 |
| EUR | IBS | Iberian | 0.35±0.03 | 0.81±0.01 | 0.35±0.04 | 0.74±0.02 |
| AFR | YRI | Yoruba in Nigeria | 0.30±0.02 | 0.69±0.00 | 0.15±0.03 | 0.66±0.03 |
| AFR | LWK | Luhya | 0.67±0.01 | 1.00±0.00 | 0.59±0.01 | 1.00±0.00 |
| AFR | GWD | Gambian | 0.26±0.02 | 0.94±0.02 | 0.23±0.02 | 0.78±0.07 |
| AFR | MSL | Mende | 0.25±0.03 | 0.93±0.02 | 0.35±0.03 | 0.81±0.04 |
| AFR | ESN | Esan in Nigeria | 0.28±0.02 | 0.00±0.01 | 0.19±0.05 | 0.28±0.13 |
| AMR | PUR | Puerto Ricans | 0.90±0.01 | 0.86±0.02 | 0.90±0.01 | 0.87±0.03 |
| AMR | CLM | Colombians | 0.69±0.01 | 0.85±0.01 | 0.84±0.01 | 0.82±0.02 |
| AMR | PEL | Peruvians | 0.88±0.01 | 0.97±0.01 | 0.94±0.01 | 0.95±0.01 |
| SAS | PJL | Punjabi | 0.89±0.01 | 0.96±0.01 | 0.96±0.01 | 0.96±0.00 |
| SAS | BEB | Bengali | 0.95±0.01 | 0.96±0.01 | 0.96±0.01 | 0.96±0.01 |
| Overall | 0.54±0.00 | 0.80±0.00 | 0.61±0.01 | 0.80±0.01 |
SVM 10 PCs = support vector machine classification model using 10 principal components, PCA 8-NN = k-nearest neighbours model based on 2D PCA map (k = 8), GTM 3 or 10 PCs = bayesian classification model based on generative topographic mapping using 3 or 10 principal components. Ancestry codes: EAS = East Asians, EUR = Europeans, AFR = Africans, AMR = Admixed Americans, SAS = South Asians. CEU and GBR were merged into one class. Each value is an average with 95% confidence interval
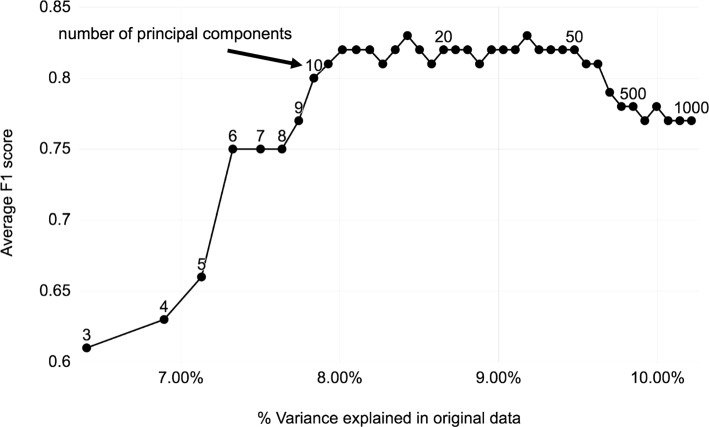
To investigate how the performance of 19 populations classification models (with CEU and GBR populations merged into one class) is changing depending on the percentage of variance explained, the cross-validated performance of GTM maps was evaluated by varying the number of principal components included in the model (Fig. 4). The F1 score increases until it reaches a plateau around 0.80 at 10-12 principal components accounting for around 8% variance explained. Interestingly, beyond 100-200 principal components the performance starts decreasing. This could be due to including more individual-level variance, which would disperse population clusters, or to the curse of dimensionality, which occurs when the number of variables increases but not enough data points are provided to populate the high-dimensional space. This indicates that the number of principal components should be optimized - our curve suggests to use 10-12 components for this pruned genotype matrix.
Fig. 4.
