Abstract
Polyglutamine expansions in the transcriptional co-repressor Atrophin-1, encoded by ATN1, cause the neurodegenerative condition dentatorubral-pallidoluysian atrophy (DRPLA) via a proposed novel toxic gain of function. We present detailed phenotypic information on eight unrelated individuals who have de novo missense and insertion variants within a conserved 16-amino-acid “HX repeat” motif of ATN1. Each of the affected individuals has severe cognitive impairment and hypotonia, a recognizable facial gestalt, and variable congenital anomalies. However, they lack the progressive symptoms typical of DRPLA neurodegeneration. To distinguish this subset of affected individuals from the DRPLA diagnosis, we suggest using the term CHEDDA (congenital hypotonia, epilepsy, developmental delay, digit abnormalities) to classify the condition. CHEDDA-related variants alter the particular structural features of the HX repeat motif, suggesting that CHEDDA results from perturbation of the structural and functional integrity of the HX repeat. We found several non-homologous human genes containing similar motifs of eight to 10 HX repeat sequences, including RERE, where disruptive variants in this motif have also been linked to a separate condition that causes neurocognitive and congenital anomalies. These findings suggest that perturbation of the HX motif might explain other Mendelian human conditions.
Keywords: allelic disorders, HX repeat, intellectual disability, developmental delay, dysmorphic
Main Text
The combination of unbiased chromosomal analysis (chromosomal microarray) and next-generation sequencing approaches (exome sequencing [ES] and whole-genome sequencing [WGS]), along with the use of databases that promote sharing of information on genotype and phenotype, is enabling the identification and validation of genetic conditions and improved diagnostic rates for complex congenital conditions.1 Such unbiased genetic approaches can also unveil the complexity of how different types of genetic variation in a particular gene can result in varied and sometimes distinct phenotypic presentations.2, 3, 4, 5, 6
ATN1 (MIM: 607462), located at chromosomal region 12p13.31, comprises 10 exons and has two transcript variants (GenBank: NM_001007026.1, NM_001940.3) that differ only in their untranslated exons. ATN1 encodes atrophin-1 (ATN1), a member of a class of evolutionarily conserved transcriptional corepressors involved in nuclear signaling.7 The normal roles of ATN1 are incompletely understood; however, converging evidence supports a role for this protein as a nuclear transcriptional regulator important in the control of brain and other organ system development.8, 9, 10 Although Atn1−/− mice are neurologically normal,8 Zhang et al.9 demonstrated that knockdown of Atn1 in rat neuronal progenitor cells (NPCs) led to significant abnormalities in brain development; these abnormalities could be largely rescued by co-transfection with a human ATN1 construct. That study also demonstrated that ATN1 is a direct target of the lysine-specific histone demethylase 1A (LSD1), a protein known to have key developmental roles, including controlling embryonic stem cell differentiation, cortical neuronal migration, and adult NPC proliferation.9 ATN1 transcripts are widely expressed, including in brain, heart, lung, kidney, and skeletal muscle; expression is higher in fetal tissues, especially in the brain.11 In the human adult brain, ATN1 is broadly expressed in multiple regions, including the amygdala, corpus callosum, hippocampus, hypothalamus, caudate nucleus, substantia nigra, subthalamic nucleus, and thalamus, consistent with a role for ATN1 in central nervous system development and function.11
The only human condition definitively associated with ATN1 to date is the autosomal-dominant neurodegenerative condition dentatorubral-pallidoluysian atrophy (DRPLA, MIM: 125370)12, 13 caused by a polyglutamine expansion in exon 5. DRPLA is characterized by the progressive neurological features of choreoathetosis, myoclonus, epilepsy, ataxia, and dementia. Age of onset ranges from infancy to late adulthood, dependent on size of the expansion.14, 15 Congenital anomalies are not a feature. The underlying pathogenic mechanism whereby polyglutamine expansion of ATN1 causes DRPLA is incompletely understood: it is postulated that a toxic gain-of-function effect of the expanded polyglutamine tract causes neurotoxicity rather than simple loss of function. These toxic effects might include formation of peri- and intranuclear inclusions; abnormal protein cleavage or abnormal phosphorylation of ATN1; and downstream suppression of cAMP-response-element-binding protein (CREB)-dependent transcriptional activation, which is required for neuronal plasticity and survival.16, 17, 18, 19
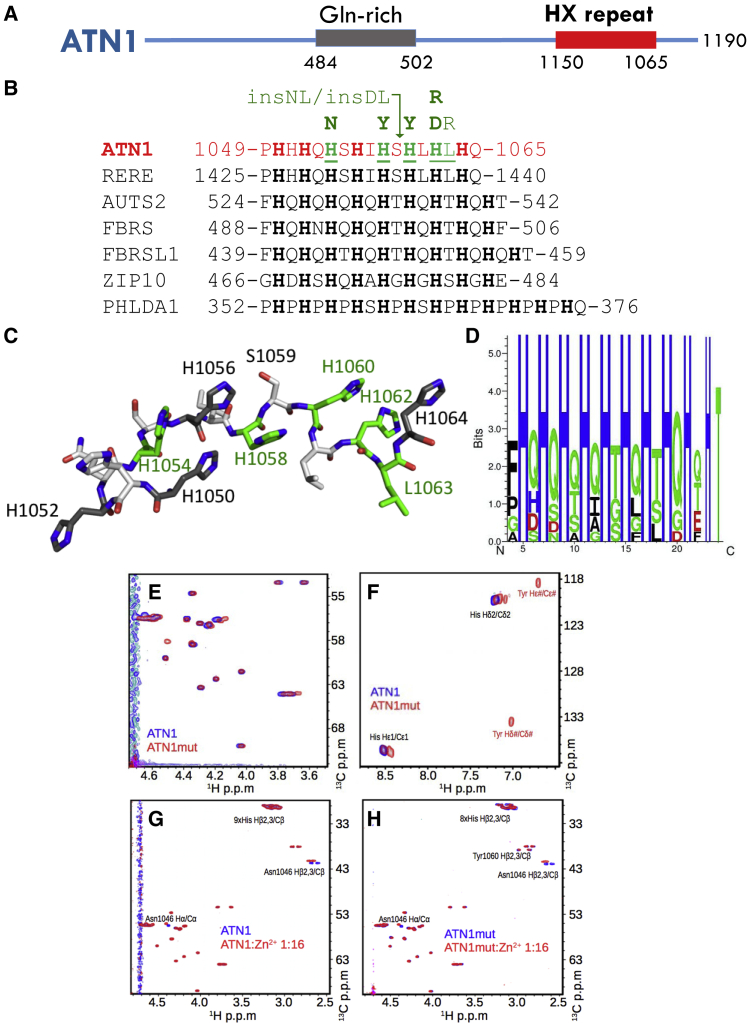
We report here on a cohort of eight individuals affected with overlapping severe primarily neurocognitive phenotypes. All of these individuals harbored de novo variants in a specific and highly evolutionarily conserved and invariant 16 amino acid motif, consisting of a histidine-rich 16 amino acid motif encoded by exon 7 of ATN1 (Figure 2). This motif is distal to the Gln-rich region involved in DRPLA, and the affected individuals lacked the progressive neurodegenerative features characteristic of DRPLA.14
Figure 2.
