Abstract
Objective
Insulin action in the brain controls metabolism and brain function, which is linked to proper mitochondrial function. Conversely, brain insulin resistance associates with mitochondrial stress and metabolic and neurodegenerative diseases. In the present study, we aimed to decipher the impact of hypothalamic insulin action on mitochondrial stress responses, function and metabolism.
Methods
To investigate the crosstalk of insulin action and mitochondrial stress responses (MSR), namely the mitochondrial unfolded protein response (UPRmt) and integrated stress response (ISR), qPCR, western blotting, and mitochondrial activity assays were performed. These methods were used to analyze the hypothalamic cell line CLU183 treated with insulin in the presence or absence of the insulin receptor as well as in mice fed a high fat diet (HFD) for three days and STZ-treated mice without or with insulin therapy. Intranasal insulin treatment was used to investigate the effect of acute brain insulin action on metabolism and mitochondrial stress responses.
Results
Acute HFD feeding reduces hypothalamic mitochondrial stress responsive gene expression of Atf4, Chop, Hsp60, Hsp10, ClpP, and Lonp1 in C57BL/6N mice. We show that insulin via ERK activation increases the expression of MSR genes in vitro as well as in the hypothalamus of streptozotocin-treated mice. This regulation propagates mitochondrial function by controlling mitochondrial proteostasis and prevents excessive autophagy under serum deprivation. Finally, short-term intranasal insulin treatment activates MSR gene expression in the hypothalamus of HFD-fed C57BL/6N mice and reduces food intake and body weight development.
Conclusions
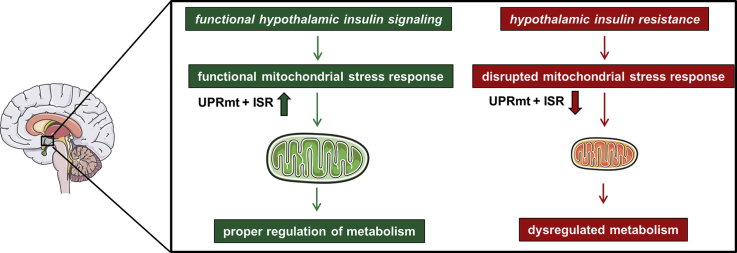
We define hypothalamic insulin action as a novel master regulator of MSR, ensuring proper mitochondrial function by controlling mitochondrial proteostasis and regulating metabolism.
Keywords: Brain insulin signaling, Mitochondrial function, Mitochondrial stress response, Autophagy, Intranasal insulin
Graphical abstract
Highlights
-
•
Hypothalamic insulin regulates mitochondrial stress responses.
-
•
Insulin controls mitochondrial function by regulating mitochondrial proteostasis.
-
•
Insulin via ERK signaling regulates MSR activation and prevents starvation-induced autophagy.
-
•
Intranasal insulin reduces HFD-induced food intake and activates the MSR.
1. Introduction
The brain relies on a constant energy supply to exert its function and consumes about 20% of energy though it represents only 2% of total body mass. The primary generators of energy are mitochondria, which facilitate brain metabolism. Thus, proper control of mitochondrial function is of utter importance for normal brain function and physiology. Not surprisingly, mitochondrial dysfunction has been observed in neurodegenerative and metabolic disorders, such as type 2 diabetes [1]. People with type 2 diabetes exhibit insulin resistance in peripheral tissues as well as in the brain. Causes for insulin resistance are multifactorial but can be a result of different stressors such as low-grade inflammation, glucotoxicity, lipotoxicity, ER stress, and mitochondrial stress [2]. Diet-induced obesity causes mitochondrial dysfunction in the rodent brain, which can be rescued with insulin sensitizers indicating that insulin can be protective for brain mitochondria [3]. Indeed, insulin is able to regulate mitochondrial membrane potential in sensory neurons of type 1 diabetic rats, and mitochondrial dynamics are altered in neurons of insulin-resistant mice [4], [5]. As neurons are especially vulnerable to mitochondrial stress due to low amounts of antioxidants, many mitochondrial diseases affect the brain and cause neurodegeneration [6], [7], [8].
The ability to sense and adapt to alterations in nutrient supply in conditions such as fasting, eating, or constant overnutrition requires cellular adaptation by precise activation of different signaling pathways. Cells have developed various adaptive responses, such as the mitochondrial unfolded protein response (UPRmt) or the integrated stress response (ISR) - here summarized as mitochondrial stress response (MSR) - to counteract cellular and mitochondrial deterioration and re-establish homeostasis. Cells respond to organelle stressors by activating transcription of cyto- and mitoprotective genes including the transcription factors Atf4 and Chop, which activate the mitochondrial chaperones and proteases such as Hsp60, Hsp10, ClpP, and Lonp1, thereby maintaining mitochondrial function [9], [10]. Prolonged fasting induces autophagy as an adaptive response, which causes the recycling of unneeded proteins allowing the synthesis of crucial proteins for cell survival or the degradation of dysfunctional organelles such as mitochondria to maintain cellular health. Conversely, these responses are interconnected [11]. Proper activation of the MSR increases lifespan and supports metabolic health [12], [13]. Interestingly, improving insulin sensitivity induces similar outcomes with enhanced longevity and improved metabolism [14], [15]. Indeed, the use of intranasal insulin, which serves as a therapeutic option to stimulate brain insulin action without affecting the periphery, enhances brain energy levels and improves metabolism and cognition even in humans [16], [17], [18].
We have previously shown that diabetic mice and patients exhibit decreased expression of the MSR gene Hsp60 in the hypothalamus, causing oxidative stress and insulin resistance [19]. Mutation of Hsp60 causes mitochondrial dysfunction and neurodegeneration, and a mutation of its co-chaperone Hsp10 is linked to a neurological disorder [20], [21], [22], [23]. Thus altered mitochondrial protein homeostasis due to Hsp60/10 dysregulation represents a link between brain insulin resistance, mitochondrial dysfunction and brain health [24].
In the present study, we investigated whether brain insulin signaling impacts mitochondrial stress responsiveness in the hypothalamus and thus affects mitochondrial function and metabolism. We find that acute insulin resistance causes a reduction of MSR gene expression and increased oxidative stress in the hypothalamus. This dysregulation is due to lack of proper brain insulin action, as insulin deficient mice exhibit a similar phenotype in the hypothalamus, which can be rescued by insulin supplementation. Brain insulin signaling controls mitochondrial function mainly via ERK activation and thereby reduces starvation-induced autophagy. Importantly, hypothalamic insulin action does not regulate mitochondrial function via enhancing mitochondrial biogenesis, as described for peripheral tissues, but controls mitochondrial function via regulation of mitochondrial proteostasis. Finally, we used intranasal insulin application, which is also used in humans, as a translational methodological approach to activate brain insulin action and investigate its effect on MSR regulation. Short-term intranasal insulin application of HFD-fed C57BL/6N mice increases levels of MSR genes in the hypothalamus and reduces food intake and weight gain, describing the insulin-MSR axis as a novel neuroprotective signaling pathway for hypothalamic function and metabolism.
2. Methods
2.1. Animal studies
Animals were kept in a temperature-controlled room (22+/− 1 °C) on a 12 h light/dark cycle with free access to food and water. All animal studies were conducted in accordance with the NIH guidelines for the care and use of laboratory animals, and all experiments were approved by the ethics committee of the State Agency of Environment, Health and Consumer Protection (State of Brandenburg, Germany).
4 week old male and female C57BL/6NCrl wildtype mice were fed with normal chow diet (NCD) (Rodent Diet with 10% kcal fat, ssniff Spezialdiäten GmbH, V1124-3) or high fed diet containing high fat and sugar (HFD) (Rodent Diet with 60% kcal fat, D12492, Research Diet inc.) over 3 days. Body weight was measured daily, and blood glucose was taken at the beginning and at the end of the study.
2.2. Streptozotocin-induced diabetes and insulin pellet implantation
8 week old male C57BL/6J mice (The Jackson Laboratory) were fasted overnight followed by i.p. injections of fresh prepared streptozotocin (150 mg/kg body weight) dissolved in citrate buffer (100 mM, pH4.5); the control group was injected with citrate buffer. Blood glucose levels of each mouse were measured every 3–4 days after the initial streptozotocin injection. To normalize streptozotocin-induced hyperglycemia, a subgroup of diabetic mice was treated at day 7 by subcutaneous implantation of LinBit Insulin pellets (LinShin Canada, Inc.) following manufacturer's manual.
2.3. Intranasal insulin application
6 week old male C57BL/6NCrl wildtype mice were fed a HFD over three days. Food intake was measured daily 9- and 24 h after intranasal saline or insulin application along with daily body weight. To apply intranasal saline or insulin, mice were fixed in the supine position and slightly overstretched, so that the tip of the nose points towards the ceiling. 1.75U/17.5 μl insulin (Actrapid 100IE/ml; Novo Nordisk Pharma GmbH Mainz) solution or saline solution in a maximum of 4.4 μl drops was alternately added at intervals of 30 s to each nostril to prevent suffocation. Attention was always paid to swallowing. After three days of HFD feeding, mice were starved overnight (16 h), followed by a final intranasal application of saline or insulin. Mice had free access to HFD and water, food intake and body weight was measured after one hour.
2.4. Analytical procedures
Blood glucose was measured with a Glucometer–Contour XT (Bayer, Leverkusen, Germany). Insulin and Leptin were measured with ELISAs from Alpco (Alpco Salem, BioCatGmbH, Switzerland) and R&D Systems (R&D Systems/Bio-Techne GmbH, Germany) respectively, according to manufacturer's guidelines. Triacylglycerols were measured with Triglyceride Reagent from ABX and NEFA were measured with NEFA-HR(2) Assay according to manufacturer's guidelines (FUJIFILM Wako Chemicals Europe GmbH, Germany). Measurements were performed in a blinded manner.
2.5. RNA isolation from tissue/cells
Total RNA was extracted from 5 to 20 mg tissue or 4 × 105–7 × 105 cells using a ReliaPrep RNA Tissue Miniprep System (Promega) or RNeasy Mini Kit (Qiagen) following manufacturer's manual including DNase I treatment.
2.6. Analysis of gene expression by quantitative PCR
Overall, between 100 ng and 1 μg of RNA from tissue were reverse transcribed in 20 μl using Oligo(dT)15 primers (Promega), Random primers (Promega), Thermo Scientific™ dNTP-Set, and M-MLV Reverse Transcriptase (Promega). Real time PCR was performed using the GoTaq qPCR master mix (Promega), gene-specific primers (200 nM each, obtained from Sigma) (Supplementary Table 1), and 10 ng cDNA. Fluorescence was monitored and analyzed in the ViiA 7 Real-Time PCR System (Applied Biosystems). Gene expression was calculated according to the ΔΔCT method using TBP (TATA-box binding protein) as reference gene. The specificity of SYBRGreen primers was confirmed by melting curve analysis.
