Main Text

Thank you, Dave Valle, for that kind introduction and thanks to our Society for this terrific honor of the McKusick Leadership Award, which has very special meaning coming from the Society in which I grew up professionally. Experiential advice for the students, postdocs, and junior faculty that are present here today is that often some of your most important mentors are not at your institution, but rather sitting next to you here at the American Society of Human Genetics Annual Meeting; introduce yourself and get involved in your Society.
Genetic and Genomic Variation of the Human Genome
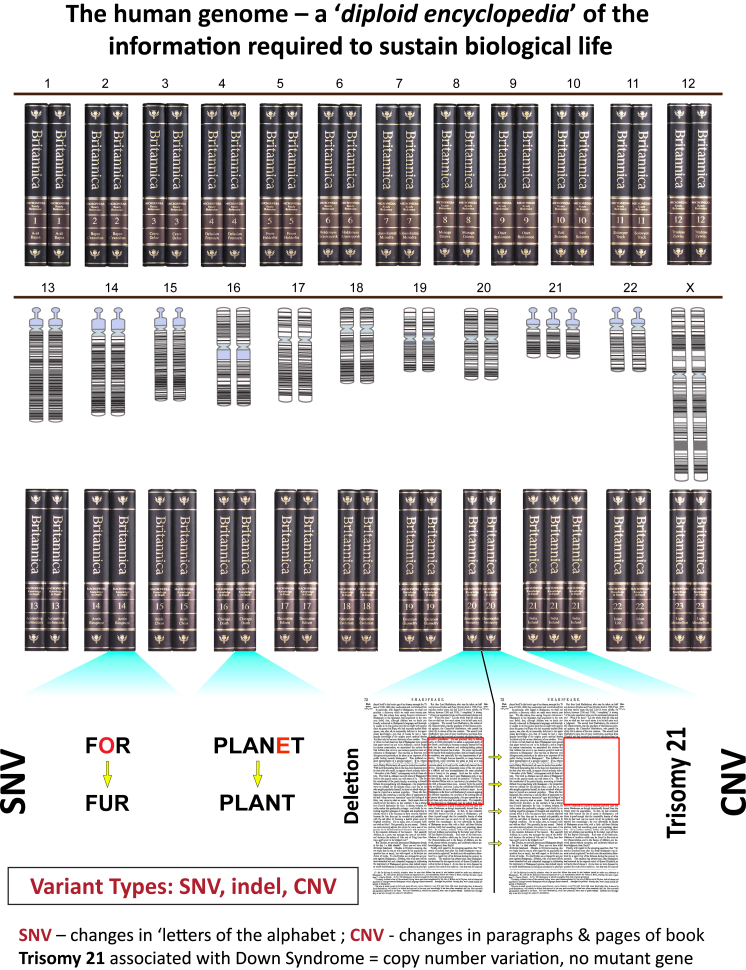
When we think of the human genome and how specific variation at a locus may result in different phenotypic outcomes, it is perhaps a good analogy to think of a diploid (i.e., two copies of each volume) 23-volume encyclopedia of the information required to sustain biological life (Figure 1). In this encyclopedia one has several “variant types:” a chromosome might be thought of as one volume of the encyclopedia. Single, simple, or small nucleotide variants (SNVs; Watson-Crick base pair changes), a type of variation that has also been referred to as a SNP (single-nucleotide polymorphism), could be analogized as a change in the alphabet that could change the meaning of a word. Likewise, indels, or small (<50 bp) insertion/deletions of Watson-Crick base pairs, could also result in a change in the meaning of the language of genetics by altering a word (codon), sentence (exon), or paragraph (gene) (Figure 1). However, there is another type or category of genetic and genomic variation, of a size between that which can be visualized microscopically by changes to a chromosome and that readily discerned molecularly by short read nucleotide sequencing—i.e., structural variants (SVs), including copy number variants (CNVs) (Figure 1). These CNVs can alter the gene copy-number and dosage at a genetic locus, causing deviations from the normal diploid state. Such deviations can have implications for haploinsufficiency and triplosensitivity disease traits; affect the interpretation of marker genotypes at a locus, sometimes distorting the results of linkage analyses;4, 5, 6 potentially inflate the significance of association studies;7 and if involving genes or even just single exons may contribute to disease gene discovery8 and molecular diagnoses (CNV + SNV).9, 10
Figure 1.
The Diploid Human Genome Can Be Analogized by a Duplicate Copy of a 23-Volume Encyclopedia Representing the Two-Copy “Diploid” Encyclopedia of Information Required to Sustain Human Biological Life
Volumes 1 through 12 of the encyclopedia; directly underneath are shown the chromosome karyograms and books for volumes numbered 13 through 23 (i.e., chromosome X in 46,XX females). The bottom of the page shows the different variant types and how they affect information flow for the words, sentences, paragraphs, pages, and volumes of the analogized human genome encyclopedia. Note, a typical SNV or Watson-Crick base pair changes that can alter the reading of the genetic code at that locus can completely distort the meaning of a word. In this analogy, trisomy 21 is shown to represent a large copy-number variation with three copies of volume 21 rather than the usual two in the diploid genome. Of note, the current version of the human genome is a haploid reference genome that does not account for copy-number variant information or SV haplotypes.1, 2, 3
In some respects, trisomy 21 (47,XY,+21), which is the variation of the human genome that causes one of the most commonly observed clinical phenotypes that we see as pediatricians and clinical geneticists, Down syndrome, is due to one large CNV. In trisomy 21, there are not really changes to specific genes, in terms of what you might observe as de novo DNA sequence changes specifying the sporadic trait, but instead three rather than the usual two copies of all genes on chromosome 21; i.e., at the chromosome 21 locus. From an alternative viewpoint, Down syndrome might be considered a multigenic trait with all the dosage-sensitive, i.e., triplosensitive, genes that map to chromosome 21 contributing to trait manifestation.
Structural Variants, Gene Dosage, and Genomic Disorders: The Evolution of a Human and Medical Genetics Concept
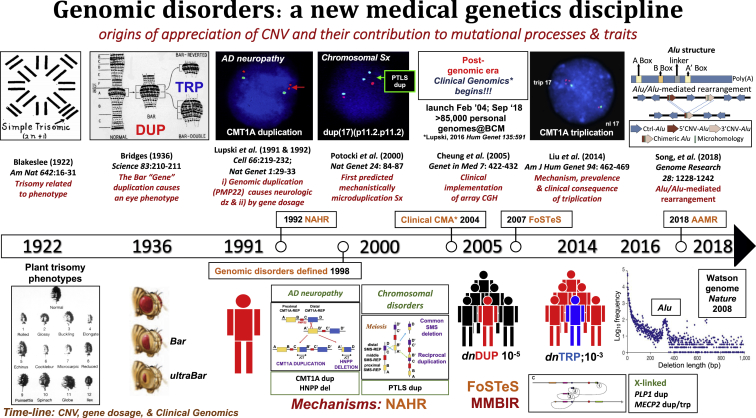
In genetics and genomics, it is of interest to trace the origins of the appreciation of CNVs, their contribution to gene dosage-dependent phenotypes or trait manifestations, and the evolution of our thinking regarding SV mutagenesis mechanisms (Figure 2). One of the first seminal papers that documented dosage, i.e., trisomy related to phenotype, was a study published almost a century ago in 1922 by Albert Blakeslee at the Cold Spring Harbor Laboratories in New York.12 He showed that the plant Datura, which has 12 chromosomes, could be found to have an extra chromosome of each one of the individual chromosomes (he coined the term simple trisomy, 2n+1), and there was an observed different plant phenotype for each of the different chromosomal trisomies (Figure 2). In 1936, Calvin Bridges used a relatively new genome technology, polytene chromosomes, to demonstrate that duplication of the Bar gene locus causes the bar eye phenotype. Bridges was part of the “fly group” at Columbia University but also performed some of these experiments at CSHL. Importantly, he showed that reversion of the duplication genotype results in a wild-type phenotype whereas triplication at the locus gave a more severe phenotype termed ultraBar13 (Figure 2). It is interesting to note his discussion and interpretation of his data as the concept of “position effect” was perhaps prominent in the genetic thinking of the time: “The respective shares attributable in the total effect to the genic balance change (i.e., dosage) and to the position-effect change seems to be at present a matter of taste.”13
Figure 2.
Timeline of Genetic Observations and Genetic and Genomic Variation Tied to Trait Manifestation Regarding Genomic Disorders and Medical Genetics/Genomics Practice
Above, key findings and papers. Below, phenotypes and traits. Also noted on the timeline, with the genome “phenotype” below, are the three major SV mutagenesis mechanisms: NAHR, FoSTeS/MMBIR, and AAMR and the de novo mutation frequencies for CMT1A duplication and triplication. The CMT1A duplication discovery opened a new field of genomic disorders “diseases caused by rearrangements of the genome incited by a genomic architecture that conveys instability.” The conceptualization and mechanistic understanding (NAHR, FoSTeS/MMBIR, AluAlu-mediated rearrangements) of genomic disorders have prompted some medical geneticists to maintain that the clinical implementation of chromosomal microarray analysis (CMA) to detect genomic disorders, including their enormous role in causing developmental, cognitive, and behavioral disabilities, was the greatest clinical benefit to emanate from the human genome project—this was until 2013 when personal genome analyses by clinical ES became a reality. A timeline of the origins of appreciation of structural variation, copy-number variation, and dosage effects and their contribution to mutational processes and trait manifestations is shown. This timeline chronicles the development of the concept of genomic disorders and the implementation of such conceptual information into clinical genomics - an emerging new medical genetics discipline. (Figure modified, expanded, and updated from Lupski.11)
In 1991, we documented that a 1.4 Mb duplication at the CMT1A locus resulted in Charcot-Marie-Tooth disease type 1A.4 CMT1A (MIM: 118220) is a distal symmetric polyneuropathy (DSP)14, 15 usually of adult onset and observed as an autosomal-dominant (AD) trait with an age-dependent penetrance. This disease phenotype is caused by a gene dosage effect as shown by experimental evidence in a paper that appeared in the first issue of the new journal Nature Genetics.16 Initial evidence that showed the specific dosage-sensitive gene was PMP22 came from four groups simultaneously showing that this peripheral myelin gene maps within the CMT1A duplication genomic interval.17, 18, 19, 20 Other evidence included rare patients who did not have a duplication, but instead had point mutation, gain-of-function (GoF) missense alleles, of PMP22.21
Further mechanistic studies of the CMT1A duplication showed that PMP22 was flanked by a unique human genome architecture of directly oriented low-copy repeats (LCRs), the 24 kb LCRs that were termed CMT1A-REP, and these could act as homologous recombination substrates and cause duplication by unequal crossing over and ectopic recombination,22 a mechanism later termed by Pawel Stankiewicz as non-allelic homologous recombination (NAHR).23 The mechanism predicted a reciprocal recombination product of deletion and this predicted deletion was shown in 1993 by Phil Chance’s group to cause hereditary neuropathy with liability to pressure palsies (HNPP [MIM: 162500]).24 Together our groups demonstrated experimentally that the CMT1A duplication and HNPP deletion were reciprocal NAHR products.25 Consistent with a HNPP disease mechanism of PMP22 haploinsufficiency was the finding of frameshift mutation, i.e., a loss-of-function (LoF) null allele, in association with rare non-deletion neuropathy in patients with HNPP.26
In 1997, we showed the same homologous recombination mechanism, NAHR, for the Smith-Magenis chromosome microdeletion syndrome (MIM: 182290) and suggested that reciprocal duplication could cause a chromosomal microduplication syndrome;27 3 years later, the predicted reciprocal recombination product was identified and it was shown to be responsible for the condition that bears the eponym Potocki-Lupski syndrome (PTLS [MIM: 610883]).28, 29 This paper that utilized chromosomal, molecular biological, and molecular cytogenetic studies of human subjects appeared in the first Nature Genetics issue of the beginning of the year 2000 and the new 21st century28 (Figure 2), 100 years after Marcella and Theodor Boveri’s work helped establish the chromosome theory of inheritance and the distinction of nuclear and cytoplasmic inheritance.30 It established that SV mutagenesis mechanisms acting on genomes could generate reciprocal recombination rearrangement end products that explain chromosomal abnormalities.28 The concept of genomic disorders was delineated in the early 1990s and formally summarized in 1998; it was predicated on two major premises: (1) disease phenotypes are due to DNA rearrangements, not DNA sequence changes, and (2) genome architecture incites genome instability.31
The first SV mutagenesis mechanism of NAHR causing genomic disorders was firmly established by experimentally finding the predicted reciprocal recombination products at several loci and defining the clinical conditions associated with them. Moreover, by establishing the NAHR mechanism as underlying chromosomal microdeletion/microduplication syndromes, it became clear that SV mutagenesis mechanisms were genomic DNA rearrangements underlying chromosomal abnormalities. Whereas the mechanism for aneuploidies in humans were established as the cellular mechanism of meiotic nondisjunction, i.e., explained by perturbations in chromosome biology, little information was known regarding mechanisms for other types of chromosomal abnormalities. However, the evidence from studies of the SMS deletion27 and reciprocal PTLS duplication28 implicated perturbations in genome biology as underlying some types of chromosomal abnormalities.