De novo Variants in ATN1 Affect a Highly Invariant Motif
(A) Schematic overview of ATN1; amino acid positions of the Gln-repeat region expanded in DRPLA and the HX repeat motif are illustrated.
(B) Human proteins with their (HX)n repeat motifs, with n ≥ 8. Start and end residue numbers are given and histidines are highlighted. For ATN1, residues found mutated in this study are underlined. The variant substitutions present in affected individuals are indicated above the sequence alignment (green). The arrow head marks the position of the two amino acid insertions Asn-Leu (insNL) and Asp-Leu (insDL). Proteins and database accession numbers are: ATN1: atrophin-1, NP_001007027.1; RERE: arginine-glutamic acid dipeptide repeats protein isoform a, NP_001036146.1; AUTS2: autism susceptibility gene 2 protein, NP_056385.1; FBRS: probable fibrosin-1, NP_001098549.2; FBRSL1: fibrosin-1-like protein, NP_001136113.1; ZIP10: zinc transporter ZIP10 precursor, NP_001120729.1; PHLADA1: pleckstrin homology-like domain family A member 1, NP_031376.3.
(C) 3D structural visualization of the ATN1 HX repeat motif. The 3D structure is not derived experimentally, but only chosen to illustrate the localization of the histidines.
(D) Amino acid enrichment within the HX regions of the 51 human sequences most similar to the ATN1 HX repeat motif (excluding His-only or His-Pro motifs). Figure was produced with Seq2Logo 2.0.20
(E-F) 2D heteronuclear 1H - 13C HSQC correlation spectra. Peptides were recorded using natural abundance of 1H and 13C in synthesized peptides. ATN1: ATN11046-1067; ATN1mut: ATN11046-1067His1060Tyr. C-D: “1:16” indicates a peptide: Zn2+ ion ratio of 1:16. pD corresponds to the pH in D2O.
For all affected individuals, within the first three months of life, there arose concerns regarding significant hypotonia, feeding difficulties, seizures, congenital malformations, and distinctive facial features, and all have severe to profound global developmental delay and/or intellectual disability, truncal hypotonia, global motor disability, and very limited verbal communication. (See Table 1 for an overview of the clinical data and Table S2 for further clinical details.) Five have a seizure disorder; for four of these, the seizure disorder could be described as a neonatal or infantile-onset developmental encephalopathy. Hearing and visual impairments and functional gastrointestinal disorders were common and frequently severe; four individuals required orogastric feeding or total parenteral nutrition. Other than suboptimal weight gain in those with more significant feeding difficulties, growth parameters were within the normal range. Individual 8 was born prematurely at 33 weeks and died at 2 months of age as a result of respiratory distress in the setting of severe multiple congenital anomalies.
Table 1.
Comparison of Clinical Features of Affected Individuals with Missense Variants in the Poly HX Domain of ATN1
| Affected Individual | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 822 |
|---|---|---|---|---|---|---|---|---|
| Current Age | 3 years | 1 year | 5 years | 7 years | 9 years | 4 years | 5 years | 2 months |
| Gender | M | M | F | F | F | F | F | F |
| Ethnicity | Argentinian | Hispanic | Hispanic | Hispanic | Saudi | Mexican | Australian | French |
| Variant cDNA1 | c.3160C>A | c.3172C>T | c.3177_3178insAACCTG | c.3177_3178insGACCTG | c.3178C>T | c.3184C>G | c.3188T>G | c.3185A>G |
| AA Change2 | p.His1054Asn | p.His1058Tyr | p.Ser1059_His1060insAsnLeu | p.Ser1059_His1060insAspLeu | p.His1060Tyr | p.His1062Asp | p.Leu1063Arg | p.His1062Arg |
| Antenatal Findings | increased nuchal translucency (karyotype N) | oligohydramnios and partial urinary obstruction | normal antenatal USS | normal antenatal USS | no | ambiguous genitalia and cardiac malformation | normal antenatal USS | normal antenatal USS, breech, PROM |
| Gestation | term | term | term | 31+6 weeks | term | term | term | 33 weeks |
| Birth Centiles: Length | NA | >90% | NA | 50%–75% | NA | 50% | 15% | 50% |
| Birth Centiles: Weight | 3%–15% | 50% | 3% | 25%–50% | NA | 15% | 3% | 50% |
| Birth Centiles: Head Circumference | NA | NA | 15%–50% | 50% | NA | 50% | 15%–50% | 75% |
| Current Centiles: Length | 3% | 85% | 25% | 5% | 25% | 30% | 3%–10% | NK |
| Current Centiles: Weight | 3% | 40% | 85% | 3% | 2% | 85% | 3% | NK |
| Current Centiles: Head Circumference | 50% | 45% | 60% | 85% | 75% | 20% | 25% | NK |
| Infantile Hypotonia | yes | yes | yes | yes | yes | yes | yes | yes |
| Current Neurology | global hypotonia | central hypotonia, appendicular spasticity | global hypotonia | global hypotonia, hyperkinetic UL movts |
central hypotonia and appendicular spasticity | global hypotonia | global hypotonia | axial hypotonia and appendicular hypertonia |
| Overt Seizure Disorder | yes; Lennox Gestaut onset 1yr |
no; no clinical seizures | no | yes; inf spasms controlled 2 AED |
yes; controlled MT |
yes; intractable neonatal onset EE (mixed types) | yes; EE onset 7 mos controlled MT |
no |
| EEG | MEA with slow spike and wave and periods of voltage attenuation | bitemporal epileptiform discharges | focal theta slowing | hypsarrhythmia + bg slowing | diffuse slowing | diffuse slowing | MEA | ND |
