Abstract
One of the debilitating diseases affecting the central nervous system is multiple sclerosis (MS). As there is no definitive treatment for MS, researchers have mainly consented with optimization of strategies which slows down the progression of the disease such as specific auto-antigens tolerance induction. In this regard, the aim of this study was design of a new double-epitope protective vaccine based on interleukin (IL)-16-neuroantigens fusion proteins. First, we selected highly antigenic epitopes of myelin basic protein (MBP) (aa 84-104) and myelin oligodendrocyte glycoprotein (MOG) (aa 99-107) from available literature and our bioinformatics analysis. The correct cleavage of our constructs and major histocompatibility complex class II binding affinities of cleaved epitopes were checked and evaluated using Pepcleave and IEDB servers, respectively. Then, different combination of MOG and MBP epitopes with or without fusion to C-terminal active part of IL-16 were designed as constructs. Afterward, Modeller and Gromacs softwares used for the investigation of the MBP, and MOG epitopes antigenicity in these constructs. The results of molecular dynamics simulations showed that IL-16 in MOG + linker + MBP + IL-16 construct does not interfere with final epitopes antigenicity of MOG + linker + MBP construct. To sum up, the construct with IL-16 is suggested as a new double-epitope tolerogenic vaccine for prevention and amelioration of MS in human.
Keywords: Fusion proteins, Molecular dynamic simulation, Multiple sclerosis, Neuroantigen, Tolerogenic vaccine
INTRODUCTION
One of the debilitating diseases that affect the central nervous system is multiple sclerosis (MS). In this disease, the target of immune attacks is myelin membrane of oligodendrocytes. This disease associated with tissue inflammation, apoptosis of neurons and autoimmune protests, atrophy of brain, and spinal cord as well as destruction of myelin sheath but its exact etiology is still unknown (1). According to the recent research, autoimmunity produced by autoreactive T cells, specific for self-epitopes of myelin proteins, plays a fundamental role in progression of the disease (2). These autoreactive T cells may recognize myelin basic protein (MBP), myelin oligodendrocyte glycoprotein (MOG), and proteolipid protein (PLP) as neuro-antigens (3,4).
There is no known definitive therapy for MS but there are currently several unspecific treatments for this disease, such as using immunosuppressive drugs to slow down the progression of the disease (4). Therefore, many researchers have considered optimization of strategies for induction of self-tolerance to the related auto-antigens (3). In this regard tolerogenic vaccines developed based on cytokine-neuroantigen fusion proteins to restore the immune homeostasis and decreases unwanted autoimmune responses (5). In this strategy, the function of cytokine domain is presumably submission of neuroantigens to antigen presenting cells (APCs) for further presentation, and stimulating the T cells population to produce more regulatory rather effector T cells (4).
Different researchers benefit from several cytokines in tolerogenic vaccines such as interleukin (IL)-16, IL-2, IL-1RA, IL-4, IL-13, IL-10, IFNβ, and granulocyte-macrophage colony-stimulating factor (GM-CSF) (4,5,6). Also, various neuroantigens or some parts of neuroantigens (as their functional epitopes) were used in these kinds of tolerogenic vaccines. For example, MBP (aa 69-87 and aa 73-87), MOG (aa 35-55), and PLP (aa 139-151) were fused with different cytokines and tested for tolerogenicity in animal models of MS, experimental autoimmune encephalomyelitis (EAE) (4). The MBP (aa 84-104) is known to be the best epitope of MBP which is recognized by T cell clones in individuals with the human leukocyte antigen (HLA)-DR2 haplotype, such as HLADRB1* 1501 which is associated with susceptibility to MS (7,8). MBP (aa 69-89) was also suggested by Mannie et al., and Blanchfield et al. (6,9,10).
