Abstract
Lassa fever virus (LASV) causes acute viral haemorrhagic fever with symptoms similar to those seen with Ebola virus infections. LASV is endemic to West Africa and is transmitted through contact with excretions of infected Mastomys natalensis rodents and other rodent species. Due to a high fatality rate, lack of treatment options and difficulties with prevention and control, LASV is one of the high-priority pathogens included in the WHO R&D Blueprint. The WHO LASV vaccine strategy relies on availability of effective diagnostic tests. Current diagnostics for LASV include in-house and commercial (primarily research-only) laboratory-based serological and nucleic acid amplification tests. There are two commercially available (for research use only) rapid diagnostic tests (RDTs), and a number of multiplex panels for differential detection of LASV infection from other endemic diseases with similar symptoms have been evaluated. However, a number of diagnostic gaps remain. Lineage detection is a challenge due to the genomic diversity of LASV, as pan-lineage sensitivity for both molecular and immunological detection is necessary for surveillance and outbreak response. While pan-lineage ELISA and RDTs are commercially available (for research use only), validation and external quality assessment (EQA) is needed to confirm detection sensitivity for all known or relevant strains. Variable sensitivity of LASV PCR tests also highlights the need for improved validation and EQA. Given that LASV outbreaks typically occur in low-resource settings, more options for point-of-care testing would be valuable. These requirements should be taken into account in target product profiles for improved LASV diagnostics.
Keywords: Lassa fever virus, LASV, in-vitro diagnostics, outbreak
Summary box.
Diagnostics are key to effective prevention and control of Lassa fever virus (LASV), a WHO R&D Blueprint priority pathogen that causes acute viral haemorrhagic fever.
Current diagnostics include laboratory-based serological and nucleic acid amplification tests as well as rapid diagnostic tests.
Challenges to LASV diagnostics include commercial availability of clinically validated pan-lineage tests, few options for point-of-care testing, differentiation from other agents that cause similar symptoms and a need for improvements to test validation, regulation and external quality assessment; target product profiles for LASV diagnostics should be refined to take into account these needs.
Introduction
Lassa fever virus (LASV) causes acute viral haemorrhagic fever (VHF) and is endemic to West Africa. Every year, approximately 100 000–300 000 people contract LASV, and 5000 people die from the infection.1–3 The recent 2018 outbreak in Nigeria saw 423 confirmed cases with a case fatality rate of 25%,4 5 and higher case fatality rates of up to 50%–70% have been reported.6 7 Because of its potential for zoonotic and human transmission as well as difficulties in treatment and prevention, LASV is one of the high-priority pathogens identified on the WHO R&D Blueprint.1 2 8 9 In June 2017, WHO finalised a LASV vaccine strategy,10 which relies on improved diagnostic tests as well as enhanced surveillance capacity in endemic countries. Here, we summarise existing LASV diagnostics and highlight remaining research and development needs.
Epidemiology
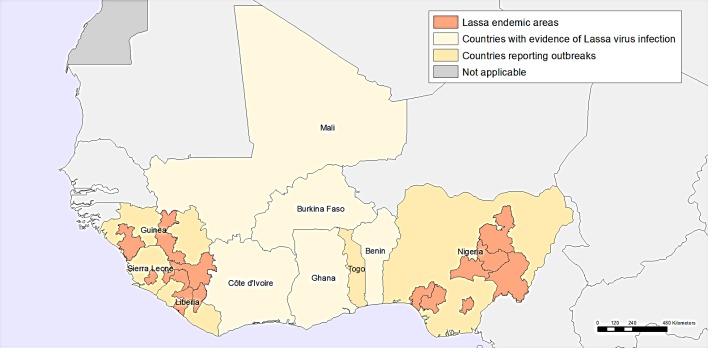
LASV is a single-stranded RNA virus of the Arenaviridae family. First identified in 1969 in Nigeria,11 Lassa fever is now endemic in West Africa including Nigeria, Sierra Leone, Guinea, Liberia, Benin, Ghana and Mali and has spread to neighbouring countries (figure 1).12–15 In some areas, 10%–16% of people admitted to hospitals every year have LASV.1 Cases have also been identified in Germany,13 15 16 the Netherlands,17 18 Sweden,19 the USA,20–22 the UK23 24 and Japan,25 largely imported after travel in West Africa.17 26–28 The long incubation period of LASV (~7–10 days) makes it one of the most commonly exported VHFs to countries outside its endemic range.
Figure 1.
Geographic distribution of Lassa fever in West Africa. Adapted from Emergencies—Lassa fever, WHO, Geographic distribution of Lassa fever in west African affected countries, 1969–2018, Copyright 2018.
Reservoir
Mastomys natalensis multimammate rodents are the most common rodent across the African continent, found predominantly in rural areas and human dwellings.29–32 These rodents show persistent LASV infection but are largely unaffected by the disease and shed the virus in their excrement.33 Seroprevalence has been reported to be as high as 60%–80% in M. natalensis populations.29 34 35 More recently, other rodent species including Hylomyscus pamfi and Mastomys erythroleucus have been shown to host LASV.36 37 Transmission to humans occurs primarily through contact with infected rodent urine or faeces; handling and consumption of infected rodents is also a pathway to infection.32 38 Airborne transmission may occur from aerosolised rodent excretions (dust) during cleaning activities.1 2 M. natalensis rodents readily colonise human areas where food is stored, contributing a significant risk for spillover, especially in communities with poor sanitation or crowded living conditions.1 2
Human-to-human transmission is less common, but LASV can be spread through direct contact with bodily secretions of persons infected with Lassa fever, presenting a higher risk for healthcare and humanitarian personnel,39–42 which increases with progression of disease and increasing viral load.41 43 There are suspected sexual transmission risks, as LASV can be detected in semen for 3 months past symptomatic infection.32 44 45
Prevention and control
Prevention of Lassa fever relies on promoting good community hygiene to reduce the potential for human-rodent contact. Measures to discourage rodents include storing grain and other food in rodent-proof containers, good hand and food hygiene, disposing of garbage away from the home, maintaining clean households and trapping rodents or employing cats as a natural deterrent.1 2 46 Regular and sustainable environmental sanitation is also needed to reduce rodent activity. Although rodents are a food source for a high percentage of some communities, consumption should be discouraged.47
Healthcare settings should employ standard infection prevention and control precautions when caring for patients.40 42 45 48–51 Healthcare and laboratory workers should handle LASV specimens under maximum biosafety level 4 (BSL-4) biological containment conditions where possible.52 53 If BSL-4 precautions are not available, samples may be handled in a class II/III biosafety cabinet under BSL-2 precautions.1 2
Early detection is critical for LASV containment, and a strong surveillance system is necessary to support interventions in endemic or ‘hot spot’ areas for LASV and other VHF.53 54 WHO and partners support national authorities in affected countries for outbreak preparedness and emergency response once an outbreak has been detected,55 as shown in recent outbreaks in Nigeria.56–60
Clinical indications and management
Early detection of LASV infection is difficult as the clinical course is highly variable, with symptoms ranging from 2 to 21 days postinfection. Lassa fever symptoms can mimic other endemic diseases such as malaria, typhoid fever and other VHFs.1 2 Although bleeding may help to discriminate VHF from other febrile illness, only 30% of patients with Lassa fever present with visible bleeding.7 42 61 62
Around 80% of people who become infected with LASV are asymptomatic or have mild symptoms of gradual fever, weakness and malaise, which often go unreported.32 63 After a few days, roughly 20% of infections progress to headache, sore throat, cough, muscle/joint pain, chest/abdominal pain, nausea, vomiting or diarrhoea. Indications of severe infection include facial swelling, fluid in the lung cavity, low blood pressure, petechiae and bruising, hepatitis and haemorrhaging of the conjunctival, gastrointestinal or mucosal tissue. Critically ill cases often present acute respiratory distress, shock, seizures, tremor, disorientation and coma. Death occurs within 14 days of onset for 15%–20% of severe cases. Young people and pregnant women are disproportionately impacted by LASV; the disease is especially serious in the third trimester, with maternal and fetal mortality reaching rates of 80% and 95%, respectively.7 32 64
In non-fatal cases, the fever subsides and the patient’s condition improves over 1–3 weeks, although renal damage65 along with neurological effects and fatigue66 can persist for several weeks. Deafness is a common side effect during the convalescent phase, typically accompanied by neurological dysfunction and vertigo.27 34 67 In half of these cases, hearing returns partially after 1–3 months.