| Level of DD or ID | severe-profound GDD | GDD | severe GDD | profound | severe | severe-profound | severe | severe |
| Visual Impairment | yes; does not fix or follow, corneal leukoma |
yes; CVI | no | yes; CVI | no | yes; CVI | yes; CVI | yes; microphthalmia |
| Hearing Impairment | yes | yes; bl mod (hearing aids) | yes; OME (grommets) | no; OME (grommets) | no; OME (grommets) | yes; bl sn | yes; bl mod (hearing aids and grommets) | NK |
| Verbal Ability | non-verbal | coos | single words | non-verbal | non-verbal | non-verbal | babbles | none |
| Gross Motor Ability | no head control, cannot roll | rolls to side | walks few steps unsupported | sits with support | immobile | sits with support | sits with support | none |
| Fine Motor Ability | holds small objects | grasps objects | grasps objects | |||||
| MRI Brain | parenchymal atrophy, unilateral PVL, left cerebellar hyperintensity | peri-sylvian polymicrogyria, parenchymal atrophy, thin CC, absent falx cerebri | normal | normal | peri-sylvian polymicrogryri, thin CC, partial absence falx cerebri, parenchymal atrophy | vermian hypoplasia | peri-sylvian polymicrogyria, thin CC, absent falx cerebri | polymicrogyria of the rt Sylvian fissure, vermian hypoplasia, thin CC |
| MRI Cervical Spine | ND | craniocervical stenosis | craniocervical stenosis | ND | ND | normal | normal | ND |
| Respiratory Symptoms | no | yes; severe OSA | no | no | yes; asthma | yes; ul choanal stenosis, O+CSA (tracheostomy) |
yes; OSA (CPAP) |
yes; respiratory distress |
| Orofacial Clefting | no; high, narrow palate | yes; small, hard palate cleft | no | no | no | no | no; high, narrow palate | yes; cleft palate and gingiva |
| GI Abnormalities | yes; pyloric hypertrophy, dysphagia GERD |
yes; GERD NEC |
yes; dysphagia constipation |
no | yes; GERD | yes; dysphagia GERD, ant anus |
yes; dysphagia GERD, constipation |
yes; ant. anus |
| Nutrition | oral feeding | TPN | self feeds | oral feeding | G tube | G tube | oral feeding | orogastric feeding |
| Congenital Heart Disease | no | yes; ASD+ | no | no | yes; VSD + ASD | yes; CoA + hypoplasia LV and AA | no | yes; large foramen ovale, persistence left SVC |
| Genitourinary Disease | right renal agenesis | cryptorchidism, VUR, ul hydrouteronephrosis rUTI |
no | no; normal USS | yes; r UTI normal USS |
no; normal USS | no; normal USS | yes; left non-dysplastic renal hypoplasia |
| Skeletal System | joint hypermobility, scoliosis | hip dysplasia | short trunk DDH, joint laxity |
mild scoliosis | DDH | sagittal craniosynostosis | normal | phalangeal hypoplasia |
| Facial Gestalt | yes | yes | yes | yes | yes | yes | yes | yes |
| Hands and Feet | overlapping digits | overlapping digits | overlapping digits | overlapping toes | abnormal PC | overlapping toes, single PC, fetal pads |
overlapping toes, fetal pads |
overlapping toes, proximally implanted thumbs, single PC |
| Others | - | - | - | - | - | inv nipples | inv nipples |
Abbreviations are as follows: AA, aortic arch; abn, abnormal; AED, antiepileptic drug; Ant. anus, anteriorly placed anus; AmA, amino acid; ASD, atrial septal defect; bg, background; bl, bilateral; CC, corpus callosum; C+OSA, central and obstructive sleep apnoea; CVI, cortical visual impairment; CoA, coarctation of the aorta; DDH, developmental disorder of the hips; EE, epileptic encephalopathy; F, female; GDD, global developmental delay; GERD, gastresophageal reflux disease; G tube, gastrostomy tube; inf spasms, infantile spasms; inv, inverted; LV, left ventricle; MEA, multifocal epileptiform activity; MT, monotherapy (i.e., controlled on monotherapy); M: male; mo: month; mod: moderate; movts: movements; NA: not available; ND: not done; NK: not known; NEC: acute necrotising enterocolitis; PC: palmar crease; PROM: premature rupture of membranes; OSA: obstructive sleep apnoea; OME: otitis media with effusions PVL: periventricular leukomalacia; sn: sensorineural; SVC: superior vena cava; TPN: total parenteral nutrition; rUTI: recurrent urinary tract infections; r: recurrent; UL: upper limb; ul: unilateral; USS: ultrasound; VUR: vesicoureteric reflux; VSD: ventriculoseptal defect; year: year.
Based on transcript GenBank: NM_001007026.1.
Protein ID GenBank: NP_001007027.1.
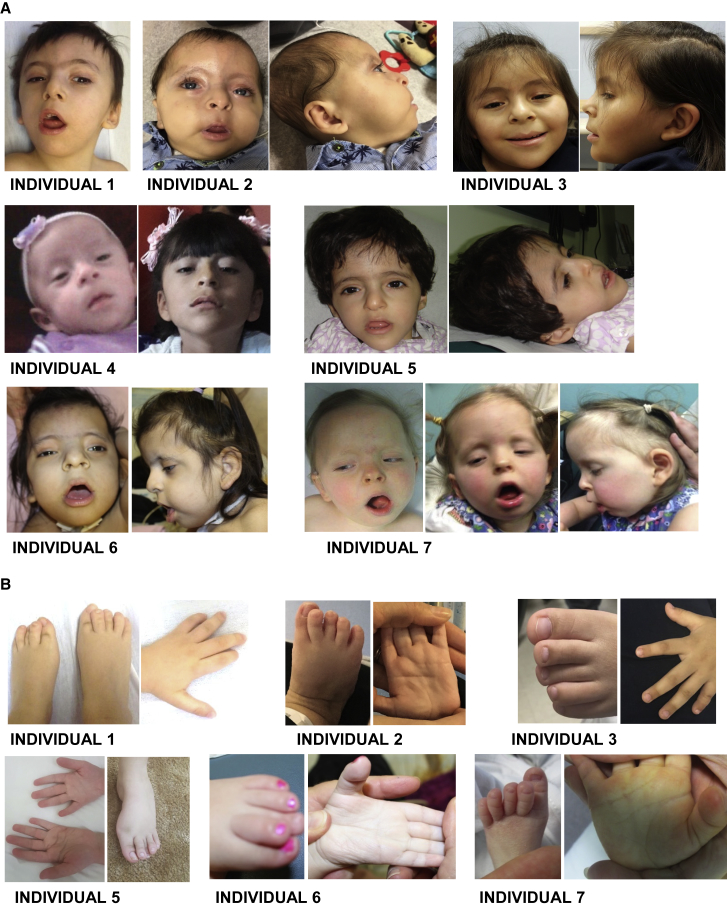
Congenital structural anomalies were common but variable between individuals: four individuals had cardiac malformations, including atrial and ventricular septal defects, plus abnormalities of the aorta and superior vena cava; two individuals had palatal clefts; three individuals had congenital renal anomalies; and two had an anteriorly placed anus. Common neuroanatomical abnormalities were evident on examination of available MRI in one center (individuals 2, 5, 7, and 8) (Figure S1). Several individuals had cranio-skeletal abnormalities: in particular, two individuals had stenosis of the craniocervical junction, which prompted screening for this complication in all individuals. When the individuals were assessed as a group, a similarity in facial features was apparent (Figure 1), a particularly striking feature being sparsity of the lateral forehead hair and low-set, posteriorly rotated ears. Characteristic hand and foot features were overlapping toes, camptodactyly, persistent fetal fingertip pads, and abnormalities of the palmar creases (Figure 1).
