Abstract
Isoprenoids are an attractive class of metabolites for enzymatic synthesis from renewable substrates. However, metabolic engineering of microorganisms for monoterpenoid production is limited by the need for time-consuming, and often non-intuitive, combinatorial tuning of biosynthetic pathway variations to meet design criteria. Towards alleviating this limitation, the goal of this work was to build a modular, cell-free platform for construction and testing of monoterpenoid pathways, using the fragrance and flavoring molecule limonene as a model. In this platform, multiple Escherichia coli lysates, each enriched with a single overexpressed pathway enzyme, are mixed to construct the full biosynthetic pathway. First, we show the ability to synthesize limonene from six enriched lysates with mevalonate substrate, an adenosine triphosphate (ATP) source, and cofactors. Next, we extend the pathway to use glucose as a substrate, which relies on native metabolism in the extract to convert glucose to acetyl-CoA along with three additional enzymes to convert acetyl-CoA to mevalonate. We find that the native E. coli farnesyl diphosphate synthase (IspA) is active in the lysate and diverts flux from the pathway intermediate geranyl pyrophospahte to farnesyl pyrophsophate and the byproduct farnesol. By adjusting the relative levels of cofactors NAD+, ATP and CoA, the system can synthesize 0.66 mM (90.2 mg l−1) limonene over 24 h, a productivity of 3.8 mg l−1 h−1. Our results highlight the flexibility of crude lysates to sustain complex metabolism and, by activating a glucose-to-limonene pathway with 9 heterologous enzymes encompassing 20 biosynthetic steps, expands an approach of using enzyme-enriched lysates for constructing, characterizing and prototyping enzymatic pathways.
Keywords: cell-free metabolic engineering, limonene, metabolic pathway prototyping, cell-free synthetic biology, E. coli crude lysate
1. Introduction
Isoprenoids are a valuable class of compounds with applications as pharmaceuticals, flavorings, fragrances, pesticides, disinfectants and chemical feedstocks.1,2 Metabolic engineering efforts have successfully endowed microorganisms the ability to synthesize a number of isoprenoids,3,4 the most notable being the anti-malarial drug precursor artemisinin.5 While the broader metabolic engineering field has multiple established success stories,6–9 efforts to construct and characterize new biosynthetic pathways remain time-consuming and often require non-intuitive optimization of enzyme expression levels.10–15
Cell-free metabolic engineering offers a unique approach for constructing metabolic pathways in vitro to accelerate biological design.16–23 Cell-free systems permit easy access to reaction conditions, can circumvent competing byproduct pathways, offer fewer toxicity constraints and avoid the need to divert energy and carbon resources to supporting cell growth.16 This exquisite control provides a high degree of flexibility to model the kinetics and stability of individual enzymes, measure metabolite fluxes in multistep pathways, study the effects of redox potential on pathway performance and experimentally isolate many other parameters confounded in living organisms. Given these features, several groups have used purified enzymes to construct components of the isoprenoid pathway including phosphoenolpyruvate (PEP) to isoprene,24 acetyl-CoA to farnesene,25 mevalonate to amorphadiene,26 acetate to dimethylallyl pyrophosphate (DMAPP),27 glucose to limonene28 and pulegone to menthol isomers.29 Alternatively, crude extract systems are capable of supporting complex metabolism without needing to purify enzymes. Prominent examples include biosynthesis of proteins,30 2,3-butanediol,31 butanol32 and others.16,33,34 However, no one to our knowledge has yet built a crude extract-based system for studying the complete isoprenoid pathway. Moreover, crude extract pathways have so far been limited to 5 enriched enzymes encompassing 17 biosynthetic steps in total.32
In this work, we sought to establish a modular cell-free platform to build monoterpene biosynthetic pathways. We constructed a cell-free pathway to the monoterpenoid limonene, a valuable fragrance and flavoring agent as well as precursor for insecticides, biofuels and medicines.35 While limonene has been successfully synthesized in cells including Escherichia coli,14,36,37 yeast38,39 and cyanobacteria,40,41 titers are low and toxicity of pathway intermediates may confound efforts to prototype highly active enzymes.42 In previous work, we used mixtures of enzyme-enriched lysates to synthesize the isoprenoid precursor mevalonate from glucose43 and quickly tested an unoptimized pathway from mevalonate to limonene.44 Building on these efforts, this manuscript details the construction of the full glucose-to-limonene pathway. In addition, we identified and characterized a competing byproduct pathway toward farnesol and tuned cofactor levels to enable sustained adenosine triphosphate (ATP) production and higher limonene titers. We anticipate this platform could have future utility as a method for prototyping enzymatic monoterpenoid pathways (e.g. studying the impact of multiple enzyme homologs) since cell-free systems avoid the need to reengineer an organism for each pathway iteration.45–52
2. Materials and methods
2.1 Strains and plasmids
Strains and plasmids used in this study are listed in Supplementary Table S1. Pathway gene sequences were codon optimized, synthesized by Gen9 (Cambridge, MA) and cloned into the modified pET-22b (Novagen/EMD Millipore, Darmstadt, Germany) high copy expression vector pETBCS31 using the Berkeley Cloning Standard (BglBrick) restriction sites.53 Strong ribosome binding site (RBS) sequences were calculated using the Salis RBS calculator (v1.1).54 The sequence encoding limonene synthase (LS) was truncated to remove a plastid signal peptide and cloned using Gibson assembly into the plasmid pJL1 behind an N-terminal tag known to improve in vitro expression. The N-terminal tag is a 15 nucleotide, AT-rich sequence which encodes the first five amino acids (Met-Glu-Lys-Lys-Ile, MEKKI) of chloramphenicol acetyl transferase, which has been used to increase expression in E. coli cell-free protein synthesis (CFPS) systems.55,56 Gene sequences (Supplementary Appendix S1) and plasmid maps are included in the supplementary data and all plasmids are available upon request. Routine cloning was performed using the E. coli strain NEB Turbo™ and all DNA-modifying enzymes were purchased from NEB (New England BioLabs, Ipswich, MA). Sequenced-confirmed plasmids were transformed into BL21(DE3) purchased from NEB for protein expression.
2.2 Cell-free proteins synthesis of LS
LS was expressed in vitro using a modified PANOx-SP CFPS system described previously32,56,57 with extract prepared using an Avestin EmulsiFlex-B15 homogenizer. After incubating the 15 µl reaction for 3 h at 30°C, geranyl pyrophospahte (GPP) was added to a final concentration of 360 µM and final volume of 25 µl. A dodecane overlay of 25 µl was added and limonene measured over time. After validation of synthase activity, the sequence was cloned into pETBCS for preparation of enriched lysates.
