Abstract
Crimean-Congo haemorrhagic fever (CCHF) is a widespread tickborne disease that circulates in wild and domestic animal hosts, and causes severe and often fatal haemorrhagic fever in infected humans. Due to the lack of treatment options or vaccines, and a high fatality rate, CCHF virus (CCHFV) is considered a high-priority pathogen according to the WHO R&D Blueprint. Several commercial reverse transcriptase PCR (RT-PCR) and serological diagnostic assays for CCHFV are already available, including febrile agent panels to distinguish CCHFV from other viral haemorrhagic fever agents; however, the majority of international laboratories use inhouse assays. As CCHFV has numerous amplifying animal hosts, a cross-sectoral ‘One Health’ approach to outbreak prevention is recommended to enhance notifications and enable early warning for genetic and epidemiological shifts in the human, animal and tick populations. However, a lack of guidance for surveillance in animals, harmonisation of case identification and validated serodiagnostic kits for animal testing hinders efforts to strengthen surveillance systems. Additionally, as RT-PCR tests tend to be lineage-specific for regional circulating strains, there is a need for pan-lineage sensitive diagnostics. Adaptation of existing tests to point-of-care molecular diagnostic platforms that can be implemented in clinic or field-based settings would be of value given the potential for CCHFV outbreaks in remote or low-resource areas. Finally, improved access to clinical specimens for validation of diagnostics would help to accelerate development of new tests. These gaps should be addressed by updated target product profiles for CCHFV diagnostics.
Keywords: crimean-congo haemorrhagic fever, CCHF, in vitro diagnostics, outbreak
Summary box.
Diagnostic tests for Crimean-Congo haemorrhagic fever virus (CCHFV), a WHO R&D Blueprint priority pathogen, include commercial reverse transcriptase PCR and serological diagnostic assays and multiplex panels to distinguish from other viral haemorrhagic fever agents.
Despite the extensive range of tests available, diagnostic gaps remain, including a need for improved surveillance for early detection, a lack of point-of-care testing options and issues with limited availability of clinical specimens for test validation.
Refinement of target product profiles for CCHFV diagnostics to include these needs will help to enhance surveillance, prevention and management of CCHFV in both human and animal hosts.
Introduction
Crimean-Congo haemorrhagic fever (CCHF) is one of the high-priority pathogens identified on the WHO R&D Blueprint because of its high case fatality rate, potential for nosocomial outbreaks and difficulties in treatment and prevention.1–3 CCHF is widespread, now found in Europe, Asia, Africa, the Middle East and the Indian subcontinent, with currently no vaccine available for widespread human or animal use. This landscape analysis is intended to provide a view to the current state of CCHF diagnostics, with particular emphasis on human diagnostics for screening, diagnosis and surveillance.
History and epidemiology of CCHF
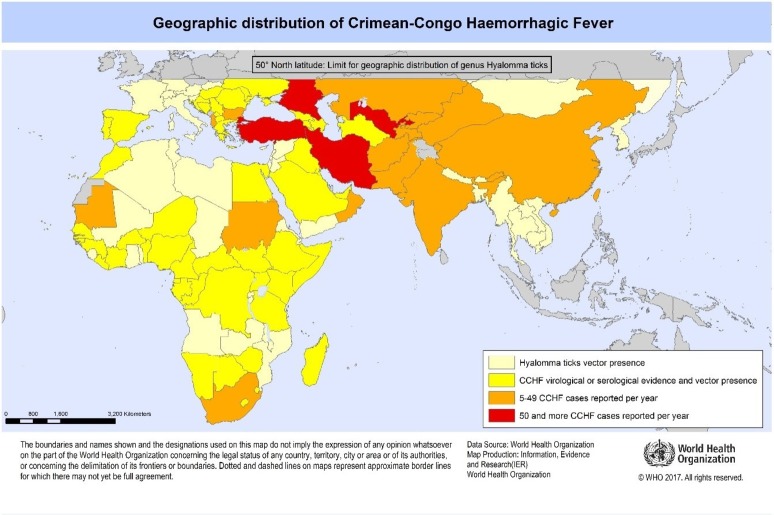
CCHF virus (CCHFV) is an orthonairovirus of the family Nairoviridae causing severe and often fatal haemorrhagic fever in humans. The disease was first described in the Crimea in 1944 and became known as ‘Crimean haemorrhagic fever’, but the virus was first isolated in Congo in 1956 and was named ‘Congo virus’; the two names converged as CCHF in 1969.2 4 5 CCHF is considered an ‘emerging’ disease across the globe, with many countries reporting new infections in recent decades.6–8 CCHF is now found in Europe, the Mediterranean, China and Central Asia, Africa, the Middle East and India (figure 1), and has been reported after long periods of absence.9 10 Cases are typically sporadic and seasonal, and occur in remote or agricultural regions.
Figure 1.
Geographical distribution of Crimean-Congo haemorrhagic fever (CCHF) (http://www.who.int/emergencies/diseases/crimean-congo-haemorrhagic-fever/en/).
CCHF reservoir and mode of transmission
Ixodid (hard) ticks are a reservoir and a vector for CCHFV, now commonly found in dry or semiarid environments across Africa, Europe, the Middle East and Asia.7 11 12 Numerous wild and domestic animals, including cattle, goats, sheep, camels and hares, serve as amplifying hosts after being bitten by infected ticks, but animal infections are difficult to detect as the tick–animal–tick cycle is asymptomatic.13 CCHFV persists throughout the tick life-cycle, enabling reservoirs of infection over long periods without vertebrate hosts.
Transmission to humans occurs most frequently among agricultural workers following the bite of an infected tick, and among slaughterhouse or veterinary workers exposed to the blood and tissue of infected livestock. Human-to-human transmission is infrequent, but has arisen from contact with infected patients or contaminated medical equipment.14–17
Clinical presentation
CCHFV infection typically has four distinct phases—incubation, prehaemorrhagic, haemorrhagic and convalescence. The incubation period is in the range of 1–5 days following a tick bite, and 5–7 days following contact with infected blood or tissues.4 11 16 The prehaemorrhagic phase is characterised by the sudden onset of a wide range of non-specific symptoms that mimic other diseases and lasts for 4–5 days. CCHFV infections may be subclinical or asymptomatic in some people; for example, high levels of seroprevalence have been detected in specific regions of Turkey and Greece.18 19
The haemorrhagic phase is generally 2 weeks in duration with rapid progressive haemorrhage. Severely ill patients may experience rapid deterioration, shock and multiorgan failure.20 In documented outbreaks, fatality rates in hospitalised patients have ranged as high as 50%, with an average mortality of 30% occurring in the second week of illness.1 2 For survivors, recovery generally begins 10–20 days after the onset of illness; however, full recovery may take up to a year.11 16 The long-term effects of CCHFV infection have not been studied well enough in survivors to determine whether or not specific complications exist.
Molecular epidemiology
CCHFV displays a high degree of sequence diversity, with divergence of 20%, 31% and 22% for the S, M and L segments among isolates in GenBank. Based on an analysis of the viral S segment, six or seven viral lineages/clades have been identified,6 11 16 21 22 suggesting a lengthy history of geographical dispersion of the virus from Africa to Europe, the Middle East and then Asia. Genome diversity can be greater in the tick than in the mammalian host.23–25 The extensive sequence diversity of CCHFV is likely due to genetic reassortment, enhanced by circulation and adaptation of strains into new geographical regions.22 26–29
Therapeutic efforts
Treatment for confirmed cases of CCHF is general supportive care with management of symptoms.2 There is no approved antiviral treatment. Although the antiviral drug ribavirin is widely used based on its in vitro activity, clinical evidence is inconclusive.30–36 Postexposure prophylaxis using ribavirin is controversial but recommended as a safe option for healthcare providers with high-risk/direct exposure to CCHF-infected patient fluids or tissue.15 17 37 38
Development of new therapies has been slow in part due to the requirement for a high-containment laboratory and the lack of a suitable animal model.6 39 40 Favipiravir is a broad-spectrum RNA inhibitor that has shown promising results.41 Tetracyclines, cationic amphiphilic drugs and ubiquitin variants are under investigation as potential antiviral therapies42–44; however, none are in clinical-stage development.