Figure 1.
Clinical Images of Affected Individuals with CHEDDA
(A) Facial images of affected individuals 1–7. Common facial features include tall foreheads with bitemporal narrowing; deeply set eyes; sparsity of the lateral forehead hair; low-set posteriorly rotated ears; a bulbous, slightly overhanging nasal tip; longer philtrum; prominent columella; and a thin upper lip.
(B) Hand and foot images of affected individuals 1, 2, 3, 5, 6 and 7. Common features include abnormalities of the palmar creases, bulbous endings to the fingers and toes, and overlapping toes.
This clinical cohort was collated through the identification of individuals with de novo ATN1 variants listed in the ClinVar database, as well as via contact with individual diagnostic laboratories and networking at human genetics conferences. Prior to ES or WGS, all individuals with a de novo ATN1 variant were undiagnosed despite clinical genetic assessment and prior genetic screening, which in all individuals included chromosomal microarray. (Further details of prior genetic studies are provided in Table S1.) Individual 5 was previously included in a large exome sequencing study,21 and individual 8 was previously described clinically without a molecular diagnosis.22 All ATN1 variants in the clinical cohort were rare in that they were absent from the 125,748 exomes and 15,708 genomes listed in the gnomAD 2.1 database (a database depleted of individuals with severe pediatric disease23) and from the BRAVO database of 62,785 healthy individuals. All missense variants were predicted to be pathogenic by the majority of in silico pathogenicity scoring tools (see Table 2). No affected individual had another plausible cause for their neurocognitive condition or congenital anomalies after ES or WGS variant filtering and prioritization (see Table S2). Segregation analysis was consistent with the variant’s being de novo for all individuals and provided no evidence of mosaicism. Details of the methods for sequencing, variant filtering, and prioritization are provided in the Supplemental Data. Genetic studies were approved by local ethics committees, and written informed consent for molecular genetic analysis and for the publication of clinical and radiological data and photographs, which were obtained as part of standard diagnostic procedures, was obtained from the participants’ legal guardians.
Table 2.
De novo ATN1 Variants Reported in this Clinical Cohort
| Affected Individual | Genomic Location (GRCh37) | Variant cDNA Change (NM_001007026.1) | Amino Acid Change (GenBank:NP_001007027.1) | Present in gnomAD Database? | Segregation | SIFT24Score | PROVEAN25Score | DANN26Score | CADD27Score | Classification as per ACMG Guidelines28 |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | GenBank: NC_000012.11: g.7048286C>A | c.3160C>A | p.His1054Asn | no | de novo | damaging (0) | damaging (−6.1) | 0.994 | 29.6 | Variant of uncertain significance. PM2: Pathogenic Moderate (absent GnomAD despite good coverage) PP3: Pathogenic Supporting (8/8 pathogenic predictions) |
| 2 | GenBank: NC_000012.11: g.7048298C>T | c.3172C>T | p.His1058Tyr | no | de novo | damaging (0) | damaging (−5.26) | 0.992 | 23 | Variant of uncertain significance. PM2:Pathogenic Moderate (absent GnomAD despite good coverage) PP3: Pathogenic Supporting (6/8 pathogenic predictions) |
| 3 | GenBank: NC_000012.11: g.7048303-7048304insAACCTG | c.3177_3178insAACCTG | p.Ser1059_His1060insAsnLeu | no | de novo | NA | NA | NA | NA | Variant of uncertain significance. PM2: Pathogenic Moderate (absent GnomAD despite good coverage) PM4: Pathogenic Moderate (in frame variant in ATN1, and is not in a repeat region.) PP3: Pathogenic Supporting: 1 pathogenic prediction from GERP (versus no benign predictions). |
| 4 | GenBank: NC_000012.11: g.7048303-7048304insGACCTG | c.3177_3178insGACCTG | p.Ser1059_His1060insAspLeu | no | de novo | NA | NA | NA | NA | Variant of uncertain significance. PM2: Pathogenic Moderate (absent GnomAD despite good coverage) PM4: Pathogenic Moderate (in frame variant in ATN1, and is not in a repeat region.) PP3: Pathogenic Supporting: 1 pathogenic prediction from GERP (versus no benign predictions). |
| 5 | GenBank: NC_000012.11: g.7048304C>T | c.3178C>T | p.His1060Tyr | no | de novo | damaging (0) | damaging (−5.21) | 0.998 | 28.7 | Variant of uncertain significance. PM2: Pathogenic Moderate (absent GnomAD despite good coverage) PP3: Pathogenic Supporting (8/8 pathogenic predictions) |
| 6 | GenBank: NC_000012.11: g.7048310C>G | c.3184C>G | p.His1062Asp | no | de novo | damaging (0) | damaging (−7.86) | 0.992 | 26.2 | Variant of uncertain significance. PM2: Pathogenic Moderate (absent GnomAD despite good coverage) PP3: Pathogenic Supporting (8/9 pathogenic predictions) |
| 7 | GenBank: NC_000012.11: g.7048314T>G | c.3188T>G | p.Leu1063Arg | no | de novo | damaging (0) | damaging (−5.33) | 0.997 | 29.6 | Variant of uncertain significance. PM2: Pathogenic Moderate (absent GnomAD despite good coverage) |
| 8 | GenBank: NC_000012.11: g.7048311A>G | c.3185A>G | p.His1062Arg | no | de novo | damaging (0) | damaging (−6.89) | 0.9938 | 24.1 | Variant of uncertain significance. PM2: Pathogenic Moderate (absent GnomAD despite good coverage) PP3: Pathogenic Supporting (8/8 pathogenic predictions) |
The histidine-rich motif that is perturbed in all individuals of our cohort is located in the C-terminal part of ATN1 (residues 1049–1065). It consists of eight HX repeats, where H is a histidine and X is any amino acid (Figures 2A–2C). The DNA region encoding for this HX repeat motif was covered in the sequencing of all individuals to a depth of at least 20 reads (Table S2). A search for the (HX)8 pattern in the human proteome (via PatternSearch29) yielded 71 sequences (see Supplemental Data), 20 of which appeared to be distinct from ATN1’s HX repeat in that the “X” position only contained histidines or prolines (see PHLDA1, Figure 2B). The remaining 51 sequences were isoforms of ATN1 and its paralogous arginine-glutamic-acid dipeptide-repeat protein RERE (which is also a transcriptional repressor); were isoforms of the paralogous autism-susceptibility gene 2 protein (AUTS2, a component of the PRC1-like complex involved in maintaining the transcriptional repressive state of many genes during development), fibrosin (FBRS), and fibrosin-like proteins (FBRSL1); or belonged to the ZIP family of zinc transporters (Figure 2). Within these 51 sequences, the “X” position showed limited variability and mostly consisted of Gln and Thr (Figure 2D).