In other studies it was shown that MBP (aa 80-105) (11) and MBP (aa 82-100) (12) are immunodominant epitopes and MBP (aa 86-95) is the central sequence of MBP for T cell clones which are specific for HLA-DR2 (11). MOG (aa 99-107) has been defined as the immune dominant HLA-DR4-restricted T cell epitope (13).
In this study, different epitopes of two neuroantigens in combination with C-terminal of IL-16 were studied and then best cytokineneuroantigen combination was selected using the bioinformatic tools. Further analysis were then carried out by molecular dynamics (MD) simulation.
MATERIALS AND METHODS
The sequence of MBP and MOG epitopes were selected according to their highly antigenic properties by literature review, checked using bioinformatics tools (IEDB web server)(http://www.iedb.org/)(14), and HLAPred (15) and considered for further analysis.
The sequence of the common isoform of human MBP 18.5 KD (Accession: NP_ 001020 261, UniProtKB: P02686-5), isoform alpha1 precursor of human MOG (Accession: NP_996532, UniProtKB: Q16653) and pro-IL-16 isoform 1 of human (Accession: NP_004504, UniProtKB: Q14005-3) were obtained from NCBI and UniProt. We designed two fusion protein vaccines (constructs 1 and 2) whose corresponding peptide sequences are summarized in Fig. 1.
Fig. 1.
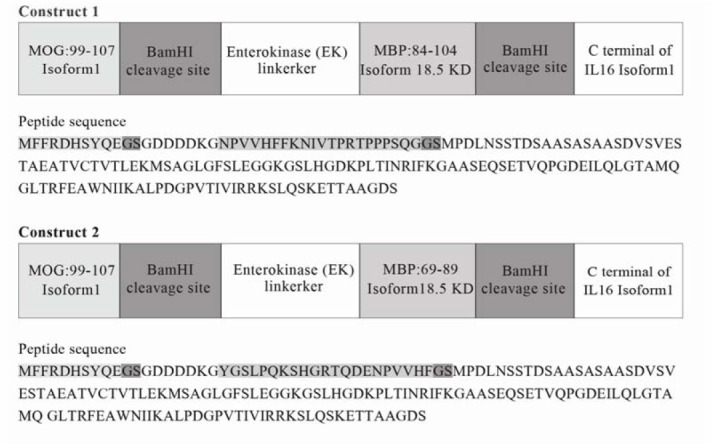
Peptide sequence of designed tolerogenic vaccine.
Among the investigated cytokines, IL-16 showed acceptable tolerogenic activity (4,6) as well as specific targeting and attachment to the CD4+ T cells (Th2) via receptor of IL-16, CD4 (6). Albeit, the N-terminal of this protein is translocated into the nucleus but its C-terminal part, a ligand for CD4, is secreted as active IL-16 (6,16). Therefore, we selected C-terminal part of IL-16 as the functional part of this cytokine to design fusion proteins.
In our fusion protein designs, we employed enterokinase (EK) linker between neuroantigens because its protease-recognition site may facilitate cleavage of constructs and therefore, better antigen processing with major histocompatibility complex (MHC) class II and more efficient presentation by APCs (4).
PepCleave server (http://peptibase.cs.biu. ac.il/PepCleave_II/) (17) was used for prediction of cleavage properties of our fusion proteins for producing intact epitopes by enzymes play roles after endocytosis of the fusion protein by APCs (18).
Modeller 9v2 was used for homology modelling of MBP (aa 84-104), MOG (aa 99-107) and whole antigenic domain (MOG (aa 99-107) + linker + MBP (aa 84-104)) (18,19). The template structure for MOG antigen sequence 99-107 was chain A of the human -MOG with 1.45 angstroms resolution (PDB code: 1PKO) (max score: 35, total score: 35, query coverage: 100%, and E value: 3e-04). The template for MBP (aa 84-104) antigen was chain A of 2LUG (max score: 71.5, total score: 71.5, query cover: 100%, and E value: 3e-17).