2.3 Cell growth and extract preparation
Cells were grown and lysed in a manner consistent with prior work.31,43,58Escherichia coli BL21(DE3) containing a pETBCS plasmid (each encoding one or more pathway enzymes) were grown in 1-l Tunair™ shake flasks at 37°C (250 rpm) in 2xYTPG media (16 g·l−1 tryptone, 10 g·l−1 yeast extract, 5 g·l−1 NaCl, 7 g·l−1 potassium phosphate monobasic, 3 g·l−1 potassium phosphate dibasic and 18 g·l−1 glucose) containing 100 µg·l−1 carbenicillin (IBI Scientific, Peosta, IA). Tryptone and yeast extract (BD Bacto™) were obtained from BD Biosciences (San Jose, CA). Cultures were induced with 0.1 mM IPTG (Santa Cruz Biotech, Dallas, TX) at 0.6 OD600nm and growth continued at 30°C for four more hours. Cells were centrifuged (8000 × g for 15 min) and the resulting pellet was rinsed twice in S30 buffer (10 mM tris acetate pH 8.2, 14 mM magnesium acetate and 60 mM potassium acetate). Cells were then flash-frozen for storage at −80°C.
To make crude extracts, cell pellets were thawed on ice, suspended in S30 buffer (0.8 ml per gram cell pellet), and lysed at 20 000 psi (homogenizing pressure) using an EmulsiFlex-B15 homogenizer (Avestin, Ottawa, ON). To remove insoluble protein and cell debris, lysed cells were centrifuged twice at 30 000 × g for 30 min and the supernatant (i.e. lysate) was transferred to a new container without disturbing the pellet. Lysate was aliquoted into 100–400 µl volumes and flash-frozen for storage at −80°C. Total protein was quantified by Bradford assay with bovine serum albumin as the standard using a microplate protocol (Bio-Rad, Hercules, CA). Overexpression of enzymes was confirmed by NuPAGE® SDS-PAGE protein gels using Coomassie stain (Life Technologies/Thermo Fisher Scientific, Waltham, MA). When making new batches of lysate, each was compared to the previous batch and found to have a relatively small variation (typically less than 5–10% of mevalonate or limonene titer, Supplementary Figure S1).
2.4 Cell-free biosynthesis of limonene
Mevalonate to limonene reactions contained glutamate salts (8 mM magnesium glutamate, 10 mM ammonium glutamate and 134 mM potassium glutamate), 100 mM Bis Tris buffer, 1 mM CoA and mixed extracts, as well as 45 mM ATP, 45 mM PEP and/or 15 mM mevalonate (pH 7.2) as noted. Mevalonate was prepared by alkaline hydrolysis of 1 M (±)-mevalonolactone with equal volume 1 M potassium hydroxide for 1.5 h at 37°C and then neutralized to pH 7.2 by addition of 18.5% hydrochloric acid.26 Six lysates, each at 1 mg·ml−1 total protein, were mixed together; each lysate is enriched with a single enzyme [mevalonate kinase (MK), phosphomevalonate kinase (PMK), pyrophosphomevalonate decarboxylase (PMD), isopentenyl pyrophosphate isomerase (IDI), geranyl diphosphate synthase (GPPS) and LS], see Supplementary Table S1 and Appendix S1. Glucose to limonene reactions contained 200 mM glucose, glutamate salts, 100 mM Bis Tris buffer, 1 mM oxidized nicotinamide adenine dinucleotide (NAD+), 1 mM ATP, 1 mM Coenzyme A (CoA) and 10 mM potassium phosphate59 (K2HPO4, pH 7.2) plus seven lysates. Six lysates, each enriched in a single enzyme (MK, PMK, PMD, IDI, GPPS or LS), are added at 1 mg·ml−1 total protein concentration to the glucose-to-limonene reaction in addition to a single lysate43 enriched with three enzymes: acetyl-CoA acetyltransferase (ACAT), hydroxymethylglutaryl-CoA synthase (HMGS) and hydroxymethylglutaryl-CoA reductase (HMGR) at a concentration of 4 mg·ml−1 total protein. Subsequent reactions substituted acetate salts (8 mM magnesium acetate, 10 mM ammonium acetate and 134 mM potassium acetate) for glutamate salts and adjusted levels of NAD+, ATP, CoA and potassium phosphate. All reagents and chemicals were purchased from Sigma Aldrich (St. Louis, MO).
All cell-free reactions (25 µl total volume) were incubated at 30°C in a 1.5 ml Eppendorf tube and were overlaid by 25 µl of dodecane to capture volatile products (i.e. limonene). At each time point, the dodecane overlay was removed for limonene analysis while the reaction itself was quenched by precipitating proteins using 25 µl of 5% trichloroacetic acid (TCA) and then centrifuging at 15 000 × g for 10 min at 4°C. The supernatant was collected and stored at −80°C until analysis. Methods to quantify mevalonate (gas chromatography-mass spectrometry) and other metabolites (high pressure liquid chromatography) are described previously.43 All measurements were completed as technical replicates since we observed that biological replicates were within 5–10% error (Supplementary Figure S1).
2.5 Quantification of limonene and other metabolites
Limonene was quantified by diluting 18 µl of dodecane overlay into 180 µl of ethyl acetate containing 20 µg·ml−1 trans-caryophyllene (Sigma) as an internal standard. This mixture was injected (1 µl) into an Agilent 7890 A Gas Chromatograph with 5977 A MSD (Agilent, Santa Clara, CA) using an Agilent HP-5MS (30 m length × 0.25 mm i.d. × 0.25 µm film) column with helium carrier gas at constant flow of 1 ml·min−1. The inlet temperature was 120°C and initial column temperature held at 120°C for 0.25 min, then increased at 40°C min−1 to 200°C, and finally increased at 30°C·min−1 to 250°C which was maintained for 0.32 more minutes. Injection volume was 1 µl with a split ratio of 20:1. Extract ion chromatograms for 93.1 m/z (limonene, peak at 2.81 min), 69.1 m/z (farnesol, peaks at 5.13 and 5.21 min) and 133.1 m/z (caryophyllene, peak at 4.31 min) were integrated using Agilent MassHunter Quantitation Analysis software. Farnesol elutes with two peaks, both of which tail slightly towards longer retention times; the second peak at 5.21 min was used for quantification. Concentration was determined by comparison to standards prepared by mixing dilutions of (R)-(+)-limonene and farnesol (Sigma) in dodecane. This mixture is pipetted into the aqueous phase of a mock cell-free reaction containing 152 mM acetate salts, 10 mM K2HPO4 and 10 mg·ml−1 of BL21 extract (which contains no expressed proteins with catalytic activity to mevalonate or limonene) and incubated for 3 h at 30°C.
3. Results
3.1 Construction of a glucose to limonene cell-free system
A set of six enzymes is known to convert mevalonate to limonene (Figure 1A). To generate enzyme-enriched lysates, well-characterized enzyme sequences were codon-optimized for E. coli and cloned into a high copy expression vector. Specifically, genes sequences for MK, PMK and PMD were obtained from Saccharomyces cerevisiae (baker’s yeast), while IDI, GPPS and LS were taken from E. coli, Abies grandis (grand fir) and Mentha spicata (spearmint), respectively. Additional information regarding strains and plasmids (Supplementary Table S1), known kinetic parameters (Supplementary Table S2), and gene sequences (Supplementary Appendix S1) are provided.