During the next decade, much was learned about genomic disorders32 and more was learned about downstream mechanisms for clinical manifestations in the decade that followed to the present day.33 I have to admit I was quite excited to read Victor McKusick’s comments published in 2007 in our Society’s journal that stated: “Molecular cytogenetics and molecular genetics have narrowed the gap between Mendelian genetics and the classic cytogenetics of clinical disorders. This is illustrated by the conditions termed genomic disorders by Lupski, many of which show Mendelian patterns of inheritance.”34
By 2004, there was a very good reference haploid genome enabling one to array different probes that mapped throughout the human genome and interrogate a personal genome for structural variation that might cause the disease trait ascertained in the clinic. The clinical genomics35 assay that was initially implemented utilized bacterial artificial chromosomes (BAC clones) as probes for the array comparative genomic hybridization (aCGH) assay. Among the BACs arrayed were those that were used clinically in different fluorescence in situ hybridization (FISH) molecular cytogenetics assays to screen for microdeletion syndromes. Rapid expansion of that clinical genomics technology took place by having oligonucleotide arrays interrogating the human genome with more and more “pixels” allowing greater and greater resolution of the human genome and the detection in patients of more and more pathogenic rare variant SVs/CNVs in the human genome.35
In 2007 (Figure 2), after mapping the size, extent, and gene content of genomic CNVs, and extensive breakpoint junction analyses of the nonrecurrent rearrangements observed in patients with Pelizaeus-Merzbacher disease (PMD [MIM: 312080]), we proposed a new SV mutagenesis mechanism. This genomic rearrangement mechanism appeared to be a DNA replication mechanism for generating structural variation associated with genomic disorders.36 The name proposed was fork stalling template switching (FoSTeS), emphasizing the interpretation of the data that showed evidence for long-distance template switching (TS) in a template-driven manner resulting in juxtaposition of genomic sequence from different human genome map positions and potential breakpoint complexity. Thus, rather than two pieces of DNA simply recombining or joining together, the breakpoint junction results from the bringing together or recombining of non-contiguous haploid reference genome sequence from two different map positions. At the breakpoint, one could often observe microhomology at the substrate human genome sequences that were joined, i.e., join-points. Thus, this was not a homologous recombination reaction, but rather the microhomology observed was interpreted as reflecting “primer binding” for the replicative recombination reaction. Moreover, this replicative recombination was “subtractive” for the microhomology versus “additive” for the microhomology,37 the latter additive microhomology as evidenced by base pair duplications at the site of insertion for replicative transposition by the Shapiro model.38
Thus, from these data generated from studying nonrecurrent PLP1 duplications associated with the genomic disorder PMD, experimental evidence showed that one was not dealing with simple end-joining. Instead, there was microhomology-mediated joining which was proposed to reflect the priming of DNA replication (that is, breakpoint can demarcate one or multiple join point(s) of discontinuous sequences). The evidence suggested long-distance TS facilitating template-driven juxtaposition of DNA sequences separated by large genomic distances in the reference haploid genome. Furthermore, breakpoint complexity could take place because of iterative TS. This idea of iterative template switching could explain breakpoint complexity but also could result in tremendous complexity to the genomic rearrangement or a rearrangement end product that revealed a complex genomic rearrangement (CGR).37
The FoSTeS concept of replicative rearrangements was further refined by combining observations and experimental data from bacteria and yeast to describe the mechanistic details of the DNA chemistry involved and this mechanism was termed microhomology-mediated break-induced replication, MMBIR.39 The precise molecular mechanistic details that were delineated put forward the idea that a collapsed replication fork (likely resulting from replication proceeding through a nick in the DNA) generates a single-ended, double-stranded DNA, seDNA—this is distinct from a double-strand break which results in a two-ended, double-stranded DNA molecule. In the MMBIR mechanism, the 3′ end of one strand was exposed after extensive exonuclease degradation (5′ to 3′) that generates a long ssDNA with a 3′ end which can become a flexible primer for initiating DNA replication. This is followed by a new low processivity replisome that can disassociate repeatedly and reform with a different template, i.e., TS, and the replication then becomes a more well established and processive replisome that continues to copy the DNA. Such iterative TS can result in the breakpoint complexity.
More recent work from yeast has further supported some of the details of the MMBIR model including the TS mediated by microhomology of human Alu repetitive sequences placed in a yeast experimental system that allows induction of seDNA40 and evidence for involvement of translesion polymerases.41 As noted, seDNA can be generated at collapsed replication forks, but such DNA ends can also be generated at the end of a chromosome, i.e., telomeres, as part of the end-replication problem or terminal deletions.42, 43 This mechanism might also generate an interrupted inverted duplication as can be observed with a breakage-fusion-bridge cycle accompanying telomere healing.44
For human genetics studies, the MMBIR model could readily explain nonrecurrent duplications and deletions, inversion, translocation, triplication, and higher order amplification—the latter from a proposed rolling circle replication. Furthermore, the MMBIR mutagenesis mechanism could begin to also explain a multitude of rearrangement end-products that were perceived as CGR, such as duplication-triplication-duplication (DUP-TRP-DUP) and DUP-NML-DUP array CGH patterns observed in humans with associated genomic disorders. Moreover, the MMBIR model made several predictions including: (1) increased SNV mutagenesis concomitant with CGR, (2) copy number neutral AOH when template switch occurs to the chromosome homolog versus the sister chromatid, and (3) higher order amplification, e.g., triplication, quadruplication, and beyond, might take place via a rolling circle type mechanism. All three of these predictions were borne out by experimental studies in human subjects.45, 46, 47, 48
Perhaps one of the most interesting rearrangement end-product structures delineated was shown to result from just two iterative template switches (TSs): the initial breakpoint junction 1 (jct1) occurring by recombination between inverted repeats and junction 2 by a microhomology-mediated TS. This rearrangement end-product structure was termed DUP-TRP/INV-DUP.45 Interestingly, by inverting a segment of DNA, new genes can be formed at the junction by now reading the reverse complement strand; and indeed the predicted transcripts were identified.49 Thus, with only two TSs, complex structural variants could be derived and new genes generated. Moreover, studies on DUP-TRP/INV-DUP demonstrated that inverted repeats45, 50 and smaller-sized LCR (<1,000 bp) were important architectural features for human genome instability.
CGR can involve only a single exon, a single gene, or multiple genes.51 They may include more than a single dosage-sensitive gene that can contribute to the disease phenotype due to triplosensitivity and as manifestations of a multigenic trait as shown for the Yuan-Harel-Lupski (YUHAL [MIM: 616652]) syndrome.52 For deletions, multigenic haploinsufficiency traits53 perhaps extend the Schmickel54 contiguous gene deletion syndrome concept. With deletion CNV, the complexity of gene variants that may contribute to clinical phenotypic traits is sometimes greater than duplication CNV as the deletions cause a locus to be hemizygous and may unmask rare variant recessive alleles55, 56 or regulatory SNPs.57 The multilocus complexity of a CGR can include an interstitial fragment of a chromosome or only its terminal end43, 58 or involve an entire chromosomal arm.51 Multigenic contributions to the disease phenotype may also relate to gene interruptions at the breakpoint junctions and/or altered gene regulation due to position effects resulting from a rearranged genome or chromosomes.59
The MMBIR mechanism could explain insertional translocations,60 small marker chromosomes (SMCs),61 and some apparently balanced translocations.62 It may also explain some cases of the somatic mutagenesis resulting in the phenomena of chromothripsis63 originally described in cancer genomes and also such complexity observed as chromoanasynthesis64 in association with developmental disorders. The extent of chromosomal/genomic structural changes that accompany chromothripsis/chromoanasynthesis/chromoanagenesis is staggering.65 Finally, the perizygotic mutagenesis observed as a potential SV mutator phenotype appears to occur by MMBIR.66
By 2014, after almost a decade of extensive experimental analyses and implementation of clinical genetics testing for the CMT1A duplication in thousands of patients with neuropathy,67 we determined the mechanism, prevalence, and clinical consequence of triplication in association with a more severe CMT1A peripheral neuropathy phenotype.68 The NAHR mechanism driven triplication at the PMP22 locus occurs as a 100-fold more frequent event from duplication to triplication than the frequency for de novo CMT1A duplication69, 70 (Figure 2). It was notable that after three quarters of a century we were given a chance in our Society’s journal to illuminate some of the questions raised by the observations of Bridges and brought up in the discussion of his 1936 one-page paper,13 although by genetics work from a completely different model organism, Homo sapiens.68
Consistent with the dosage hypothesis, triplication at the PMP22 locus and the MECP2 locus caused a more severe neurological disease phenotype than that which was observed with the duplication.45, 68, 71 As anticipated by the gene-dosage hypothesis, CMT1A triplication gave a severe DSP and with similar severity to that observed with homozygous CMT1A duplication;72 both instances have four copies of PMP22, the former with three copies in cis (i.e., heterozygous triplication) and the latter with two copies each in trans (i.e., homozygous duplication) having different implications for transmission genetics.73
The gene-dosage hypothesis for phenotypes, i.e., triplosensitivity and haploinsufficiency as mechanisms for disease traits, was strongly supported by data from the CMT1A duplication and HNPP deletion studies that solidified the concept of gene dosage. However, it is perhaps of heuristic interest that earlier human genetics data on gene dosage and disease were not appreciated and perhaps even misunderstood; the latter possibility might relate to understanding the difference between a disease population (i.e., a group of patients with the same diagnostic classification) and a disease trait. In 1987 French investigators demonstrated APP gene duplication in early-onset Alzheimer families and karyotypically normal (46,XY with likely submicroscopic duplication on chromosome 21) Down syndrome.74 This was of interest because of the known clinical association of early-onset Alzheimer dementia in patients with Down syndrome due to trisomy 21. Two reports then argued against APP duplication in Alzheimer disease.75, 76 Of note, these arguments were based on negative data studies in fewer than ten patients with Alzheimer dementia! It would take 20 years to overcome these negative data. Seven pages of negative data published in two papers, more than twice the three pages of positive data in the original paper. Twenty years later APP duplication was definitively shown to cause early-onset autosomal-dominant (i.e., AD) Alzheimer disease in five French kindreds.77 I often wondered what would have happened if we had found the PMP22 point mutation,21, 78 initially in 1 of about 100 CMT families studied, and not the CMT1A duplication?4, 79
Finally, to bring the timeline (Figure 2) up to date from an SV mutagenesis mechanism perspective, this year we have provided evidence for perhaps a different mechanistic way to think about CNV of smaller sizes (e.g., <10 kb), particularly with respect to single exon drop out alleles. Evidence from the Watson genome, the first personal genome determined by massively parallel next generation sequencing technology,80 documented an intriguing SV mutagenesis finding. Interestingly, allele frequency spectrum data for CNV deletion size showed that, for those deletion alleles <1,000 bp in size, there is a logarithmic increase in the frequency for which deletion CNVs are found in a personal genome. Remarkably, there is a large peak frequency observed at the ∼300 base pair size that corresponds to the Alu repetitive sequence element (Figure 2). Alu is a repetitive sequence initially described by re-association kinetics, not genomic sequencing; it is distributed in ∼1,000,000 copies as an interspersed repetitive element throughout the human haploid reference. Specifically, this ∼300 bp peak allele frequency represents dimorphic Alu in the Watson personal diploid genome at a given map position whereby on one chromosome the Alu is present and on the other chromosome it is not present. It suggests that Alu transposition, or Alu-mediated rearrangement, might be responsible for a tremendous amount of evolution of the human genome.