Molecular epidemiology
Members of the arenavirus family are composed of an ambisense RNA genome and a nucleoprotein (NP), surrounded by a lipid envelope and a glycoprotein. The LASV genome consists of two single-stranded RNA segments: the small segment (S, 3.5 kb) encodes the NP and the glycoprotein precursor (GPC); the large segment (L, 7.2 kb) encodes the RNA-dependent RNA polymerase (LP) and the matrix protein (Z).
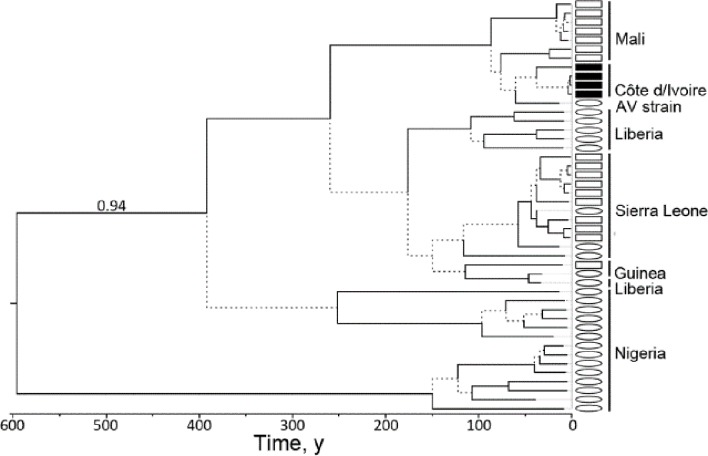
Once thought to be genetically stable, considerable genomic variation has been detected for geographically distant LASV strains, with phenotypic differences even among closely related isolates. LASV phylogeny is far more genetically diverse than Ebola virus in that <75% sequence is conserved for LASV compared with >97% for Ebola.68 69 LASV comprises at least four lineages: the prototype LASV strain isolated from Eastern Nigeria is lineage I; strains isolated from Southern Central and Northern Central Nigeria are lineage II and III and the large group from Guinea, Liberia and Sierra Leone are lineage IV (with three distinct subclades including the Josiah strain). A fifth ‘strain AV’ lineage from Mali/Cote D’Ivoire has been proposed (figure 2)16 70 71 as well as a newly designated lineage VI strain linked to Togo.72
Figure 2.
Bayesian chronogram of LASV based on the genomic L segment. LASV sequences of human origin indicated by ovals and sequences of Mastomys natalensis indicated by squares (black squares indicate ‘strain AV’ sequences). This tree was built under the assumption of a molecular clock and is therefore rooted. Source: Kouadio et al. 151 LASV, Lassa fever virus.
Therapeutic efforts
The recommended course for clinical management of LASV is general supportive care with management of symptoms. There is no approved antiviral treatment; however, evidence indicates that Lassa responds to ribavirin if administered at early onset of symptoms. Oral ribavirin has been used for postexposure prophylaxis for persons at high risk of secondary infection.15 41 42 48 While ribavirin studies have demonstrated a noticeable decrease in mortality for severe cases, high fatality rates have been observed for both ribavirin-treated and untreated patients, which underscores the need for better Lassa therapeutics.7 73
Favipiravir is another broad-spectrum RNA inhibitor (licensed for influenza) that has broad-spectrum activity against RNA viruses and has been shown to decrease Lassa viremia in animal models.74–78 Small molecules with benzimidazole-related scaffolds possess activity against a variety of arenaviruses and can interfere with LASV processing or viral entry. One such compound (ST-193) has been assessed in an animal model of Lassa fever, where Lassa-treated guinea pigs exhibited fewer signs of disease and enhanced survival.79 80 A similar optimised analogue (ST-161) had subnanomolar activity against LASV using a plaque reduction assay with live virus.81
Small interfering RNA particles (siRNA) can been shown to inhibit LASV replication system for in vitro studies.82 NP-directed and L-directed siRNAs demonstrated antiviral activity in a Lassa vero cell assay against five Lassa isolates. Both siRNAs inhibited replication of virus strains by up to 1 log unit with no apparent effect on cell viability. However, siRNA candidates must perfectly base pair to the target sequence, which could be a significant roadblock given the amount of genomic variation with LASV.
Monoclonal antibodies specific for LASV neutralisation cloned from West African Lassa fever survivors83 appear to bind to individual or combined Lassa GP protein subunits, which can potently neutralise all four LASV lineages—an early start to immunotherapeutic development and vaccine design.84 In particular, in the recent elucidation of the crystal structure of the trimeric, prefusion ectodomain of LASV GP bound to human neutralising antibody may accelerate development of antibody-based LASV therapeutics.85
Vaccine efforts
Currently, there is no vaccine that protects against LASV, and there are no vaccines available for use in animals to reduce zoonotic transmission.1 2 To stimulate vaccine development, WHO finalised a Lassa fever vaccine Target Product Profile (TPP) in June 2017, which was made available to target vaccine scientists, product developers, manufacturers and funding agencies after public consultation.10 In response, LASV was made one of the priorities for vaccine development funding by the multiagency Coalition for Epidemic Preparedness Innovations (CEPI), using a multisector partnership approach to finance and develop vaccines for infections of epidemic potential.86 87
A number of different platform technologies have been evaluated as potential Lassa fever vaccines, including virus-like particles,88 89 DNA vaccines,90 91 non-replicating (inactivated) viruses such as Lassa92 and Yellow Fever (17D),93 94 Chimpanzee adenoviral vectors,95 recombinant Vesicular stomatitis virus (VSV/LASV)96–98 and reassortment viral vectors such MOPV/LASV.99–101 For the vaccine platforms, recombinant VSV/LASV and reassortant ML29 are considered the most advanced vaccine candidates in clinical development.102 103 Despite promising preclinical evaluation, none of these vaccine candidates have yet advanced to human clinical trials.
Lassa diagnostics
Laboratory diagnosis of LASV infection is made by detection of the virus (culture), LASV RNA, LASV-specific IgG or IgM antibody response or LASV antigens shed during replication. LASV RNA is detected using a nucleic acid amplification test, which can include techniques such as PCR, loop-mediated isothermal amplification (LAMP) and strand displacement assays. Antibodies and antigens can be detected by indirect immunofluorescence assay test (IFA or IIFT), western blot (WB), ELISA or rapid diagnostic test (RDT) formats. Active infections can be diagnosed by virus isolation, PCR, LASV antigen positivity or IgM, along with clinical symptoms consistent with Lassa fever.
Diagnostic test types differ in complexity, infrastructure requirements and appropriateness for a rapid response. An overview of the implementation requirements for the range of diagnostics available for LASV is shown in table 1.
Table 1.
Diagnostics infrastructure comparison
| Test type | Lab infrastructure requirements (example) |
Training requirements (example) |
Turnaround time | Typical cost | In-house test | Commercial source |
| Virus isolation, neutralisation | HIGH (BSL-3/4) (reference laboratory) |
HIGH (advanced lab technician) |
7–10 days | – | >3 | – |
| NAAT reference (includes multiplex) |
HIGH (BSL-3/4) (reference laboratory) |
HIGH to MODERATE (advanced lab technician) |
2–3 hours 1–2 hours (prep) |
$30–$100 | >10 | 3–5 |
| NAAT POC | MODERATE/BSL-2 (district hospital) |
MODERATE (lab technician) |
1–2 hours | $15–$30 | – | – |
| ELISA, IFA/IIFT, WB | HIGH to MODERATE (regional lab, district hospital) |
MODERATE (lab technician) |
3–4 hours | $5–$15 | >10 | 1–2 |
| RDT | LOW (clinic, health centre, field settings) |
LOW (nurse, healthcare worker) |
<30 min | $1–$20 | – | 1 |
IFA/IIFT, indirect immunofluorescence assay test; NAAT, nucleic acid amplification test; POC, point of care; RDT, rapid diagnostic test; WB, western blot.
The majority of international laboratories use in-house LASV assays (see tables 2 and 3), with reports suggesting an equal number of published and unpublished protocols.104 Commercial assays for PCR and serology are available but are primarily labelled for research use only (see online supplementary tables S1 and S2).
Table 2.