Ancestry classification performance vs. variance explained Generative Topographic Mapping (GTM) ancestry classification model performance as a function of number of principal components used to train the model
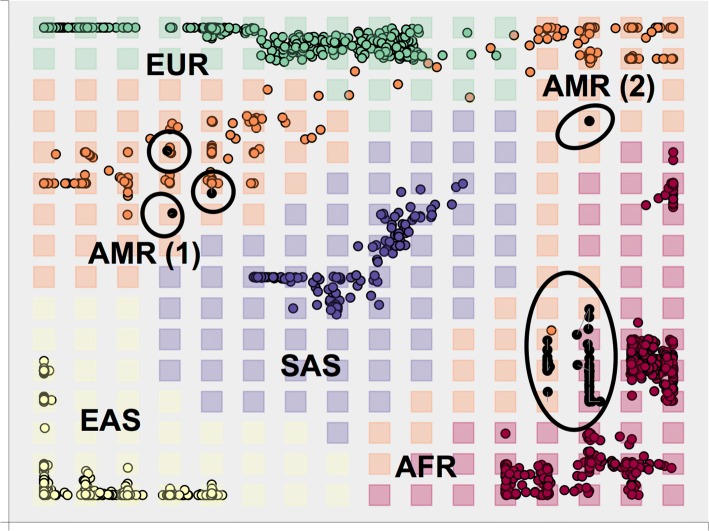
A final map was built with 10 principal components and the complete training set of 20 populations (cf. Fig. 5). The six populations that were not used to build the GTM map were used to generate posterior probabilities of superpopulation membership, which can be interpreted as the probability for a tested population pop to belong to a superpopulation: P(AFR|pop) would be the probability of African ancestry for tested population pop. Results are presented in Table 3. Indian Telugu from the UK (ITU), Sri Lankan Tamil from the UK (STU), and Gujarati Indian from Houston (GIH) are all predicted as South Asians with P(SAS|pop)=1 - none of them is mapped to another ancestry group. Individuals with Mexican ancestry from Los Angeles (MXL) are mostly mapped as Admixed Americans with a small European membership probability, whereas Americans of African ancestry in Southwest USA (ASW) and African Caribbeans in Barbados (ACB) show more mixed results - with high probabilities for both African and Admixed American superpopulations. Figure 5 shows how Americans of African ancestry in Southwest USA are distributed on the map: most of them are mapped near the African ancestry group but are assigned to empty nodes, where no African individual in the training set was mapped; some others are close to the Colombian/Peruvian group (AMR 1) and others to the Puerto Rican group (AMR 2).
Fig. 5.
Projected Americans of African ancestry in Southwest USA (ASW) on a GTM map. Generative Topographic Map (GTM) trained with 10 principal components. Coloured points represent individuals coloured by ancestry or superpopulation (AFR, AMR, EAS, EUR, SAS). Squares represent GTM nodes coloured by most probable ancestry. The highlighted black points represent mean positions of ASW individuals projected onto the map. Grey lines map mean positions of individuals on the map to their most probable node. Ancestry codes: EAS = East Asians, EUR = Europeans, AFR = Africans, AMR = Admixed Americans, SAS = South Asians
Table 3.
Posterior probabilities of superpopulation memberships in 6 test populations obtained by a GTM model trained with all superpopulations
| Population (pop) | P(AFR|pop) | P(AMR|pop) | P(EAS|pop) | P(EUR|pop) | P(SAS|pop) |
|---|---|---|---|---|---|
| ASW | 0.55 | 0.45 | 0 | 0 | 0 |
| ACB | 0.89 | 0.11 | 0 | 0 | 0 |
| MXL | 0 | 0.98 | 0 | 0.02 | 0 |
| GIH | 0 | 0 | 0 | 0 | 1 |
| STU | 0 | 0 | 0 | 0 | 1 |
| ITU | 0 | 0 | 0 | 0 | 1 |
NB: GTM classification models are restricted by an applicability domain defined by the training set. Here, the training set contains twenty 1000 Genomes Project, excluding [ASW, ACB, MXL, GIH, STU, ITU]. These posterior probabilities should be considered as a similarity measure between test populations and populations used to build the map, and not as an absolute measure of population admixture. Abbreviations: ASW = Americans of African Ancestry in SW USA; ACB = African Caribbeans in Barbados; MXL = Mexican Ancestry from Los Angeles USA; GIH = Gujarati Indian from Houston, Texas; STU = Sri Lankan Tamil from the UK; ITU = Indian Telugu from the UK; EUR = Europeans; EAS = East Asians; AMR = Admixed Americans; SAS = South Asians
Additional analysis 1: African-only GTM
A separate GTM was built with African populations exclusively (cf. Additional file 15). Americans of African ancestry in Southwest USA (ASW) and Africans Caribbeans in Barbados (ACB) were excluded from the training set, which included: Esan in Nigeria (ESN); Yoruba in Ibadan, Nigeria (YRI); Gambian in Western Divisions in The Gambia (GWD); Luhya in Webuye, Kenya (LWK); and Mende in Sierra Leone (MSL). We projected onto this African-only map ASW and ACB populations, but also other superpopulations (EUR, EAS, SAS, AMR), in order to distinguish populations based on their African variation. ASW and ACB are both mapped near Nigerian populations, whereas all other superpopulations (EUR, EAS, SAS, and AMR) are mapped in the same approximate location near the Luhya (LWK) - posterior probabilities of ancestry membership are provided in Table 4. However, these superpopulations are mapped in locations that are not populated by the training set; no strong conclusion should be inferred from these results. Moreover, the 1000 Genomes Project does not contain many African ethnic groups. Constructing an African-only map with more ethnic groups would be an interesting follow-up to this analysis.
