Abstract
The increasing incidences of cancer at the global scale have recently resulted in the invention of various biotechnology approaches among which the oncolytic virotherapy is a new strategy for the treatment of multiple tumors. Herpes simplex virus (HSV) based vectors are one of the most studied oncolytic agents, worldwide. Moreover, syngeneic animal models are the principal parts of the oncolytic virotherapies investigation. The effects of a dual fluorescent γ34.5 deleted vector-HSV-GR- on three mouse tumor cell lines were studied in this work. We previously generated a dual fluorescent labeled oncolytic HSV-HSV-GR- (both copies of γ34.5 were inactivated by insertion of two distinct fluorescent dyes, GFP and mCherry) in our laboratory; subsequently, they were used as oncolytic viruses. The three 4T1, TC-1, and CT26 cell lines were infected with HSV-GR. The infection efficacy and the elimination potency of HSV-GR were analyzed by photomicrography and flow cytometry methods. HSV-GR showed a significant efficiency to infect the cell lines examined. Flow cytometry analyses demonstrated that HSV-GR infected 89.3%, 86.1%, and 92.4% of 4T1, TC-1, and CT26 cells, respectively. Moreover, propidium iodide (PI) staining of infected cells indicated that HSV-GR could kill 27.9%, 21.2%, and 21.3% of 4T1, TC-1, and CT26 cells, respectively. Interestingly, HSV-GR infected cells were capable of expressing both GFP and mCherry at the same time. The promising effects of the oncolytic virus HSV-GR in the mouse syngeneic tumor cell system have shed more light on the therapeutic potential of this anti-cancer approach.
Keywords: Flow cytometry, Oncolytic HSV, Oncolytic virotherapy, Syngeneic tumor cell lines
INTRODUCTION
Cancer is a leading cause of death, worldwide (1). There are many types of cancer treatment including surgery, chemotherapy, and radiotherapy (2). However, most of these methods are either insufficient to treat cancers or exhibit side effects with highly damaging potential on proliferative tissues (3). New cancer treatment methods are needed due to limitations or side effects of conventional strategies of cancer treatment. Oncolytic virotherapy is a new strategy for targeting and elimination of different tumors. Currently, different types of viruses have been developed as oncolytic viruses (OVs), worldwide (4,5). These viruses are promising tools designed to both replicate in and lyse tumor cells by direct cytocidal effects while leaving normal cells unscathed (6,7). Such viruses exhibit the potential of targeting various tumor types and induce durable systemic antitumor immune responses with minimal side effects, as compared with conventional therapies (1).
Herpes simplex virus (HSV) is one of the most popular viruses in the OVs era. Properties including the ability to infect a diverse range of tumor cell lines, well-known life cycles, and genomic structure, as well as large genome space for transgene insertion, have made HSV-1 a perfect tool for cancer therapy (8). HSV-1 has a rapid infection cycle and can lyse cells within 10-h of infection, followed by the release of new viral particles (9).
HSV-1 vectors that are deficient in both copies of γ34.5 genes are safe enough for application as oncolytic HSV (10,11). The γ34.5 gene is one of the virulence factors of HSV and has been demonstrated as a critical determinant in the selective targeting of tumor cells in herpes-mediated virotherapy (11). HSV infection induces protein kinase R activation, the host defense mechanism against viral infection, and subsequently shuts off host protein synthesis (12). The HSV-γ34.5 gene reverses this phenomenon and reactivates host protein synthesis by dephosphorylation of translation factors (12).
On the other hand, preclinical characterization and validation of new cancer treatments require laboratory models. In this way, in vitro and ex vivo examination of novel therapeutic anti-cancer agents lead to remarkable progress in cancer therapy and applied as primary tools for the investigation of efficacy and safety of therapeutic approaches (13). The 4T1 (mouse breast tumor cell line) (14,15,16), CT26 (a mouse colon tumor cell line) (15,17), and TC-1 (a mouse lung cell) are three most well-studied mouse tumor models successfully treated with OVs (14). These three cell lines are considered as the counterparts of the cells causing three major human cancers. In addition, these cells have been used reputedly in many previous similar studies as target cell lines (9,13,14).
Considering the importance of newly developed agent efficacy investigations, we here evaluated our previously-developed double fluorescent oncolytic HSV (green-red) (HSV-GR) activity on three well-studied mouse tumor cells.
