Abstract
Background
Malignant melanoma is recalcitrant to most existing chemotherapies, and aberrant expression of miR-211 plays prominent roles in progression of melanoma. However, the trigger mechanism of aberrant miR-211 expression in melanoma is still elusive.
Material/Methods
We used qRT-PCR to test miR-211 expression. Cell viability assay and mouse xenograft assay were performed to examine the role of miR-211 on the sensitivity of melanoma cells to cisplatin. The epigenetic modification of miR-211 promoter was assess by DNA methylation analysis and DAC treatment.
Results
In this study, decreased miR-211 expression was detected. Bisulfite sequencing PCR showed that DNA hypermethylation contributed to the downregulation of miR-211 in melanoma tissues. In melanoma cells, overexpressed 211 could enhance the anticancer effect of cisplatin and restoration of miR-211 rendered susceptibility to cisplatin in cisplatin-resistant cells. And the same result was showed in vivo by mouse xenograft assay. What is more, DAC treatment could increase miR-211 expression and EZH2 expression was increased in cisplatin-resistant cells. MiR-211 could be transcriptionally repressed by EZH2 mediated promoter methylation.
Conclusions
Taken together, our findings revealed that epigenetic modification of miR-211 governed melanoma cell chemosensitivity and were involved in the progression of tumorigenesis.
MeSH Keywords: Abnormalities, Drug-Induced; Epigenomics
Background
Melanoma is a malignant tumor, arising from melanocytes, whose incidence has a drastic increase during the past 30 years [1]. Of note, disseminating to distant sites and visceral organs is the key transition for melanoma to be aggressive and life-threatening [2], which in not suitable for surgical excision and intrinsic or acquired resistance frequently occur with biological, chemical, or physical therapies [3,4]. Melanoma is common in Caucasians with a median survival time of only 6–9 months and a 3-year survival of 15% [3]. However, knowledge of melanoma in Asian patients is scant. In Chinese patients, mucosal and acral melanomas account for the majority of melanoma cases among the 4 subtypes [5]. A retrospective analysis showed that the 5-year overall and disease-free survival (DFS) rates and median survival time were 41.6% and 43 months, and 12.3% and 20 months in 522 ethnic Chinese patients respectively, which were substantially worse than those in Caucasian patients [6]. Cisplatin (CDDP), an anticancer drug, is widely used in cancers [7,8], however, without a very effective in melanoma owing to the strong drug resistance [9]. Thus, this is urgent for melanoma patients to clarify the mechanisms underlying the resistance of melanoma cells to chemotherapeutic drugs such as CDDP, especially for those in advanced stages.
Increasing evidences show that epigenetic regulation involved in aberrant expression of non-coding genes, such as microRNAs, which cause significant impact in disease occurrence and cure. DNA methylation, as one main style of the epigenetic modification, has been implicated in various biological processes, including tumorigenesis and chemosensitivity [10]. In GBM (glioblastoma multiforme), decreased microRNA-211 (miR-211) caused by methylation-mediated could enhance cell viability and EMT (epithelial-mesenchymal transition) via AKT/β-catenin pathway [11]. In melanoma, DNMT1 could mediate the methylation level of miR-211 promoter to inhibit EMT [12]. MMP9 downregulation increase miR-211 promoter methylation to manage glioma cell biological behavior [13]. However, a precise understanding of the mechanism between aberrantly expressed miR-211 and drug resistance in melanoma remain uncharacterized.
DNA is predominantly methylated at the CpG island by concerted action of 3 DNA methyltransferases, including de novo methyltransferases DNMT3A and DNMT3B and DNA methylation maintenance methyltransferase DNMT1 [14]. Recent study had revealed the effect of DNMT1 on miR-211 in melanoma [12]. What is more, the methylation of the chromatin core group protein also can mediate the silencing of miRNAs, such as EZH2, which is the catalytic component of polycomb repressor complex 2, administers tri-methylation of H3K27. In some cancers, overexpressed EZH2 caused miRNAs silencing, such as, miR-31 in prostate cancer [15] and miR-622 in HCC (hepatocellular carcinoma) [16]. Hypothesis was made that methylation-mediated silencing of miR-211 take part in chemosensitivity and patients’ cure.
Evidence presented in this study points at the possible connection between the methylation level of miR-211 and drug-sensitivity and patient outcomes. In this study, we firstly revealed that EZH2 mediated methylation of miR-211 is a mechanism implicated in melanoma. In short, theoretical basis was provided that the epigenetic modification of miR-211 serves as unrealized potential targets of cancer therapy.
Material and Methods
Cell culture
Melanoma cell lines, including A375 and SK-MEL-28 were purchased from ATCC and cultured in DMEM medium (Hyclone) supplemented with 10% FBS at 37°C and 5% CO2.
Cell viability assay
A cell count kit-8 (CCK-8) assay (Dojindo, Kumamoto, Japan) was employed to quantitatively evaluate cell viability at the indicated time points and the absorbance was measured at 450 nm in a microplate reader.