Table 4.
Posterior probabilities of African ethnicity membership in 6 test populations obtained by a GTM model trained on African populations exclusively
| Population (pop) | P(ESN|pop) | P(YRI|pop) | P(GWD|pop) | P(LWK|pop) | P(MSL|pop) |
|---|---|---|---|---|---|
| ASW | 0.24 | 0.37 | 0.11 | 0.13 | 0.14 |
| ACB | 0.29 | 0.42 | 0.07 | 0.07 | 0.15 |
| EUR | 0.04 | 0.10 | 0.21 | 0.62 | 0.04 |
| EAS | 0.09 | 0.19 | 0.21 | 0.44 | 0.07 |
| AMR | 0.07 | 0.15 | 0.23 | 0.49 | 0.06 |
| SAS | 0.06 | 0.13 | 0.21 | 0.53 | 0.05 |
NB: GTM classification models are restricted by an applicability domain defined by the training set. Here, the training set contains only African populations, excluding ASW and ACB subsets. These posterior probabilities should be considered as a similarity measure between test populations and populations used to build the map, and not as an absolute measure of population admixture. Abbreviations: ASW = Americans of African Ancestry in SW USA; ACB = African Caribbeans in Barbados; ESN = Esan in Nigeria; YRI = Yoruba in Ibadan; Nigeria; GWD = Gambian in Western Divisions in the Gambia; LWK = Luhya in Webuye, Kenya; MSL = Mende in Sierra Leone; EUR = Europeans; EAS = East Asians; AMR = Admixed Americans; SAS = South Asians
Additional analysis 2: Arabidopsis thaliana
To test our methods on non-human genomes, we generated GTM, t-SNE and PCA maps for 1135 Arabidopsis thaliana genomes (a model plant organism) from the 1001 Genomes Consortium [14]. Visualizations are available in Additional files 16 and 17. PCA can separate the strains by continent but not by individual countries, as opposed to GTM and t-SNE, which find more fine-grained clusters corresponding to individual countries or regions, such as Spain, Southern Sweden, Northern Sweden, Southern Italy, or Northern Italy.
Discussion
Defining the training set
Our classification models were trained using known ancestry labels and a reference population (1000 Genomes Project). However, any other reference population could be used as a training set. In this application, populations expected to be more homogeneous were included in the training set. The choice of training set populations could also depend on the goal of the study, such as distinguishing between African populations in an African-only dataset, in which case a better classification model could be built using exclusively African samples.
Testing new data
To predict the ancestry of new individuals (test set) using a model trained on a reference population (training set), SNPs in the test matrix should correspond to the SNPs in the train matrix. This was not an issue in this paper, where populations from 1000 Genomes Project were used for both training and test. But in the more general case, many of the SNPs in the training set will be missing from the test set. Missing values in the test matrix should be imputed using the reference population, which can be achieved using genome imputation softwares such as MaCH [15] or IMPUTE2 [16].
Outliers
GTM or t-SNE maps can also be used to identify ancestry outliers, i.e. mislabeled individuals. Outliers are typically mapped to single points far away from their expected clusters. These data points should be removed from the training set used to build the classification model. By observing t-SNE and GTM maps, outliers can readily be identified in the 1000 Genomes Project.
Hyperparameter optimization
One major drawback of GTM and t-SNE is hyperparameter optimization. GTM has at least four hyperparameters to optimize, and t-SNE at least three. The maps presented in this paper have fixed hyperparameters (cf. Methods). However, hyperparameters might have a significant impact on the shape of the map, and can be optimized to obtain better visualization and classification performance. For GTM classification models, typical performance measures such as the F1 score, balanced accuracy, area under the curve (AUC) or Matthews correlation coefficient (MCC) can be used to select the optimal values for these hyperparameters.
Conclusion
PCA provides a good visualization of the superpopulations in the 1000 Genomes Project (AFR, AMR, EUR, EAS, SAS), but is not ideal for more fine-grained clustering and does not provide probabilistic models for admixed populations. On the other hand, both t-SNE and GTM provide a way to cluster and visualize more complex population substructures. GTM, as opposed to t-SNE, can be harnessed to generate comprehensive ancestry classification models. Moreover, new individuals can be directly projected onto a pre-constructed GTM map - which makes it the ideal choice to cluster individuals based on pre-defined panels. We showed how to assess ancestry membership probabilities using GTM and interpret them through visualization. By generating t-SNE or GTM maps with increasing number of principal components, we can estimate the percentage of variance explained to identify population substructures - this could also be useful to account for population stratification in genome-wide association studies.