To probe the impact of a pathological variant on the molecular behavior of the ATN1 protein, we studied two ATN1-derived polypeptides containing the HX repeat motif in solution by nuclear magnetic resonance (NMR). The peptides 1046-NVTPHHHQHSHIHSHLHLHQQD-1067 (ATN11046–1067) and another bearing the variant p.His1060Tyr 1046-NVTPHHHQHSHIHSYLHLHQQD-1067 (ATN11046–1067p.His1060Tyr) were commercially synthesized and dissolved in 500 μL of 100% D2O at a concentration of 2.2 mg/mL. NMR experiments were performed on a 700 MHz Bruker spectrometer at 25°C and a pD (the pH for D2O) between 5.51 and 6.4. The NMR data were processed by NMRPipe30 and analyzed with SPARKY. 1H and 13C resonances for the two peptides were analyzed via standard procedures31 on the basis of 2D homonuclear 2D 1H-1H TOCSY (mixing times were 10 and 80 ms) and 2D 1H-1H ROESY, as well as 2D 1H-1H NOESY (mixing times were 300 and 500 ms); these analyses were based on 2D 1H-13C HSQC experiments, which made use of the natural abundance of 13C, were carried out using separate tuning for the aliphatic and aromatic regions. For details, see Supplemental Data.
For all histidines, the cross peaks corresponding to the Hδ2/Cδ2 atoms and those corresponding to the Hε1/Cε1 atoms were each clustered at indistinguishably close 1H and 13C chemical shifts (Figure 2E). Moreover, 2D homonuclear 1H -1H ROESY and 2D 1H -1H spectra showed that the trivial (|j − i| = 0) and short-range (|j − i| < 2) NOE cross peaks were absent or of very weak intensity (Figure 2F). Both types of experiments indicated that the regularly spaced occurrence of histidines introduces a spatial and dynamical synchronization of the histidines. This synchronization was lost in ATN11046–1067p.His1060Tyr, which displayed dispersed cross peaks of the side chain aromatic region and stronger intensity of the NOE cross peaks.
Side-chain imidazole rings of histidines are known to coordinate metal ions.32, 33 Zn2+ binding was assessed by the stepwise addition of ZnCl2 stock solution (500 mM in 100% D2O) to the solution of peptides reaching the peptide: Zn2+ molar ratios of 1:0.5, 1:1, 1:4, 1:8, 1:16, 1:32, and 1:48. For each peptide:Zn2+ ratio, the pD was checked and corrected if required, and the same 1D and 2D NMR spectra as for the free peptides were recorded (see Supplemental Data). We found that only the histidines of the p.His1060Tyr mutant, but not of the wild-type peptide, bound Zn2+ at a pD of 5.5, as demonstrated by the histidine 1Hβ2,3/13Cβ chemical shift and signal intensity changes upon addition of Zn2+ (Figures 2G and 2H). This particular feature was lost upon deprotonation of the histidines at higher pD (Figures S2A and S2B). Thus, under the conditions used, the HX -repeat motif created specific and unusual pH-dependent zinc-binding properties for the histidine-rich sequence, and those properties were abolished by the variant.
Herein, we describe a recognizable constellation of severe neurocognitive impairment, distinctive facial features, and pleiotropic but overlapping congenital anomalies in eight affected individuals with de novo variants in the HX repeat motif encoded within exon 7 of ATN1. This static (non-progressive) syndromic phenotype is distinct from the neurodegenerative condition of DRPLA, which is caused by a triplet repeat expansion in exon 5 of ATN1. This repeat expansion is thought to result in a toxic gain of function. We propose the name CHEDDA (congenital hypotonia, epilepsy, developmental delay, digit abnormalities) to distinguish this previously unreported condition from ATN1-related DRPLA. Our clinical observations are consistent with a role for ATN1 as a key nuclear transcriptional regulator involved in the regulation of organ development, including development of the brain and heart,9, 10, 34 and hint at a critical role of the ATN1 HX repeat motif in the control of human embryonic development. That the variants cause a simple haploinsufficiency of ATN1 is unlikely given the presence of a number (albeit a very small number) of healthy individuals with heterozygous stop-gain, frameshift, and canonical splice variants in gnomAD and BRAVO and the clustering of the variants in CHEDDA within a specific restricted protein motif.
The HX repeat motif was only briefly described in 1991 in a purely bioinformatics-based “hypothesis” publication.35 The authors reported the presence of (HX)n repeat motifs (where n is the number of times the motif is repeated) in certain Drosophila transcription factors and suggested that this motif might be used for coordinating zinc binding. Interestingly, the wild-type ATN1 HX repeat sequence showed a strong pH dependency for zinc binding in vitro. This feature appears to be linked to the regular histidine spacing, which introduces a specific synchronization of the histidine side chains. The introduction of the p.His1060Tyr variant present in an affected individual in the cohort disrupted this synchronization and allowed zinc binding, as expected for poly-histidine motifs, thus endowing ATN1 with a novel property, albeit with unclear molecular consequences. On a molecular level, the ATN1 HX repeat motif appears therefore to give rise to specific features that distinguish it from other poly-histidine motifs. The ATN1 HX repeat might serve as a specific pH-dependent interaction motif for ions and/or proteins or other biomolecules.