The starting structure for IL-16 called 1I16. Because IL-16 is ubiquitinated in the cell the 3D structure of ubiquitinated IL-16 was made via connecting the ubiquitin (PDB code: 2ZNV) to IL-16 via Discovery Studio 3.5 Client. According to Phosphosite plus site (20), three ubiquitin were connected to three lysine residues (aa 22,57,103) of IL-16. PDB code “2ZNV” is related to the crystal structure of human AMSH-LP DUB domain in complex with lysine 63-linked ubiquitin dimer. In this structure, ubiquitin is linked to lysine and is similar to ubiquitinated IL-16. Only in this PDB structure, ubiquitin was connected to lysine residue. In addition, this structure has the best resolution (1.6 Å), among all ubiquitin PDB structures. We also added one methionine residue to N-terminal of the fusion protein and three methionines to N-terminal chain of ubiquitin chains. Of the 1000 models generated with Modeller, the one corresponding to the lowest value of discrete optimized protein energy (DOPE) and the fewest restraints violations was selected for further analysis. This structure was verified via RAMPAGE server (http://mordred.bioc .cam.ac.uk/~rapper/rampage.php) (21) (98% of residues were in favored region and 2% were in allowed region for both construct 1 and antigenic domain) (data not shown). In addition, the structure of MBP (aa 84-104) has deamidated glutamine and phosphorylated threonines (22). These modifications were applied after modeling by Modeller software via Hyperchem7 software (http://www.chemistrysoftware. com/hyperchem/hyperchem 7.5.htm).
At first, free MBP (aa 84-104) and MOG (aa 99-107) epitopes, ubiquitinated IL-16 domain and antigenic domains (MOG (aa 99-107) + linker + MBP (aa 84-104)) (four structures) were separately utilized for MD simulation by Gromacs 5.1.1 package under G43A1 force field with spc216 water model and the time step of 1 fs, for 20 ns (23,24) with the same procedure as mentioned in our previous works (25,26,27). The charge of phosphorylated threonine was set to -2e (28), and the charge of deamidated glutamine was set to zero. The total charge of the antigenic domain was -17e, then 74 Na+ and 57 Cl- ions were added to the system for reaching to ionic strength 140 mM by replacing with water molecules in random positions. The ionic strength of 140 mM is similar to the ionic strength of cells (29). Then the obtained final structures of the antigenic domain (MOG + linker + MBP) and ubiquitinated IL-16 were connected to each other via Discovery studio software and 3D structure of construct 1 (MOG + linker + MBP + C-terminal part of IL16) was produced. Construct 1 system contained 30 Na+ and 24 Cl- ions and it was simulated for 20 ns with the same previous mentioned conditions. In brief, we only mention the results of the antigenic domain and construct 1 in this paper. Prediction of aggregation propensity of construct 1 structure and assessment of solubility were investigated with the Aggrescan3D server (30).
Also, for cloning of construct 1 in Saccharomyces cerevisiae for future researches, its coding sequence was optimized with JCat web server (31).
RESULTS
Bioinformatics results
According to previous studies and bioinformatic tools the sequence of MBP (aa 84-104), MBP (aa 69-89) and MOG (aa 99-107) epitopes were selected for further analysis.
The results of the PepCleave server showed that MBP (aa 84-104) epitope in construct 1 is cleaved with the score of 1.08 and MBP (aa 69-89) epitope in construct 2 is cleaved with the score of 0.02. This was because of differences in the orders of epitopes in our designed constructs. Therefore, MBP (aa 84-104) in construct 1 can be cleaved with more specificity than construct 2. Also according to the PepCleave server MOG (aa 99-107) epitope in both constructs is cleaved with score 0.23 which is in an acceptable interval, therefore construct 1 was selected for modeling and MD simulation. The result of aggrescan 3D server indicated negative values for the average and total score. It means this protein structure is soluble; average and total score are respectively -0.93 and -373.21.
