Abstract
Objective
Mitochondria are organelles primarily responsible for energy production, and recent evidence indicates that alterations in size, shape, location, and quantity occur in response to fluctuations in energy supply and demand. We tested the impact of acute and chronic exercise on mitochondrial dynamics signaling and determined the impact of the mitochondrial fission regulator Dynamin related protein (Drp)1 on exercise performance and muscle adaptations to training.
Methods
Wildtype and muscle-specific Drp1 heterozygote (mDrp1+/−) mice, as well as dysglycemic (DG) and healthy normoglycemic men (control) performed acute and chronic exercise. The Hybrid Mouse Diversity Panel, including 100 murine strains of recombinant inbred mice, was used to identify muscle Dnm1L (encodes Drp1)-gene relationships.
Results
Endurance exercise impacted all aspects of the mitochondrial life cycle, i.e. fission-fusion, biogenesis, and mitophagy. Dnm1L gene expression and Drp1Ser616 phosphorylation were markedly increased by acute exercise and declined to baseline during post-exercise recovery. Dnm1L expression was strongly associated with transcripts known to regulate mitochondrial metabolism and adaptations to exercise. Exercise increased the expression of DNM1L in skeletal muscle of healthy control and DG subjects, despite a 15% ↓(P = 0.01) in muscle DNM1L expression in DG at baseline. To interrogate the role of Dnm1L further, we exercise trained male mDrp1+/− mice and found that Drp1 deficiency reduced muscle endurance and running performance, and altered muscle adaptations in response to exercise training.
Conclusion
Our findings highlight the importance of mitochondrial dynamics, specifically Drp1 signaling, in the regulation of exercise performance and adaptations to endurance exercise training.
Keywords: Mitochondrial dynamics, Drp1, Exercise performance, Exercise training
Highlights
-
•
Skeletal muscle expression of the mitochondrial fission regulator Dnm1L (encodes Drp1) is increased in mice and men during acute exercise.
-
•
DNM1L expression is reduced in muscle from dysglycemic vs. normoglycemic men at rest and during exercise.
-
•
Phospho-Drp1Ser616 (activation) is elevated in muscle from male and female mice during acute exercise and returns to baseline during post-exercise recovery.
-
•
Dnm1L expression is critical for exercise performance and muscle adaptations in response to endurance exercise training.
1. Introduction
Mitochondria are intracellular organelles responsible for the production of ATP by oxidative phosphorylation and contain maternally inherited DNA (16,569 base pairs) that code for two rRNAs, 22 tRNAs, and 13 polypeptides [1], [2], [3], [4]. The mtDNA-encoded polypeptides are all subunits of enzyme complexes of oxidative phosphorylation. However, the vast majority of mitochondrial proteins (numbering approximately 1500 in mammals) are encoded by the nuclear genome. Although originally viewed as solitary organelles, recent findings show a highly dynamic and connected mitochondrial network that adapts in size and morphology to changes in cellular environment [5], [6], [7]. This dynamic mitochondrial network possesses a life cycle including biogenesis, rearrangement of the network via fission-fusion, and removal of damaged or unneeded mitochondria by autophagic turnover, or mitophagy [8], [9]. These lifecycle processes appear interdependent and critical to overall mitochondrial health, as chronic alterations in a single aspect of the lifecycle contribute to network dysmorphology, impaired oxidative phosphorylation, and global metabolic dysfunction [8], [10], [11], [12], [13], [14], [15], [16], [17], [18], [19], [20], [21], [22], [23].
Although the process of fission-fusion dynamics, occurs on the order of minutes [24], [25] and is linked with longer-term mitochondrial quality control [26], [27], [28], [29] and mtDNA replication [30], more recent evidence shows that mitochondrial remodeling is also requisite for rapid shifts in substrate metabolism [31], [32], [33], [34], [35]. Signal transduction controlling mitochondrial remodeling to promote acute coupling of energy demand and supply is an area of intense investigation [36]. No one study has systematically interrogated the impact of acute exercise and long-term training on all three phases of the mitochondrial lifecycle (biogenesis, fission/fusion, and mitophagy). To fill these gaps in our knowledge, we examined how the mitochondrial life cycle responds to three different endurance exercise interventions: acute exercise, chronic exercise training, and acute exercise after chronic exercise training. This research led to the identification of a novel role for Drp1 during acute exercise which we explored further in a mouse model harboring a skeletal muscle-specific Drp1 heterozygous (mDrp1+/−) deletion. Our findings show that Drp1 is critical for regulating exercise performance and mitochondrial adaptations to chronic training.
2. Materials and methods
2.1. Human studies
Between the years of 2011–2012, 26 sedentary (<1 exercise session/week) men of Scandinavian origin from Oslo, Norway (aged 40–65 years), were recruited into the Skeletal Muscles, Myokines and Glucose Metabolism (MyoGlu) intervention trial and divided into the following groups: 1) normoglycemic (control) with BMI <27 kg/m2 (n = 13), or 2) dysglycemic (DG) with BMI 27–32 kg/m2 and either impaired fasting plasma glucose, impaired glucose tolerance or insulin resistance (based on HOMA-IR) (n = 13). Exclusion criteria included family history of diabetes (for controls only), known hypertension, liver or kidney disease, chronic inflammatory diseases, or on any medication expected to affect glucose metabolism (lipid lowering, anti-hypertensive, ASA, corticosteroids, etc.). More details have been described previously [37], [38]. Skeletal muscle biopsies (n = 154) were obtained from the vastus lateralis muscle during the bicycle tests. The 1st and 2nd skeletal muscle biopsies were obtained from the same incision site, but from opposite directions. After sterilization, the subcutaneous and superficial tissues were injected with Xylocain-adrenaline 10 mg/mL + 5 μg/mL. A 6 mm muscle biopsy needle (Pelomi, Albertslund, Denmark) was used with a 50 mL syringe for vacuum generation. Biopsies were quickly rinsed in cold PBS and dissected on a cold aluminum plate to remove blood etc. before freezing. The MyoGlu study design is presented in Figure 1A.
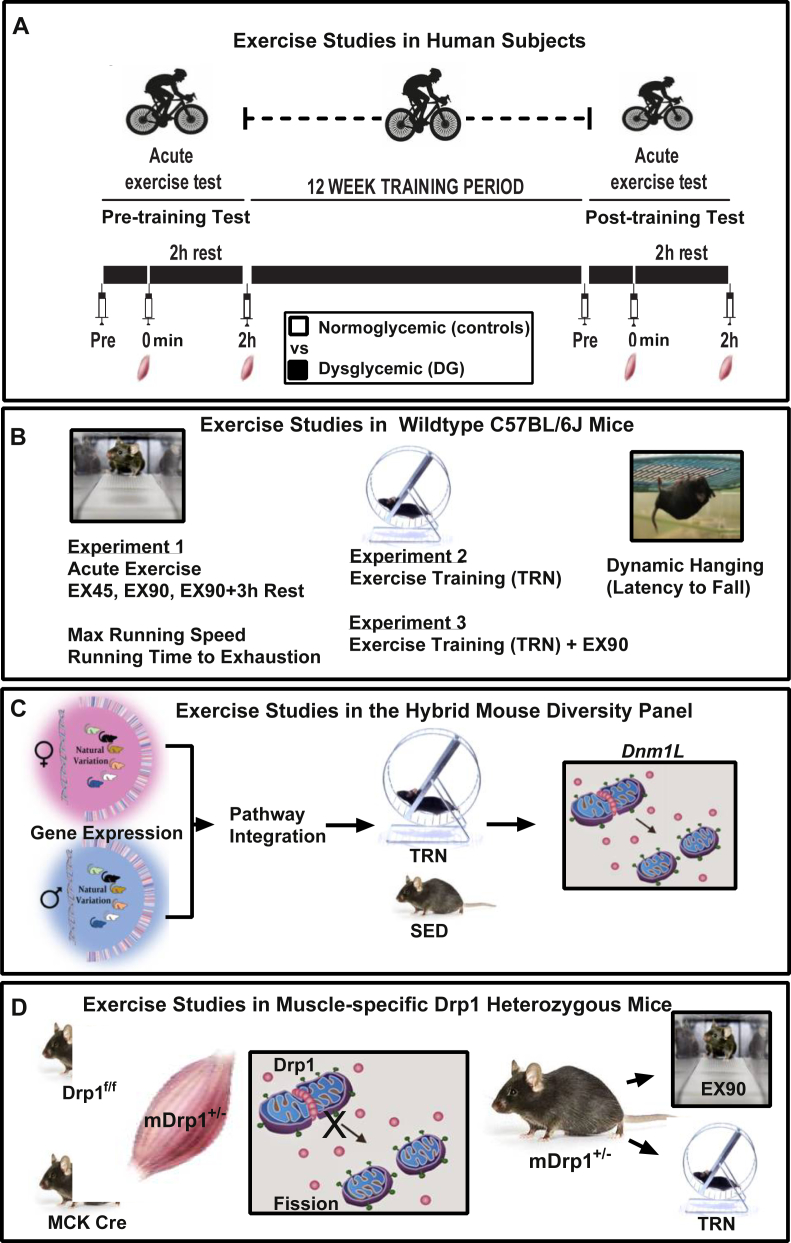
Figure 1.
Schematic depicting exercise protocols for humans and mice. (A) Acute and chronic cyclying exercise was performed by healthy normoglycemic (control) and Dysglycemic (DG) men, age 40–65 years (N = 13/group). (B) Wildtype mice were studied during acute (EX45, EX90, EX90 + 3 h Rest) and chronic exercise (TRN, TRN + EX90) experiments 1–3. Exercise training (TRN) was achieved using voluntary running wheels for 30 days. Muscle was harvested 30 h after the last bout of exercise. An additional cohort of TRN mice performed an acute bout of moderate intensity treadmill running (TRN + EX90). (C) A panel of recombinant inbred strains of mice (84 strains of male and female mice, N = 4 mice/strain and a 15-strain subset of female mice, N = 4 mice/strain), the UCLA HMDP, were studied to determine the relationship between sex and Dnm1L expression and Dnm1L gene–gene relationships. (D) Muscle-specific Drp1 heterozygous (mDrp1+/−) mice were studied at rest and following EX90 and TRN.
2.2. Animal studies
2.2.1. Exercise protocols
Female wildtype C57BL/6J mice 4–6 months of age were obtained from Jackson Laboratories and separated into one of three groups prior to experimentation to ensure no statistical differences in body weight between groups. An additional cohort of male C57BL/6J mice (N = 6 mice/group, SED vs. EX90) and three cohorts of female mice representing mouse strains of varying running capacity BALB/CJ (high capacity running), A/J (medium capacity running), and C3H/HeJ (low capacity running) were studied (N = 6 mice/group, SED vs. EX90) to confirm the absence of a sex and strain dependence on acute exercise signaling, with specific focus on Drp1 phosphorylation status. Mice were group housed and had free access to standard laboratory food (Teklad 8604, Calories: 25% protein, 14% fat, 54% carbohydrate) and water unless otherwise indicated. Each exercise group was matched with an independent control sedentary group (SED) (Figure 1B).
2.2.1.1. Experiment one (acute endurance exercise)
Experimental groups consisted of: sedentary (SED), 45 min exercise (EX45), 90 min exercise (EX90 cohort 1), and 90 min exercise + 3 h sedentary recovery (EX90+3 h Rest) (Figure 1). The moderate intensity endurance exercise protocol included treadmill running (5% grade) at 15 m/min for the indicated times (Figure 1B) [39], [40]. All mice were acclimated to the treadmill by running for 10 min at 5–10 m/min on two separate occasions during the 2–4 days prior to experimentation. Mice in the SED groups were fasted for 6 h prior to tissue harvest. Mice in the EX45, EX90, and EX90 + 3 h Rest groups were fasted for 3 h prior to exercise. Mice in the EX90 + 3 h Rest group were given ad libitum access to water during the 3 h post exercise recovery period. All mice were given ad libitum access to water during the fasting period. Tissues were removed immediately following exercise completion or at the indicated post-exercise time, snap-frozen in liquid nitrogen, and stored at −80 °C until analysis.
2.2.1.2. Experiment two (TRN)
Experimental groups: sedentary control (SED) or 30 days of in cage voluntary wheel running (TRN). All animals were individually housed during the exercise training period. Wheels in cages of SED mice were locked throughout the experiment. Daily running activity and weekly body weights were recorded for 30 consecutive days. After 30 days of TRN, wheels were locked for 30 h prior to euthanasia and tissue harvest (after 6 h fasting). Tissues were snap-frozen in liquid nitrogen and stored at −80 °C until analysis (Figure 1B).