The hypothesis that disruption in the spacing of the histidines in HX motifs will affect critical functioning is supported by our observation that nine of the 19 reported individuals with neurodevelopmental disorder with or without anomalies of the brain, eye, and heart (NEDBEH, MIM: 616975) have de novo variants disrupting the HX motif of RERE and that those individuals with variants in the HX motif, as opposed to the rest of the protein, are more likely to have congenital anomalies, including septal cardiac, eye, and brain anomalies.36 We also note that three rare de novo variants (SCV000571291.3, SCV000837721.1, and SCV000493076.1) in the HX motif of AUTS2 are listed in ClinVar as likely pathogenic and occurred in individuals with an intellectual-disability and congenital-anomaly phenotype.
A possible link between dysregulation of ATN1 expression and another congenital syndromic neurocognitive condition, Pallister Killian syndrome (PKS, mosaic tetrasomy 12p, MIM: 601803), has also been previously postulated by Kaur et al.37 ATN1 lies within the PKS critical region on 12p13.31, and dysregulation of the expression of ATN1, among other genes, was demonstrated in fibroblasts from affected individuals with PKS. Kaur et al., postulated that ATN1 overexpression could be a key driver of the phenotype of PKS via dysregulation of the key developmental HOX genes through the action of the master transcriptional regulator CREBBP, although direct evidence was lacking. This speculation is intriguing, given certain similarities in phenotype between individuals in our cohort and individuals affected by PKS; such similarities included severe cognitive impairment, hypotonia, distinctive facial features including high forehead and sparse fronto-temporal hair at birth (the latter representing a relatively rare clinical finding), and variable congenital anomalies, including high arched or cleft palate, polymicrogyria, limb and genitourinary anomalies, and congenital heart defects.38 Indeed, PKS was considered as a differential diagnosis in individual 7 of our cohort.
The de novo variants in the HX motif of ATN1 reported here account for one out of 6,100 individuals who had a neurological phenotype and had exome sequencing performed through Baylor Genetics and for five out of 13,640 individuals who had neurodevelopmental delay and had exome sequencing performed through GeneDx; these latter individuals included another affected individual who had the variant ATN1 (c.3178C>T [p.His1060Tyr] GenBank: NM_001007026.1) (ClinVar: SCV000620232.1) and whose family did not give consent for the inclusion of clinical data. These data suggests a frequency of CHEDDA between 1.6 × 10−4 and 3.7 × 10−4 individuals with neurocognitive and/or neurological disorders. Despite this apparent rarity of CHEDDA, we postulate that more affected individuals might have already had a variant detected in this region through diagnostic ES and WGS, but the lack of the progressive neurological phenotype characteristic of DRPLA might have caused the variants to be unreported or classified as variants of uncertain clinical significance. This situation is similar to those of other clinically distinct conditions our groups have recently described,2, 3, 4 and it highlights genotype-phenotype complexity and the importance of rigorous evaluation of the possible pathogenicity of unreported variants through international clinical and basic science collaborations.39, 40 To assist with the dissemination of accessible information about CHEDDA to clinicians and families of affected individuals, we have adopted ATN1 on the Human Disease Gene Webseries.
Clarifying the biological role of the HX repeat motif in the ATN1 and AUTS2 protein families and understanding how this function might be altered in CHEDDA and potentially linked conditions such as PKS will require further work. Such work might lead to the development of targeted therapies. For example, Zhang et al. demonstrated that the clinical LSD1 inhibitor, tranylcypromine, could suppress ATN1 expression, and they suggested that this agent might have potential therapeutic implications for conditions resultant from aberrant ATN1 expression.39, 41 Further studies into the primary gene-regulatory functions of ATN1 and how this might be altered in CHEDDA and potentially linked conditions such as PKS are warranted.
Declaration of Interests
The authors declare no competing interests.
Acknowledgments
The authors thank the affected individuals and their families for their participation in this study and also thank the clinical neurology, general pediatric, and clinical genetics teams involved with their clinical care and support. We thank Toby Baldwin, Toni Saville, and Michael Buckley at SEALS genetics laboratory for their technical assistance. The research was supported by the National Health and Medical Research Council (GNT11149630 to E.E.P.; GNT0512123 to T.R.), Office of Health and Medical Research, NSW Health (to E.E.P.), King Abdulaziz City for Science and Technology (13-BIO1113-20 and 15-BIO3688-20 to F.S.A.), King Salman Center for Disability Research (F.S.A.), Saudi Human Genome Program (F.S.A.), and the King Abdullah University of Science and Technology through baseline funds (S.T.A., S.H., J.M., H.A., F.A., L.J., M.J.) and the Award No. FCC1/1976-25 from the Office of Sponsored Research (S.T.A.). The Department of Molecular and Human Genetics at Baylor College of Medicine receives revenue from clinical genetic testing done at Baylor Genetics Laboratory.
Published: February 28, 2019
Footnotes
Supplemental Data can be found with this article online at https://doi.org/10.1016/j.ajhg.2019.01.013.
Contributor Information
Fowzan S. Alkuraya, Email: falkuraya@kfshrc.edu.sa.
Stefan T. Arold, Email: stefan.arold@kaust.edu.sa.