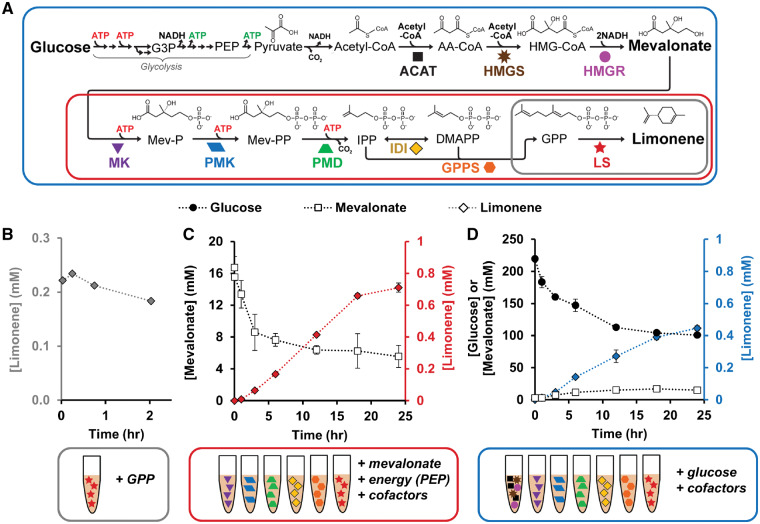
Figure 1.
Mixing of extracts containing overexpressed enzymes recapitulates limonene biosynthesis in vitro. (A) Enzymatic route for limonene synthesis via the mevalonate pathway. (B) Validation of LS activity. (C) Conversion of mevalonate to limonene via six enzyme-enriched lysates plus cofactors and ATP source. (D) Conversion of glucose to limonene via seven enzyme-enriched lysates plus cofactors. G3P, glyceraldehyde 3-phosphate; PEP, phosphoenolpyruvate; AA-CoA, acetoacetyl-CoA; HMG-CoA, 3-hydroxy-3-methylglutaryl-CoA; Mev-P, mevalonate-5-phosphate; Mev-PP, mevalonate pyrophosphate; IPP, isopentenyl pyrophosphate; DMAPP, dimethylallyl pyrophosphate; GPP, geranyl pyrophosphate. Values represent averages (n = 3) and error bars represent 1 SD.
We grew multiple E. coli cultures, each containing a different enzyme-encoding plasmid, induced gene expression and harvested cells containing overexpressed protein. While most enzymes expressed with a high percentage of soluble protein (Supplementary Figure S2), we found that GPPS was mostly insoluble (GPPS1.0, Supplementary Figure S3). By decreasing the strength of the RBS, we produced a strain with lowered, but soluble expression of GPPS (GPPS2.1, Supplementary Figure S3), and this strain was used in subsequent experiments. With six strains each overexpressing a soluble enzyme (Supplementary Figure S4), we then made crude cell lysates of each by disrupting the cells using high-pressure homogenization and removing cellular debris by centrifugation.
We leveraged the plug-and-play nature of cell-free systems to build a complete limonene biosynthesis pathway in vitro in a step-wise manner. As a first step, we validated the activity of the final enzyme (LS) in isolation. We synthesized LS in vitro using cell-free protein synthesis and added substrate GPP directly to the reaction and observed limonene at concentrations around 0.2 mM (Figure 1B), a similar titer compared to a previous in vitro study.36 We conducted preliminary experiments to use cell-free protein synthesis to assemble the full pathway32 but found that low GPPS protein yields limited this approach. Therefore, to extend the pathway to use mevalonate as substrate, six extracts (each enriched in a single enzyme via heterologous overexpression in vivo) were mixed at equal ratios with mevalonate and cofactors. Since the first three reactions of the pathway are ATP-driven, we tested several energy sources including glutamate, ATP and PEP (Supplementary Figure S5). The highest limonene titer occurred when 45 mM PEP was added to 15 mM mevalonate, generating 0.71 ± 0.028 mM limonene (Figure 1C). PEP and ADP are converted to pyruvate and ATP, respectively, by pyruvate kinase present in the cell lysate and this maintains a consistent level of ATP (above 100 μM over 24 h) rather than adding ATP directly (Supplementary Figure S5). Given that the full pathway produced limonene and these enzymes have been previously characterized, we did not individually assess the activity of each individual enzyme. After completing a mevalonate-to-limonene module, we wanted to expand the pathway to generate the mevalonate and ATP substrates from glucose instead of needing to add expensive, high-energy molecules like PEP.
To build the complete glucose-to-limonene pathway, we coupled multiple modules of metabolism together—glycolysis, an acetyl-CoA-to-mevalonate module, and our mevalonate-to-limonene module. Endogenous metabolism in E. coli crude lysates contains all the glycolytic enzymes necessary to convert glucose to acetyl-CoA. A previously developed module for acetyl-CoA-to-mevalonate conversion consists of a single extract enriched with three enzymes, ACAT, HMGS and HMGR.43 By mixing this ACAT/HMGS/HMGR enriched lysate capable of converting glucose to mevalonate with the six mevalonate-to-limonene lysates plus glucose and cofactors, we are able to construct the complete limonene biosynthetic pathway from glucose. Conveniently, glycolysis generates six net ATP from three glucose molecules, these six ATP are available to convert two mevalonate to one limonene molecule. However, glycolysis generates eight excess reduced nicotinamide adenine dinucleotide (NADH) for every three glucose, which must be recycled to NAD+ by native metabolism in the lysate (Supplementary Figure S6). Using cofactor concentrations established in our prior work,43 the system generates 0.45 ± 0.015 mM limonene over 24 h (Figure 1D). Glucose concentration decreases as the reaction progresses and mevalonate accumulates as well as limonene. In total, we extended the metabolic pathway six steps from our previous work43 which requires 20 total metabolic steps from glucose to limonene. This result highlights the ability of cell-free biology to recapitulate long pathways, as has also been shown in other reports with even more demanding chemistry like polyketides.23,60–63
3.2 Characterization of farnesyl pyrophosphate synthase activity (IspA) in the crude lysate
Since E. coli naturally produces isoprenoids using the non-mevalonate DXP/MEP pathway, we next wanted to determine if any endogenous enzymes in the lysate—such as natively expressed idi and farnesyl diphosphate synthase (ispA) (Figure 2A)—were accelerating or competing with limonene production.
Figure 2.
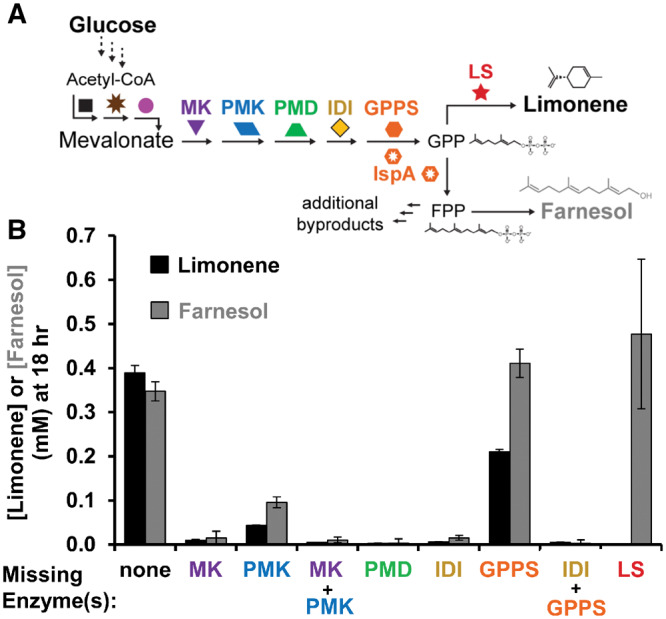
Removing lysates from a complete reaction demonstrates activity of E. coli’s native geranyl pyrophosphate synthase IspA. (A) Metabolic pathway describing IspA’s dual activity to generate FPP in addition to GPP. (B) Limonene synthesis from glucose using seven lysates (black bar). Each subsequent reaction is missing one or more enriched lysates. Limonene is generated in the condition in which the geranyl pyrophosphate synthase-enriched lysate is removed indicating IspA is active. Values represent averages (n = 3) and error bars represent 1 SD.