In-house NAAT tests for LASV
| Assay type | Target | Multiplex | Detection | Detection limit | Reference |
| RT-PCR | S segment (GPC) | – | Acrylamide gel | 102 TCID50 | 110 |
| RT-PCR | S segment (GPC) | – | Agarose gel/ Southern blot | 101.6 TCID50 | 118 |
| RT-PCR | S segment (GPC) | – | Standard | 1–10 copies/assay | 45 109 144 |
| real-time RT-PCR (qRT-PCR) |
S segment (GPC) | – | SYBR-green | 8.6–16 copies/assay 1545–2835 copies/mL | 45 144 152 |
| RT-PCR | S segment (NP) | 10-plex | Mass spectrometry | 20 copies/assay | 138 |
| RT-PCR | S segment (NP) | >20-plex | Microarray | 1900 copies/assay | 139 |
| RT-PCR | L segment (LP) | – | Standard | 4290 copies/mL | 144 153 |
| RT-PCR | L segment (LP) | 8-plex | Standard | 1200 copies/assay 105 copies/mL | 137 |
| RT-PCR | S segment (GPC) | – | Standard | 4–30 copies/assay 342–2560 copies/mL | 45 107 144 |
| qRT-PCR | S segment (GPC), L segment (LP), | – | SYBR-green | n/a | 45 123 |
| One-step qRT-PCR | S segment (GPC, NP) | – | Standard | 234–583 copies | 45 141 |
| RT-PCR | S segment (GPC), L segment (LP) | – | Standard | 4290 copies/mL (serial assays) | 122 144 |
| RT-LAMP | S segment (GPC) | – | Turbidity | 100 copies | 154 |
| One-step RT-PCR | S segment (GPC) | – | Microarray | 1540 copies/mL | 45 155 |
| One-step qRT-PCR | S segment (GPC) | 4-plex | Standard | 45–150 copies/assay | 136 |
| One-step RT-PCR/LDR | L segment (LP) | 11-plex | Microarray | 100 copies/mL | 140 |
LAMP, loop-mediated isothermal amplification; LASV, Lassa fever virus; LDR, ligase detection reaction; NAAT, nucleic acid amplification test; RT, reverse transcriptase; q, quantitative.
Table 3.
In-house serological and antigen tests for LASV
| Assay type | Target | Lineage | Multiplex | Reference |
| ELISA | IgG, IgM, Ag | Lineage IV | – | 128 |
| Immunoblot | IgG, IgM | Lineage IV (NP) | – | 38 |
| ELISA | IgM, NP Ag | Lineage IV | – | 41 131 |
| ELISA | IgG, IgM | Lineage IV | – | 129 |
| ELISA | IgG, NP Ag | Strain AV (NP) | – | 133 |
| ELISA | IgG, IgM | Lineage III, IV, V | – | 106 |
| ELISA | NP Ag | Lineage IV | – | 7 |
| ELISA | IgG, IgM | Strain AV (NP) | – | 132 |
| RDT | NP Ag | Lineage IV | – | 7 45 |
| RDT | NP Ag | Lineage II, III, IV | – | 108 |
| Bead assay | IgG | Lineage IV (GP, NP) | 7-plex | 45 143 |
| Bead assay | IgM, NP, GP | Lineage IV | LASV/EBOV | 45 142 |
EBOV, Ebola virus; LASV, Lassa fever virus; RDT, rapid diagnostic test.
Lineage detection
Lassa lineage diversity must be considered in the choice of PCR primers and serology antigens, as test sensitivity relies on homology between the unknown specimen and the molecular probes or proteins used for detection. As more lineages and clades are revealed, and as mutations occur, tests will need to be revalidated and enriched to encompass variations observed at the nucleotide (20%–25%) and amino acid (8%–12%) levels.66 For some use cases, tests developed for a specific lineage or region may be appropriate; other cases may require detection of all known strains.62 105–108
Specimen diversity
LASV can be found in many body fluid compartments during infection besides blood, including urine,19 24 109 110 semen,11 32 44 45 cerebrospinal fluid,66 111 with evidence from throat swabs.41 111 There is evidence that acute infections detected in the CNS can report as negative in blood.112 LASV can persist in the central nervous system, urine and semen long after viral clearance in the blood, possibly in immunologically protected compartments.66 113 114
Viral titre is highly predictive of disease outcome.115 LASV RNA was detected as early as 3 days after infection in monkeys, and viral load peaked between 6 and 12 days in survivors but increased until death in fatally infected animals.116 For patients ill enough to be admitted to hospital, the average viral load in serum is 103–104 copies/mL, which clears within 21 days for survivors.66 109 117 118 Patients with viral load>108 copies/mL typically progress to multiorgan failure and shock, with viremia increasing to 108–109 copies/mL days before death;119 similar levels were found for cultured cell titres >103 TCID50/mL.111 120
Fatality has also been linked to suppressed or diminished IgM response throughout infection, supporting the hypothesis that an early and vigorous humoral response is critical to surviving infection.114 121 In monkeys, IgG response can appear as late as 12 days postinfection for both fatal and surviving cases, but titres were much higher in the survivors.116
Molecular diagnostics
Molecular diagnostic tests (PCR, LAMP and similar assays) are designed to detect highly conserved regions of the pathogen genome and are commonly used as the most sensitive method to detect active infection. Molecular diagnostic tests for LASV generally target the S genome segment encoding the GPC or the NP regions (table 2). Tests targeting the LP genome segment are non-specific for Old World arenavirus detection. Given the high mutation rate and genetic diversity of LASV, molecular diagnostic tests must encompass this sequence diversity or otherwise risk false negatives in mutated or ‘uncovered’ strains. Amplicons produced by the older tests can be sequenced to evaluate homology to known strains and new probes can be generated from sequencing data to improve lineage sensitivity.19 122 123 In some cases, multiple PCR assays could be performed to enhance coverage (see online supplementary table S1).
bmjgh-2018-001116supp002.pdf (411.8KB, pdf)
Serological and antigen detection assays
Serological tests can be used to detect IgM and IgG antibodies raised against LASV antigens as well as direct capture and detection of LASV antigens. LASV NP, GP and Z proteins have been shown to be immunogenic.124 125 IgM and antigen tests can be used to detect active infection, although not all patients have detectable IgM at the acute stage113 126 and both IgM and IgG antibodies can be immunosuppressed in severe cases.113 127 128 Particularly for endemic settings, IgG is generally used only for surveillance.11 129 130
LASV antigen detection has been demonstrated to be a robust method for detection of active infection. While genetic sequence diversity and minor genetic mutations typically have less of an impact on protein sequence, early antigen tests have shown some variation in LASV lineage sensitivity. In tandem, detection of LASV NP antigen and anti-LASV IgM demonstrated 88% sensitivity and 90% specificity for early stage infection, sufficient for diagnosis in ~90% of PCR-positive cases.108 131
Serology test platforms for LASV include IFA/IIFT, ELISA, WB, multiplex bead assays and one RDT (table 3). IFA tests using infected Vero cells were the traditional method for LASV serodiagnosis, but have been largely displaced by ELISA due to time and biosafety constraints.45 Most serological tests were developed using antigens from LASV lineage IV (Josiah strain), though the AV strain has also been used.132 133 A small number of ELISA kits are commercially available, although primarily marketed as research-use only (see online supplementary table S2).
bmjgh-2018-001116supp003.pdf (401KB, pdf)
Rapid tests
LASV RDTs, which leverage the same antibody/antigen capture agents as an ELISA but packaged in a stripped-down lateral flow format, can play an important role for patient care and outbreak response in outlying laboratories and clinics. A dipstick-based RDT for LASV has been developed that detects NP from fingerstick whole blood specimens.7 108 134 135 For LASV lineage IV (Josiah strain), the dipstick LASV RDT performed with good sensitivity (91% sensitivity, 86% specificity) compared with its progenitor ELISA (94% sensitivity, 84% specificity; both relative to qPCR). The monoclonal capture agents developed for this LASV RDT showed reduced sensitivity to LASV lineages II and III; improvements to the assay using a polyclonal approach suggest increased pan-lineage antigen sensitivity.108
Syndromic approach
At the early stages, the symptoms of Lassa fever can mimic other endemic diseases such as malaria, typhoid fever and other VHFs. A syndromic approach that tests for pathogens based on a syndrome such as VHF, using multiplex panels to quickly identify or eliminate likely pathogens from a single specimen, could be more effective in expediting LASV outbreak detection.
Several groups have demonstrated multiplex PCR assays for differential detection of VHF including Lassa. Real-time RT-PCR assays were developed for 28 VHFs to be processed as 4-plex reactions136 and 8-plex reactions.137 To overcome the limitations of fluorescence detection, combining RT-PCR with tagged primers can enable detection of 64 distinct species using mass spectrometry.138 Others have paired conventional assays with multiplex detection formats such as bead-based platforms139 140 and microarrays.140 A comprehensive set of 48 TaqMan-based PCR assays has been developed that can enable large-scale parallel processing of VHF agents.141 In addition, PCR panels for febrile agent and biothreat panels (5, 20 and 26-member panels including Lassa) are commercially available in bead-based and real-time TaqMan format (see online supplementary table S1).