2.7. Cell culture
CLU183 cells (mHypoA-2/23 CLU183) were cultivated with DMEM GlutaMAX (Gibco) supplemented with 1 mM sodium pyruvate (Sigma–Aldrich), 10% fetal bovine serum (FBS) (Pan, South Africa) and 1% penicillin-streptomycin (Gibco). All cell cultures were maintained at 37 °C with 5% CO2. For all experiments, the cells were seeded one day before the stimulation.
CLU183 cells were serum-starved for 3 h with DMEM GlutaMAX, 1 mM sodium pyruvate, 1% penicillin-streptomycin, and 0.1% BSA prior to any stimulations. Next, CLU183 cells were either serum-deprived (control) or stimulated with 100 nM insulin (Sigma–Aldrich) for 6 h or 16 h. Note, for western blot analysis, 1x amino acids (Gibco) have been added to cultivating medium. For MAPK/ERK kinase inhibition, CLU183 cells were serum-starved and at the same time pre-treated with 10 μM UO126 (InvivoGen) for 3 h and afterwards stimulated with insulin (100 nM) for indicated time points. For JNK-activation, CLU183 cells were serum-starved and at the same time pre-treated with 1 μM anisomycin (Sigma–Aldrich) for 3 h and afterwards stimulated with insulin (100 nM) for indicated time points. To inhibit IGF-1 receptor (IGF-1R), cells were pre-treated with 400 nM JB-1 (Sigma) for 3 h during serum-starvation and afterwards stimulated with insulin (10 nM) for indicated time points.
To generate Hsp60 knockdown (KD) neurons, CLU183 were transfected with either siGENOME non-targeting siRNA (Dharmacon) or SMARTpool: siGENOME Hspd1 (Hsp60) siRNA (M-062629-01-0005, Dharmacon) using DharmaFECT 1 Transfection Reagent (Dharmacon). For this, 2.2 × 106 cells were plated one day prior to transfection. Solution A contained 25 pM siRNA in Opti-MEM™ I reduced serum medium (Gibco) and transfection reagent was diluted 1:80 in Opti-MEM (Solution B). Both solutions were incubated at room temperature for 5 min separately before mixing and incubating them for 20 min at room temperature. Cells were then incubated with the mixture and penicillin-streptomycin free full medium for 6–8 h and medium was changed the following day. 3 days after transfection seahorse and western blot analysis were performed.
2.8. Western blot analysis
Western blots were performed on 10–15 μg total protein lysates loaded on 10% SDS-PAGE gels and transferred to PVDF membranes (GE HealthCare Life Science, Amersham) for 3 h at 90 V. After transfer, membranes were blocked in Starting Block (StartingBlockTM T20 (TBS) Blocking Buffer Thermo Scientific #37543) for 1 h. Following blocking, membranes were probed with primary antibody overnight at 4 °C. Antibodies used were IRβ (Cell Signaling, #3025, 1:1000), pIKKαβSer176/180 (Cell Signaling #2697, 1:1000), IKKβ (Cell Signaling #2370 1:1000) pSAPK/pJNKThr183/Tyr185 (Cell Signaling #9251, 1:1000), SAPK/JNK (Cell Signaling #9258, 1:1000), p-p38 MAPKThr180/Tyr182 (Cell Signaling #9211, 1:1000), p38 MAPK (Cell Signaling #9212, 1:1000), Hsp60 (Cell Signaling #4870, 1:1000), LONP1 (Sigma SAB1411647, 1:1000), ClpP(Santa Cruz sc-271284, 1:500), LC3 A/B (Cell Signaling #12741, 1:1000), p62 (abcam ab56416, 1:2000), ATF4 (Cell Signaling, #11815, 1:1000), CHOP (Cell Signaling, #2895, 1:1000), Total OXPHOS Rodent WB Antibody Cocktail (Abcam, #ab110413, 1:2000), fumarate hydratase (Santa Cruz sc-100743, 1:1000), VDAC (Cell Signaling #4866, 1:1000), β-Actin + HRP (Sigma–Aldrich A3854 1:10000). Membranes were then incubated with peroxidase-conjugated secondary antibodies (anti-rabbit Dianova #711-065-152, 1:10000, anti-mouse Dianova #715-065-150, 1:10000) at room temperature for 1 h. Specific bands were detected with chemiluminescence assay (WesternBright ECL Biozym 541005X) using the ChemiDoc Touch Imaging System (BioRad). To remove the phospho-epitope of a protein and run the total antibody, membranes were incubated in stripping buffer (RestoreTM PLUS Western Blot Stripping Buffer Thermo Scientific #46430) for 20 min at room temperature, re-blocked for 30 min and re-probed. In the end, membranes were re-probed with β-Actin, which served as an internal loading control for western blotting or stained with fast green. Band intensities were quantified via densitometric analysis using Image Lab 5.2.1 software.
2.9. Protein carbonylation assay
Protein carbonyl groups were detected with an antibody after protein derivatization as previously described [25]. Briefly, after ponceau staining, the membrane was washed with water and then equilibrated in 1X TBS +20% methanol for 5 min. Afterwards, the membrane was washed for 5 min with 10% HCl and then incubated for derivatization in 5 mM 2,4-dinitrophenylhydrazine (DNPH) in 10% HCl for 10 min. Following this, the excess of DNPH was removed by washing twice for 5 min with 10% HCl. The final step consisted of washing the membrane five times for 5 min with 50% methanol and washing one time for 5 min with 1X TBS. After briefly washing with MQ water, the membrane was blocked for 1 h in starting block, then washed three times with 1X TBS-T, and probed with anti-DNP primary antibody (Sigma, D9656, 1:10000) at 4 °C.
2.10. Seahorse assay
Changes in mitochondrial respiration were monitored using Seahorse XF Mito Stress Test Kit and the Seahorse XF96 extracellular flux analyzer, measuring oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) of adherent cells to test mitochondrial functionality. All compound concentrations were tested and optimized before and were consistent through all Seahorse runs for all experimental setups. Final concentrations for the compounds used were 2 μM for Oligomycin (Port A), 1 μM for FCCP (Port B), 1 μM for Rotenone/2 μM for Antimycin A (Port C). Cells were seeded at a density of 5000 cells/well two days prior to experiment within a 96 well microplate in full medium. Four wells were prepared without cells and used as background signal. On the following day full medium was removed, cells were washed with PBS, and starvation medium was added, with or without MAPK Kinase Inhibitor (UO126) at a final concentration of 10 μM. 3 h later, insulin was added to the designated wells to a final concentration of 100 nM. Cells were incubated with insulin for 16 h. On the assay day, cells were washed twice with Seahorse Medium (XF base Minimum DMEM supplemented with 1 mM sodium pyruvate, 2 mM glutamine and 10 mM glucose) and incubated in the final amount of assay medium in a non-CO2 incubator at 37 °C to maintain pH levels. After calibration, the microplate was placed in the Seahorse XF96 flux analyzer and the experiment was performed according to protocol. Data were normalized to total protein level due to the 96 well setting of the Seahorse XF96 flux analyzer and were analyzed using Wave 2.4.0 software.
2.11. Insulin receptor knockout generation
In order to delete IRs specifically in murine hypothalamic neurons, we used pSpCas9(BB)-2A-GFP (PX458) vector, which contains guide RNA (gRNA) against IR. To generate the insulin receptor knockout cell line, the CLU183 were transfected using 10 μg of gRNA-expressing plasmid. One day before transfection, the cells were plated at a density of 1 × 105 cells. For the transfection, the FuGENE® HD Transfection Reagent (Promega) was used and performed according to manufacturer's guidelines. Forty-eight hours post-transfection, GFP-positive cells were selected by flow cytometry. IR knockout single clones were confirmed based on the absence of IR protein as detected by Western blot technique using anti-IRβ Antibody.
guide: INSR_469_fw CACCGATACGGGACCAGTCGATAG
INSR_469_rv AAACCTATCGACTGGTCCCGTATC
2.12. Electron microscopy
CLU183 cells were serum-starved for 3 h with DMEM GlutaMAX, 1 mM sodium pyruvate, 1% penicillin-streptomycin and 0.1% BSA prior to any stimulation. Following CLU183 cells were either serum-deprived (control) or stimulated with 100 nM insulin (Sigma–Aldrich) for 16 h. Confluent neurons were fixed by immersion in 2.5% glutaraldehyde in 0.19 M sodium cacodylate buffer at pH 7.4, postfixed in 1% reduced osmium tetroxide [26] in aqua bidest, and subsequently stained with 2% uranyl acetate in maleate buffer, pH 4.7. The specimens were dehydrated in graded ethanol and embedded in epoxy resin. Embedding of neurons in Epon 812 was performed as described by Luft [27] and modified by Reale [26]. Ultrathin sections were picked up onto Formvarcarbon-coated grids, stained with lead citrate, and viewed in a transmission electron microscope (TEM 910; Zeiss Elektronenmikroskopie, Oberkochen, Germany).
2.13. Statistical analysis
All the data were presented as mean ± SEM. Two groups were compared using unpaired two-tailed Student's t-test or Mann–Whitney when necessary. 1-way ANOVA followed by Dunnett's post hoc test for multiple comparisons was used to compare more than two groups. 2-way ANOVA was performed to detect the interactions between e.g. genotype and treatment (insulin, IR KO), and Tukey's post hoc analysis was performed when appropriate.