Vaccine efforts
There is currently no safe and effective CCHF vaccine widely available for human use.1 2 A vaccine developed in 1970 was based on CCHFV cultivated in suckling mouse brain. Vaccinated individuals developed anti-CCHFV antibodies, but the neutralising activity was low.45 The vaccine is licensed in Bulgaria and used on a small scale in eastern Europe, but it is unlikely to gain international regulatory approval due to concerns with efficacy and allergic responses.46 Several studies indicate that neutralising antibodies alone are insufficient protection against CCHFV challenge,39 47 48 and it is unclear whether conserved neutralising epitopes are present for all CCHFV isolates.49
Newly developed mouse models that mimic human CCHF disease are useful to study vaccine candidates. A recent study vaccinated mice with a DNA vaccine (encoding a ubiquitin-linked version of CCHFV Gc, Gn and NP) and demonstrated 100% efficient preventive immunity against lethal CCHFV challenge.39 Using this mouse model, vaccination with a cell culture-based vaccine from the CCHFV Turkey-Kelkit06 strain provided partial protection (80%) against a high-dose challenge.47 A recombinant vaccine expressing CCHFV glycoproteins (GP, M segment) using a modified vaccinia virus Ankara (MVA) protected 100% of recipient animals up to 14 days postchallenge in a lethal challenge model adapted to represent infection via a tick bite.50 However, a similar study using MVA to express the CCHFV nucleoprotein (NP, S segment) generated an immune response but failed to protect animals from lethal disease.51
Veterinary vaccines may be an alternative or complementary approach to human vaccines. Livestock vaccines against CCHF could play an important role in preventing human infection by controlling exposure during animal slaughter, as well as interrupting the vector cycle during tick feeding.8 46 Infection control of CCHFV in wildlife populations will remain a challenge, however. Currently there are no vaccines available for use in animals,1 2 although the MVA GP vaccine is currently being evaluated in sheep.52
CCHF diagnostics
Laboratory tests used to diagnose CCHF include reverse transcriptase (RT)-PCR, immunofluorescence assay (IFA), antibody (IgG, IgM) and antigen-capture ELISA, and virus isolation. Patients suspected of CCHF are primarily diagnosed by RT-PCR as these assays provide the highest detection sensitivity to active infection at the earliest time point. Lineage detection may be challenged by the high diversity and in situ evolution of CCHFV, particularly for RT-PCR assays which rely on a conserved genomic sequence for detection.53–57 Serological detection is less impacted by minor genomic variations. Given CCHFV strain variations, it is recommended that nucleic acid amplification tests (NAAT, eg, RT-PCR) be used in combination with immunological assays for highest detection sensitivity;8 58 59 however many low-resource settings may not have the capacity for PCR testing, especially at the early stages of an outbreak. Virus isolation is rarely used as a diagnostic tool because of the stringent biosafety containment level (BSL-4) required.
NAAT typically requires the highest laboratory infrastructure, including biosafety hoods and a clean room or PCR workstation, while most serological tests (ELISA, IFA) can be run on the benchtop in a more modest laboratory environment (table 1).60 61 Ideally, point-of-care (POC) NAAT tests are fully automated, with samples delivered to an integrated cartridge that contains all the reagents necessary for sample processing and analysis; this process can be performed without a biosafety hood, depending on the sample preparation requirements (here defined as BSL-2 for human disease).60 61 Once the cartridge is inserted into the instrument, no further manual steps are required. Rapid diagnostic tests (RDTs) are typically designed for field or home use. Turnaround time for each test is specified by the manufacturer; turnaround time per result can include additional time (days to weeks) for sample transport and processing at the reference lab.62–64
Table 1.
Diagnostics infrastructure comparison
| Test type | Infrastructure requirements | Training requirements | Turnaround time | Inhouse or prototype | Commercial source | Target population |
| Virus isolation, neutralisation | High (BSL-4) (reference laboratory) |
High (advanced lab technician) | 3–7 days | Several | – | Human, animal |
| NAAT reference (including multiplex) | High (BSL-3/4) (regional lab, reference laboratory) |
High to moderate (advanced lab technician) |
2.5 hours 1–2 hours prep |
>10 | >5 | Human, animal, ticks |
| NAAT POC | Moderate/BSL-2 (district hospital) |
Moderate (laboratory technician) |
1–2 hours | 1 | – | Human, ticks, culture |
| ELISA, IFA | High to moderate (regional lab, district hospital) |
Moderate (laboratory technician) |
3–4 hours | >10 | 6 | Human, animal, culture |
| RDTs | Low (clinic, health centre, field settings) |
Low (nurse, healthcare worker) |
<30 min | – | – | – |
BSL, biosafety containment level; IFA, immunofluorescence assay; NAAT, nucleic acid amplification test; POC, point of care; RDT, rapid diagnostic test.
Several commercial assays for PCR and serology are available (online supplementary table S1 and S2), although the majority of international laboratories use inhouse assays, likely due to an investment in tests developed from regional CCHV strains.7 8 53 59 It has also been suggested that commercial tests may be too expensive, difficult to order or not available to international customers.8 65 The majority of the inhouse assays have a publication history, several with published data on diagnostic performance or external quality assessment (tables 2 and 3).
Table 2.
Inhouse NAAT tests for CCHF: references, reviews, EQA
| Test type | Reference | Review/EQA | Labs using the method (n)65 | CCHFV target54 |
| qRT-PCR | 73 | 54 65 | 3 | 24 genomic targets |
| qRT-PCR | 113 | 54 | – | 19 strains worldwide |
| qRT-PCR | 76 78 | 53 54 59 65 | 2 | Kosovo Hoti and Drosdov strains |
| qRT-PCR | 114, updated 57 | 54 | – | 18 strains worldwide (updated 26) |
| qRT-PCR | 115 116 | 65 | 3 | – |
| qRT-PCR | 117 118 | 53 54 59 65 | 12 | 17 strains worldwide |
| qRT-PCR | 119 | 53 59 65 | 2 | – |
| qRT-PCR | 120 | 54 | – | 19 Southern African strains |
| qRT-PCR | 121 | 54 | – | All known worldwide strains, including the AP92 strain |
| qRT-PCR | 122 | – | – | – |
| Nested RT-PCR | 123 | 53 59 65 | 2 | – |
| Nested RT-PCR | 74 | 53 65 | 2 | – |
| Nested RT-PCR | 22 | 65 | 1 | – |
| Nested RT-PCR | 124 | – | – | – |
| Conventional RT-PCR | 72 125 | 54 65 | 2 | 7 geographically diverse strains |
| Isothermal RPA | 82 | – | – | 7 geographically diverse strains |
| PCR multiplex | 64 92 | – | 2 | – |
| Low density array | 118 | 53 54 | – | Strains worldwide |
| High density array | 90 | 54 | – | Nigerian strain |
| Multiplex RT-PCR/NGS | 84 | – | – | 46 VHF species |
CCHF, Crimean-Congo haemorrhagic fever; CCHFV, CCHF virus; EQA, external quality assessment; NAAT, nucleic acid amplification test; NGS, next-generation sequencing; qRT-PCR, quantitative real-time RT-PCR; RPA, recombinase polymerase amplification; VHF, viral haemorrhagic fever.
Table 3.
Inhouse serological tests for CCHF: references, reviews, EQA
| Test type | Reference source | Review/EQA |
| ELISA (Ag, IgG, IgM) | 87 126 | 8 |
| ELISA (IgG, Ag) | 104 127 | 53/8 |
| ELISA (IgG) | 128 | 8 |
| ELISA (IgG, IgM) | 129 | 8 |
| ELISA (IgG, IgM) | 130 | 53/8 |
| ELISA (IgG) | 131 | 53/16 |
| ELISA (IgG) | 132 | 8 16 |
| ELISA (IgM) | 133 | 8 |
| ELISA (IgG) | 134 | – |
| Competitive ELISA | 134 | |
| Double-Ab ELISA | 100 | – |
| Immune complex ELISA | 135 | – |
| Multiplex ELISA | 91 | (7 viral species) |
| IFA (IgG, IgM) | 87 | 8 |
| IFA (IgG) | 136 | 53/8 |
| IFA (IgG) | 137 | 8 |
CCHF, Crimean-Congo haemorrhagic fever; EQA, external quality assessment; IFA, immunofluorescence assay.