Transfection assay
MiR-211 mimics (50 nM), miRNA control (miR-NC) (GenePharma, Shanghai, China) and 20 nM si-EZH2 were transfected into cells by Lipofectamine 2000 (Invitrogen) following the manufacturer’s instructions.
Quantitative RT–PCR analysis
These experiments were conducted according to the previously mentioned [17]. The primers used as follow, miR-211, forward, 5′-GCTCGTCGGGTCGGGCCTATTG-3′ and reverse, 5′-CCGCCCCTATTGCTTAAGCCCACG-3′; U6 gene as a normalizing control. U6, forward, 5′-CTCGCTTCGGCAGCACA-3′ and reverse, 5′-AACGCTTCACGAATTTGCGT-3′. EZH2, forward, 5′-CAGTAAAAATGTGTCCTGCAAGAA-3′ and reverse, 5′-TCAAGGGATTTCCATTTCTCTTTCGA-3′; GAPDH, forward, 5′-ACCACAGTCCATGCCATCAC-3′ and reverse, 5′-TCCACCACCCTGTTGCTGTA-3.
DAC treatment
Cells were treatment with 5 or 10 μM of 5-aza-2′-deoxycytidine (DAC, Sigma-Aldrich, St. Louis, MO, USA) for 3 days and drug in medium were replaced every 24 hours. Control cells were incubated with same volume DMSO.
DNA methylation analysis
This assay was performed as previously reported [18]. DNA (1 μg) was carried out according to the manufacturer’s instruction (Epitect Bisulfite Kit, Qiagen). Pre-treated DNA with bisulfite was amplified with designed sequence-specific primers binding to the miR-211 promoter: 5′-GTTATTGAAGTTAATAACGGTGATTGATA-3′ (forward) and 5′-CTTCCTCGGAATTAACTATTACTGCG-3′ (reverse).
Western blot
Immunoblot analysis was conducted as previously mentioned [19]. The primary antibodies used include anti-DNMT1 (1: 1000, ab87654, Abcam), anti-EZH2 (1: 1000; BD), anti-GAPDH (1: 1000, CST).
Mouse xenograft assay
All protocols using animals were approved by the Medical Ethics Committee of Second Affiliated Hospital of Harbin Medical University. All animal care followed institutional guidelines. Male BALB/c nude mice, 6-week-old, were implanted subcutaneously with 1×106 SK-MEL-28 cells. When the tumor volume reached 100 mm3, mice were randomly assigned to 4 groups; the time was defined as day 1, which was the starting point for treatment. CDDP (3.0 mg/kg body weight) were intraperitoneally injected, twice-weekly for 4 weeks. Once each week, mice were weighed, and tumor volume was measured using the following formula: Tumor volume=1/2×(width)2×length.
Clinical tissue samples
The 10 normal skin and 38 primary melanomas samples used in this study were gathered from patients who had tumor resection performed at Department of Surgery, Department of Plastic and Reconstructive Surgery, Second Affiliated Hospital of Harbin Medical University, from January 2006 to May 2014. All human samples with informed consent were enjoyed supports by the Medical Ethics Committee of Second Affiliated Hospital of Harbin Medical University. The ethical approval number, ky2018-156.
Statistical analysis
Two-tailed Student’s t-test was conducted to determine the statistical significance using Prism GraphPad or SPSS 20.0 (Chicago, IL, USA). Kaplan-Meier and log-rank t-test were used to analyze the survival. P<0.05 was considered statistically significant.
Results
Downregulated miR-211 in melanoma patients
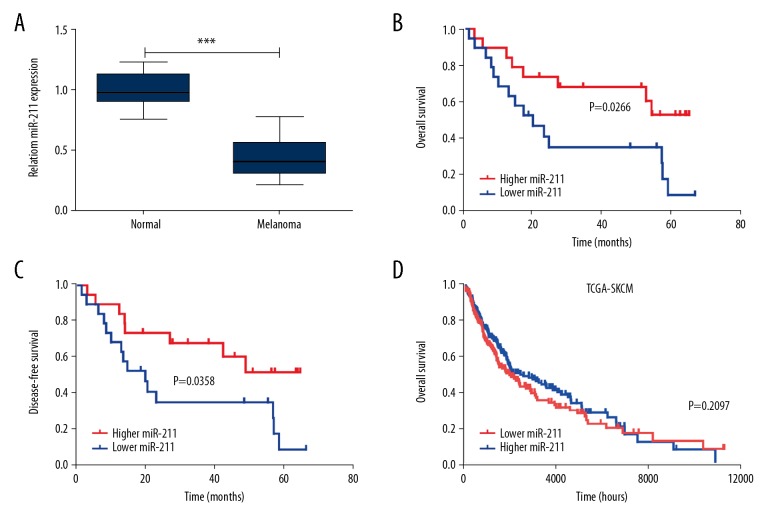
To clarify miR-211 expression pattern in the process of melanoma, we used 10 normal skin samples and 38 primary melanoma samples to conduct qRT-PCR. We noticed that miR-211 was decreased in primary melanomas compared with normal skin (Figure 1A). In the clinicopathological parameters analysis within 38 melanoma samples (Table 1), significant correlation was presented between miR-211 expression with tumor thickness and AJCC stage, while no statistical significance in age, gender, ulceration metastasis, and solar elastosis. Collectively, these findings suggested that miRNA-211 was downregulated in melanomas and might be involved in the carcinogenesis.