3. Results
3.1. Acute HFD feeding reduces MSR activation in the hypothalamus
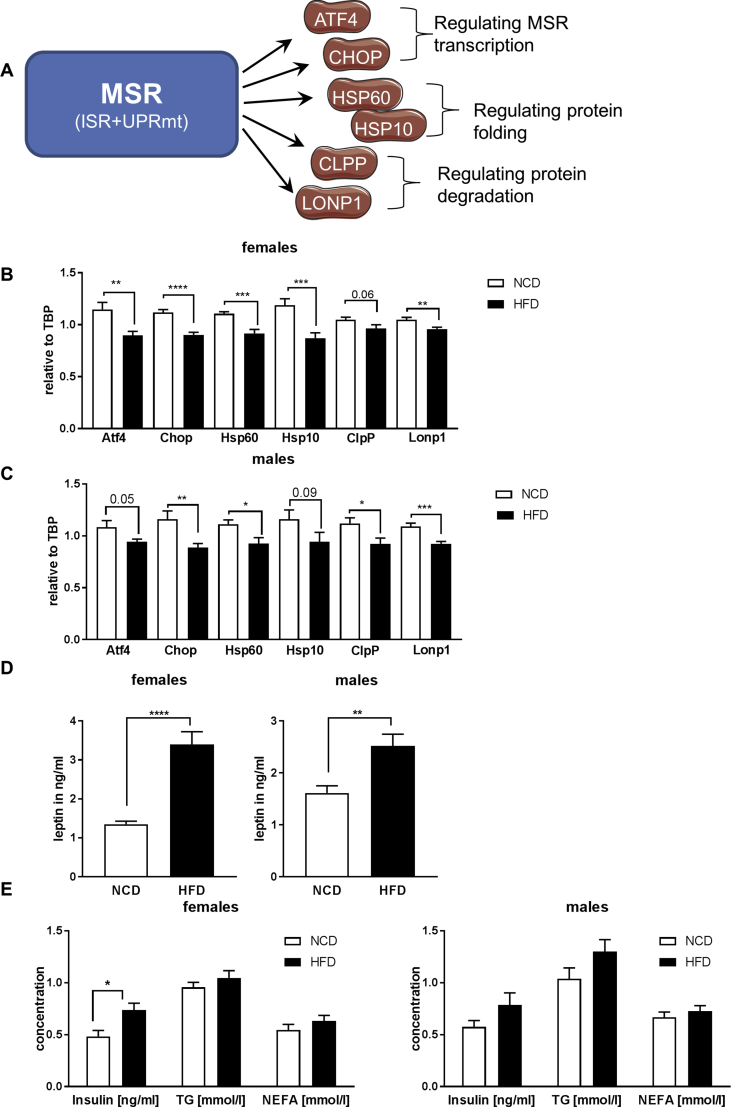
To determine whether acute high fat diet (HFD) affects the MSR in the hypothalamus, we fed 4 week old male and female C57BL/6N mice a normal chow diet (NCD) or a HFD for only 3 days. Quantitative PCR (qPCR) analysis of hypothalamic samples revealed a significant 10–27% reduction of mRNA levels of Atf4, Chop, Hsp60, Hsp10, ClpP, and Lonp1 in HFD-fed mice compared to NCD, with a more prominent phenotype in females (Figure 1A–C). This coincided with increased oxidative stress exemplified by an increase in carbonylation of high molecular proteins in the hypothalamus (Suppl. Fig. 1A, B). To test whether acute HFD feeding also affected other mitochondrial parameters in the hypothalamus, gene expression of key players of mitochondrial dynamics and biogenesis were assessed. This analysis revealed unaltered expression of the master regulator of mitochondrial biogenesis Pgc1α. Further acute HFD feeding did not affect gene expression of the mitochondrial fission gene Drp1, but caused a significant downregulation of the mitochondrial fusion gene Mfn2, which has been previously reported. A reduction of Mfn2 in hypothalamic neurons can cause hyperleptinemia [28], which was also present in our analyzed mouse cohorts (Suppl. Fig. 1C). This response was specific to the hypothalamus, as other investigated brain regions and tissues, such as hippocampus and gonadal adipose tissue, did not exhibit this regulation (Suppl. Fig. 1D,E). Acute HFD-feeding did not affect body weight, plasma insulin, triglycerides and non-esterified fatty acid levels in males compared to NCD-fed controls, but resulted in marked hyperglycemia, as a sign of early insulin resistance. In comparison, HFD-fed females exhibited increased body weight, normal glycemia, unaltered triglycerides and non-esterfied fatty acid levels compared to NCD-fed control, but were hyperinsulinemic, also a sign of early insulin resistance (Figure 1E, Suppl. Fig. 1F,G). As short term HFD feeding has been shown to induce activation of serine/threonine protein kinases which cause insulin resistance, we assessed activation of c-Jun kinase (JNK) and IκB kinase (IKK), as well as inhibitory serine-phosphorylation of insulin receptor substrate 1 (IRS-1). This analysis revealed a considerable increase of IRS-1 Ser1101 phosphorylation, as a further marker of insulin resistance, in addition to enhanced activation of JNK and IKK in males fed a HFD, whereas females were unaffected (Suppl. Fig. 1H,I).
Figure 1.
Acute high fat diet feeding reduces the mitochondrial stress response (MSR) in the hypothalamus. A: Schematic presentation of key genes of the MSR: ATF4 (Activating Transcription Factor 4), CHOP (C/EBP Homologous Protein), HSP60 (Heat Shock Protein 60), HSP10 (Heat Shock Protein 10), CLPP (Clp Protease), LONP1 (Lon Protease). B, C: (B) mRNA expression of Atf4, Chop, Hsp60, Hsp10, ClpP, and Lonp1 in hypothalamic samples of 4 weeks old female and (C) male C57BL/6N mice fed a NCD or HFD for 3 days n = 12 for NCD, n = 12 for HFD. D, E: (D) Leptin, (E) Insulin, triglycerides (TG) and non-esterified fatty acid (NEFA) concentration of 4 weeks old (left) female and (right) male C57BL/6N mice fed a NCD or HFD for 3 days n = 12 for NCD, n = 12 for HFD. *, P < 0.05, **, P < 0.01, ***, P < 0.001, ****, P < 0.0001. All the data are presented as mean ± SEM.
3.2. Hypothalamic insulin action controls MSR activation
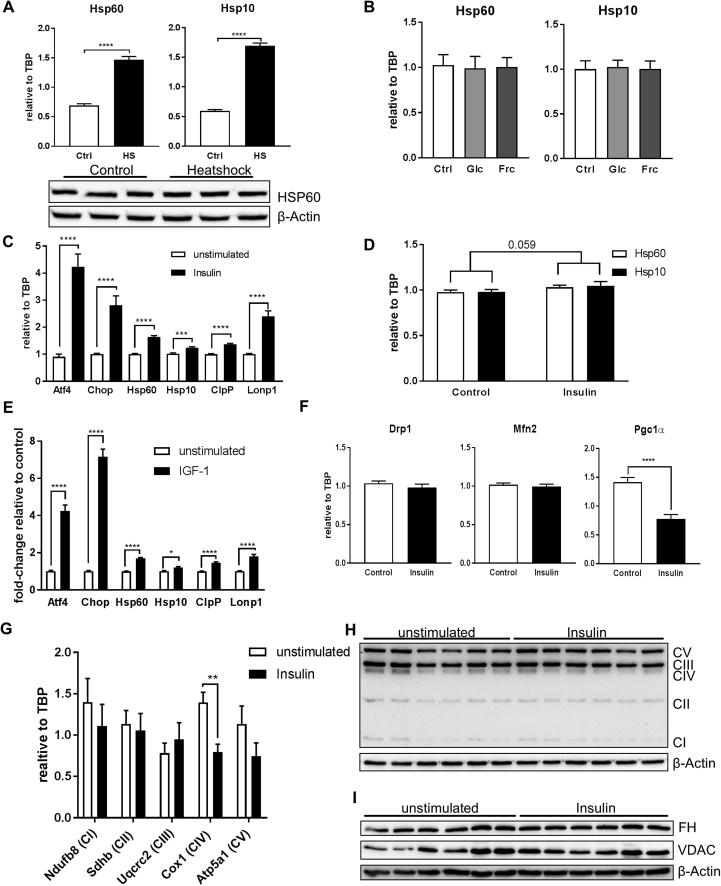
Next, we investigated causes for the observed downregulation of MSR gene expression in the hypothalamus of HFD-fed animals. To this end, we tested the metabolic regulation of Hsp60 and Hsp10 using the hypothalamic cell line CLU183. First, we confirmed the classical regulation by heat shock of the mitochondrial chaperones Hsp60 and Hsp10 in these cells resulting in a 3-fold increase in mRNA levels. The increased expression of HSP60 was not detectable using western blotting technique due to high basal expression of HSP60 (Suppl. Fig. 2A), confirming data that only chronically elevated levels of HSPs can be determined by western blot and highly quantitative methods such as mass spectrometry are needed to assess transient increases of heat shock protein levels [29], [30]. To this end, we assessed Hsp60 and Hsp10 regulations by qPCR throughout this study, as it represents so far the common standard methodology to analyze their regulation and induction [30]. As HFD-fed males were hyperglycemic, we assessed the effect of elevated glucose concentrations on mitochondrial chaperone expression. Neither high concentrations of glucose nor high concentrations of fructose affected gene expression of Hsp60 and Hsp10 indicating that hyperglycemia per se is not causal for the observed reduction of mitochondrial chaperones in vivo (Suppl. Fig. 2B).
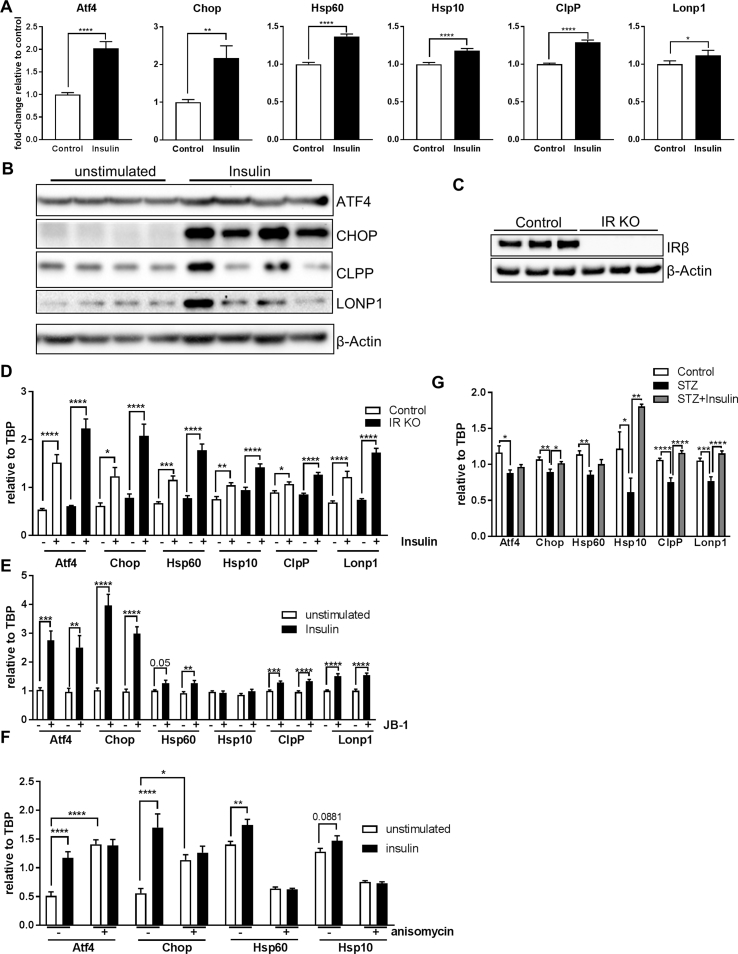
As HFD-fed mice exhibited early insulin resistance, we tested whether insulin was able to regulate the MSR. We assessed mRNA levels of these genes in CLU183 cells in serum-starved and serum-starved conditions supplemented with 100 nM insulin for 6 h. This analysis revealed that 6 h insulin treatment caused a significant upregulation of all tested genes ranging from an increase of 17% for Lonp1 up to 217% for Chop in CLU183 cells compared to unstimulated control (Figure 2A). This response was still apparent after 16 h of insulin stimulation as evidenced by gene and protein expression, indicating a long-term effect of insulin on MSR activation (Figure 2B, Suppl. Fig. 2C). To investigate these data in a more physiological cell model, we treated primary cortical neurons with 100 nM insulin for 16 h. Insulin also increased the expression of Hsp60 and Hsp10 in primary neurons (p = 0.059) confirming the insulin-induced control of Hsp60/10 transcription (Suppl. Fig. 2D). Insulin and IGF-1 use the common canonical signaling pathway to exert their function. So we tested whether IGF-1 signaling was also able to induce gene expression of MSR genes. 6 h of IGF-1 (10 nM) stimulation of serum-starved CLU183 cells caused a significant increase of all tested MSR genes compared to control (Suppl. Fig. 2E). To test whether insulin specifically increases expression of MSR genes and not overall gene expression of mitochondria-related genes, we analyzed expression levels of mitochondrial fission and fusion genes Drp1 and Mfn2 as well as a regulator of mitochondrial biogenesis Pgc1α. Stimulating serum-starved CLU183 cells with 100 nM insulin for 6 h did not affect Drp1 or Mfn2 mRNA levels but significantly reduced Pgc1α (Suppl. Fig. 2F). As insulin and IGF-1 are able to cross-activate IR and IGF-1R, we analyzed the effect of the respective receptors in controlling the MSR. Stimulating CRISPR/Cas9-mediated IR knockout CLU183 cells with 100 nM insulin, a concentration which also activates IGF-1R, was not only sufficient to activate the MSR, but also had a greater regulatory effect on MSR as evidenced by a significant genotype effect (p < 0,0001) (Figure 2C,D). To inhibit IGF-1R activation, we used the IGF-1R inhibitor JB-1. To this end, we stimulated cells with only 10 nM insulin in the absence and presence of JB-1. Insulin was still able to enhance mRNA levels of Atf4, Chop, Hsp60, Hsp10, ClpP, and Lonp1 in IGF-1R inhibited conditions. Overall, these data clearly demonstrate that the MSR is engaged by both insulin and IGF-1 signaling cascades in hypothalamic neurons (Figure 2E).