Extensive analyses in patients suggest that for some loci a predominant mutational mechanism is exon deletion driving alleles within the disease gene and that these exonic deletions can be the result of Alu-Alu-mediated rearrangement.50, 81, 82 Thus, in a recently reported paper we provided computational and experimental evidence that Alu-Alu-mediated rearrangement occurs by MMBIR.83 In this Alu-Alu-mediated rearrangement (AAMR; Figure 2) mechanism, a TS occurs by using the Alu microhomology as a substrate sequence to form Watson-Crick base pairing of the primer and after TS initiate the replicative repair.40, 83 Of note, some of the first intragenic exonic deletion and duplication CNV alleles reported were found at the LDLR locus causing familial hypercholesterolemia with susceptibility to AD familial hypercholesterolemia (MIM: 143890) and atherosclerosis with early myocardial infarction as a complex trait.84, 85
SV Mutagenesis Mechanisms, Mirror Traits, and Gene and Genome Evolution
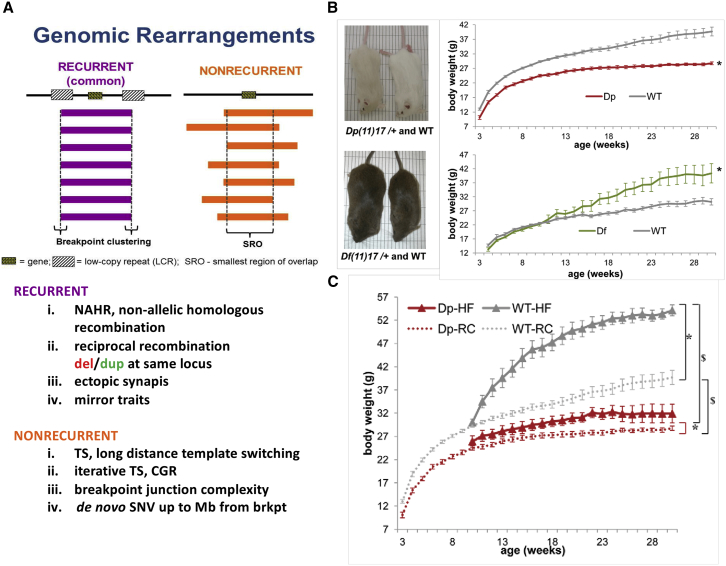
In general, from experimental studies of personal genomes from individuals with genomic disorders, two types of simple deletion and duplication rearrangements have been delineated (Figure 3). One is recurrent rearrangements, sometimes also referred to as the “commonly found” rearrangement at a given locus, and the second is nonrecurrent rearrangements (Figure 3). What we have learned from the study of recurrent rearrangements is that the flanking LCRs represent a genome architecture, or intrinsic property of the human genome, rendering genomic instability. Moreover, these flanking LCRs can be used as homologous recombination (HR) substrates to mediate the rearrangement via NAHR. We have also learned that reciprocal recombination results in either deletion or duplication at the same locus.88, 89 The mechanism has further been shown to represent an end product of ectopic synapses and it is these ectopic synapses that then mediate an apparent ectopic recombination or an NAHR.90
Figure 3.
Two Major Categories of Simple Deletion and Duplication Rearrangement End Products: Recurrent and Nonrecurrent, and Lesson Learned from Studies of These Rearrangement End Products
(A) Definition of recurrent or common rearrangements where breakpoint clustering occurs in flanking low-copy repeat (LCR) sequences and a dosage-sensitive gene maps within the rearranged interval of the human genome. For nonrecurrent rearrangements, the breakpoints can be different in different patients, but the smallest region of overlap (SRO) identifies a potential dosage-sensitive gene(s).
(B) Lessons learned from recurrent rearrangements include the concept of mirror traits whereby copy-number gain of a specific interval results in one phenotypic extreme or copy-number loss results in the mirror image trait phenotypic extreme as illustrated here for weight and the animal models for PTLS [Dp(11)17] and SMS [Df(11)17].86
(C) Documents that a mirror trait is truly a genetic trait at the PTLS locus in that there is protection from weight gain, obesity, and the metabolic syndrome, when the environment changes to that of a high fat diet fed to the animal model. Note, the wild-type littermates gain a tremendous amount of weight due to this environmental change and the G × E interaction, but heterozygous duplication animals are protected from weight gain and metabolic syndrome. Parts of figure modified from Lacaria et al.87
Of note, from a human and medical genetics standpoint, for reciprocal recombination products the phenotypic features conveyed can sometimes be observed to represent mirror traits. The term mirror traits refers to opposite extremes of phenotype observed in association with copy-number loss versus gain at the locus (e.g., reciprocal duplication/deletion) with a copy number variant locus contributing to a quantitative trait.91, 92 Examples of mirror traits include obesity (metabolic syndrome) versus underweight (lean), microcephaly versus macrocephaly,93, 94 and short stature versus tall stature. Complex neurobehavioral traits, such as licking behavior in the chromosome engineered mouse models for PTLS and SMS,95 can even represent mirror trait phenotypes. Some neurobehavioral and neurocognitive traits in human have been proposed to represent mirror phenotype extremes in a population distribution96—macrocephaly and microcephaly with autism and schizophrenia CNV, respectively, in association with reciprocal deletion or duplication at 1q21.1 and 16p11.2 provide further evidence for mirror traits.97, 98, 99, 100, 101
A number of mirror traits were described at the SMS/PTLS locus86 including a mirror trait for weight/BMI (Figure 3B) also shown to contribute to metabolic syndrome in chromosome engineered mice.87 Moreover, the mirror trait of “leanness” associated with duplication of this locus is resistant to environmental influences, such as a high fat diet (Figure 3C), on weight (gene by environment, G × E,87 interactions).
NAHR can be the mechanism underlying some chromosome inversions, isodicentric isochromosome102 formation [isodicentric17q, isoXq, and isoYq] (by nonsister chromatid recombination of inverted LCR on the short arm), or recurrent translocations.103 Based on this mechanistic thinking of NAHR-driven reciprocal recombination, the reciprocal recombination products, and genomic architecture of flanking LCR driving rearrangements associated with several genomic disorders, the specific loci underlying neurocognitive phenotypes/syndromes due to reciprocal recombination were found at many human genome map positions.104, 105 Evidence for new genomic disorders and CNV loci affecting neurocognition provide an opportunity to better describe and further characterize neurodevelopmental traits,106, 107 enabling potential biological insights allowing potential avenues for variant tailored therapeutic interventions and educational programs.
The second major class or type of observed rearrangement end product is nonrecurrent rearrangements (Figure 3A). The major mechanism involved here seems to be long-distance TS. Iterative TS can result in breakpoint junction complexity and can also result in CGR.37 Because the replicative repair mechanism may use a polymerase that has reduced processivity, one may see breakpoint complexity as the fork collapses and undergoes another TS before the replisome is stabilized and a processive replisome is established. This processive replisome may replicate large genomic distances. Moreover, in this replicative repair mechanism, evidence suggest that the polymerase utilized has less fidelity than the intergenerational DNA polymerases and can generate SNV during replicative repair. Thus, one can create SNV hypermutation in proximity to the breakpoint junctions because of de novo SNV up to a megabase from the breakpoint junctions or join points formed during the TS in the MMBIR mechanism.108
This FoSTeS/MMBIR replicative repair mechanism appears also to play a critical role in the evolutionary processes as it may contribute to a host of observations regarding the evolution of genes and genomes, including: (1) gene duplication and divergence, (2) gene amplification (e.g., methotrexate resistance in cancer, via rolling circle replication), (3) exon shuffling, (4) new gene creation by inversion, reverse complementary strand read at SV breakpoint junctions, (5) locus-specific mutation rates for CNV versus SNV at the locus = between 10−4 and 10−6 versus 10−8, respectively, (6) one can regulate mutation rates via the FoSTeS/MMBIR mechanism. Thus, regarding the latter two points (5 and 6), structural variant mutagenesis mechanisms may contribute to the “mutational rheostat” essential to evolution—NAHR (meiotic) will require one or two generations to respond to environmental change whereas for FoSTeS/MMBIR this can happen during early embryogenesis and it may perhaps be that this mechanism is particularly prone to occurring during a perizyogotic mutagenesis period with rapid replication/cell division during early embryogenesis.66
Rare Variants, Clan Genomics, and Disease Traits
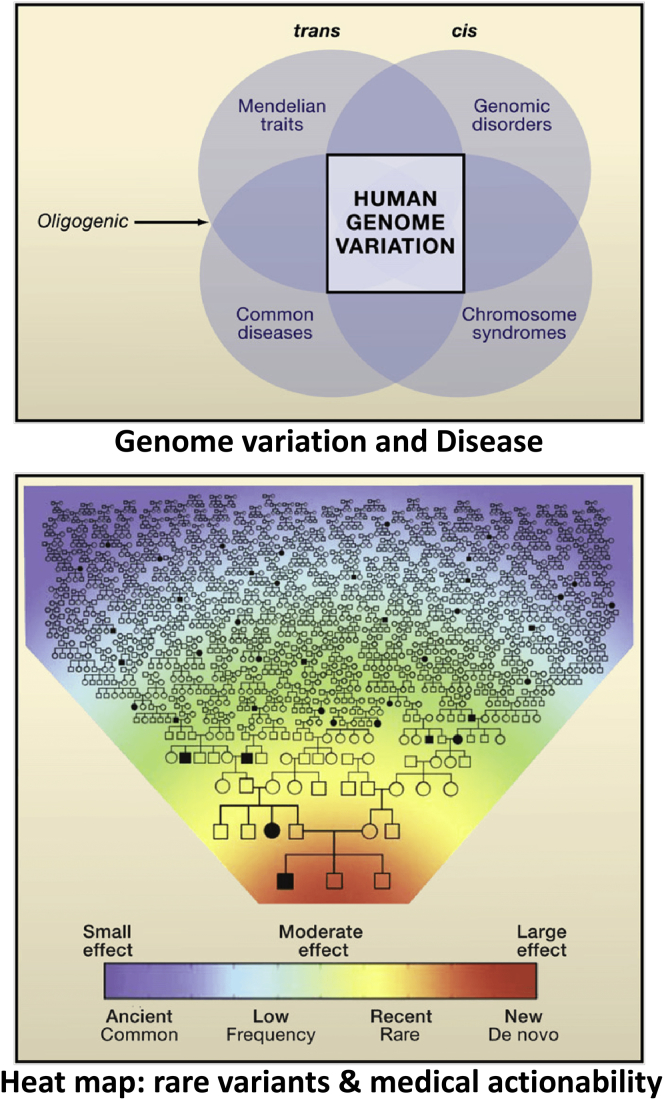
SV mutagenesis and the genomic disorders caused by such rare variant CNVs have taught us more about human genome variation in the context of clearly observed clinical phenotypes. In general, four major types of disorders are observed in the clinic for which there are genetic influences. These include chromosomal syndromes, genomic disorders, Mendelian traits, and common diseases that may be complex traits. The latter complex traits are those disease traits for which there are clear genetic influences, but the phenotype does not show a Mendelian segregation pattern consistent with a single locus effect. Such traits may be multi-factorial traits with both genetic and environmental influences (Figure 4A). As DNA sequencing technologies were applied to personal genomes from many individuals, and one could begin to study variation of individual loci and personal genomes at scale, it has become apparent that the genome of any one individual has a tremendous amount of rare variation for which one did not begin to see its existence until the personal genomes from many different individuals and populations were studied.