Figure 1.
Melanoma patients with downregulated miR-211 has a poor prognosis. (A) qRT-PCR measurements of miR-211 level in 10 normal skin and 38 primary melanoma samples (MM). Data represent the mean ± standard deviation. (B, C) Patients were subdivided into 2 groups by median of miR-211 level of all samples. Kaplan-Meier curves representing the overall survival and disease-free survival of melanoma patients was stratified. P value was determined by log-rank test. (D) Kaplan-Meier was performed to analyze the overall survival of melanoma patients according to the miR-211 expression in TCGA cohort, the median value of expression was utilized as a cutoff (log-rank test, P=0.2097).
Table 1.
Correlations between miR-211 and clinicopathologic features in melanoma patients.
| Clinicopathological feature | Total 62 | Expression of miR-211 | p Value | |
|---|---|---|---|---|
| Low (%) | High (%) | |||
| Age (years) | ||||
| ≤50 | 21 | 11 (52.4) | 10 (47.6) | 0.744 |
| >50 | 17 | 8 (47.1) | 9 (52.9) | |
| Gender | ||||
| Male | 24 | 13 (54.2) | 11 (45.8) | 0.501 |
| Female | 14 | 6 (42.9) | 8 (57.1) | |
| Ulceration | ||||
| Yes | 23 | 14 (60.9) | 9 (39.1) | 0.097 |
| No | 15 | 5 (33.3) | 10 (66.7) | |
| Tumor thickness | ||||
| ≤2 mm | 16 | 5 (31.3) | 11 (61.7) | 0.049 |
| >2 mm | 22 | 14 (63.6) | 8 (36.4) | |
| Solar elastosis | ||||
| Yes | 19 | 11 (57.9) | 8 (42.1) | 0.330 |
| No | 19 | 8 (42.1) | 11 (57.9) | |
| Metastasis | ||||
| Yes | 17 | 10 | 7 | 0.328 |
| No | 21 | 9 | 12 | |
| AJCC stage | ||||
| I + II | 19 | 6 (31.6) | 13 (68.4) | 0.023 |
| III + iv | 19 | 13 (68.4) | 6 (31.6) | |
Meanwhile, Kaplan-Meier analysis was conducted in the 38 melanomas, which were bisected by the median expression of miR-211, and the results revealed that worse overall survival and disease-free survival (DFS) were existed in low miR-211 group (Figure 1B, P=0.0266 and Figure 1C, P=0.0358). However, no significant statistical significance was presented in the TCGA database (Figure 1D, P=0.2097). Thereafter, Cox proportional hazards regression analysis was performed, and univariate Cox proportional hazards regression analysis indicated that miR-211, ulceration, tumor thickness, and AJCC stage were hazardous prognostic factors for the overall survival of melanomas. Nevertheless, multivariate Cox proportional hazards regression analysis showed that miR-211 cannot be used as independent predictors of the overall survival (Table 2). Taken together, the analysis suggested that miR-211 was not an independent prognostic factor in melanoma.
Table 2.
Univariate and multivariate analyses of prognostic parameters for survival in melanomas.
| Prognostic parameter | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|
| HR (95% CI) | P value | HR (95% CI) | P value | |
| MiR-211 expression (high vs. low) | 0.389 (0.164–0.923) | 0.032 | 1.219 (0.457–3.251) | 0.692 |
| Age (≤50 vs. >50) | 1.049 (0.459–2.396) | 0.910 | ||
| Gender (Female vs. Male) | 1.362 (0.595–3.115) | 0.465 | ||
| Ulceration (no vs. yes) | 4.421 (1.574–12.418) | 0.005 | 2.899 (0.971–8.657) | 0.057 |
| Tumor thickness (≤2 mm vs. >2 mm) | 4.092 (1.533–10.920) | 0.005 | 3.218 (1.064–9.733) | 0.038 |
| Solar elastosis (no vs. yes) | 2.121 (0.908–4.953) | 0.082 | ||
| AJCC stage (I + II vs. III + IV) | 6.103 (2.199–16.943) | 0.001 | 4.812 (1.585–14.605) | 0.006 |
CI – confidence interval; HR – hazard ratio.