MATERIALS AND METHODS
Cells and virus
African green monkey kidney cell line (Vero) (NCBI-C101), 4T1 (NCBI-C604), TC-1 (mouse mammary carcinoma cell line) (NCBI-C569), CT26 (NCBI-C532) and BHK 21 (baby hamster kidney cell line) (NCBIC107) were purchased from National Cell Bank of Iran (NCBI, Pasteur Institute of Iran, Tehran, I.R. Iran). Vero and 4T1 cells were cultured in RPMI 1640 (Thermo Fisher Scientific, Gibco™, USA) supplemented with 10% fetal bovine serum (FBS) (Thermo Fisher Scientific, Gibco™, USA) and incubated at 37 °C. The above-mentioned cell lines were cultured in Dulbecco modified Eagle’s medium (DMEM; Thermo Fisher Scientific, Gibco™, USA) supplemented with 10% FBS. The cell cultures were incubated at 37 °C in a humidified atmosphere of 5% CO2. HSV-1 was kindly provided as a gift by Dr. Houriyeh Soleimanjahi (Tarbiat Modarres University, Tehran, I.R. Iran). Virus stocks were generated from low-multiplicity infections.
Herpes simplex virus propagation
Vero cells were used for HSV propagations. The day before infection, Vero cells were plated into 10-cm culture dishes and incubated at 37 °C, 5% CO2. After 24 h of incubation, cells were infected with HSV1 at a multiplicity of infection (MOI) of 1. The supernatant was harvested, aliquoted, titrated (18,19) and stored at -70 °C when the total cytopathic effect observed.
Titration of progeny viruses
The plaque assay technique was used to determine viral titers (20). In brief, precultured Vero cells were seeded into 6-well plates and infected with serial dilutions (up to 10 logs) of the virus samples. After 2 h of incubation at 37 °C, virus inoculum was removed, and freshly-prepared RPMI (supplemented with 2% FBS and 0.1% pooled human immune globulin, Sigma chemical Co., Germany) was added to the cells. The plates were incubated at 37 °C for 3 to 4 days until plaques were visible. The infected cells were then fixed for 5 min with methanol and stained with Giemsa for 20 min to visualize plaques. Afterward, plaques were counted, and the average number of plaques was determined. The mCherry (in context of BleCherry, bleomycin-mCherry fusion protein) and green fluorescent protein (GFP) positive plaques were observed with an inverted fluorescence microscope. (CETI, Inverso TC100 Medline, UK) (20).
Recombinant virus generation
The two recombinant HSV based viruses (HSV-GFP and HSV-GR) were generated using conventional homologous recombination methods using specific recombination shuttle vectors as described previously (20). Briefly, two distinct shuttle vectors were made with the PCR-cloning technique for inactivation of γ34.5 and insertion of two fluorescent dyes, GFP and mCherry into γ34.5-deleted sites. The pSL-ICP34.5L-CMV-GFP-pA-ICP34.5R was constructed by amplification of two right and left flanking regions with PCR and subsequently the GFP expression cassette was inserted between them. For the construction of HSV-GFP, cultured BHK cells were transfected by designed shuttle vector and subsequently infected with wild-type HSV-1. Afterward, recombinant viruses were isolated by plaque purification technique using GFP as a reporter signal. A similar procedure was applied for the generation of HSV-GR. The 6-well plated BHK cells were transfected with pSL-HomoF1-CMV-BleCherry-pA-HomoF2 and subsequently infected by HSV-GFP. Afterward, the recombinant virus was isolated and purified by plaque purification method. Finally, the isolated recombinant viruses were cultured, titrated and stored at -70 °C for further applications (20). The schematic structure of HSV-GR is depicted in Fig. 1.
Fig. 1.
Schematic structure of herpes simplex virus green-red (HSV-GR). The oncolytic HSV-GR was developed from a wild type strain. Modifications include deletion of both γ34.5 genes and insertion of a GFP and BleCherry expression cassette. CMVp, Cytomegalovirus promoter; BleCherry, bleomycin-mCherry fusion protein; pAs, polyadenylation signal; GFP, green fluorescent protein.
Fluorescent microscopy
Direct observation of HSV-GR infection potency was conducted using fluorescent potency was conducted using fluorescent microscopy. Cell lines 4T1, TC-1, and CT26 were plated into 6-well plates and were subsequently infected by of HSV-GR at MOI of 1. Forty-four h later, the infected cells were observed with invert fluorescent microscope (Motic, Scientific Imaging Company, China) (21).