Figure 4.
Clan Genomics, Rare Variants, and Medical Actionability
Above shows the types of conditions that can be influenced by genetic variation and human genome variation. These include chromosomal syndromes as analogized in Figure 1 and genomic disorders delineated and defined in Figure 2. Mendelian disease traits are due to single locus variation while some of complex diseases and multi-factorial traits may result from digenic and oligogenic inheritance or multi-locus pathogenic variation. Below analogizes the concept of a clan genomics whereby rare variants that have arisen recently in the patient, family, or clan are more likely to be the medically actionable variants and provide insight into the genetic influences on disease phenotypes as this variation is tied to disease biology and disease trait manifestation. Figure modified from Lupski et al.109
This rare variation for SNV has similar observations to that which was discerned for CNV in clinical populations. The variant causing pathology might either be de novo or recently inherited within the family or clan structure. But perhaps more important and pragmatic for the individual and for clinical medicine was the conceptual realization of what rare variation and medical actionability actually might mean. It is the conceptualization of this realization that lead to the clan genomics hypothesis,109 a term coined by Richard Gibbs. Simply stated, the clan genomics hypothesis argues that genome variation with respect to human disease and medical actionability has to do with rare variants (SNVs and/or CNVs) that have large effect and which have arose recently in the population history. Therefore, new mutation in you and your recent ancestors, and novel combinations of mutations you inherit from your parents at a locus through transmission genetics, account for many medically actionable variants.
The clan genomics model provides a powerful framework to contrast the importance of rare variants versus common variants in disease. It is distinct from the rare variant/rare disease (RVRD) hypothesis. Thus, clan genomics provides a theoretical framework for aligning human disease biology with evolutionary biology and genetics, the latter of which deals with per locus variation and trait manifestation. Thus, if the phenotype that is observed clinically is treated as a disease trait, the medically actionable variants will be the rare variants that represent potentially pathogenic variation contributing to disease trait manifestations. Moreover, one can utilize a series of rare variant filtering strategies to discern this potentially pathogenic rare variation from the tremendous amount of variation found in each personal genome. Using such rare-variant, family-based genomic studies, one can narrow to a potentially small group of rare variants those which might contribute to the disease phenotype; this has ramifications for disease gene discoveries and for molecular diagnosis.
From one perspective, one might state that this concept of clan genomics is nothing new. We already knew that common variants have a small effect and rare variants have a large effect, and that is why Mendelian diseases perhaps are rare and that these were highly penetrant allelic variants in association with Mendelian disease. Nevertheless, the clan genomics hypothesis back in 2011 had profound implications for family-based genomics and rare variant filtering strategies to examine per locus variation in the context of the patient, family, or clan. Clan genomics is about patient, family, and clan—disease traits not disease populations.
This concept of clan genomics is further illustrated by case reports including, for example, one documenting cousins with Charcot-Marie-Tooth disease, in whom testing revealed one cousin had recessive Charcot-Marie-Tooth disease due to MTMR2 homozygous mutation while the other cousin, one of fraternal twins, had a de novo PMP22 duplication, the CMT1A duplication, causing the disease.110 Of note, case/family study reports of molecular cytogenetic testing111 and gene panel testing applied clinically112 also resulted in some of the first clinical observations of multi-locus pathogenic variants in a patient. Using a family-based genomics approach and a trio analysis (proband with sporadic disease trait and unaffected parents) one can readily observe the new CMT1A duplication mutation associated with the sporadic trait in the nuclear family. Also, a quad approach in the cousin’s nuclear family will find the homozygous mutation or biallelic variation associated with the recessive CMT trait in the cousin; homozygosity due to an identity-by-descent (IBD) of a rare variant that might be private to the clan. Thus, every time a search for a molecular diagnosis by clinical exome is performed and the rare-variant family-based analysis undertaken, one is testing the clan genomics hypothesis. No evidence has been obtained to reject the hypothesis, whereas a substantial amount of evidence in tens of thousands of exomes applied in research studies113, 114 and clinical studies9 support the clan genomics hypothesis.
Family-based genomics can also be approached by determining whether there is evidence for a potential IBD due to consanguineous relationship between the parents. This can be performed on the exome data by determining a B allele frequency and using BafCalulator to find genomic intervals with absence of heterozygosity (AOH).115 Whereas genetics is the study of single locus variation in the context of a family history, genomics takes into account both the family history and de novo mutation as well as the aggregation of pathogenic alleles at a locus by transmission genetics. Moreover, genomics allows direct assay of potential multilocus pathogenic variation to be assessed. Clan genomics studies the individual, the family, and the clan, which is a different emphasis from what is investigated by a genome-wide association study (GWAS) approach, which concentrates on a disease population. Thus, for medical actionability the most important thing for individuals regarding clinical implementation of their personal genome is what their nearest relatives (e.g., parents through transmission genetics) gave them and de novo events at that locus. The population from which the subject comes is not that relevant.
The clan genomics hypothesis has been tested in a broad study of the Centers for Mendelian Genomics (CMG) and collaborators. To date, about 60,000 samples have been collected from more than 22,000 families and the CMG and collaborators work to date includes almost 4,000 collaborators with subject samples coming from 80 countries worldwide. The CMG have begun to analyze more than 2,200 different phenotypes. Thus far, about 47,000 exomes and more than 1,000 whole-genome sequencing studies have been performed. The work of the CMG and collaborators continues to be disseminated through different means of making the information public. This includes more than 500 publications with 5,832 contributing co-authors from 1,540 organizations in 77 countries.114 About 1,700 candidate disease genes have been identified, with the level of evidence in support of these disease candidates being quite extensive for about 600 of them. The remaining candidates will need verification in other families or with other variants being tied to the same or similar phenotype before being clearly established as a “disease gene.” About 400 genes were known disease genes, but the phenotype observed expands beyond what the known phenotype had been associated with that gene to date as detailed in the clinical synopsis of the disease trait in the OMIM entry. This could be due to different allelic variants and an allelic series establishing the extent and variability of phenotypes that one may see with pathogenic variation at that locus. Alternatively, observed phenotypic expansion in an individual could be because there is a second locus for pathogenic variation and this makes it appear that there is phenotypic expansion.115 Currently, the CMG and collaborators identify a novel candidate disease gene in about 1 in every 30 exomes that are performed on different subject samples.114
The clan genomics hypothesis has also been tested in the clinic where indeed a high rate of identifying patients with sporadic disease traits due to new mutations has been observed.9 Current molecular diagnostic yield of clinical exomes is approximately 25%–30% of cases. Notably, research strategies for optimizing family-based analyses of exomes could be employed for molecular diagnosis.116 Re-analyses of exome data116 once or twice a year increases the molecular diagnostic rate. In a study following up the original publication of a pilot clinical genomics study using exome sequencing (ES),117 it was shown that after a 5-year period, the molecular diagnostic rate had increased from 24.8% to 46.8% (unpublished data). Likewise, a follow up of the original 2,000 studied subjects who underwent clinical ES9 documents that 3–4 years later, the diagnostic rate increased from 25.2% to 36.7%. Notably, in both of these re-analysis cohorts, the biggest contributor to new molecular diagnoses was new disease gene discovery. Other contributors were clinical updated information and utilization of additional family members for genomic studies (e.g., unaffected parents for trio analysis, affected siblings for analysis of shared rare variants). In the follow up of the JAMA study117 a significant contributor besides variant re-classification was actual CNV detection.
An interesting finding to come from extensive analyses of thousands of clinical exomes is that the molecular diagnostic rate is approximately 30%. Of note, about 5% of the molecularly diagnosed cases actually have pathogenic variation at more than one locus.118 Thus, about 1 in 20 families with a “solveable” clinical exome, i.e., productive for a molecular diagnosis, have a disease phenotype that results from genomic multi-locus pathogenic variation. Each variant at each individual locus may cause a dominant, recessive, or X-linked disease trait for the trait associated with each individual locus. For the loci including a monoallelic variant causing an autosomal-dominant (AD) trait, 68% were found to be due to de novo variants whereas of those causing an X-linked (XL) trait, 52% were found to be de novo. Remarkably, both variants were de novo in 44.7% (17/38) of cases diagnosed by mono-allelic variants (i.e., causing an AD+AD or AD+XL trait) at both loci.118
Patients having multiple molecular diagnoses can be challenging to clinicians because the patient can manifest a blended phenotype. These blended phenotypes can be further dissected into those whereby each locus causes distinct parts of the blended phenotype versus the blended phenotypes in which the clinical phenotype caused by each locus can have some overlap and it may just look like a more severe, for example, neurological disease if both of the gene loci affect the nervous system and cause cognitive phenotypes.119 Dual diagnosis with overlapping phenotypes may represent a mutational burden within an interactome or pathway, biological structure/unit (e.g., cilia, neuron), or organ system.118
Recently, we documented that apparent phenotypic expansion can sometimes illuminate multi-locus pathogenic variation. Studies of intra-familial genotypic and phenotypic variability are able to show that a more severely affected individual in a family had multi-locus pathogenic variation while the less severely affected sibling inherited pathogenic variation at only one locus.115 We have also recently obtained preliminary evidence that suggest the clan genomics hypothesis may extend to multi-locus pathogenic variation. In studies of arthrogryposis, a complex trait, we show that within the Turkish population there is a high rate of multi-locus variation for which two or more loci both encode an AR disease trait and that the biallelic variants have come together because of IBD (unpublished data). Whereas in North American populations clinical diagnostic exomes have multi-locus pathogenic variation in about 1:20 solved cases, in the Turkish population of patients with arthrogryposis about 11% (11/101) of subjects had two or more loci with pathogenic variation. Remarkably, 88.9% (16/18) were AR+AR whereas the frequency of such cases in the North American cohort was 22% (p < 0.0001) (unpublished observation).