Methods
Data and quality control
Genotypes of 2504 people in the 1000 Genomes Project Phase III were downloaded from ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/release/20130502 [12]. Variants were removed based on a Hardy-Weinberg equilibrium exact test p-value filter (< 0.001) and genotyping rate filter (> 0.02). The Hardy-Weinberg equilibrium test measures whether the ratio between homozygous and heterozygous genotypes differs significantly from prediction under HWE assumptions. SNPs from the major histocompatibility complex (MHC) on chromosome 6 and in the chromosome 8 inversion region were excluded. The remaining SNPs were pruned twice using plink 1.9 [17, 18] with windows of 1000 variants and step size 10, pair-wise squared correlation threshold = 0.02, and minor allele frequency > 0.05. The pruning operation deals with linkage desequilibrium or non-random association of alleles at different loci: it reduces the number of SNPs, keeps SNPs in linkage equilibrium, and thereby reduces data dimensionality. A training set was built by removing the following populations: Americans of African ancestry in Southwest USA (code = ASW); African Caribbeans in Barbados (ACB); Mexican ancestry from Los Angeles USA (MXL); Gujarati Indian from Houston, Texas (GIH); Sri Lankan Tamil from the UK (STU); and Indian Telugu from the UK (ITU). We used these populations as an external test set to predict the degree of relative admixture in individuals and populations. For the classification models, we also merged British in England and Scotland (GBR) and Utah Residents with Northern and Western European Ancestry (CEU) to obtain a single category for Northern and Western European Ancestry.
Additional dataset: Arabidopsis thaliana
We used an additional dataset of 1135 Arabidopsis thaliana genomes extracted from the 1001 Genomes Project [14]; the genotypes and an imputed SNP matrix could be downloaded from 1001genomes.org. Arabidopsis thaliana was the first plant genome to be sequenced and is a commonly used model organism. Variants were removed using a permissive genotyping rate filter (> 0.2). SNPs were pruned using plink 1.9 [17, 18] with windows of 100 variants and step size 10, pair-wise squared correlation threshold = 0.1, and minor allele frequency > 0.05. We merged the imputed SNP matrix with our filtered list of SNPs to obtain a filtered imputed SNP matrix.
Visualization of ancestry clusters using t-SNE and GTM
t-SNE [5] translates similarities between points into probabilities; Gaussian joint probabilities in the original input space and Student’s t-distributions in the latent space. The Kullback-Leibler divergence between data distributions in the input and latent space is minimized with gradient descent. t-SNE has several parameters to optimize: the learning rate for gradient descent, the perplexity of distributions in the initial space, and the early exaggeration. In this paper, we used the scikit-learn v0.19.1 implementation for t-SNE [19], with default learning rate = 200, perplexity = 30, and early exaggeration = 12. The main disadvantage of t-SNE is its lack of a framework to project new points onto a pre-trained map - a feature available in PCA and GTM.
The core principle of GTM [6] is to fit a manifold into the high-dimensional initial space. The points yk on the manifold Y in the initial space are the centers of normal probability distributions of g, which here are individuals described by the genotype matrix G:
| 1 |
where β is the common inverse variance of these distributions and W is the parameters matrix of the mapping function y(x;W) which maps nodes xk in the latent space to yk: y(xk;W)=Wϕ(xk), where ϕ(xk) is a set of radial basis functions. W and β are optimized with an expectation-maximization (EM) algorithm maximizing the overall data likelihood. The responsibility or posterior probability that the individual gn in the original genotype space is generated from the kth node in the latent space is computed using Bayes theorem:
| 2 |
These responsibilities are used to compute the mean position of an individual on the map x(gn), by averaging over all nodes on the map:
| 3 |
We used the python package ugtm v1.1.4 [20] for generative topographic mapping, and scripts used for ancestry classification are available online (https://github.com/hagax8/ancestry_viz). GTM has several hyperparameters to tune, which might have a high impact on the shape of the map: the number of radial basis functions, a width factor for these functions, map grid size, and a regularization parameter.