Flow cytometry assay
Infection efficiency and cytotoxicity of HSV-GR were analyzed using flow cytometry. To this end, the 4T1, TC-1, and CT26 cells were seeded into 6-well plate and subsequently were infected with MOI 1 of the recombinant virus. After 22 h, the infected cells were treated with trypsin, collected and then washed twice with PBS. Finally, cells were resuspended in PBS and analyzed with flow cytometry (Cyflow, Partec, Germany). To evaluate the cytopathic effects of HSV-GR, the collected cells were stained with PI (22) and then analyzed with flow cytometry (23). In each assay, fluorescence was detected by flow cytometry, using the FL1 channel to detect GFP or the FL3 channel to detect PI staining of the DNA. Forward and side scatter gates were set to exclude cell debris.
RESULTS
Effect of inactivation of both copies of γ34.5 genes on HSV-GR titer
Titer of wild-type HSV-1 and HSV-GR were determined using standard plaque assay methods (detailed procedures are described in the materials and methods). HSV-1 and the materials and methods). HSV-1 and HSV-GR yielded 2 × 108 and 3 × 107 PFU/mL, respectively (Fig. 2). Hence, we observed that inactivation of γ34.5 has affected the viral replication and has led to a 10-fold decrease in recombinant virus titers. This finding emphasizes once again on the critical role of the γ34.5 gene in the viral replication. Constant expression of fluorescent dye was observed after several infection cycles in Vero and other three cell lines.
Fig. 2.
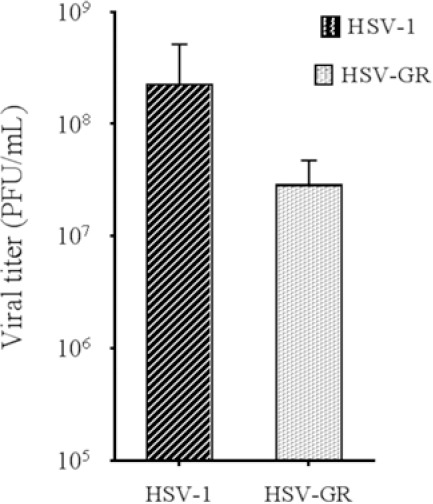
Titration of wild type and recombinant viruses. Vero cells were inoculated with wild type or recombinant virus. The virus titer was determined by titration on Vero cells. Herpes simplex virus green-red yielded 3 × 107 and HSV-1 yielded 2 × 108 PFU/mL.
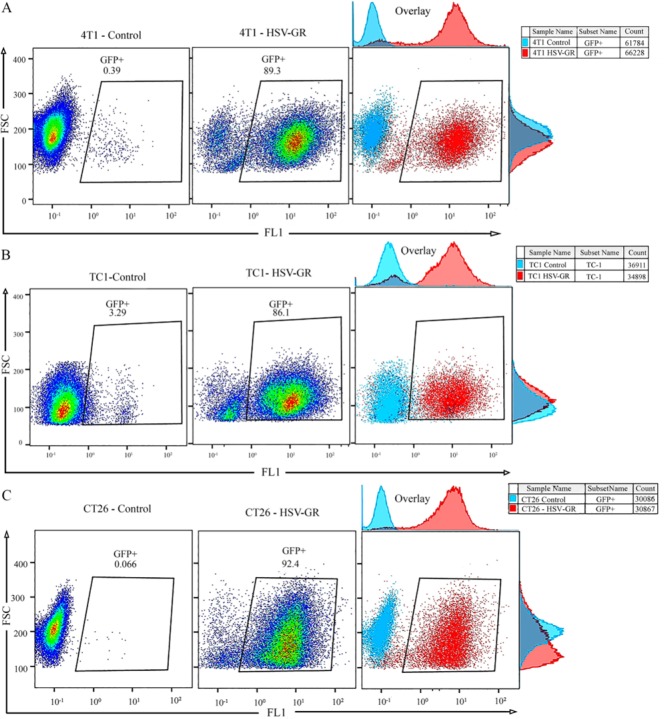
Determination of HSV-GR infection potency on 4T1, TC-1, and CT26 cell lines
After infection with HSV-GR, the cultured cells were analyzed by the fluorescence microscopy. Fig. 3 shows the target cells after 24-h of infection. Nearly all the infected cells could express both GFP and mCherry. Our findings indicated that the cell lines 4T1, TC-1, and CT26 are susceptible to HSV infections and HSV-GR has efficiently infected them (Fig. 3). These findings represented that all examined cell lines are susceptible to HSV infections.