A multiplex bead-based immunoassay platform for differential diagnosis using antigens and IgM for Lassa and Ebola demonstrated greater detection sensitivity for LASV GP antigen and IgM (25× and 5×, respectively) than conventional ELISA.142 A similar approach was used to survey pathogen exposure in West Africa for multiplex IgG detection of Lassa, Ebola, Marburg, Rift Valley fever and Crimean-Congo Haemorrhagic Fever as well as pan-flavivirus and pan-alphavirus.143
Challenges for Lassa diagnostics
As laboratory-developed protocols are generally not manufactured for distribution or regulated by an international agency, there remains a need for commercial pan-LASV tests with assured and reproducible quality that can be procured by any international laboratory or agency. The ideal LASV diagnostic test would detect all known LASV lineages with high sensitivity. While PCR enables the most sensitive platform for early detection, the high genetic diversity of LASV may result in false negatives for older tests, and to date, only a few attempts at pan-LASV PCR probe sets have been demonstrated. And while antigen tests are typically less dependent on minor genetic mutations, early results have shown some variation in lineage sensitivity, with further work underway to demonstrate pan-Lassa validation.108
Validation and quality assessment can be valuable in evaluating the entire testing process, including sample preparation, method of amplification or capture, detection methods and lineage sensitivity. Even for identical protocols, laboratory proficiency can significantly impact the quality of results. In 2004 and 2015, external quality assessments (EQAs) were conducted by the European Network for Diagnostics of Imported Viral Diseases using well-characterised Lassa panels for 24 international laboratories.104 144 Early LASV detection rates varied from 50% to 85.7%, with later improvement of 58% to 100%; however, 7 of 24 labs fell below good-to-acceptable performance and 11 labs reported false-negative results.
Sourcing specimens for clinical validation can be a major roadblock for both diagnostic development and quality control. International reference standards and proficiency panels could assist development and validation of diagnostic tests and also help improve laboratory proficiency and EQA efforts towards quality. Several international reference institutes, including the WHO International Biological Reference custodian laboratories, could be sources for specimens for development and validation (see online supplementary list S1). As new diagnostic tests are validated, these agencies could assist in routine EQA monitoring of test performance using up-to-date clinical specimen panels and reference standards.
bmjgh-2018-001116supp001.pdf (399.4KB, pdf)
Conclusion
Diagnostics are essential for the recognition and control of outbreaks of LASV, one of the most widespread and genetically diverse agents of VHF. Improved LASV tests are needed for endemic clinical management, outbreak response and vaccine and therapeutic clinical trials. Surveillance across all LASV lineages and regions will continue to be important to rapidly identify ‘hot spots’ for intervention and containment, and to monitor genetic and geographical shifts in both human and animal reservoir populations. For vaccine development, diagnostics are fundamental to detecting and differentiating an infection challenge from vaccine immunity. Finally, there is strong consensus that early diagnosis and treatment increases the survival rate from LASV infection.
This review has identified test resources for Lassa molecular diagnostics and serology and further described a large number of in-house LASV tests used across the international community. However, several gaps identified in the 2016 WHO R&D Blueprint remain.8 145 Efforts should be made to advance existing diagnostic platforms towards clinical validation and regulatory approval. Implementation of diagnostics could be further refined with more detailed understanding of LASV kinetics across a range of sample types, and a more robust point-of-care or field-appropriate design where appropriate. Tests that are currently available could benefit from EQA with the goal of standardisation of test sensitivity, specificity and lineage/regional coverage.
Given the challenges identified for LASV detection in endemic and often low-resourced settings, rapid on-site diagnosis of suspect cases may bridge the gap. A range of assays have been already developed for commercial point-of-care platforms, thus it is possible that current LASV assays could be adapted into a cartridge-based format. The combination of rapid, point-of-care antigen detection with a point-of-care confirmatory test has been suggested as an ideal implementation for early case detection and outbreak response.108 146 147
Each of the aforementioned diagnostic use scenarios have different performance requirements for text complexity, sensitivity, specificity and turnaround time. Development of new and improved LASV diagnostics could be facilitated by a clearly defined set of use cases to describe where and how diagnostic tests are most needed, coupled with TPPs with detailed performance characteristics for the highest priority molecular and immunodiagnostics.148 While numerous barriers to achieving regulatory approval exist,142 initiatives from WHO and other organisations such as the Foundation for Innovative New Diagnostics (FIND) and CEPI are intended to make development and commercialisation of LASV diagnostics more feasible.86 148–150
Acknowledgments
We gratefully acknowledge all those who attended the WHO R&D Blueprint Roadmaps: Consultation on Lassa fever in January 2018. Editorial assistance for later drafts was provided by Rachel Wright, PhD, funded by FIND, according to Good Publication Practice guidelines (http://annals.org/aim/fullarticle/2424869/good-publication-practice-communicating-company-sponsored-medical-research-gpp3).
Footnotes
Handling editor: Seye Abimbola
Contributors: LTM contributed to drafting the manuscript and provided background research for the manuscript. CK-C contributed insight into diagnostic needs for outbreak pathogens. All authors reviewed, edited and approved the final version of the manuscript.
Funding: FIND was funded for this work by UK aid from the UK Government.
Disclaimer: The opinions expressed in this article are those of the authors and do not necessarily reflect those of the institutions or organisations with which they are affiliated.
Competing interests: None declared.
Patient consent: Not required.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data sharing statement: No additional data are available.
References
- 1. CDC , 2017. Lassa fever. Available from: https://www.cdc.gov/vhf/lassa/index.html [accessed 26 Oct 2018].
- 2. WHO , 2017. Lassa fever. Available from: http://www.who.int/csr/disease/lassafever/en/ [accessed 26 Oct 2018].
- 3. WHO , 2017. Lassa fact sheet. Available from: http://www.who.int/mediacentre/factsheets/fs179/en/ [accessed 26 Oct 2018].
- 4. Nigeria Centre for Disease Control , 2018. Available from: http://www.ncdc.gov.ng/diseases/sitreps/?cat=5&name=An%20update%20of%20Lassa%20fever%20outbreak%20in%20Nigeria [accessed 26 Oct 2018].
- 5. WHO , 2018. Lassa fever outbreak news. Available from: http://www.who.int/csr/don/archive/disease/lassa_fever/en/ [accessed 26 Oct 2018].
- 6. WHO , 2017. Lassa Fever – Nigeria. Available from: http://www.who.int/csr/don/28-june-2017-lassa-fever-nigeria/en/ [accessed 26 Oct 2018].
- 7. Shaffer JG, Grant DS, Schieffelin JS, et al. . Lassa fever in post-conflict sierra leone. PLoS Negl Trop Dis 2014;8:e2748 10.1371/journal.pntd.0002748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. WHO , 2017. R&D Blueprint for action to prevent epidemics. Available from: http://www.who.int/blueprint/en/ [accessed 26 Oct 2018].
- 9. Mehand MS, Millett P, Al-Shorbaji F, et al. . World Health Organization Methodology to Prioritize Emerging Infectious Diseases in Need of Research and Development. Emerg Infect Dis 2018;24 10.3201/eid2409.171427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. WHO , 2017. Target Product Profiles (TPPs). Available from: http://www.who.int/immunization/research/ppc-tpp/target_product_profiles/en/ [accessed 26 Oct 2018].
- 11. Frame JD. Surveillance of Lassa fever in missionaries stationed in West Africa. Bull World Health Organ 1975;52:593–8. [PMC free article] [PubMed] [Google Scholar]
- 12. Patassi AA, Landoh DE, Mebiny-Essoh Tchalla A, et al. . Emergence of Lassa Fever Disease in Northern Togo: Report of Two Cases in Oti District in 2016. Case Rep Infect Dis 2017;2017:1–5. 10.1155/2017/8242313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lehmann C, Kochanek M, Abdulla D, et al. . Control measures following a case of imported Lassa fever from Togo, North Rhine Westphalia, Germany, 2016. Euro Surveill 2017;22 10.2807/1560-7917.ES.2017.22.39.17-00088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Patassi AA, Landoh DE, Mebiny-Essoh Tchalla A, et al. . Emergence of lassa fever disease in northern togo: report of two cases in oti district in 2016. Case Rep Infect Dis 2017;2017:1–5. 10.1155/2017/8242313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ehlkes L, George M, Samosny G, et al. . Management of a Lassa fever outbreak, Rhineland-Palatinate, Germany, 2016. Euro Surveill 2017;22 10.2807/1560-7917.ES.2017.22.39.16-00728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Günther S, et al. Imported lassa fever in Germany: molecular characterization of a new lassa virus strain. Emerg Infect Dis 2000;6:466–76. 10.3201/eid0605.000504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. van Paassen J, Bauer MP, van Dissel JV, et al. . Viral haemorrhagic fever in returned travellers; a review on clinical symptoms, management, and outbreak prevention 2010.