Epigenetically silenced cause downregulated miR-211
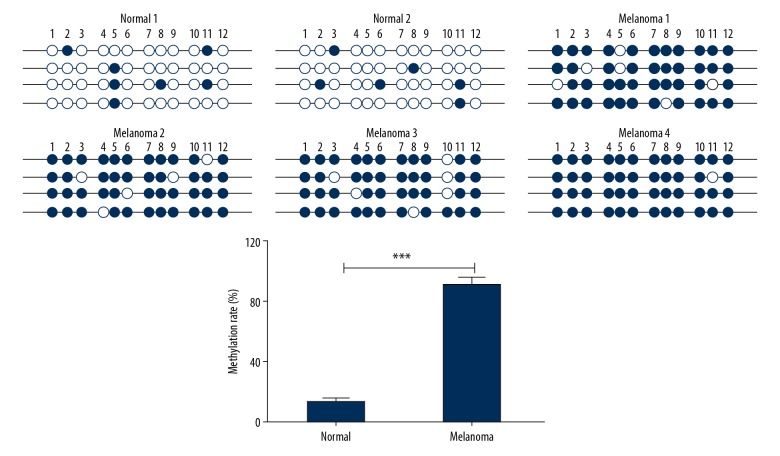
Previous publications have revealed that epigenetic modification on the promoter, as a typical trigger mechanism, cause aberrant gene expression. To further asked the reason of miR-211 downregulated in melanoma, the methylation level of miR-211 in melanoma was detected by bisulfite sequencing PCR (BSP) in 4 melanoma tissues and 2 normal skin samples. The results revealed that the melanoma tissues had higher methylation rates in miR-211 promoter compared with normal skin (Figure 2). This means that miR-211 is epigenetic regulation via DNA methylation.
Figure 2.
MiR-211 is downregulated by hypermethylation of CpG island in DNA promoter. Hypermethylation of the miR-211 promoter and relevant methylation rates were detected in 4 melanoma tissues and 2 normal skin, *** P<0.001.
Upregulation of miR-211 enhances in vitro sensitivity of melanoma cells to CDDP
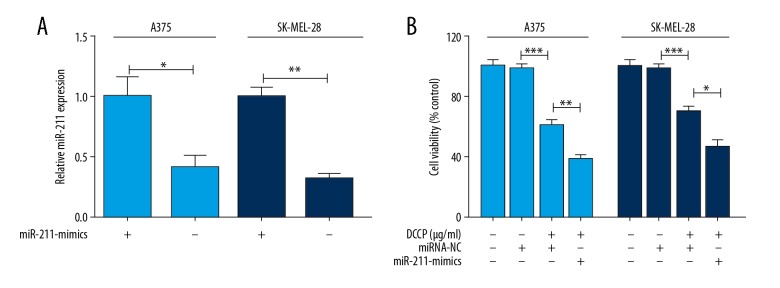
Dysregulation of miRNAs has been reported to be associated with chemoresistance of human cancers. However, whether miR-211 expression affects the sensitivity of melanoma cells is not fully understood. To determine this, the stably transfected melanoma cells were treated with 5 μg/mL CDDP for 48 hours [20]. The results indicated that upregulation of miR-211 (Figure 3A) effectively enhance the sensitivity of melanoma cells to CDDP (Figure 3B).
Figure 3.
Effect of miR-211 upregulation on the sensitivity of melanoma cells to cisplatin (CDDP) in vitro. (A) Quantitative RT-PCR analysis of miR-211 expression in miR-NC or miR-211-mimics cells. (B) MiR-211-mimics obviously enhanced CDDP’s effect on cell viability. Data represent the mean ±SEM, * P<0.05, ** P<0.01, *** P<0.001.
Downregulated miR-211 is correlated with CDDP resistance in melanoma cells
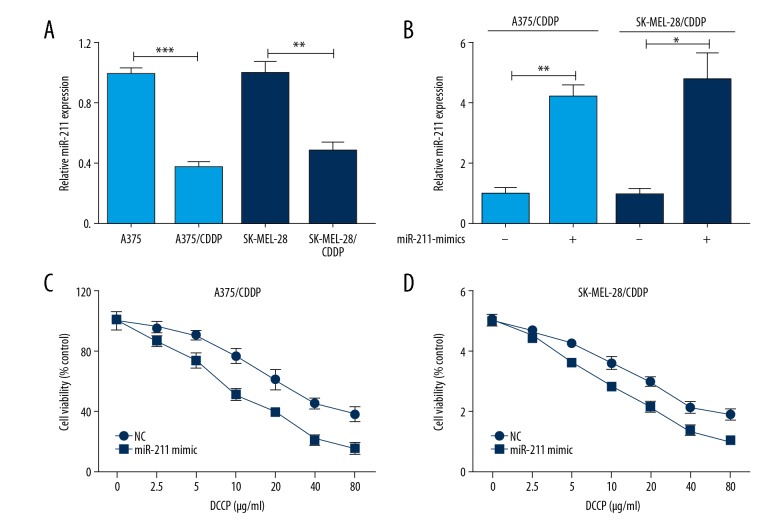
To better understand the role of miR-211 in CDDP resistance, 2 CDDP-resistant melanoma cell lines, termed A375/CDDP and SK-MEL-28/CDDP, were established by treated with CDDP (1 μg/mL) for 24 hours and then incubated with increasing concentrations of CDDP for additional 30 days for sufficient acquisition of resistance. Interestingly, striking decreased miR-211 was found in CDDP-resistant cells (Figure 4A). And overexpression of miR-211 (Figure 4B) could render chemosensitive of CDDP-resistant cells to CDDP (Figure 4C, 4D). All data suggested that low miR-211 could contribute to CDDP-resistance in human melanoma cells.
Figure 4.