Fig. 3.
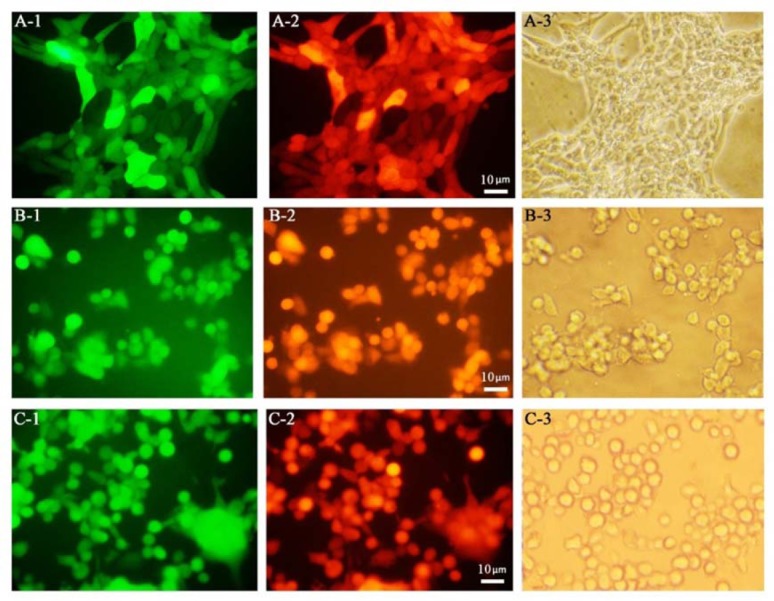
Fluorescent microscopy analyses of herpes simplex virus green-red infected cells. Preparation of infected cells are described in material and method section. Fluorescent imaging was done 24 h post infection. (A) 4T1 tumor cells; A-1, GFP positive; A-2, BleCherry positive; A-3, Bright field. (B) TC-1 tumor cells; B-1, GFP positive; B-2, BleCherry positive; B-3, bright field; (magnification × 200).
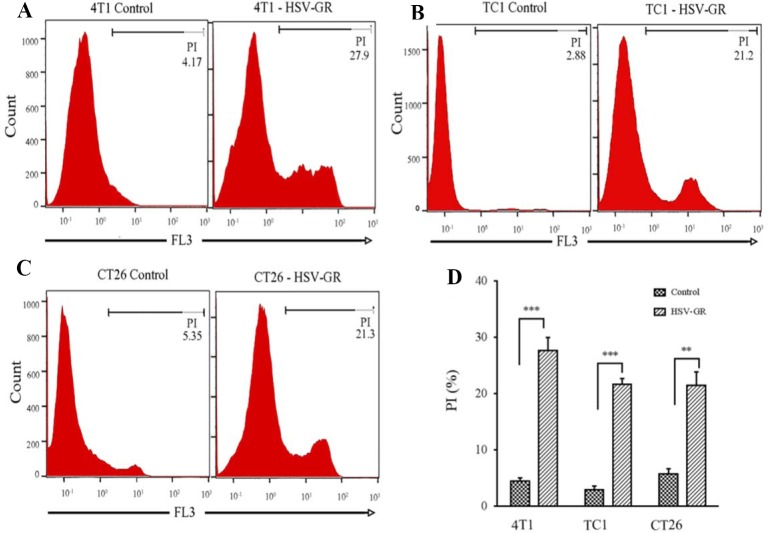
In vitro characterization of HSV-GR on syngeneic tumor cell lines
4T1 and TC-1 cells were seeded into the 6-well plate, and sub-confluent cells were infected with HSV-GR at an MOI of 1. Twenty-four h post infections, cells were harvested and three wells were stained with PI for cell death analysis. All samples were analyzed with flow cytometry instrument. (Fig. 4).
Fig. 4.
In vitro analysis of the infection potency of herpes simplex virus green-red (HSV-GR) in 4T1, TC-1 and CT26 cells using flow cytometry. HSV-GR was used to infect three mouse tumor cells at the indicated multiplicity of infection (MOIs) and times. Infected cells have been analyzed as described using the flow cytometry. GFP fluorescence signal was gated at FL1 channel to determine infection potency. (A) The representative dot plot demonstrates the GFP expression of HSV-GR infected 4T1 cells at a MOI of 1; 89.3% of infected cells demonstrate GFP expression. (B) Dot plot showing GFP expression coordinates of TC-1 cells infected with HSV-GR at MOI of 1; 86.1 percent of infected cells show GFP expression. (C) The representative dot plot showing GFP expression of CT26 cells infected with HSVGR at MOI of 1; 92.4% of infected cells demonstrate GFP expression.