- 18. Swaan CM, van den Broek PJ, Wijnands S, et al. . Management of viral haemorrhagic fever in the Netherlands. Euro Surveill 2002;7:48–50. 10.2807/esm.07.03.00338-en [DOI] [PubMed] [Google Scholar]
- 19. Grahn A, Bråve A, Lagging M, et al. . Imported Case of Lassa Fever in Sweden With Encephalopathy and Sensorineural Hearing Deficit. Open Forum Infect Dis 2016;3:ofw198 10.1093/ofid/ofw198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Amorosa V, MacNeil A, McConnell R, et al. . Imported Lassa Fever, Pennsylvania, USA, 2010. Emerg Infect Dis 2010;16:1598–600. 10.3201/eid1610.100774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Centers for Disease Control and Prevention (CDC) Imported Lassa fever--New Jersey, 2004. MMWR Morb Mortal Wkly Rep 2004;53:894–7. [PubMed] [Google Scholar]
- 22. Holmes GP, McCormick JB, Trock SC, et al. . Lassa fever in the United States. Investigation of a case and new guidelines for management. N Engl J Med 1990;323:1120–3. [DOI] [PubMed] [Google Scholar]
- 23. Atkin S, Anaraki S, Gothard P, et al. . The first case of Lassa fever imported from Mali to the United Kingdom, February 2009. Euro Surveill 2009;14:19145. [PubMed] [Google Scholar]
- 24. Kitching A, Addiman S, Cathcart S, et al. . A fatal case of Lassa fever in London, January 2009. Euro Surveill 2009;14:19117. [PubMed] [Google Scholar]
- 25. Hirabayashi Y, Oka S, Goto H, et al. . An imported case of Lassa fever with late appearance of polyserositis. J Infect Dis 1988;158:872–5. 10.1093/infdis/158.4.872 [DOI] [PubMed] [Google Scholar]
- 26. Hewson R. Lessons learnt from imported cases and onward transmission of Lassa fever in Europe support broader management of viral haemorrhagic fevers. Euro Surveill 2017;22 10.2807/1560-7917.ES.2017.22.39.17-00661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Macher AM, Wolfe MS. Historical Lassa fever reports and 30-year clinical update. Emerg Infect Dis 2006;12:835–7. 10.3201/eid1205.050052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Spira AM. Assessment of travellers who return home ill. The Lancet 2003;361:1459–69. 10.1016/S0140-6736(03)13141-8 [DOI] [PubMed] [Google Scholar]
- 29. Demby AH, Inapogui A, Kargbo K, et al. . Lassa fever in Guinea: II. Distribution and prevalence of Lassa virus infection in small mammals. Vector-Borne and Zoonotic Diseases 2001;1:283–97. 10.1089/15303660160025912 [DOI] [PubMed] [Google Scholar]
- 30. McCormick JB, Webb PA, Krebs JW, et al. . A prospective study of the epidemiology and ecology of Lassa fever. J Infect Dis 1987;155:437–44. 10.1093/infdis/155.3.437 [DOI] [PubMed] [Google Scholar]
- 31. Monath TP, Newhouse VF, Kemp GE, et al. . Lassa virus isolation from Mastomys natalensis rodents during an epidemic in Sierra Leone. Science 1974;185:263–5. 10.1126/science.185.4147.263 [DOI] [PubMed] [Google Scholar]
- 32. Richmond JK, Baglole DJ. Lassa fever: epidemiology, clinical features, and social consequences. BMJ 2003;327:1271–5. 10.1136/bmj.327.7426.1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mariën J, Borremans B, Gryseels S, et al. . No measurable adverse effects of Lassa, Morogoro and Gairo arenaviruses on their rodent reservoir host in natural conditions. Parasit Vectors 2017;10:210 10.1186/s13071-017-2146-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. McCormick JB, King IJ, Webb PA, et al. . A case-control study of the clinical diagnosis and course of Lassa fever. J Infect Dis 1987;155:445–55. 10.1093/infdis/155.3.445 [DOI] [PubMed] [Google Scholar]
- 35. Mylne AQN, Pigott DM, Longbottom J, et al. . Mapping the zoonotic niche of Lassa fever in Africa. Trans R Soc Trop Med Hyg 2015;109:483–92. 10.1093/trstmh/trv047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Olayemi A, Cadar D, Magassouba N, et al. . New Hosts of The Lassa Virus. Sci Rep 2016;6:25280 10.1038/srep25280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Olayemi A, Oyeyiola A, Obadare A, et al. . Widespread arenavirus occurrence and seroprevalence in small mammals, Nigeria. Parasit Vectors 2018;11:416 10.1186/s13071-018-2991-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. ter Meulen J, Koulemou K, Wittekindt T, et al. . Detection of Lassa virus antinucleoprotein immunoglobulin G (IgG) and IgM antibodies by a simple recombinant immunoblot assay for field use. J Clin Microbiol 1998;36:3143–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bonney JHK, Nyarko EO, Ohene S-A, et al. . Molecular confirmation of Lassa fever imported into Ghana. Afr J Lab Med 2016;5:288 10.4102/ajlm.v5i1.288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Fisher-Hoch SP, Tomori O, Nasidi A, et al. . Review of cases of nosocomial Lassa fever in Nigeria: the high price of poor medical practice. BMJ 1995;311:857–9. 10.1136/bmj.311.7009.857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Haas WH, Breuer T, Pfaff G, et al. . Imported Lassa fever in Germany: surveillance and management of contact persons. Clin Infect Dis 2003;36:1254–8. 10.1086/374853 [DOI] [PubMed] [Google Scholar]
- 42. Iroezindu MO, Unigwe US, Okwara CC, et al. . Lessons learnt from the management of a case of Lassa fever and follow-up of nosocomial primary contacts in Nigeria during Ebola virus disease outbreak in West Africa. Trop Med Int Health 2015;20:1424–30. 10.1111/tmi.12565 [DOI] [PubMed] [Google Scholar]
- 43. Johnson KM, Monath TP. Imported Lassa fever-reexamining the algorithms. N Engl J Med Overseas Ed 1990;323:1139–41. 10.1056/NEJM199010183231611 [DOI] [PubMed] [Google Scholar]
- 44. McElroy AK, Akondy RS, Harmon JR, et al. . A case of human Lassa virus infection with robust acute T-cell activation and long-term virus-specific t-cell responses. J Infect Dis 2017;215:1862–72. 10.1093/infdis/jix201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Raabe V, Koehler J. Laboratory diagnosis of Lassa fever. J Clin Microbiol 2017;55:1629–37. 10.1128/JCM.00170-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Eisen RJ, Atiku LA, Boegler KA, et al. . An evaluation of removal trapping to control rodents inside homes in a plague-endemic region of rural northwestern uganda. Vector-Borne and Zoonotic Diseases 2018;18:458–63. 10.1089/vbz.2018.2276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Inegbenebor U, Okosun J, Inegbenebor J. Prevention of lassa fever in Nigeria. Trans R Soc Trop Med Hyg 2010;104:51–4. 10.1016/j.trstmh.2009.07.008 [DOI] [PubMed] [Google Scholar]
- 48. Ajayi NA, Nwigwe CG, Azuogu BN, et al. . Containing a Lassa fever epidemic in a resource-limited setting: outbreak description and lessons learned from Abakaliki, Nigeria (January–March 2012). Int J Infect Dis 2013;17:e1011–6. 10.1016/j.ijid.2013.05.015 [DOI] [PubMed] [Google Scholar]
- 49. Ehichioya DU, Hass M, Ölschläger S, et al. . Lassa fever, Nigeria, 2005–2008. Emerg Infect Dis 2010;16:1040–1. 10.3201/eid1606.100080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ijarotimi IT, Ilesanmi OS, Aderinwale A, et al. . Knowledge of Lassa fever and use of infection prevention and control facilities among health care workers during Lassa fever outbreak in Ondo State, Nigeria. Pan Afr Med J 2018;30:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Grahn A, Bråve A, Tolfvenstam T, et al. . Absence of Nosocomial Transmission of Imported Lassa Fever during Use of Standard Barrier Nursing Methods. Emerg Infect Dis 2018;24:978–87. 10.3201/eid2406.172097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. CDC , 2017. Ebola hemorrhagic fever. Available from: https://www.cdc.gov/vhf/ebola/index.html [accessed 26 Oct 2018].