bmjgh-2018-001114supp001.pdf (450.8KB, pdf)
Specimens and sampling
CCHFV is classified as BSL-4 in some countries, but if serum samples have been inactivated (eg, with virucides, gamma rays, formaldehyde, heat and so on), they can be manipulated in a basic biosafety environment.1 2 Otherwise, CCHF patient samples present an extreme biohazard risk and should only be conducted under maximum biological containment conditions.8 11 15
CCHFV can be detected in the saliva and urine from prehaemorrhagic stage patients, consistent with other viral haemorrhagic fevers (VHF).66–69 CCHFV has also been detected in nasal, conjunctival, rectal and vaginal swabs of severe cases.70 A more thorough study of CCHF viral kinetics is needed to determine whether viral RNA peaks in serum during early-stage infection, then later in urine during the recovery stage as with other VHF.71 In particular, urine and saliva samples may be desirable for easy collection and handling for CCHF diagnosis. Culture of CCHFV from urine indicates the potential for prolonged viraemia up to 30 days, as well as potential transmission risk in recovering patients.34 37
Molecular diagnostics
RT-PCR-based techniques typically target the nucleoprotein gene region in the S segment, which is the more conserved region of the CCHFV genome across geographical isolates.72–75 CCHFV RNA peaks in the first week after symptom onset and can be detected for up to 3 weeks.58 72 75 76 Viral load, which varies considerably among patients with CCHF, can be an indicator of severity.73 77–80 For moderate cases, serum viral loads are initially 102–104 copies/mL, while severe cases have initial viral loads typically 104–107 copies/mL, with 108–1010 copies/mL predictive of fatal outcome.77 78 81
Quantitative real-time RT-PCR (qRT-PCR) has better performance over conventional RT-PCR or nested RT-PCR, with a lower contamination rate, higher sensitivity and specificity, and better time-effectiveness59 65; laboratories performing only nested or conventional RT-PCR have been advised to implement qRT-PCR to improve assay performance and viral load determination. An isothermal NAAT test has been developed based on recombinase polymerase amplification, enabling amplification at a single temperature in a more ‘crude’ sample which may prove more amenable as a field diagnostic or in low-resource laboratories.82 Next-generation sequencing (NGS) enables comprehensive genome analysis and has been used for CCHFV phylogeny83 84; however, this complex and expensive approach is not currently practical for diagnostic screening.
There are many inhouse laboratory tests, some with primers designed for regional circulating strains (table 2).53 59 85 86 Several commercial RT-PCR kits are available, typically with primers designed to target broad consensus sequences within the S segment, including several European Commission marked (CE) diagnostics and research use-only (RUO)-labelled products in stand-alone and multiplex test formats (online supplementary table S1).
Serological assays
Serological assays are sensitive to antigenic variation, but generally less impacted by genetic variation.53 Most assays target the CCHFV N protein, which induces an early, strong and long-lasting immune response in humans.86 Active CCHFV infection can be detected by IgM or a significant increase in IgG titre following the acute phase of infection 4–9 days after symptom onset; however, severe and fatal cases often do not mount a detectable antibody response.79 87 88 Detection of anti-CCHFV IgG can indicate current or resolved infection (often years after infection) and can be useful in surveillance epidemiological studies. For CCHFV, capture ELISA has been shown to be more sensitive than IFA or neutralisation assay.87–89 Virus neutralisation assays are less useful for diagnosis, since CCHFV elicits relatively low levels of neutralising antibodies, but can be useful for epidemiology and vaccine research. CCHFV neutralisation is generally performed using plaque reduction neutralisation, with 5–7 days for results.
As with RT-PCR, many of the CCHFV serological tests in use were developed as inhouse assays with limited validation (table 3).8 53 59 Several commercial ELISA (IgG, IgM) and IFA kits are available, although primarily marketed as RUO (online supplementary table S2).
bmjgh-2018-001114supp003.pdf (420.7KB, pdf)
Rapid diagnostic tests
RDTs can leverage the same antibody/antigen capture agents as an ELISA but in a lateral flow strip format, with minimal specimen processing (blood, plasma, swabs). This enables a faster time to result (10–30 min, however with a lower detection sensitivity than ELISA, due in part to reduced sample volume).90 RDTs are ideal screening tests, suitable for field testing and low infrastructure settings,87 although follow-up confirmatory testing is often required. RDTs have been used to effectively screen and triage suspected high-risk cases of diseases such as Ebola and dengue88 89; however, the literature shows no evidence of CCHF RDT development. The primary challenge is detection sensitivity, as the IgG/IgM serological response is typically detectable only 5 days postinfection, and often undetectable in severe and fatal infections.
Syndromic multiplex approach
CCHF screening tests need to be able to distinguish CCHF from other types of VHF, particularly in regions where VHF viruses may be endemic and maintained in the region in natural reservoirs. In some instances, a multiplex approach may be the better option for definitive identification, or at least to rule in or rule out more virulent pathogens.4 7 58
For analysis of circulating reservoirs in Sierra Leone, a bead-based immunoassay was used to detect IgG antibodies for multiple pathogens, which included Lassa, Ebola, Marburg and Rift Valley fever viruses, CCHFV, and pan-assays for flaviviruses and alphaviruses.91 In 675 human serum samples, 50% were positive for Lassa, 5% for Ebola, 11% for Marburg, 2% for Rift Valley fever, 2% for CCHF, 53% for flaviviruses and 56% for alphaviruses. A multiplex PCR approach developed as a universal array for simultaneous identification of Ebola, Marburg, CCHF, Lassa fever, Rift Valley fever, dengue and Yellow fever, as well as Variola and Vaccinia virus for smallpox, detected all viruses in 32 different isolates, with no cross-reactivity with other emerging viruses.64 Finally, a qRT-PCR-based card-based platform developed for 26 acute febrile illnesses,92 including 15 viruses, 8 bacteria and 3 protozoa, achieved an overall 88% sensitivity and 99% specificity compared with individual real-time RT-PCR assays.73
In addition, febrile agent panels (20 and 10 member panels including CCHF) are recently commercially available using bead-based and real-time TaqMan assays with a limit of detection of 10 copies/mL (online supplementary table S1).
Challenges for CCHF diagnostics
Surveillance
Surveillance programmes for humans, animals and ticks in endemic and bordering non-endemic areas can be used to monitor the spread of disease.8 93 As infected animals are usually asymptomatic, only active surveillance or human case detection will reveal CCHFV in circulation. Seroconversion in animals is a good indicator of CCHFV prevalence; when domestic animals in Turkey and Bulgaria were tested for CCHFV-specific IgG antibodies, the mean seroprevalence was 26% for Bulgaria and 57% for Turkey, with some provinces reporting seroprevalence of almost 90%.94 In both rural and urban settings, similar ‘random sampling’ surveillance programmes have been employed for ticks85 95–97 and other ruminants.98 99 However, routine reservoir/host monitoring is not broadly implemented, and surveillance is challenged by a lack of serodiagnostic tests suitable for large-scale animal testing,100 no clear guidance for standardised surveillance of CCHFV in the animal health sector, and the cost of routine implementation.6 For human surveillance, high prevalence endemic countries (Iran, Iraq, Pakistan and Turkey) report human cases annually through health surveillance systems, although not uniformly effective.101 Other countries (Afghanistan, Egypt, Oman, Saudi Arabia and United Arab Emirates) have occasional human cases reported; these and surrounding non-endemic countries would benefit from active surveillance systems for early identification of hot spots.102
Harmonisation of case definition
For CCHF surveillance, harmonisation of case identification is necessary to enhance notifications and estimate disease burden, as well as to enable early warning for genetic and epidemiological shifts in the human, animal and tick populations.6 59 95 103 National CCHF prevention and control programmes should be strengthened and supported by the respective Ministries of Health and international agencies.6 63 102 To assist these goals, a guideline development group for CCHF has been established by WHO to formulate recommendations, evaluate optimal implementation and develop guidelines on clinical management,104 as well as ongoing efforts towards the WHO Roadmap to prioritise research and product development for CCHF.105
Clinical validation
During the early stage of an outbreak, diagnostic tests are often evaluated using the strains most relevant to that region. Diagnostic test development could be accelerated through validation and external quality control (EQA) using up-to-date clinical specimen panels and reference standards, particularly since prior EQA performance indicated a wide range in laboratory test sensitivity. While the majority of laboratories received high marks, the observed sensitivities ranged from 75% to 100% for serological assays and from 43% to 100% for molecular assays (with outliers as low as 25% for older test methods).53 65 Specifically, routine EQA studies should include a range of CCHFV genotypes and concentrations to accurately evaluate and compare diagnostic performance. To the extent possible, patient specimens could be characterised and maintained for diagnostic test evaluation and quality assurance. In the absence of clinical specimens, a recombinant approach may be needed to generate sufficient quantities of quality control material.106 107
For CCHF diagnostic test developers, sourcing clinical specimens has been a major roadblock to both molecular and serological assay validation.8 The manufacturing process requires a substantial amount of reference material, and often companies develop inhouse calibration standards to control supply and lot-to-lot variability. There is little incentive to seek international regulatory approval; even for commercial suppliers, the investment for regulatory approval is often subject to market demand. Some international reference institutes, including the WHO International Biological Reference custodian laboratories, provide reference materials or specimen panels for validation or EQA/proficiency (online supplementary list S1). A specimen bank would be beneficial to CCHFV diagnostic development; however, this effort faces significant challenges given the sporadic nature of human cases which typically occur in remote agricultural regions across 30 countries, with only several hundred cases confirmed each year.6
bmjgh-2018-001114supp002.pdf (399.2KB, pdf)
Turnaround time and POC testing
As RT-PCR testing requires a high infrastructure laboratory and a turnaround time of 2–5 days, a more flexible approach is needed for an outbreak, with options that serve both animal and human populations. POC and ‘near-POC’ molecular diagnostic platforms have significantly lower infrastructure requirements and have been implemented in decentralised laboratories.108–110 These POC instruments are compact and self-contained, with automated sample preparation, and most healthcare workers can be trained for operation in clinic or field-based settings. Given the range of assays already developed for these commercial platforms, it is likely that current CCHFV RT-PCR assays could be readily adapted to the POC cartridge-based format.