Molecular dynamics simulation results
The root mean square deviation (RMSD) of backbone atoms relative to starting structure as a reference was calculated for construct 1 and antigenic domain (Fig. 2). The kinetics and potential energy fluctuation were in the equal and opposite direction in construct 1 and antigenic domain (data not shown). This figure shows that proteins reach equilibrium after about 10 ns MD simulation and simulation times were adequate.
Fig. 2.
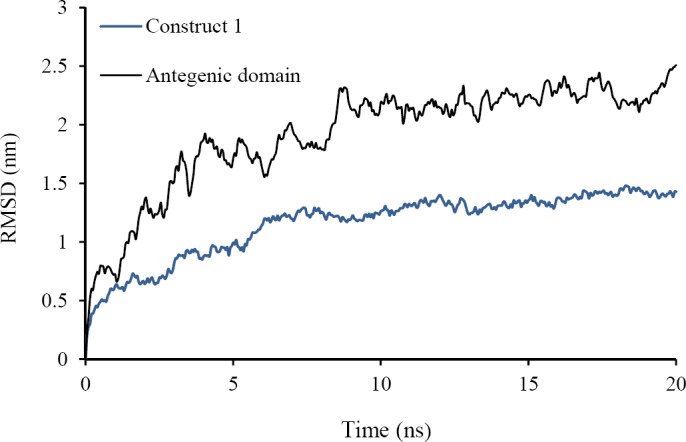
Root mean square deviation (RMSD) of the backbone of the fusion protein and fusion protein without interleukin 16 (free) during 20 ns molecular dynamic simulation. Blue line relates to fusion protein and black line relates to fusion protein without interleukin-16 (free).
Table 1 shows the average of temperature, potential energy, radius of gyration (Rg), and the distance between center of mass of MOG or MBP and IL-16 in construct 1. Also in this table the average of accessible surface area of MBP and MOG and the average of root mean square fluctuation (RMSF) of backbone of MOG + MBP residues in construct 1 and antigenic domain during the last 5 of 20 ns MD simulation were mentioned. Small standard deviation of RMSD and Rg and temperature in both proteins show that the systems reach to stable structure and thermal equilibrium. It should be noted there were a significant number of hydrogen bonds between water and construct 1 or antigenic domain and within them; therefore construct 1 and antigenic domain is soluble and stable.
Table 1.
The results of the last 5 ns of molecular dynamic simulation for fusion protein and free fusion protein.
| Criterion | Fusion protein | Free fusion protein |
|---|---|---|
| RMSD (nm) | 1.4 ± 0.04 | 2.28 ± 0.1 |
| Temperature (K) | 300.1 ± 1.06 | 300.3 ± 1.9 |
| Potential energy (kj/mol) | -1499736 | -401091 |
| Radius of gyration (nm) | 2.48 ± 0.02 | 1.2 ± 0.07 |
| Distance between center of mass MOG and IL-16 (nm) | 3.03 ± 0.2 | - |
| Distance between center of mass MOG and MBP (nm) | 2.5 ± 0.1 | 1.64 ± 0.1 |
| Distance between center of mass MBP and IL-16(nm) | 1.3 ± 0.05 | - |
| Total area of MOG (nm2) | 14.6 ± 1.06 | 14.4 ± 0.6 |
| Total area of MBP 84-104 (nm2) | 28.8 ± 0.6 | 23.58 ± 0.8 |
| h-bonds protein-protein | 274.5 ± 9.5 | - |
| h-bonds protein-water | 795.34 ± 19.48 | - |
RMSD, root mean square deviation; MOG, myelin oligodendrocyte glycoprotein; IL-16, interleukin-16; MBP, myelin basic protein; (-), absence of IL-16 in the free fusion protein construct.
The distance between the center of mass MOG epitope and IL-16 in construct 1 was 3.03 ± 0.17 nm (Table 1), it means that the EK linker can separate MOG antigen from IL-16 and IL-16 does not have intervention with antigenicity of MOG.