Ancestry classification models
PCA does not provide a comprehensive framework to build a probabilistic classification model. However, a simple classification model associated with the 2-dimensional plot can be built using the k-NN approach in three steps: (1) a PCA plot is constructed from a training set, (2) a test set is projected on the plot, and (3) each test individual is assigned the predominant ancestry amongst its k nearest neighbors in the training set. We did not construct k-NN models for t-SNE since it is not straightforward to project new points onto a t-SNE map. On the other hand, GTM provides a probabilistic framework which can be used to build classification models and generate class membership probabilities [10]. GTM responsibilities can be seen as feature vectors: they encode individuals depending on their position on the map, which is discretized into a finite number of nodes (positions). They can be used to estimate the probability of a specific ancestry given the position on map, using Bayes’ theorem
| 4 |
where P(xk|a) is computed as follows:
| 5 |
where Rkn is the responsibility of node xk for an individual belonging to population a, which counts Na individuals. It is then possible to predict the ancestry profile P(a|gi) of a new individual with associated responsibilities {Rki}
| 6 |
GTM nodes xk can be represented as points coloured by most probable ancestry amax using P(amax|xk). We compared performances of visual classifications (PCA and GTM) with linear support vector machine classification (SVM), a classical machine learning algorithm. Linear SVM is only dependent on C, the penalty hyperparameter. Increasing C increases the variance of the model and decreases its bias. In this application, classification performance is estimated by the average F1 score over all ancestry classes in a 5-fold cross-validation experiment (5-CV) repeated 10 times. The F1 score is a harmonic mean of precision and recall. For each of the 10 repetitions, labels are predicted for 5 partitions of the data, which are concatenated to obtain predicted values for the entire dataset. From these, F1 scores are computed for each class a and repetition j. The per-class performance measure is computed across the 10 repetitions:
| 7 |
The overall model performance measure is a weighted average across per-class F1 scores, with weights equal to the number of individuals in the class:
| 8 |
This procedure is performed for each parameter combination and for each algorithm (PCA, GTM, SVM). The best model for each algorithm is defined as having the largest overall F1 score. Only the performance of the best model is reported in the Results section. For PCA, we vary k (the number of neighbours) from 1 to 10. For GTM, we set the map grid size (number of nodes) = 16*16, the number of RBFs = 4*4, regularization = 0.1 and rbf width factor = 0.3. For linear SVM, the penalty parameter is set to C=2r where r runs from -5 to 10.
Posterior probabilities of ancestry membership for whole populations
All our models are trained with only twenty 1000 Genomes Project populations. Six populations are used as an external test set (cf. foregoing section Data and quality control). Posterior probabilities of ancestry membership are estimated for all individuals in these test populations (Eq. 6) based on observed superpopulation distributions (Eq. 5). We also generate probabilities of belonging to a superpopulation for each population as a whole, by replacing individual responsibilities {Rki} in equation 6 by an overall population responsibility {Rkp}
| 9 |
It should be noted that these responsibilities {Rkp} correspond to the averaged distribution of the population on the map, and can be used to compare populations and estimate their diversity.
Additional files
GTM map of twenty 1000 Genomes Project populations. Interactive GTM map of twenty 1000 Genomes Project populations. File name: 1000G_GTM_20populations.html. The file can be viewed in a web browser with internet access. (HTML 2416 kb)
t-SNE map of twenty 1000 Genomes Project populations. Interactive t-SNE map of twenty 1000 Genomes Project populations. File name: 1000G_t-SNE_20populations.html. The file can be viewed in a web browser with internet access. (HTML 589 kb)
GTM projection, test set 1: Americans of African ancestry in SW USA (ASW). Projection of Americans of African ancestry in SW USA (black points) onto a GTM map trained with 10 principal components. File name: 1000G_GTM_projection_ASW.html. The file can be viewed in a web browser with internet access. (HTML 437 kb)
GTM projection, test set 2: African Caribbeans in Barbados (ACB). Projection of African Caribbeans in Barbados (black points) onto a GTM map trained with 10 principal components. File name: 1000G_GTM_projection_ACB.html. The file can be viewed in a web browser with internet access. (HTML 471 kb)
GTM projection, test set 3: Mexican Ancestry from Los Angeles USA (MXL). Projection of individuals of Mexican ancestry from Los Angeles USA (black points) onto a GTM map trained with 10 principal components. File name: 1000G_GTM_projection_MXL.html. The file can be viewed in a web browser with internet access. (HTML 439 kb)
GTM projection, test set 4: Gujarati Indian from Houston, Texas (GIH). Projection of Gujarati Indian from Houston (black points) onto a GTM map trained with 10 principal components. File name: 1000G_GTM_projection_GIH.html. The file can be viewed in a web browser with internet access. (HTML 483 kb)