Conclusion
This analysis identified several commercial sources for CCHF molecular diagnostics and serology, as well as a large number of inhouse tests. Despite this, several of the gaps identified in the 2016 WHO R&D Blueprint remain.3 111 A more detailed understanding of CCHF viral and antibody kinetics is needed across the broad range of sample types. Routine EQA, using well-characterised and up-to-date specimen panels, would be valuable for both clinical validation and proficiency testing. Surveillance is currently limited by lack of harmonisation and availability of validated serological tests.
Development of novel and next-generation diagnostic technologies for CCHF would benefit from a refined set of target product profiles (TPP) with detailed clinical and operational design specifications, including a range of minimal to optimal performance characteristics. Application-driven TPPs can be designed to support the development of CCHF diagnostics that have been identified here to accelerate care and minimise transmission risk: POC diagnostics for patient triage, screening and field testing; syndromic PCR panels for expediting differential diagnosis of CCHF from other VHF pathogens; and NGS to monitor circulating strains and viral mutations, particularly to assess the sensitivity of probe design used in molecular diagnostics. Ongoing initiatives include a CCHF-specific TPP currently being developed by WHO as part of their roadmap.112
Acknowledgments
We gratefully acknowledge all those who attended the WHO R&D Blueprint Roadmaps: Consultation on Crimean-Congo Fever in March 2018. Editorial assistance for later drafts was provided by Rachel Wright, PhD, funded by FIND, according to Good Publication Practice guidelines.
Footnotes
Handling editor: Seye Abimbola
Contributors: LTM contributed to drafting the manuscript and provided background research for the manuscript. CK-C contributed insight into the diagnostic needs for outbreak pathogens. Both authors reviewed, edited and approved the final version of the manuscript.
Funding: Publication of this article was funded by FIND. FIND was funded for this work by UK Aid from the UK Government.
Competing interests: None declared.
Patient consent: Not required.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data sharing statement: No additional data are available.
References
- 1. CDC , 2017. Crimean-Congo Hemorrhagic Fever (CCHF).. Available from: https://www.cdc.gov/vhf/crimean-congo/index.html [Accessed 12 Oct 2017].
- 2. WHO , 2017. a. Crimean-Congo haemorrhagic fever (CCHF).. Available from: http://www.who.int/csr/disease/crimean_congoHF/en/ [Accessed 12 Oct 2017].
- 3. WHO , 2017. R&D Blueprint for action to prevent epidemics.. Available from: http://www.who.int/blueprint/en/ [Accessed 12 Sep 2017].
- 4. Chinikar S, Mirahmadi R, Moradi M. Crimean-Congo Hemorrhagic Fever (CCHF) : Lorenzo-Morales J, Zoonosis. IntechOpen, 2012:193–212. 10.5772/2125 [DOI] [Google Scholar]
- 5. Whitehouse CA. Crimean–Congo hemorrhagic fever. Antiviral Res 2004;64:145–60. 10.1016/S0166-3542(04)00163-9 [DOI] [PubMed] [Google Scholar]
- 6. Al-Abri SS, Abaidani IA, Fazlalipour M, et al. Current status of crimean-congo haemorrhagic fever in the world health organization eastern mediterranean region: issues, challenges, and future directions. Int J Infect Dis 2017;58:82–9. 10.1016/j.ijid.2017.02.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ergonul O. Crimean-Congo hemorrhagic fever virus: new outbreaks, new discoveries. Curr Opin Virol 2012;2:215–20. 10.1016/j.coviro.2012.03.001 [DOI] [PubMed] [Google Scholar]
- 8. Mertens M, Schmidt K, Ozkul A, et al. The impact of Crimean-Congo hemorrhagic fever virus on public health. Antiviral Res 2013;98:248–60. 10.1016/j.antiviral.2013.02.007 [DOI] [PubMed] [Google Scholar]
- 9. Messina JP, Pigott DM, Golding N, et al. The global distribution of Crimean-Congo hemorrhagic fever. Trans R Soc Trop Med Hyg 2015;109:503–13. 10.1093/trstmh/trv050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kizito S, Okello PE, Kwesiga B, et al. Notes from the field: Crimean-Congo Hemorrhagic fever outbreak — central Uganda, august–september 2017. MMWR Morb Mortal Wkly Rep;67:646–7. 10.15585/mmwr.mm6722a6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ergönül O. Crimean-Congo haemorrhagic fever. Lancet Infect Dis 2006;6:203–14. 10.1016/S1473-3099(06)70435-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Charrel RN, Attoui H, Butenko AM, et al. Tick-borne virus diseases of human interest in Europe. Clin Microbiol Infect 2004;10:1040–55. 10.1111/j.1469-0691.2004.01022.x [DOI] [PubMed] [Google Scholar]
- 13. Spengler JR, Bergeron É, Rollin PE. Seroepidemiological studies of Crimean-Congo hemorrhagic fever virus in domestic and wild animals. PLoS Negl Trop Dis 2016;10:e0004210 10.1371/journal.pntd.0004210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Appannanavar SB, Mishra B. An update on crimean congo hemorrhagic fever. J Glob Infect Dis 2011;3:285–92. 10.4103/0974-777X.83537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Aslam S, Latif MS, Daud M, et al. Crimean-Congo hemorrhagic fever: risk factors and control measures for the infection abatement. Biomed Rep 2016;4:15–20. 10.3892/br.2015.545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bente DA, Forrester NL, Watts DM, et al. Crimean-Congo hemorrhagic fever: history, epidemiology, pathogenesis, clinical syndrome and genetic diversity. Antiviral Res 2013;100:159–89. 10.1016/j.antiviral.2013.07.006 [DOI] [PubMed] [Google Scholar]
- 17. Guven G, Talan L, Altintas ND, et al. An Unexpected fatal CCHF case and management of exposed health care workers. Int J Infect Dis 2017;55:118–21. 10.1016/j.ijid.2016.12.026 [DOI] [PubMed] [Google Scholar]
- 18. Papa A, Sidira P, Tsatsaris A. Spatial cluster analysis of Crimean-Congo hemorrhagic fever virus seroprevalence in humans, Greece. Parasite Epidemiol Control 2016;1:211–8. 10.1016/j.parepi.2016.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bodur H, Akinci E, Ascioglu S, et al. Subclinical infections with Crimean-Congo hemorrhagic fever virus, Turkey. Emerg Infect Dis 2012;18:640-2 10.3201/eid1804.111374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Spengler JR, Bente DA. Crimean–Congo hemorrhagic fever in Spain — new arrival or silent resident? N Engl J Med Overseas Ed 2017;377:106–8. 10.1056/NEJMp1707436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chinikar S, Bouzari S, Shokrgozar MA, et al. Genetic diversity of Crimean Congo hemorrhagic fever virus strains from Iran. J Arthropod Borne Dis 2016;10:127–40. [PMC free article] [PubMed] [Google Scholar]
- 22. Deyde VM, Khristova ML, Rollin PE, et al. Crimean-Congo hemorrhagic fever virus genomics and global diversity. J Virol 2006;80:8834–42. 10.1128/JVI.00752-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Xia H, Beck AS, Gargili A, et al. Transstadial transmission and long-term association of Crimean-Congo hemorrhagic fever virus in ticks shapes genome plasticity. Sci Rep 2016;6:35819 10.1038/srep35819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gargili A, Estrada-Peña A, Spengler JR, et al. The role of ticks in the maintenance and transmission of Crimean-Congo hemorrhagic fever virus: a review of published field and laboratory studies. Antiviral Res 2017;144:93–119. 10.1016/j.antiviral.2017.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Brackney DE, Armstrong PM. Transmission and evolution of tick-borne viruses. Curr Opin Virol 2016;21:67–74. 10.1016/j.coviro.2016.08.005 [DOI] [PubMed] [Google Scholar]