Accession Numbers
ATN1 c.3178C>T (p.His1060Tyr): GenBank: NM_001007026.1 and ClinVar: SCV000221666.1
ATN1 c.3177_3178insGACCTG (p.Ser1059_His1060insAspLeu): GenBank: NM_001007026.1 and ClinVar: SCV000619648.1
ATN1 c.3177_3178insAACCTG (p.Ser1059_His1060insAsnLeu): GenBank: NM_001007026.1 and ClinVar: SCV000571353.3
ATN1 c.3184C>G (p.His1062Asp): GenBank: NM_001007026.1 and ClinVar: SCV000528073.3
ATN1 c.3160C>A (p.His1054Asn): GenBank: NM_001007026.1 and ClinVar: SCV000678263.1
ATN1 c.3172C>T (p.His1058Tyr): GenBank: NM_001007026.1 and ClinVar: SCV000678264.1
ATN1: c.3188T>G (p.Leu1063Arg): GenBank: NM_001007026.1 and ClinVar: SCV000678265.1
ATN1 c.3185A>G (p.His1062Arg): GenBank: NM_001007026.1 and ClinVar: SCV000853264
Web Resources
DECIPHER, https://decipher.sanger.ac.uk/
Ensembl, https://www.ensembl.org/index.html
ExAC database, http://exac.broadinstitute.org
gnomAD database, http://gnomad.broadinstitute.org/
Human Disease Gene Webseries, http://humandiseasegenes.nl/
Mendelian Inheritance in Man, http://www.omim.org
PROVEAN, http://provean.jcvi.org
UCSC Genome Browser, http://genome.ucsc.edu
Supplemental Data
References
- 1.Boycott K.M., Rath A., Chong J.X., Hartley T., Alkuraya F.S., Baynam G., Brookes A.J., Brudno M., Carracedo A., den Dunnen J.T. International cooperation to enable the diagnosis of all rare genetic diseases. Am. J. Hum. Genet. 2017;100:695–705. doi: 10.1016/j.ajhg.2017.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Palmer E.E., Kumar R., Gordon C.T., Shaw M., Hubert L., Carroll R., Rio M., Murray L., Leffler M., Dudding-Byth T., DDD Study A Recurrent de novo nonsense variant in ZSWIM6 results in severe intellectual disability without frontonasal or limb malformations. Am. J. Hum. Genet. 2017;101:995–1005. doi: 10.1016/j.ajhg.2017.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Patel N., Faqeih E., Anazi S., Alfawareh M., Wakil S.M., Colak D., Alkuraya F.S. A novel APC mutation defines a second locus for Cenani-Lenz syndrome. J. Med. Genet. 2015;52:317–321. doi: 10.1136/jmedgenet-2014-102850. [DOI] [PubMed] [Google Scholar]
- 4.Aldahmesh M.A., Mohamed J.Y., Alkuraya H.S., Verma I.C., Puri R.D., Alaiya A.A., Rizzo W.B., Alkuraya F.S. Recessive mutations in ELOVL4 cause ichthyosis, intellectual disability, and spastic quadriplegia. Am. J. Hum. Genet. 2011;89:745–750. doi: 10.1016/j.ajhg.2011.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alkuraya F.S. Discovery of mutations for Mendelian disorders. Hum. Genet. 2016;135:615–623. doi: 10.1007/s00439-016-1664-8. [DOI] [PubMed] [Google Scholar]
- 6.Monies D., Maddirevula S., Kurdi W., Alanazy M.H., Alkhalidi H., Al-Owain M., Sulaiman R.A., Faqeih E., Goljan E., Ibrahim N. Autozygosity reveals recessive mutations and novel mechanisms in dominant genes: implications in variant interpretation. Genet. Med. 2017;19:1144–1150. doi: 10.1038/gim.2017.22. [DOI] [PubMed] [Google Scholar]
- 7.Wang L., Tsai C.C. Atrophin proteins: An overview of a new class of nuclear receptor corepressors. Nucl. Recept. Signal. 2008;6:e009. doi: 10.1621/nrs.06009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shen Y., Lee G., Choe Y., Zoltewicz J.S., Peterson A.S. Functional architecture of atrophins. J. Biol. Chem. 2007;282:5037–5044. doi: 10.1074/jbc.M610274200. [DOI] [PubMed] [Google Scholar]
- 9.Zhang F., Xu D., Yuan L., Sun Y., Xu Z. Epigenetic regulation of Atrophin1 by lysine-specific demethylase 1 is required for cortical progenitor maintenance. Nat. Commun. 2014;5:5815. doi: 10.1038/ncomms6815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wood J.D., Nucifora F.C., Jr., Duan K., Zhang C., Wang J., Kim Y., Schilling G., Sacchi N., Liu J.M., Ross C.A. Atrophin-1, the dentato-rubral and pallido-luysian atrophy gene product, interacts with ETO/MTG8 in the nuclear matrix and represses transcription. J. Cell Biol. 2000;150:939–948. doi: 10.1083/jcb.150.5.939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Onodera O., Oyake M., Takano H., Ikeuchi T., Igarashi S., Tsuji S. Molecular cloning of a full-length cDNA for dentatorubral-pallidoluysian atrophy and regional expressions of the expanded alleles in the CNS. Am. J. Hum. Genet. 1995;57:1050–1060. [PMC free article] [PubMed] [Google Scholar]
- 12.Nagafuchi S., Yanagisawa H., Ohsaki E., Shirayama T., Tadokoro K., Inoue T., Yamada M. Structure and expression of the gene responsible for the triplet repeat disorder, dentatorubral and pallidoluysian atrophy (DRPLA) Nat. Genet. 1994;8:177–182. doi: 10.1038/ng1094-177. [DOI] [PubMed] [Google Scholar]
- 13.Koide R., Ikeuchi T., Onodera O., Tanaka H., Igarashi S., Endo K., Takahashi H., Kondo R., Ishikawa A., Hayashi T. Unstable expansion of CAG repeat in hereditary dentatorubral-pallidoluysian atrophy (DRPLA) Nat. Genet. 1994;6:9–13. doi: 10.1038/ng0194-9. [DOI] [PubMed] [Google Scholar]
- 14.Hasegawa A., Ikeuchi T., Koike R., Matsubara N., Tsuchiya M., Nozaki H., Homma A., Idezuka J., Nishizawa M., Onodera O. Long-term disability and prognosis in dentatorubral-pallidoluysian atrophy: a correlation with CAG repeat length. Mov. Disord. 2010;25:1694–1700. doi: 10.1002/mds.23167. [DOI] [PubMed] [Google Scholar]
- 15.Veneziano L., Frontali M. Drpla. In: Adam M.P., Ardinger H.H., Pagon R.A., Wallace S.E., Bean L.J.H., Stephens K., Amemiya A., editors. GeneReviews. University of Washington; Seattle, WA: 1993. [Google Scholar]
- 16.Ross C.A., Poirier M.A. Protein aggregation and neurodegenerative disease. Nat. Med. 2004;10(Suppl):S10–S17. doi: 10.1038/nm1066. [DOI] [PubMed] [Google Scholar]
- 17.Nucifora F.C., Jr., Ellerby L.M., Wellington C.L., Wood J.D., Herring W.J., Sawa A., Hayden M.R., Dawson V.L., Dawson T.M., Ross C.A. Nuclear localization of a non-caspase truncation product of atrophin-1, with an expanded polyglutamine repeat, increases cellular toxicity. J. Biol. Chem. 2003;278:13047–13055. doi: 10.1074/jbc.M211224200. [DOI] [PubMed] [Google Scholar]