- 53. WHO , 2017. Integrated disease surveillance and response in the African Region. Available from: http://www.who.int/csr/resources/publications/ebola/disease-surveillance-afro/en/ [accessed 26 Oct 2018].
- 54. Gautret P, Okolo S, Elachola H, et al. . Mitigating the risks of global spread of Lassa fever at the 2018 Hajj pilgrimage. Travel Med Infect Dis 2018;23:99–100. 10.1016/j.tmaid.2018.04.016 [DOI] [PubMed] [Google Scholar]
- 55. WHO , 2017. Regional Office for Africa. WHO calls for early diagnostic tests for Lassa fever. Available from: http://www.afro.who.int/news/who-calls-early-diagnostic-tests-lassa-fever [accessed 26 Oct 2018].
- 56. Isere EE, Fatiregun AA, Ilesanmi O, et al. . Lessons learnt from epidemiological investigation of Lassa Fever outbreak in a southwest state of Nigeria December 2015 to April 2016. PLoS Curr 2018;10 10.1371/currents.outbreaks.bc4396a6650d0ed1985d731583bf5ded [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Fatiregun A, Isere E. Epidemic preparedness and management: a guide on Lassa fever outbreak preparedness plan. Niger Med J 2017;58:1–6. 10.4103/0300-1652.218414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Tambo E, Adetunde OT, Olalubi OA. Re-emerging Lassa fever outbreaks in Nigeria: Re-enforcing “One Health” community surveillance and emergency response practice. Infect Dis Poverty 2018;7:37 10.1186/s40249-018-0421-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Akpede GO, Asogun DA, Okogbenin SA, et al. . Lassa fever outbreaks in Nigeria. Expert Rev Anti Infect Ther 2018;16:663–6. 10.1080/14787210.2018.1512856 [DOI] [PubMed] [Google Scholar]
- 60. The Lancet Infectious Diseases Lassa fever and global health security. Lancet Infect Dis 2018;18:357 10.1016/S1473-3099(18)30179-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lamunu M, Lutwama JJ, Kamugisha J, et al. . Containing a haemorrhagic fever epidemic: the Ebola experience in Uganda (October 2000–January 2001). Int J Infect Dis 2004;8:27–37. 10.1016/j.ijid.2003.04.001 [DOI] [PubMed] [Google Scholar]
- 62. Ogbu O, Ajuluchukwu E, Uneke CJ. Lassa fever in West African sub-region: an overview. J Vector Borne Dis 2007;44:1–11. [PubMed] [Google Scholar]
- 63. Asogun DA, Adomeh DI, Ehimuan J, et al. . Molecular diagnostics for lassa fever at Irrua specialist teaching hospital, Nigeria: lessons learnt from two years of laboratory operation. PLoS Negl Trop Dis;6:e1839 10.1371/journal.pntd.0001839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Price ME, Fisher-Hoch SP, Craven RB, et al. . A prospective study of maternal and fetal outcome in acute Lassa fever infection during pregnancy. BMJ 1988;297:584–7. 10.1136/bmj.297.6648.584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Okokhere P, Colubri A, Azubike C, et al. . Clinical and laboratory predictors of Lassa fever outcome in a dedicated treatment facility in Nigeria: a retrospective, observational cohort study. Lancet Infect Dis 2018;18:684–95. 10.1016/S1473-3099(18)30121-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Günther S, Weisner B, Roth A, et al. . Lassa fever encephalopathy: Lassa virus in cerebrospinal fluid but not in serum. J Infect Dis 2001;184:345–9. 10.1086/322033 [DOI] [PubMed] [Google Scholar]
- 67. White HA. Lassa fever. A study of 23 hospital cases. Trans R Soc Trop Med Hyg 1972;66:390–8. 10.1016/0035-9203(72)90269-6 [DOI] [PubMed] [Google Scholar]
- 68. Andersen KG, Shapiro BJ, Matranga CB, et al. . Clinical Sequencing uncovers origins and evolution of Lassa Virus. Cell 2015;162:738–50. 10.1016/j.cell.2015.07.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Attar N. Viral evolution: the history of Lassa virus. Nat Rev Microbiol 2015;13:600. [DOI] [PubMed] [Google Scholar]
- 70. Leski TA, Stockelman MG, Moses LM, et al. . Sequence variability and geographic distribution of Lassa virus, Sierra Leone. Emerg Infect Dis 2015;21:609–18. 10.3201/eid2104.141469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Manning JT, Forrester N, Paessler S. Lassa virus isolates from Mali and the Ivory Coast represent an emerging fifth lineage. Front Microbiol 2015;6:1037 10.3389/fmicb.2015.01037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Whitmer SLM, Strecker T, Cadar D, et al. . New lineage of Lassa virus, Togo, 2016. Emerg Infect Dis 2018;24:599–602. 10.3201/eid2403.171905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Cross RW, Mire CE, Branco LM, et al. . Treatment of Lassa virus infection in outbred guinea pigs with first-in-class human monoclonal antibodies. Antiviral Res 2016;133:218–22. 10.1016/j.antiviral.2016.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Oestereich L, Rieger T, Lüdtke A, et al. . Efficacy of favipiravir alone and in combination with ribavirin in a Lethal, immunocompetent mouse model of Lassa fever. J Infect Dis 2016;213:934–8. 10.1093/infdis/jiv522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Raabe VN, Kann G, Ribner BS, et al. . Favipiravir and ribavirin treatment of epidemiologically linked cases of Lassa fever. Clin Infect Dis 2017;65:855–9. 10.1093/cid/cix406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Safronetz D, Rosenke K, Westover JB, et al. . The broad-spectrum antiviral favipiravir protects guinea pigs from lethal Lassa virus infection post-disease onset. Sci Rep 2015;5:14775 10.1038/srep14775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Sepúlveda CS, García CC, Damonte EB. Determining the virus life-cycle stage blocked by an antiviral. Methods Mol Biol Clifton NJ 2018;1604:371–92. [DOI] [PubMed] [Google Scholar]
- 78. Rosenke K, Feldmann H, Westover JB, et al. . Use of favipiravir to treat Lassa virus infection in Macaques. Emerg Infect Dis 2018;24:1696–9. 10.3201/eid2409.180233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Cashman KA, Smith MA, Twenhafel NA, et al. . Evaluation of Lassa antiviral compound ST-193 in a guinea pig model. Antiviral Res 2011;90:70–9. 10.1016/j.antiviral.2011.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Larson RA, Dai D, Hosack VT, et al. . Identification of a broad-spectrum arenavirus entry inhibitor. J Virol 2008;82:10768–75. 10.1128/JVI.00941-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Burgeson JR, Gharaibeh DN, Moore AL, et al. . Lead optimization of an acylhydrazone scaffold possessing antiviral activity against Lassa virus. Bioorg Med Chem Lett 2013;23:5840–3. 10.1016/j.bmcl.2013.08.103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Müller S, Günther S. Broad-spectrum antiviral activity of small interfering RNA targeting the conserved RNA termini of Lassa virus. Antimicrob Agents Chemother 2007;51:2215–8. 10.1128/AAC.01368-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Robinson JE, Hastie KM, Cross RW, et al. . Most neutralizing human monoclonal antibodies target novel epitopes requiring both Lassa virus glycoprotein subunits. Nat Commun 2016;7:11544 10.1038/ncomms11544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Mire CE, Cross RW, Geisbert JB, et al. . Human-monoclonal-antibody therapy protects nonhuman primates against advanced Lassa fever. Nat Med 2017;23:1146–9. 10.1038/nm.4396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Hastie KM, Zandonatti MA, Kleinfelter LM, et al. . Structural basis for antibody-mediated neutralization of Lassa virus. Science 2017;356:923–8. 10.1126/science.aam7260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Plotkin SA. Vaccines for epidemic infections and the role of CEPI. Hum Vaccin Immunother 2017;13:2755–62. 10.1080/21645515.2017.1306615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Røttingen J-A, Gouglas D, Feinberg M, et al. . New Vaccines against Epidemic Infectious Diseases. N Engl J Med Overseas Ed 2017;376:610–3. 10.1056/NEJMp1613577 [DOI] [PubMed] [Google Scholar]