Increased miR-211 level renders chemosensitive to cisplatin (CDDP) in CDDP-resistant melanoma cells. (A) miR-211 was significantly down-regulated in CDDP-resistant cells (A375/CDDP and SK-MEL-28/CDDP). (B) Upregulated miR-211 was in CDDP-resistant cells (A375/CDDP and SK-MEL-28/CDDP). (C, D) The representative curves of growth inhibition of A375/CDDP and SK-MEL-28/CDDP cells after transfecting miR-211 mimics or control, respectively. * P<0.05; ** P<0.01; *** P<0.001.
Methylation-mediated silencing of miR-211 mediated melanoma cell chemosensitivity
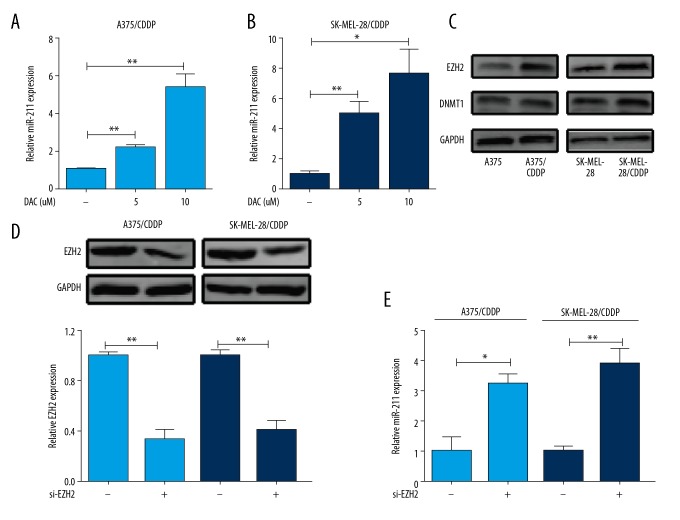
To further investigate the reason of decreased miR-211 in CDDP-resistance cells, we considered an alternative mechanism of miRNA downregulation by epigenetic modification silencing, which including DNA methylation and histone modification, the main regulatory mechanisms involved gene aberrant expression [21]. First, a demethylating agent 5-aza-2′-deoxycytidine (DAC) was used and significantly increased miR-211 was reproduced in A375/CDDP (Figure 5A) and SK-MEL-28/CDDP (Figure 5B). Following, we checked the expression of DNMT1 and EZH2, which had been reported in the methylation-mediated silencing of miR-211 [12,11]. The results showed that higher EZH2 expression was emerged in A375/CDDP and SK-MEL-28/CDDP compared to wildtypes, respectively. While no significant change in DNMT1 (Figure 5C). EZH2 downregulation (Figure 5D) accelerated miR-211 expression (Figure 5E). These data indicated that epigenetically silenced miR-211 by DNA methylation was closely associated with CDDP-resistant.
Figure 5.
Methylation-mediated silencing of miR-211 mediated melanoma cell chemosensitivity. (A, B) MiR-211expression was increased with demethylating agent DAC treatment in A375/CDDP and SK-MEL-28/cisplatin (CDDP) cells, U6 was used as a loading control. Values are means ±SEM. (C). Higher EZH2 expression was in A375/CDDP and SK-MEL-28/CDDP cells compared with wildtype cells, while no markedly change in DNMT1. GAPDH served as the loading control. (D). EZH2 was knockdown in A375/CDDP and SK-MEL-28/CDDP cells. (E). Knockdown of EZH2 could increase miR-211 expression. * P<0.05; ** P<0.01.
Targeting miR-211 mediated the chemosensitivity of CDDP in vivo
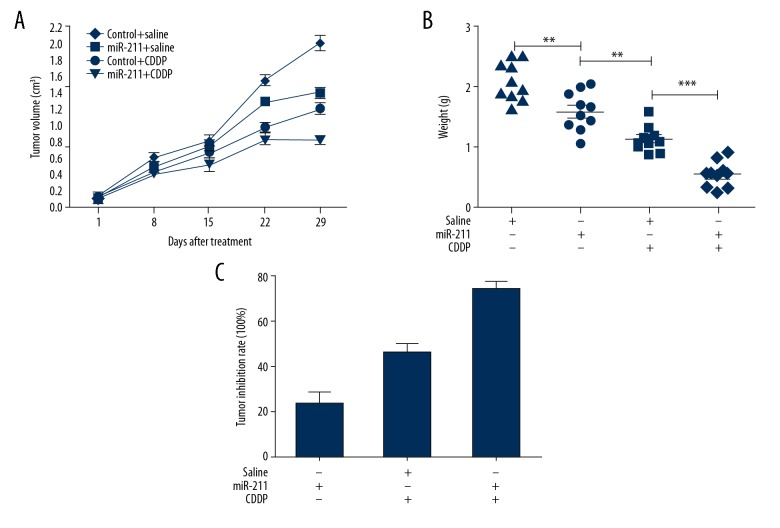
To further confirm our in vitro findings, in vivo experiments were employed. As shown in Figure 6A, the tumors formed from miR-211 upregulated cells grew significantly slower than those from control groups after the treatment with CDDP. At 29 days after inoculation, the average tumor weight of miR-211 upregulated was significantly lower than that of control groups following CDDP treatment (P<0.05; Figure 6B). As a consequence, higher inhibition rate was presented due to miR-211 upregulated (Figure 6C). From the above, increasing miR-211 could enhance cytotoxic effects of CDDP on tumors.