Figure 2.
Insulin controls the mitochondrial stress response. A: mRNA expression of Atf4, Chop, Hsp60, Hsp10, ClpP and Lonp1 after 6 h insulin stimulation of CLU183 hypothalamic neurons. Data of three independent experiments with a total of n = 18. The term ‘unstimulated’ or ‘Control’ always represents serum-starved conditions. B: Protein expression of ATF4, CHOP, CLPP, and LONP1 after 16 h of insulin stimulation of CLU183 hypothalamic cells, n = 4. β-Actin served as a loading control. Data from three independent experiments. C, D: (C) Protein expression of insulin receptor (IR) for verification of IR knockout (KO) CLU183 cells. β-Actin served as a loading control. (D) mRNA expression of Atf4, Chop, Hsp60, Hsp10, ClpP, and Lonp1 after 6 h of insulin stimulation of control or IR KO CLU183 cells. Data of three independent experiments with a total of n = 9. E: mRNA expression of Atf4, Chop, Hsp60, Hsp10, ClpP, and Lonp1 after 6 h of insulin stimulation of vehicle treated or 400 nM JB-1 treated CLU183 cells. Data of three independent experiments with a total of n = 12. F: mRNA expression of Atf4, Chop, Hsp60, Hsp10, ClpP, and Lonp1 after 16 h of insulin stimulation of vehicle treated or 1 μM anisomycin treated CLU183 cells. Data of three independent experiments with a total of n = 12. G: mRNA expression of Atf4, Chop, Hsp60, Hsp10, ClpP, and Lonp1 in hypothalamic samples of control, STZ-treated and STZ-treated and insulin-supplemented male mice, n = 11 for control, n = 7 for STZ, n = 10 for STZ + Ins treated male mice. *, P < 0.05, **, P < 0.01, ***, P < 0.001, ****, P < 0.0001. All the data are presented as mean ± SEM.
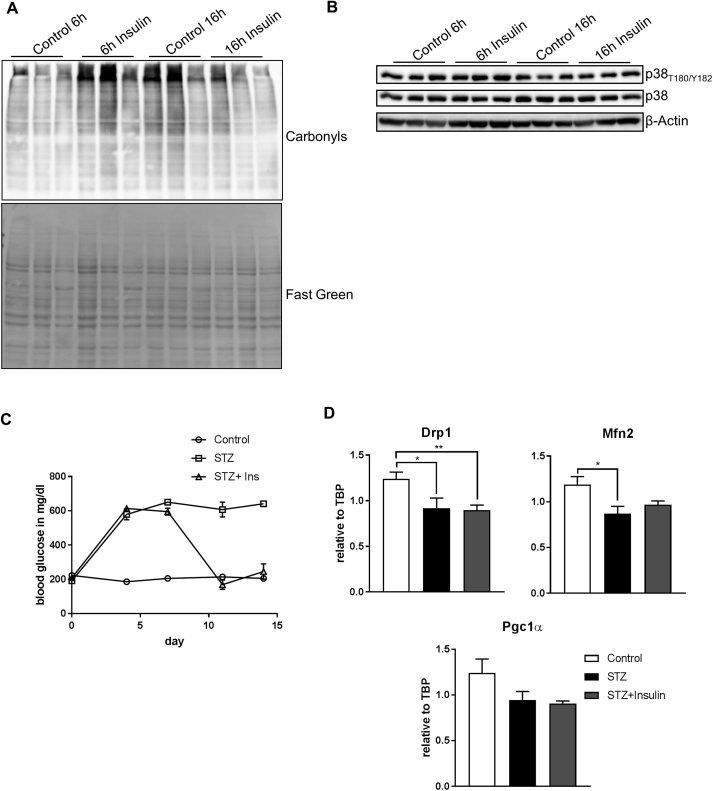
To rule out that prolonged insulin treatment was detrimental to neurons and caused cellular stress, MAPK activation and oxidative stress were investigated. This analysis showed that 6 h and 16 h insulin treatment caused neither oxidative stress nor p38 activation, indicating that this treatment was not detrimental in this experimental setup (Suppl. Fig. 3A,B). Prolonged insulin treatment has been shown to cause insulin resistance [31]. To show that proper insulin signaling and not insulin resistance was responsible for the observed effect on MSR regulation, we induced insulin resistance in vitro by stimulating cells with the JNK activator anisomycin [32]. 16 h co-treatment of insulin with the JNK activator anisomycin was sufficient to blunt completely insulin's effect in regulating MSR gene expression, highlighting that insulin signaling and not resistance causes regulation of the MSR (Figure 2F). Interestingly, sole activation of JNK was sufficient to activate Atf4 and Chop as part of the ISR but did not affect UPRmt gene expression.
To substantiate the transcriptional control of MSR genes by insulin, we analyzed MSR mRNA levels in hypothalamic samples of control, streptozotocin-treated/insulin deficient (STZ) mice along with STZ-treated mice treated with subcutaneous insulin pellets. As expected STZ treatment caused acute hyperglycemia, while insulin treatment for one week corrected hyperglycemia in STZ-treated mice (Suppl. Fig. 3C). Strikingly, STZ-treated animals exhibited an overall reduction of MSR mRNA levels in the hypothalamus, which was rescued by the insulin treatment, confirming the crucial role of hypothalamic insulin action in regulating the MSR (Figure 2G). Conversely with our in vitro studies (Suppl. Fig. 2F), insulin was not able to increase gene expression of Drp1 and Mfn2 as well as Pgc1α in STZ-treated animals (Suppl. Fig. 3D). These data demonstrate that insulin/IGF signaling induces specifically MSR gene expression in vitro and in vivo, while activation of JNK inhibits insulin's effect on the MSR expression in these experimental setups.
3.3. Insulin signaling regulates mitochondrial function and autophagy in hypothalamic neurons
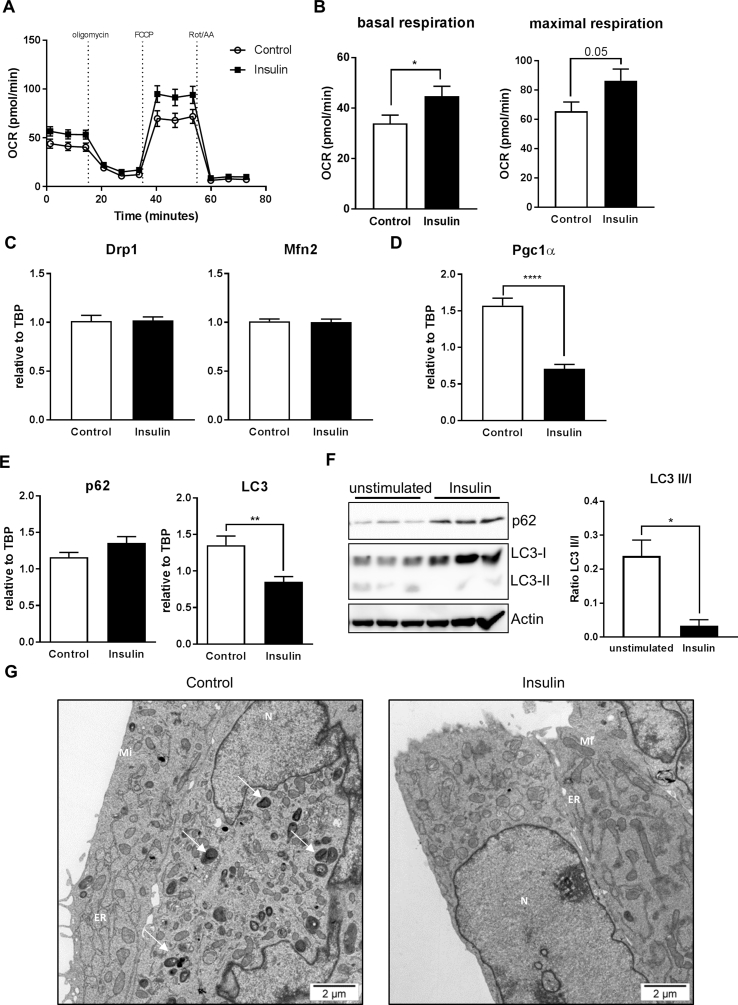
Next we tested whether insulin dependent control of MSR genes influences mitochondrial function and cell homeostasis. Serum-starved CLU183 cells were stimulated for 16 h with 100 nM insulin and the oxygen consumption rate was determined using a Seahorse XF analyzer. This analysis revealed an insulin-induced 32% increase in basal respiration and a concomitant 32% increase in maximal respiration (p = 0.05) compared to serum-starved control (Figure 3A,B). This enhanced mitochondrial respiration was not due to an increase in mitochondrial biogenesis or altered mitochondrial dynamics as evidenced by unaltered expression of mitochondrial fission and fusion genes Drp1 and Mfn2 with a reduction of the mitochondrial biogenesis gene Pgc1α on gene expression level after 16 h of Insulin stimulation (Figure 3C,D). Furthermore, this effect on the mitochondrial function was also not associated with a major regulation of key mitochondrial enzymes such complexes of the electron transport chain (ETC), shown by the unregulated protein expression of subunits of the ETC (Suppl. Fig 2H,I)
Figure 3.