- 88. Branco LM, Grove JN, Geske FJ, et al. . Lassa virus-like particles displaying all major immunological determinants as a vaccine candidate for Lassa hemorrhagic fever. Virol J 2010;7:279 10.1186/1743-422X-7-279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Kainulainen MH, Spengler JR, Welch SR, et al. . Use of a Scalable Replicon-Particle Vaccine to Protect Against Lethal Lassa Virus Infection in the Guinea Pig Model. J Infect Dis 2018;217:1957–66. 10.1093/infdis/jiy123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Cashman KA, Wilkinson ER, Shaia CI, et al. . A DNA vaccine delivered by dermal electroporation fully protects cynomolgus macaques against Lassa fever. Hum Vaccin Immunother 2017;13:2902–11. 10.1080/21645515.2017.1356500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Rodriguez-Carreno MP, Nelson MS, Botten J, et al. . Evaluating the immunogenicity and protective efficacy of a DNA vaccine encoding Lassa virus nucleoprotein. Virology 2005;335:87–98. 10.1016/j.virol.2005.01.019 [DOI] [PubMed] [Google Scholar]
- 92. McCormick JB, Mitchell SW, Kiley MP, et al. . Inactivated Lassa virus elicits a non protective immune response in rhesus monkeys. J Med Virol 1992;37:1–7. 10.1002/jmv.1890370102 [DOI] [PubMed] [Google Scholar]
- 93. Bredenbeek PJ, Molenkamp R, Spaan WJM, et al. . A recombinant Yellow Fever 17D vaccine expressing Lassa virus glycoproteins. Virology 2006;345:299–304. 10.1016/j.virol.2005.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Jiang X, Dalebout TJ, Bredenbeek PJ, et al. . Yellow fever 17D-vectored vaccines expressing Lassa virus GP1 and GP2 glycoproteins provide protection against fatal disease in guinea pigs. Vaccine 2011;29:1248–57. 10.1016/j.vaccine.2010.11.079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Ewer K, Sebastian S, Spencer AJ, et al. . Chimpanzee adenoviral vectors as vaccines for outbreak pathogens. Hum Vaccin Immunother 2017;13:3020–32. 10.1080/21645515.2017.1383575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Fisher-Hoch SP, McCormick JB, Auperin D, et al. . Protection of rhesus monkeys from fatal Lassa fever by vaccination with a recombinant vaccinia virus containing the Lassa virus glycoprotein gene. Proc Natl Acad Sci U S A 1989;86:317–21. 10.1073/pnas.86.1.317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Geisbert TW, Jones S, Fritz EA, et al. . Development of a new vaccine for the prevention of Lassa fever. PLoS Med 2005;2:e183 10.1371/journal.pmed.0020183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Safronetz D, Mire C, Rosenke K, et al. . A recombinant vesicular stomatitis virus-based Lassa fever vaccine protects guinea pigs and macaques against challenge with geographically and genetically distinct Lassa viruses. PLoS Negl Trop Dis 2015;9:e0003736 10.1371/journal.pntd.0003736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Carnec X, Matéo M, Page A, et al. . A vaccine platform against arenaviruses based on a recombinant hyper-attenuated Mopeia virus expressing heterologous glycoproteins. J Virol 2018;92 10.1128/JVI.02230-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Lukashevich IS, Patterson J, Carrion R, et al. . A live attenuated vaccine for Lassa fever made by reassortment of Lassa and Mopeia viruses. J Virol 2005;79:13934–42. 10.1128/JVI.79.22.13934-13942.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Zapata JC, Goicochea M, Nadai Y, et al. . Genetic variation in vitro and in vivo of an attenuated Lassa vaccine candidate. J Virol 2014;88:3058–66. 10.1128/JVI.03035-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Lukashevich IS, Pushko P. Vaccine platforms to control Lassa fever. Expert Rev Vaccines 2016;15:1135–50. 10.1080/14760584.2016.1184575 [DOI] [PubMed] [Google Scholar]
- 103. Warner BM, Safronetz D, Stein DR. Current research for a vaccine against Lassa hemorrhagic fever virus. Drug Des Devel Ther 2018;12:2519–27. 10.2147/DDDT.S147276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Niedrig M, Schmitz H, Becker S, et al. . First international quality assurance study on the rapid detection of viral agents of bioterrorism. J Clin Microbiol 2004;42:1753–5. 10.1128/JCM.42.4.1753-1755.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Bowen MD, Rollin PE, Ksiazek TG, et al. . Genetic diversity among Lassa virus strains. J Virol 2000;74:6992–7004. 10.1128/JVI.74.15.6992-7004.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Emmerich P, Günther S, Schmitz H. Strain-specific antibody response to Lassa virus in the local population of west Africa. J Clin Virol 2008;42:40–4. 10.1016/j.jcv.2007.11.019 [DOI] [PubMed] [Google Scholar]
- 107. Olschläger S, Lelke M, Emmerich P, et al. . Improved detection of Lassa virus by reverse transcription-PCR targeting the 5' region of S RNA. J Clin Microbiol 2010;48:2009–13. 10.1128/JCM.02351-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Boisen ML, Hartnett JN, Shaffer JG, et al. . Field validation of recombinant antigen immunoassays for diagnosis of Lassa fever. Sci Rep 2018;8:5939 10.1038/s41598-018-24246-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Demby AH, Chamberlain J, Brown DW, et al. . Early diagnosis of Lassa fever by reverse transcription-PCR. J Clin Microbiol 1994;32:2898–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Lunkenheimer K, Hufert FT, Schmitz H. Detection of Lassa virus RNA in specimens from patients with Lassa fever by using the polymerase chain reaction. J Clin Microbiol 1990;28:2689–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Johnson KM, McCormick JB, Webb PA, et al. . Clinical virology of Lassa fever in hospitalized patients. J Infect Dis 1987;155:456–64. 10.1093/infdis/155.3.456 [DOI] [PubMed] [Google Scholar]
- 112. Okokhere PO, Erameh CO, Alikah F, et al. . Acute Lassa Virus encephalitis with Lassa virus in the cerebrospinal fluid but absent in the blood: a case report with a positive outcome. Case Rep Neurol 2018;10:150–8. 10.1159/000490374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Branco LM, Grove JN, Boisen ML, et al. . Emerging trends in Lassa fever: redefining the role of immunoglobulin M and inflammation in diagnosing acute infection. Virol J 2011;8:478 10.1186/1743-422X-8-478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Fischer WA, Wohl DA. Moving Lassa fever research and care into the 21st century. J Infect Dis 2017;215:1779–81. 10.1093/infdis/jix206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Yun NE, Walker DH. Pathogenesis of Lassa fever. Viruses 2012;4:2031–48. 10.3390/v4102031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Baize S, Marianneau P, Loth P, et al. . Early and strong immune responses are associated with control of viral replication and recovery in lassa virus-infected cynomolgus monkeys. J Virol 2009;83:5890–903. 10.1128/JVI.01948-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Panning M, Emmerich P, Olschläger S, et al. . Laboratory diagnosis of Lassa fever, liberia. Emerg Infect Dis 2010;16:1041–3. 10.3201/eid1606.100040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Trappier SG, Conaty AL, Farrar BB, et al. . Evaluation of the polymerase chain reaction for diagnosis of Lassa virus infection. Am J Trop Med Hyg 1993;49:214–21. 10.4269/ajtmh.1993.49.214 [DOI] [PubMed] [Google Scholar]
- 119. Schmitz H, Köhler B, Laue T, et al. . Monitoring of clinical and laboratory data in two cases of imported Lassa fever. Microbes Infect 2002;4:43–50. 10.1016/S1286-4579(01)01508-8 [DOI] [PubMed] [Google Scholar]
- 120. McCormick JB, King IJ, Webb PA, et al. . Lassa fever. Effective therapy with ribavirin. N Engl J Med 1986;314:20–6. 10.1056/NEJM198601023140104 [DOI] [PubMed] [Google Scholar]
- 121. Prescott JB, Marzi A, Safronetz D, et al. . Immunobiology of Ebola and Lassa virus infections. Nat Rev Immunol 2017;17:195–207. 10.1038/nri.2016.138 [DOI] [PubMed] [Google Scholar]
- 122. Coulibaly-N'Golo D, Allali B, Kouassi SK, et al. . Novel arenavirus sequences in Hylomyscus sp. and Mus (Nannomys) setulosus from Côte d'Ivoire: implications for evolution of arenaviruses in Africa. PLoS One 2011;6:e20893 10.1371/journal.pone.0020893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Safronetz D, Lopez JE, Sogoba N, et al. . Detection of Lassa virus, Mali. Emerg Infect Dis 2010;16:1123–6. 10.3201/eid1607.100146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Günther S, Lenz O, virus L. Lassa virus. Crit Rev Clin Lab Sci 2004;41:339–90. 10.1080/10408360490497456 [DOI] [PubMed] [Google Scholar]