2.2.1.3. Experiment three (TRN + EX90)
Experimental groups included: sedentary control (SED), endurance exercise for 90 min (EX90 cohort 2), and 30 days in cage voluntary wheel running followed by 90 min of acute endurance exercise (TRN + EX90). All mice involved in experiment 3 followed the same protocol as mice in experiment 2. Five days prior to completion of the 30-day voluntary wheel running program, mice in the EX90 and TRN + EX90 groups performed a maximum treadmill running test. Prior to the maximum running test, mice were acclimated to the treadmill by running for 10 min at 5–15 m/min on two separate occasions. After 30 days voluntary wheel running, wheels were locked for the TRN + EX90 group. On the following day, SED mice fasted for 6 h, whereas EX90 and TRN + EX90 mice fasted for 3 h before completion of a single bout of moderate intensity endurance exercise. The moderate intensity endurance exercise protocol consisted of 90 min of treadmill running (5° incline) at 15 m/min or 18–20 m/min for the EX90 and TRN + EX90 groups respectively, so as to maintain the same relative exercise intensity between the groups [40], [41]. At the completion of the exercise study, animals were euthanized and tissues immediately removed, snap-frozen in liquid nitrogen, and stored at −80 °C until analysis (Figure 1B).
2.2.2. Hybrid Mouse Diversity Panel (HMDP)
All mice were obtained from The Jackson Laboratory and bred at the University of California, Los Angeles. Mice were maintained on a chow diet (Ralston Purina Company) until 8 weeks of age when they either continued on the chow diet or were provided a high fat/high sucrose diet (HF/HS Research Diets D12266B; 8 weeks) with the following composition: 16.8% kcal protein, 51.4% kcal carbohydrate, 31.8% kcal fat. A complete list of the strains included in our study is included in Supplemental Table 1. This resource was established to provide a platform for high resolution genome wide mapping and systems level analysis of gene–gene and gene–trait relationships. Because DNM1L expression levels were induced in human subjects following exercise training, herein and in previous reports [42], we determined Dnm1L gene correlations from muscle of male and female high fat/high sucrose-fed HMDP animals [43]. In addition, a 15-strain subset of female HMDP mice underwent an exercise training protocol as described in experiment 2 (Figure 1C).
2.2.3. Muscle-specific Drp1 heterozygous mice (mDrp1+/−)
Floxed Dnm1L mice were a generous gift from Hiromi Sesaki, Johns Hopkins University [44]. Floxed mice were mated with a transgenic line in which Cre recombinase was driven by the muscle creatine kinase (MCK) promoter as previously described [45]. Female f/f Control and muscle-specific heterozygote (mDrp1+/−) mice were fed a chow diet and studied at basal/sedentary (SED) and following acute exercise (EX90) and chronic exercise (TRN) (Figure 1D). Muscle heterozygotes were utilized because the reduction in Drp1 total protein is more physiologic recapitulating findings for mouse models of insulin resistance that we have studied previously [46]. Moreover, in contrast to the muscle Drp1 homozygotes, the heterozygotes maintain muscle mass with aging. Experimental groups included pre-training (pre-TRN) vs. post-training (post-TRN) for both genotypes, f/f control vs. mDrp1+/− (Figure 1D). Mice completed a treadmill run to exhaustion both prior to start and after completion of the training protocol. Before each run to exhaustion test, mice were acclimated to treadmill running as described previously. The remainder of the experimental period was the same as described for experiment 2 (Exercise training).
Animal studies were approved by the University of California, Los Angeles Institutional Animal Care and Use Committee. All animal care, maintenance, surgeries, and euthanasia were conducted in accordance with this Institutional Animal Care and Use Committee and the National Institute of Health.
2.3. Maximum running speed, dynamic hanging, and run to exhaustion testing
Mouse genotypes were blinded to the experimenter for all tests. Maximal running speed test was performed as described previously [47]. Briefly, mice were acclimated to treadmill running on two separate occasions prior to the maximum running speed test. On testing day, mice were given a 5-min warm-up at 5–10 m/min. The treadmill speed was increased by 3 m/min until mice were unable to maintain the speed for 10 consecutive sec following gentle encouragement if required. Mice were given three attempts at each speed and approximately 60 s of rest after each increase in treadmill speed. Latency to fall test, used as an index of grip strength and muscle endurance, was performed as previously described [48]. Mice were acclimated to the wire grid on two separate occasions prior to testing. Mice performed three trials and the data from the three trials were averaged and reported as means ± SEM. Mice were given five min of rest between each trail. Run to exhaustion was performed as described previously [49]. Briefly, mice were acclimated to treadmill running on two separate occasions, and prior to the test a 5 min warm-up was followed by an incremental increase in treadmill speed (3 m/min) starting from 10 m/min (fixed 5° incline). Running speed intervals were 3 min in length, and the test was terminated when mice could no longer perform the running exercise (indicated by >5 s of inactivity on the resting grid).
2.4. Immunoblot analysis
Whole quadriceps muscle from both legs were pulverized together into a powder while frozen in liquid nitrogen. A homogenous sample of pulverized muscle was used for immunoblotting [50]. Proteins from each individual whole cell homogenate were normalized (expressed relative to the pixel densitometry) to glyceraldehyde 3-phosphate dehydrogenase (GAPDH, AM4300, Ambion). Phosphorylation-specific proteins were normalized (expressed relative to pixel densitometry) to the same unphosphorylated protein (e.g. phosphorylated Drp1 at Ser 616 was expressed relative to the pixel densitometry of Drp1 for each individual sample). Primary antibodies included: Mitochondrial Fission Protein 1 (Fis1, GTX111010, GeneTex), Mitochondrial fission factor (MFF, ab81127, Abcam), Mitofusin 1 (MFN1, 75–162, NeuroMab), Mitofusin2 (MFN2, ab56889, Abcam), Transcription factor A, Mitochondrial (TFAM, ab131607, Abcam), Dynamin-related protein 1 (Drp1, 8570, Cell Signaling), phosphorylated Drp1Ser616 (4494, Cell Signaling), phosphorylated Drp1Ser637 (6319, Cell Signaling), OxPhos Complex I to V (ab110413, Abcam), Optic atrophy 1 (Opa1, 612606, BD Biosciences), PTEN Induced Putative Kinase 1 (Pink1, 10006283, Cayman Chemical), Parkin (2132, Cell Signaling), phosphorylated ParkinSer65 (ab154995, Abcam), Microtubule-associated proteins 1A/1B light chain 3B (LC3B, 2775, Cell Signaling), Sequestosome 1 (p62, 5114, Cell Signaling), Beclin-1 (3738, Cell Signaling), Autophagy Related 7 (ATG7, 8558, Cell Signaling), Protein Kinase A (PKA, 4782, Cell Signaling), AMP-Activated Protein Kinase α (AMPKα, 2793, Cell Signaling), Unc-51 Like Autophagy Activating Kinase 1 (Ulk1, 8054, Cell Signaling), Regulator of Calcineurin (RCAN, SAB2101967, Sigma–Aldrich), Calcineurin A (CalA, 2614, Cell Signaling), Calcineurin B (CalB, ab94535, Abcam), phospho-Ulk1Ser555 (5869, Cell Signaling), Autophagy Related 10 (ATG10, PA5-20454, ThermoFisher), phospho-AMPKαThr172 (50081, Cell Signaling), DJ1 (2134, Cell Signaling), PGC1α (2178, Cell Signaling), and phospho-PKAThr197 (5661, Cell Signaling). In certain instances, membranes probed for phospho-proteins were stripped and reprobed for total protein. In Figure 2B top panel, the Drp1 immunoblot is duplicated in Supplemental Figure 1 to confirm equal loading of protein for the p-Drp1Ser637 immunoblot. In the interest of space, most representative immunoblots are presented in the supplement.
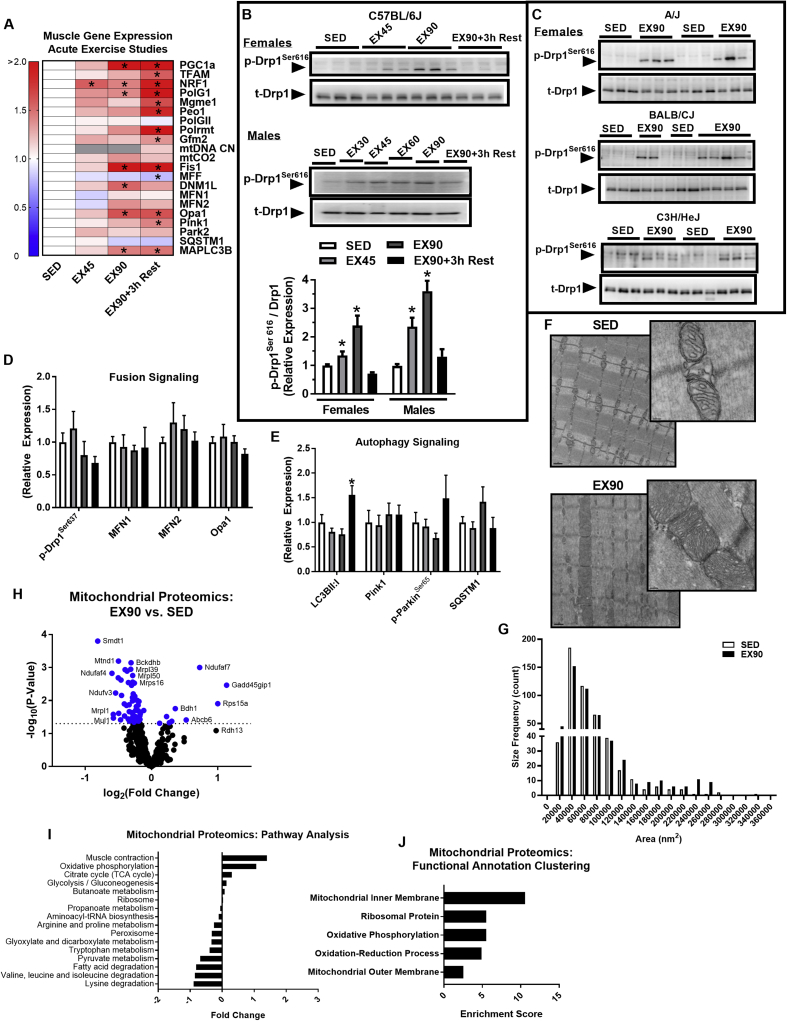
Figure 2.
A single session of moderate intensity exercise increases pro-fission phosphorylation of Drp1 in a time dependent manner and alters the mitochondrial proteome. (A) Quadriceps mRNA expression (presented in a heat map), representing mitochondrial fission, fusion, and autophagy signaling, in SED, EX45, EX90, and EX90+3 h Rest (N = 6/group; one-way ANOVA with Tukey post hoc analysis). (B–E) Immunoblot densitometry of quadriceps protein and phospho-protein abundance of mitochondrial fission, fusion, and autophagy signaling, in SED (open bars), EX45 (light gray bars), EX90 (dark gray bars), and EX90+3 h Rest (closed black bars) for C57BL/6J (N = 6/group; additional representative immunoblots presented in Supplemental Figure 1; one-way ANOVA with Tukey's post hoc analysis). Muscle Drp1Ser616 phosphorylation in (A) male and female C57BL/6J (N = 6 mice/time point), and (C) female A/J, BALB/CJ, and C3H/HeJ mice (N = 6 mice/group). (F) Transmission electron micrograph (TEM) images of mouse soleus muscle from SED and EX90 groups. (G) Frequency of mitochondrial area (nm2) determined from the TEMs obtained from SED and EX90 muscle (494 mitochondria visualized/group; Kolmogorov–Smirnov test for cumulative distribution comparison, P = 0.52). (H) Mitochondrial proteomics of gastrocnemius muscle represented in a volcano plot for EX90 vs. SED groups. Dashed line indicates significance threshold and blue dots indicate significance SED vs. EX90 (two-tailed unpaired t-test with Bonferroni correction). (I) Mitochondrial proteomics pathway analysis. (J) Mitochondrial proteomics functional annotation of significantly impacted protein clusters. Data are means ± SEM (N = 6/group). *, P < 0.05 vs. SED.
2.5. DNA & RNA extraction, cDNA synthesis, quantitative RT-PCR, and microarrays
DNA and RNA were extracted from a homogenous portion of frozen quadriceps muscle homogenate using DNeasy/RNeasy Isolation kits (Qiagen) as described by the manufacturer. Isolated DNA and RNA was tested for concentration and purity using a NanoDrop Spectrophotometer (Thermo Scientific). Isolated RNA was converted into cDNA, checked for purity, and qPCR of the resulting cDNA levels was performed as previously described [50]. All genes were normalized to the housekeeping gene Ppia or 18S. Mitochondrial DNA content was assessed as a ratio of mitochondrial DNA (mtCO2) to nuclear DNA (SDHA). See Supplemental Table 2 for a list of qPCR primers.