Figure 6.
Effect of miR-211 upregulation on the in vivo sensitivity of melanoma cells to cisplatin (CDDP). (A) Much smaller tumors were appeared in mice co-treated with increased miR-211 and CDDP compared with other groups. (B, C) Mice from increased miR-211 and CDDP showed lowest tumor burdens and highest inhibition rate. Tumor inhibition rate was calculated on the base of tumor weight, ** P<0.01; *** P<0.001.
Discussion
Poor outcome often occurs in late-stage melanomas with a median survival time of only 6 to 9 months and a 3-year survival of 15% [3,22]. Whereas surgical treatments for thin melanomas have achieved a good prognosis, drugs commonly used to treat advanced melanoma rarely achieve durable responses. Even though remarkable antitumor effects have emerged from immunotherapy and targeted therapy in melanoma, CDDP-based treatment is still classic treatments, in most cases, in combination with other therapies [23]. However, the strong resistance to platinum-based drugs results in a major obstacle in melanoma successful therapy. As known to us, aberrant miRNAs frequently occur in various types of cancers through regulating many cancer-pertinent cellular processes, including chemoresistance [24,25]. So, miRNAs may be used as biomarkers for predicting chemo-responsiveness to maximize therapeutic effect and minimize treatment toxicity.
In the beginning, miR-211 expression was assessed at mRNA level. The data confirmed that miR-211 expression was decreased in melanomas compared with normal. Furthermore, we detected the relationship between miR-211 expression and prognosis in 38 melanomas. The results demonstrated that poor prognosis was existed in lower miR-211 expression samples. The remarkable thing is that no significant statistical significance between miR-211 and prognosis was presented in the TCGA database, which may be due to the different race origin of the patient and too small sample size in this study. Notably, multivariate Cox proportional hazards regression analysis showed no statistical significance between miR-211 expression and the overall survival. Thus, we concluded that miR-211 cannot be as an independent prognostic factor in melanomas. And, the credibility of these conclusions may be discounted due to such a small sample size. In future studies, much more samples should be collected to verify the phenomenon above.
Extensive reports suggested that the miR-211 was abnormal in multiple kinds of malignant tumors. In melanoma, reduced miR-211 was found to enhance melanoma invasiveness [26]. Upregulation of miR-211 decreased the invasive potential of melanoma by directly suppresses NFAT5 and TFGBR2 and indirectly suppresses IGF2R [26]. Besides, miR-211 overexpression could result in decreased BRN2 protein [27] and the ion channel KCNMA1 [28] expression to inhibit cellular invasion. NUAK1 could be mediated by miR-211 in the regulation of cellular adhesion pathways necessary for metastasis [29]. However, very little was known in the effect of miR-211 on CDDP-resistance. In this study, we comprehensively confirmed the expression pattern of miR-211 in CDDP-resistant melanoma cells and firstly showed that miR-211 could mediate the sensitivity of melanoma cells to CDDP.
Epigenetic modification could regulate chromatin state and gene expression through DNA methylation state, histone modifications, ncRNA expression, chromatin remodeling, and nucleosome positioning without altering DNA sequences [30]. Of these, aberrant DNA methylation and histone modifications have been most intensively studied. So, we assume that epigenetic mechanism may be undergone during CDDP-long treating. Previous studies had demonstrated that miR-211 could be epigenetic regulated by DNMT1 [12] and EZH2 [11]. In the current research, EZH2 was increased in CDDP-resistant cells, with no visible difference in DNMT1. As well as, decreased EZH2 could significantly upregulated miR-211. Herein, we speculated that CDDP tolerance mainly due to tri-methylation of histone H3 at Lys 27 (H3K27me3) catalyzed by EZH2. Over the past decades, the role of EZH2 in cancer progression attracted much attention. EZH2 is frequently overexpressed in multi malignant tumors, such as prostate cancer, breast cancer, and ovarian cancer. Accumulating studies indicate that EZH2 could promote malignant phenotype in vivo and in vitro [31]. Besides, emerging researches showed that EZH2 could methylate miRNAs in a PRC2-independent manner [15,16,32], as well as be mediated by miRNAs [33,34]. Thus, as a key regulator, EZH2 is a potential target for cancer therapy, and much more studies within pre-clinical and clinical should be performed.