Insulin increases mitochondrial function and inhibits autophagy. A, B: (A) Representative bioenergetics profile and its (B) analysis of unstimulated and 16 h insulin treated CLU183 cells using the Seahorse Bioflux Analyzer XF96e. n = 10–12 per treatment. This experiment has been independently repeated at least three times. C: mRNA expression of Drp1 and Mfn2 as marker for mitochondrial dynamics after 16 h of insulin stimulation of CLU183 cells. Data of three independent experiments with a total of n = 16–18. D: mRNA expression of Pgc-1α as a mitochondrial biogenesis marker after 16 h of insulin stimulation of CLU183 cells. Data of three independent experiments with a total of n = 16–18. E: mRNA expression of p62 and LC3 after 6 h insulin stimulation of CLU183 hypothalamic cells. Data of three independent experiments with a total of n = 13. F: Protein expression of p62 and LC3 and densitometric analysis after 16 h insulin stimulation of CLU183 hypothalamic cells, n = 3. β-Actin served as a loading control. Data from three independent experiments. G: Representative electron microscopy analysis of serum-starved and 16 h insulin treated CLU183 cells. N = Nucleus, Mi = Mitochondria, ER = Endoplasmatic reticulum. White arrows indicate autophagosomes. In total 77–92 pictures per group were taken. *, P < 0.05, **, P < 0.01, ***, P < 0.001, ****, P < 0.0001. All the data are presented as mean ± SEM.
Mitochondrial dysfunction and prolonged starvation induces autophagy as a stress-induced response to nutrient deprivation [33]. We hypothesized that insulin was able to maintain mitochondrial function and thus counteracts starvation-induced autophagy. 100 nM insulin stimulation for 16 h did not affect p62 gene expression but was sufficient to reduce LC3 expression by 53% indicating decreased autophagy (Figure 3E). Confirming this, insulin was able to inhibit serum-starvation induced autophagy as evidenced by increased p62 protein levels and a reduction in LC3II/I conversion compared to serum-starvation control (Figure 3F). In agreement with data from gene and protein expression analysis, electron microscopy experiments revealed the occurrence of autophagosomes in 16 h serum-deprived CLU183 cells, which were almost completely absent in insulin-stimulated cells (Figure 3G).
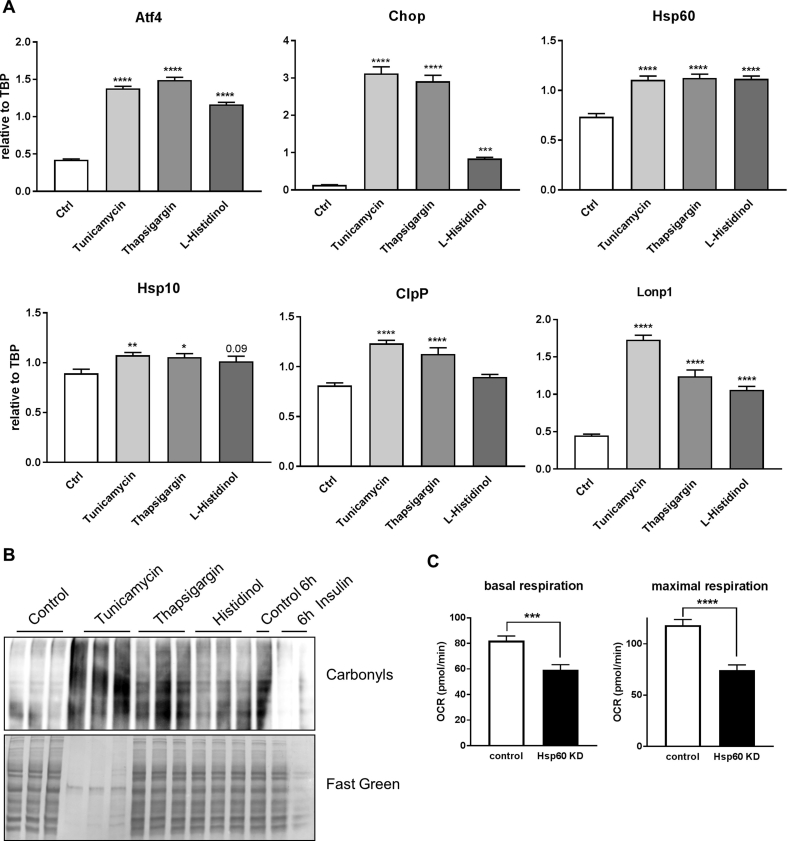
Our data show that insulin modestly increases expression of MSR genes and does not cause cellular stress. Yet, excessive activation of the MSR causes cellular death [34]. To further investigate this, we treated CLU183 cells with known activators of endoplasmatic reticulum (ER) stress and ISR and compared it to insulin's effect on oxidative stress as marker of cell stress. Treating cells with 5 μg/ml thapsigargin, 1 μM tunicamycin or 2 mM L-histidinol for 6 h increased mRNA levels of Atf4, Chop as well as Hsp60 and Hsp10 beyond normal values (Suppl. Fig. 4A). These treatments caused massive oxidative stress as evidenced by increased protein carbonylation, which was not apparent after insulin stimulation (Suppl. Fig. 4B) further highlighting that insulin's modest effect on MSR does not harm the cell.
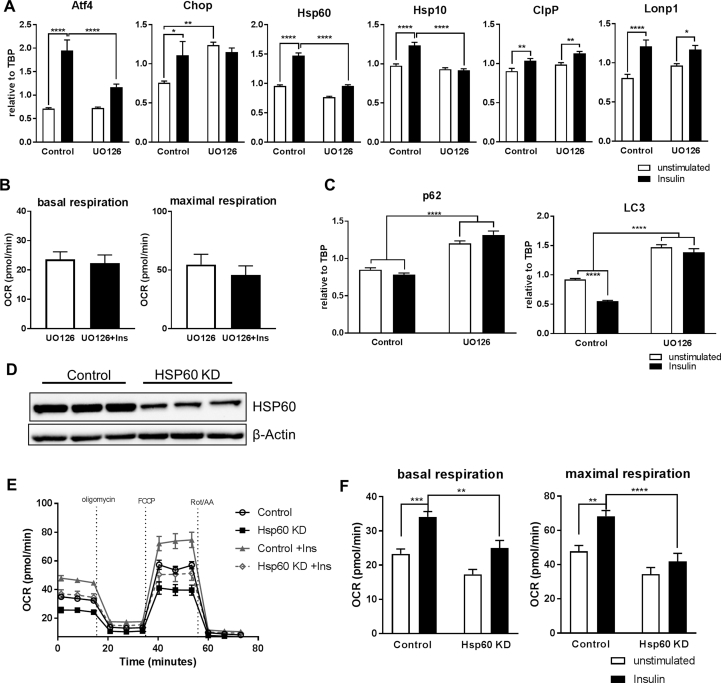
3.4. Insulin signaling controls MSR, mitochondrial function and autophagy via ERK activation
The obtained data suggests that hypothalamic insulin action can regulate the MSR in a concerted effort. In silico analysis of the promoters of Atf4, Chop, Hsp60, Hsp10, ClpP, and Lonp1 showed that these genes might be regulated via MAPK signaling (http://opossum.cisreg.ca/oPOSSUM3/). As insulin regulates the MAP-kinase pathway, we next inhibited ERK signaling using UO126. Pretreating cells with 10 μM UO126 followed by 16 h insulin stimulation abolished the insulin-induced increase in MSR gene expression of Atf4, Chop, Hsp60, and Hsp10. Unexpectedly, inhibition of ERK signaling increased Chop mRNA levels which was not further upregulated by insulin whereas gene expressions of ClpP and Lonp1 were independent of insulin-induced ERK activation (Figure 4A). As ERK-dependent insulin signaling upregulates the MSR genes Hsp60 and 10, which are key genes of mitochondrial protein folding and function, we investigated the influence of ERK inhibition on mitochondrial function using the Seahorse XF analyzer. UO126 treatment abrogated the insulin-induced increase of basal and maximal respiration in serum-deprived CLU183 cells (Figure 4B). Next, we investigated the autophagic response in insulin-treated CLU183 cells in the absence or presence of the ERK inhibitor UO126. ERK inhibition caused a significant 42% increase of p62 expression, which was not further influenced by insulin. Confirmatory, while insulin was able to decrease LC3 mRNA levels, LC3 expression was significantly upregulated in ERK inhibited cells and was unresponsive to insulin (Figure 4C). These data show that insulin via ERK signaling improves mitochondrial function and counteracts excessive autophagy under serum-deprived conditions.
Figure 4.
Insulin regulation of mitochondrial chaperone expression is ERK dependent, and the mitochondrial chaperone complex Hsp60/Hsp10 is crucial to convey beneficial effects of insulin on mitochondrial function. A: mRNA expression of Atf4, Chop, Hsp60, Hsp10, ClpP and Lonp1 of unstimulated or UO126 pre-treated CLU183 cells followed by 100 nM insulin stimulation for 16 h. Data of three independent experiments with a total of n = 11–12. B: Analysis of unstimulated or UO126 pre-treated CLU183 cells followed by 16 h of 100 nM insulin stimulation using the Seahorse Bioflux Analyzer XF96e of three independent experiments with an n = 18–24 per treatment. C: mRNA expression of p62 and LC3 of unstimulated and UO126 pre-treated CLU183 cells followed by 16 h of 100 nM insulin stimulation. Data of three independent experiments with a total of n = 25–27. D–F: (D) Verification of successful generated siRNA-mediated Hsp60 knockdown assessing protein expression in CLU183 cells. (E, F) Representative bioenergetics profile and its analysis of control and Hsp60 KD CLU183 cells and after 16 h of insulin stimulation and 16 h insulin treated CLU183 cells using the Seahorse Bioflux Analyzer XF96e. n = 15–16 per treatment. Combined data of three independent experiments. *, P < 0.05, **, P < 0.01, ***, P < 0.001, ****, P < 0.0001. All the data are presented as mean ± SEM.
We have recently shown that Hsp60 is a key mitochondrial chaperone and is crucial for mitochondrial homeostasis. In addition, insulin regulates Hsp60 expression via ERK activation and ERK inhibition causes mitochondrial dysfunction [35]. Thus, we investigated whether insulin's ability to propagate mitochondrial health depends on HSP60 expression. siRNA mediated knockdown (KD) of HSP60 caused a 79% reduction in protein levels (Figure 4D). KD of HSP60 in CLU183 cells exhibited an expected 28% decrease in basal and 37% decrease in maximal respiration compared to control under basal conditions (Suppl. Fig. 4C) [19]. Importantly, while 100 nM insulin stimulation for 16 h increased basal respiration by 36% and maximal respiration by 28% in serum-starved control cells, insulin's effect was severely blunted to regulate mitochondrial respiration in HSP60 knockdown cells (Figure 4E,F). These data demonstrate that insulin's ability to regulate mitochondrial function in serum deprivation depends on the presence of HSP60, underlining the importance of HSP60 to convey insulin's effects on mitochondrial function in the hypothalamus.