Total RNA from HMDP mouse muscle (211 females, 228 males) was hybridized to Affymetrix HT_MG-430A arrays and scanned using standard Affymetrix protocols. To reduce the chances of spurious association results, RMA normalization was performed after removing all individual probes with SNPs and all probesets containing 8 or more SNP-containing probes, which resulted in 22,416 remaining probesets. To determine the accuracy of our microarray data, we tested by qPCR the expression of a dozen genes and found an r = 0.7 between qPCR and microarray.
Frozen human muscle biopsies were cooled in liquid nitrogen and crushed to powder by a pestle in a liquid nitrogen-cooled mortar as described by Langleite et al. [51]. Frozen biopsies were transferred into 1 mL QIAzol Lysis Reagent (Qiagen, Hilden, Germany) and homogenized using TissueRuptor (Qiagen) at full speed for 15 s, twice. Total RNA was isolated from the homogenate using RNeasy Mini Kit (Qiagen). RNA integrity and concentration were determined using Agilent RNA 6000 Nano Chips on a Bioanalyzer 2100 (Agilent Technologies Inc, Santa Clara, CA). Using High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster, CA), 200 ng of total RNA was converted to cDNA for TaqMan real-time RT-PCR. The cDNA reaction mixture was diluted in water and cDNA equivalent of 25 ng RNA used for each sample. Quantitative real-time PCR was performed with reagents and instruments from Applied Biosystems in the 96-well format using a 7900HT Fast instrument and the SDS 2.3 software (Applied Biosystems). A predeveloped primer and probe set (TaqMan assays; Applied Biosystems) was used to analyze mRNA levels of Dynamin Related Protein 1 (DNM1L) and control gene Peroxisome Proliferator-Activated Receptor Gamma, Coactivator 1 Alpha (PPARGC1A, Hs01016719_m1). Relative target mRNA expression levels were calculated as 2−ΔCt, and normalized to beta-2 microglobulin (B2M, Hs00984230_m1) [52]. mRNA sequencing was performed using the Illumina HiSeq 2000 system (Illumina, San Diego, CA). cDNA sequenced reads alignment was carried out using Tophat v2.0.8. Reads counted by gene feature were performed by featureCounts in Rsubread 1.14.2.
2.6. Plasma lactate
Plasma lactate was determined using the Eton Bioscience (San Diego, CA) l-Lactate Assay Kit I protocol Version 7. Briefly, immediately following euthanasia and prior to tissue removal, whole blood was removed via 27-gauge needle from the abdominal aorta and centrifuged at 2,000 x G for 2 min in EDTA-coated tubes. Plasma was removed and a portion was diluted 1:10 in MiliQ H2O before performing the lactate assay as per manufacturer's instructions.
2.7. Muscle glycogen
Muscle glycogen was assessed in excised quadriceps muscle using the following protocol adapted from [53]. Briefly, ∼100 mg of gastrocnemius muscle was weighed and cut into small pieces. 1 mL of 30% KOH was added and boiled for ∼25 min. Tubes were cooled to room temperature after which 2 mL of 95% ethanol was added. Tubes were incubated for 30 min on ice, centrifuged at 550 x G for 30 min at 4 °C, and the resulting supernatant was removed. Pellets were dissolved in 1 mL of MiliQ H2O after which 1 mL of 5% phenol was added. Samples were further diluted 2:3 in MiliQ H2O and 5 mL of 96–98% H2SO4 was added. Following incubation on ice for 30 min, the optical density was measured at 490 nm. Concentrations were determined using an eight-point glycogen standard curve and normalized to mg of tissue weight assayed.
2.8. Tissue histology
Gastrocnemius muscles from both Control and mDrp1+/− SED and TRN animals were sectioned and stained for hematoxylin and eosin or cytochrome c oxidase (COX) as previously described [54]. Muscle fiber area, nuclei number, and fiber number were counted or measured using ImageJ software.
2.9. Mitochondrial isolation & proteomic analysis
Mitochondria were isolated from gastrocnemius muscle using a Dounce homogenizer and Mitochondria Isolation Kit for Tissue (Thermo Scientific) with a Percoll density method for added purification [55]. Isolated mitochondria were lysed using bath sonication and a buffer containing: 0.5% sodium deoxycholate, 12 mM sodium lauroyl sarcosine, and 50 mM triethyl ammonium bicarbonate (TEAB). Samples were treated with tris (2-carboxyethyl) phosphine, chloroacetamide, and incubated overnight with Sequencing Grade Modified Trypsin (Promega, Madison, WI) following addition of equal volume ethyl acetate/trifluoroacetic acid. Supernatants were discarded after centrifugation and the resulting phase was desalted on a C18-silica disk (3M, Maplewood, MN) according to Rappsilber's protocol [56]. The collected eluent was chemically modified using a TMT10plex Isobaric Label Reagent Set (Thermo Fisher Scientific) as per the manufacturer's protocol and an aliquot was taken for measurement of total peptide concentration (Pierce Quantitative Colorimetric Peptide, Thermo Fisher Scientific). The samples were then pooled according to protein content and fractionated via high pH reversed-phase chromatography using a 1260 Infinity LC System (Agilent Technologies, Santa Clara, CA) and a ZORBAX 300 Extend-C18 column (Agilent Technologies, 0.3 × 150 mm, 3.5 μm). Twelve fractions were injected onto a reverse phase nanobore HPLC column (AcuTech Scientific) using an Eksigent NanoLC-2D system (Sciex, Framingham, MA). The effluent from the column was directed to a nanospray ionization source connected to a hybrid quadrupole-Orbitrap mass spectrometer (Q Exactive Plus, Thermo Fisher Scientific) acquiring mass spectra in a data-dependent mode alternating between a full scan (m/z 350–1700, automated gain control (AGC) target 3 × 106, 50 ms maximum injection time, FWHM resolution 70,000 at m/z 200) and up to 10 MS/MS scans (quadrupole isolation of charge states ≥ 2, isolation width 1.2 Th) with previously optimized fragmentation conditions (normalized collision energy of 32, dynamic exclusion of 30 s, AGC target 1 × 105, 100 ms maximum injection time, FWHM resolution 35,000 at m/z 200). The raw data were analyzed in Proteome Discoverer 2.2, providing measurements of peptide relative abundance. Downstream analysis was performed using DAVID Bioinformatics Resources and Ingenuity Pathway Analysis.
2.10. Statistical analysis
Values are presented as means ± SEM and expressed relative to the respective control group. Group differences were assessed by one-way ANOVA followed by Tukey's Honest Significant Difference post hoc test, Student's t-test, or two-way ANOVA where appropriate. Histogram representing mitochondrial area distribution between SED vs. EX90 was assessed by Kolmogorov–Smirnov test. Statistical significance was established a priori at P < 0.05 (Graph Pad Prism 7.0).
3. Results
3.1. The impact of acute exercise on markers of the mitochondrial life cycle
We used a variety of exercise protocols to challenge mouse muscle in order to determine the impact on acute mitochondrial signaling and chronic mitochondrial adaptation. As expected, acute exercise for 90 min (EX90) induced expression of regulators of mitochondrial biogenesis and this elevation was sustained into the 3 h rest period following acute exercise (EX90+3h Rest; P < 0.05, Figure 2A). Pgc1a, NRF1, and Polg1 were all significantly induced during acute exercise and this robust expression profile was durable into the 3 h recovery period. By comparison, muscle expression of Tfam, Peo1, Polrmt, and Gfm2 genes associated specifically with mtDNA replication and transcription, were significantly induced in the recovery period. Despite the increase in expression of the mitochondrial biogenesis gene profile, we did not detect an increase mtDNA copy number or mtCO2 expression, a gene transcribed by the mitochondrial genome (P > 0.05, Figure 2A), until the exercise stimulus was chronic as with training.
Similar to markers of mitochondrial biogenesis, regulators of mitochondrial fission, Fis1 (outer mitochondrial membrane bound Drp1 docking protein) and Dnm1L (encodes the protein Drp1) were upregulated in muscle of mice after EX90, although only Fis1 remained elevated during the 3 h recovery period, Dnm1L returned to baseline (EX90+3 h Rest) (P < 0.05, Figure 2A). In contrast to Fis1, Mff expression was reduced during acute exercise and this reduction in expression reached statistical significance during the 3 h recovery period (EX90+3 h Rest) (P < 0.05, Figure 2A). Expression of outer mitochondrial membrane fusion markers Mfn1 and Mfn2 were unchanged by exercise (P > 0.05, Figure 2A). However, muscle expression of the internal mitochondrial membrane cristae remodeling regulator Opa1 was increased by EX90, and robustly maintained during exercise recovery (EX90+3h Rest; P < 0.05, Figure 2A). General markers of mitophagy and autophagy, Pink1 and Mapcl3b, respectively, were both increased only during the recovery period from EX90 (EX90+3h Rest) (P < 0.05, Figure 2A).
In contrast to the mRNA response to exercise, the protein level of all regulators of the mitochondrial life cycle remained unchanged during exercise and during the acute post-exercise recovery period (P > 0.05, Figure 2B–D, Supplemental Figure 1A). Because we observed a sharp contrast in the response of Dnm1L mRNA vs. Drp1 protein during exercise, we investigated the activation state of the protein. We immunoblotted the phosphorylation status at Ser616, (purported activation site), and found that it was robustly elevated in muscle from both female and male mice during EX45-EX90, but returned to baseline SED levels during post-exercise recovery (EX90 + 3h Rest) (P < 0.05, Figure 2B). To confirm that the exercise-induced phosphorylation of Drp1Ser616 is robust and reproducible, we immunoblotted muscle homogenates from 3 additional strains of female mice with varied running capacity (BALB/CJ high, A/J medium, and C3H/HeJ low) following 90 min of treadmill running (EX90). In each of the 3 strains, Drp1Ser616 was markedly elevated over sedentary control (Figure 2C). In contrast to the increase in fission signaling, we detected no change in pro-fusion markers including Drp1Ser637 (purported inhibitory site [57], [58]), Mfn 1/2, and OPA1 during exercise or after 3 h exercise recovery (Figure 2D; Supplemental Figure 1A).
Because damaged or unneeded mitochondria are eliminated by autophagic-lysosomal degradation, we assessed macro- and micro-autophagy (specifically mitophagy) signaling. Macroautophagy, commonly reflected as a ratio of LC3BII:LC3BI (LC3B processing includes protein cleavage and lipidation), was elevated in muscle only during the 3 h recovery period (EX90+3h Rest) (P < 0.05, Figure 2E and Supplemental Figure 1A). Although we observed a shift in the molecular weight distribution of the outer mitochondrial membrane (OMM) potential-sensitive Ser–Thr kinase PINK1 at EX90 and during the recovery period, the difference from SED did not reach statistical significance (Supplemental Figure 1A). No difference in Parkin or p-Parkin was detected during exercise or after a 3 h exercise recovery period (Figure 2E and Supplemental Figure 1A).
Electron micrograph images show that acute exercise induces a remarkably diverse response in mitochondrial morphology by location, presumably the consequence of regional energy demand (Figure 2F). After 90 min of exercise, some mitochondria along specific areas of the muscle z-line (intermyofibrillar mitochondria comprise 80% of the total network) appear smaller and more round with dense cristae architecture, whereas in other areas, mitochondria are enlarged with enhanced mitochondria–mitochondria contacts, or electron dense contacts (EDCs) as previously observed by Picard et al. [59]. The diversity in response may indicate that there are mitochondrial sub-populations in skeletal muscle with altered morphology to enable specific processes in response to changing energy supply and demand that occurs during activity [31], [60]. In aggregate and congruent with findings by Picard et al. [59], we observed no difference in the distribution of mitochondrial area between the groups, SED vs. EX90 (Figure 2G; P = 0.52).
To obtain a global picture of the mitochondrial proteome during acute exercise, we performed mass spectroscopy analysis on mitochondria from gastrocnemius muscle from female C57BL/6J mice (SED vs. EX90). Interestingly, the expression of the majority of proteins altered by EX90 were reduced, 69 of 550 (P < 0.05, Figure 2H). Although analysis revealed alterations in a number of pathways, none reached statistical significance (P > 0.05, Figure 2I). Functional annotation classification revealed proteins involved in basic mitochondrial processes and sub-cellular localization as significantly impacted by acute treadmill running, EX90 (Figure 2J). Mitochondrial proteins significantly reduced by acute exercise (N = 60) included Smdt1 (calcium uniporter, Single-Pass Membrane Protein With Aspartate Rich Tail 1) and mtND1 (NADH Dehydrogenase, Subunit 1, Complex 1). Of the 9 mitochondrial proteins significantly increased by EX90, Ndufaf7 (NADH:Ubiquinone Oxidoreductase Complex 1 Assembly Factor 7) and Bdh1 (3-Hydroxybutyrate Dehydrogenase 1) were most notable (Figure 2H).