What is more, we found that miR-211 was frequently downregulated in melanoma tissues. As known to us, epigenetic modified silencing in CpG island on promoter, as a typical trigger mechanism, cause aberrant gene expression [10]. Previous publications have revealed that methylation-mediated decrease of miR-211 facilitates cells proliferation and EMT [11] and this epigenetic control can be mediated by MMP-9 in GBM [13]. MiR-211 could be regulated by DNMT1 in melanoma cells [12]. Inspired by this, BSP was performed and the results exhibited higher methylation lever of miR-211 in melanoma tissues. All results agree with this view that methylation of promoter caused reduced miR-211. And all above firstly determine the action of the miR-211 in response to CDDP in melanoma. More profoundly, recent studies shed light on the epigenetic therapy including numerous drugs that target specific enzymes involved in the epigenetic regulation of gene expression for cancers. Hence, together with our research data prompted that utilizing demethylating agents or combined with EZH2 inhibitor to enhance miR-211 expression may have a potential value for melanoma treatment. And the epigenetic state of miR-211 could rich diagnostic indicators for precision medicine, as well as, offer criterion for adjuvant therapy and drug-resistance, such as CDDP. In summary, our results provide new evidence for epigenetic therapy and it is worthy of further in-depth study.
Conclusions
In conclusion, the data above firstly demonstrated that methylation in miR-211 promoter decreases the toxic effect of CDDP on melanoma cells, as well as involved in the carcinogenesis, which might be valuable for provides a therapeutic potential for melanoma treatment in the future.
Footnotes
Source of support: This work was supported by grants from Fund of Health and Family Planning Commission of Heilongjiang Province and Fund of The Second Affiliated Hospital of Harbin Medical University Middle-Young Innovation
Conflicts of interest
None.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. Cancer J Clin. 2015;65(1):5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 2.Orgaz JL, Sanz-Moreno V. Emerging molecular targets in melanoma invasion and metastasis. Pigment Cell Melanoma Res. 2013;26(1):39–57. doi: 10.1111/pcmr.12041. [DOI] [PubMed] [Google Scholar]
- 3.Eggermont AM, Spatz A, Robert C. Cutaneous melanoma. Lancet. 2014;383(9919):816–27. doi: 10.1016/S0140-6736(13)60802-8. [DOI] [PubMed] [Google Scholar]
- 4.Burris HA., 3rd Overcoming acquired resistance to anticancer therapy: Focus on the PI3K/AKT/mTOR pathway. Cancer Chemother Pharmacol. 2013;71(4):829–42. doi: 10.1007/s00280-012-2043-3. [DOI] [PubMed] [Google Scholar]
- 5.Kang XJ, Shi XH, Chen WJ, et al. Analysis of KIT mutations and c-KIT expression in Chinese Uyghur and Han patients with melanoma. Clin Exp Dermatol. 2016;41(1):81–87. doi: 10.1111/ced.12659. [DOI] [PubMed] [Google Scholar]
- 6.Chi Z, Li S, Sheng X, et al. Clinical presentation, histology, and prognoses of malignant melanoma in ethnic Chinese: A study of 522 consecutive cases. BMC Cancer. 2011;11:85. doi: 10.1186/1471-2407-11-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dilruba S, Kalayda GV. Platinum-based drugs: Past, present and future. Cancer Chemother Pharmacol. 2016;77(6):1103–24. doi: 10.1007/s00280-016-2976-z. [DOI] [PubMed] [Google Scholar]
- 8.Galluzzi L, Vitale I, Michels J, et al. Systems biology of cisplatin resistance: Past, present and future. Cell Death Dis. 2014;5(5):e1257. doi: 10.1038/cddis.2013.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Megahed AI, Koon HB. What is the role of chemotherapy in the treatment of melanoma? Curr Treat Options Oncol. 2014;15(2):321–35. doi: 10.1007/s11864-014-0277-5. [DOI] [PubMed] [Google Scholar]
- 10.Baylin SB, Jones PA. A decade of exploring the cancer epigenome – biological and translational implications. Nat Rev Cancer. 2011;11(10):726–34. doi: 10.1038/nrc3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li W, Miao X, Liu L, et al. Methylation-mediated silencing of microRNA-211 promotes cell growth and epithelial to mesenchymal transition through activation of the AKT/β-catenin pathway in GBM. Oncotarget. 2017;8(15):25167. doi: 10.18632/oncotarget.15531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu H, Yang W. MiR-211 is epigenetically regulated by DNMT1 mediated methylation and inhibits EMT of melanoma cells by targeting RAB22A. Biochem Biophys Res Commun. 2016;476(4):400–5. doi: 10.1016/j.bbrc.2016.05.133. [DOI] [PubMed] [Google Scholar]