- 26. Lukashevich IS, Patterson J, Carrion R, et al. A live attenuated vaccine for Lassa fever made by reassortment of Lassa and Mopeia viruses. J Virol 2005;79:13934–42. 10.1128/JVI.79.22.13934-13942.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lukashev AN, Klimentov AS, Smirnova SE, et al. Phylogeography of Crimean Congo hemorrhagic fever virus. PLoS One 2016;11:e0166744 10.1371/journal.pone.0166744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chinikar S, Shah-Hosseini N, Bouzari S, et al. Assessment of recombination in the s-segment genome of Crimean-Congo hemorrhagic fever virus in Iran. J Arthropod Borne Dis 2016;10:12–23. [PMC free article] [PubMed] [Google Scholar]
- 29. Zhou Z, Deng F, Han N, et al. Reassortment and migration analysis of Crimean-Congo haemorrhagic fever virus. J Gen Virol 2013;94 2536–48. 10.1099/vir.0.056374-0 [DOI] [PubMed] [Google Scholar]
- 30. Johnson S, Henschke N, Maayan N, et al. Ribavirin for treating Crimean Congo haemorrhagic fever. Cochrane Database Syst Rev 2018;6:CD012713 10.1002/14651858.CD012713.pub2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ascioglu S, Leblebicioglu H, Vahaboglu H, et al. Ribavirin for patients with Crimean-Congo haemorrhagic fever: a systematic review and meta-analysis. J Antimicrob Chemother 2011;66:1215–22. 10.1093/jac/dkr136 [DOI] [PubMed] [Google Scholar]
- 32. Dokuzoguz B, Celikbas AK, Gök ŞE, et al. Severity scoring index for Crimean-Congo hemorrhagic fever and the impact of ribavirin and corticosteroids on fatality. Clin Infect Dis 2013;57:1270–4. 10.1093/cid/cit527 [DOI] [PubMed] [Google Scholar]
- 33. Ozbey SB, Kader Ç, Erbay A, et al. Early use of ribavirin is beneficial in Crimean-Congo hemorrhagic fever. Vector Borne Zoonotic Dis 2014;14:300–2. 10.1089/vbz.2013.1421 [DOI] [PubMed] [Google Scholar]
- 34. Soares-Weiser K, Thomas S, Thomson G, et al. Ribavirin for Crimean-Congo hemorrhagic fever: systematic review and meta-analysis. BMC Infect Dis 2010;10:207 10.1186/1471-2334-10-207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bodur H, Erbay A, Akıncı E, et al. Effect of oral ribavirin treatment on the viral load and disease progression in Crimean-Congo hemorrhagic fever. Int J Infect Dis 2011;15:e44–47. 10.1016/j.ijid.2010.09.009 [DOI] [PubMed] [Google Scholar]
- 36. Ertem G, Sönmezer MÇ, Temoçin F, et al. The efficacy of oral ribavirin on clinical and laboratory parameters inCrimean-Congo hemorrhagic fever: an observational study from Turkey. Turk J Med Sci 2016;46:1407–14. 10.3906/sag-1506-92 [DOI] [PubMed] [Google Scholar]
- 37. Conger NG, Paolino KM, Osborn EC, et al. Health care response to CCHF in US soldier and nosocomial transmission to health care providers, Germany, 2009. Emerg Infect Dis 2015;21:23–31. 10.3201/eid2101.141413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Guner R, Hasanoglu I, Tasyaran MA, et al. Is ribavirin prophylaxis effective for nosocomial transmission of Crimean-Congo hemorrhagic fever? Vector Borne Zoonotic Dis 2014;14:601–5. 10.1089/vbz.2013.1546 [DOI] [PubMed] [Google Scholar]
- 39. Hinkula J, Devignot S, Åkerström S, et al. Immunization with DNA Plasmids Coding for Crimean-Congo hemorrhagic fever virus capsid and envelope proteins and/or virus-like particles induces protection and survival in challenged mice. J Virol 2017;91 10.1128/JVI.02076-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Spengler JR, Bente DA. Therapeutic intervention in Crimean-Congo hemorrhagic fever: where are we now? Future Virol 2015;10:203–6. 10.2217/fvl.14.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Oestereich L, Rieger T, Neumann M, et al. Evaluation of antiviral efficacy of ribavirin, arbidol, and T-705 (favipiravir) in a mouse model for Crimean-Congo hemorrhagic fever. PLoS Negl Trop Dis 2014;8:e2804 10.1371/journal.pntd.0002804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Salata C, Calistri A, Parolin C, et al. Antiviral activity of cationic amphiphilic drugs. Expert Rev Anti Infect Ther 2017;15:483–92. 10.1080/14787210.2017.1305888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sharifi A, Amanlou A, Moosavi-Movahedi F, et al. Tetracyclines as a potential antiviral therapy against Crimean Congo hemorrhagic fever virus: Docking and molecular dynamic studies. Comput Biol Chem 2017;70:1–6. 10.1016/j.compbiolchem.2017.06.003 [DOI] [PubMed] [Google Scholar]
- 44. Zhang W, Bailey-Elkin BA, Knaap RCM, et al. Potent and selective inhibition of pathogenic viruses by engineered ubiquitin variants. PLoS Pathog 2017;13:e1006372 10.1371/journal.ppat.1006372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mousavi-Jazi M, Karlberg H, Papa A, et al. Healthy individuals' immune response to the bulgarian Crimean-Congo hemorrhagic fever virus vaccine. Vaccine 2012;30:6225–9. 10.1016/j.vaccine.2012.08.003 [DOI] [PubMed] [Google Scholar]
- 46. Dowall SD, Carroll MW, Hewson R. Development of vaccines against Crimean-Congo haemorrhagic fever virus. Vaccine 2017;35:6015–23. 10.1016/j.vaccine.2017.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Canakoglu N, Berber E, Tonbak S, et al. Immunization of knock-out α/β interferon receptor mice against high lethal dose of Crimean-Congo hemorrhagic fever virus with a cell culture based vaccine. PLoS Negl Trop Dis 2015;9:e0003579 10.1371/journal.pntd.0003579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kortekaas J, Vloet RP, McAuley AJ, et al. Crimean-Congo hemorrhagic fever virus subunit vaccines induce high levels of neutralizing antibodies but no protection in STAT1 knockout mice. Vector Borne Zoonotic Dis 2015;15:759–64. 10.1089/vbz.2015.1855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ahmed AA, McFalls JM, Hoffmann C, et al. Presence of broadly reactive and group-specific neutralizing epitopes on newly described isolates of Crimean-Congo hemorrhagic fever virus. J Gen Virol 2005;86 3327–36. 10.1099/vir.0.81175-0 [DOI] [PubMed] [Google Scholar]
- 50. Buttigieg KR, Dowall SD, Findlay-Wilson S, et al. A novel vaccine against Crimean-Congo haemorrhagic fever protects 100% of animals against lethal challenge in a mouse model. PLoS One 2014;9:e91516 10.1371/journal.pone.0091516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Dowall SD, Buttigieg KR, Findlay-Wilson SJ, et al. A Crimean-Congo Hemorrhagic Fever (CCHF) viral vaccine expressing nucleoprotein is immunogenic but fails to confer protection against lethal disease. Hum Vaccin Immunother 2016;12:519–27. 10.1080/21645515.2015.1078045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Pirbright Institute , 2018. Crimean-Congo haemorrhagic fever.. Available from: https://www.pirbright.ac.uk/viruses/crimean-congo-haemorrhagic-fever [Accessed 27 Sep 2018].