Studies using family-based genomics analyses from ES and whole-genome sequencing (WGS) have begun to elucidate genetic models that may apply to common disease and complex traits. These models include multi-locus pathogenic variation,118 mutational burden,120 and the compound inheritance gene dosage model.121, 122, 123 The latter compound inheritance gene dosage model ties common variant alleles to disease biology and may be important at expression quantitative trait loci, eQTL. Compound inheritance may have particular relevance to birth defects as complex traits that result from perturbations in developmental pathways. Extensive studies in humans and mice on a spinal disease, congenital scoliosis, has elucidated this genetic model at the TBX6/Tbx6 locus.121, 122, 123 Recent work supports a role for compound inheritance in another birth defect—congenital anomalies of the kidney and urinary tract (CAKUT)124 involving TBX6—and in lethal developmental lung disorders due to disruption of the TBX4-FGF10 pathway.125
In summary, clan genomics provides the theoretical framework for a rare-variant, family-based genomics approach in research and the clinic. Clan genomics also ties disease biology to evolutionary theory. SV mutagenesis11 contributes to chromosomal syndromes, genomic disorders, and the allelic architecture (exon dropout alleles; CNV + dnSNV) of Mendelian disease traits. Of the molecularly diagnosed cases from a clinical referral population, about 1 in 20 individuals have two or more pathogenic variant alleles. Multi-locus pathogenic variation provides opportunities for exploring genetic models of complex traits. The compound inheritance gene dosage model121, 122, 123 ties common variant alleles to disease traits and disease biology and may have particular relevance to birth defects where homozygosity for severe LoF alleles, or mutational burden in a developmental pathway, may result in embryonic lethality—a lot more of this to come during the next few years of study.
In closing, I would like to thank the students and postdocs who have passed through the Lupski laboratory for their hard-earned data and all that they have taught me, many thanks also to my terrific faculty colleagues and outstanding leadership of our Department and the Baylor College of Medicine Human Genome Sequencing Center (Figure 5). A special thank you to my family for taking this journey through life in science with me, and finally Victor McKusick for teaching us to focus on patients, families, and clans—disease traits not disease populations.
Figure 5.
Many People to Thank and Apologize for Those Missing from the Collage
Declaration of Interests
J.R.L. has stock ownership in 23andMe and Lasergen, Inc., is a paid consultant for Regeneron Pharmaceuticals and the Regeneron Genetics Center, and is a coinventor on multiple United States and European patents related to molecular diagnostics for inherited neuropathies, eye diseases, and bacterial genomic fingerprinting. The Department of Molecular and Human Genetics at Baylor College of Medicine derives revenue from chromosomal microarray analysis (CMA) and clinical exome sequencing (ES) offered in the Baylor Genetics (BG) Laboratory. J.R.L. is a member of the Scientific Advisory Board (SAB) of BG.
Web Resources
Baylor Genetics (BG) Laboratory, http://baylorgenetics.com
Centers for Mendelian Genomics (CMG), http://mendelian.org/
OMIM, https://www.omim.org/
References
- 1.Lupski J.R. Genome structural variation and sporadic disease traits. Nat. Genet. 2006;38:974–976. doi: 10.1038/ng0906-974. [DOI] [PubMed] [Google Scholar]
- 2.Carvalho C.M., Lupski J.R. Copy number variation at the breakpoint region of isochromosome 17q. Genome Res. 2008;18:1724–1732. doi: 10.1101/gr.080697.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yuan B., Liu P., Gupta A., Beck C.R., Tejomurtula A., Campbell I.M., Gambin T., Simmons A.D., Withers M.A., Harris R.A. Comparative genomic analyses of the human NPHP1 locus reveal complex genomic architecture and its regional evolution in primates. PLoS Genet. 2015;11:e1005686. doi: 10.1371/journal.pgen.1005686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lupski J.R., de Oca-Luna R.M., Slaugenhaupt S., Pentao L., Guzzetta V., Trask B.J., Saucedo-Cardenas O., Barker D.F., Killian J.M., Garcia C.A. DNA duplication associated with Charcot-Marie-Tooth disease type 1A. Cell. 1991;66:219–232. doi: 10.1016/0092-8674(91)90613-4. [DOI] [PubMed] [Google Scholar]
- 5.Matise T.C., Chakravarti A., Patel P.I., Lupski J.R., Nelis E., Timmerman V., Van Broeckhoven C., Weeks D.E. Detection of tandem duplications and implications for linkage analysis. Am. J. Hum. Genet. 1994;54:1110–1121. [PMC free article] [PubMed] [Google Scholar]
- 6.Lupski J.R. 2002 Curt Stern Award Address. Genomic disorders recombination-based disease resulting from genomic architecture. Am. J. Hum. Genet. 2003;72:246–252. doi: 10.1086/346217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu J., Zhou Y., Liu S., Song X., Yang X.Z., Fan Y., Chen W., Akdemir Z.C., Yan Z., Zuo Y., DISCO (Deciphering disorders Involving Scoliosis and COmorbidities) Study The coexistence of copy number variations (CNVs) and single nucleotide polymorphisms (SNPs) at a locus can result in distorted calculations of the significance in associating SNPs to disease. Hum. Genet. 2018;137:553–567. doi: 10.1007/s00439-018-1910-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gambin T., Yuan B., Bi W., Liu P., Rosenfeld J.A., Coban-Akdemir Z., Pursley A.N., Nagamani S.C.S., Marom R., Golla S. Identification of novel candidate disease genes from de novo exonic copy number variants. Genome Med. 2017;9:83. doi: 10.1186/s13073-017-0472-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang Y., Muzny D.M., Xia F., Niu Z., Person R., Ding Y., Ward P., Braxton A., Wang M., Buhay C. Molecular findings among patients referred for clinical whole-exome sequencing. JAMA. 2014;312:1870–1879. doi: 10.1001/jama.2014.14601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bayer D.K., Martinez C.A., Sorte H.S., Forbes L.R., Demmler-Harrison G.J., Hanson I.C., Pearson N.M., Noroski L.M., Zaki S.R., Bellini W.J. Vaccine-associated varicella and rubella infections in severe combined immunodeficiency with isolated CD4 lymphocytopenia and mutations in IL7R detected by tandem whole exome sequencing and chromosomal microarray. Clin. Exp. Immunol. 2014;178:459–469. doi: 10.1111/cei.12421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lupski J.R. Structural variation mutagenesis of the human genome: Impact on disease and evolution. Environ. Mol. Mutagen. 2015;56:419–436. doi: 10.1002/em.21943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blakeslee A.F. Variations in Datura due to changes in chromosome number. Am. Nat. 1922;642:16–31. [Google Scholar]
- 13.Bridges C.B. The Bar “Gene” a Duplication. Science. 1936;83:210–211. doi: 10.1126/science.83.2148.210. [DOI] [PubMed] [Google Scholar]
- 14.England J.D., Gronseth G.S., Franklin G., Carter G.T., Kinsella L.J., Cohen J.A., Asbury A.K., Szigeti K., Lupski J.R., Latov N., American Academy of Neurology Practice Parameter: evaluation of distal symmetric polyneuropathy: role of autonomic testing, nerve biopsy, and skin biopsy (an evidence-based review). Report of the American Academy of Neurology, American Association of Neuromuscular and Electrodiagnostic Medicine, and American Academy of Physical Medicine and Rehabilitation. Neurology. 2009;72:177–184. doi: 10.1212/01.wnl.0000336345.70511.0f. [DOI] [PubMed] [Google Scholar]
- 15.England J.D., Gronseth G.S., Franklin G., Carter G.T., Kinsella L.J., Cohen J.A., Asbury A.K., Szigeti K., Lupski J.R., Latov N., American Academy of Neurology Practice Parameter: evaluation of distal symmetric polyneuropathy: role of laboratory and genetic testing (an evidence-based review). Report of the American Academy of Neurology, American Association of Neuromuscular and Electrodiagnostic Medicine, and American Academy of Physical Medicine and Rehabilitation. Neurology. 2009;72:185–192. doi: 10.1212/01.wnl.0000336370.51010.a1. [DOI] [PubMed] [Google Scholar]
- 16.Lupski J.R., Wise C.A., Kuwano A., Pentao L., Parke J.T., Glaze D.G., Ledbetter D.H., Greenberg F., Patel P.I. Gene dosage is a mechanism for Charcot-Marie-Tooth disease type 1A. Nat. Genet. 1992;1:29–33. doi: 10.1038/ng0492-29. [DOI] [PubMed] [Google Scholar]
- 17.Patel P.I., Roa B.B., Welcher A.A., Schoener-Scott R., Trask B.J., Pentao L., Snipes G.J., Garcia C.A., Francke U., Shooter E.M. The gene for the peripheral myelin protein PMP-22 is a candidate for Charcot-Marie-Tooth disease type 1A. Nat. Genet. 1992;1:159–165. doi: 10.1038/ng0692-159. [DOI] [PubMed] [Google Scholar]
- 18.Valentijn L.J., Bolhuis P.A., Zorn I., Hoogendijk J.E., van den Bosch N., Hensels G.W., Stanton V.P., Jr., Housman D.E., Fischbeck K.H., Ross D.A. The peripheral myelin gene PMP-22/GAS-3 is duplicated in Charcot-Marie-Tooth disease type 1A. Nat. Genet. 1992;1:166–170. doi: 10.1038/ng0692-166. [DOI] [PubMed] [Google Scholar]
- 19.Timmerman V., Nelis E., Van Hul W., Nieuwenhuijsen B.W., Chen K.L., Wang S., Ben Othman K., Cullen B., Leach R.J., Hanemann C.O. The peripheral myelin protein gene PMP-22 is contained within the Charcot-Marie-Tooth disease type 1A duplication. Nat. Genet. 1992;1:171–175. doi: 10.1038/ng0692-171. [DOI] [PubMed] [Google Scholar]
- 20.Matsunami N., Smith B., Ballard L., Lensch M.W., Robertson M., Albertsen H., Hanemann C.O., Müller H.W., Bird T.D., White R. Peripheral myelin protein-22 gene maps in the duplication in chromosome 17p11.2 associated with Charcot-Marie-Tooth 1A. Nat. Genet. 1992;1:176–179. doi: 10.1038/ng0692-176. [DOI] [PubMed] [Google Scholar]
- 21.Roa B.B., Garcia C.A., Suter U., Kulpa D.A., Wise C.A., Mueller J., Welcher A.A., Snipes G.J., Shooter E.M., Patel P.I., Lupski J.R. Charcot-Marie-Tooth disease type 1A. Association with a spontaneous point mutation in the PMP22 gene. N. Engl. J. Med. 1993;329:96–101. doi: 10.1056/NEJM199307083290205. [DOI] [PubMed] [Google Scholar]
- 22.Pentao L., Wise C.A., Chinault A.C., Patel P.I., Lupski J.R. Charcot-Marie-Tooth type 1A duplication appears to arise from recombination at repeat sequences flanking the 1.5 Mb monomer unit. Nat. Genet. 1992;2:292–300. doi: 10.1038/ng1292-292. [DOI] [PubMed] [Google Scholar]