Using the glucose-to-limonene pathway configuration, we tested the necessity of each heterologous pathway enzyme to produce limonene by removing each enzyme-enriched extract one at a time. This leave-one-out type of experiment is quite straightforward with the cell-free approach. When measuring limonene, we found that the system does indeed produce limonene without supplementation of GPPS (Figure 2B) suggesting that IspA is active. The small amount of limonene produced in the condition lacking PMK suggests that MK may promiscuously catalyze a second phosphorylation event. Notably, levels of IDI are not high enough in the lysate to catalyze limonene biosynthesis without supplementation.
We hypothesized that other isoprenoid byproducts might exist because previous literature has characterized IspA to have bifunctional catalytic activity as a geranyl/farnesyl diphosphate synthase generating farnesyl pyrophsophate (FPP) (C15) as the primary product.64 We indeed found that FPP is converted to farnesol, which accumulates in the cell-free reaction at a similar concentration to limonene (Figure 2B). This indicates that GPPS, IspA and LS are competing for isopentenyl pyrophosphate (IPP), DMAPP and GPP substrates, particularly since farnesol accumulates highest in the condition where LS is absent from the reaction (Figure 2B). Future iterations of this work could consider incorporating an S80F mutation of IspA that favors GPP65,66 or implementing transient knockdown strategies67,68 to conditionally inactivate this essential gene and thus increase limonene concentration in the reaction. However, as the goal of this work was to establish a modular cell-free platform to build monoterpene biosynthetic pathways in crude extracts, we next sought to validate that tuning small molecule cofactors and energy substrates would impact pathway performance.
3.3 Tuning limonene production by modulating cofactor concentrations
Within enzymatic systems, the concentrations of small molecule cofactors and energy substrates can have a significant impact on pathway activity.32,43,69 To demonstrate the ability of cell-free systems to rapidly tune biosynthetic pathway performance, and with an eye towards improving the glucose-to-limonene titer, multiple cofactor conditions were explored. Similar to our recent effort,69 we focused on physiochemical salts that are used to mimic the cytoplasm. We also targeted molecules directly involved in the limonene pathways itself. We first adjusted the amount of potassium phosphate and found that increasing concentrations are deleterious to limonene titer (Supplementary Figure S7A). This is consistent with other results showing phosphate levels higher than 30 mM are inhibitory69,70; however, previous studies had suggested a small concentrations (10 mM) are beneficial for glycolytic activity.43,59 We next tested combinations of supplemental cofactors. NAD+, ATP and CoA were removed one-by-one, in pairs, and all together and we found that a system without any supplemented cofactors catalyzed the same limonene titer as the initial condition with 1 mM each (Supplementary Figure S7B). This is not altogether surprising since lysates are not dialyzed during preparation and are known to contain catalytic amounts of NAD+, ATP and CoA which we have previously measured.43 While not explored in this study, our previous work suggests that dialysis of crude lysates may be insufficient to remove all cofactors and other small molecules (such as nucleotides).71
In order to better characterize the complex reaction environment, we next considered a panel of metabolites over the course of the cell-free reaction under four different conditions (Figure 3). We compared glutamate to acetate as an alternative counterion for K+, NH4+ and Mg2+ ions added to the reaction; acetate salts had improved the titer of mevalonate in our previous work.43 We also tested the presence or absence of supplemental cofactors (1 mM ATP/CoA/NAD+ and 10 mM phosphate). The addition of cofactors to the reaction accelerates the rate of glucose breakdown and lactate production (Figure 3A, B). However, the highest limonene titer utilizes glutamate salts without added cofactors and generates 0.66 ± 0.009 mM (90.2 mg·l−1) limonene over 24 h (Figure 3F and Supplementary Figure S8). We hypothesize that this condition may be favorable for limonene production because the cofactor limitation slows the rate of byproduct lactate. While the metabolism of acetate to acetyl-CoA provides a potentially beneficial secondary carbon source (Figure 3C), it does so at the expense of ATP which may constrain the ATP-dependent enzymes MK, PMK and PMD. For example, mevalonate, the substrate for MK, accumulates to over 40 mM in acetate salt conditions (Figure 3D) indicating that ATP is potentially limiting. By measuring ATP concentrations in the reaction, we see that glutamate conditions have a more consistent level of ATP—above 0.4 mM (with cofactors) and 0.01 mM (without cofactors)—compared to acetate salts (Supplementary Figure S9A). This validates the notion that consistent levels of ATP are important to maintain the reaction. We also see evidence of glutamate metabolism in the accumulation of TCA cycle intermediates succinate (no cofactors) and citrate (with cofactors) (Supplementary Figure S9B, C). Finally, 10 mM ethanol accumulates across all conditions where it is an additional sink for excess NADH (Supplementary Figure S9D).
Figure 3.
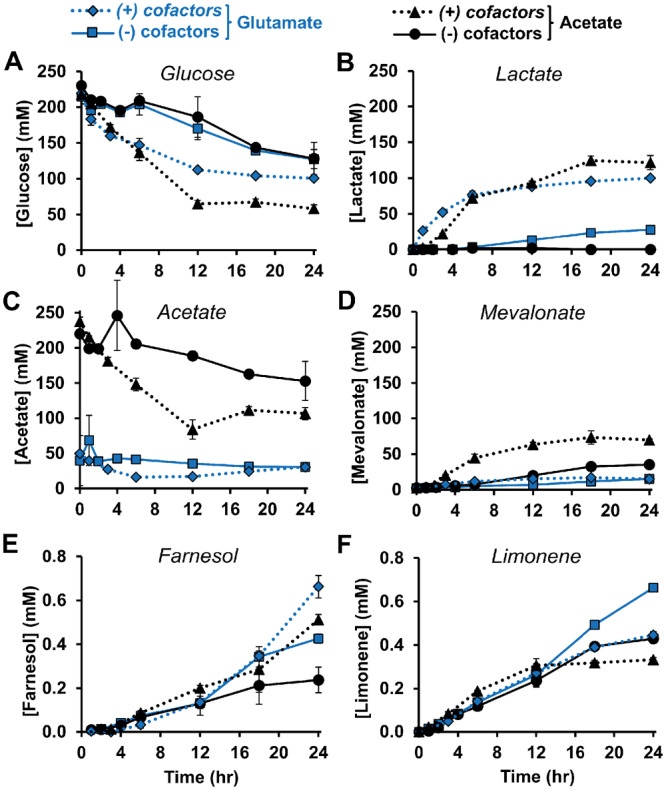
Concentrations of metabolites under four different reaction conditions. Glucose (A), lactate (B), acetate (C), mevalonate (D), farnesol (E) and limonene (F). The (−) condition has no added NAD+, ATP, CoA or potassium phosphate while the (+) condition has 1 mM NAD+, 1 mM ATP, 1 mM CoA and 10 mM potassium phosphate added. Glutamate and acetate are two possible counterion molecules for added Mg2+, K+ and NH4+. Values represent averages (n = 3) and error bars represent 1 SD.