GTM projection, test set 5: Sri Lankan Tamil from the UK (STU). Projection of Sri Lankan Tamil from the UK (black points) onto a GTM map trained with 10 principal components. File name: 1000G_GTM_projection_STU.html. The file can be viewed in a web browser with internet access. (HTML 482 kb)
GTM projection, test set 6: Indian Telugu from the UK (ITU). Projection of Indian Telugu from the UK (black points) onto a GTM map trained with 10 principal components. File name: 1000G_GTM_projection_ITU.html. The file can be viewed in a web browser with internet access. (HTML 482 kb)
1000 Genomes Project populations. Table of 1000 Genomes Project populations and superpopulations and the number of individuals in each category. File name: 1000G_populations.html. (HTML 7 kb)
Variance explained in first principal components of genotype matrix. Variance explained in 100 first principal components of the genotype matrix for twenty 1000 Genomes Projects Populations, which were used as a training set to build our models. File name: varianceExplained.html. (HTML 13 kb)
5-fold cross-validated precision for twenty 1000 Genomes Project populations (19 classes) using SVM, PCA or GTM. Precision of optimized models for the following algorithms: SVM 10 PCs = support vector machine classification model using 10 principal components, PCA 8-NN = k-nearest neighbours model based on 2D PCA map (k = 8), GTM 3 or 10 PCs = bayesian classification model based on generative topographic mapping using 3 or 10 principal components. File name: precision_crossvalidation_19classes.html. (HTML 7 kb)
5-fold cross-validated recall for twenty 1000 Genomes Project populations (19 classes) using SVM, PCA or GTM. Recall of optimized models for the following algorithms: SVM 10 PCs = support vector machine classification model using 10 principal components, PCA 8-NN = k-nearest neighbours model based on 2D PCA map (k = 8), GTM 3 or 10 PCs = bayesian classification model based on generative topographic mapping using 3 or 10 principal components. File name: recall_crossvalidation_19classes.html. (HTML 8 kb)
5-fold cross-validated precision for five 1000 Genomes Project superpopulations (5 classes). Precision of optimized models for the following algorithms: SVM 10 PCs = support vector machine classification model using 10 principal components, PCA 8-NN = k-nearest neighbours model based on 2D PCA map (k = 8), GTM 3 or 10 PCs = bayesian classification model based on generative topographic mapping using 3 or 10 principal components. File name: precision_crossvalidation_5classes.html. (HTML 3 kb)
5-fold cross-validated recall for five 1000 Genomes Project superpopulations (5 classes). Recall of optimized models for the following algorithms: SVM 10 PCs = support vector machine classification model using 10 principal components, PCA 8-NN = k-nearest neighbours model based on 2D PCA map (k = 8), GTM 3 or 10 PCs = bayesian classification model based on generative topographic mapping using 3 or 10 principal components. File name: recall_crossvalidation_5classes.html. (HTML 3 kb)
African-only GTM map. Interactive GTM map for AFR superpopulation (1000 Genomes Project), excluding ASW and ACB populations, and projections of following test sets: two African ancestry populations (ASW and ACB), and 1000 Genomes superpopulations (EUR, EAS, AMR, and SAS) on the AFR map).File name: AFR_maps.pdf. (PDF 1414 kb)
Arabidopsis map coloured by country. Interactive map of 1135 Arabidopsis thaliana genomes from the 1001 Genomes project. File name: worldmap_arabidopsis_countries.html. The file can be viewed in a web browser with internet access. (HTML 571 kb)
Arabidopsis map coloured by admixture group. Interactive map of 1135 Arabidopsis thaliana genomes from the 1001 Genomes project, coloured by admixture group. File name: worldmap_arabidopsis_admixed.html. The file can be viewed in a web browser with internet access. (HTML 571 kb)
Acknowledgements
We thank Dr. Jonathan Coleman from our team at King’s College London for his advice on preparing 1000 Genomes Project data. Arabidopsis thaliana sequence data were produced by the Weigel laboratory at the Max Planck Institute for Developmental Biology.
Funding
HG and GB acknowledge funding from the US National Institute of Mental Health (PGC3: U01 MH109528). This work was also supported in part by the National Institute for Health Research (NIHR) Biomedical Research Centre at South London and Maudsley NHS Foundation Trust and King’s College London. The views expressed are those of the authors and not necessarily those of the NHS, the NIHR or the Department of Health. High performance computing facilities were funded with capital equipment grants from the GSTT Charity (STR130505) and Maudsley Charity (980). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Availability of data and materials
Results supporting the conclusions of this article are included within the article and its additional files. Visualizations can also be accessed on a dedicated web platefrom (https://lovingscience.com/ancestries). We provide the code to reproduce our results for both 1000 Genomes Project (https://github.com/hagax8/ancestry_viz) and Arabidopsis thaliana (https://github.com/hagax8/arabidopsis_viz). The ugtm python package used to build the GTM models is also accessible online (https://github.com/hagax8/ugtm).