- 125. Günther S, Kühle O, Rehder D, et al. . Antibodies to Lassa virus Z protein and nucleoprotein co-occur in human sera from Lassa fever endemic regions. Med Microbiol Immunol 2001;189:225–9. 10.1007/s004300100061 [DOI] [PubMed] [Google Scholar]
- 126. Wulff H, Johnson KM. Immunoglobulin M and G responses measured by immunofluorescence in patients with Lassa or Marburg virus infections. Bull World Health Organ 1979;57:631–5. [PMC free article] [PubMed] [Google Scholar]
- 127. Bond N, Schieffelin JS, Moses LM, et al. . A historical look at the first reported cases of Lassa fever: IgG antibodies 40 years after acute infection. Am J Trop Med Hyg 2013;88:241–4. 10.4269/ajtmh.12-0466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Niklasson BS, Jahrling PB, Peters CJ. Detection of Lassa virus antigens and Lassa virus-specific immunoglobulins G and M by enzyme-linked immunosorbent assay. J Clin Microbiol 1984;20:239–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Emmerich P, Thome-Bolduan C, Drosten C, et al. . Reverse ELISA for IgG and IgM antibodies to detect Lassa virus infections in Africa. J Clin Virol 2006;37:277–81. 10.1016/j.jcv.2006.08.015 [DOI] [PubMed] [Google Scholar]
- 130. Sogoba N, Rosenke K, Adjemian J, et al. . Lassa virus seroprevalence in sibirilia commune, Bougouni District, Southern Mali. Emerg Infect Dis 2016;22:657–63. 10.3201/eid2204.151814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Bausch DG, Rollin PE, Demby AH, et al. . Diagnosis and clinical virology of Lassa fever as evaluated by enzyme-linked immunosorbent assay, indirect fluorescent-antibody test, and virus isolation. J Clin Microbiol 2000;38:2670–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Gabriel M, Adomeh DI, Ehimuan J, et al. . Development and evaluation of antibody-capture immunoassays for detection of Lassa virus nucleoprotein-specific immunoglobulin M and G. PLoS Negl Trop Dis 2018;12:e0006361 10.1371/journal.pntd.0006361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Saijo M, Georges-Courbot MC, Marianneau P, et al. . Development of recombinant nucleoprotein-based diagnostic systems for Lassa fever. Clin Vaccine Immunol 2007;14:1182–9. 10.1128/CVI.00101-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Boisen ML, Oottamasathien D, Jones AB, et al. . Development of prototype filovirus recombinant antigen immunoassays. J Infect Dis 2015;212(Suppl 2):S359–67. 10.1093/infdis/jiv353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Boisen ML, Cross RW, Hartnett JN, et al. . Field Validation of the ReEBOV antigen rapid test for point-of-care diagnosis of ebola virus infection. J Infect Dis 2016;214(suppl 3):S203–9. 10.1093/infdis/jiw261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Pang Z, Li A, Li J, et al. . Comprehensive multiplex one-step real-time TaqMan qRT-PCR assays for detection and quantification of hemorrhagic fever viruses. PLoS One 2014;9:e95635 10.1371/journal.pone.0095635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. He J, Kraft AJ, Fan J, et al. . Simultaneous detection of CDC category "A" DNA and RNA Bioterrorism agents by use of multiplex PCR & RT-PCR enzyme hybridization assays. Viruses 2009;1:441–59. 10.3390/v1030441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Palacios G, Briese T, Kapoor V, et al. . MassTag polymerase chain reaction for differential diagnosis of viral hemorrhagic fever. Emerg Infect Dis 2006;12:692–5. 10.3201/eid1204.051515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Palacios G, Quan PL, Jabado OJ, et al. . Panmicrobial oligonucleotide array for diagnosis of infectious diseases. Emerg Infect Dis 2007;13:73–81. 10.3201/eid1301.060837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Das S, Rundell MS, Mirza AH, et al. . A multiplex PCR/LDR assay for the simultaneous identification of category a infectious pathogens: agents of viral hemorrhagic fever and variola virus. PLoS One 2015;10:e0138484 10.1371/journal.pone.0138484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Trombley AR, Wachter L, Garrison J, et al. . Comprehensive panel of real-time TaqMan polymerase chain reaction assays for detection and absolute quantification of filoviruses, arenaviruses, and New World hantaviruses. Am J Trop Med Hyg 2010;82:954–60. 10.4269/ajtmh.2010.09-0636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Satterly NG, Voorhees MA, Ames AD, et al. . Comparison of MagPix Assays and Enzyme-Linked Immunosorbent Assay for Detection of Hemorrhagic Fever Viruses. J Clin Microbiol 2017;55:68–78. 10.1128/JCM.01693-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. O'Hearn AE, Voorhees MA, Fetterer DP, et al. . Serosurveillance of viral pathogens circulating in West Africa. Virol J 2016;13:163 10.1186/s12985-016-0621-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Nikisins S, Rieger T, Patel P, et al. . International external quality assessment study for molecular detection of Lassa virus. PLoS Negl Trop Dis 2015;9:e0003793 10.1371/journal.pntd.0003793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. WHO , 2017. Accelerating R&D processes. Available from: http://www.who.int/blueprint/what/research-development/en/ [12 Sep 2018].
- 146. Peeling RW, Boeras DI, Nkengasong J. Re-imagining the future of diagnosis of Neglected Tropical Diseases. Comput Struct Biotechnol J 2017;15:271–4. 10.1016/j.csbj.2017.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Modjarrad K, Moorthy VS, Ben Embarek P, et al. . A roadmap for MERS-CoV research and product development: report from a World Health Organization consultation. Nat Med 2016;22:701–5. 10.1038/nm.4131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. WHO , 2018. Lassa Fever Research and Development (R&D) Roadmap. Available from: http://www.who.int/blueprint/priority-diseases/key-action/LassaFever_Draft_Roadmap_publiccomment_MAY2018.pdf [accessed 24 Sep 2018].
- 149. FIND , 2018. FIND pilots partnership-based business model for outbreak response as first investment in new epidemic preparedness strategy. Available from: https://www.finddx.org/news/find-pilots-partnership-based-business-model-outbreak-response-first-investment-new-epidemic-preparedness-strategy/ [accessed 24 Sep 2018].
- 150. FIND , 2018. Target product profiles. Available from: https://www.finddx.org/target-product-profiles [accessed 28 Sep 2018].
- 151. Kouadio L, Nowak K, Akoua-Koffi C, et al. . Lassa Virus in multimammate rats, Côte d'Ivoire, 2013. Emerg Infect Dis 2015;21:1481–3. 10.3201/eid2108.150312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Drosten C, Göttig S, Schilling S, et al. . Rapid detection and quantification of RNA of Ebola and Marburg viruses, Lassa virus, Crimean-Congo hemorrhagic fever virus, Rift Valley fever virus, dengue virus, and yellow fever virus by real-time reverse transcription-PCR. J Clin Microbiol 2002;40:2323–30. 10.1128/JCM.40.7.2323-2330.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Vieth S, Drosten C, Lenz O, et al. . RT-PCR assay for detection of Lassa virus and related Old World arenaviruses targeting the L gene. Trans R Soc Trop Med Hyg 2007;101:1253–64. 10.1016/j.trstmh.2005.03.018 [DOI] [PubMed] [Google Scholar]
- 154. Fukuma A, Kurosaki Y, Morikawa Y, et al. . Rapid detection of Lassa virus by reverse transcription-loop-mediated isothermal amplification. Microbiol Immunol 2011;55:44–50. 10.1111/j.1348-0421.2010.00286.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Olschläger S, Günther S. Rapid and specific detection of Lassa virus by reverse transcription-PCR coupled with oligonucleotide array hybridization. J Clin Microbiol 2012;50:2496–9. 10.1128/JCM.00998-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
bmjgh-2018-001116supp002.pdf (411.8KB, pdf)
bmjgh-2018-001116supp003.pdf (401KB, pdf)
bmjgh-2018-001116supp001.pdf (399.4KB, pdf)