4. Discussion
In this study, we have validated the ability to mix seven enzyme-enriched lysates to construct a 20-step enzymatic pathway from glucose to limonene containing 9 heterologous enzyme steps. The system generates 0.66 mM (90.2 mg·l−1) limonene over 24 h with a productivity of 3.8 mg·l−1·h−1. While the goal of the study was to validate the cell-free platform for characterizing and building monoterpenoid pathways and not for biomanufacturing, cell-free production is similar (2-fold less) to a recently published in vivo pathway using glucose (8.4 mg·l−1·h−1).14 However, our approach does not require DNA manipulation to balance pathway flux or tune promoters and RBSs10–13; this decreases the cycle time between experiments. Additionally, a cell-free biomanufacturing system would have fewer toxicity constraints72 since there is no membrane to be disrupted by limonene and fewer barriers to limonene extraction into the overlay or headspace; such a system would perhaps be well suited for natural product34 or antimicrobial peptide synthesis.73
While previous efforts have recapitulated portions of isoprenoid biosynthesis using purified enzymes,24–27 the crude extract system presented here reconstitutes the entire pathway from glucose to limonene. This highlights the flexibility that comes with crude extract systems to recycle excess cofactors (Supplementary Figure S6).31,32,43 One benefit of purified systems, though, is that there is greater control over pathway operation due to a lack of competing pathways. However, these systems can be limited in their ability to regenerate cofactors.16,22 Recently, the development of an innovative molecular purge valve system74 led to the establishment of a high-yielding (>15 g·l−1, >95% conversion) glucose to limonene system from purified enzymes that balances cofactors.28 Such valves will be crucial in further expanding cell-free technologies to modulate metabolism in both crude extract and purified enzyme systems.
While there are many compelling benefits for using cell-free systems, there are also potential complications. First, our assumption that each enriched lysate is a modular component different only in its enriched enzyme may not be accurate. The overexpression of different enzymes may cause the host cells to have dissimilar proteomes due to cell adaptation and gene expression changes during growth. These differences in the proteome could provide a confounding effect to analysis of data when lysates are mixed. Using CFPS to generate pathway enzymes can limit this effect as there will be no dynamic host response but differences in protein yield could still result in small differences in metabolite concentration. Second, the approach as described is limited to proteins that can be expressed in E. coli and expression optimization does occasionally require time and effort (Supplementary Figure S3); however, mixing lysates from multiple organisms has been shown to address this challenge23,75 Third, modeling frameworks need to be developed; however, some efforts have already begun to adapt genome-scale metabolic models to cell-free metabolic engineering.76
Looking forward, the cell-free platform described here sets the stage for studying the performance of monoterpenoid metabolic enzymes. In this approach, cell-free lysates, each selectively enriched with an overexpressed enzyme homolog, could be generated in parallel and then combinatorially mixed to construct a full biosynthetic pathway. This would enable the identification of novel sets of enzymes which enable the high limonene titer. For example, the MK from S. cerevisiae used in this work is known to be feedback inhibited by pathway intermediates DMAPP, IPP, GPP, FPP and GGPP and its substrate (mevalonate); other homologs are reported to be less feedback inhibited and could be tested.77–79 Though we used well-characterized enzymes with known kinetic parameters (Supplementary Table S2), such a prototyping environment could also be used to characterize new enzyme homologs and compliment efforts to implement kinetics-guided optimization of metabolic pathways in cells.80 Since GPP is a substrate for many different enzymes, the lessons learned from this testing strategy could be adapted to study other monoterpene products by exchanging the LS extract for one enriched in a different synthase; others have highlighted this possibility in cells by swapping plasmids encoding a monoterpene synthase.81 Further, we could study pathway operation in novel metabolic contexts by making made genomic modifications to the extract source strain. An obvious target would be to mutate the native IspA to favor GPP production;65,66 previous in vivo efforts have tried this with mixed results.82–85 Each of these could be exciting directions, enabled by the rapid nature of prototyping in cell-free systems.45–52 Additionally, integration of cell-free protein synthesis would enable swifter design-build-test cycle times, reducing the time to build pathways to hours rather than days.32 For instance, a recent method utilizing synthetic consortia to produce purified translation machinery could enable added control over reaction conditions for cost-effective, high-throughput analysis.86
In summary, we expect this work to help advance a methodology of using enzyme-enriched lysates for constructing, characterizing and prototyping enzymatic pathways. This could ultimately prove to be a useful tool in streamlining and accelerating metabolic engineering in living cells.25
Supplementary Material
Acknowledgements
Special thanks to Ashty Karim for helpful comments on the manuscript. We would also like to thank Jay Keasling87 for graciously sharing plasmids pGW322, pGW350 and pGW360, which were the basis for our codon-optimized plasmids.
Funding
We gratefully acknowledge the Department of Energy (BER grant: DE-SC0018249), the Joint Genome Institute Community Science Program (Project 503280), the David and Lucile Packard Foundation (2011–37152) and the Dreyfus Teacher-Scholar Program for funding and support. QMD is supported, in part, by the Northwestern Molecular Biophysics Training Program funded by the National Institute of General Medical Sciences (NIGMS) of the National Institutes of Health (5T32 GM008382).
Conflict of interest statement. None declared.