3.2. Impact of exercise training on mitochondrial life cycle markers and proteome
As expected, exercise training (TRN, average distance 6.81 ± 0.32 km/day) promoted body weight reduction compared to pre-training body weight (P < 0.05, Pre-TRN = 22.3 ± 0.4 g, Post-TRN = 20.4 ± 0.7 g; Figure 3A). Inguinal and ovarian (gonadal) adipose tissue weight was reduced following TRN as compared to SED, whereas heart weight was increased by TRN as previously shown (P < 0.05, Figure 3A right panel). No difference in muscle weight was detected between the SED control and TRN groups, although these were two independent cohorts of littermate mice.
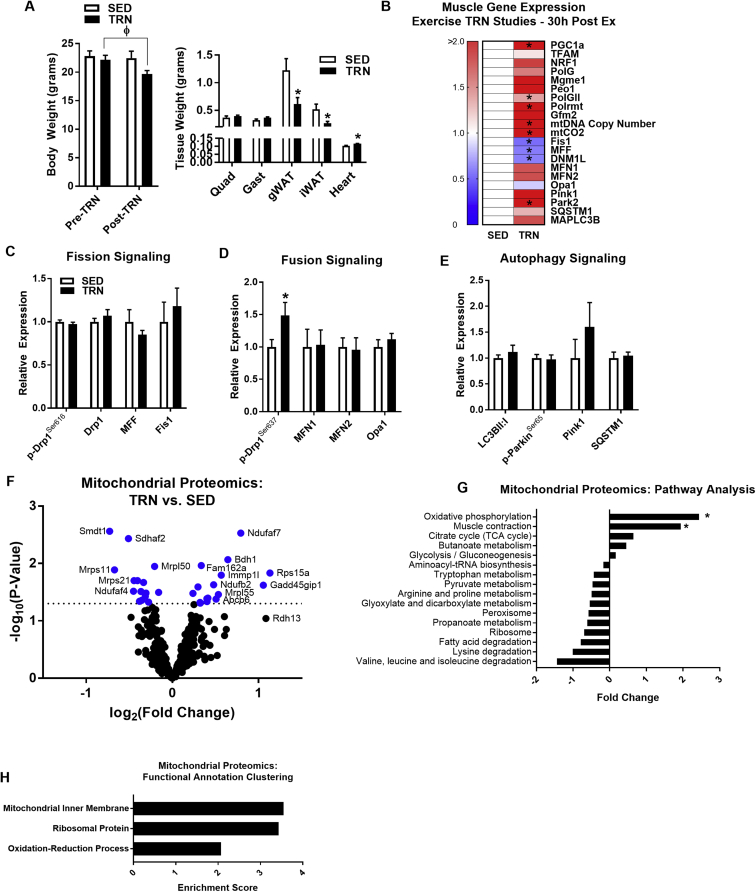
Figure 3.
Chronic exercise training increases pro-fusion phosphorylation of Drp1 in post-exercise recovery period (Experiment 2). (A) Body and tissue weights (quadriceps, gastrocnemius, gonadal WAT, inguinal WAT, and heart) in SED and TRN female mice (N = 6/group; two-way repeated measures ANOVA with Tukey post hoc analysis for left panel and; two-tailed unpaired t-test for right panel). (B) Quadriceps mRNA abundance (presented in a heat map), representing mitochondrial fission, fusion, and autophagy signaling, in SED vs. TRN (N = 6/group; two-tailed unpaired t-test). Muscles were harvested from TRN mice 30 h following the cessation of exercise including a 6 h fast. (C–E) Immunoblot densitometry of quadriceps protein and phospho-protein abundance representing mitochondrial fission, fusion, and autophagy signaling, in SED (open bars) and TRN (closed bars) mice (N = 6/group) (representative immunoblots presented in Supplemental Figure 1). (F) Mitochondrial proteomics determined by mass spectrometry analysis of gastrocnemius muscle and presented as a volcano plot (dashed line indicates significance threshold; blue dots indicate significance, N = 4/group; two-tailed unpaired t-test with Bonferroni correction). (G) Mitochondrial proteomics pathway analysis (two-tailed unpaired t-test), and (H) mitochondrial functional annotation of significantly impacted protein clusters. Data are means ± SEM. *, P < 0.05 vs. SED.
At the end of the 30 d exercise training period, running wheels were locked. Animals were euthanized 30 h after the wheels were locked (6 h after food removal), so as to avoid the effects of the last exercise bout. In muscle, several, but not all, transcripts associated with mitochondrial biogenesis were robustly upregulated following TRN (Figure 3B) including Pgc1a (↑2.7-fold), PolgII (↑1.4-fold), and Polrmt (↑12.3-fold) compared with SED (P < 0.05, Figure 3B). The copy number of mtDNA (↑86%; P = 0.026) and the expression of the mitochondrial transcribed gene mtCO2 were also significantly higher in TRN vs SED animals (P < 0.05, Figure 3B). Gene expression levels of regulators of mitochondrial fission were reduced 30 h after exercise in TRN animals (P < 0.05, Figure 3B), whereas expression levels of mitochondrial fusion, mitophagy, and autophagy genes were elevated but most failed to reach significance (P > 0.05, Figure 3B). The protein levels of mitochondrial fission and fusion markers were also assessed by immunoblotting. Similar to acute exercise, the protein levels of regulators of the mitochondrial life cycle were unchanged by exercise training (P > 0.05, Figure 3C–E, Supplemental Figure 1B). However, we observed that the phosphorylation state of Drp1 at Ser637, a post-translational modification purported to reduce Drp1 translocation and mitochondrial fission, was elevated in TRN animals (P < 0.05, Figure 3D), suggesting a possible TRN-induced shift in mitochondrial dynamics in the post-exercise recovery phase.
The mitochondrial proteome in gastrocnemius muscle from female mice was assessed following training, and in contrast to acute treadmill running, long-term volitional wheel running promoted a marked upregulation of mitochondrial proteins over that observed during acute exercise (P < 0.05, Figure 3F–H). Overall, fewer proteins were diminished after acute exercise following TRN compared to acute exercise of previously SED animals (Figure 2, Figure 3). Thus, our findings support the notion that TRN may enhance mitochondrial proteostasis [61]. Pathway analysis revealed a number of functional categories altered by TRN, including oxidative phosphorylation as the most highly induced by TRN (P < 0.05, Figure 3G). As would be predicted, functional annotation classification revealed that chronic exercise impacted proteins involved in basic mitochondrial processes and sub-cellular localization (Figure 3H).
3.3. Impact of TRN on acute exercise performance and mitochondrial dynamics signaling
To determine the effect of TRN on acute exercise performance, TRN mice performed an acute about of treadmill running (TRN + EX90) at the same relative intensity performed by control EX90 (cohort 2). Consistent with TRN alone group, TRN + EX90 mice ran 6.47 ± 0.24 km per day and showed a similar reduction in body weight (P < 0.05, Pre-TRN + EX90 = 22.5 ± 0.8 g, Post-TRN + EX90 = 20.9 ± 0.8 g). Moreover, inguinal and gonadal white adipose tissue fat pad weights were reduced in TRN + EX90 when compared to SED and EX90 animals (cohort 2) (P < 0.05). Maximum running speed was assessed one week prior to completion of the experimental protocol in all animals. A significantly higher maximum running speed (51.7 ± 1.4 m/min) during acute treadmill running was achieved by TRN female mice compared with untrained animals (40.7 ± 2.9 m/min, P < 0.05).
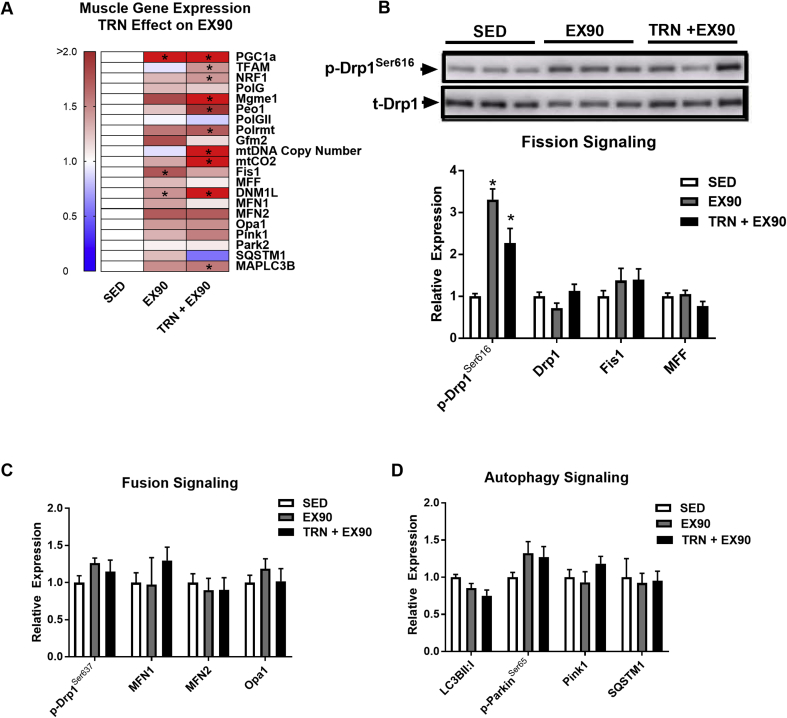
Regardless of training status, acute exercise (EX90) induced a similar increase in expression of PGC1α (identical expression levels observed between EX90 and TRN + EX90 groups; P < 0.05, Figure 4A). Our findings show that alterations to the mitochondrial genome including regulators of DNA replication, translation, and copy number, require a repeated exercise stimulus delivered by training. Whereas TRN appears to heighten the expression response of Dnm1L to acute exercise (EX90) (Figure 4A; EX90 vs. TRN + EX90, P = 0.018), Drp1Ser616 was similar between the EX groups (TRN + EX90 vs. EX90; Figure 4B). Drp1 total protein levels were unchanged by exercise regardless of training status, although localization of the protein and its macromolecular structure are critical for understanding its action upon the mitochondrion. Additionally, we observed no change in other fission-fusion-autophagy markers in EX90 or TRN + EX90 vs. SED (Figure 4B–D, Supplemental Figure 1C).
Figure 4.
Drp1 pro-fission signaling is increased during acute exercise in muscle from trained female mice. (A) Quadriceps mRNA abundance (presented in a heat map), representing mitochondrial fission, fusion, and autophagy signaling, in SED, EX90, and TRN + EX90 (N = 6/group; one-way ANOVA with Tukey post hoc analysis). Muscles were harvested from both EX90 groups immediately following the cessation of exercise. (B–D) Quadriceps protein and phospho-protein representing mitochondrial fission, fusion, and autophagy signaling, in SED (open bars), EX90 (gray bars), and TRN + EX90 (black bars) (N = 6/group; representative immunoblots presented in Figures. 4B and S1; one-way ANOVA with Tukey post hoc analysis). Data are means ± SEM (N = 6/group). *, P < 0.05 vs. SED.
Although the protein level of mitophagy or macroautophagy regulators were not different between SED, EX90, and TRN + EX90 (P > 0.05, Figure 4D), we observed an increase in expression of the macroautophagy regulator LC3B, Maplc3b in TRN + EX90 compared with SED animals, a finding similar to the increase in EX90 alone (P < 0.05, Figure 4A). A limitation of this study is that we did not measure autophagic flux by inhibiting lysosomal function or LC3B processing.