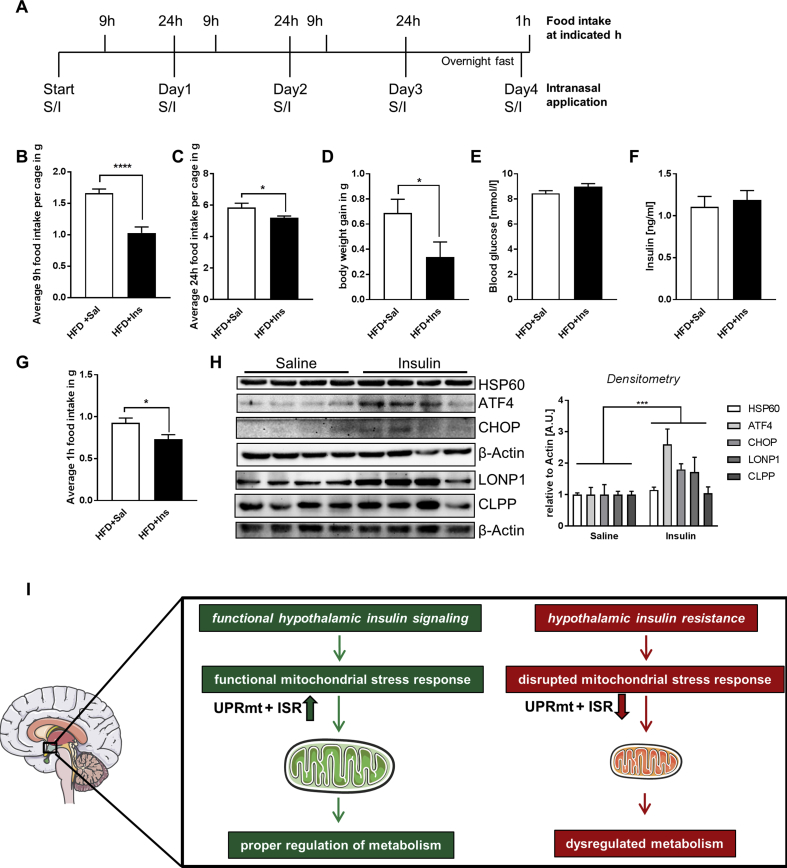
3.5. Intranasal insulin treatment decreases HFD-induced food intake and weight gain with an increased hypothalamic MSR activation
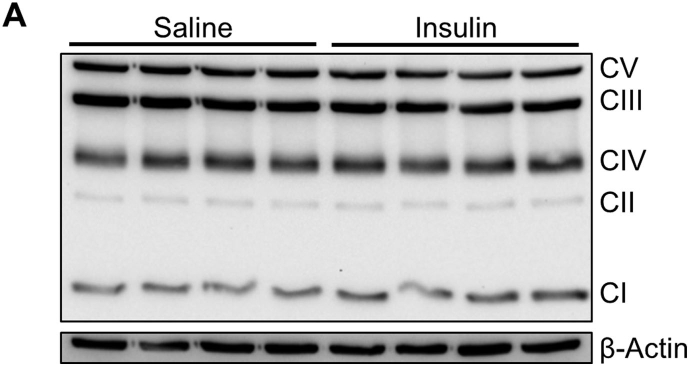
To test whether increased brain insulin action counteracts the negative consequences of diet-induced weight gain and regulates the MSR, we treated C57BL/6N mice with intranasal saline or 1.75U insulin for four consecutive days while feeding them a HFD (Figure 5A). Intranasal insulin treatment reduced food intake by maximal 38% after 9 h of treatment (Figure 5B), a time when insulin increases the expression of MSR genes (Figure 2A). Daily intranasal insulin application caused a 12% reduction of average daily food intake and a 50% reduction in body weight gain after three days of HFD while not affecting blood glucose or insulin levels (Figure 5C–F). To also assess the effect of intranasal insulin treatment on starvation-induced food intake after three days of HFD, mice were fasted overnight, followed by intranasal saline or insulin treatment and food intake was determined after 1 h. This analysis revealed that intranasal insulin treatment not only reduced food intake of palatable food, but also reduced starvation-induced food intake of HFD, showing the impact of intranasal insulin application on food intake regulation under HFD conditions (Figure 5G). Importantly, intranasal insulin application resulted specifically in elevated hypothalamic protein levels of MSR protein expression in the hypothalamus while not influencing the expression of other functional important mitochondrial proteins like the electron transport chain complexes (Figure 5H, Suppl. Fig. 5A), confirming our in vitro data (Figure 2B) and showing that insulin counteracts acute metabolic effects of HFD consumption with an increased hypothalamic MSR activation.
Figure 5.
Intranasal insulin application reduces food intake and weight development in HFD-fed mice. A: Experimental setup of intranasal insulin treatment of 6 week old male C57BL/6N mice feeding a HFD for a total of four days. S = saline, I = insulin. B: Average 9 h food intake per cage after intranasal insulin application of 6 week-old C57BL/6N mice fed a HFD. Two male mice were housed per cage with a total of 16 saline and 16 insulin treated mice. n = 8 (per treatment). C: Average 24 h food intake per cage after intranasal insulin application of mice fed a HFD. Two male mice were housed per cage with a total of 16 saline and 16 insulin treated mice. n = 8 (per treatment). D: Body weight gain of intranasal saline and insulin treated mice fed a HFD for three consecutive days. n = 16 (per treatment). E, F: (D) Blood glucose and (E) plasma insulin concentrations of mice fed a HFD and treated with intranasal saline or insulin for four consecutive days. Each group n = 15–16 (per treatment). G: Average 1 h HFD food intake after overnight fast of mice treated with intranasal saline or insulin. Each group n = 15–16. H: Hypothalamic protein expression of HSP60, ATF4, CHOP, LONP1, and CLPP and densitometric analysis of 6 week-old C57BL/6N mice fed a HFD after intranasal saline or insulin treatment, n = 4–8. β-Actin served as a loading control. two-way ANOVA, insulin's effect. All the data are presented as mean ± SEM. *, P < 0.05, **, P < 0.01, ***, P < 0.001, ****, P < 0.0001. I: Model of hypothalamic insulin regulation of the MSR.
4. Discussion
Brain insulin action has a crucial function in regulating metabolism ranging from controlling food intake over hepatic glucose production to behavior [36]. Conversely, brain insulin resistance is a hallmark of type 2 diabetes and is present in neurodegenerative diseases such as Alzheimer's disease. These disorders are also characterized by mitochondrial dysfunction. Thus, the ability of mitochondria to adapt to differences in nutrient supply and cellular stressors is of crucial importance for optimal metabolism. Deteriorated mitochondria cause excessive oxidative stress, inducing several adaptive stress responses such as the ISR and UPRmt or, in more severe cases, autophagy to reduce the burden of cellular stress. Insulin has been shown to exert neuro-protective functions and regulate mitochondrial function. By using multiple cell lines and mouse models, we identify hypothalamic insulin signaling as a major integrator of mitochondrial stress responsiveness. We demonstrate that insulin signaling controls the ISR and UPRmt - summarized as MSR - and as a result inhibits serum starvation-induced autophagy. Hypothalamic insulin resistance is linked to the reduction the expression of key genes of the MSR, which seems to dependent on proper insulin signaling, as this response can be rescued by insulin treatment in STZ-treated, insulin-deficient mice. As insulin was given peripherally, this effect may also be secondary due to peripheral effects. Yet, we show that the MSR is indeed controlled by insulin/IGF-1, activating their receptors and downstream ERK signaling in vitro and in vivo (Figure 2, Figure 5H) indicating that brain insulin action regulates hypothalamic MSR. This regulation improves mitochondrial function and inhibits autophagy under metabolic, serum-deprived stress and thus propagates neuronal health. We show that the mitochondrial chaperone HSP60 is a key target in this response as knockdown of HSP60 ablates insulin's protective effect on neuronal mitochondrial function. Finally, we demonstrate that intranasal insulin counteracts HFD-induced metabolic alterations by reducing food intake and concomitantly activating the MSR in the hypothalamus.
It has been shown that long-term HFD feeding causes disrupted mitochondria and impaired mitochondrial biogenesis with reduced expression of Mfn2 and Pgc1α in the hypothalamus [28], [37]. Our data confirm the reduction of Mfn2 after acute HFD treatment, but reveal unaltered Pgc1α levels. From a mechanistic standpoint, it is unlikely that insulin directly regulates Mfn2 expression in the hypothalamus as insulin does not affect its expression in vitro or in vivo but it is rather a consequence of altered metabolism (Figure 3C, Suppl. Fig. 3D). In addition, we do not observe a reduction of Pgc1α after acute HFD feeding, indicating Pgc1α regulation is only observed after long-term obesity or severe hyperglycemia (Suppl. Fig. 3D) [37]. Further, insulin decreases Pgc1α gene expression in vitro but not in vivo, suggesting that Pgc1α gene expression in the hypothalamus is under complex regulation and further research is needed [38]. Thus, we show for the first time that hypothalamic insulin action improves mitochondrial function not by affecting mitochondrial biogenesis but by controlling mitochondrial proteostasis. In addition, intranasal insulin treatment regulates food intake with activation of the hypothalamic MSR.
We show that insulin/IGF-1 action in the hypothalamus can act as a master regulator of the UPRmt and ISR in healthy conditions, thereby controlling adaptive responses to ensure neuronal health. Neuronal insulin receptor deficiency results in mitochondrial dysfunction with smaller mitochondria showing that insulin receptor signaling is important for mitochondrial health in the brain [39]. The fact that loss of IR expression does not affect basal expression rates of the MSR (Figure 2D) suggests either that loss of IR can be compensated by IGF-1R expression or that basal expression is not controlled by insulin. Consistent with this hypothesis, the promoter of Hsp60 and 10 does not contain a TATA-Box, which enables constant, basal transcription [40], but several Sp1 binding sites. Interestingly, insulin activates Sp1, which may facilitate insulin-induced regulation of at least the mitochondrial chaperones Hsp60 and Hsp10 [41].
In vitro experiments using mammalian cells have revealed that mitochondrial protein misfolding activates the serine/threonine protein kinase JNK, which is crucial for the UPRmt [42]. This response causes increased expression of Hsp60 and Hsp10 [10]. Yet, in hypothalami of insulin resistant, deficient and type 2 diabetic mice, Hsp60 and Hsp10 are downregulated and JNK is activated (Figure 1C, Suppl. Fig. 1I) (see also Attie lab diabetes database, http://diabetes.wisc.edu/) [19]. Our data show that an activation of JNK inhibits insulin-induced gene expression of the UPRmt in the hypothalamus, yet is sufficient to induce the ISR (Figure 2F). These data highlight that in metabolic stressed conditions the regulation of the different MSR genes is more complex than previously anticipated and warrants further investigation.
Intranasal insulin application reduces food intake and increases ATP levels in the brain in humans which relates to improved mitochondrial function, phenotypes we also observed in mice after intranasal insulin application [18]. It will be of interest to test whether our proposed insulin regulatory mechanism is also present in humans.