Photo-micrography and flow cytometry data analysis demonstrated that all examined cell lines in this study are susceptible to HSV infections. After infection with HSV-GR, nearly all of the infected cells showed expression of both green (GFP) and red (BleCherry) colors, constantly (Fig. 3). As shown in Fig. 4A, 89.3% of the 4T1 cells representing high-level GFP expression in infection with recombinant virus at MOI of 1.
The similar pattern observed for TC-1 cells. Figure 3B shows the photo-microscopy of the infected TC-1 cells. As shown in Fig. 3B, both GFP and mCherry were continuously expressed in TC-1 cells. Flow cytometry analysis of the infected cells revealed that more than 86% of TC-1 cells were infected with HSV-GR at MOI of 1 (Fig. 4B). The CT26 cell demonstrated the same infection patterns. Photo-micrograph of infected CT26 cells shows that both GFP and mCherry were expressed in nearly all of the infected cells (Fig. 3C). Flow cytometry analysis demonstrates that more than 92% of CT26 cells expressed that more than 92% of CT26 cells expressed GFP 24 h after infections (Fig. 4C). Collectively, these findings indicated that our recombinant virus could efficiently infect and replicate in target cells. Finally, HSV-GR’s killing potency was measured by PI staining of infected cells (Fig. 5A-C). We observed that 27.9% of 4T1 cells (Fig. 5A), 21.2% of TC-1 cells (Fig. 5B) and 21.3% of CT26 (Fig. 5C) cells were stained with PI. The data analysis was performed with GraphPad PRISM and Statistical significance was determined with ANOVA (P < 0.05) (Fig. 5D).
Fig. 5.
In vitro analysis of the killing efficacy of herpes simplex virus green-red (HSV-GR) in 4T1, TC-1 and CT26 cells using flow cytometry. The percentage of dead cells was measured by PI staining of HSV-GR infected cells. PI fluorescence signal was gated at FL3 channel. (A) Histogram plot of PI stained control and HSV-GR infected 4T1 cells, 27.9% of infected cells were PI positive. (B) Histogram plot of control and HSV-GR infected TC-1 cells, 21.2% of infected cells were stained with PI. (C) CT26 control and infected cells were stained with PI, 21.3% of infected cells are PI positive. (D) The percentage of necrotic cells after HSV-GR infection was assayed. Each value represents the mean ± SD of three independent samples. ** And *** indicate significant differences (P ≤ 0.01, P ≤ 0.001, respectively) between 2 groups.
DISCUSSION
Currently, many kinds of cancers have remained as incurable diseases. Oncolytic viruses are emerging tools for eliminating tumor cells by replication within and inducing cell death without any adverse effects to normal cells. HSV is one of the most studied OVs approved by the Food and Drug Administration (FDA) for melanoma treatment lately (24,25).
Application of replication-competent oncolytic HSVs vectors for cancer therapy has been well documented in previous studies (13,26).
In the present study, we used a double fluorescent expressing oncolytic HSV-HSV-GR which was constructed through the deletion of both copies of γ34.5 copies and insertion of GFP and BleCherry coding sequence into deletion sites. This vector could be isolated easily by fluorescent microscopy or fluorescence-activated cell sorting for rapid isolation or enrichment of recombinant viruses (27).
HSV-GR could be applied as a platform for the construction of new generation of recombinant HSV, whereas one color could be replaced by therapeutic or immunostimulatory genes, the other one could serve as a tracking dye. Up to our best knowledge, this is the first report of replacing both copies of γ34.5 with two distinct fluorescent dyes which is used in three different murine tumor cell lines.
Our results are in line with those of previous reports that attenuated, replicationcompetent oncolytic HSVs can infect and destroy various tumor cell lines in vitro (10,20,28,29). It has been recently demonstrated that the deletion of γ34.5 genes attenuates the HSV-GR recombinant virus. Our recombinant HSV-GR yielded one log lower than wild-type HSV-1 on Vero cells due to inactivation of both copies of γ34.5 genes (Fig. 2).