The distance between centers of mass of two antigens (MOG and MBP) was 2.50 ± 0.10 nm in construct 1, and then two antigens do not interfere with each other. Comparison of the accessible surface area of MOG antigen in the antigenic domain and in construct 1 presented that accessible surface area of MOG antigen was similar in the antigenic domain and in both proteins (about 14.4 nm2). However, the accessible surface area of MBP in construct 1 is more than the antigenic domain. In the antigenic domain, MOG epitope bends and cover MPB surface, but in construct 1, MBP is contiguous to IL-16 and MOG cannot bend and cover MBP (Table 1). The average of RMSF of backbone in MOG + MBP residues during the last 5 ns of MD simulation in construct 1, and antigenic domain shows that average of RMSF in MOG + MBP residues in construct 1 is more than antigenic domain in the construct 2 (Table 1). Then flexibility and functionality and binding power of antigenic domain in construct 1 beside to IL-16 is more than antigenic domain without IL-16. Conformational change or flexibility in the protein, results in a series of rearrangements that lead to a complex with tighter binding (32).
DISCUSSION
In the pathogenesis of MS myelin proteins are recognized as autoantigens by autoreactive T cells (3,4). The main objective of recent research is self-tolerance induction to auto-antigens for diseases control. Cytokineneuroantigen fusion proteins are known as the newest strategy in animal models of MS (4).
Our suggested therapeutic vaccine encompasses some new features. First, both epitopes of two different neuroantigens MOG (aa 99-107) and MBP (aa 84-104), which were reported in previous studies (11,12) as immunodominant epitopes (not used in tolerogenic vaccines), were subsequently confirmed by IEDB and HLAPred in this study and included at once in our construct. Firstly, other researchers designed vaccines that just contained one kind of myelin epitope in combination with one cytokine, such as MBP (aa 69-88) (6,17,19), MBP (aa 73-87) (33,34), the epitopes in guinea pig or murine MOG (aa 35-55), or the PLP (aa 139-151) (34). Secondly, these epitopes MBP (aa 84-104), MOG (aa99-107) are highly conserved among rat, mouse, and human. Suitable cleavage of our recombinant fusion protein was checked by using the Pepcleave server and their appropriate binding to MHC class II molecules accomplished by IEDB server.
Interestingly, our considered epitopes bind with a high affinity to two different alleles of MHC-class II which both are associated with risk of MS. In this case, DRB1*1501 and DRB1*0401 show a better affinity for binding to MBP (aa 84-104) and MOG (aa 99-107), respectively (16,19). Therefore, fusion of both MBP and MOG epitopes in a single vaccine can help us to produce a multifunctional tolerogenic vaccine. Second, a nonimmunogenic linker with a known proteaserecognition site, EK, was used between two epitopes to enhance the possibility of their correct cleavage (4). Third, BamHI restriction sites are located on both sides of MBP epitope to ensure the possibility of producing a single-epitope-IL-16 fusion protein if required. Forth, our tolerogenic vaccine consisted of IL-16 which has been an effective cytokine in other animal model studies (6). Therefore, in our study, C-terminal IL-16 was chosen as the suitable cytokine for our new vaccine, double epitope tolerogenic fusion protein. This vaccine may transform extracted lymphocytes to tolerogenic T cells (2).
The construct (construct 1) with MBP (aa 84-104) was chosen and its structural properties was investigated by MD simulation method. The results of MD simulation showed that surface areas of MOG antigen in the construct 1 are similar to antigenic domain in the construct 2 but the surface areas of MBP in construct 1 are greater than the antigenic domain of construct 2. Also flexibility and binding power of MOG + MBP residues are higher in construct 1. As a result, it can be concluded that construct 1 sustain their antigenic property for vaccination.