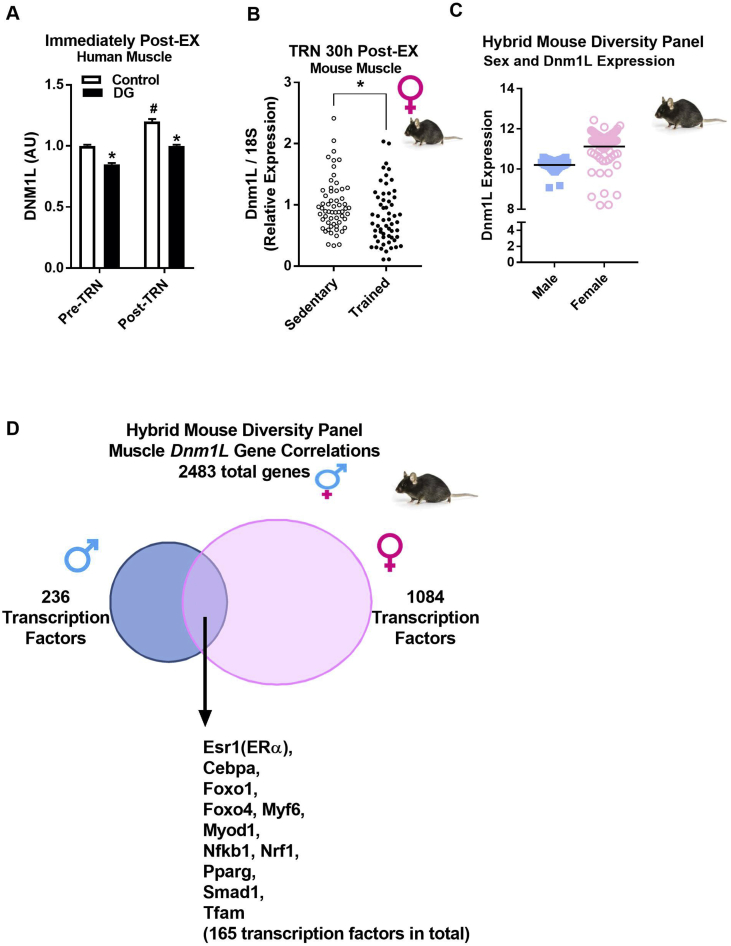
3.4. Muscle DNM1L expression in healthy normoglycemic and dysglycemic men, and in a panel of recombinant inbred mice
Because Dnm1L expression was significantly induced by acute and chronic exercise in female mice, we assessed the effect of exercise on muscle DNM1L expression in sedentary normoglycemic and dysglycemic men. We found that basal DNM1L expression was reduced by 15% (P = 0.01) in muscle of dysglycemic men compared with healthy normoglycemic controls (Figure 5A). Following 12 weeks of endurance and strength training (TRN), DNM1L expression was increased in both control (P = 0.029) and dysglycemic (P = 0.002) men; however, due to the differences in baseline expression, levels in DG men were similar to untrained controls following TRN (Figure 5A). Moreover, we examined Dnm1L gene expression in a 15-strain subset of the HMDP following 30 d of long-term exercise training. Similar to findings for TRN in C57BL/6J, a reduction in Dnm1L gene expression was observed in muscle harvested 30 h after the last training bout in recombinant inbred mice of the HMDP (P < 0.05, Figure 5B). To determine the genes most highly associated with Dnm1L expression, we mined the muscle expression data from the HMDP (Figure 5B). We found that muscle Dnm1L expression is higher in female mice vs. males, and that the variance about the mean was larger for females compared with male mice (Figure 5C). Interestingly, specific transcription factors, transcription co-factors, and chromatin remodeling elements known to play a role in muscle and mitochondrial metabolism were the most highly correlated transcripts with Dnm1L (P < 0.05, Figure 5D). These findings are consistent with our prior studies showing that Dnm1L is induced by estrogen receptor agonists [46].
Figure 5.
DNM1L gene expression and gene correlations in muscle from normoglycemic and dysglycemic men at rest and during exercise, and in 15 strains of recombinant inbred mice from the Hybrid Mouse Diversity Panel (HMDP). (A) Muscle DNM1L expression levels at basal and immediately following cycling training in N = 13 normoglycemic control and N = 13 dysglycemic (DG) men aged 40–65 years (two-way repeated measures ANOVA with Tukey's post hoc analysis). (B) Dnm1L muscle gene expression (presented as a dot plot) in a fifteen-strain subset (N = 4 mice/strain; two-tailed unpaired t-test) female HMDP mice, SED vs. TRN (normal chow fed mice 12 weeks of age). Muscles were harvested from TRN mice 30 h following the cessation of exercise (including a 6 h fast). (C) Dnm1L expression levels were elevated in female mouse muscle and showed greater variability over males. (D) Genes significantly correlated with Dnm1L expression (cut off criteria P < 0.0001; yield = 2483 genes significantly correlated including a high number of transcription factors) in skeletal muscle of male and female HMDP mice (12 weeks of age) fed a HF/HS diet for eight weeks (N = 84 strains/sex, 4 mice/strain). Muscles were harvested from HMDP mice following a 4–6 h fast. *, P < 0.05 vs. SED. #, P < 0.05 Pre-TRN vs. Post-TRN human muscle DNM1L expression.
3.5. Dnm1L is critical for exercise performance and training adaptations in muscle
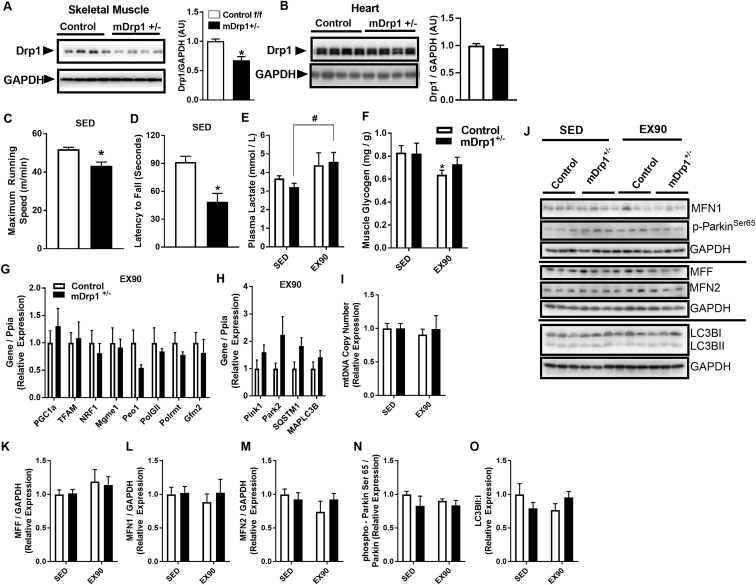
To determine the role Drp1 in acute exercise performance and exercise training-induced mitochondrial adaptations, mice with a conditional muscle-specific heterozygous deletion (mDrp1+/−; Figure 6A) performed 90 min treadmill running (EX90) or 30 d of volitional wheel running (TRN). Because we used the MCK Cre mouse line to achieve deletion of the Drp1 floxed allele, we confirmed a normal expression and protein abundance of Drp1 in cardiac muscle compared with Control (f/f) (Figure 6B). Prior to chronic exercise training, we performed acute exercise studies including tests of maximal running speed and dynamic hanging (latency to fall). Both maximal running speed and hanging time were reduced in mDrp1+/− mice vs. Controls (P < 0.05, Figure 6C–D). Following 90 min of moderate intensity exercise, plasma lactate was similarly elevated in both Control and mDrp1+/− animals (P < 0.05, Figure 6E), although the increase in EX90 over SED was only statistically significant for mDrp1+/−. Interestingly, muscle glycogen was significantly reduced following acute exercise (EX90) for f/f Control, but was not statistically different between SED vs. EX90 for mDrp1+/− animals (P < 0.05, Figure 6F).
Figure 6.
Muscle-specific reduction in Drp1 reduces maximal running speed and muscle endurance capacity in female mice. (A–B) Immunoblots and densitometric analysis of skeletal and cardiac muscle Drp1 total protein in Control and mDrp1+/− mice (representative immunoblots N = 4 per genotype; densitometry N = 8; two-way unpaired t-test). (C) Maximum running speed (m/min), and (D) latency to fall (seconds; N = 3 with three independent trials/per animal) in SED Control (open bars) and mDrp1+/− (closed bars) mice (N = 7–9; two-tailed unpaired t-test). (E) Plasma lactate (mmol/l) and (F) muscle glycogen (mg/g) from quadriceps muscle of control (open bars) and mDrp1+/− (closed bars), SED vs. EX90 mice using repeated measures ANOVA and two-way ANOVA with Tukey's post hoc analysis respectively. (G–H) Quadriceps gene expression in Control (open bars) and mDrp1+/− (closed bars) mice after EX90 (two-tailed unpaired t-test). (I) Mitochondrial DNA copy number in quadriceps muscle of SED and EX90, Control (open bars) and mDrp1+/− (closed bars) mice (two-way ANOVA with Tukey post hoc analysis). (J–O) Inmmunoblots and densitometric analysis of protein and phospho-protein expression of select fission, fusion, and autophagy markers from quadriceps muscle of SED and EX90, Control (open bars) and mDrp1+/− (closed bars) mice (two-way ANOVA with Tukey's post hoc analysis). Data are means ± SEM. N = 6–8/group. *, P < 0.05 vs. control SED. #, P < 0.05 SED vs. EX90 for mDrp1+/−.
Gene expression of mitochondrial biogenesis and autophagy markers were examined for Control and mDrp1+/− animals following acute exercise (P > 0.05, Figure 6G–I). Similar to our observations for wildtype C57BL/6J mice (SED vs. EX90), all markers of mitochondrial biogenesis as well as mtDNA copy number were identical between the genotypes (f/f Control vs. mDrp1+/−) at rest and following EX90 (P > 0.05, Figure 6G,I). We also evaluated protein and mRNA expression of select markers of mitochondrial fission, fusion, mitophagy, and autophagy, which were comparable between Control and mDrp1+/− animals (P > 0.05, Figure 6H, J-P).
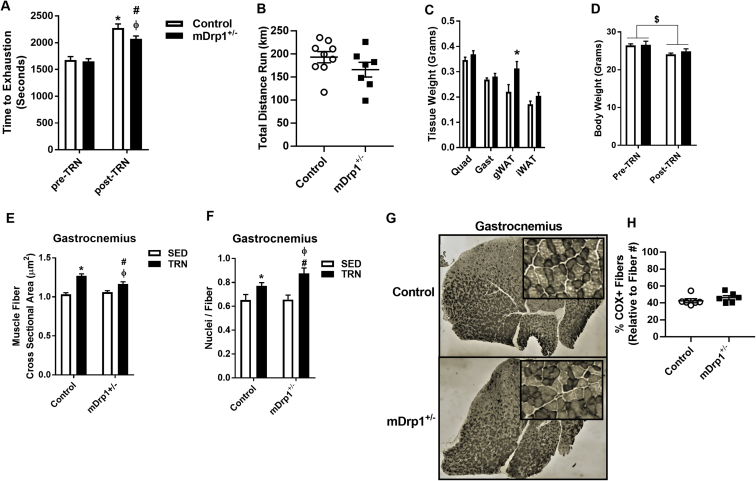
Prior to training we determined whether muscle Drp1 deletion impacted exercise capacity, and found that time to exhaustion was unaffected by Drp1 expression in untrained animals (P > 0.05, Figure 7A). By contrast, running time to exhaustion increased by 20–40% in Control floxed animals following training (Figure 7A, P < 0.05); however, the training improvement in exercise capacity was significantly blunted in mDrp1+/− mice. This difference in training-induced performance occurred despite identical body weight loss and total distance run during the 30-day training protocol between the genotypes (Figure 7A–B, D). Although we detected no difference in body weight between the genotypes, gonadal white adipose tissue (gWAT) weight was significantly higher in mDrp1+/− animals post-training (P < 0.05, Figure 7C). Muscle fiber cross sectional area in gastrocnemius muscle was unchanged in SED Control and mDrp1+/− animals (P < 0.05, Figure 7E). However, although muscle cross-sectional area increased in both genotypes of mice post-training, a larger increase in muscle cross-sectional area was observed for f/f Control vs. mDrp1+/− (P < 0.05, Figure 7E). Nuclei per muscle fiber was also increased in both genotypes post-training, although a larger increase in nuclei number was observed in mDrp1+/− mice (P < 0.05, Figure 7F). COX staining was similar between the genotypes following exercise training (Figure 7G–H).
Figure 7.
Muscle-specific reduction in Drp1 alters response to exercise training (TRN). (A) Time to treadmill running exhaustion displayed in seconds and (B) cumulative distance run in kilometers for female Control (open bars) vs. mDrp1+/− (closed bars) mice (N = 7–9; two-tailed unpaired t-test). (C) Tissue weights in age-matched littermates of SED and TRN Control vs. mDrp1+/− (two-tailed unpaired t-test). (D) Body weights of Control vs. mDrp1+/−, pre- and post-exercise training (two-way repeated measures ANOVA with Tukey post hoc analysis). (E) Muscle fiber cross sectional area and (F) nuclei number/fiber from gastrocnemius muscle of SED (open bars) and TRN (closed bars) Control vs. mDrp1+/− (two-tailed unpaired t-test). (G–H) Cytochrome C oxidase staining in gastrocnemius muscle from TRN Control and mDrp1+/− animals (representative image for each genotype; COX + staining between the genotypes compared using two-tailed unpaired t-test). Data are means ± SEM (N = 7–9/group). *, P < 0.05, vs. Control. #, P < 0.05, from pre-training or SED mDrp1+/−. Φ, P < 0.05 vs. post-training or TRN Control. $, P < 0.05 main affect from pre-training.