In the present study, we evaluated the infectivity and cytotoxicity of our recombinant viruses -HSV-GR- on three mice syngeneic tumor cell lines, 4T1, TC-1, and CT26. We found that HSV-GR could efficiently infect and kill these cell lines within 24 h after infection. The infection potency of HSV-GR was evaluated with fluorescent microscopy and flow cytometry, as we described in methods section. Our data showed that nearly all infected cells expressed both GFP (green color) and mCherry (red color) constantly (Fig. 3). Afterwards, the percentage of infected cells was determined with flow cytometery. We observed that more than 90% of mentioned cells were infected with HSV-GR (Fig. 4). These findings demonstrate that all three examined cell lines were efficiently infected with HSV-GR. Moreover, we investigated the cytopathic effects of HSV-GR in three mentioned cell lines using PI staining. Consequently, our data demonstrated that HSV-GR could kill 27.9%, 22.1%, and 21.3% of 4T1, TC-1, and CT26 cells, respectively (Fig. 5).
Previous studies highlighted the fact that the growth and metastatic properties of 4T1 tumor models are similar to characteristics of the human breast cancers (16,28). We showed that HSV-GR could infect (more than 90%) and kill (27.9%) 4T1 cells 24 h post infection with HSV-GR at an MOI of 1. The results of this study support previous findings that oncolytic HSV can infect and kill 4T1 tumor cell lines. Thomas and Fraser demonstrated that HSV1716 (a double deleted γ34.5 inactivated oncolytic HSV) could infect and kill 4T1 tumor cells in vitro and in vivo (16).
Zhao et al. demonstrated that both oncolytic HSV-1 and oncolytic HSV-2 could efficiently infect and lyse 4T1 and TC-1 cells (14). They concluded that oncolytic HSV is a safe and effective therapeutic agent for 4T1 breast cancer. In agreement with them, we demonstrated that our HSV-GR could infect and lyse 4T1 and TC-1 cells. We observed that 27.9 % of 4T1 and 21.2% of TC-1 cells were killed 24 h after infection with oncolytic HSV-GR at an MOI of 1 (Fig. 4). Zhao et al. reported that oncolytic HSV1 and oncolytic HSV2 could infect about 50% of 4T1 and 100% of TC-1 cells. Consistent with their study, we demonstrated that HSV-GR could infect more than 86% of TC-1 cells. In our experiments, 89.3% of 4T1 cells were infected with HSV-GR that is higher than that was reported by Zhao et al. (14).
Nakamori et al. reported that Baco-1 (a GFP positive HSV) could only infect 20% of 4T1 cells at an MOI of 1 (30). In contrast with their study, our microscopic and flowcytometric data demonstrated that about 90% of 4T1 cells were infected with HSV-GR at an MOI of 1 (Figs. 3 and 4).
Toda et al. demonstrated that G207 (both γ34.5 genes and ICP6 genes were inactivated) could efficiently infect and kill CT26 cells at an MOI of 1 (31). The results of Toda et al. confirm our results since we demonstrated that HSV-GR could infect CT26 cells and express both GFP and BleCherry, respectively (Fig. 3). Moreover, HSV-GR killed 21.3% of infected CT26 cells (Fig. 5C).
Bennett et al. showed that NV1023 (a multi-mutant HSV-strain F) could replicate inside and kill CT26 cell in vitro. They concluded that NV1023 has the oncolytic potential to eliminate colon cancer cells (17). Consistent with their study, we showed that HSV-GR could infect and kill CT26 cell lines properly (Fig. 5C)..
Esaki et al. showed that the combination of HF10 (a highly attenuated HSV-1) and gemcitabine resulted in a considerable reduction of the growth of CT26 tumor cells in comparison with each of them, separately (32). They demonstrated that HSV could infect and lyse the CT26 cell. In accordance with Esaki et al., our work showed that HSV-GR could infect and lyse the CT26 cells, efficiently (Figs. 4 and 5).
CONCLUSION
As a conclusion, mouse tumor cell lines provide significant tools for investigation of oncolytic virotherapy efficacy. In this study, we showed that the HSV-GR recombinant virus could efficiently infect and eliminate three mice syngeneic cell lines. We have demonstrated that HSV-GR infects and replicates in 4T-1, CT26, and TC-1 cells. Moreover, PI staining results represented that HSV-GR could lyse infected cells within 24 h after infection. Further studies concerning HSV-GR oncolytic properties in vivo can shed more light on the potential of this anticancer approach.
ACKNOWLEDGEMENT
This study was a part of Ph.D thesis submitted by Shahriyar Abdoli, which was financially supported by Pasteur Institute of Iran, Tehran, I.R. Iran. The authors thank all the members of virology lab for their helpful consultation and technical assistance.