Results of JCat server showed that codon adaptation index of fusion protein before optimization in Saccharomyces cerevisiae is 0.050 and after optimization reached to 1.0 and the GC content of this fusion protein before optimization is 55.23% and after optimization becomes 44.38%, then GC content reduced to suitable content for construct 1 (i.e. 38.148).
Since our construct was optimized for expression in yeast host may lead to an easier and more precise production in high scale. However, other researchers selected the expression system of Baculovirus in human embryonic kidney cells for different fusion proteins (4,6,10,33,35).
CONCLUSION
To sum up, as the construct 1 obtained better scores by different predictive servers as well as by MD simulation, this fusion protein is suggested for further functional experiments as a tolerogenic vaccine for MS. We hope that this work could suggest a better tolerogenic vaccine for amelioration of MS disease.
ACKNOWLEDGMENTS
This work was financially supported (Grant No. 141/375) by post-graduate office of Shahrekord University, Shahrekord, I.R. Iran. We would also like to thank Mr. Gholamreza Banisharif-Dehkordi for his technical assistance.
REFERENCES
- 1.Koriem KMM. Multiple sclerosis: New insights and trends. Asian Pac J Trop Biomed. 2016;6(5):429–440. [Google Scholar]
- 2.Correale J, Farez M, Gilmore W. Vaccines for multiple sclerosis: progress to date. CNS Drugs. 2008;22(3):175–198. doi: 10.2165/00023210-200822030-00001. [DOI] [PubMed] [Google Scholar]
- 3.Fissolo N, Montalban X, Comabella M. DNA-based vaccines for multiple sclerosis: current status and future directions. Clin Immunol. 2012;142(1):76–83. doi: 10.1016/j.clim.2010.11.011. [DOI] [PubMed] [Google Scholar]
- 4.Mannie MD, Blanchfield JL, Islam SM, Abbott DJ. Cytokine-neuroantigen fusion proteins as a new class of tolerogenic, therapeutic vaccines for the treatment of inflammatory demyelinating disease in rodent models of multiple sclerosis. Front Immunol. 2012;3:255–270. doi: 10.3389/fimmu.2012.00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mannie MD, Curtis AD. Tolerogenic vaccines for Multiple Sclerosis. Hum Vaccin Immunother. 2013;9(5):1032–1038. doi: 10.4161/hv.23685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mannie MD, Abbott DJ. A Fusion protein consisting of IL-16 and the encephalitogenic peptide of myelin basic protein constitutes an antigen-specific tolerogenic vaccine that inhibits experimental autoimmune encephalomyelitis. J Immunol. 2007;179(3):1458–1465. doi: 10.4049/jimmunol.179.3.1458. [DOI] [PubMed] [Google Scholar]
- 7.Alcina A, Abad-Grau MM, Fedetz M, Izquierdo G, Lucas M, Fernández O, et al. Multiple sclerosis risk variant HLA-DRB1* 1501 associates with high expression of DRB1 gene in different human populations. PLoS One. 2012;7(1):e29819. doi: 10.1371/journal.pone.0029819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krogsgaard M, Wucherpfennig KW, Cannella B, Hansen BE, Svejgaard A, Pyrdol J, et al. Visualization of myelin basic protein (MBP) T cell epitopes in multiple sclerosis lesions using a monoclonal antibody specific for the human histocompatibility leukocyte antigen (HLA)-DR2-MBP 85-99 complex. J Exp Med. 2000;191(8):1395–1412. doi: 10.1084/jem.191.8.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blanchfield JL, Mannie MD. A GMCSFneuroantigen fusion protein is a potent tolerogen in experimental autoimmune encephalomyelitis (EAE) that is associated with efficient targeting of neuroantigen to APC. J Leukoc Biol. 2010;87(3):509–521. doi: 10.1189/jlb.0709520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mannie MD, Abbott DJ, Blanchfield JL. Experimental autoimmune encephalomyelitis in Lewis rats: IFN-beta acts as a tolerogenic adjuvant for induction of neuroantigen-dependent tolerance. J Immunol. 2009;182(9):5331–5341. doi: 10.4049/jimmunol.0803756. [DOI] [PubMed] [Google Scholar]