- 23.Stankiewicz P., Lupski J.R. Genome architecture, rearrangements and genomic disorders. Trends Genet. 2002;18:74–82. doi: 10.1016/s0168-9525(02)02592-1. [DOI] [PubMed] [Google Scholar]
- 24.Chance P.F., Alderson M.K., Leppig K.A., Lensch M.W., Matsunami N., Smith B., Swanson P.D., Odelberg S.J., Disteche C.M., Bird T.D. DNA deletion associated with hereditary neuropathy with liability to pressure palsies. Cell. 1993;72:143–151. doi: 10.1016/0092-8674(93)90058-x. [DOI] [PubMed] [Google Scholar]
- 25.Chance P.F., Abbas N., Lensch M.W., Pentao L., Roa B.B., Patel P.I., Lupski J.R. Two autosomal dominant neuropathies result from reciprocal DNA duplication/deletion of a region on chromosome 17. Hum. Mol. Genet. 1994;3:223–228. doi: 10.1093/hmg/3.2.223. [DOI] [PubMed] [Google Scholar]
- 26.Nicholson G.A., Valentijn L.J., Cherryson A.K., Kennerson M.L., Bragg T.L., DeKroon R.M., Ross D.A., Pollard J.D., McLeod J.G., Bolhuis P.A. A frame shift mutation in the PMP22 gene in hereditary neuropathy with liability to pressure palsies. Nat. Genet. 1994;6:263–266. doi: 10.1038/ng0394-263. [DOI] [PubMed] [Google Scholar]
- 27.Chen K.S., Manian P., Koeuth T., Potocki L., Zhao Q., Chinault A.C., Lee C.C., Lupski J.R. Homologous recombination of a flanking repeat gene cluster is a mechanism for a common contiguous gene deletion syndrome. Nat. Genet. 1997;17:154–163. doi: 10.1038/ng1097-154. [DOI] [PubMed] [Google Scholar]
- 28.Potocki L., Chen K.S., Park S.S., Osterholm D.E., Withers M.A., Kimonis V., Summers A.M., Meschino W.S., Anyane-Yeboa K., Kashork C.D. Molecular mechanism for duplication 17p11.2- the homologous recombination reciprocal of the Smith-Magenis microdeletion. Nat. Genet. 2000;24:84–87. doi: 10.1038/71743. [DOI] [PubMed] [Google Scholar]
- 29.Potocki L., Bi W., Treadwell-Deering D., Carvalho C.M., Eifert A., Friedman E.M., Glaze D., Krull K., Lee J.A., Lewis R.A. Characterization of Potocki-Lupski syndrome (dup(17)(p11.2p11.2)) and delineation of a dosage-sensitive critical interval that can convey an autism phenotype. Am. J. Hum. Genet. 2007;80:633–649. doi: 10.1086/512864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Satzinger H. Theodor and Marcella Boveri: chromosomes and cytoplasm in heredity and development. Nat. Rev. Genet. 2008;9:231–238. doi: 10.1038/nrg2311. [DOI] [PubMed] [Google Scholar]
- 31.Lupski J.R. Genomic disorders: structural features of the genome can lead to DNA rearrangements and human disease traits. Trends Genet. 1998;14:417–422. doi: 10.1016/s0168-9525(98)01555-8. [DOI] [PubMed] [Google Scholar]
- 32.Lupski J.R. Genomic disorders ten years on. Genome Med. 2009;1:42. doi: 10.1186/gm42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harel T., Lupski J.R. Genomic disorders 20 years on-mechanisms for clinical manifestations. Clin. Genet. 2018;93:439–449. doi: 10.1111/cge.13146. [DOI] [PubMed] [Google Scholar]
- 34.McKusick V.A. Mendelian Inheritance in Man and its online version, OMIM. Am. J. Hum. Genet. 2007;80:588–604. doi: 10.1086/514346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lupski J.R. Clinical genomics: from a truly personal genome viewpoint. Hum. Genet. 2016;135:591–601. doi: 10.1007/s00439-016-1682-6. [DOI] [PubMed] [Google Scholar]
- 36.Lee J.A., Carvalho C.M., Lupski J.R. A DNA replication mechanism for generating nonrecurrent rearrangements associated with genomic disorders. Cell. 2007;131:1235–1247. doi: 10.1016/j.cell.2007.11.037. [DOI] [PubMed] [Google Scholar]
- 37.Carvalho C.M., Lupski J.R. Mechanisms underlying structural variant formation in genomic disorders. Nat. Rev. Genet. 2016;17:224–238. doi: 10.1038/nrg.2015.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shapiro J.A. Molecular model for the transposition and replication of bacteriophage Mu and other transposable elements. Proc. Natl. Acad. Sci. USA. 1979;76:1933–1937. doi: 10.1073/pnas.76.4.1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hastings P.J., Ira G., Lupski J.R. A microhomology-mediated break-induced replication model for the origin of human copy number variation. PLoS Genet. 2009;5:e1000327. doi: 10.1371/journal.pgen.1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mayle R., Campbell I.M., Beck C.R., Yu Y., Wilson M., Shaw C.A., Bjergbaek L., Lupski J.R., Ira G. DNA REPAIR. Mus81 and converging forks limit the mutagenicity of replication fork breakage. Science. 2015;349:742–747. doi: 10.1126/science.aaa8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sakofsky C.J., Ayyar S., Deem A.K., Chung W.H., Ira G., Malkova A. Translesion polymerases drive microhomology-mediated break-induced replication leading to complex chromosomal rearrangements. Mol. Cell. 2015;60:860–872. doi: 10.1016/j.molcel.2015.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lowden M.R., Flibotte S., Moerman D.G., Ahmed S. DNA synthesis generates terminal duplications that seal end-to-end chromosome fusions. Science. 2011;332:468–471. doi: 10.1126/science.1199022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yatsenko S.A., Hixson P., Roney E.K., Scott D.A., Schaaf C.P., Ng Y.T., Palmer R., Fisher R.B., Patel A., Cheung S.W., Lupski J.R. Human subtelomeric copy number gains suggest a DNA replication mechanism for formation: beyond breakage-fusion-bridge for telomere stabilization. Hum. Genet. 2012;131:1895–1910. doi: 10.1007/s00439-012-1216-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McClintock B. The stability of broken ends of chromosomes in Zea Mays. Genetics. 1941;26:234–282. doi: 10.1093/genetics/26.2.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carvalho C.M., Ramocki M.B., Pehlivan D., Franco L.M., Gonzaga-Jauregui C., Fang P., McCall A., Pivnick E.K., Hines-Dowell S., Seaver L.H. Inverted genomic segments and complex triplication rearrangements are mediated by inverted repeats in the human genome. Nat. Genet. 2011;43:1074–1081. doi: 10.1038/ng.944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Carvalho C.M., Pehlivan D., Ramocki M.B., Fang P., Alleva B., Franco L.M., Belmont J.W., Hastings P.J., Lupski J.R. Replicative mechanisms for CNV formation are error prone. Nat. Genet. 2013;45:1319–1326. doi: 10.1038/ng.2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carvalho C.M., Pfundt R., King D.A., Lindsay S.J., Zuccherato L.W., Macville M.V., Liu P., Johnson D., Stankiewicz P., Brown C.W., DDD Study Absence of heterozygosity due to template switching during replicative rearrangements. Am. J. Hum. Genet. 2015;96:555–564. doi: 10.1016/j.ajhg.2015.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Beck C.R., Carvalho C.M., Banser L., Gambin T., Stubbolo D., Yuan B., Sperle K., McCahan S.M., Henneke M., Seeman P. Complex genomic rearrangements at the PLP1 locus include triplication and quadruplication. PLoS Genet. 2015;11:e1005050. doi: 10.1371/journal.pgen.1005050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zuccherato L.W., Alleva B., Whiters M.A., Carvalho C.M., Lupski J.R. Chimeric transcripts resulting from complex duplications in chromosome Xq28. Hum. Genet. 2016;135:253–256. doi: 10.1007/s00439-015-1614-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gu S., Yuan B., Campbell I.M., Beck C.R., Carvalho C.M., Nagamani S.C., Erez A., Patel A., Bacino C.A., Shaw C.A. Alu-mediated diverse and complex pathogenic copy-number variants within human chromosome 17 at p13.3. Hum. Mol. Genet. 2015;24:4061–4077. doi: 10.1093/hmg/ddv146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang F., Khajavi M., Connolly A.M., Towne C.F., Batish S.D., Lupski J.R. The DNA replication FoSTeS/MMBIR mechanism can generate genomic, genic and exonic complex rearrangements in humans. Nat. Genet. 2009;41:849–853. doi: 10.1038/ng.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yuan B., Harel T., Gu S., Liu P., Burglen L., Chantot-Bastaraud S., Gelowani V., Beck C.R., Carvalho C.M., Cheung S.W. Nonrecurrent 17p11.2p12 rearrangement events that result in two concomitant genomic disorders: The PMP22-RAI1 contiguous gene duplication syndrome. Am. J. Hum. Genet. 2015;97:691–707. doi: 10.1016/j.ajhg.2015.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yuan B., Neira J., Gu S., Harel T., Liu P., Briceño I., Elsea S.H., Gómez A., Potocki L., Lupski J.R. Nonrecurrent PMP22-RAI1 contiguous gene deletions arise from replication-based mechanisms and result in Smith-Magenis syndrome with evident peripheral neuropathy. Hum. Genet. 2016;135:1161–1174. doi: 10.1007/s00439-016-1703-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schmickel R.D. Contiguous gene syndromes: a component of recognizable syndromes. J. Pediatr. 1986;109:231–241. doi: 10.1016/s0022-3476(86)80377-8. [DOI] [PubMed] [Google Scholar]
- 55.Liburd N., Ghosh M., Riazuddin S., Naz S., Khan S., Ahmed Z., Riazuddin S., Liang Y., Menon P.S., Smith T. Novel mutations of MYO15A associated with profound deafness in consanguineous families and moderately severe hearing loss in a patient with Smith-Magenis syndrome. Hum. Genet. 2001;109:535–541. doi: 10.1007/s004390100604. [DOI] [PubMed] [Google Scholar]
- 56.Shiow L.R., Paris K., Akana M.C., Cyster J.G., Sorensen R.U., Puck J.M. Severe combined immunodeficiency (SCID) and attention deficit hyperactivity disorder (ADHD) associated with a Coronin-1A mutation and a chromosome 16p11.2 deletion. Clin. Immunol. 2009;131:24–30. doi: 10.1016/j.clim.2008.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kurotaki N., Shen J.J., Touyama M., Kondoh T., Visser R., Ozaki T., Nishimoto J., Shiihara T., Uetake K., Makita Y. Phenotypic consequences of genetic variation at hemizygous alleles: Sotos syndrome is a contiguous gene syndrome incorporating coagulation factor twelve (FXII) deficiency. Genet. Med. 2005;7:479–483. doi: 10.1097/01.gim.0000177419.43309.37. [DOI] [PubMed] [Google Scholar]