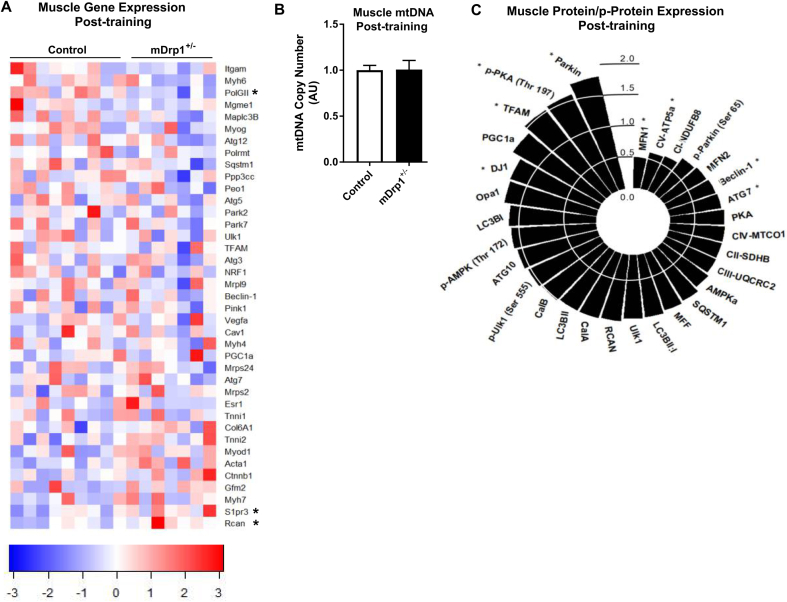
Several genes related to mitochondrial biogenesis were measured in muscle of both genotypes following TRN, and we identified PolGII as the only gene reduced in mDrp1+/− vs. f/f Control (P < 0.05, Figure 8A). No differences between the genotypes for other genes related to muscle structure, autophagy, mitophagy, muscle size, mitochondrial protein synthesis, vascularization, and cellular signaling were detected following TRN (P > 0.05, Figure 8A). In contrast to PolGII, expression of Sphingosine-1-Phosphate Receptor 3 (S1pr3), and Regulator of Calcineurin (RCAN) 1 were increased in muscle of mDrp1+/− following TRN vs. f/f Control (P < 0.05, Figure 8A). mtDNA copy number was not different between f/f Control and mDrp1+/− groups following TRN (P > 0.05, Figure 8B). We evaluated protein expression of regulators of the mitochondrial lifecycle and found alterations in MFN1, Beclin-1, ATG7, DJ1, TFAM, and Parkin (P < 0.05, Figure 8C, Supplemental Figure 2). An increase in the phosphorylation of Protein Kinase A (PKA) at Threonine 197 in TRN mDrp1+/− over f/f Control was also observed (P < 0.05, Figure 8C). We have previously shown that activation of the RCAN1-PKA axis impairs mitochondrial fission and promotes a hyperfused mitochondrial phenotype [46]. We hypothesize, that the capacity to rapidly remodel mitochondria dictates metabolic flexibility, and our findings suggest that Drp1 action, underlies in part, the enhancement in mitochondrial dynamics and improvements in metabolic health and performance associated with exercise training.
Figure 8.
Alterations in gene and protein expression following chronic exercise training in female Control and mDrp1+/− animals. (A) Quadriceps gene expression expressed as fold-change for mDrp1+/− TRN relative to Control TRN animals (two-tailed unpaired t-test; normalized to 18S). (B) Mitochondrial DNA copy number in quadriceps muscle of female Control (open bars) and mDrp1+/− (closed bars) animals following TRN (two-tailed unpaired t-test). (C) Muscle protein expression in mDrp1+/− relative to Control following TRN (two-tailed unpaired t-test; representative immunoblots presented in Supplemental Figure 2). Muscles were harvested from TRN mice 30 h following the cessation of exercise including a 6 h fast. Data are means ± SEM. N = 7–9/group. *, P < 0.05 mDrp1+/− vs. Control.
4. Discussion
Exercise imposes energetic demand upon skeletal muscle cells; however, the mechanisms by which mitochondria remodel and adapt to meet the metabolic challenge remains inadequately understood [62]. Because mitochondria are responsible for energy production within skeletal muscle cells, we determined how endurance exercise impacts mitochondrial dynamics signaling. To accomplish this we utilized three exercise protocols and examined more than 20 regulators of mitochondrial function. Our findings show that regardless of training status, all aspects of the mitochondrial lifecycle are impacted by acute exercise and chronic endurance exercise training.
Acute treadmill running promoted increased expression of genes related to mitochondrial biogenesis including Pgc1a, TFAM, NRF1, and Polg1. Additionally, expression levels of fission markers, Fis1 and Dnm1L, and cristae remodeling gene Opa1, were increased by 90 min of exercise and expression of these genes remained elevated during the 3hr post-recovery period. Similar to mice, acute exercise increased DNM1L gene expression in healthy normoglycemic untrained and trained men, as well as dysglycemic men who displayed reduced basal pre-exercise DNM1L expression levels compared with metabolically healthy individuals. Our findings in human subjects are in line with previous work by Kruse et al. [63]. When the post-exercise recovery period was extended to 30 h in SED and TRN mice, only Pgc1a and regulators of the mitochondrial genome, PolgII and Polrmt, as well as mtDNA copy number remained elevated. Genes regulating mitochondrial fission including Fis1, Mff, and Dnm1L were all significantly reduced in TRN vs. SED muscle when measured 30 h after the last exercise bout.
Although no alteration in total Drp1 protein from whole cell lysates was detected for any of the exercise interventions, we observed a robust increase in phosphorylation of Drp1 at its purported activation site Ser616, and we have previously shown that Ser616:Ser637 phosphorylation status correlates with Drp1 abundance on the OMM [64]. The increase in phosphorylation status occurred as early as 45 min and was sustained through 90 min of treadmill running, the longest duration of acute exercise performed. Our findings of an exercise-induced increase in muscle Drp1Ser616 in both male (C57BL/6J) and female (C57BL/6J, BALB/CJ, A/J, and C3H/HeJ) mice, is consistent with findings published by Kruse et al. [63]. During the 3 h post-exercise recovery period, Drp1Ser616 levels returned to basal phosphorylation status. Collectively, our findings suggest that the timing of measurement during exercise, post-exercise, and following training is critical because mitochondrial remodeling (reflected by signal transduction in this study) is a rapid and dynamic process.
Since Drp1Ser616 is previously associated with enhanced mitochondrial fission [65], we have been intrigued by the preponderance of electron micrograph images, including those generated by our laboratory, showing no to modest increase in mitochondrial size, but enhanced electron dense contacts (EDCs) between mitochondria [59]. Findings by Liu et al. [66], using high resolution microscopy, provide some resolution to these apparently conflicting observations by providing evidence for two modes of fusion-fission in mammalian cells. These modes include the well-described complete mitochondrial fusion vs. transient fusion. In contrast to complete fusion events, transient fusion is characterized by rapid kiss-and-run dynamics, in which mitochondria undergo heightened fission-fusion cycling with fission occurring proximal to the site of mitochondria–mitochondria contact. By comparison to complete fusion, which markedly alters mitochondrial morphology, kiss-and-run fusion permits architectural maintenance. Of interest, transient kiss-and-run fusion events are highly dependent upon rapid Drp1 action [66]. Microscopy studies elegantly show that genetic Drp1 inactivation reduces transient fusion flux, but elevates the proportion of complete fusion events whereby all four membranes are fused, full exchange of mitochondrial contents is achieved, and long-term alteration of mitochondrial topology is observed [66]. Functionally these distinct fusion events appear to serve different purposes; complete fusion is shown to be critical for mtDNA maintenance [20], [67], whereas transient fusion is thought to be essential for mitochondrial metabolism and motility [33], [66], [68]. Of interest, kiss-and-run dynamics require specific IMM fusion-independent actions of OPA1 on the cristae and cytoskeletal anchorage [66], and these events may be influenced by muscle tension and motor protein force generation. Indeed, an increase in electron dense mitochondrial contacts characterized as transient tethers have been previously observed during acute exercise [59]. Similar to our findings, exercise-induced EDCs were observed in the absence of change in OPA1 and MFN2 protein abundance [59].
A major limitation in our understanding of mitochondrial dynamics during exercise is our heavy reliance on signaling and electron microscopy (static observations), and our inability to visualize individual mitochondria in vivo in real time. Moreover, a limitation of the present study is that we did not discretely measure mitochondrial dynamics signaling in the subsarcolemmal and intermyofibrillar compartments. Considering that it is now well-accepted that several distinct mitochondrial pools reside in skeletal muscle and serve vastly different functions, it could be that selective mitochondrial remodeling paradigms are engaged to drive specific mitochondrial activities including oxidative metabolism, uncoupling of oxidative phosphorylation, calcium buffering, retrograde signaling, and DNA replication. Case in point, subsarcolemmal (SS) mitochondria are thought to provide energy for membrane related events including cell signaling, substrate and ion transport, while intermyofibrillar (IMF) mitochondria supply ATP to contracting myofibrils [69], [70], [71]. SS mitochondria represent only 20–30% of the total skeletal muscle mitochondrial pool, but appear by 3D analysis to have a different morphology from IMF (including form factor, aspect ratio, and circularity) [72], and to be more responsive to physical activity, but markedly reduced in abundance in aging, obesity and type 2 diabetes [69], [73], [74]. Moreover, mitochondrial ultrastructural analyses by Picard and Turnbull [72] suggest that there are active processes involved in mitochondrial shaping that maintain selective morphology to preserve mitochondrial function in these distinct mitochondrial populations. In addition to the two distinct mitochondrial pools previously characterized by [69], [70], [74], more recent evidence has emerged describing a unique mitochondrial pool that is intimately associated with the lipid droplet and involved in the regulation of lipid storage and metabolism [60]. Considering the robust phenotype of lipid accumulation in muscle lacking Drp1, mitochondrial dynamics signaling may very well play an important role in substrate storage and utilization [60]. Additional studies are required to determine how these specific mitochondrial pools are regulated so as to better understand structure–function relationships at rest and during physical activity.
Since sex is shown to play a key role in the regulation of metabolism and mitochondrial function, an important aspect of our study was the assessment of sex in controlling the natural variation in muscle Dnm1L expression of a large panel of recombinant inbred mice. We observed that muscle Dnm1L expression levels were elevated in females compared with males, and that there was significantly greater variability in muscle Dnm1L expression for females. These findings suggest a possible sex difference in the regulation of mitochondrial dynamics supporting known differences in mitochondrial density, substrate metabolism, and insulin sensitivity between males and females [75], [76], [77]. Muscle Dnm1L expression levels in both sexes correlated with a variety of factors that affect muscle metabolism and mitochondrial function including Esr1, Pparg, Tfam, Nrf1, Myod1, and Myf6. We exercise trained a 15-strain subset of the HMDP and found a similar acute exercise-induced increase in Dnm1L expression compared to C57BL/6J; moreover, also similar to our findings for C57BL/6J, Dnm1L expression declined to 20% below that of SED untrained for all TRN HMDP strains studied during a 30h post-exercise recovery period. These data are consistent with findings by Fealy et al. showing reduced DNM1L expression as well as Drp1Ser616 phosphorylation in human muscle during longer duration post-exercise recovery [42].
Taken together, our findings from the study of several cohorts of mice and human subjects under different paradigms of acute and chronic exercise show internal consistency with respect to an acute exercise-induced increase in Dnm1L expression and Drp1 phosphorylation, followed by a return to baseline in Dnm1L expression and Drp1 phospho-protein levels during post-exercise recovery. In addition, we found that muscle Drp1 deletion reduced maximal running speed and muscular endurance, and blunted the improvement in running time to exhaustion following training. These impairments in performance are in line with our previous findings of reduced oxidative metabolism, increased lactic acid production, muscle lipid accumulation and insulin resistance in muscle cells with Drp1 knockdown [46]. The precise actions of Drp1 during exercise and recovery require further investigation, especially since this mechanoenzyme has been implicated not only in mitochondrial fission, but also mtDNA replication, mitophagy, mitochondrial motility, and substrate metabolism [7], [27], [30], [31], [78]. Targeted studies focused on cellular distribution and enzymatic actions as a consequence of post-translational modification are warranted.
5. Conclusions
In conclusion, our findings indicate that endurance exercise is sufficient to induce changes in the mitochondrial life cycle including mitochondrial fission signaling through Drp1. Furthermore, lacking Drp1 reduced exercise performance and altered training-induced adaptations. Here we report that DNM1L expression levels are reduced in muscle from dysglycemic men and this could be an underlying contributor of metabolic dysfunction in these individuals. In mouse models of fission-fusion incompetence, we and others have shown that impaired dynamic flux of mitochondrial remodeling is associated with derangements in metabolism and insulin sensitivity. Because the processes of mitochondrial fission, fusion, biogenesis, and quality control are interdependent, strategies aimed at enhancing mitochondrial lifecycle flux capacity may be effective in combating diseases associated with metabolic dysfunction.