- 18.Okamura-Oho Y., Miyashita T., Nagao K., Shima S., Ogata Y., Katada T., Nishina H., Yamada M. Dentatorubral-pallidoluysian atrophy protein is phosphorylated by c-Jun NH2-terminal kinase. Hum. Mol. Genet. 2003;12:1535–1542. doi: 10.1093/hmg/ddg168. [DOI] [PubMed] [Google Scholar]
- 19.Fischbeck K.H. Kennedy disease. J. Inherit. Metab. Dis. 1997;20:152–158. doi: 10.1023/a:1005344403603. [DOI] [PubMed] [Google Scholar]
- 20.Thomsen M.C., Nielsen M. Seq2Logo: a method for construction and visualization of amino acid binding motifs and sequence profiles including sequence weighting, pseudo counts and two-sided representation of amino acid enrichment and depletion. Nucleic Acids Res. 2012;40 doi: 10.1093/nar/gks469. W281-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anazi S., Maddirevula S., Faqeih E., Alsedairy H., Alzahrani F., Shamseldin H.E., Patel N., Hashem M., Ibrahim N., Abdulwahab F. Clinical genomics expands the morbid genome of intellectual disability and offers a high diagnostic yield. Mol. Psychiatry. 2017;22:615–624. doi: 10.1038/mp.2016.113. [DOI] [PubMed] [Google Scholar]
- 22.Mosca A.L., Laurent N., Guibaud L., Callier P., Thauvin-Robinet C., Mugneret F., Huet F., Grimaldi M., Gouyon J.B., Sandre D., Faivre L. Polymicrogyria, cerebellar vermis hypoplasia, severe facial dysmorphism and cleft palate: a new syndrome? Eur. J. Med. Genet. 2007;50:48–53. doi: 10.1016/j.ejmg.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 23.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ng P.C., Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Choi Y., Sims G.E., Murphy S., Miller J.R., Chan A.P. Predicting the functional effect of amino acid substitutions and indels. PLoS ONE. 2012;7:e46688. doi: 10.1371/journal.pone.0046688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Quang D., Chen Y., Xie X. DANN: A deep learning approach for annotating the pathogenicity of genetic variants. Bioinformatics. 2015;31:761–763. doi: 10.1093/bioinformatics/btu703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rentzsch P., Witten D., Cooper G.M., Shendure J., Kircher M. CADD: Predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 2019;47(D1):D886–D894. doi: 10.1093/nar/gky1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E., ACMG Laboratory Quality Assurance Committee Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zimmermann L., Stephens A., Nam S.Z., Rau D., Kübler J., Lozajic M., Gabler F., Söding J., Lupas A.N., Alva V. A completely reimplemented MPI bioinformatics toolkit with a new HHpred server at its core. J. Mol. Biol. 2018;430:2237–2243. doi: 10.1016/j.jmb.2017.12.007. [DOI] [PubMed] [Google Scholar]
- 30.Delaglio F., Grzesiek S., Vuister G.W., Zhu G., Pfeifer J., Bax A. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 31.Wüthrich K. NMR with proteins and nucleic acids. Europhys. News. 1986;17:11–13. [Google Scholar]
- 32.Witkowska D., Rowinska-Zyrek M., Valensin G., Kozlowski H. Specific poly-histidyl and poly-cysteil protein sites involved in Ni2+ homeostasis in Helicobacter pylori. Impact of Bi3+ ions on Ni2+ binding to proteins. Structural and thermodynamic aspects. Coord. Chem. Rev. 2012;256:133–148. [Google Scholar]
- 33.Remelli M., Brasili D., Guerrini R., Pontecchiani F., Potocki S., Rowinska-Zyrek M., Watly J., Kozlowski H. Zn(II) and Ni(II) complexes with poly-histidyl peptides derived from a snake venom. Inorg. Chim. Acta. 2017;472:149–156. [Google Scholar]
- 34.Zhang C.L., Zou Y., Yu R.T., Gage F.H., Evans R.M. Nuclear receptor TLX prevents retinal dystrophy and recruits the corepressor atrophin1. Genes Dev. 2006;20:1308–1320. doi: 10.1101/gad.1413606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Janknecht R., Sander C., Pongs O. (HX)n repeats: a pH-controlled protein-protein interaction motif of eukaryotic transcription factors? FEBS Lett. 1991;295:1–2. doi: 10.1016/0014-5793(91)81369-j. [DOI] [PubMed] [Google Scholar]
- 36.Fregeau B., Kim B.J., Hernández-García A., Jordan V.K., Cho M.T., Schnur R.E., Monaghan K.G., Juusola J., Rosenfeld J.A., Bhoj E. De Novo mutations of RERE cause a genetic syndrome with features that overlap those associated with proximal 1p36 deletions. Am. J. Hum. Genet. 2016;98:963–970. doi: 10.1016/j.ajhg.2016.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kaur M., Izumi K., Wilkens A.B., Chatfield K.C., Spinner N.B., Conlin L.K., Zhang Z., Krantz I.D. Genome-wide expression analysis in fibroblast cell lines from probands with Pallister Killian syndrome. PLoS ONE. 2014;9:e108853. doi: 10.1371/journal.pone.0108853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Izumi K., Krantz I.D. Pallister-Killian syndrome. Am. J. Med. Genet. C. Semin. Med. Genet. 2014;166C:406–413. doi: 10.1002/ajmg.c.31423. [DOI] [PubMed] [Google Scholar]
- 39.Shi Z., Fujii K., Kovary K.M., Genuth N.R., Röst H.L., Teruel M.N., Barna M. Heterogeneous ribosomes preferentially translate distinct subpools of mRNAs genome-wide. Mol. Cell. 2017;67:71–83.e7. doi: 10.1016/j.molcel.2017.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Galan A., Lozano G., Piñeiro D., Martinez-Salas E. G3BP1 interacts directly with the FMDV IRES and negatively regulates translation. FEBS J. 2017;284:3202–3217. doi: 10.1111/febs.14184. [DOI] [PubMed] [Google Scholar]
- 41.Kondrashov N., Pusic A., Stumpf C.R., Shimizu K., Hsieh A.C., Ishijima J., Shiroishi T., Barna M. Ribosome-mediated specificity in Hox mRNA translation and vertebrate tissue patterning. Cell. 2011;145:383–397. doi: 10.1016/j.cell.2011.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.