- 58.Yatsenko S.A., Brundage E.K., Roney E.K., Cheung S.W., Chinault A.C., Lupski J.R. Molecular mechanisms for subtelomeric rearrangements associated with the 9q34.3 microdeletion syndrome. Hum. Mol. Genet. 2009;18:1924–1936. doi: 10.1093/hmg/ddp114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lupski J.R., Stankiewicz P. Genomic disorders: molecular mechanisms for rearrangements and conveyed phenotypes. PLoS Genet. 2005;1:e49. doi: 10.1371/journal.pgen.0010049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gu S., Szafranski P., Akdemir Z.C., Yuan B., Cooper M.L., Magriñá M.A., Bacino C.A., Lalani S.R., Breman A.M., Smith J.L. Mechanisms for complex chromosomal insertions. PLoS Genet. 2016;12:e1006446. doi: 10.1371/journal.pgen.1006446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Grochowski C.M., Gu S., Yuan B., Tcw J., Brennand K.J., Sebat J., Malhotra D., McCarthy S., Rudolph U., Lindstrand A. Marker chromosome genomic structure and temporal origin implicate a chromoanasynthesis event in a family with pleiotropic psychiatric phenotypes. Hum. Mutat. 2018;39:939–946. doi: 10.1002/humu.23537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nilsson D., Pettersson M., Gustavsson P., Förster A., Hofmeister W., Wincent J., Zachariadis V., Anderlid B.M., Nordgren A., Mäkitie O. Whole-genome sequencing of cytogenetically balanced chromosome translocations identifies potentially pathological gene disruptions and highlights the importance of microhomology in the mechanism of formation. Hum. Mutat. 2017;38:180–192. doi: 10.1002/humu.23146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stephens P.J., Greenman C.D., Fu B., Yang F., Bignell G.R., Mudie L.J., Pleasance E.D., Lau K.W., Beare D., Stebbings L.A. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 2011;144:27–40. doi: 10.1016/j.cell.2010.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu P., Erez A., Nagamani S.C., Dhar S.U., Kołodziejska K.E., Dharmadhikari A.V., Cooper M.L., Wiszniewska J., Zhang F., Withers M.A. Chromosome catastrophes involve replication mechanisms generating complex genomic rearrangements. Cell. 2011;146:889–903. doi: 10.1016/j.cell.2011.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Maher C.A., Wilson R.K. Chromothripsis and human disease: piecing together the shattering process. Cell. 2012;148:29–32. doi: 10.1016/j.cell.2012.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu P., Yuan B., Carvalho C.M.B., Wuster A., Walter K., Zhang L., Gambin T., Chong Z., Campbell I.M., Coban Akdemir Z. An organismal CNV mutator phenotype restricted to early human development. Cell. 2017;168:830–842.e7. doi: 10.1016/j.cell.2017.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.DiVincenzo C., Elzinga C.D., Medeiros A.C., Karbassi I., Jones J.R., Evans M.C., Braastad C.D., Bishop C.M., Jaremko M., Wang Z. The allelic spectrum of Charcot-Marie-Tooth disease in over 17,000 individuals with neuropathy. Mol. Genet. Genomic Med. 2014;2:522–529. doi: 10.1002/mgg3.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liu P., Gelowani V., Zhang F., Drory V.E., Ben-Shachar S., Roney E., Medeiros A.C., Moore R.J., DiVincenzo C., Burnette W.B. Mechanism, prevalence, and more severe neuropathy phenotype of the Charcot-Marie-Tooth type 1A triplication. Am. J. Hum. Genet. 2014;94:462–469. doi: 10.1016/j.ajhg.2014.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lupski J.R. Genomic rearrangements and sporadic disease. Nat. Genet. 2007;39(7, Suppl):S43–S47. doi: 10.1038/ng2084. [DOI] [PubMed] [Google Scholar]
- 70.Turner D.J., Miretti M., Rajan D., Fiegler H., Carter N.P., Blayney M.L., Beck S., Hurles M.E. Germline rates of de novo meiotic deletions and duplications causing several genomic disorders. Nat. Genet. 2008;40:90–95. doi: 10.1038/ng.2007.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ramocki M.B., Peters S.U., Tavyev Y.J., Zhang F., Carvalho C.M., Schaaf C.P., Richman R., Fang P., Glaze D.G., Lupski J.R., Zoghbi H.Y. Autism and other neuropsychiatric symptoms are prevalent in individuals with MeCP2 duplication syndrome. Ann. Neurol. 2009;66:771–782. doi: 10.1002/ana.21715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kaku D.A., Parry G.J., Malamut R., Lupski J.R., Garcia C.A. Nerve conduction studies in Charcot-Marie-Tooth polyneuropathy associated with a segmental duplication of chromosome 17. Neurology. 1993;43:1806–1808. doi: 10.1212/wnl.43.9.1806. [DOI] [PubMed] [Google Scholar]
- 73.Riccardi V.M., Lupski J.R. Duplications, deletions, and single-nucleotide variations: the complexity of genetic arithmetic. Genet. Med. 2013;15:172–173. doi: 10.1038/gim.2012.124. [DOI] [PubMed] [Google Scholar]
- 74.Delabar J.M., Goldgaber D., Lamour Y., Nicole A., Huret J.L., de Grouchy J., Brown P., Gajdusek D.C., Sinet P.M. Beta amyloid gene duplication in Alzheimer’s disease and karyotypically normal Down syndrome. Science. 1987;235:1390–1392. doi: 10.1126/science.2950593. [DOI] [PubMed] [Google Scholar]
- 75.Podlisny M.B., Lee G., Selkoe D.J. Gene dosage of the amyloid beta precursor protein in Alzheimer’s disease. Science. 1987;238:669–671. doi: 10.1126/science.2960019. [DOI] [PubMed] [Google Scholar]
- 76.Tanzi R.E., Bird E.D., Latt S.A., Neve R.L. The amyloid beta protein gene is not duplicated in brains from patients with Alzheimer’s disease. Science. 1987;238:666–669. doi: 10.1126/science.2890207. [DOI] [PubMed] [Google Scholar]
- 77.Rovelet-Lecrux A., Hannequin D., Raux G., Le Meur N., Laquerrière A., Vital A., Dumanchin C., Feuillette S., Brice A., Vercelletto M. APP locus duplication causes autosomal dominant early-onset Alzheimer disease with cerebral amyloid angiopathy. Nat. Genet. 2006;38:24–26. doi: 10.1038/ng1718. [DOI] [PubMed] [Google Scholar]
- 78.Roa B.B., Garcia C.A., Pentao L., Killian J.M., Trask B.J., Suter U., Snipes G.J., Ortiz-Lopez R., Shooter E.M., Patel P.I., Lupski J.R. Evidence for a recessive PMP22 point mutation in Charcot-Marie-Tooth disease type 1A. Nat. Genet. 1993;5:189–194. doi: 10.1038/ng1093-189. [DOI] [PubMed] [Google Scholar]
- 79.Warner L.E., Roa B.B., Lupski J.R. Absence of PMP22 coding region mutations in CMT1A duplication patients: further evidence supporting gene dosage as a mechanism for Charcot-Marie-Tooth disease type 1A. Hum. Mutat. 1996;8:362–365. doi: 10.1002/(SICI)1098-1004(1996)8:4<362::AID-HUMU10>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 80.Wheeler D.A., Srinivasan M., Egholm M., Shen Y., Chen L., McGuire A., He W., Chen Y.J., Makhijani V., Roth G.T. The complete genome of an individual by massively parallel DNA sequencing. Nature. 2008;452:872–876. doi: 10.1038/nature06884. [DOI] [PubMed] [Google Scholar]
- 81.Boone P.M., Liu P., Zhang F., Carvalho C.M., Towne C.F., Batish S.D., Lupski J.R. Alu-specific microhomology-mediated deletion of the final exon of SPAST in three unrelated subjects with hereditary spastic paraplegia. Genet. Med. 2011;13:582–592. doi: 10.1097/GIM.0b013e3182106775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Boone P.M., Yuan B., Campbell I.M., Scull J.C., Withers M.A., Baggett B.C., Beck C.R., Shaw C.J., Stankiewicz P., Moretti P. The Alu-rich genomic architecture of SPAST predisposes to diverse and functionally distinct disease-associated CNV alleles. Am. J. Hum. Genet. 2014;95:143–161. doi: 10.1016/j.ajhg.2014.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Song X., Beck C.R., Du R., Campbell I.M., Coban-Akdemir Z., Gu S., Breman A.M., Stankiewicz P., Ira G., Shaw C.A., Lupski J.R. Predicting human genes susceptible to genomic instability associated with Alu/Alu-mediated rearrangements. Genome Res. 2018;28:1228–1242. doi: 10.1101/gr.229401.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lehrman M.A., Goldstein J.L., Russell D.W., Brown M.S. Duplication of seven exons in LDL receptor gene caused by Alu-Alu recombination in a subject with familial hypercholesterolemia. Cell. 1987;48:827–835. doi: 10.1016/0092-8674(87)90079-1. [DOI] [PubMed] [Google Scholar]
- 85.Lehrman M.A., Schneider W.J., Südhof T.C., Brown M.S., Goldstein J.L., Russell D.W. Mutation in LDL receptor: Alu-Alu recombination deletes exons encoding transmembrane and cytoplasmic domains. Science. 1985;227:140–146. doi: 10.1126/science.3155573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ricard G., Molina J., Chrast J., Gu W., Gheldof N., Pradervand S., Schütz F., Young J.I., Lupski J.R., Reymond A., Walz K. Phenotypic consequences of copy number variation: insights from Smith-Magenis and Potocki-Lupski syndrome mouse models. PLoS Biol. 2010;8:e1000543. doi: 10.1371/journal.pbio.1000543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lacaria M., Saha P., Potocki L., Bi W., Yan J., Girirajan S., Burns B., Elsea S., Walz K., Chan L. A duplication CNV that conveys traits reciprocal to metabolic syndrome and protects against diet-induced obesity in mice and men. PLoS Genet. 2012;8:e1002713. doi: 10.1371/journal.pgen.1002713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bi W., Park S.S., Shaw C.J., Withers M.A., Patel P.I., Lupski J.R. Reciprocal crossovers and a positional preference for strand exchange in recombination events resulting in deletion or duplication of chromosome 17p11.2. Am. J. Hum. Genet. 2003;73:1302–1315. doi: 10.1086/379979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Reiter L.T., Murakami T., Koeuth T., Pentao L., Muzny D.M., Gibbs R.A., Lupski J.R. A recombination hotspot responsible for two inherited peripheral neuropathies is located near a mariner transposon-like element. Nat. Genet. 1996;12:288–297. doi: 10.1038/ng0396-288. [DOI] [PubMed] [Google Scholar]