Acknowledgments
We would like to take this opportunity to thank Hiromi Sesaki for the generous gift of the floxed Drp1 mouse model. This research was supported in part by the UCLA Department of Medicine, The UCLA Iris Cantor Women’s Health Center Research Foundation and UCLA CTSI (ULTR000124), The UCLA Claude D. Pepper Older Americans Independence Center, and the National Institutes of Health (DK109724, P30DK063491, and NURSA NDSP to ALH under parent award U24DK097748). TMM was supported by a Kirschstein-NRSA predoctoral fellowship (F31DK108657), a Carl V. Gisolfi Memorial Research grant from the American College of Sports Medicine, and a predoctoral graduate student award from the Dornsife College at the University of Southern California. AJL2 was supported by an NIH T32 Neuroendocrinology, Sex Differences, and Reproduction Training Grant (5T32HD007228), Center for Duchenne Muscular Dystrophy Training Grant (T32AR065972), Molecular, Cellular, and Integrative Physiology Training Grant (T32GM065823), and the American College of Sports Medicine - NASA Space Physiology Research Grant. ZZ was supported by UCLA Claude Pepper Older Americans Independence Center funded by the National Institute on Aging (5P30AG028748), NIH/NCATS UCLA CTSI Grant (UL1TR000124), and UCLA Center for Duchenne Muscular Dystrophy-NIH NIAMS (U54 AR052646) Wellstone Center of Excellence Training Fellowship. SKM was supported by a grant from the Department of Veterans Affairs (I01BX000323). AJL3,4 was supported by NIH grants HL28481 and HL30568.
The MyoGlu trial is registered at ClinicalTrail.gov; NCT01803568. This work was supported by grants from the Institute of Basic Medical Sciences, UiO, Johan Throne-Holst Foundation for Nutrition Research, Freia Medical Research Foundation, the “Functional Genomics” and “Infrastructure” programs of the Research Council of Norway and the Southeastern Regional Health Authorities and EU-financed FP7 project (NutriTech grant agreement no: 289511). We thank Anne Randi Enget, Ansgar Heck and Birgitte Nellemann for taking the biopsies, and Tor I Gloppen, Torstein Dalen, Håvard Moen, Marius A Dahl, Guro Grøthe, Egil Johansen, Katrine A Krog, Øyvind Skattebo, Daniel S Tangen, Kristoffer K Jensen, Hans K Stadheim, and Eirin Rise for conducting the human strength and endurance intervention (MyoGlu) under supervision of Professor Jørgen Jensen. Sequencing was performed by PhD Gregor Gilfilan at the Norwegian Sequencing Centre (www.sequencing.uio.no) in collaboration with Torgeir Holen supported by the Research Council of Norway and the South-eastern Regional Health Authorities, Norway.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2018.11.012.
Conflict of interest
None declared.
Abbreviations
- 18S
18S ribosomal RNA
- Acta1
Actin1 Alpha 1
- AMPKA
Protein Kinase AMPK-Activated Catalytic Subunit Alpha 1
- Atg10
Autophagy Related 10
- Atg12
Autophagy Related 12
- Atg3
Autophagy Related 3
- Atg5
Autophagy Related 5
- Atg7
Autophagy Related 7
- Cav1
Caveolin 1
- Cebpa
CCAAT/Enhancer Binding Protein Alpha
- CII-SDHB
Succinate Dehydrogenase Complex Iron Sulfur Subunit B
- CI-NDUFB8
NADH: Ubiquinone Oxidoreductase Subunit B8
- CIV-mtCO1
Mitochondrially Encoded Cytochrome C Oxidase I
- Col6A1
Collagen Type VI Alpha 1 Chain
- Ctnnb1
Catenin Beta 1
- CV-ATP5a
ATP Synthase F1 Subunit Alpha
- DJ1
Parkinson Protein 7 (Park7)
- Drp1
Dynamin Related Protein 1 (DNM1L)
- Esr1
Estrogen Receptor 1
- Fis1
Fission, Mitochondrial 1
- Foxo1
Forkhead Box O1
- Foxo4
Forkhead Box O4
- Gast
Gastrocnemius Muscle
- Gfm2
G Elongation Factor Mitochondrial 2
- gWAT
Gonadal White Adipose Tissue
- Itgam
Integrin Subunit Alpha M
- iWAT
Inguinal White Adipose Tissue
- Kat2b
Lysine Acetyltransferase 2B
- LC3B
Mirotubule Associated Protein 1 Light Chain 3 Beta (MAPLC3B)
- MFF
Mitochondrial Fission Factor
- MFN1
Mitofusin 1
- MFN2
Mitofusin 2
- Mgme1
Mitochondrial Genome Maintenance Exonuclease 1
- Mrpl9
Mitochondrial Ribosomal Protein L9
- Mrps2
Mitochondrial Ribosomal Protein S2
- Mrps24
Mitochondrial Ribosomal Protein S24
- mtCO2
Mitochondrially Encoded Cytochrome C Oxidase II
- mtDNA
Mitochondrial DNA
- Myf6
Myogenic Factor 6
- Myh4
Myosin Heavy Chain 4
- Myh6
Myosin Heavy Chain 6
- Myh7
Myosin Heavy Chain 7
- Myod1
Myogenic Differentiation 1
- Myog
Myogenin
- Nfkb1
Nuclear Factor Kappa B Subunit 1
- Nfkbia
NFKB Inhibitor Alpha
- NRF1
Nuclear Respiratory Factor 1
- NRF2
Nuclear Respiratory Factor 2
- Opa1
Optic Atrophy Protein 1
- Park2
Parkinson Disease 2 (Parkin)
- Peo1
Twinkle mtDNA Helicase
- PGC1α
Peroxisome proliferator-activated receptor gamma coactivator 1-alpha
- Pink1
PTEN Induced Putative Kinase 1
- PKA
Protein Kinase CAMPK-Activated Catalytic Subunit Alpha
- PolG
DNA Polymerase Gamma, Catalytic Subunit
- PolGII
DNA Polymerase Gamma 2, Accessory Subunit
- Polrmt
RNA Polymerase Mitochondrial
- Pparg
Peroxisome Proliferator Activated Receptor Gamma
- Ppia
Peptidylprolyl isomerase A
- Ppp3cc
Protein Phosphatase 3 Catalytic Subunit Gamma (CalA)
- Quad
Quadriceps Muscle
- RCAN
Regulator of Calcineurin
- S1pr3
Sphingosine-1-Phosphate Receptor 3
- SIII-UQCRC2
Ubiquinol-Cytochrome C Reductase Core Protein 2
- Smad1
SMAD Family Member 1
- SQSTM1
Sequestosome 1 (p62)
- TFAM
Mitochondrial Transcription Factor A
- Tgfb1
Transforming Growth Factor Beta 1
- Tnni1
Troponin I1
- Tnni2
Troponin I2
- Ulk1
Unc-51 Llike Autophagy Activating Kinase 1
- Vegfa
Vascular Endothelial Growth Factor A
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Bo H., Zhang Y., Ji L.L. Redefining the role of mitochondria in exercise: a dynamic remodeling. Annals of the New York Academy of Sciences. 2010;1201:121–128. doi: 10.1111/j.1749-6632.2010.05618.x. [DOI] [PubMed] [Google Scholar]
- 2.Anderson S. Sequence and organization of the human mitochondrial genome. Nature. 1981;290(5806):457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 3.Bibb M.J. Sequence and gene organization of mouse mitochondrial DNA. Cell. 1981;26(2 Pt 2):167–180. doi: 10.1016/0092-8674(81)90300-7. [DOI] [PubMed] [Google Scholar]
- 4.Montoya J. Identification of initiation sites for heavy-strand and light-strand transcription in human mitochondrial DNA. Proceedings of the National Academy of Sciences of the United State of America. 1982;79(23):7195–7199. doi: 10.1073/pnas.79.23.7195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lackner L.L. Shaping the dynamic mitochondrial network. BMC Biology. 2014;12:35. doi: 10.1186/1741-7007-12-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liesa M., Palacín M., Zorzano A. Mitochondrial dynamics in mammalian health and disease. Physiological Reviews. 2009;89(3):799–845. doi: 10.1152/physrev.00030.2008. [DOI] [PubMed] [Google Scholar]
- 7.Twig G., Shirihai O.S. The interplay between mitochondrial dynamics and mitophagy. Antioxidants and Redox Signaling. 2011;14(10):1939–1951. doi: 10.1089/ars.2010.3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Twig G., Hyde B., Shirihai O.S. Mitochondrial fusion, fission and autophagy as a quality control axis: the bioenergetic view. Biochimica et Biophysica Acta. 2008;1777(9):1092–1097. doi: 10.1016/j.bbabio.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Narendra D. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. The Journal of Cell Biology. 2008;183(5):795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pejznochova M. Mitochondrial DNA content and expression of genes involved in mtDNA transcription, regulation and maintenance during human fetal development. Mitochondrion. 2010;10(4):321–329. doi: 10.1016/j.mito.2010.01.006. [DOI] [PubMed] [Google Scholar]
- 11.Jornayvaz F.R., Shulman G.I. Regulation of mitochondrial biogenesis. Essays in Biochemistry. 2010;47:69–84. doi: 10.1042/bse0470069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miller F.J. Precise determination of mitochondrial DNA copy number in human skeletal and cardiac muscle by a PCR-based assay: lack of change of copy number with age. Nucleic Acids Research. 2003;31(11):e61. doi: 10.1093/nar/gng060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dickinson A. The regulation of mitochondrial DNA copy number in glioblastoma cells. Cell Death and Differentiation. 2013;20(12):1644–1653. doi: 10.1038/cdd.2013.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taanman J.W. The mitochondrial genome: structure, transcription, translation and replication. Biochimica et Biophysica Acta. 1999;1410(2):103–123. doi: 10.1016/s0005-2728(98)00161-3. [DOI] [PubMed] [Google Scholar]
- 15.Scarpulla R.C. Metabolic control of mitochondrial biogenesis through the PGC-1 family regulatory network. Biochimica et Biophysica Acta. 2011;1813(7):1269–1278. doi: 10.1016/j.bbamcr.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Montgomery M.K., Turner N. Mitochondrial dysfunction and insulin resistance: an update. Endocrine in Connectivity. 2015;4(1):R1–R15. doi: 10.1530/EC-14-0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Westermann B. Mitochondrial fusion and fission in cell life and death. Nature Reviews Molecular Cell Biology. 2010;11(12):872–884. doi: 10.1038/nrm3013. [DOI] [PubMed] [Google Scholar]
- 18.Seo A.Y. New insights into the role of mitochondria in aging: mitochondrial dynamics and more. Journal of Cell Science. 2010;123(Pt 15):2533–2542. doi: 10.1242/jcs.070490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chan D.C. Fusion and fission: interlinked processes critical for mitochondrial health. Annual Review of Genetics. 2012;46:265–287. doi: 10.1146/annurev-genet-110410-132529. [DOI] [PubMed] [Google Scholar]
- 20.Chen H. Mitochondrial fusion is required for mtDNA stability in skeletal muscle and tolerance of mtDNA mutations. Cell. 2010;141(2):280–289. doi: 10.1016/j.cell.2010.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nochez Y. Acute and late-onset optic atrophy due to a novel OPA1 mutation leading to a mitochondrial coupling defect. Molecular Vision. 2009;15:598–608. [PMC free article] [PubMed] [Google Scholar]
- 22.Bach D. Mitofusin-2 determines mitochondrial network architecture and mitochondrial metabolism. A novel regulatory mechanism altered in obesity. Journal of Biological Chemistry. 2003;278(19):17190–17197. doi: 10.1074/jbc.M212754200. [DOI] [PubMed] [Google Scholar]
- 23.Shen Q. Mutations in Fis1 disrupt orderly disposal of defective mitochondria. Molecular Biology of the Cell. 2014;25(1):145–159. doi: 10.1091/mbc.E13-09-0525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haigh S.E., GFP P.A.- A window into the subcellular adventures of the individual mitochondrion. Novartis Foundation symposium. 2007;287:21–36. doi: 10.1002/9780470725207.ch3. Discussion 36–46. [DOI] [PubMed] [Google Scholar]
- 25.Twig G. Tagging and tracking individual networks within a complex mitochondrial web with photoactivatable GFP. American Journal of Physiology Cell Physiology. 2006;291(1):C176–C184. doi: 10.1152/ajpcell.00348.2005. [DOI] [PubMed] [Google Scholar]
- 26.Iglewski M. Mitochondrial fission and autophagy in the normal and diseased heart. Current Hypertension Reports. 2010;12(6):418–425. doi: 10.1007/s11906-010-0147-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Twig G. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. The EMBO Journal. 2008;27(2):433–446. doi: 10.1038/sj.emboj.7601963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee Y. Mitochondrial autophagy by Bnip3 involves Drp1-mediated mitochondrial fission and recruitment of Parkin in cardiac myocytes. American Journal of Physiology Heart and Circulatory Physiology. 2011;301(5):H1924–H1931. doi: 10.1152/ajpheart.00368.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ashrafi G., Schwarz T.L. The pathways of mitophagy for quality control and clearance of mitochondria. Cell Death and Differentiation. 2013;20(1):31–42. doi: 10.1038/cdd.2012.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lewis S.C., Uchiyama L.F., Nunnari J. ER-mitochondria contacts couple mtDNA synthesis with mitochondrial division in human cells. Science. 2016;353(6296):aaf5549. doi: 10.1126/science.aaf5549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wikstrom J.D., Twig G., Shirihai O.S. What can mitochondrial heterogeneity tell us about mitochondrial dynamics and autophagy? The International Journal of Biochemistry and Cell Biology. 2009;41(10):1914–1927. doi: 10.1016/j.biocel.2009.06.006. [DOI] [PubMed] [Google Scholar]