- 11.Blanchfield JL, Mannie MD. A GMCSFneuroantigen fusion protein is a potent tolerogen in experimental autoimmune encephalomyelitis (EAE) that is associated with efficient targeting of neuroantigen to APC. J Leukoc Biol. 2010;87(3):509–521. doi: 10.1189/jlb.0709520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wucherpfennig KW, Catz I, Hausmann S, Strominger JL, Steinman L, Warren KG. Recognition of the immunodominant myelin basic protein peptide by autoantibodies and HLA-DR2- restricted T cell clones from multiple sclerosis patients. Identity of key contact residues in the Bcell and T-cell epitopes. J Clin Invest. 1997;100(5):1114–1122. doi: 10.1172/JCI119622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Forsthuber TG, Shive CL, Wienhold W, de Graaf K, Spack EG, Sublett R, et al. T cell epitopes of human myelin oligodendrocyte glycoprotein identified in HLA-DR4 (DRB1*0401) transgenic mice are encephalitogenic and are presented by human B cells. J Immunol. 2001;167(12):7119–7125. doi: 10.4049/jimmunol.167.12.7119. [DOI] [PubMed] [Google Scholar]
- 14.Vita R, Overton JA, Greenbaum JA, Ponomarenko J, Clark JD, Cantrell JR, et al. The immune epitope database (IEDB) 3.0. Nucleic Acids Res. 2015;43(Database issue):D405–D412. doi: 10.1093/nar/gku938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Adams HP, Koziol JA. Prediction of binding to MHC class I molecules. J Immunol Methods. 1995;185(2):181–190. doi: 10.1016/0022-1759(95)00111-m. [DOI] [PubMed] [Google Scholar]
- 16.Cruikshank WW, Kornfeld H, Center DM. Interleukin-16. J Leukoc Biol. 2000;67(6):757–766. doi: 10.1002/jlb.67.6.757. [DOI] [PubMed] [Google Scholar]
- 17.Hoze E, Tsaban L, Maman Y, Louzoun Y. Predictor for the effect of amino acid composition on CD4+ T cell epitopes preprocessing. J Immunol Methods. 2013;391(1-2):163–173. doi: 10.1016/j.jim.2013.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Payab N, Mahnam K, Shakhsi-Niaei M. Computational comparison of two new fusion proteins for multiple Sclerosis. Res Pharm Sci. 2018;13(5):394–403. doi: 10.4103/1735-5362.236832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eswar N, Webb B, Marti-Renom MA, Madhusudhan MS, Eramian D, Shen MY, et al. Comparative protein structure modeling using Modeller. Curr Protoc Bioinformatics. 2006;Chapter 5:Unit–5. doi: 10.1002/0471250953.bi0506s15. 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hornbeck PV, Zhang B, Murray B, Kornhauser JM, Latham V, Skrzypek E. PhosphoSitePlus, 2014: mutations, PTMs and recalibrations. Nucleic Acids Res. 2015;43(Database issue):D512–D520. doi: 10.1093/nar/gku1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lovell SC, Davis IW, Arendall WB, 3rd, de Bakker PI, Word JM. Structure validation by Calpha geometry: phi,psi and Cbeta deviation. Proteins: Structure, Function & Genetics. 2003;50(3):437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- 22.Zhang C, Walker AK, Zand R, Moscarello MA, Yan JM, Andrews PC. Myelin basic protein undergoes a broader range of modifications in mammals than in lower vertebrates. J Proteome Res. 2012;11(10):4791–4802. doi: 10.1021/pr201196e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Van Der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJ. GROMACS: fast, flexible, and free. J Comput Chem. 2005;26(16):1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 24.Darden T, York D, Pedersen L. Particle mesh Ewald: An N log (N) method for Ewald sums in large systems. J Chem Phys. 1998;98(12):10089–10092. [Google Scholar]