References
- 1. Bohlmann J., Keeling C.I. (2008) Terpenoid biomaterials. Plant J., 54, 656–669. [DOI] [PubMed] [Google Scholar]
- 2. Leavell M.D., McPhee D.J., Paddon C.J. (2016) Developing fermentative terpenoid production for commercial usage. Curr. Opin. Biotechnol., 37, 114–119. [DOI] [PubMed] [Google Scholar]
- 3. George K.W., Alonso-Gutierrez J., Keasling J.D., Lee T.S. (2015) Isoprenoid drugs, biofuels, and chemicals—artemisinin, farnesene, and beyond. Adv. Biochem. Eng. Biotechnol., 148, 355–389. [DOI] [PubMed] [Google Scholar]
- 4. Li Y., Pfeifer B.A. (2014) Heterologous production of plant-derived isoprenoid products in microbes and the application of metabolic engineering and synthetic biology. Curr. Opin. Plant Biol., 19, 8–13. [DOI] [PubMed] [Google Scholar]
- 5. Paddon C.J., Westfall P.J., Pitera D.J., Benjamin K., Fisher K., McPhee D., Leavell M.D., Tai A., Main A., Eng D., et al. (2013) High-level semi-synthetic production of the potent antimalarial artemisinin. Nature, 496, 528–532. [DOI] [PubMed] [Google Scholar]
- 6. Nielsen J., Fussenegger M., Keasling J., Lee S.Y., Liao J.C., Prather K., Palsson B. (2014) Engineering synergy in biotechnology. Nat. Chem. Biol., 10, 319–322. [DOI] [PubMed] [Google Scholar]
- 7. Keasling J.D. (2012) Synthetic biology and the development of tools for metabolic engineering. Metab. Eng., 14, 189–195. [DOI] [PubMed] [Google Scholar]
- 8. Lee J.W., Na D., Park J.M., Lee J., Choi S., Lee S.Y. (2012) Systems metabolic engineering of microorganisms for natural and non-natural chemicals. Nat. Chem. Biol., 8, 536–546. [DOI] [PubMed] [Google Scholar]
- 9. Nielsen J., Keasling J.D. (2016) Engineering cellular metabolism. Cell, 164, 1185–1197. [DOI] [PubMed] [Google Scholar]
- 10. Blazeck J., Liu L., Redden H., Alper H. (2011) Tuning gene expression in Yarrowia lipolytica by a hybrid promoter approach. Appl. Environ. Microbiol., 77, 7905–7914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ajikumar P.K., Xiao W.-H., Tyo K.E., Wang Y., Simeon F., Leonard E., Mucha O., Phon T.H., Pfeifer B., Stephanopoulos G. (2010) Isoprenoid pathway optimization for Taxol precursor overproduction in Escherichia coli. Science, 330, 70–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Du J., Yuan Y., Si T., Lian J., Zhao H. (2012) Customized optimization of metabolic pathways by combinatorial transcriptional engineering. Nucleic Acids Res., 40, e142–e142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Biggs B.W., De Paepe B., Santos C.N.S., De Mey M., Ajikumar P.K. (2014) Multivariate modular metabolic engineering for pathway and strain optimization. Curr. Opin. Biotechnol., 29, 156–162. [DOI] [PubMed] [Google Scholar]
- 14. Alonso-Gutierrez J., Kim E.-M., Batth T.S., Cho N., Hu Q., Chan L.J.G., Petzold C.J., Hillson N.J., Adams P.D., Keasling J.D., et al. (2015) Principal component analysis of proteomics (PCAP) as a tool to direct metabolic engineering. Metab. Eng., 28, 123–133. [DOI] [PubMed] [Google Scholar]
- 15. Smanski M.J., Bhatia S., Zhao D., Park YJin., B A Woodruff L., Giannoukos G., Ciulla D., Busby M., Calderon J., Nicol R., et al. (2014) Functional optimization of gene clusters by combinatorial design and assembly. Nat. Biotechnol., 32, 1241–1249. [DOI] [PubMed] [Google Scholar]
- 16. Dudley Q.M., Karim A.S., Jewett M.C. (2015) Cell‐free metabolic engineering: biomanufacturing beyond the cell. Biotechnol. J., 10, 69–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hodgman C.E., Jewett M.C. (2012) Cell-free synthetic biology: thinking outside the cell. Metab. Eng., 14, 261–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Guterl J.K., Sieber V. (2013) Biosynthesis “debugged”: novel bioproduction strategies. Eng. Life Sci., 13, 4–18. [Google Scholar]
- 19. Rollin J.A., Tam T.K., Zhang Y.-H.P. (2013) New biotechnology paradigm: cell-free biosystems for biomanufacturing. Green Chem., 15, 1708–1719. [Google Scholar]
- 20. Swartz J.R. (2012) Transforming biochemical engineering with cell‐free biology. AIChE J., 58, 5–13. [Google Scholar]
- 21. Morgado G., Gerngross D., Roberts T.M., Panke S. (2016) Synthetic biology for cell-free biosynthesis: fundamentals of designing novel in vitro multi-enzyme reaction networks. In: H Zhao, A-P Zeng (eds). Synthetic Biology – Metabolic Engineering. Springer, Cham, pp. 117–146. [DOI] [PubMed] [Google Scholar]
- 22. Karim A.S., Dudley Q.M., Jewett M.C. (2017) Cell‐free synthetic systems for metabolic engineering and biosynthetic pathway prototyping. In: C Wittmann, JC Liao (eds). Industrial Biotechnology: Microorganisms, Vol. 1 Weinheim, Germany: Wiley-VCH, pp. 125–148. [Google Scholar]
- 23. Santander P.J., Roessner C.A., Stolowich N.J., Holderman M.T., Scott A.I. (1997) How corrinoids are synthesized without oxygen: nature’s first pathway to vitamin B12. Chem. Biol., 4, 659–666. [DOI] [PubMed] [Google Scholar]
- 24. Korman T.P., Sahachartsiri B., Li D., Vinokur J.M., Eisenberg D., Bowie J.U. (2014) A synthetic biochemistry system for the in vitro production of isoprene from glycolysis intermediates. Protein Sci., 23, 576–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhu F., Zhong X., Hu M., Lu L., Deng Z., Liu T. (2014) In vitro reconstitution of mevalonate pathway and targeted engineering of farnesene overproduction in Escherichia coli. Biotechnol. Bioeng., 111, 1396–1405. [DOI] [PubMed] [Google Scholar]
- 26. Chen X., Zhang C., Zou R., Zhou K., Stephanopoulos G., Too H.P. (2013) Statistical experimental design guided optimization of a one-pot biphasic multienzyme total synthesis of amorpha-4,11-diene. PLoS One, 8, e79650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rodriguez S.B., Leyh T.S. (2014) An enzymatic platform for the synthesis of isoprenoid precursors. PLoS One, 9, e105594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Korman T.P., Opgenorth P.H., Bowie J.U. (2017) A synthetic biochemistry platform for cell free production of monoterpenes from glucose. Nat. Commun., 8, 15526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Toogood H.S., Cheallaigh A.N., Tait S., Mansell D.J., Jervis A., Lygidakis A., Humphreys L., Takano E., Gardiner J.M., Scrutton N.S. (2015) Enzymatic menthol production: one-pot approach using engineered Escherichia coli. ACS Synth. Biol., 4, 1112–1123. [DOI] [PubMed] [Google Scholar]
- 30. Carlson E.D., Gan R., Hodgman C.E., Jewett M.C. (2012) Cell-free protein synthesis: applications come of age. Biotechnol. Adv., 30, 1185–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kay J.E., Jewett M.C. (2015) Lysate of engineered Escherichia coli supports high-level conversion of glucose to 2,3-butanediol. Metab. Eng., 32, 133–142. [DOI] [PubMed] [Google Scholar]