REFERENCES
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA: a cancer journal for clinicians. 2016;66(1):7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.DeSantis CE, Lin CC, Mariotto AB, Siegel RL, Stein KD, Kramer JL, et al. Cancer treatment and survivorship statistics, 2014. CA: a cancer journal for clinicians. 2014;64(4):252–71. doi: 10.3322/caac.21235. [DOI] [PubMed] [Google Scholar]
- 3.Friedman GK, Pressey JG, Reddy AT, Markert JM, Gillespie GY. Herpes Simplex Virus Oncolytic Therapy for Pediatric Malignancies. Mol Ther. 2009;17(7):1125–35. doi: 10.1038/mt.2009.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Singh PK, Doley J, Kumar GR, Sahoo A, Tiwari AK. Oncolytic viruses & their specific targeting to tumour cells. The Indian journal of medical research. 2012;136(4):571. [PMC free article] [PubMed] [Google Scholar]
- 5.Seymour LW, Fisher KD. Oncolytic viruses: finally delivering. Br J Cancer. 2016;114(4):357–61. doi: 10.1038/bjc.2015.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang F, Wang B-R, Wu Y-Q, Wang F-C, Zhang J, Wang Y-G. Oncolytic viruses against cancer stem cells: A promising approach for gastrointestinal cancer. World Journal of Gastroenterology. 2016;22(35):7999–8009. doi: 10.3748/wjg.v22.i35.7999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meerani S, Yao Y. Oncolytic viruses in cancer therapy. Eur J Sci Res. 2010;40(156171.3) [Google Scholar]
- 8.Argnani R, Lufino M, Manservigi M, Manservigi R. Replication-competent herpes simplex vectors: design and applications. Gene Ther. 2005;12(S1):S170–S7. doi: 10.1038/sj.gt.3302622. [DOI] [PubMed] [Google Scholar]
- 9.Speranza MC, Kasai K, Lawler SE. Preclinical Mouse Models for Analysis of the Therapeutic Potential of Engineered Oncolytic Herpes Viruses. ILAR journal. 2016;57(1):63–72. doi: 10.1093/ilar/ilw002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kanai R, Zaupa C, Sgubin D, Antoszczyk SJ, Martuza RL, Wakimoto H, et al. Effect of γ34. 5 deletions on oncolytic herpes simplex virus activity in brain tumors. Journal of virology. 2012;86(8):4420–31. doi: 10.1128/JVI.00017-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Y, Zhang C, Chen X, Yu J, Wang Y, Yang Y, et al. ICP34. 5 protein of herpes simplex virus facilitates the initiation of protein translation by bridging eukaryotic initiation factor 2α (eIF2a) and protein phosphatase 1. Journal of biological chemistry. 2011;286(28):24785–92. doi: 10.1074/jbc.M111.232439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Campadelli-Fiume G, De Giovanni C, Gatta V, Nanni P, Lollini PL, Menotti L. Rethinking herpes simplex virus: the way to oncolytic agents. Reviews in medical virology. 2011;21(4):213–26. doi: 10.1002/rmv.691. [DOI] [PubMed] [Google Scholar]
- 13.Falls T, Roy DG, Bell JC, Bourgeois-Daigneault M-C. Murine Tumor Models for Oncolytic Rhabdo-Virotherapy. ILAR journal. 2016;57(1):73–85. doi: 10.1093/ilar/ilv048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao Q, Zhang W, Ning Z, Zhuang X, Lu H, Liang J, et al. A Novel Oncolytic Herpes Simplex Virus Type 2 Has Potent Anti-Tumor Activity. PLOS ONE. 2014;9(3):e93103. doi: 10.1371/journal.pone.0093103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arulanandam R, Batenchuk C, Varette O, Zakaria C, Garcia V, Forbes NE, et al. Microtubule disruption synergizes with oncolytic virotherapy by inhibiting interferon translation and potentiating bystander killing. 2015;6:6410. doi: 10.1038/ncomms7410. [DOI] [PubMed] [Google Scholar]
- 16.Thomas DL, Fraser NW. HSV-1 therapy of primary tumors reduces the number of metastases in an immune-competent model of metastatic breast cancer. Molecular Therapy. 2003;8(4):543–51. doi: 10.1016/s1525-0016(03)00236-3. [DOI] [PubMed] [Google Scholar]