- 90.Liu P., Lacaria M., Zhang F., Withers M., Hastings P.J., Lupski J.R. Frequency of nonallelic homologous recombination is correlated with length of homology: evidence that ectopic synapsis precedes ectopic crossing-over. Am. J. Hum. Genet. 2011;89:580–588. doi: 10.1016/j.ajhg.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jacquemont S., Reymond A., Zufferey F., Harewood L., Walters R.G., Kutalik Z., Martinet D., Shen Y., Valsesia A., Beckmann N.D. Mirror extreme BMI phenotypes associated with gene dosage at the chromosome 16p11.2 locus. Nature. 2011;478:97–102. doi: 10.1038/nature10406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Owen D., Bracher-Smith M., Kendall K.M., Rees E., Einon M., Escott-Price V., Owen M.J., O’Donovan M.C., Kirov G. Effects of pathogenic CNVs on physical traits in participants of the UK Biobank. BMC Genomics. 2018;19:867. doi: 10.1186/s12864-018-5292-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Brunetti-Pierri N., Berg J.S., Scaglia F., Belmont J., Bacino C.A., Sahoo T., Lalani S.R., Graham B., Lee B., Shinawi M. Recurrent reciprocal 1q21.1 deletions and duplications associated with microcephaly or macrocephaly and developmental and behavioral abnormalities. Nat. Genet. 2008;40:1466–1471. doi: 10.1038/ng.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Dumas L.J., O’Bleness M.S., Davis J.M., Dickens C.M., Anderson N., Keeney J.G., Jackson J., Sikela M., Raznahan A., Giedd J. DUF1220-domain copy number implicated in human brain-size pathology and evolution. Am. J. Hum. Genet. 2012;91:444–454. doi: 10.1016/j.ajhg.2012.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Heck D.H., Gu W., Cao Y., Qi S., Lacaria M., Lupski J.R. Opposing phenotypes in mice with Smith-Magenis deletion and Potocki-Lupski duplication syndromes suggest gene dosage effects on fluid consumption behavior. Am. J. Med. Genet. A. 2012;158A:2807–2814. doi: 10.1002/ajmg.a.35601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Crespi B., Stead P., Elliot M. Evolution in health and medicine Sackler colloquium: Comparative genomics of autism and schizophrenia. Proc. Natl. Acad. Sci. USA. 2010;107(Suppl 1):1736–1741. doi: 10.1073/pnas.0906080106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.International Schizophrenia Consortium Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008;455:237–241. doi: 10.1038/nature07239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lupski J.R. Schizophrenia: Incriminating genomic evidence. Nature. 2008;455:178–179. doi: 10.1038/455178a. [DOI] [PubMed] [Google Scholar]
- 99.Stefansson H., Rujescu D., Cichon S., Pietiläinen O.P., Ingason A., Steinberg S., Fossdal R., Sigurdsson E., Sigmundsson T., Buizer-Voskamp J.E., GROUP Large recurrent microdeletions associated with schizophrenia. Nature. 2008;455:232–236. doi: 10.1038/nature07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Weiss L.A., Shen Y., Korn J.M., Arking D.E., Miller D.T., Fossdal R., Saemundsen E., Stefansson H., Ferreira M.A., Green T., Autism Consortium Association between microdeletion and microduplication at 16p11.2 and autism. N. Engl. J. Med. 2008;358:667–675. doi: 10.1056/NEJMoa075974. [DOI] [PubMed] [Google Scholar]
- 101.Shinawi M., Liu P., Kang S.H., Shen J., Belmont J.W., Scott D.A., Probst F.J., Craigen W.J., Graham B.H., Pursley A. Recurrent reciprocal 16p11.2 rearrangements associated with global developmental delay, behavioural problems, dysmorphism, epilepsy, and abnormal head size. J. Med. Genet. 2010;47:332–341. doi: 10.1136/jmg.2009.073015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Barbouti A., Stankiewicz P., Nusbaum C., Cuomo C., Cook A., Höglund M., Johansson B., Hagemeijer A., Park S.S., Mitelman F. The breakpoint region of the most common isochromosome, i(17q), in human neoplasia is characterized by a complex genomic architecture with large, palindromic, low-copy repeats. Am. J. Hum. Genet. 2004;74:1–10. doi: 10.1086/380648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Liu P., Carvalho C.M., Hastings P.J., Lupski J.R. Mechanisms for recurrent and complex human genomic rearrangements. Curr. Opin. Genet. Dev. 2012;22:211–220. doi: 10.1016/j.gde.2012.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sharp A.J., Hansen S., Selzer R.R., Cheng Z., Regan R., Hurst J.A., Stewart H., Price S.M., Blair E., Hennekam R.C. Discovery of previously unidentified genomic disorders from the duplication architecture of the human genome. Nat. Genet. 2006;38:1038–1042. doi: 10.1038/ng1862. [DOI] [PubMed] [Google Scholar]
- 105.Dittwald P., Gambin T., Szafranski P., Li J., Amato S., Divon M.Y., Rodríguez Rojas L.X., Elton L.E., Scott D.A., Schaaf C.P. NAHR-mediated copy-number variants in a clinical population: mechanistic insights into both genomic disorders and Mendelizing traits. Genome Res. 2013;23:1395–1409. doi: 10.1101/gr.152454.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Männik K., Mägi R., Macé A., Cole B., Guyatt A.L., Shihab H.A., Maillard A.M., Alavere H., Kolk A., Reigo A. Copy number variations and cognitive phenotypes in unselected populations. JAMA. 2015;313:2044–2054. doi: 10.1001/jama.2015.4845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lupski J.R. Cognitive phenotypes and genomic copy number variations. JAMA. 2015;313:2029–2030. doi: 10.1001/jama.2015.4846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Beck C.R., Carvalho C.M.B., Coban Akdemir Z., Sedlazeck F.J., Song X., Meng Q., Hu J., Doddapaneni H., Chong Z., Chen E.S. Megabase length hypermutation accompanies human structural variation at 17p11.2. Cell. 2018;176 doi: 10.1016/j.cell.2019.01.045. Published online February 28, 3019. 10.1016/j.cell.2019.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lupski J.R., Belmont J.W., Boerwinkle E., Gibbs R.A. Clan genomics and the complex architecture of human disease. Cell. 2011;147:32–43. doi: 10.1016/j.cell.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Verny C., Ravisé N., Leutenegger A.L., Pouplard F., Dubourg O., Tardieu S., Dubas F., Brice A., Genin E., LeGuern E. Coincidence of two genetic forms of Charcot-Marie-Tooth disease in a single family. Neurology. 2004;63:1527–1529. doi: 10.1212/01.wnl.0000142082.65144.ee. [DOI] [PubMed] [Google Scholar]
- 111.Potocki L., Chen K.S., Koeuth T., Killian J., Iannaccone S.T., Shapira S.K., Kashork C.D., Spikes A.S., Shaffer L.G., Lupski J.R. DNA rearrangements on both homologues of chromosome 17 in a mildly delayed individual with a family history of autosomal dominant carpal tunnel syndrome. Am. J. Hum. Genet. 1999;64:471–478. doi: 10.1086/302240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Saporta A.S., Sottile S.L., Miller L.J., Feely S.M., Siskind C.E., Shy M.E. Charcot-Marie-Tooth disease subtypes and genetic testing strategies. Ann. Neurol. 2011;69:22–33. doi: 10.1002/ana.22166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Chong J.X., Buckingham K.J., Jhangiani S.N., Boehm C., Sobreira N., Smith J.D., Harrell T.M., McMillin M.J., Wiszniewski W., Gambin T., Centers for Mendelian Genomics The genetic basis of Mendelian phenotypes: discoveries, challenges, and opportunities. Am. J. Hum. Genet. 2015;97:199–215. doi: 10.1016/j.ajhg.2015.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Posey J.E., O’Donnell-Luria A.H., Chong J.X., Harel T., Jhangiani S.N., Coban Akdemir Z.H., Buyske S., Pehlivan D., Carvalho C.M.B., Baxter S. Insights into genetics, human biology and disease gleaned from family based genomic studies. Genet. Med. 2019 doi: 10.1038/s41436-018-0408-7. Published online January 18, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Karaca E., Posey J.E., Coban Akdemir Z., Pehlivan D., Harel T., Jhangiani S.N., Bayram Y., Song X., Bahrambeigi V., Yuregir O.O. Phenotypic expansion illuminates multilocus pathogenic variation. Genet. Med. 2018;20:1528–1537. doi: 10.1038/gim.2018.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Eldomery M.K., Coban-Akdemir Z., Harel T., Rosenfeld J.A., Gambin T., Stray-Pedersen A., Küry S., Mercier S., Lessel D., Denecke J. Lessons learned from additional research analyses of unsolved clinical exome cases. Genome Med. 2017;9:26. doi: 10.1186/s13073-017-0412-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Yang Y., Muzny D.M., Reid J.G., Bainbridge M.N., Willis A., Ward P.A., Braxton A., Beuten J., Xia F., Niu Z. Clinical whole-exome sequencing for the diagnosis of mendelian disorders. N. Engl. J. Med. 2013;369:1502–1511. doi: 10.1056/NEJMoa1306555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Posey J.E., Harel T., Liu P., Rosenfeld J.A., James R.A., Coban Akdemir Z.H., Walkiewicz M., Bi W., Xiao R., Ding Y. Resolution of disease phenotypes resulting from multilocus genomic variation. N. Engl. J. Med. 2017;376:21–31. doi: 10.1056/NEJMoa1516767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Girirajan S., Rosenfeld J.A., Coe B.P., Parikh S., Friedman N., Goldstein A., Filipink R.A., McConnell J.S., Angle B., Meschino W.S. Phenotypic heterogeneity of genomic disorders and rare copy-number variants. N. Engl. J. Med. 2012;367:1321–1331. doi: 10.1056/NEJMoa1200395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Gonzaga-Jauregui C., Harel T., Gambin T., Kousi M., Griffin L.B., Francescatto L., Ozes B., Karaca E., Jhangiani S.N., Bainbridge M.N., Baylor-Hopkins Center for Mendelian Genomics Exome sequence analysis suggests that genetic burden contributes to phenotypic variability and complex neuropathy. Cell Rep. 2015;12:1169–1183. doi: 10.1016/j.celrep.2015.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wu N., Ming X., Xiao J., Wu Z., Chen X., Shinawi M., Shen Y., Yu G., Liu J., Xie H. TBX6 null variants and a common hypomorphic allele in congenital scoliosis. N. Engl. J. Med. 2015;372:341–350. doi: 10.1056/NEJMoa1406829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Yang N., Wu N., Zhang L., Zhao Y., Liu J., Liang X., Ren X., Li W., Chen W., Dong S. TBX6 compound inheritance leads to congenital vertebral malformations in humans and mice. Hum. Mol. Genet. 2019;28:539–547. doi: 10.1093/hmg/ddy358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Liu J., Wu N., Yang N., Takeda K., Chen W., Li W., Du R., Liu S., Zhou Y., Liu Z. TBX6-associated congenital scoliosis (TACS) as a novel clinically uniform subtype of congenital scoliosis. Genet. Med. 2018 doi: 10.1038/s41436-018-0377-x. Published online January 14, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Verbitsky M., Westland R., Perez A., Kiryluk K., Liu Q., Krithivasan P., Mitrotti A., Fasel D.A., Batourina E., Sampson M.G. The copy number variation landscape of congenital anomalies of the kidney and urinary tract. Nat. Genet. 2018;51:117–127. doi: 10.1038/s41588-018-0281-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Karolak J.A., Vincent M., Deutsch G., Gambin T., Cogné B., Pichon O., Vetrini F., Mefford H.C., Dines J.N., Golden-Grant K. Complex compound inheritance of lethal lung developmental disorders due to disruption of the TBX-FGF pathway. Am. J. Hum. Genet. 2019;104:213–228. doi: 10.1016/j.ajhg.2018.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]