- 32.Wikstrom J.D. A novel high-throughput assay for islet respiration reveals uncoupling of rodent and human islets. PLoS One. 2012;7(5):e33023. doi: 10.1371/journal.pone.0033023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mishra P., Chan D.C. Metabolic regulation of mitochondrial dynamics. The Journal of Cell Biology. 2016;212(4):379–387. doi: 10.1083/jcb.201511036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mahdaviani K. Mfn2 deletion in brown adipose tissue protects from insulin resistance and impairs thermogenesis. EMBO Reports. 2017;18(7):1123–1138. doi: 10.15252/embr.201643827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pisani D.F. Mitochondrial fission is associated with UCP1 activity in human brite/beige adipocytes. Molecular Metabolism. 2018;7:35–44. doi: 10.1016/j.molmet.2017.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yan Z., Lira V.A., Greene N.P. Exercise training-induced regulation of mitochondrial quality. Exercise and Sport Sciences Reviews. 2012;40(3):159–164. doi: 10.1097/JES.0b013e3182575599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee S. Skeletal muscle phosphatidylcholine and phosphatidylethanolamine respond to exercise and influence insulin sensitivity in men. Scientific Reports. 2018;8(1):6531. doi: 10.1038/s41598-018-24976-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li Y. Subsarcolemmal lipid droplet responses to a combined endurance and strength exercise intervention. Physics Reports. 2014;2(11) doi: 10.14814/phy2.12187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Davis J.M. Exercise, alveolar macrophage function, and susceptibility to respiratory infection. Journal of Applied Physiology (1985) 1997;83(5):1461–1466. doi: 10.1152/jappl.1997.83.5.1461. [DOI] [PubMed] [Google Scholar]
- 40.Ghosh S. Selecting exercise regimens and strains to modify obesity and diabetes in rodents: an overview. Clinical Science (London) 2010;119(2):57–74. doi: 10.1042/CS20090389. [DOI] [PubMed] [Google Scholar]
- 41.O'Neill H.M. AMP-activated protein kinase (AMPK) beta1beta2 muscle null mice reveal an essential role for AMPK in maintaining mitochondrial content and glucose uptake during exercise. Proceedings of the National Academy of Sciences of the United Statesof America. 2011;108(38):16092–16097. doi: 10.1073/pnas.1105062108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fealy C.E. Exercise training decreases activation of the mitochondrial fission protein dynamin-related protein-1 in insulin-resistant human skeletal muscle. Journal of Applied Physiology (1985) 2014;117(3):239–245. doi: 10.1152/japplphysiol.01064.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Parks B.W. Genetic control of obesity and gut microbiota composition in response to high-fat, high-sucrose diet in mice. Cell Metabolism. 2013;17(1):141–152. doi: 10.1016/j.cmet.2012.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wakabayashi J. The dynamin-related GTPase Drp1 is required for embryonic and brain development in mice. The Journal of Cell Biology. 2009;186(6):805–816. doi: 10.1083/jcb.200903065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hevener A.L. Muscle-specific Pparg deletion causes insulin resistance. Nature Medicine. 2003;9(12):1491–1497. doi: 10.1038/nm956. [DOI] [PubMed] [Google Scholar]
- 46.Ribas V. Skeletal muscle action of estrogen receptor alpha is critical for the maintenance of mitochondrial function and metabolic homeostasis in females. Science Translational Medicine. 2016;8(334):334ra54. doi: 10.1126/scitranslmed.aad3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lerman I. Genetic variability in forced and voluntary endurance exercise performance in seven inbred mouse strains. Journal of Applied Physiology (1985) 2002;92(6):2245–2255. doi: 10.1152/japplphysiol.01045.2001. [DOI] [PubMed] [Google Scholar]
- 48.Mandillo S. Early motor deficits in mouse disease models are reliably uncovered using an automated home-cage wheel-running system: a cross-laboratory validation. Disease Models and Mechanisms. 2014;7(3):397–407. doi: 10.1242/dmm.013946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fan W. PPARdelta promotes running endurance by preserving glucose. Cell Metabolism. 2017;25(5):1186–1193 e4. doi: 10.1016/j.cmet.2017.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Drew B.G. HSP72 is a mitochondrial stress sensor critical for Parkin action, oxidative metabolism, and insulin sensitivity in skeletal muscle. Diabetes. 2014;63(5):1488–1505. doi: 10.2337/db13-0665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Langleite T.M. Insulin sensitivity, body composition and adipose depots following 12 w combined endurance and strength training in dysglycemic and normoglycemic sedentary men. Archives of Physiology and Biochemistry. 2016;122(4):167–179. doi: 10.1080/13813455.2016.1202985. [DOI] [PubMed] [Google Scholar]
- 52.Lee S. Effect of energy restriction and physical exercise intervention on phenotypic flexibility as examined by transcriptomics analyses of mRNA from adipose tissue and whole body magnetic resonance imaging. Physics Reports. 2016;4(21) doi: 10.14814/phy2.13019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dubois M. A colorimetric method for the determination of sugars. Nature. 1951;168(4265):167. doi: 10.1038/168167a0. [DOI] [PubMed] [Google Scholar]
- 54.Wanagat J. Mitochondrial DNA deletion mutations colocalize with segmental electron transport system abnormalities, muscle fiber atrophy, fiber splitting, and oxidative damage in sarcopenia. The FASEB Journal. 2001;15(2):322–332. doi: 10.1096/fj.00-0320com. [DOI] [PubMed] [Google Scholar]
- 55.Graham J.M. Purification of a crude mitochondrial fraction by density-gradient centrifugation. Current Protocol in Cell Biology. 2001 doi: 10.1002/0471143030.cb0304s04. [Chapter 3]: p. Unit 3 4. [DOI] [PubMed] [Google Scholar]
- 56.Rappsilber J., Mann M., Ishihama Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nature Protocols. 2007;2(8):1896–1906. doi: 10.1038/nprot.2007.261. [DOI] [PubMed] [Google Scholar]
- 57.Smirnova E. Dynamin-related protein Drp1 is required for mitochondrial division in mammalian cells. Molecular Biology of the Cell. 2001;12(8):2245–2256. doi: 10.1091/mbc.12.8.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cereghetti G.M. Dephosphorylation by calcineurin regulates translocation of Drp1 to mitochondria. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(41):15803–15808. doi: 10.1073/pnas.0808249105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Picard M. Acute exercise remodels mitochondrial membrane interactions in mouse skeletal muscle. Journal of Applied Physiology (1985) 2013;115(10):1562–1571. doi: 10.1152/japplphysiol.00819.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Benador I.Y. Mitochondria bound to lipid droplets have unique bioenergetics, composition, and dynamics that support lipid droplet expansion. Cell Metabolism. 2018;27(4):869–885 e6. doi: 10.1016/j.cmet.2018.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Robinson M.M. Enhanced protein translation underlies improved metabolic and physical adaptations to different exercise training modes in young and old humans. Cell Metabolism. 2017;25(3):581–592. doi: 10.1016/j.cmet.2017.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Green H.J. Mechanisms of muscle fatigue in intense exercise. Journal of Sports Science. 1997;15(3):247–256. doi: 10.1080/026404197367254. [DOI] [PubMed] [Google Scholar]
- 63.Kruse R. Intact initiation of autophagy and mitochondrial fission by acute exercise in skeletal muscle of patients with Type 2 diabetes. Clinical Science. 2017;131(1):37–47. doi: 10.1042/CS20160736. [DOI] [PubMed] [Google Scholar]
- 64.Archer S.L. Mitochondrial dynamics--mitochondrial fission and fusion in human diseases. New England Journal of Medicine. 2013;369(23):2236–2251. doi: 10.1056/NEJMra1215233. [DOI] [PubMed] [Google Scholar]
- 65.Chang C.R., Blackstone C. Dynamic regulation of mitochondrial fission through modification of the dynamin-related protein Drp1. Annals of the New York Academy of Sciences. 2010;1201:34–39. doi: 10.1111/j.1749-6632.2010.05629.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu X. Mitochondrial 'kiss-and-run': interplay between mitochondrial motility and fusion-fission dynamics. The EMBO Journal. 2009;28(20):3074–3089. doi: 10.1038/emboj.2009.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Clayton D.A. Transcription and replication of mitochondrial DNA. Human Reproduction. 2000;15(Suppl. 2):11–17. doi: 10.1093/humrep/15.suppl_2.11. [DOI] [PubMed] [Google Scholar]
- 68.Cogliati S. Mitochondrial cristae shape determines respiratory chain supercomplexes assembly and respiratory efficiency. Cell. 2013;155(1):160–171. doi: 10.1016/j.cell.2013.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ritov V.B. Deficiency of subsarcolemmal mitochondria in obesity and type 2 diabetes. Diabetes. 2005;54(1):8–14. doi: 10.2337/diabetes.54.1.8. [DOI] [PubMed] [Google Scholar]
- 70.Koves T.R. Subsarcolemmal and intermyofibrillar mitochondria play distinct roles in regulating skeletal muscle fatty acid metabolism. American Journal of Physiology Cell Physiology. 2005;288(5):C1074–C1082. doi: 10.1152/ajpcell.00391.2004. [DOI] [PubMed] [Google Scholar]
- 71.Cogswell A.M., Stevens R.J., Hood D.A. Properties of skeletal muscle mitochondria isolated from subsarcolemmal and intermyofibrillar regions. American Journal of Physiology. 1993;264(2 Pt 1):C383–C389. doi: 10.1152/ajpcell.1993.264.2.C383. [DOI] [PubMed] [Google Scholar]
- 72.Picard M., White K., Turnbull D.M. Mitochondrial morphology, topology, and membrane interactions in skeletal muscle: a quantitative three-dimensional electron microscopy study. Journal of Applied Physiology (1985) 2013;114(2):161–171. doi: 10.1152/japplphysiol.01096.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bizeau M.E., Willis W.T., Hazel J.R. Differential responses to endurance training in subsarcolemmal and intermyofibrillar mitochondria. Journal of Applied Physiology (1985) 1998;85(4):1279–1284. doi: 10.1152/jappl.1998.85.4.1279. [DOI] [PubMed] [Google Scholar]
- 74.Menshikova E.V. Effects of exercise on mitochondrial content and function in aging human skeletal muscle. Journal of Gerontology A Biological Science in Medical Science. 2006;61(6):534–540. doi: 10.1093/gerona/61.6.534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yki-Jarvinen H. Sex and insulin sensitivity. Metabolism. 1984;33(11):1011–1015. doi: 10.1016/0026-0495(84)90229-4. [DOI] [PubMed] [Google Scholar]
- 76.Hevener A. Female rats do not exhibit free fatty acid-induced insulin resistance. Diabetes. 2002;51(6):1907–1912. doi: 10.2337/diabetes.51.6.1907. [DOI] [PubMed] [Google Scholar]
- 77.Montero D. Sexual dimorphism of substrate utilization: differences in skeletal muscle mitochondrial volume density and function. Experimental Physiology. 2018;103(6):851–859. doi: 10.1113/EP087007. [DOI] [PubMed] [Google Scholar]
- 78.Wikstrom J.D. Hormone-induced mitochondrial fission is utilized by brown adipocytes as an amplification pathway for energy expenditure. The EMBO Journal. 2014;33(5):418–436. doi: 10.1002/embj.201385014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.