- 32. Karim A.S., Jewett M.C. (2016) A cell-free framework for rapid biosynthetic pathway prototyping and enzyme discovery. Metab. Eng., 36, 116–126. [DOI] [PubMed] [Google Scholar]
- 33. Bujara M., Schümperli M., Billerbeck S., Heinemann M., Panke S. (2010) Exploiting cell‐free systems: implementation and debugging of a system of biotransformations. Biotechnol. Bioeng., 106, 376–389. [DOI] [PubMed] [Google Scholar]
- 34. Goering A.W., Li J., McClure R.A., Thomson R.J., Jewett M.C., Kelleher N.L. (2017) In vitro reconstruction of nonribosomal peptide biosynthesis directly from DNA using cell-free protein synthesis. ACS Synth. Biol., 6, 39–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jongedijk E., Cankar K., Buchhaupt M., Schrader J., Bouwmeester H., Beekwilder J. (2016) Biotechnological production of limonene in microorganisms. Appl. Microbiol. Biotechnol., 100, 2927–2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Willrodt C., David C., Cornelissen S., Bühler B., Julsing M.K., Schmid A. (2014) Engineering the productivity of recombinant Escherichia coli for limonene formation from glycerol in minimal media. Biotechnol. J., 9, 1000–1012. [DOI] [PubMed] [Google Scholar]
- 37. Alonso-Gutierrez J., Chan R., Batth T.S., Adams P.D., Keasling J.D., Petzold C.J., Lee T.S. (2013) Metabolic engineering of Escherichia coli for limonene and perillyl alcohol production. Metab. Eng., 19, 33–41. [DOI] [PubMed] [Google Scholar]
- 38. Jongedijk E., Cankar K., Ranzijn J., Krol S., Bouwmeester H., Beekwilder J. (2015) Capturing of the monoterpene olefin limonene produced in Saccharomyces cerevisiae. Yeast, 32, 159–171. [DOI] [PubMed] [Google Scholar]
- 39. Cao X., Lv Y.-B., Chen J., Imanaka T., Wei L.-J., Hua Q. (2016) Metabolic engineering of oleaginous yeast Yarrowia lipolytica for limonene overproduction. Biotechnol. Biofuels, 9, 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Davies F.K., Work V.H., Beliaev A.S., Posewitz M.C. (2014) Engineering limonene and bisabolene production in wild type and a glycogen-deficient mutant of Synechococcus sp. PCC 7002. Front. Bioeng. Biotechnol., 2, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wang X., Liu W., Xin C., Zheng Y., Cheng Y., Sun S., Li R., Zhu X.-G., Dai S.Y., Rentzepis P.M., et al. (2016) Enhanced limonene production in cyanobacteria reveals photosynthesis limitations. Proc. Natl. Acad. Sci. USA, 113, 14225–14230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. George K.W., Thompson M.G., Kim J., Baidoo E.E.K., Wang G., Benites V.T., Petzold C.J., Chan L.J.G., Yilmaz S., Turhanen P., et al. (2018) Integrated analysis of isopentenyl pyrophosphate (IPP) toxicity in isoprenoid-producing Escherichia coli. Metab. Eng., 47, 60–72. [DOI] [PubMed] [Google Scholar]
- 43. Dudley Q.M., Anderson K.C., Jewett M.C. (2016) Cell-free mixing of Escherichia coli crude extracts to prototype and rationally engineer high-titer mevalonate synthesis. ACS Synth. Biol., 5, 1578–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Casini A., Chang F.-Y., Eluere R., King A.M., Young E.M., Dudley Q.M., Karim A., Pratt K., Bristol C., Forget A., et al. (2018) A pressure test to make 10 molecules in 90 days: external evaluation of methods to engineer biology. J. Am. Chem. Soc., 140, 4302–4316. [DOI] [PubMed] [Google Scholar]
- 45. Wu Y.Y., Culler S., Khandurina J., Van Dien S., Murray R.M. (2015) Prototyping 1,4-butanediol (BDO) biosynthesis pathway in a cell-free transcription-translation (TX-TL) system. bioRxiv 017814. doi: 10.1101/017814. [Google Scholar]
- 46. Bogorad I.W., Lin T.-S., Liao J.C. (2013) Synthetic non-oxidative glycolysis enables complete carbon conservation. Nature, 502, 693–697. [DOI] [PubMed] [Google Scholar]
- 47. Siegal-Gaskins D., Tuza Z.A., Kim J., Noireaux V., Murray R.M. (2014) Gene circuit performance characterization and resource usage in a cell-free “breadboard”. ACS Synth. Biol., 3, 416–425. [DOI] [PubMed] [Google Scholar]
- 48. Takahashi M.K., Chappell J., Hayes C.A., Sun Z.Z., Kim J., Singhal V., Spring K.J., Al-Khabouri S., Fall C.P., Noireaux V., et al. , (2015) Rapidly characterizing the fast dynamics of RNA genetic circuitry with cell-free transcription–translation (TX-TL) systems. ACS Synth. Biol., 4, 503–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Karig D.K., Iyer S., Simpson M.L., Doktycz M.J. (2012) Expression optimization and synthetic gene networks in cell-free systems. Nucleic Acids Res., 40, 3763–3774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhang Y., Meng Q., Ma H., Liu Y., Cao G., Zhang X., Zheng P., Sun J., Zhang D., Jiang W., et al. (2015) Determination of key enzymes for threonine synthesis through in vitro metabolic pathway analysis. Microb. Cell Fact., 14, 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tan G.-Y., Zhu F., Deng Z., Liu T. (2016) In vitro reconstitution guide for targeted synthetic metabolism of chemicals, nutraceuticals and drug precursors. Synth. Syst. Biotechnol., 1, 25–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kelwick R., Ricci L., Chee S.M., Bell D., Webb A.J., Freemont P.S. (2018) Cell-free prototyping strategies for enhancing the sustainable production of polyhydroxyalkanoates bioplastics. Synth. Biol., 3, ysy016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Anderson J.C., Dueber J.E., Leguia M., Wu G.C., Goler J.A., Arkin A.P., Keasling J.D. (2010) BglBricks: a flexible standard for biological part assembly. J. Biol. Eng., 4, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Salis H.M., Mirsky E.A., Voigt C.A. (2009) Automated design of synthetic ribosome binding sites to control protein expression. Nat. Biotechnol., 27, 946–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Swartz J.R., Jewett M.C., Woodrow K.A. (2004) Cell-free protein synthesis with prokaryotic combined transcription-translation. In: P Balbás, A Lorence (eds). Recombinant Gene Expression. Methods in Molecular Biology, vol. 267. Totowa, NJ: Humana Press, pp. 169–182. [DOI] [PubMed] [Google Scholar]
- 56. Jewett M.C., Swartz J.R. (2004) Mimicking the Escherichia coli cytoplasmic environment activates long‐lived and efficient cell‐free protein synthesis. Biotechnol. Bioeng., 86, 19–26. [DOI] [PubMed] [Google Scholar]
- 57. Kwon Y.-C., Jewett M.C. (2015) High-throughput preparation methods of crude extract for robust cell-free protein synthesis. Sci. Rep., 5, Article ID: 8663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Jewett M.C., Calhoun K.A., Voloshin A., Wuu J.J., Swartz J.R. (2008) An integrated cell‐free metabolic platform for protein production and synthetic biology. Mol. Syst. Biol., 4, 220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Calhoun K.A., Swartz J.R. (2005) Energizing cell‐free protein synthesis with glucose metabolism. Biotechnol. Bioeng., 90, 606–613. [DOI] [PubMed] [Google Scholar]