- 53. Vanhomwegen J, Alves MJ, Županc TA, et al. Diagnostic assays for Crimean-Congo hemorrhagic fever. Emerg Infect Dis 2012;18:1958–65. 10.3201/eid1812.120710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Tezer H, Polat M. Diagnosis of Crimean-Congo hemorrhagic fever. Expert Rev Anti Infect Ther 2015;13:555–66. 10.1586/14787210.2015.1021782 [DOI] [PubMed] [Google Scholar]
- 55. Zivcec M, Albarino CG, Guerrero LIW, et al. Genome sequences of Crimean-Congo hemorrhagic fever virus strains isolated in South Africa, Namibia, and Turkey. Genome Announc 2017;5:e01060–17. 10.1128/genomeA.01060-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Emmerich P, Jakupi X, von Possel R, et al. Viral metagenomics, genetic and evolutionary characteristics of Crimean-Congo hemorrhagic fever orthonairovirus in humans, Kosovo. Infect Genet Evol 2018;65:6–11. 10.1016/j.meegid.2018.07.010 [DOI] [PubMed] [Google Scholar]
- 57. Koehler JW, Delp KL, Hall AT, et al. Sequence optimized real-time reverse transcription polymerase chain reaction assay for detection of Crimean-Congo hemorrhagic fever virus. Am J Trop Med Hyg 2018;98:211–5. 10.4269/ajtmh.17-0165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Drosten C, Kümmerer BM, Schmitz H, et al. Molecular diagnostics of viral hemorrhagic fevers. Antiviral Res 2003;57 61–87. 10.1016/S0166-3542(02)00201-2 [DOI] [PubMed] [Google Scholar]
- 59. Fernandez-Garcia MD, Negredo A, Papa A, et al. European survey on laboratory preparedness, response and diagnostic capacity for Crimean-Congo haemorrhagic fever, 2012. Euro Surveill 2014;19:20844 10.2807/1560-7917.ES2014.19.26.20844 [DOI] [PubMed] [Google Scholar]
- 60. Roberts T, Bygrave H, Fajardo E, et al. Challenges and opportunities for the implementation of virological testing in resource-limited settings. J Int AIDS Soc 2012;15:17324 10.7448/IAS.15.2.17324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wang S, Lifson MA, Inci F, et al. Advances in addressing technical challenges of point-of-care diagnostics in resource-limited settings. Expert Rev Mol Diagn 2016;16:449–59. 10.1586/14737159.2016.1142877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Al Johani S, Hajeer AH. MERS-CoV diagnosis: an update. J Infect Public Health 2016;9:216–9. 10.1016/j.jiph.2016.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Fletcher TE, Gulzhan A, Ahmeti S, et al. Infection prevention and control practice for Crimean-Congo hemorrhagic fever-a multi-center cross-sectional survey in Eurasia. PLoS One 2017;12:e0182315 10.1371/journal.pone.0182315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Das S, Rundell MS, Mirza AH, et al. A Multiplex PCR/LDR assay for the simultaneous identification of category a infectious pathogens: agents of viral hemorrhagic fever and variola virus. PLoS One 2015;10:e0138484 10.1371/journal.pone.0138484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Escadafal C, Olschläger S, Avšič-Županc T, et al. First international external quality assessment of molecular detection of Crimean-Congo hemorrhagic fever virus. PLoS Negl Trop Dis 2012;6:e1706 10.1371/journal.pntd.0001706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Formenty P, Leroy EM, Epelboin A, et al. Detection of Ebola virus in oral fluid specimens during outbreaks of Ebola virus hemorrhagic fever in the Republic of Congo. Clin Infect Dis 2006;42:1521–6. 10.1086/503836 [DOI] [PubMed] [Google Scholar]
- 67. Lunkenheimer K, Hufert FT, Schmitz H. Detection of lassa virus RNA in specimens from patients with Lassa fever by using the polymerase chain reaction. J Clin Microbiol 1990;28:2689–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Mizuno Y, Kotaki A, Harada F, et al. Confirmation of dengue virus infection by detection of dengue virus type 1 genome in urine and saliva but not in plasma. Trans R Soc Trop Med Hyg 2007;101:738–9. 10.1016/j.trstmh.2007.02.007 [DOI] [PubMed] [Google Scholar]
- 69. Pettersson L, Klingström J, Hardestam J, et al. Hantavirus RNA in saliva from patients with hemorrhagic fever with renal syndrome. Emerg Infect Dis 2008;14:406–11. 10.3201/eid1403.071242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Negredo A, de la Calle-Prieto F, Palencia-Herrejón E, et al. Autochthonous Crimean–Congo Hemorrhagic Fever in Spain. N Engl J Med Overseas Ed 2017;377:154–61. 10.1056/NEJMoa1615162 [DOI] [PubMed] [Google Scholar]
- 71. Van den Bossche D, Cnops L, Van Esbroeck M. Recovery of dengue virus from urine samples by real-time RT-PCR. Eur J Clin Microbiol Infect Dis 2015;34:1361–7. 10.1007/s10096-015-2359-0 [DOI] [PubMed] [Google Scholar]
- 72. Burt FJ, Leman PA, Smith JF, et al. The use of a reverse transcription-polymerase chain reaction for the detection of viral nucleic acid in the diagnosis of Crimean-Congo haemorrhagic fever. J Virol Methods 1998;70:129–37. 10.1016/S0166-0934(97)00182-1 [DOI] [PubMed] [Google Scholar]
- 73. Drosten C, Göttig S, Schilling S, et al. Rapid detection and quantification of RNA of Ebola and Marburg viruses, Lassa virus, Crimean-Congo hemorrhagic fever virus, Rift Valley fever virus, dengue virus, and yellow fever virus by real-time reverse transcription-PCR. J Clin Microbiol 2002;40:2323–30. 10.1128/JCM.40.7.2323-2330.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Rodriguez LL, Maupin GO, Ksiazek TG, et al. Molecular investigation of a multisource outbreak of Crimean-Congo hemorrhagic fever in the United Arab Emirates. Am J Trop Med Hyg 1997;57:512–8. 10.4269/ajtmh.1997.57.512 [DOI] [PubMed] [Google Scholar]
- 75. Bodur H, Akıncı E, Ongürü P, et al. Detection of Crimean-Congo hemorrhagic fever virus genome in saliva and urine. Int J Infect Dis 2010;14:e247–49. 10.1016/j.ijid.2009.04.018 [DOI] [PubMed] [Google Scholar]
- 76. Duh D, Saksida A, Petrovec M, et al. Novel one-step real-time RT-PCR assay for rapid and specific diagnosis of Crimean-Congo hemorrhagic fever encountered in the Balkans. J Virol Methods 2006;133:175–9. 10.1016/j.jviromet.2005.11.006 [DOI] [PubMed] [Google Scholar]
- 77. Cevik MA, Erbay A, Bodur H, et al. Viral load as a predictor of outcome in Crimean-Congo hemorrhagic fever. Clin Infect Dis 2007;45:e96–e100. 10.1086/521244 [DOI] [PubMed] [Google Scholar]
- 78. Duh D, Saksida A, Petrovec M, et al. Viral load as predictor of Crimean-Congo hemorrhagic fever outcome. Emerg Infect Dis 2007;13:1769–72. 10.3201/eid1311.070222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Ergonul O, Celikbas A, Baykam N, et al. Analysis of risk-factors among patients with Crimean-Congo haemorrhagic fever virus infection: severity criteria revisited. Clin Microbiol Infect 2006;12:551–4. 10.1111/j.1469-0691.2006.01445.x [DOI] [PubMed] [Google Scholar]
- 80. Hasanoglu I, Guner R, Carhan A, et al. Dynamics of viral load in Crimean Congo hemorrhagic fever. J Med Virol 2018;90:639–43. 10.1002/jmv.24990 [DOI] [PubMed] [Google Scholar]
- 81. Akinci E, Bodur H, Sunbul M, et al. Prognostic factors, pathophysiology and novel biomarkers in Crimean-Congo hemorrhagic fever. Antiviral Res 2016;132:233–43. 10.1016/j.antiviral.2016.06.011 [DOI] [PubMed] [Google Scholar]
- 82. Bonney LC, Watson RJ, Afrough B, et al. A recombinase polymerase amplification assay for rapid detection of Crimean-Congo haemorrhagic fever virus infection. PLoS Negl Trop Dis 2017;11:e0006013 10.1371/journal.pntd.0006013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Papa A, Papadopoulou E, Tsioka K, et al. Isolation and whole-genome sequencing of a Crimean-Congo hemorrhagic fever virus strain, Greece. Ticks Tick Borne Dis 2018;9:788–91. 10.1016/j.ttbdis.2018.02.024 [DOI] [PubMed] [Google Scholar]