- 25.Mahnam K, Hoghoughi A. In silico studies on fingolimod and cladribine binding to p53 gene and its implication in prediction of their carcinogenicity potential. MBD. 2014;1(2):105–122. [Google Scholar]
- 26.Mansourian M, Madadkar-Sobhani A, Mahnam K, Fassihi A, Saghaie L. Characterization of adenosine receptor in its native environment: insights from molecular dynamics simulations of palmitoylated/glycosylated, membrane-integrated human A(2B) adenosine receptor. J Mol Model. 2012;18(9):4309–4324. doi: 10.1007/s00894-012-1427-y. [DOI] [PubMed] [Google Scholar]
- 27.Mansouri A, Mahnam K. Designing new surfactant peptides for binding to carbon nanotubes via computational approaches. J Mol Graph Model. 2017;74:61–72. doi: 10.1016/j.jmgm.2017.02.016. [DOI] [PubMed] [Google Scholar]
- 28.Polverini E, Coll EP, Tieleman DP, Harauz G. Conformational choreography of a molecular switch region in myelin basic protein-molecular dynamics shows induced folding and secondary structure type conversion upon threonyl phosphorylation in both aqueous and membrane-associated environments. Biochim Biophys Acta. 2011;1808(3):674–683. doi: 10.1016/j.bbamem.2010.11.030. [DOI] [PubMed] [Google Scholar]
- 29.The Properties of Water and their Role in Colloidal and Biological Systems. In: van Oss CJ, editor; Hubbard A, editor. Interface Science and Technology. Vol. 16. Amsterdam: Academic Press; 2008. pp. 31–48. [Google Scholar]
- 30.Zambrano R, Jamroz M, Szczasiuk A, Pujols J, Kmiecik S, Ventura S. AGGRESCAN3D (A3D): server for prediction of aggregation properties of protein structures. Nucleic Acids Res. 2015;43(W1):W306–W313. doi: 10.1093/nar/gkv359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grote A, Hiller K, Scheer M, Münch R, Nörtemann B, Hempel DC, et al. JCat: a novel tool to adapt codon usage of a target gene to its potential expression host. Nucleic Acids Res. 2005;33(Web Server issue):W526–W531. doi: 10.1093/nar/gki376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koshland DE. Application of a theory of enzyme specificity to protein synthesis. Proc Natl Acad Sci U S A. 1958;44(2):98–104. doi: 10.1073/pnas.44.2.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mannie MD, Devine JL, Clayson BA, Lewis LT, Abbott DJ. Cytokine-neuroantigen fusion proteins: new tools for modulation of myelin basic protein (MBP)-specific T cell responses in experimental autoimmune encephalomyelitis. J Immunol Methods. 2007;319(1-2):118–132. doi: 10.1016/j.jim.2006.11.005. [DOI] [PubMed] [Google Scholar]
- 34.Mannie MD, Clayson BA, Buskirk EJ, DeVine JL, Hernandez JJ, Abbott DJ. IL-2/neuroantigen fusion proteins as antigen-specific tolerogens in experimental autoimmune encephalomyelitis (EAE): correlation of T cell-mediated antigen presentation and tolerance induction. J Immunol. 2007;178(5):2835–2843. doi: 10.4049/jimmunol.178.5.2835. [DOI] [PubMed] [Google Scholar]
- 35.Abbott DJ, Blanchfield JL, Martinson DA, Russell SC, Taslim N, Curtis AD, et al. Neuroantigen-specific, tolerogenic vaccines: GMCSF is a fusion partner that facilitates tolerance rather than immunity to dominant self-epitopes of myelin in murine models of experimental autoimmune encephalomyelitis (EAE) BMC Immunol. 2011;12:72–89. doi: 10.1186/1471-2172-12-72. [DOI] [PMC free article] [PubMed] [Google Scholar]