- 13.Asuthkar S, Velpula KK, Chetty C, et al. Epigenetic regulation of miRNA-211 by MMP-9 governs glioma cell apoptosis, chemosensitivity and radiosensitivity. Oncotarget. 2012;3(11):1439–54. doi: 10.18632/oncotarget.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Robaina MC, Mazzoccoli L, Arruda VO, et al. Deregulation of DNMT1, DNMT3B and miR-29s in Burkitt lymphoma suggests novel contribution for disease pathogenesis. Exp Mol Pathol. 2015;98(2):200–7. doi: 10.1016/j.yexmp.2015.03.006. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Q, Padi SK, Tindall DJ, Guo B. Polycomb protein EZH2 suppresses apoptosis by silencing the proapoptotic miR-31. Cell Death Dis. 2014;5:e1486. doi: 10.1038/cddis.2014.454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu H, Liu Y, Liu W, et al. EZH2-mediated loss of miR-622 determines CXCR4 activation in hepatocellular carcinoma. Nat Commun. 2015;6:8494. doi: 10.1038/ncomms9494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang J, Lv J, Zhang F, et al. MicroRNA-211 expression is downregulated and associated with poor prognosis in human glioma. J Neurooncol. 2017;133(3):553–59. doi: 10.1007/s11060-017-2464-2. [DOI] [PubMed] [Google Scholar]
- 18.Fu Y, Feng MX, Yu J, et al. DNA methylation-mediated silencing of matricellular protein dermatopontin promotes hepatocellular carcinoma metastasis by alpha3beta1 integrin-Rho GTPase signaling. Oncotarget. 2014;5(16):6701–15. doi: 10.18632/oncotarget.2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tian GA, Zhu CC, Zhang XX, et al. CCBE1 promotes GIST development through enhancing angiogenesis and mediating resistance to imatinib. Sci Rep. 2016;6:31071. doi: 10.1038/srep31071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang HL, Si LB, Zeng A, et al. MicroRNA-21 antisense oligonucleotide improves the sensitivity of A375 human melanoma cell to Cisplatin: An in vitro study. J Cell Biochem. 2018;119(4):3129–41. doi: 10.1002/jcb.26455. [DOI] [PubMed] [Google Scholar]
- 21.Sarkar D, Leung EY, Baguley BC, et al. Epigenetic regulation in human melanoma: Past and future. Epigenetics. 2015;10(2):103–21. doi: 10.1080/15592294.2014.1003746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Balch CM, Gershenwald JE, Soong S, et al. Final version of 2009 AJCC Melanoma Staging and Classification. J Clin Oncol. 2009;27(36):6199–206. doi: 10.1200/JCO.2009.23.4799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Flaherty KT, Lee SJ, Zhao F, et al. Phase III trial of carboplatin and paclitaxel with or without sorafenib in metastatic melanoma. J Clin Oncol. 2013;31(3):373–79. doi: 10.1200/JCO.2012.42.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Du Z, Niu S, Xu X, Xu Q. MicroRNA31-NDRG3 regulation axes are essential for hepatocellular carcinoma survival and drug resistance. Cancer Biomark. 2017;19(2):221–30. doi: 10.3233/CBM-170568. [DOI] [PubMed] [Google Scholar]
- 25.Ma J, Dong C, Ji C. MicroRNA and drug resistance. Cancer Gene Ther. 2010;17(8):523–31. doi: 10.1038/cgt.2010.18. [DOI] [PubMed] [Google Scholar]
- 26.Levy C, Khaled M, Iliopoulos D, et al. Intronic miR-211 assumes the tumor suppressive function of its host gene in melanoma. Mol Cell. 2010;40(5):841–49. doi: 10.1016/j.molcel.2010.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boyle GM, Woods SL, Bonazzi VF, et al. Melanoma cell invasiveness is regulated by miR-211 suppression of the BRN2 transcription factor. Pigment Cell Melanoma Res. 2011;24(3):525–37. doi: 10.1111/j.1755-148X.2011.00849.x. [DOI] [PubMed] [Google Scholar]
- 28.Mazar J, DeYoung K, Khaitan D, et al. The regulation of miRNA-211 expression and its role in melanoma cell invasiveness. PLoS One. 2010;5(11):e13779. doi: 10.1371/journal.pone.0013779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bell RE, Khaled M, Netanely D, et al. Transcription factor/microRNA axis blocks melanoma invasion program by miR-211 targeting NUAK1. J Invest Dermatol. 2014;134(2):441–51. doi: 10.1038/jid.2013.340. [DOI] [PubMed] [Google Scholar]
- 30.Rodriguez-Paredes M, Esteller M. Cancer epigenetics reaches mainstream oncology. Nat Med. 2011;17(3):330–39. doi: 10.1038/nm.2305. [DOI] [PubMed] [Google Scholar]
- 31.Gan L, Yang Y, Li Q, et al. Epigenetic regulation of cancer progression by EZH2: From biological insights to therapeutic potential. Biomark Res. 2018;6:10. doi: 10.1186/s40364-018-0122-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Golbabapour S, Majid NA, Hassandarvish P, et al. Gene silencing and polycomb group proteins: An overview of their structure, mechanisms and phylogenetics. OMICS. 2013;17(6):283–96. doi: 10.1089/omi.2012.0105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu L, Beckebaum S, Iacob S, et al. MicroRNA-101 inhibits human hepatocellular carcinoma progression through EZH2 downregulation and increased cytostatic drug sensitivity. J Hepatol. 2014;60(3):590–98. doi: 10.1016/j.jhep.2013.10.028. [DOI] [PubMed] [Google Scholar]
- 34.Konno Y, Dong P, Xiong Y, et al. MicroRNA-101 targets EZH2, MCL-1 and FOS to suppress proliferation, invasion and stem cell-like phenotype of aggressive endometrial cancer cells. Oncotarget. 2014;5(15):6049–62. doi: 10.18632/oncotarget.2157. [DOI] [PMC free article] [PubMed] [Google Scholar]