- 84. Brinkmann A, Ergünay K, Radonić A, et al. Development and preliminary evaluation of a multiplexed amplification and next generation sequencing method for viral hemorrhagic fever diagnostics. PLoS Negl Trop Dis 2017;11:e0006075 10.1371/journal.pntd.0006075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Fernández de Mera IG, Chaligiannis I, Hernández-Jarguín A, et al. Combination of RT-PCR and proteomics for the identification of Crimean-Congo hemorrhagic fever virus in ticks. Heliyon 2017;3:e00353 10.1016/j.heliyon.2017.e00353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Papa A, Mirazimi A, Köksal I, et al. Recent advances in research on Crimean-Congo hemorrhagic fever. J Clin Virol 2015;64:137–43. 10.1016/j.jcv.2014.08.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Burt FJ, Leman PA, Abbott JC, et al. Serodiagnosis of Crimean-Congo haemorrhagic fever. Epidemiol Infect 1994;113:551–62. 10.1017/S0950268800068576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Shepherd AJ, Swanepoel R, Gill DE. Evaluation of enzyme-linked immunosorbent assay and reversed passive hemagglutination for detection of Crimean-Congo hemorrhagic fever virus antigen. J Clin Microbiol 1988;26:347–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Saluzzo JF, Le Guenno B. Rapid diagnosis of human Crimean-Congo hemorrhagic fever and detection of the virus in naturally infected ticks. J Clin Microbiol 1987;25:922–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Filippone C, Marianneau P, Murri S, et al. Molecular diagnostic and genetic characterization of highly pathogenic viruses: application during Crimean-Congo haemorrhagic fever virus outbreaks in Eastern Europe and the Middle East. Clin Microbiol Infect 2013;19:E118–28. 10.1111/1469-0691.12075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. O'Hearn AE, Voorhees MA, Fetterer DP, et al. Serosurveillance of viral pathogens circulating in West Africa. Virol J 2016;13:163 10.1186/s12985-016-0621-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Liu J, Ochieng C, Wiersma S, et al. Development of a taqman array card for acute-febrile-illness outbreak investigation and surveillance of emerging pathogens, including ebola virus. J Clin Microbiol 2016;54:49–58. 10.1128/JCM.02257-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Mild M, Simon M, Albert J, et al. Towards an understanding of the migration of Crimean-Congo hemorrhagic fever virus. J Gen Virol 2010;91 199–207. 10.1099/vir.0.014878-0 [DOI] [PubMed] [Google Scholar]
- 94. Mertens M, Schuster I, Sas MA, et al. Crimean-Congo hemorrhagic fever virus in bulgaria and Turkey. Vector Borne Zoonotic Dis 2016;16:619–23. 10.1089/vbz.2016.1944 [DOI] [PubMed] [Google Scholar]
- 95. Orkun Ö, Karaer Z, Çakmak A, et al. Crimean-Congo hemorrhagic fever virus in ticks in Turkey: a broad range tick surveillance study. Infect Genet Evol 2017;52:59–66. 10.1016/j.meegid.2017.04.017 [DOI] [PubMed] [Google Scholar]
- 96. Sherifi K, Rexhepi A, Berxholi K, et al. Crimean-Congo hemorrhagic fever virus and Borrelia burgdorferi sensu lato in Ticks from Kosovo and Albania. Front Vet Sci 2018;5:38 10.3389/fvets.2018.00038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Moming A, Yue X, Shen S, et al. Prevalence and phylogenetic analysis of Crimean-Congo hemorrhagic fever virus in ticks from different ecosystems in Xinjiang, China. Virol Sin 2018;33:67–73. 10.1007/s12250-018-0016-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. De Liberato C, Frontoso R, Magliano A, et al. Monitoring for the possible introduction of Crimean-Congo haemorrhagic fever virus in Italy based on tick sampling on migratory birds and serological survey of sheep flocks. Prev Vet Med 2018;149:47–52. 10.1016/j.prevetmed.2017.10.014 [DOI] [PubMed] [Google Scholar]
- 99. Christova I, Panayotova E, Groschup MH, et al. High seroprevalence for Crimean-Congo haemorrhagic fever virus in ruminants in the absence of reported human cases in many regions of Bulgaria. Exp Appl Acarol 2018;75:227–34. 10.1007/s10493-018-0258-7 [DOI] [PubMed] [Google Scholar]
- 100. Sas MA, Comtet L, Donnet F, et al. A novel double-antigen sandwich ELISA for the species-independent detection of Crimean-Congo hemorrhagic fever virus-specific antibodies. Antiviral Res 2018;151:24–6. 10.1016/j.antiviral.2018.01.006 [DOI] [PubMed] [Google Scholar]
- 101. Yousaf MZ, Ashfaq UA, Anjum KM, et al. Crimean-Congo Hemorrhagic Fever (CCHF) in Pakistan: The "Bell" is Ringing Silently. Crit Rev Eukaryot Gene Expr 2018;28:93–100. 10.1615/CritRevEukaryotGeneExpr.2018020593 [DOI] [PubMed] [Google Scholar]
- 102. Spengler JR, Bente DA, Bray M, et al. Second international conference on Crimean-Congo hemorrhagic fever. Antiviral Res 2018;150:137–47. 10.1016/j.antiviral.2017.11.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Papa A, Pappa S, Panayotova E, et al. Molecular epidemiology of Crimean-Congo hemorrhagic fever in Bulgaria–an update. J Med Virol 2016;88:769–73. 10.1002/jmv.24400 [DOI] [PubMed] [Google Scholar]
- 104. Saijo M, Tang Q, Shimayi B, et al. Antigen-capture enzyme-linked immunosorbent assay for the diagnosis of crimean-congo hemorrhagic fever using a novel monoclonal antibody. J Med Virol 2005;77:83–8. 10.1002/jmv.20417 [DOI] [PubMed] [Google Scholar]
- 105. WHO , 2018. Crimean-Congo haemorrhagic fever (CCHF). RD Bluepr. Available from: http://www.who.int/blueprint/priority-diseases/key-action/crimean-congo-haemorrhagic-fever/en/ [Accessed 24 Sep 2018].
- 106. Chan M, Jiang B, Tan TY. Using pooled recombinant plasmids as control materials for diagnostic real-time PCR. Clin Lab 2016;62:1893–901. 10.7754/Clin.Lab.2016.160114 [DOI] [PubMed] [Google Scholar]
- 107. Baadenhuijsen H, Kuypers A, Weykamp C, et al. External Quality Assessment in The Netherlands: time to introduce commutable survey specimens. Lessons from the Dutch "Calibration 2000" project. Clin Chem Lab Med 2005;43:304-7 10.1515/CCLM.2005.052 [DOI] [PubMed] [Google Scholar]
- 108. Goel N, Ritchie AV, Mtapuri-Zinyowera S, et al. Performance of the SAMBA I and II HIV-1 Semi-Q Tests for viral load monitoring at the point-of-care. J Virol Methods 2017;244:39–45. 10.1016/j.jviromet.2017.03.001 [DOI] [PubMed] [Google Scholar]
- 109. Jani IV, Meggi B, Vubil A, et al. Evaluation of the whole-blood alere Q NAT point-of-care RNA Assay for HIV-1 viral load monitoring in a primary health care setting in Mozambique. J Clin Microbiol 2016;54:2104–8. 10.1128/JCM.00362-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Moyo S, Mohammed T, Wirth KE, et al. Point-of-care cepheid xpert HIV-1 viral load test in Rural African communities is feasible and reliable. J Clin Microbiol 2016;54:3050–5. 10.1128/JCM.01594-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Mehand MS, Millett P, Al-Shorbaji F, et al. World health organization methodology to prioritize emerging infectious diseases in need of research and development. Emerg Infect Dis 2018;24 10.3201/eid2409.171427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. WHO , 2018. Roadmap for Research and Product Development against Crimean-Congo Haemorrhagic Fever (CCHF). Available from: http://www.who.int/blueprint/priority-diseases/key-action/cchf-draft-r-and-d-roadmap.pdf [Accessed 24 Sep 2018].