Abbreviations
- AFR
African
- AMR
Admixed American
- EAS
East Asian
- EUR
European
- GTM
Generative topographic mapping
- GWAS
Genome-wide assocation study
- PCA
Principal component analysis
- SAS
South Asian
- SNP
Single nucleotide polymorphism
- SVM
Support vector machine
- t-SNE
t-distributed stochastic neighbor embedding
Authors’ contributions
HG designed the study, conducted the analyses, and wrote the original draft. GB contributed to the writing of the manuscript. Both authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Héléna A. Gaspar, Email: helena.gaspar@kcl.ac.uk
Gerome Breen, Email: gerome.breen@kcl.ac.uk.
References
- 1.Tian C, Gregersen PK, Seldin MF. Accounting for ancestry: population substructure and genome-wide association studies. Hum Mol Genet. 2008;17(R2):143–50. doi: 10.1093/hmg/ddn268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38(8):904–9. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 3.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155(2):945–59. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19(9):1655–64. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maaten L. Visualizing High-Dimensional data using t-SNE. J Mach Learn Res. 2008;9:2579–605. [Google Scholar]
- 6.Bishop CM, Svensén M, Williams CKI. GTM: The generative topographic mapping. Neural Comput. 1998;10(1):215–34. [Google Scholar]
- 7.Li W, Cerise JE, Yang Y, Han H. Application of t-SNE to human genetic data. J Bioinform Comput Biol. 2017;15(4):1750017. doi: 10.1142/S0219720017500172. [DOI] [PubMed] [Google Scholar]
- 8.Bushati N, Smith J, Briscoe J, Watkins C. An intuitive graphical visualization technique for the interrogation of transcriptome data. Nucleic Acids Res. 2011;39(17):7380–9. doi: 10.1093/nar/gkr462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Amir E-AD, Davis KL, Tadmor MD, Simonds EF, Levine JH, Bendall SC, Shenfeld DK, Krishnaswamy S, Nolan GP, Pe’er D. viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nat Biotechnol. 2013;31(6):545–52. doi: 10.1038/nbt.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gaspar HA, Marcou G, Horvath D, Arault A, Lozano S, Vayer P, Varnek A. Generative topographic mapping-based classification models and their applicability domain: application to the biopharmaceutics drug disposition classification system (BDDCS) J Chem Inf Model. 2013;53(12):3318–25. doi: 10.1021/ci400423c. [DOI] [PubMed] [Google Scholar]
- 11.Gaspar HA, Baskin II, Marcou G, Horvath D, Varnek A. Chemical data visualization and analysis with incremental generative topographic mapping: big data challenge. J Chem Inf Model. 2015;55(1):84–94. doi: 10.1021/ci500575y. [DOI] [PubMed] [Google Scholar]
- 12.1000 Genomes Project Consortium. Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini JL, McCarthy S, McVean GA, Abecasis GR. A global reference for human genetic variation. Nature. 2015;526(7571):68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cortes C, Vapnik V. Support-vector networks. Mach Learn. 1995;20(3):273–97. [Google Scholar]
- 14.1001 Genomes Consortium. Electronic address: magnus.nordborg@gmi.oeaw.ac.at. 1001 Genomes Consortium 1135 genomes reveal the global pattern of polymorphism in arabidopsis thaliana. Cell. 2016;166(2):481–91. doi: 10.1016/j.cell.2016.05.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol. 2010;34(8):816–34. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5(6):1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, de Bakker PIW, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, et al. Scikitlearn: Machine Learning in Python. J Mach Learn Res. 2011;12:2825–30. [Google Scholar]
- 20.Gaspar HA. ugtm: A Python Package for Data Modeling and Visualization Using Generative Topographic Mapping. J Open Res Softw. 2018;6:21 5. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
GTM map of twenty 1000 Genomes Project populations. Interactive GTM map of twenty 1000 Genomes Project populations. File name: 1000G_GTM_20populations.html. The file can be viewed in a web browser with internet access. (HTML 2416 kb)
t-SNE map of twenty 1000 Genomes Project populations. Interactive t-SNE map of twenty 1000 Genomes Project populations. File name: 1000G_t-SNE_20populations.html. The file can be viewed in a web browser with internet access. (HTML 589 kb)
GTM projection, test set 1: Americans of African ancestry in SW USA (ASW). Projection of Americans of African ancestry in SW USA (black points) onto a GTM map trained with 10 principal components. File name: 1000G_GTM_projection_ASW.html. The file can be viewed in a web browser with internet access. (HTML 437 kb)
GTM projection, test set 2: African Caribbeans in Barbados (ACB). Projection of African Caribbeans in Barbados (black points) onto a GTM map trained with 10 principal components. File name: 1000G_GTM_projection_ACB.html. The file can be viewed in a web browser with internet access. (HTML 471 kb)