- 17.Bennett JJ, Malhotra S, Wong RJ, Delman K, Zager J, St-Louis M, et al. Interleukin 12 Secretion Enhances Antitumor Efficacy of Oncolytic Herpes Simplex Viral Therapy for Colorectal Cancer. Annals of Surgery. 2001;233(6):819–26. doi: 10.1097/00000658-200106000-00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fabiani M, Limongi D, Palamara AT, De Chiara G, Marcocci ME. A Novel Method to Titrate Herpes Simplex Virus-1 (HSV-1) Using Laser-Based Scanning of Near-Infrared Fluorophores Conjugated Antibodies. Frontiers in microbiology. 2017;8:1085. doi: 10.3389/fmicb.2017.01085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blaho JA, Morton ER, Yedowitz JC. Herpes simplex virus: propagation, quantification, and storage. Current protocols in microbiology. 2005;(Chapter 14) doi: 10.1002/9780471729259.mc14e01s00. Unit 14E.1. [DOI] [PubMed] [Google Scholar]
- 20.Abdoli S, Roohvand F, Teimoori-Toolabi L, Shokrgozar MA, Bahrololoumi M, Azadmanesh K. Construction of Various gamma34.5 Deleted Fluorescent-Expressing Oncolytic herpes Simplex type 1 (oHSV) for Generation and Isolation of HSV-Based Vectors. Iranian biomedical journal. 2017;21(4):206–17. doi: 10.18869/acadpub.ibj.21.4.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sanderson MJ, Smith I, Parker I, Bootman MD. Fluorescence Microscopy. Cold Spring Harbor Protocols. 2014;2014(10):pdb. doi: 10.1101/pdb.top071795. top071795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu H. Cell Cycle Analysis Using Propidium Iodide Staining with GFP Detection. Bio-protocol. 2012;2(7):e199. [Google Scholar]
- 23.Riccardi C, Nicoletti I. Analysis of apoptosis by propidium iodide staining and flow cytometry. Nature Protocols. 2006;1:1458. doi: 10.1038/nprot.2006.238. [DOI] [PubMed] [Google Scholar]
- 24.Yura Y. Presage of oncolytic virotherapy for oral cancer with herpes simplex virus. The Japanese dental science review. 2017;53(2):53–60. doi: 10.1016/j.jdsr.2016.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.DiGiulio S. FDA Approves First Oncolytic Virus Therapy—Imlygic for Melanoma. Oncology Times. 2015 [Google Scholar]
- 26.Peters C, Rabkin SD. Designing herpes viruses as oncolytics. Molecular Therapy—Oncolytics. 2015;2 doi: 10.1038/mto.2015.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Popov S, Mirshahidi S, Essono S, Song R, Wang X, Ruprecht RM. Generation of Recombinant Vaccinia Viruses via Green Fluorescent Protein Selection. DNA and Cell Biology. 2009;28(3):103–8. doi: 10.1089/dna.2008.0792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martuza RL, Malick A, Markert JM, Ruffner KL, Coen DM. Science. 5007. Vol. 252. New York, NY: 1991. Experimental therapy of human glioma by means of a genetically engineered virus mutant; pp. 854–6. [DOI] [PubMed] [Google Scholar]
- 29.Learmonth K, Braidwood L, Woll P, Conner J. Immune responses following intrapleural administration of oncolytic SEPREHVIR in patients with malignant pleural mesothelioma. Journal for immunotherapy of cancer. 2015;3(Suppl 2):P335. [Google Scholar]
- 30.Nakamori M, Fu X, Rousseau R, Chen S-Y, Zhang X. Destruction of nonimmunogenic mammary tumor cells by a fusogenic oncolytic herpes simplex virus induces potent antitumor immunity. Molecular Therapy. 9(5):658–65. doi: 10.1016/j.ymthe.2004.02.019. [DOI] [PubMed] [Google Scholar]
- 31.Toda M, Rabkin SD, Kojima H, Martuza RL. Herpes simplex virus as an in situ cancer vaccine for the induction of specific anti-tumor immunity. Human gene therapy. 1999;10(3):385–93. doi: 10.1089/10430349950018832. [DOI] [PubMed] [Google Scholar]
- 32.Esaki S, Goshima F, Kimura H, Murakami S, Nishiyama Y. Enhanced antitumoral activity of oncolytic herpes simplex virus with gemcitabine using colorectal tumor models. International Journal of Cancer. 2013;132(7):1592–601. doi: 10.1002/ijc.27823. [DOI] [PubMed] [Google Scholar]