- 60. Cheng Q., Xiang L., Izumikawa M., Meluzzi D., Moore B.S. (2007) Enzymatic total synthesis of enterocin polyketides. Nat. Chem. Biol., 3, 557. [DOI] [PubMed] [Google Scholar]
- 61. Lowry B., Robbins T., Weng C.-H., O’Brien R.V., Cane D.E., Khosla C. (2013) In vitro reconstitution and analysis of the 6-deoxyerythronolide B synthase. J. Am. Chem. Soc., 135, 16809–16812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Lowry B., Walsh C.T., Khosla C. (2015) In vitro reconstitution of metabolic pathways: insights into nature’s chemical logic. Synlett, 26, 1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Li J., Zhang L., Liu W. (2018) Cell-free synthetic biology for in vitro biosynthesis of pharmaceutical natural products. Synth. Syst. Biotechnol., 3, 83–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Fujisaki S., Nishino T., Katsuki H. (1986) Isoprenoid synthesis in Escherichia coli. Separation and partial purification of four enzymes involved in the synthesis. J. Biochem., 99, 1327–1337. [DOI] [PubMed] [Google Scholar]
- 65. Reiling K.K., Yoshikuni Y., Martin V.J., Newman J., Bohlmann J., Keasling J.D. (2004) Mono and diterpene production in Escherichia coli. Biotechnol. Bioeng., 87, 200–212. [DOI] [PubMed] [Google Scholar]
- 66. Lee P.C., Petri R., Mijts B.N., Watts K.T., Schmidt-Dannert C. (2005) Directed evolution of Escherichia coli farnesyl diphosphate synthase (IspA) reveals novel structural determinants of chain length specificity. Metab. Eng., 7, 18–26. [DOI] [PubMed] [Google Scholar]
- 67. Kim S.K., Han G.H., Seong W., Kim H., Kim S.-W., Lee D.-H., Lee S.-G. (2016) CRISPR interference-guided balancing of a biosynthetic mevalonate pathway increases terpenoid production. Metab. Eng., 38, 228–240. [DOI] [PubMed] [Google Scholar]
- 68. Li S., Jendresen C.B., Grünberger A., Ronda C., Jensen S.I., Noack S., Nielsen A.T. (2016) Enhanced protein and biochemical production using CRISPRi-based growth switches. Metab. Eng., 38, 274–284. [DOI] [PubMed] [Google Scholar]
- 69. Karim A.S., Heggestad J.T., Crowe S.A., Jewett M.C. (2018) Controlling cell-free metabolism through physiochemical perturbations. Metab. Eng., 45, 86–94. [DOI] [PubMed] [Google Scholar]
- 70. Kim D.M., Swartz J.R. (1999) Prolonging cell‐free protein synthesis with a novel ATP regeneration system. Biotechnol. Bioeng., 66, 180–188. [PubMed] [Google Scholar]
- 71. Caschera F., Karim A.S., Gazzola G., d’Aquino A.E., Packard N.H., Jewett M.C. (2018) High-throughput optimization cycle of a cell-free ribosome assembly and protein synthesis system. ACS Synth. Biol., 7, 2841–2853. [DOI] [PubMed] [Google Scholar]
- 72. Chubukov V., Mingardon F., Schackwitz W., Baidoo E.E., Alonso-Gutierrez J., Hu Q., Lee T.S., Keasling J.D., Mukhopadhyay A. (2015) Acute limonene toxicity in Escherichia coli is caused by limonene hydroperoxide and alleviated by a point mutation in alkyl hydroperoxidase AhpC. Appl. Environ. Microbiol., 81, 4690–4696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Des Soye B.J., Davidson S.R., Weinstock M.T., Gibson D.G., Jewett M.C. (2018) Establishing a high-yielding cell-free protein synthesis platform derived from Vibrio natriegens. ACS Synth. Biol., 7, 2245–2255. [DOI] [PubMed] [Google Scholar]
- 74. Opgenorth P.H., Korman T.P., Bowie J.U. (2014) A synthetic biochemistry molecular purge valve module that maintains redox balance. Nat. Commun, 5, 4113. [DOI] [PubMed] [Google Scholar]
- 75. Yi T., Lim H.J., Lee S.J., Lee K.-H., Kim D.-M. (2018) Synthesis of (R, R)-2, 3-butanediol from starch in a hybrid cell-free reaction system. J. Ind. Eng. Chem., 67, 231–235. [Google Scholar]
- 76. Bujara M., Panke S. (2012) In silico assessment of cell‐free systems. Biotechnol. Bioeng., 109, 2620–2629. [DOI] [PubMed] [Google Scholar]
- 77. Primak Y.A., Du M., Miller M.C., Wells D.H., Nielsen A.T., Weyler W., Beck Z.Q. (2011) Characterization of a feedback-resistant mevalonate kinase from the archaeon Methanosarcina mazei. Appl. Environ. Microbiol., 77, 7772–7778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Dorsey J.K., Porter J.W. (1968) The inhibition of mevalonic kinase by geranyl and farnesyl pyrophosphates. J. Biol. Chem., 243, 4667–4670. [PubMed] [Google Scholar]
- 79. Garcia D.E. (2013) The in vitro characterization of heterologously expressed enzymes to inform in vivo biofuel production optimization. Thesis. University of California-Berkeley. [Google Scholar]
- 80. Weaver L.J., Sousa M.M., Wang G., Baidoo E., Petzold C.J., Keasling J.D. (2015) A kinetic‐based approach to understanding heterologous mevalonate pathway function in E. coli. Biotechnol . Bioeng., 112, 111–119. [DOI] [PubMed] [Google Scholar]
- 81. Leferink N.G., Jervis A.J., Zebec Z., Toogood H.S., Hay S., Takano E., Scrutton N.S. (2016) A ‘Plug and Play’ platform for the production of diverse monoterpene hydrocarbon scaffolds in Escherichia coli. Chem. Select, 1, 1893–1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Mendez-Perez D., Alonso-Gutierrez J., Hu Q., Molinas M., Baidoo E.E.K., Wang G., Chan L.J.G., Adams P.D., Petzold C.J., Keasling J.D., et al. (2017) Production of jet fuel precursor monoterpenoids from engineered Escherichia coli. Biotechnol. Bioeng., 114, 1703–1712. [DOI] [PubMed] [Google Scholar]
- 83. Du F.-L., Yu H.-L., Xu J.-H., Li C.-X. (2014) Enhanced limonene production by optimizing the expression of limonene biosynthesis and MEP pathway genes in E. coli. Biores. Bioproc., 1, 10. [Google Scholar]
- 84. Tashiro M., Kiyota H., Kawai-Noma S., Saito K., Ikeuchi M., IIjima Y., Umeno D. (2016) Bacterial production of pinene by a laboratory-evolved pinene-synthase. ACS Synth. Biol., 5, 1011–1020. [DOI] [PubMed] [Google Scholar]
- 85. Chen F., Li W., Jiang L., Pu X., Yang Y., Zhang G., Luo Y. (2016) Functional characterization of a geraniol synthase-encoding gene from Camptotheca acuminata and its application in production of geraniol in Escherichia coli. J. Ind. Microbiol. Biotechnol., 43, 1281–1292. [DOI] [PubMed] [Google Scholar]
- 86. Villarreal F., Contreras-Llano L.E., Chavez M., Ding Y., Fan J., Pan T., Tan C. (2018) Synthetic microbial consortia enable rapid assembly of pure translation machinery. Nat. Chem. Biol., 14, 29. [DOI] [PubMed] [Google Scholar]
- 87. Dueber J.E., Wu G.C., Malmirchegini G.R., Moon T.S., Petzold C.J., Ullal A.V., Prather K.L., Keasling J.D. (2009) Synthetic protein scaffolds provide modular control over metabolic flux. Nat. Biotechnol., 27, 753–759. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.