In addition, intranasal insulin treatment attenuates the cognitive decline in some patients with Alzheimer's disease, showing that insulin action is also neuro-protective in humans [43]. It seems that patients who do not profit from these intranasal insulin interventions show a stronger link between mitochondrial damage and cognitive decline, suggesting that this effect is difficult to regulate by insulin therapy [44], [45], [46]. Interestingly, these ApoE4 allele carrying patients exhibit already abnormal, elevated levels of MSR genes such as ATF4, presumably due to ERK activation, suggesting that insulin cannot further regulate this response [47], [48]. It will be interesting to see whether patients who benefit from intranasal insulin therapy have low levels of MSR genes in basal conditions. Here, whole genome biomarker expression analysis in blood and brain samples have shown good correlations for gene expression of the MSR gene ATF4 in PBMC compared to brain, indicating that this method may be useful for clarifying the effect of intranasal insulin treatment on the MSR in the brain [49]. Thus, further research is needed to identify these potential treatment-susceptible patient cohorts.
As mitochondrial dysfunction and insulin resistance represent common factors in brain diseases, dysregulation of MSR might be a crucial part for the development of brain diseases. Indeed, defective expression and/or mutations of tested MSR genes are linked to brain alterations, behavioral abnormalities and cognitive impairments [20], [21], [50], [51], [52].
As hyperinsulinemia can cause per se brain insulin resistance, improving insulin sensitivity by use of insulin sensitizers such as metformin might help to improve the MSR in insulin resistant brains, since metformin has been shown to increases MSR genes, reduce oxidative stress, and improve mitochondrial as well as brain function in HFD-fed animals [53], [54]. There is a current clinical trial listed to assess metformin on brain insulin action on elderly patients. It will be interesting to see how this treatment affects brain energy levels and insulin action in the brain of these patients.
The fact that hypothalami of male HFD fed mice exhibit increased protein carbonylation suggests that also proteasomal activity might be affected which counteracts protein toxicity. Insulin is able to regulate proteasomal activity and it has been demonstrated that proteasomal activity via Nrf1 can act as a complementary mechanism of MSR in brown fat [55]. But whether this holds true for the brain is so far unknown.
We and others have previously shown that the reduction of MSR gene Hsp60 causes hypothalamic insulin resistance and neurodegeneration pointing to the importance of this response for metabolism and brain health [19], [21], [22], [56]. Here, we show that insulin-induced MSR activation is crucial to antagonize serum-deprivation induced mitochondrial dysfunction and autophagy unveiling a novel mechanism of how insulin is able to regulate neuronal metabolism. The fact that elevated Chop levels are linked to improvement of mitochondrial function and inhibition of autophagy reveals that induction of Chop does not constitute a death signal but can be beneficial for neurons as in the case of neuronal survival after seizures [57]. In addition, it has been shown that starvation-induced hypothalamic autophagy is crucial to increase Agrp expression and food intake [58]. Our finding that insulin is able to inhibit starvation-induced autophagy in hypothalamic neurons points to a novel mechanism how hypothalamic insulin action is able to reduce food intake. Indeed, we observe that daily intranasal insulin treatment reduces food intake and increases MSR activation in the hypothalamus of mice fed a HFD diet, indicating that this regulation is metabolically relevant.
In summary, we define hypothalamic insulin/IGF-1 signaling as a new, crucial regulator of mitochondrial stress responsiveness via ERK activation and with this, the propagation of mitochondrial function and inhibition of serum-deprived induced autophagy. With functional hypothalamic insulin signaling the metabolism is flexible and able to adapt to different nutrient supply. In the case of insulin resistance in the hypothalamus, the mitochondrial stress response is disrupted, leading to mitochondrial dysfunction, excessive autophagy, and increased weight gain (Model in Figure 5I). In extreme cases, this mitochondrial dysfunction can even lead to neuronal cell death, as previously described in cases of age-associated neurodegenerative diseases like Alzheimer's disease [59]. Our data offer a novel insight into how brain insulin action via controlling adaptive stress responses regulates neuronal health, acts as a neuroprotective hormone, and regulates metabolism.
Author contributions
K.W., S.B., and M.R. designed the study, researched data, and wrote the manuscript. E.A., C.C., M.S., R.H., W.C., K.W., T.F., K.R., J.W., and C.R.K. researched data and helped design experiments. A.K. designed the study, supervised all work, and wrote the manuscript.
Funding
We thank Dr. Michael Schupp for providing plasmids (insulin receptor gRNA plasmid). This work was supported by the Deutsche Forschungsgemeinschaft (DFG) grant project KL 2399/4-1 (to A.K.) and by a grant from the German Ministry of Education and Research (BMBF) and the State of Brandenburg (DZD grant 82DZD00302). This work was also supported by NIH grants R37 DK031036 and R01 DK033201 (to C.R.K.). The authors declare no conflict of interest or competing financial interests.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2019.01.001.
Conflict of interest
None declared.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Fig S1.
Fig S2.
Fig S3.
Fig S4.
Fig S5.
References
- 1.Chan D.C. Mitochondria: dynamic organelles in disease. Aging and Development Cell. 2006;125:1241–1252. doi: 10.1016/j.cell.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 2.Boucher J., Kleinridders A., Kahn C.R. Insulin receptor signaling in normal and insulin-resistant states. Cold Spring Harbor Perspectives in Biology. 2014;6 doi: 10.1101/cshperspect.a009191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pipatpiboon N., Pratchayasakul W., Chattipakorn N., Chattipakorn S.C. PPARgamma agonist improves neuronal insulin receptor function in hippocampus and brain mitochondria function in rats with insulin resistance induced by long term high-fat diets. Endocrinology. 2012;153:329–338. doi: 10.1210/en.2011-1502. [DOI] [PubMed] [Google Scholar]
- 4.Edwards J.L., Quattrini A., Lentz S.I., Figueroa-Romero C., Cerri F., Backus C. Diabetes regulates mitochondrial biogenesis and fission in mouse neurons. Diabetologia. 2010;53:160–169. doi: 10.1007/s00125-009-1553-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang T.J., Price S.A., Chilton L., Calcutt N.A., Tomlinson D.R., Verkhratsky A. Insulin prevents depolarization of the mitochondrial inner membrane in sensory neurons of type 1 diabetic rats in the presence of sustained hyperglycemia. Diabetes. 2003;52:2129–2136. doi: 10.2337/diabetes.52.8.2129. [DOI] [PubMed] [Google Scholar]
- 6.Friedman J. Why is the nervous System vulnerable to oxidative stress? In: Gadoth N., Göbel H.H., editors. Oxidative stress and free radical damage in neurology. Humana Press; Totowa, NJ: 2011. pp. 19–27. [Google Scholar]
- 7.Miyazaki I., Asanuma M. Dopaminergic neuron-specific oxidative stress caused by dopamine itself. Acta Medica Okayama. 2008;62:141–150. doi: 10.18926/AMO/30942. [DOI] [PubMed] [Google Scholar]
- 8.Chaturvedi R.K., Flint Beal M. Mitochondrial diseases of the brain. Free Radical Biology and Medicine. 2013;63:1–29. doi: 10.1016/j.freeradbiomed.2013.03.018. [DOI] [PubMed] [Google Scholar]
- 9.Quiros P.M., Prado M.A., Zamboni N., D'Amico D., Williams R.W., Finley D. Multi-omics analysis identifies ATF4 as a key regulator of the mitochondrial stress response in mammals. The Journal of Cell Biology. 2017;216:2027–2045. doi: 10.1083/jcb.201702058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao Q., Wang J., Levichkin I.V., Stasinopoulos S., Ryan M.T., Hoogenraad N.J. A mitochondrial specific stress response in mammalian cells. The EMBO Journal. 2002;21:4411–4419. doi: 10.1093/emboj/cdf445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kroemer G., Marino G., Levine B. Autophagy and the integrated stress response. Molecular Cell. 2010;40:280–293. doi: 10.1016/j.molcel.2010.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Houtkooper R.H., Mouchiroud L., Ryu D., Moullan N., Katsyuba E., Knott G. Mitonuclear protein imbalance as a conserved longevity mechanism. Nature. 2013;497:451–457. doi: 10.1038/nature12188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mottis A., Jovaisaite V., Auwerx J. The mitochondrial unfolded protein response in mammalian physiology. Mammalian Genome. 2014;25:424–433. doi: 10.1007/s00335-014-9525-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun C., Zhang F., Ge X., Yan T., Chen X., Shi X. SIRT1 improves insulin sensitivity under insulin-resistant conditions by repressing PTP1B. Cell Metabolism. 2007;6:307–319. doi: 10.1016/j.cmet.2007.08.014. [DOI] [PubMed] [Google Scholar]
- 15.Wijsman C.A., Rozing M.P., Streefland T.C., le Cessie S., Mooijaart S.P., Slagboom P.E. Familial longevity is marked by enhanced insulin sensitivity. Aging Cell. 2011;10:114–121. doi: 10.1111/j.1474-9726.2010.00650.x. [DOI] [PubMed] [Google Scholar]
- 16.Benedict C., Hallschmid M., Hatke A., Schultes B., Fehm H.L., Born J. Intranasal insulin improves memory in humans. Psychoneuroendocrinology. 2004;29:1326–1334. doi: 10.1016/j.psyneuen.2004.04.003. [DOI] [PubMed] [Google Scholar]
- 17.Heni M., Wagner R., Kullmann S., Veit R., Mat Husin H., Linder K. Central insulin administration improves whole-body insulin sensitivity via hypothalamus and parasympathetic outputs in men. Diabetes. 2014;63:4083–4088. doi: 10.2337/db14-0477. [DOI] [PubMed] [Google Scholar]
- 18.Jauch-Chara K., Friedrich A., Rezmer M., Melchert U.H., Scholand-Engler H.G., Hallschmid M. Intranasal insulin suppresses food intake via enhancement of brain energy levels in humans. Diabetes. 2012;61:2261–2268. doi: 10.2337/db12-0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kleinridders A., Lauritzen H.P., Ussar S., Christensen J.H., Mori M.A., Bross P. Leptin regulation of Hsp60 impacts hypothalamic insulin signaling. Journal of Clinical Investigation. 2013;123:4667–4680. doi: 10.1172/JCI67615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bie A.S., Fernandez-Guerra P., Birkler R.I., Nisemblat S., Pelnena D., Lu X. Effects of a mutation in the HSPE1 gene encoding the mitochondrial Co-chaperonin HSP10 and its potential association with a neurological and developmental disorder. Frontiers in Molecular Biosciences. 2016;3:65. doi: 10.3389/fmolb.2016.00065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hansen J.J., Durr A., Cournu-Rebeix I., Georgopoulos C., Ang D., Nielsen M.N. Hereditary spastic paraplegia SPG13 is associated with a mutation in the gene encoding the mitochondrial chaperonin Hsp60. The American Journal of Human Genetics. 2002;70:1328–1332. doi: 10.1086/339935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Magnoni R., Palmfeldt J., Christensen J.H., Sand M., Maltecca F., Corydon T.J. Late onset motoneuron disorder caused by mitochondrial Hsp60 chaperone deficiency in mice. Neurobiology of Disease. 2013;54:12–23. doi: 10.1016/j.nbd.2013.02.012. [DOI] [PubMed] [Google Scholar]