- 113. Yapar M, Aydogan H, Pahsa A, et al. Rapid and quantitative detection of Crimean-Congo hemorrhagic fever virus by one-step real-time reverse transcriptase-PCR. Jpn J Infect Dis 2005;58:358–62. [PubMed] [Google Scholar]
- 114. Garrison AR, Alakbarova S, Kulesh DA, et al. Development of a TaqMan minor groove binding protein assay for the detection and quantification of Crimean-Congo hemorrhagic fever virus. Am J Trop Med Hyg 2007;77:514–20. 10.4269/ajtmh.2007.77.514 [DOI] [PubMed] [Google Scholar]
- 115. Midilli K, Gargılı A, Ergonul O, et al. Imported Crimean-Congo hemorrhagic fever cases in Istanbul. BMC Infect Dis 2007;7:54 10.1186/1471-2334-7-54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Midilli K, Gargılı A, Ergonul O, et al. The first clinical case due to AP92 like strain of Crimean-Congo hemorrhagic fever virus and a field survey. BMC Infect Dis 2009;9:90 10.1186/1471-2334-9-90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Wölfel R, Paweska JT, Petersen N, et al. Virus detection and monitoring of viral load in Crimean-Congo hemorrhagic fever virus patients. Emerg Infect Dis 2007;13:1097–100. 10.3201/eid1307.070068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Wölfel R, Paweska JT, Petersen N, et al. Low-density macroarray for rapid detection and identification of Crimean-Congo hemorrhagic fever virus. J Clin Microbiol 2009;47:1025–30. 10.1128/JCM.01920-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Papa A, Drosten C, Bino S, et al. Viral load and Crimean-Congo hemorrhagic fever. Emerg Infect Dis 2007;13:805–6. 10.3201/eid1305.061588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Kondiah K, Swanepoel R, Paweska JT, et al. A Simple-Probe real-time PCR assay for genotyping reassorted and non-reassorted isolates of Crimean-Congo hemorrhagic fever virus in southern Africa. J Virol Methods 2010;169:34–8. 10.1016/j.jviromet.2010.06.010 [DOI] [PubMed] [Google Scholar]
- 121. Atkinson B, Chamberlain J, Logue CH, et al. Development of a real-time RT-PCR assay for the detection of Crimean-Congo hemorrhagic fever virus. Vector Borne Zoonotic Dis 2012;12:786–93. 10.1089/vbz.2011.0770 [DOI] [PubMed] [Google Scholar]
- 122. Jääskeläinen AJ, Kallio-Kokko H, Ozkul A, et al. Development and evaluation of a real-time RT-qPCR for detection of Crimean-Congo hemorrhagic fever virus representing different genotypes. Vector Borne Zoonotic Dis 2014;14:870–2. 10.1089/vbz.2014.1577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Schwarz TF, Nsanze H, Longson M, et al. Polymerase chain reaction for diagnosis and identification of distinct variants of Crimean-Congo hemorrhagic fever virus in the United Arab Emirates. Am J Trop Med Hyg 1996;55:190–6. 10.4269/ajtmh.1996.55.190 [DOI] [PubMed] [Google Scholar]
- 124. Aziz TAG, Ali DJ, Jaff DO. Molecular and serological detection of Crimean-Congo hemorrhagic fever virus in sulaimani province, Iraq. J Biosci Med 2016;04:36–42. 10.4236/jbm.2016.44006 [DOI] [Google Scholar]
- 125. Burt FJ, Swanepoel R. Molecular epidemiology of African and Asian Crimean-Congo haemorrhagic fever isolates. Epidemiol Infect 2005;133:659–66. 10.1017/S0950268805003730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Burt FJ, Swanepoel R, Braack LE. Enzyme-linked immunosorbent assays for the detection of antibody to Crimean-Congo haemorrhagic fever virus in the sera of livestock and wild vertebrates. Epidemiol Infect 1993;111:547–57. 10.1017/S0950268800057277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Saijo M, Qing T, Niikura M, et al. Recombinant nucleoprotein-based enzyme-linked immunosorbent assay for detection of immunoglobulin G antibodies to Crimean-Congo hemorrhagic fever virus. J Clin Microbiol 2002;40:1587–91. 10.1128/JCM.40.5.1587-1591.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Qing T, Saijo M, Lei H, et al. Detection of immunoglobulin G to Crimean-Congo hemorrhagic fever virus in sheep sera by recombinant nucleoprotein-based enzyme-linked immunosorbent and immunofluorescence assays. J Virol Methods 2003;108:111–6. 10.1016/S0166-0934(02)00267-7 [DOI] [PubMed] [Google Scholar]
- 129. Garcia S, Chinikar S, Coudrier D, et al. Evaluation of a Crimean-Congo hemorrhagic fever virus recombinant antigen expressed by semliki forest suicide virus for IgM and IgG antibody detection in human and animal sera collected in Iran. J Clin Virol 2006;35:154–9. 10.1016/j.jcv.2005.02.016 [DOI] [PubMed] [Google Scholar]
- 130. Emmerich P, Avsic-Zupanc T, Chinikar S, et al. Early serodiagnosis of acute human Crimean-Congo hemorrhagic fever virus infections by novel capture assays. J Clin Virol 2010;48:294–5. 10.1016/j.jcv.2010.05.002 [DOI] [PubMed] [Google Scholar]
- 131. Samudzi RR, Leman PA, Paweska JT, et al. Bacterial expression of Crimean-Congo hemorrhagic fever virus nucleoprotein and its evaluation as a diagnostic reagent in an indirect ELISA. J Virol Methods 2012;179:70–6. 10.1016/j.jviromet.2011.09.023 [DOI] [PubMed] [Google Scholar]
- 132. Dowall SD, Richards KS, Graham VA, et al. Development of an indirect ELISA method for the parallel measurement of IgG and IgM antibodies against Crimean-Congo haemorrhagic fever (CCHF) virus using recombinant nucleoprotein as antigen. J Virol Methods 2012;179:335–41. 10.1016/j.jviromet.2011.11.020 [DOI] [PubMed] [Google Scholar]
- 133. Mourya DT, Yadav PD, Shete AM, et al. Detection, isolation and confirmation of Crimean-Congo hemorrhagic fever virus in human, ticks and animals in Ahmadabad, India, 2010-2011. PLoS Negl Trop Dis 2012;6:e1653 10.1371/journal.pntd.0001653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Mertens M, Vatansever Z, Mrenoshki S, et al. Circulation of Crimean-Congo hemorrhagic fever virus in the former yugoslav republic of Macedonia revealed by screening of cattle sera using a novel enzyme-linked immunosorbent assay. PLoS Negl Trop Dis 2015;9:e0003519 10.1371/journal.pntd.0003519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Emmerich P, Mika A, von Possel R, et al. Sensitive and specific detection of Crimean-Congo Hemorrhagic Fever Virus (CCHFV)-specific IgM and IgG antibodies in human sera using recombinant CCHFV nucleoprotein as antigen in μ-capture and IgG immune complex (IC) ELISA tests. PLoS Negl Trop Dis 2018;12:e0006366 10.1371/journal.pntd.0006366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Saijo M, Qing T, Niikura M, et al. Immunofluorescence technique using HeLa cells expressing recombinant nucleoprotein for detection of immunoglobulin G antibodies to Crimean-Congo hemorrhagic fever virus. J Clin Microbiol 2002;40:372–5. 10.1128/JCM.40.2.372-375.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Xia H, Li P, Yang J, et al. Epidemiological survey of Crimean-Congo hemorrhagic fever virus in Yunnan, China, 2008. Int J Infect Dis 2011;15:e459–63. 10.1016/j.ijid.2011.03.013 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
bmjgh-2018-001114supp001.pdf (450.8KB, pdf)
bmjgh-2018-001114supp003.pdf (420.7KB, pdf)
bmjgh-2018-001114supp002.pdf (399.2KB, pdf)