GTM projection, test set 3: Mexican Ancestry from Los Angeles USA (MXL). Projection of individuals of Mexican ancestry from Los Angeles USA (black points) onto a GTM map trained with 10 principal components. File name: 1000G_GTM_projection_MXL.html. The file can be viewed in a web browser with internet access. (HTML 439 kb)
GTM projection, test set 4: Gujarati Indian from Houston, Texas (GIH). Projection of Gujarati Indian from Houston (black points) onto a GTM map trained with 10 principal components. File name: 1000G_GTM_projection_GIH.html. The file can be viewed in a web browser with internet access. (HTML 483 kb)
GTM projection, test set 5: Sri Lankan Tamil from the UK (STU). Projection of Sri Lankan Tamil from the UK (black points) onto a GTM map trained with 10 principal components. File name: 1000G_GTM_projection_STU.html. The file can be viewed in a web browser with internet access. (HTML 482 kb)
GTM projection, test set 6: Indian Telugu from the UK (ITU). Projection of Indian Telugu from the UK (black points) onto a GTM map trained with 10 principal components. File name: 1000G_GTM_projection_ITU.html. The file can be viewed in a web browser with internet access. (HTML 482 kb)
1000 Genomes Project populations. Table of 1000 Genomes Project populations and superpopulations and the number of individuals in each category. File name: 1000G_populations.html. (HTML 7 kb)
Variance explained in first principal components of genotype matrix. Variance explained in 100 first principal components of the genotype matrix for twenty 1000 Genomes Projects Populations, which were used as a training set to build our models. File name: varianceExplained.html. (HTML 13 kb)
5-fold cross-validated precision for twenty 1000 Genomes Project populations (19 classes) using SVM, PCA or GTM. Precision of optimized models for the following algorithms: SVM 10 PCs = support vector machine classification model using 10 principal components, PCA 8-NN = k-nearest neighbours model based on 2D PCA map (k = 8), GTM 3 or 10 PCs = bayesian classification model based on generative topographic mapping using 3 or 10 principal components. File name: precision_crossvalidation_19classes.html. (HTML 7 kb)
5-fold cross-validated recall for twenty 1000 Genomes Project populations (19 classes) using SVM, PCA or GTM. Recall of optimized models for the following algorithms: SVM 10 PCs = support vector machine classification model using 10 principal components, PCA 8-NN = k-nearest neighbours model based on 2D PCA map (k = 8), GTM 3 or 10 PCs = bayesian classification model based on generative topographic mapping using 3 or 10 principal components. File name: recall_crossvalidation_19classes.html. (HTML 8 kb)
5-fold cross-validated precision for five 1000 Genomes Project superpopulations (5 classes). Precision of optimized models for the following algorithms: SVM 10 PCs = support vector machine classification model using 10 principal components, PCA 8-NN = k-nearest neighbours model based on 2D PCA map (k = 8), GTM 3 or 10 PCs = bayesian classification model based on generative topographic mapping using 3 or 10 principal components. File name: precision_crossvalidation_5classes.html. (HTML 3 kb)
5-fold cross-validated recall for five 1000 Genomes Project superpopulations (5 classes). Recall of optimized models for the following algorithms: SVM 10 PCs = support vector machine classification model using 10 principal components, PCA 8-NN = k-nearest neighbours model based on 2D PCA map (k = 8), GTM 3 or 10 PCs = bayesian classification model based on generative topographic mapping using 3 or 10 principal components. File name: recall_crossvalidation_5classes.html. (HTML 3 kb)
African-only GTM map. Interactive GTM map for AFR superpopulation (1000 Genomes Project), excluding ASW and ACB populations, and projections of following test sets: two African ancestry populations (ASW and ACB), and 1000 Genomes superpopulations (EUR, EAS, AMR, and SAS) on the AFR map).File name: AFR_maps.pdf. (PDF 1414 kb)
Arabidopsis map coloured by country. Interactive map of 1135 Arabidopsis thaliana genomes from the 1001 Genomes project. File name: worldmap_arabidopsis_countries.html. The file can be viewed in a web browser with internet access. (HTML 571 kb)
Arabidopsis map coloured by admixture group. Interactive map of 1135 Arabidopsis thaliana genomes from the 1001 Genomes project, coloured by admixture group. File name: worldmap_arabidopsis_admixed.html. The file can be viewed in a web browser with internet access. (HTML 571 kb)
Data Availability Statement
Results supporting the conclusions of this article are included within the article and its additional files. Visualizations can also be accessed on a dedicated web platefrom (https://lovingscience.com/ancestries). We provide the code to reproduce our results for both 1000 Genomes Project (https://github.com/hagax8/ancestry_viz) and Arabidopsis thaliana (https://github.com/hagax8/arabidopsis_viz). The ugtm python package used to build the GTM models is also accessible online (https://github.com/hagax8/ugtm).