- 23.Magnoni R., Palmfeldt J., Hansen J., Christensen J.H., Corydon T.J., Bross P. The Hsp60 folding machinery is crucial for manganese superoxide dismutase folding and function. Free Radical Research. 2014;48:168–179. doi: 10.3109/10715762.2013.858147. [DOI] [PubMed] [Google Scholar]
- 24.astro J.P., Wardelmann K., Grune T., Kleinridders A. Mitochondrial chaperones in the brain: safeguarding brain health and metabolism? Frontiers in Endocrinology. 2018;9:196. doi: 10.3389/fendo.2018.00196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Castro J.P., Ott C., Jung T., Grune T., Almeida H. Carbonylation of the cytoskeletal protein actin leads to aggregate formation. Free Radical Biology and Medicine. 2012;53:916–925. doi: 10.1016/j.freeradbiomed.2012.06.005. [DOI] [PubMed] [Google Scholar]
- 26.Reale E., Luciano L., Brandes G. Alterations in the morphology of glycoconjugate molecules caused by histochemical procedures: comparison of renal glomeruli and articular cartilage. The Histochemical Journal. 1992;24:153–165. doi: 10.1007/BF01047465. [DOI] [PubMed] [Google Scholar]
- 27.Luft J.H. Improvements in epoxy resin embedding methods. The Journal of Biophysical and Biochemical Cytology. 1961;9:409–414. doi: 10.1083/jcb.9.2.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schneeberger M., Dietrich M.O., Sebastian D., Imbernon M., Castano C., Garcia A. Mitofusin 2 in POMC neurons connects ER stress with leptin resistance and energy imbalance. Cell. 2013;155:172–187. doi: 10.1016/j.cell.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Munch C. The different axes of the mammalian mitochondrial unfolded protein response. BMC Biology. 2018;16:81. doi: 10.1186/s12915-018-0548-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Munch C., Harper J.W. Mitochondrial unfolded protein response controls matrix pre-RNA processing and translation. Nature. 2016;534:710–713. doi: 10.1038/nature18302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mayer C.M., Belsham D.D. Central insulin signaling is attenuated by long-term insulin exposure via insulin receptor substrate-1 serine phosphorylation, proteasomal degradation, and lysosomal insulin receptor degradation. Endocrinology. 2010;151:75–84. doi: 10.1210/en.2009-0838. [DOI] [PubMed] [Google Scholar]
- 32.Aguirre V., Uchida T., Yenush L., Davis R., White M.F. The c-Jun NH(2)-terminal kinase promotes insulin resistance during association with insulin receptor substrate-1 and phosphorylation of Ser(307) Journal of Biological Chemistry. 2000;275:9047–9054. doi: 10.1074/jbc.275.12.9047. [DOI] [PubMed] [Google Scholar]
- 33.Klionsky D.J., Abdelmohsen K., Abe A., Abedin M.J., Abeliovich H., Acevedo Arozena A. 3rd ed. vol. 12. 2016. pp. 1–222. (Guidelines for the use and interpretation of assays for monitoring autophagy). Autophagy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hiramatsu N., Chiang W.C., Kurt T.D., Sigurdson C.J., Lin J.H. Multiple mechanisms of unfolded protein response-induced cell death. American Journal of Pathology. 2015;185:1800–1808. doi: 10.1016/j.ajpath.2015.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ripple M.O., Kim N., Springett R. Acute mitochondrial inhibition by mitogen-activated protein kinase/extracellular signal-regulated kinase kinase (MEK) 1/2 inhibitors regulates proliferation. Journal of Biological Chemistry. 2013;288:2933–2940. doi: 10.1074/jbc.M112.430082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kleinridders A., Ferris H.A., Cai W., Kahn C.R. Insulin action in brain regulates systemic metabolism and brain function. Diabetes. 2014;63:2232–2243. doi: 10.2337/db14-0568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morselli E., Fuente-Martin E., Finan B., Kim M., Frank A., Garcia-Caceres C. Hypothalamic PGC-1alpha protects against high-fat diet exposure by regulating ERalpha. Cell Reports. 2014;9:633–645. doi: 10.1016/j.celrep.2014.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fernandez-Marcos P.J., Auwerx J. Regulation of PGC-1alpha, a nodal regulator of mitochondrial biogenesis. American Journal of Clinical Nutrition. 2011;93:884S–890S. doi: 10.3945/ajcn.110.001917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kleinridders A., Cai W., Cappellucci L., Ghazarian A., Collins W.R., Vienberg S.G. Insulin resistance in brain alters dopamine turnover and causes behavioral disorders. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:3463–3468. doi: 10.1073/pnas.1500877112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hansen J.J., Bross P., Westergaard M., Nielsen M.N., Eiberg H., Borglum A.D. Genomic structure of the human mitochondrial chaperonin genes: HSP60 and HSP10 are localised head to head on chromosome 2 separated by a bidirectional promoter. Human Genetics. 2003;112:71–77. doi: 10.1007/s00439-002-0837-9. [DOI] [PubMed] [Google Scholar]
- 41.Solomon S.S., Majumdar G., Martinez-Hernandez A., Raghow R. A critical role of Sp1 transcription factor in regulating gene expression in response to insulin and other hormones. Life Sciences. 2008;83:305–312. doi: 10.1016/j.lfs.2008.06.024. [DOI] [PubMed] [Google Scholar]
- 42.Horibe T., Hoogenraad N.J. The chop gene contains an element for the positive regulation of the mitochondrial unfolded protein response. PLoS One. 2007;2:e835. doi: 10.1371/journal.pone.0000835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Craft S., Baker L.D., Montine T.J., Minoshima S., Watson G.S., Claxton A. Intranasal insulin therapy for Alzheimer disease and amnestic mild cognitive impairment: a pilot clinical trial. Archives of Neurology. 2012;69:29–38. doi: 10.1001/archneurol.2011.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Freiherr J., Hallschmid M., Frey W.H., 2nd, Brunner Y.F., Chapman C.D., Holscher C. Intranasal insulin as a treatment for Alzheimer's disease: a review of basic research and clinical evidence. CNS Drugs. 2013;27:505–514. doi: 10.1007/s40263-013-0076-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Risner M.E., Saunders A.M., Altman J.F., Ormandy G.C., Craft S., Foley I.M. Efficacy of rosiglitazone in a genetically defined population with mild-to-moderate Alzheimer's disease. The Pharmacogenomics Journal. 2006;6:246–254. doi: 10.1038/sj.tpj.6500369. [DOI] [PubMed] [Google Scholar]
- 46.Gibson G.E., Haroutunian V., Zhang H., Park L.C., Shi Q., Lesser M. Mitochondrial damage in Alzheimer's disease varies with apolipoprotein E genotype. Annals of Neurology. 2000;48:297–303. [PubMed] [Google Scholar]
- 47.Hoe H.S., Harris D.C., Rebeck G.W. Multiple pathways of apolipoprotein E signaling in primary neurons. Journal of Neurochemistry. 2005;93:145–155. doi: 10.1111/j.1471-4159.2004.03007.x. [DOI] [PubMed] [Google Scholar]
- 48.Segev Y., Barrera I., Ounallah-Saad H., Wibrand K., Sporild I., Livne A. PKR inhibition rescues memory deficit and ATF4 overexpression in ApoE epsilon4 human replacement mice. Journal of Neuroscience. 2015;35:12986–12993. doi: 10.1523/JNEUROSCI.5241-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rollins B., Martin M.V., Morgan L., Vawter M.P. Analysis of whole genome biomarker expression in blood and brain. American Journal of Medical Genetics Part B Neuropsychiatric Genetics. 2010;153B:919–936. doi: 10.1002/ajmg.b.31062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Theunissen T.E., Szklarczyk R., Gerards M., Hellebrekers D.M., Mulder-Den Hartog E.N., Vanoevelen J. Specific MRI abnormalities reveal severe perrault syndrome due to CLPP defects. Frontiers in Neurology. 2016;7:203. doi: 10.3389/fneur.2016.00203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Strauss K.A., Jinks R.N., Puffenberger E.G., Venkatesh S., Singh K., Cheng I. CODAS syndrome is associated with mutations of LONP1, encoding mitochondrial AAA+ Lon protease. The American Journal of Human Genetics. 2015;96:121–135. doi: 10.1016/j.ajhg.2014.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pasini S., Corona C., Liu J., Greene L.A., Shelanski M.L. Specific downregulation of hippocampal ATF4 reveals a necessary role in synaptic plasticity and memory. Cell Reports. 2015;11:183–191. doi: 10.1016/j.celrep.2015.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pintana H., Apaijai N., Pratchayasakul W., Chattipakorn N., Chattipakorn S.C. Effects of metformin on learning and memory behaviors and brain mitochondrial functions in high fat diet induced insulin resistant rats. Life Sciences. 2012;91:409–414. doi: 10.1016/j.lfs.2012.08.017. [DOI] [PubMed] [Google Scholar]
- 54.Quentin T., Steinmetz M., Poppe A., Thoms S. Metformin differentially activates ER stress signaling pathways without inducing apoptosis. Disease Models & Mechanisms. 2012;5:259–269. doi: 10.1242/dmm.008110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bartelt A., Widenmaier S.B., Schlein C., Johann K., Goncalves R.L.S., Eguchi K. Brown adipose tissue thermogenic adaptation requires Nrf1-mediated proteasomal activity. Nature Medicine. 2018;24:292–303. doi: 10.1038/nm.4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Magen D., Georgopoulos C., Bross P., Ang D., Segev Y., Goldsher D. Mitochondrial hsp60 chaperonopathy causes an autosomal-recessive neurodegenerative disorder linked to brain hypomyelination and leukodystrophy. The American Journal of Human Genetics. 2008;83:30–42. doi: 10.1016/j.ajhg.2008.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Engel T., Sanz-Rodgriguez A., Jimenez-Mateos E.M., Concannon C.G., Jimenez-Pacheco A., Moran C. CHOP regulates the p53-MDM2 axis and is required for neuronal survival after seizures. Brain. 2013;136:577–592. doi: 10.1093/brain/aws337. [DOI] [PubMed] [Google Scholar]
- 58.Kaushik S., Rodriguez-Navarro J.A., Arias E., Kiffin R., Sahu S., Schwartz G.J. Autophagy in hypothalamic AgRP neurons regulates food intake and energy balance. Cell Metabolism. 2011;14:173–183. doi: 10.1016/j.cmet.2011.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gibson G.E., Starkov A., Blass J.P., Ratan R.R., Beal M.F. Cause and consequence: mitochondrial dysfunction initiates and propagates neuronal dysfunction, neuronal death and behavioral abnormalities in age-associated neurodegenerative diseases. Biochimica et Biophysica Acta 1802. 2010:122–134. doi: 10.1016/j.bbadis.2009.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.